Abstract
Thiamine pyrophosphate (TPP) is an essential cofactor for various pivotal cellular processes in all living organisms, including bacteria. Thiamine biosynthesis occurs in bacteria but not in humans; therefore, the enzymes in this pathway are attractive targets for antibiotic development. Among these enzymes, thiamine monophosphate kinase (ThiL) catalyzes the final step of this pathway, phosphorylating thiamine monophosphate to produce TPP. Here, we extensively investigated ThiL in Pseudomonas aeruginosa, a major pathogen responsible for hospital-acquired infections. We demonstrate that thiL deletion abolishes not only thiamine biosynthesis but also thiamine salvage capability and results in growth defects of the ΔthiL strain even in the presence of thiamine derivatives, except for TPP. Most importantly, the pathogenesis of the ΔthiL strain was markedly attenuated, compared with that of WT cells, with lower inflammatory cytokine induction and 103–104-fold decreased bacterial loads in an in vivo infection model in which the intracellular TPP level was in the submicromolar range. To validate P. aeruginosa ThiL (PaThiL) as a drug target, we further characterized its biochemical properties, determining a Vmax of 4.0 ± 0.2 nmol·min−1 and Km values of 111 ± 8 and 8.0 ± 3.5 μm for ATP and thiamine monophosphate, respectively. An in vitro small-molecule screening assay identified PaThiL inhibitors including WAY213613, a noncompetitive inhibitor with a Ki value of 13.4 ± 2.3 μm and potential antibacterial activity against P. aeruginosa. These comprehensive biological and biochemical results indicate that PaThiL represents a potential drug target for the development of an augmented repertoire of antibiotics against P. aeruginosa.
Keywords: thiamine, thiamine monophosphate kinase, bacterial metabolism, bacterial pathogenesis, Pseudomonas aeruginosa, infection, antibacterial target, PaThiL, virulence, drug target
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen associated with a wide range of acute and chronic infections of various body sites, including the urinary tract, skin, and respiratory tract (1). Patients with compromised immune defenses due to underlying diseases such as cancer or HIV infection or with severe burns, cystic fibrosis, bronchiectasis, or chronic obstructive pulmonary disease are particularly susceptible to P. aeruginosa infection (2–7). The mainstay of treatment for P. aeruginosa infection is antibiotics. However, the expression of multiple efflux pumps, reduced permeability of the outer membrane, the capacity to form biofilms, and the presence of persisters have rendered P. aeruginosa intrinsically resistant to many antibiotics (8, 9). Given this natural resistance, excessive use of antibiotics is required to treat P. aeruginosa infections, accelerating the development of drug-resistant strains. Indeed, multidrug-resistant and extensively drug-resistant P. aeruginosa strains are now prevalent worldwide, with the emergence of pan-drug-resistant strains threatening global public health (10–13). The World Health Organization recently listed drug-resistant P. aeruginosa as a priority pathogen necessitating urgent action to develop novel antibiotics to overcome current antibiotic resistance (14).
Thiamine is a crucial molecule in all living organisms, from microorganisms to mammals. Therefore, thiamine metabolism has attracted growing attention in the development of various drugs, including antibiotics (15–19). The physiologically active form of thiamine, thiamine pyrophosphate (TPP), plays important roles as a cofactor in various essential cellular processes, including carbohydrate, lipid, and amino acid metabolism (20–22). Microorganisms such as bacteria and fungi, as well as plants, produce TPP via de novo biosynthetic pathways that mammals lack (23, 24). The TPP biosynthetic pathway of bacteria involves the separate biosynthesis of thiazole and pyrimidine moieties, which are joined to form thiamine monophosphate (TMP) in a reaction catalyzed by thiamine phosphate synthase (ThiE) (25–27). Thiamine monophosphate kinase (ThiL) catalyzes the final step of the pathway by phosphorylating TMP to TPP, the biologically active form of the cofactor (28, 29). In addition to the TPP biosynthetic pathway, bacteria are capable of salvaging thiamine from exogenous sources to generate TPP. Some bacteria, such as Bacillus subtilis, take up exogenous thiamine and convert it to TPP in a one-step reaction catalyzed by thiamine pyrophosphate kinase (TPK), similar to the mammalian thiamine salvage pathway (30–33). Other bacteria, including Escherichia coli, first convert thiamine to TMP by thiamine kinase (ThiK) and then subsequently generate TPP by adding one additional phosphate to TMP through ThiL, the enzyme in the main TPP biosynthetic pathway (30, 34). Because TPP is indispensable for bacterial survival and humans lack the TPP biosynthetic pathway, enzymes involved in bacterial TPP biosynthesis are potential targets in the development of new antibiotics. ThiL is of particular interest, considering its key role in thiamine metabolism. Nevertheless, few studies, involving a limited number of bacterial species, have been conducted on ThiL, and it has never been validated as a target for antibacterial agents (28, 29, 35, 36).
In this work, we constructed a clean thiL deletion mutant of P. aeruginosa and investigated the impact of thiL deficiency on bacterial survival and in vivo pathogenesis. We also biochemically characterized P. aeruginosa ThiL and identified small molecules that inhibit the enzyme by using an optimized luminescent kinase assay. To the best of our knowledge, this is the first work to demonstrate the role of P. aeruginosa ThiL in bacterial physiology and pathogenesis and the first to validate ThiL as a new target for drug development, providing comprehensive biochemical characterization of ThiL and identifying its inhibitors.
Results
Role of ThiL in P. aeruginosa thiamine metabolism
To investigate the physiological role of ThiL in P. aeruginosa, we constructed an in-frame deletion mutant of the thiL gene in P. aeruginosa PAO1 by two-step allele exchange (37–39). TPP was supplied via selection medium for the last step of mutant generation, to avoid the loss of bacterial viability due to impaired thiamine biosynthesis. Deletion of thiL in the bacterial genome was confirmed by amplification of the thiL flanking region (Fig. S1). Phenotypic analysis of the ΔthiL mutant confirmed that deletion of thiL is lethal to P. aeruginosa unless TPP is exogenously provided in the medium (Fig. 1). Complementation with the plasmid expressing thiL (pthiL) relieved the growth defect of the ΔthiL mutant caused by TPP depletion, suggesting that ThiL is essential for TPP biosynthesis i0n P. aeruginosa (see Fig. 2A).sss
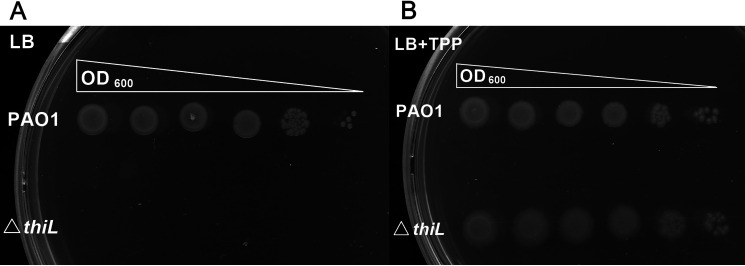
Figure 1.
Phenotypic analysis of thiL deletion in P. aeruginosa. A, a serial dilution of the thiL deletion mutant (ΔthiL) of P. aeruginosa was plated on LB agar and showed no growth after a 24-h incubation at 37 °C. B, TPP supplementation (1 mm) in LB agar rescued the growth defect of ΔthiL.
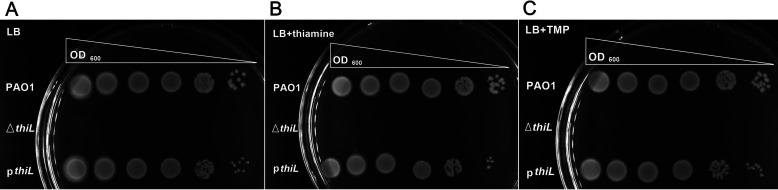
Figure 2.
Phenotypic analysis of ΔthiL and thiL complementation by the pBSPII SK(−)-thiL plasmid in the thiL deletion mutant (pthiL) with thiamine or TMP supplementation. The ΔthiL strain did not grow on LB medium (A) or LB medium supplemented with thiamine (50 μm) (B) or TMP (50 μm) (C) in a 24-h incubation at 37 °C; pthiL showed normal growth in LB medium (A) and LB medium supplemented with thiamine (B) or TMP (C).
The thiamine salvage pathway is another way to generate TPP in many organisms, including bacteria and even mammals, which cannot synthesize TPP de novo. Two types of direct thiamine salvage pathways have been identified in bacteria, namely, one-step pyrophosphorylation of thiamine to TPP by thiamine pyrophosphokinase (TPK/ThiN) and two steps of subsequent monophosphorylation of thiamine to TMP and then TPP by ThiK and ThiL, respectively. As the thiamine salvage pathway in P. aeruginosa has yet to be characterized and no thiamine transporter genes have been identified in this species (26), we first tested whether the bacteria are capable of producing TPP using extracellular thiamine. We found that the thiE mutant in which de novo TPP synthesis is impaired was able to grow in the presence of extracellular thiamine, TMP, and TPP, indicating that P. aeruginosa can take up thiamine derivatives and has a thiamine salvage pathway (Fig. 3). Unlike the thiE mutant, the ΔthiL strain exhibited a growth defect even with exogenously provided thiamine and TMP (Fig. 2). This result suggests that ThiL is involved in not only de novo TPP synthesis but also the thiamine salvage pathway in P. aeruginosa, indicating that ThiL plays a critical role in P. aeruginosa thiamine metabolism.
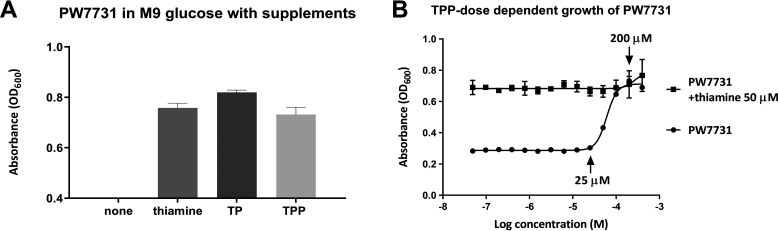
Figure 3.
Growth of the thiE mutant (PW7731) of P. aeruginosa in M9 glucose supplemented with thiamine, TMP (TP), or TPP. A, absorbance (OD600) of PW7731 showed normal growth in minimal medium supplemented with thiamine (50 μm), TMP (50 μm), or TPP (100 μm). B, TPP dose-dependent growth of PW7731 revealed the salvage concentration range (25–200 μm). Error bars are means ± S.D. of triplicate experimental groups.
Role of ThiL in the virulence of P. aeruginosa in vivo
Next, we sought to determine the role of ThiL in the virulence of P. aeruginosa in vivo. C57BL/6 mice were infected intranasally with 2 × 107 CFU of the PAO1, ΔthiL, or thiL-complemented strains. During the first 20 h, all infected mice exhibited decreased movement, compared with uninfected mice. Histopathological analysis of the lung tissues at 20 h postinfection also suggested that, unlike uninfected control mice, all infected mice showed a distinct inflammatory response composed mainly of peribronchial and alveolar neutrophilic infiltrates, with neutrophilic consolidation and smooth muscle hyperplasia in the arterioles of the lung tissues (Fig. 4). Although mice infected with the ΔthiL strain exhibited a slightly lower degree of lung neutrophil infiltration (8.2 ± 6.3%) than mice infected with the PAO1 strain (11 ± 4%), the difference was not significant, suggesting that the ΔthiL mutant is capable of recruiting host immune cells and causing lung inflammation similar to the PAO1 strain (Fig. S2). Interestingly, when the average bacterial load in the left lobe of the lungs was measured at 20 h postinfection (n = 5 for each group), over 1,000 times more bacteria were found in mice infected with the PAO1 strain (1.6 × 108 CFU) and thiL-complemented strain (1.6 × 108 CFU), compared with mice infected with the ΔthiL strain (6.7 × 104 CFU) (Fig. 5A). In addition, the survival rates of the infected mice clearly demonstrated the attenuated virulence of the ΔthiL strain, compared with the PAO1 and thiL-complemented strains. Over 50% of mice infected with the PAO1 and thiL-complemented strains died by 24 h postinfection, and the rest of the infected mice were dead by 48 h postinfection (Fig. 5B). In contrast, uninfected control mice and mice infected with the ΔthiL mutant survived as long as 72 h postinfection (Fig. 5B).
Figure 4.
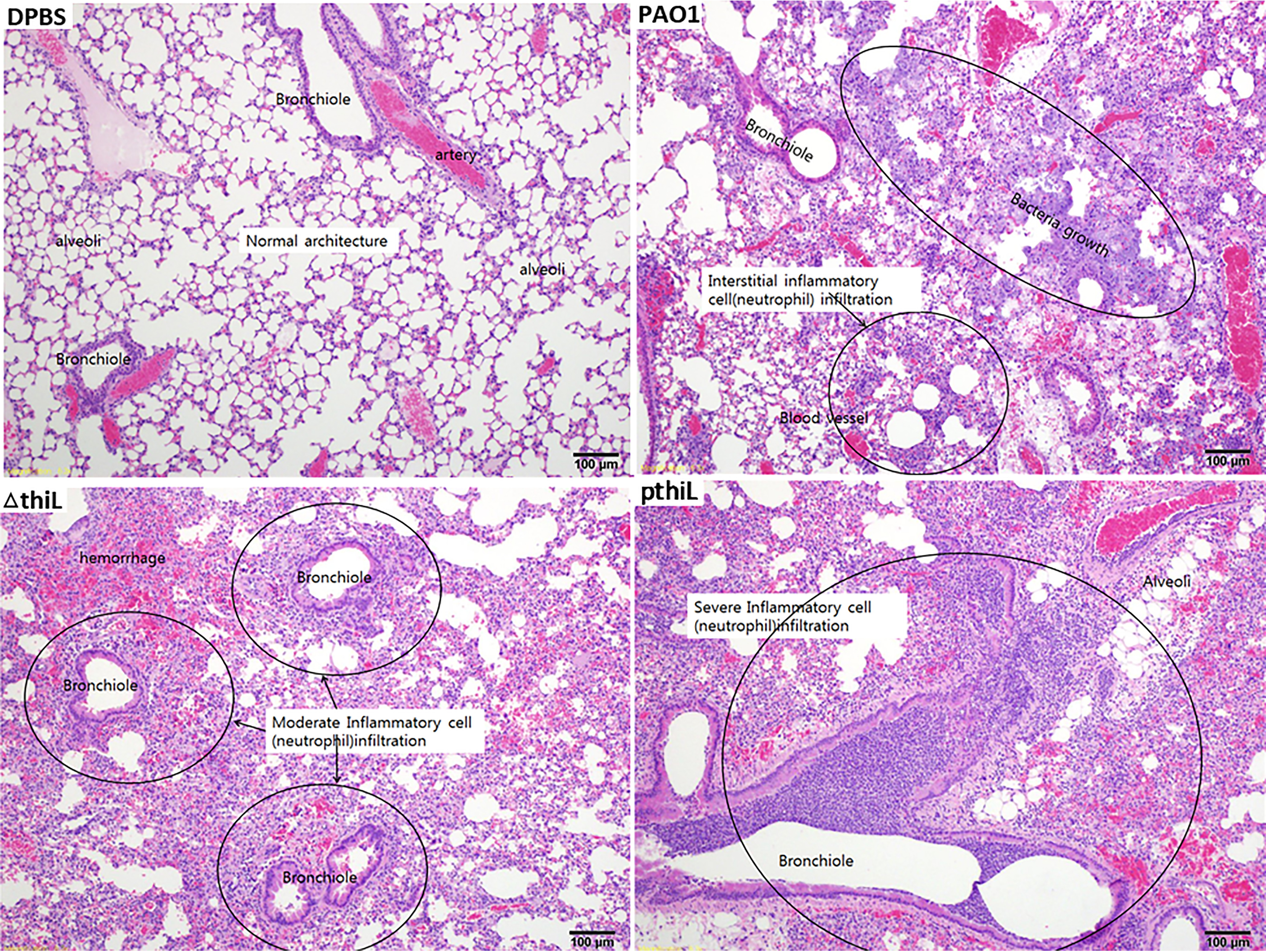
Neutrophil infiltration of the left lobe of lungs in C57BL/6 mice infected with PAO1, ΔthiL, or pthiL. Intranasal infections were applied to 6-week-old mice with DPBS, PAO1, ΔthiL, or pthiL. H&E staining showed moderate to severe neutrophil infiltration (marked in circles) in bronchioles and blood vessels of the lungs after 20 h of infection. Magnification, ×100. Representative images of triplicate experimental groups are shown.
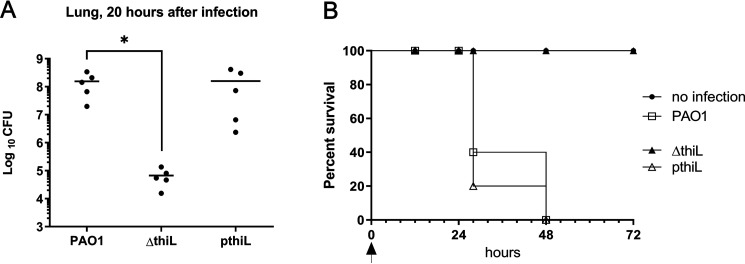
Figure 5.
Bacterial loads in mouse lungs infected with PAO1, ΔthiL, or pthiL and survival analysis of the infection groups. A, CFU analysis showed relatively low bacterial loads in the lungs of mice infected with ΔthiL (6.7 × 104 CFU), compared with PAO1 (1.6 × 108 CFU) or pthiL (1.6 × 108 CFU), after 20 h of infection. *, p < 0.05. B, a survival rate of 100% was observed in the mice infected with ΔthiL for 72 h after infection. The survival rates were reduced to below 50% in the mice infected with PAO1 or pthiL strains by 24 h after infection. The arrow indicates the time of infection (n = 5 for each experimental group, repeated three times).
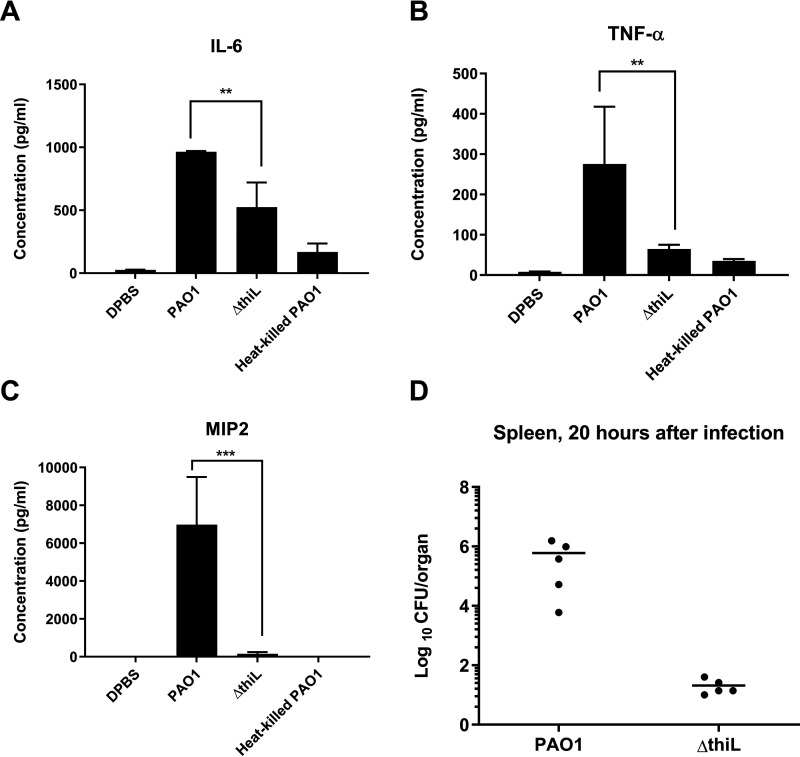
Because there was a discrepancy between the results of the lung histology and survival analyses, we further investigated pathogenesis in the infected mice by measuring the host immune response in the blood. Levels of three proinflammatory cytokines, IL-6, IL-8, and tumor necrosis factor α (TNFα), were measured in the blood of infected mice at 20 h postinfection. The level of macrophage inflammatory protein-2 (MIP-2) was measured as a murine counterpart of IL-8. Levels of all three tested cytokines were significantly higher in PAO1-infected mice than in ΔthiL-infected mice (Fig. 6, A, B, and C). Similarly, the splenic bacterial count in mice infected with the PAO1 strain (6 × 105 CFU/organ) was 4 orders of magnitude higher than that in ΔthiL-infected mice (2 × 101 CFU/organ) (Fig. 6D). Taken together, these results suggest that overall virulence is attenuated in the ΔthiL strain due to decreased severity of bacteremia and sepsis, ultimately leading to survival of the mice, demonstrating that ThiL is essential for the full virulence of P. aeruginosa.
Figure 6.
Proinflammatory cytokine (IL-6, TNFα, and MIP-2) levels in the blood of mice 20 h after infection with PAO1, ΔthiL, or heat-killed PAO1 and bacterial loads (CFU/organ) in mouse spleen 20 h after infection with PAO1 or ΔthiL. A–C, ELISAs for IL-6 (A), TNFα (B), and MIP-2 (C) in mouse plasma indicated significantly lower cytokine levels in ΔthiL groups, compared with PAO1. **, p < 0.01; ***, p < 0.001. D, CFU analysis showed 104-fold lower bacterial loads in the spleens of mice infected with ΔthiL, compared with PAO1 (n = 5 for each experimental group, repeated three times).
Biochemical characterization of P. aeruginosa ThiL
Despite its importance in bacterial physiology and virulence, to our knowledge P. aeruginosa ThiL (PaThiL) has never been studied before, at either the molecular or biochemical level. For biochemical characterization of the enzyme, the P. aeruginosa thiL gene (PA4051) was PCR amplified and cloned into pET28b. The recombinant PaThiL was purified as an ∼37-kDa His-tagged protein using a nickel-nitrilotriacetic acid affinity matrix, with >95% purity (Fig. S3). We found that purified PaThiL was highly unstable without desalting, exhibiting 80% and 56% activity loss within 24 h at 4 °C and −210 °C, respectively (Fig. S4). A further gradual decrease in activity was observed over time. Therefore, purified PaThiL was frozen and stored in liquid nitrogen (−210 °C) after immediate desalting, and activity was retained for over 1 month without significant loss (Fig. S4).
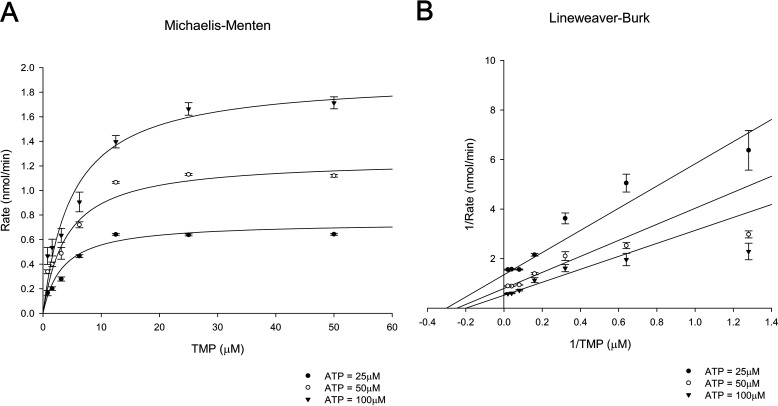
Previous studies of ThiL used a HPLC-based assay or a coupled assay using apo-carboxylase to assess ThiL activity (28, 36, 40). Because both types of assays are quite inconvenient and inefficient, we adapted a luminescent kinase assay that detects ATP consumption and optimized it to determine PaThiL enzymatic kinetics (see Experimental Procedures). The final assay was established to analyze the activity of 10 µg of PaThiL in a reaction buffer containing 0.05 mm TMP, 0.05 mm ATP, 50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, and 350 mm KCl. The Km values for TMP and ATP, as well as the Vmax value, were calculated based on Michaelis-Menten and Lineweaver-Burk plots of PaThiL activity in reactions containing several different concentrations of ATP and TMP. The Vmax value of PaThiL was 4.0 ± 0.2 nmol·min−1. The Km values for ATP and TMP were 111 ± 8 μm and 8.0 ± 3.5 μm, respectively, with a random bi-bi mechanism (Fig. 7). Similar ranges of Km values were previously reported for partially purified E. coli ThiL (270 and 1.1 μm for ATP and TMP, respectively) (36). Further characterization of PaThiL revealed that the enzyme phosphorylates oxythiamine monophosphate (a TMP analogue) with a Km value of 15.2 ± 2.0 μm (Fig. S5). Thiamine, as well as other thiamine analogues, including oxythiamine, pyrithiamine, and amprolium, were not phosphorylated by PaThiL, indicating that prior acquisition of monophosphate is a requirement for PaThiL substrates (Fig. S6). TPP was not further phosphorylated by ThiL, suggesting that PaThiL does not contribute to the generation of intracellular thiamine triphosphate.
Figure 7.
Kinetic parameters of ThiL determined by measuring the rate of kinase activity. Enzymatic assays were performed for isolated ThiL (10 µg) with various concentrations of TMP (0–50 μm). The assays were initiated with three ATP concentrations (25, 50, and 100 μm) to determine ThiL activity rates. TMP data were plotted in Michaelis-Menten (A) and Lineweaver-Burk (B) functions. The results indicated Km values of ThiL for ATP (111 ± 8 μm) and TMP (8.0 ± 3.5 μm) and Vmax (4.0 ± 0.2 nmol·min−1). Error bars are means ± S.D. of triplicate experimental groups.
Validation of PaThiL as a new antibacterial target
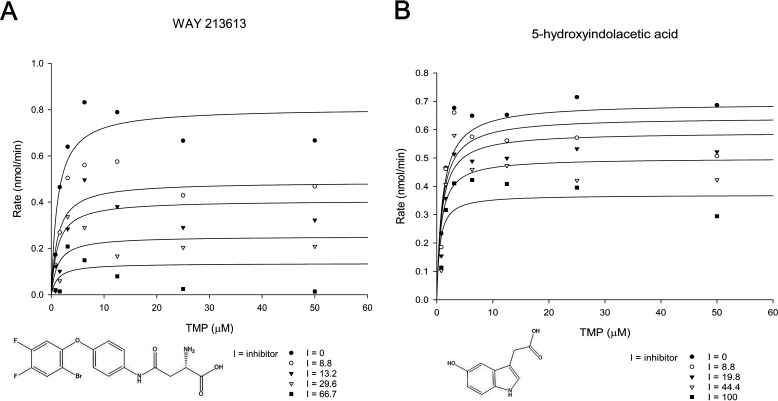
Gram-negative bacteria such as P. aeruginosa are notorious for their multidrug and pan-drug resistance, which has created an urgent need for new classes of antibiotics. Our in vivo virulence study of the ΔthiL mutant indicated that ThiL is a promising novel candidate for an antibacterial molecule, as humans have no PaThiL homologue. In order to validate the druggability of PaThiL, ∼2,800 commercially available compounds were screened for inhibitory activity against PaThiL. The screening was conducted using a slight modification of our established in vitro PaThiL assay. The screening assay was initiated upon addition of 5 µg of purified PaThiL to a reaction mixture containing 10 μm TMP and 100 μm test compound; ThiL activity was measured by monitoring ATP consumption during a 10-min reaction. WAY213613 (Tocriscreen) and 5-hydroxyindolacetic acid (Lopac) were among several compounds that inhibited PaThiL. Further biochemical studies revealed that WAY213613 was a noncompetitive inhibitor of TMP for PaThiL, with a Ki value of 13.4 ± 2.3 μm, whereas 5-hydroxyindolacetic acid was an uncompetitive inhibitor that exhibited a Ki value of 114 ± 27 μm (Fig. 8). Additional binding mode analyses of each compound with the homology modeling-derived 3D structure of PaThiL suggested energetically favorable interactions with PaThiL (data not shown).
Figure 8.
Enzymatic assay of ThiL with ThiL inhibitors for determination of inhibition kinetics and Ki values. Various concentrations of WAY213613 (0–66.7 μm) or 5-hydroxyindolacetic acid (0–100 μm) were applied to the ThiL assays with ATP (100 μm) and TMP (0–50 μm) to determine ThiL activity rates. The activity rates of ThiL with WAY213613 (A) or 5-hydroxyindolacetic acid (B) were plotted in Michaelis-Menten functions. The kinetic parameters showed that WAY213613 was a noncompetitive inhibitor with a Ki value of 13.4 ± 2.3 μm and 5-hydroxyindolacetic acid was an uncompetitive inhibitor with a Ki value of 114 ± 27 μm.
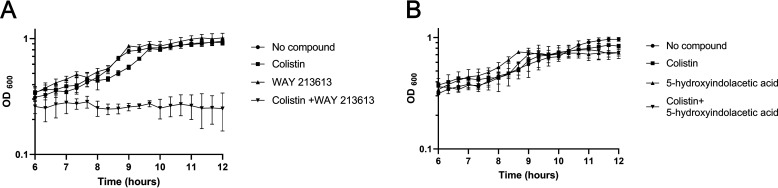
We next tested whether the identified PaThiL inhibitors exhibited antibacterial activity. Because the outer membrane of P. aeruginosa can impede the entry of drugs into the cell and thereby mask potential antibacterial activity of PaThiL inhibitors, we also tested the antibacterial activity with a low concentration of colistin (0.5 µg/ml), which can increase cell permeability by disrupting the bacterial outer membrane while not affecting bacterial viability (Fig. S7 and Table S1). Although 5-hydroxyindoleacetic acid did not exhibit antibacterial activity at 100 μm even in the presence of colistin, we found that 100 μm WAY213613 completely stopped bacterial growth not by itself but in the presence of colistin (Fig. 9). These results suggest that PaThiL is a valid target for development of new antibacterial therapeutics, demonstrating the feasibility of identification of small molecules that inhibit PaThiL.
Figure 9.
Growth kinetics of PAO1 in antibacterial effect tests with WAY213613 or 5-hydroxyindolacetic acid. No antibacterial activities were observed with 100 μm WAY213613 (A) or 5-hydroxyindolacetic acid (B). A bacterial inhibition effect for WAY213613 (100 μm) was observed with addition of a sublethal dose of colistin (0.5 µg/ml) (A). 5-Hydroxyindolacetic acid (100 μm) did not show an antibacterial effect with addition of colistin (0.5 µg/ml) (B). Control groups were treated with no compound in LB medium or LB medium with colistin (0.5 µg/ml). Error bars are means ± S.D. of triplicate experimental groups.
Discussion
TPP-dependent enzymes, including transketolase, pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, and 1-deoxy-d-xylulose-5-phosphate synthase, catalyze essential cellular reactions in bacteria, from central metabolism to biosynthesis of amino acids, cofactors, and lipids (21). Recently, TPP-dependent pyruvate and α-ketoglutarate dehydrogenase complexes in P. aeruginosa were found to intracellularly reduce phenazine derivatives such as pyocyanin (a pseudomonal toxin), thus contributing to iron acquisition and redox homeostasis of the bacteria in addition to their designated roles in central metabolism (41). This evidence indicates that TPP deficiency can have a substantial effect on the metabolic network of bacteria, thus making TPP metabolism a very attractive target for the development of new antibiotics. Indeed, inhibitors of ThiE exhibit antibacterial activity against Mycobacterium tuberculosis, a notorious human pathogen (42). Because M. tuberculosis does not harbor the genes for the thiamine salvage pathway and transporters, inhibition of ThiE can effectively deplete the TPP pool by impairing TPP biosynthesis, the only source of TPP in the bacteria, ultimately causing bacterial death.
However, inducing the death of many pathogenic bacteria via TPP deficiency is hampered by several challenges, particularly in vivo; 1) complete deficiency of TPP requires concurrent inhibition of both thiamine salvage and TPP biosynthesis, and 2) levels of thiamine derivatives in the host, specifically TPP, should not be sufficient for directly promoting bacterial survival. In this work, we addressed these two challenges by investigating ThiL in P. aeruginosa. We demonstrated that ThiL is the key enzyme for both de novo TPP synthesis and thiamine salvage in P. aeruginosa by revealing an in vitro growth defect of the thiL mutant, thus indicating a possibility to deplete the TPP pool in P. aeruginosa by targeting ThiL (Fig. 1).
Although thiamine transporter genes have not been identified in P. aeruginosa, the growth of thiE and thiL mutants in the presence of thiamine, TMP, or TPP suggests the existence of transporters that import thiamine derivatives in P. aeruginosa (26) (Figs. 2 and 3). In E. coli, intracellular TPP concentrations were measured at 150–300 μm under optimal growth conditions, which are presumably TPP levels for normal bacterial growth (43, 44). Consistent with this notion, when we determined the extracellular TPP concentration necessary to relieve the growth defect of the thiE mutant in which the TPP biosynthetic pathway is impaired, we found that a minimum TPP concentration of 25 μm was required to detect any degree of thiE mutant growth and a concentration over 200 μm was required to fully support the growth of the thiE mutant (Fig. 3). This result suggests that, if P. aeruginosa relies solely on exogenous TPP for growth, then the concentration should be higher than 25 μm. Reported TPP levels in mouse blood range from 0.3 to 1.2 μm, well below the TPP concentration sufficient to support growth of the thiL mutant (45, 46). Consistent with these data, we found that the thiL mutant exhibited markedly reduced pathogenicity in a mouse infection model, compared with the WT strain (Figs. 5 and 6). TPP levels in healthy human blood (116–138 nm) are even lower than in mouse blood, suggesting that ThiL is a viable therapeutic target in the treatment of Pseudomonas infections (47, 48).
We also validated PaThiL as a suitable target for new antibacterial agents by conducting an intensive biochemical characterization followed by identification of inhibitors of PaThiL. Considering the high intracellular concentrations of ThiL substrates (ATP, ∼1.8 mm; TMP, ∼12.5 μm) and product (TPP, ∼0.3 mm) in bacteria (44), noncompetitive ThiL inhibitors might be more beneficial than competitive inhibitors that probably require a very high affinity for ThiL. Interestingly, we found that WAY213613, a known inhibitor of the EAAT2 glutamate transporter, could inhibit PaThiL in a noncompetitive manner, with a Ki value of 13.4 ± 2.3 μm. When we tested antibacterial activity, WAY213613 was able to inhibit the growth of P. aeruginosa, although the permeability of the bacterial outer membrane had to be increased to detect the activity (Fig. 9).
Antibiotic-resistant bacteria such as pan-drug-resistant P. aeruginosa pose an immediate threat to global public health and highlight an urgent need for new antibiotics and targets. In this work, we extensively characterized PaThiL and demonstrated its roles in bacterial physiology and pathogenesis. Although this study focused on PaThiL, this enzyme is expected to play similar roles in other bacteria that have a thiamine salvage pathway composed of ThiK and ThiL, such as E. coli and Salmonella species (26). Taken together, the results of this work demonstrate that ThiL is a suitable therapeutic target for the development of new drugs to treat not only P. aeruginosa infections but potentially many other bacterial infections as well.
Experimental procedures
Bacterial strains, culture media, and chemicals
All strains and plasmids used in this study are listed in Table 1. Bacteria were cultured in Luria-Bertani (LB) broth at 37 °C with agitation. The thiE transposon mutant (PW7731) was cultured in minimal medium supplemented with TMP for normal growth. LB powder was dissolved in purified water and autoclaved at 121 °C for 15 min. As minimal medium, M9 salt powder was dissolved in purified water and autoclaved at 121 °C for 15 min. Sterile MgSO4 (2 mm) and CaCl2 (0.1 mm) were added to M9 solution for complete minimal medium. Supplements for the media, including glucose (20 mm), thiamine, TMP, and TPP, were sterilized by filtration through a 0.22-µm PVDF membrane (Millipore, Burlington, MA) before addition. Agar (1.5%) was added to the medium to prepare solid growth plates. Kanamycin (50 mg/ml) and ampicillin (100 mg/ml) were dissolved in purified water and sterilized by filtration through a 0.22-µm PVDF membrane. Antibiotic stock solutions were stored at −20 °C. Isopropyl β-d-1-thiogalactopyranoside (IPTG) (1 m) was dissolved in purified water and filtered through a 0.22-µm filter. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO), except for ATP and IPTG, which were purchased from Thermo Fisher Scientific (Waltham, MA).
Table 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Note |
|---|---|---|
| Strains | ||
| P. aeruginosa PAO1 | WT | ATCC |
| E. coli BL21(DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Invitrogen |
| E. coli SM10 λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir | Yonsei Univ. |
| PW7731 | thiE transposon mutant, lacZbpO2q3B12 | (53) |
| ΔthiL | PAO1 with thiL(PA4051) in-frame gene deletion | This study |
| Plasmidsa | ||
| pET28b(+) | E. coli expression vector, KmR | Novagen |
| pCVD442-Gm | sacB suicide vector from pUM24, GmR | Yonsei Univ. |
| pBSP II SK(−) | Broad-host-range expression vector, AmpR | (54) |
| pETthiL | PAO1 thiL gene inserted in pET28b(+) | This study |
| pthiL | PAO1 thiL gene inserted in pBSP II SK(−) | This study |
| Primersb | ||
| thiL (PA4051) | Forward, TAGGTTGGATCCCTGCGGCTGTCCACCTACGAGC | BamHI |
| Forward, TAGGTTCATATGGGTGAGTTCGAGCTGATCCGCC | NdeI | |
| Reverse, TAGGTTAAGCTTTCAGTCACGTTGGGTTCCGAAATGTTGG | HindIII | |
| nusB (PA4052) | Forward, GTTATAGCATGCGTGAGCAATCAAGACAGCGGC | SphI |
| Reverse, ATTAATCGAGCTCTCAGCGCTTGCCGCCG | SacI | |
| pgpA (PA4050) | Forward, GTTGTTGAGCTCGTGACTGAGCATCCTGATCAG | SacI |
| Reverse, GTTGTTCCCGGGTCAGACCAGCCAGTG | SmaI |
aKmR, kanamycin resistance; GmR, gentamycin resistance; AmpR, ampicillin resistance.
bRestriction sites are underlined.
Growth of the thiE transposon mutant (PW7731) with thiamine, TMP, or TPP
M9 glucose was supplemented with thiamine (50 μm), TMP (50 μm), or TPP (100 μm), and the medium was applied to PW7731 with an initial OD600 of 0.02. After a 16-h incubation with agitation, absorbance (OD600) was measured for each growth condition. The TPP salvage concentration was determined by 2-fold serial dilution of TPP in a range from 0.02 to 400 μm. Each concentration of TPP was applied to PW7731 with an initial OD600 of 0.0001, which was prepared by 1/500 dilution of the bacteria at an OD600 of 0.05, and the mutant was incubated in 37 °C for 16 h. The absorbance (OD600) was measured with a multilabel plate reader.
Overexpression and isolation of ThiL
The gene encoding thiamine monophosphate kinase (thiL) was amplified by PCR analysis using the primers listed in Table 1 and was inserted into pET28b(+) (Novagen, Madison, WI), a vector for the cloning and expression of recombinant proteins with His tags, at restriction sites between NdeI and HindIII. The plasmid was transformed into E. coli BL21 cells, which were selected on medium containing kanamycin (50 µg/ml). IPTG (1 mm) was added to the mid-log-phase culture to induce protein expression, and the cells were incubated for 2.5 h at 37 °C with agitation. The cells were centrifuged at 4,000 × g, and the resulting pellet was resuspended in 5 ml of 100 mm Tris-HCl (pH 8.0) supplemented with a protease inhibitor (Roche, Mannheim, Germany). The suspended cells were then sonicated using a Bioruptor sonication system (Diagenode, Denville, NJ) and centrifuged at 14,000 × g. ThiL protein in the supernatant was purified by affinity chromatography (GE Healthcare, Freiburg, Germany). The collected protein was desalted using a 30kDa Centrifugal Filter Unit (Millipore) and suspended in 100 mm Tris-HCl (pH 8.0). The amount of protein in the collection was quantified using the microassay protocol for the Bradford protein assay, with BSA as a standard. The protein was aliquoted in cryotubes and stored at −210 °C for long-term preservation. The purity of the isolated protein was over 95%, which was confirmed by Coomassie Blue staining.
ThiL assay
The ThiL assay was established based on previous protocols (28, 40). The assay reaction buffer contained 0.05 mm TMP, 0.05 mm ATP, 50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, and 350 mm KCl. A total of 10 µg ThiL protein was added to the reaction buffer to initiate the kinase reaction. After 10 min, the amount of ATP was determined using a luminescence assay (Promega, Madison, WI). ATP hydrolysis in the reaction buffer without TMP was less than 1% in 10 min. Km and Vmax values were determined for TMP and ATP using the ThiL assay with TMP and ATP concentrations ranging from 0.78 to 50 μm and from 25 to 100 μm, respectively, in 2-fold dilution steps. The assay data were fit to Michaelis-Menten or Lineweaver-Burk functions using the random bisubstrate module in Sigma Plot v. 14 (Systat Software Inc., San Jose, CA) to obtain the Km and Vmax values. To determine the Ki of ThiL inhibitors, ThiL assays were carried out with WAY213613 (0–66.7 μm) or 5-hydroxyindolacetic acid (0–100 μm). The activity rates of ThiL were fit to Michaelis-Menten or Lineweaver-Burk functions using the single-substrate/single-inhibitor module in SigmaPlot.
Bacterial growth kinetics with ThiL inhibitors
For measurement of PAO1 growth kinetics with ThiL inhibitors, 100 μm WAY213613 or 5-hydroxyindolacetic acid was applied to PAO1 with or without colistin at 0.5 µg/ml. The growth kinetic assays were carried out at 37 °C in 96-well plates with an initial OD600 of 0.0001. Absorbance values (OD600) were measured every 20 min for 13 h with a multilabel plate reader (SpectraMax M5; Molecular Devices, San Jose, CA). A checkerboard assay was also performed for the combination of WAY213613 and colistin in the ranges of 0–200 μm and 0–20 μm, respectively. The MIC of each antimicrobial agent alone and in combination was defined as the lowest concentration that inhibited visible growth of PAO1. The fractional inhibitory concentration (FIC) index was calculated by the formula FIC= (MIC of WAY213613 in combination/MIC of WAY213613 alone) + (MIC of colistin in combination/MIC of colistin alone), as described previously (49, 50).
Construction of thiL deletion mutants via allelic exchange
To construct a P. aeruginosa mutant lacking thiL, allelic exchange was carried out according to previous studies (37, 39). Briefly, both upstream (nusB; 480 bp) and downstream (pgpA; 516 bp) genes flanking thiL were amplified by PCR analysis using the primers listed in Table 1. Both ends of the amplified genes carried specific restriction sites (Table 1). The downstream gene and pCVD442-Gm, a suicide vector, were double-digested with SacI and SmaI and then ligated. Consecutively, the upstream gene and pCVD442-Gm-pgpA were double-digested with SphI and SmaI and then ligated to obtain the suicide vector carrying the amplified genes. The generated vector was electroporated into E. coli SM10 λpir to facilitate conjugation into P. aeruginosa. Transconjugants were selected on medium containing gentamicin (100 µg/ml), and allele exchange was induced by incubation in medium containing sucrose (6%). The deletion mutant was selected on LB medium supplemented with 1 mm TPP, and deletion of thiL was confirmed by PCR analysis.
Generation of the complemented strain of the thiL deletion mutant
To generate a complemented strain of the P. aeruginosa ΔthiL mutant, the amplified thiL gene was double-digested with BamHI and HindIII and inserted into pBSP II SK(−) to create pthiL. The constructed plasmid was electroporated into competent cells of the PAO1 ΔthiL strain using a previously reported method (51). Briefly, the bacterial culture was washed twice and suspended with 300 mm sucrose at room temperature to generate electrocompetent cells. Next, 1 µg pthiL was mixed with 100 µl electrocompetent cells, and the pulse was applied to the mixture (P. aeruginosa 2.5-kV setting; Bio-Rad). LB medium was then immediately added to the mixture and incubated for 1 h at 37 °C. Transformed cell were selected on LB agar plates supplemented with ampicillin (0.5 mg/ml). Complementation was confirmed by PCR analysis and bacterial growth without TPP supplementation.
P. aeruginosa acute murine infection model
Inbred 6-week-old female C57BL/6 mice were purchased from Orient Bio (Seongnam, Korea) and maintained in our animal facility, which cared for the animals in accordance with the institutional guidelines. All mice were subjected to a 1-week adjustment period prior to the experiment. The protocol was approved by the Animal Care and Ethics Committee of Institut Pasteur Korea (Approval number IPK-17014).
The acute infection model was established based on previously published methods (52). Bacteria were grown to mid-log phase and washed three times with Dulbecco's PBS (DPBS). Mice were anesthetized with a mixture of ketamine and xylazine and inoculated intranasally with 2 × 107 CFU bacteria suspended in 20 µl DPBS. At 20 h after inoculation, mice were sacrificed using isoflurane, and both lungs and the spleen were harvested. The left lung and spleen were homogenized in PBS and then plated separately onto LB agar plates for WT and complementation strains; the LB agar was supplemented with TPP for the ΔthiL strain. The plates were incubated for 20 h at 37 °C, and then the viable bacterial cells were counted to determine the number of bacteria in the organs. The cranial lobe of the lungs was placed in 10% formalin for histopathological analysis by H&E staining. For histopathological analysis, 4-μm-thick sections of fixed lungs were blindly analyzed for cellular infiltration using Image-Pro Plus v.4.5 (Media Cybernetics, Rockville, MD). Briefly, degrees of neutrophil infiltration were assessed as percentages of inflammatory lesions per total area in microscopic images (magnification, ×40) of the lung sections. Another set of mice (n = 5) were subjected to body weight measurements and assessments of clinical signs of infection, and results were recorded until 72 h after inoculation. At the end of the experiment, the mice were euthanized using isofluorane.
Determination of proinflammatory cytokine plasma concentrations
At the time of sacrifice, an average of 0.7 ml of whole blood was withdrawn from each mouse by cardiac puncture and collected in heparin-treated tubes. The blood was immediately centrifuged at 2,000 × g for 20 min to obtain plasma. The isolated plasma was fast-frozen on dry ice and stored at −80 °C. To measure cytokine concentrations, we used Quantikine ELISA kits for IL-6, TNFα, MIP-2, and IL-1β (R & D Systems, Minneapolis, MN). Each assay was performed according to the manufacturer's instructions.
Data availability
All data are contained within the article and the supporting information.
Supplementary Material
Acknowledgments
We thank Prof. Sangsun Yoon for invaluable advice on this research.
This article contains supporting information.
Author contributions—H. J. K. and S. J. conceptualization; H. J. K. and S. J. data curation; H. J. K., H. L., Y. L., and I. C. formal analysis; H. J. K. and S. J. supervision; H. J. K. and S. J. funding acquisition; H. J. K., H. L., Y. L., I. C., Y. K., S. L., and S. J. investigation; H. J. K. and H. L. visualization; H. J. K. and H. L. methodology; H. J. K., H. L., and S. J. writing-original draft; H. J. K. and S. J. project administration; H. J. K. and S. J. writing-review and editing; I. C., Y. K., S. L., and S. J. software.
Funding and additional information—This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (Grant NRF-2017M3A9G6068246), as well as Gyeonggi-do.
Conflict of interest—The authors declare that they have no conflicts of interests with the contents of this article.
- TPP
- thiamine pyrophosphate
- ThiL
- thiamine monophosphate kinase
- ThiE
- thiamine phosphate synthase
- ThiK
- thiamine kinase
- TMP
- thiamine monophosphate
- TPK
- thiamine pyrophosphate kinase
- TNFα
- tumor necrosis factor α
- MIP-2
- macrophage inflammatory protein 2
- PaThiL
- Pseudomonas aeruginosa thiamine monophosphate kinase
- LB
- Luria-Bertani
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- DPBS
- Dulbecco's phosphate-buffered saline.
References
- 1. Gellatly S. L., and Hancock R. E. (2013) Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173 10.1111/2049-632X.12033 [DOI] [PubMed] [Google Scholar]
- 2. Chatzinikolaou I., Abi-Said D., Bodey G. P., Rolston K. V., Tarrand J. J., and Samonis G. (2000) Recent experience with Pseudomonas aeruginosa bacteremia in patients with cancer: retrospective analysis of 245 episodes. Arch. Intern. Med. 160, 501–509 10.1001/archinte.160.4.501 [DOI] [PubMed] [Google Scholar]
- 3. Manfredi R., Nanetti A., Ferri M., and Chiodo F. (2000) Pseudomonas spp. complications in patients with HIV disease: an eight-year clinical and microbiological survey. Eur. J. Epidemiol. 16, 111–118 10.1023/A:1007626410724 [DOI] [PubMed] [Google Scholar]
- 4. Branski L. K., Al-Mousawi A., Rivero H., Jeschke M. G., Sanford A. P., and Herndon D. N. (2009) Emerging infections in burns. Surg. Infect. (Larchmt.) 10, 389–397 10.1089/sur.2009.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parkins M. D., Somayaji R., and Waters V. J. (2018) Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin. Microbiol. Rev. 31, e00019–18 10.1128/CMR.00019-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilson R., Aksamit T., Aliberti S., De Soyza A., Elborn J. S., Goeminne P., Hill A. T., Menendez R., and Polverino E. (2016) Challenges in managing Pseudomonas aeruginosa in non-cystic fibrosis bronchiectasis. Respir. Med. 117, 179–189 10.1016/j.rmed.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 7. Choi J., Oh J. Y., Lee Y. S., Hur G. Y., Lee S. Y., Shim J. J., Kang K. H., and Min K. H. (2018) Pseudomonas aeruginosa infection increases the readmission rate of COPD patients. Int. J. Chron. Obstruct. Pulmon. Dis. 13, 3077–3083 10.2147/COPD.S173759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fajardo A., Martinez-Martin N., Mercadillo M., Galan J. C., Ghysels B., Matthijs S., Cornelis P., Wiehlmann L., Tummler B., Baquero F., and Martinez J. L. (2008) The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 3, e1619 10.1371/journal.pone.0001619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moradali M. F., Ghods S., and Rehm B. H. (2017) Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7, 39 10.3389/fcimb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruiz-Garbajosa P., and Canton R. (2017) Epidemiology of antibiotic resistance in Pseudomonas aeruginosa: implications for empiric and definitive therapy. Rev. Esp. Quimioter. 30, Suppl. 1, 8–12 [PubMed] [Google Scholar]
- 11. Botelho J., Grosso F., and Peixe L. (2019) Antibiotic resistance in Pseudomonas aeruginosa: mechanisms, epidemiology and evolution. Drug Resist. Updat. 44, 100640 10.1016/j.drup.2019.07.002 [DOI] [PubMed] [Google Scholar]
- 12. Fernandes M., Vira D., Medikonda R., and Kumar N. (2016) Extensively and pan-drug resistant Pseudomonas aeruginosa keratitis: clinical features, risk factors, and outcome. Graefes. Arch. Clin. Exp. Ophthalmol. 254, 315–322 10.1007/s00417-015-3208-7 [DOI] [PubMed] [Google Scholar]
- 13. Barbier F., and Wolff M. (2010) Multi-drug resistant Pseudomonas aeruginosa: towards a therapeutic dead end? Med. Sci. (Paris) 26, 960–968 10.1051/medsci/20102611960 [DOI] [PubMed] [Google Scholar]
- 14. Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., Ouellette M., Outterson K., Patel J., Cavaleri M., Cox E. M., et al. (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 15. Wang J., Zhang X., Ma D., Lee W. P., Xiao J., Zhao Y., Go V. L., Wang Q., Yen Y., Recker R., and Xiao G. G. (2013) Inhibition of transketolase by oxythiamine altered dynamics of protein signals in pancreatic cancer cells. Exp. Hematol. Oncol. 2, 18 10.1186/2162-3619-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cho J. H., Lee M. Y., Baig I. A., Ha N. R., Kim J., and Yoon M. Y. (2013) Biochemical characterization and evaluation of potent inhibitors of the Pseudomonas aeruginosa PA01 acetohydroxyacid synthase. Biochimie (Paris) 95, 1411–1421 10.1016/j.biochi.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 17. Swier L. J., Monjas L., Guskov A., de Voogd A. R., Erkens G. B., Slotboom D. J., and Hirsch A. K. (2015) Structure-based design of potent small-molecule binders to the S-component of the ECF transporter for thiamine. ChemBioChem 16, 819–826 10.1002/cbic.201402673 [DOI] [PubMed] [Google Scholar]
- 18. Lunse C. E., Schuller A., and Mayer G. (2014) The promise of riboswitches as potential antibacterial drug targets. Int. J. Med. Microbiol. 304, 79–92 10.1016/j.ijmm.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 19. Lunse C. E., Scott F. J., Suckling C. J., and Mayer G. (2014) Novel TPP-riboswitch activators bypass metabolic enzyme dependency. Front. Chem. 2, 53 10.3389/fchem.2014.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singleton C. K., and Martin P. R. (2001) Molecular mechanisms of thiamine utilization. Curr. Mol. Med. 1, 197–207 10.2174/1566524013363870 [DOI] [PubMed] [Google Scholar]
- 21. Bunik V. I., Tylicki A., and Lukashev N. V. (2013) Thiamin diphosphate-dependent enzymes: from enzymology to metabolic regulation, drug design and disease models. FEBS J. 280, 6412–6442 10.1111/febs.12512 [DOI] [PubMed] [Google Scholar]
- 22. Frank R. A., Leeper F. J., and Luisi B. F. (2007) Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell. Mol. Life Sci. 64, 892–905 10.1007/s00018-007-6423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Settembre E., Begley T. P., and Ealick S. E. (2003) Structural biology of enzymes of the thiamin biosynthesis pathway. Curr. Opin. Struct. Biol. 13, 739–747 10.1016/j.sbi.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 24. Jurgenson C. T., Begley T. P., and Ealick S. E. (2009) The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603 10.1146/annurev.biochem.78.072407.102340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tylicki A., Lotowski Z., Siemieniuk M., and Ratkiewicz A. (2018) Thiamine and selected thiamine antivitamins: biological activity and methods of synthesis. Biosci. Rep. 38, BSR20171148 10.1042/BSR20171148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodionov D. A., Vitreschak A. G., Mironov A. A., and Gelfand M. S. (2002) Comparative genomics of thiamin biosynthesis in procaryotes: new genes and regulatory mechanisms. J. Biol. Chem. 277, 48949–48959 10.1074/jbc.M208965200 [DOI] [PubMed] [Google Scholar]
- 27. Begley T. P., Downs D. M., Ealick S. E., McLafferty F. W., Van Loon A. P., Taylor S., Campobasso N., Chiu H. J., Kinsland C., Reddick J. J., and Xi J. (1999) Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171, 293–300 10.1007/s002030050713 [DOI] [PubMed] [Google Scholar]
- 28. Webb E., and Downs D. (1997) Characterization of thiL, encoding thiamin-monophosphate kinase, in Salmonella typhimurium. J. Biol. Chem. 272, 15702–15707 10.1074/jbc.272.25.15702 [DOI] [PubMed] [Google Scholar]
- 29. McCulloch K. M., Kinsland C., Begley T. P., and Ealick S. E. (2008) Structural studies of thiamin monophosphate kinase in complex with substrates and products. Biochemistry 47, 3810–3821 10.1021/bi800041h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Melnick J., Lis E., Park J. H., Kinsland C., Mori H., Baba T., Perkins J., Schyns G., Vassieva O., Osterman A., and Begley T. P. (2004) Identification of the two missing bacterial genes involved in thiamine salvage: thiamine pyrophosphokinase and thiamine kinase. J. Bacteriol. 186, 3660–3662 10.1128/JB.186.11.3660-3662.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karunakaran R., Ebert K., Harvey S., Leonard M. E., Ramachandran V., and Poole P. S. (2006) Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J. Bacteriol. 188, 6661–6668 10.1128/JB.00641-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dhir S., Tarasenko M., Napoli E., and Giulivi C. (2019) Neurological, psychiatric, and biochemical aspects of thiamine deficiency in children and adults. Front. Psychiatry 10, 207 10.3389/fpsyt.2019.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nosaka K., Kaneko Y., Nishimura H., and Iwashima A. (1993) Isolation and characterization of a thiamin pyrophosphokinase gene, THI80, from Saccharomyces cerevisiae. J. Biol. Chem. 268, 17440–17447 [PubMed] [Google Scholar]
- 34. Imamura N., and Nakayama H. (1982) thiK and thiL loci of Escherichia coli. J. Bacteriol. 151, 708–717 10.1128/JB.151.2.708-717.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sullivan A. H., Dranow D. M., Horanyi P. S., Lorimer D. D., Edwards T. E., and Abendroth J. (2019) Crystal structures of thiamine monophosphate kinase from Acinetobacter baumannii in complex with substrates and products. Sci. Rep. 9, 4392 10.1038/s41598-019-40558-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishino H. (1972) Biogenesis of cocarboxylase in Escherichia coli: partial purification and some properties of thiamine monophosphate kinase. J. Biochem. 72, 1093–1100 10.1093/oxfordjournals.jbchem.a129996 [DOI] [PubMed] [Google Scholar]
- 37. Lee K. M., Go J., Yoon M. Y., Park Y., Kim S. C., Yong D. E., and Yoon S. S. (2012) Vitamin B12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect. Immun. 80, 1639–1649 10.1128/IAI.06161-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Donnenberg M. S., and Kaper J. B. (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59, 4310–4317 10.1128/IAI.59.12.4310-4317.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Philippe N., Alcaraz J. P., Coursange E., Geiselmann J., and Schneider D. (2004) Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255 10.1016/j.plasmid.2004.02.003 [DOI] [PubMed] [Google Scholar]
- 40. Nakayama H., and Hayashi R. (1972) Biosynthetic pathway of thiamine pyrophosphate: a special reference to the thiamine monophosphate-requiring mutant and the thiamine pyrophosphate-requiring mutant of Escherichia coli. J. Bacteriol. 112, 1118–1126 10.1128/JB.112.3.1118-1126.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Glasser N. R., Wang B. X., Hoy J. A., and Newman D. K. (2017) The pyruvate and α-ketoglutarate dehydrogenase complexes of Pseudomonas aeruginosa catalyze pyocyanin and phenazine-1-carboxylic acid reduction via the subunit dihydrolipoamide dehydrogenase. J. Biol. Chem. 292, 5593–5607 10.1074/jbc.M116.772848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khare G., Kar R., and Tyagi A. K. (2011) Identification of inhibitors against Mycobacterium tuberculosis thiamin phosphate synthase, an important target for the development of anti-TB drugs. PLoS ONE 6, e22441 10.1371/journal.pone.0022441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leonardi R., and Roach P. L. (2004) Thiamine biosynthesis in Escherichia coli: in vitro reconstitution of the thiazole synthase activity. J. Biol. Chem. 279, 17054–17062 10.1074/jbc.M312714200 [DOI] [PubMed] [Google Scholar]
- 44. Gigliobianco T., Lakaye B., Wins P., El Moualij B., Zorzi W., and Bettendorff L. (2010) Adenosine thiamine triphosphate accumulates in Escherichia coli cells in response to specific conditions of metabolic stress. BMC Microbiol. 10, 148 10.1186/1471-2180-10-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suzuki K., Yamada K., Fukuhara Y., Tsuji A., Shibata K., and Wakamatsu N. (2017) High-dose thiamine prevents brain lesions and prolongs survival of Slc19a3-deficient mice. PLoS ONE 12, e0180279 10.1371/journal.pone.0180279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Andrade J. A. A., Gayer C. R. M., Nogueira N. P. A., Paes M. C., Bastos V., Neto J., Alves S. C. Jr., Coelho R. M., da Cunha M., Gomes R. N., Aguila M. B., Mandarim-de-Lacerda C. A., Bozza P. T., and da Cunha S. (2014) The effect of thiamine deficiency on inflammation, oxidative stress and cellular migration in an experimental model of sepsis. J. Inflamm. (Lond.) 11, 11 10.1186/1476-9255-11-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gangolf M., Czerniecki J., Radermecker M., Detry O., Nisolle M., Jouan C., Martin D., Chantraine F., Lakaye B., Wins P., Grisar T., and Bettendorff L. (2010) Thiamine status in humans and content of phosphorylated thiamine derivatives in biopsies and cultured cells. PLoS ONE 5, e13616 10.1371/journal.pone.0013616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Odum E. P., and Wakwe V. C. (2012) Plasma concentrations of water-soluble vitamins in metabolic syndrome subjects. Niger. J. Clin. Pract. 15, 442–447 10.4103/1119-3077.104522 [DOI] [PubMed] [Google Scholar]
- 49. Cruz M. C., Goldstein A. L., Blankenship J. R., Del Poeta M., Davis D., Cardenas M. E., Perfect J. R., McCusker J. H., and Heitman J. (2002) Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21, 546–559 10.1093/emboj/21.4.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krucinska J., Lombardo M. N., Erlandsen H., Hazeen A., Duay S. S., Pattis J. G., Robinson V. L., May E. R., and Wright D. L. (2019) Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate. Sci. Rep. 9, 17106 10.1038/s41598-019-53301-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Choi K. H., Kumar A., and Schweizer H. P. (2006) A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 52. Filloux A., and Ramos J. L. (2014) Preface: Pseudomonas methods and protocols. Methods Mol. Biol. 1149 10.1007/978-1-4939-0473-0 [DOI] [PubMed] [Google Scholar]
- 53. Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M. V. and Manoil C. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 100, 14339–14344 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schweizer H. P. (2001) Vectors to express foreign genes and techniques to monitor gene expression in Pseudomonads. Curr Opin Biotechnol. 12, 439–445 10.1016/s0958-1669(00)00242-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the supporting information.