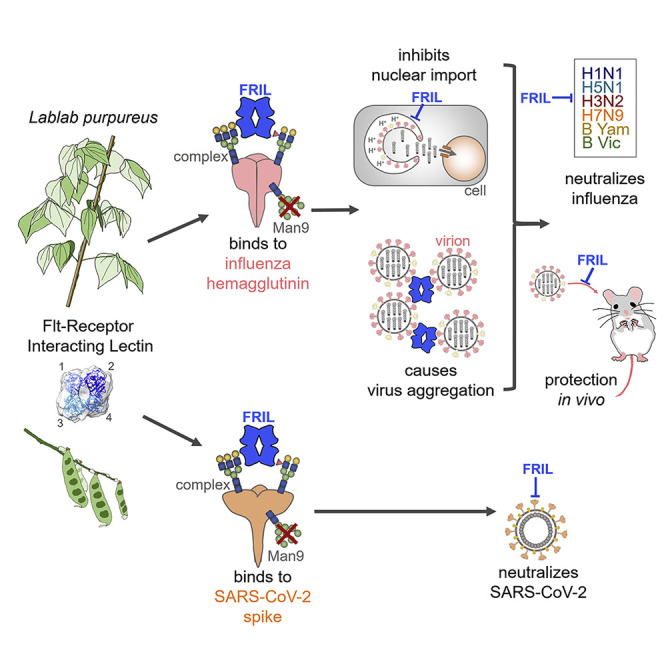
Abstract
The influenza virus hemagglutinin (HA) and coronavirus spike (S) protein mediate virus entry. HA and S proteins are heavily glycosylated, making them potential targets for carbohydrate binding agents such as lectins. Here, we show that the lectin FRIL, isolated from hyacinth beans (Lablab purpureus), has anti-influenza and anti-SARS-CoV-2 activity. FRIL can neutralize 11 representative human and avian influenza strains at low nanomolar concentrations, and intranasal administration of FRIL is protective against lethal H1N1 infection in mice. FRIL binds preferentially to complex-type N-glycans and neutralizes viruses that possess complex-type N-glycans on their envelopes. As a homotetramer, FRIL is capable of aggregating influenza particles through multivalent binding and trapping influenza virions in cytoplasmic late endosomes, preventing their nuclear entry. Remarkably, FRIL also effectively neutralizes SARS-CoV-2, preventing viral protein production and cytopathic effect in host cells. These findings suggest a potential application of FRIL for the prevention and/or treatment of influenza and COVID-19.
Keywords: lectin, influenza, coronavirus, SARS-CoV-2, antiviral, N-glycosylation, hemagglutinin, spike
Graphical Abstract
Highlights
-
•
FRIL is a plant lectin with potent anti-influenza and anti-SARS-CoV-2 activity
-
•
FRIL preferentially binds to complex-type N-glycans on viral glycoproteins
-
•
FRIL inhibits influenza virus entry by sequestering virions in late endosomes
-
•
Intranasal administration of FRIL protects against lethal H1N1 challenge in mice
Liu et al. demonstrate that FRIL, a plant lectin isolated from the hyacinth bean, has potent antiviral activity against SARS-CoV-2 and diverse influenza virus strains. FRIL is effective in vivo against H1N1. FRIL’s antiviral activity is mediated by binding to complex-type N-glycans on viral glycoproteins, interfering with viral entry.
Introduction
Each year, influenza virus infections cause more than half a million deaths worldwide (Lozano et al., 2012). Several neuraminidase (NA) inhibitors and a polymerase acidic protein (PA) inhibitor are available as therapeutics, with a third class, M2 inhibitors, obsolete due to widespread resistance (Heo, 2018). Many monoclonal antibodies that target the viral entry glycoprotein hemagglutinin (HA) are also in various stages of clinical development (Koszalka et al., 2017). However, due to the virus’ high mutational capacity, the spread of NA- and PA-inhibitor-resistant mutants are a concern (Yang et al., 2011; Imai et al., 2020), and mutations have been shown to decrease antibody binding to HA, resulting in reduced efficacy (Wu et al., 2014). Antiviral strategies that aim at components of the virus particle, such as post-translational glycosylations that incur a high fitness cost for mutation, can be advantageous for combating influenza.
A variety of complex, high-mannose and hybrid-type N-glycans are attached by cellular mechanisms to influenza surface glycoproteins. They are essential for correct protein folding and trafficking, affect HA receptor binding, and help the influenza virus evade host antibody detection through the shielding of immunogenic epitopes (Wu et al., 2017). The latter phenomenon of “glycan shield” has been proposed as to why most circulating human influenza viruses, in response to rising immunity in the population, have steadily increased the number of glycosylation sites on their HAs (Tate et al., 2014). The essential role of N-glycans on viral fitness, coupled with influenza virus’ limited ability to alter their composition, makes viral surface glycans a potential target for antiviral strategies.
Glycans are also commonly found for other enveloped viruses, of which the glycosylation profile varies among the surface proteins of each virus. The novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for the currently ongoing COVID-19 pandemic that has caused over 15 million reported cases worldwide and over 600,000 deaths as of July 2020, also contains the heavily glycosylated surface glycoprotein spike (S) protein. This glycoprotein gives the virion its namesake crown-like appearance and also mediates virus attachment and entry into the host cell. The SARS-CoV-2 S gene encodes 22 N-linked glycosylation sites per monomer, of which most feature complex- or hybrid-type glycans (Watanabe et al., 2020). Because no vaccines or therapeutics are currently available for COVID-19, studies of SARS-CoV-2 as a prime target for anti-viral lectins, especially those that bind to complex-type N-glycans, are of special interest and substantial importance.
Many lectins with antiviral properties have been discovered, with most being high-mannose-binding lectins directed against the heavily glycosylated human immunodeficiency virus (HIV) (Mitchell et al., 2017). Griffithsin (GRFT), a lectin isolated from the red algae Griffithsia sp., is currently in clinical trials as a topical vaginal gel for the prevention of HIV (Gianchecchi et al., 2019). A major obstacle for exogenous lectin treatment in vivo is their potential toxicity, as they may also recognize sugar moieties on host cells. So far, only the high-mannose-binding lectins Cyanovirin (CVN) and H84T Banlec have been shown to be protective against influenza virus challenge in mice by intranasal administration (Smee et al., 2008; Swanson et al., 2015). H84T Banlec is also intraperitoneally protective (Covés-Datson et al., 2020). Lectins that inhibit influenza virus and bind to complex-type glycans include the Nicotiana tabacum agglutinin and Urtica dioica agglutinin (Gordts et al., 2015; Balzarini et al., 1992).
Several mannose, N-acetylglucosamine (GlcNAc), and mannose/glucose-specific exogenous lectins can inhibit SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), and other mammalian and avian coronaviruses (O’Keefe et al., 2010; Millet et al., 2016; Kumaki et al., 2011; Hsieh et al., 2010; Greig and Bouillant, 1977), by interfering with coronavirus entry and egress through interactions with the viral S protein (Keyaerts et al., 2007). However, no reports on the effectiveness of anti-viral lectins against SARS-CoV-2 have been made.
Lablab purpureus, previously known as Dolichos lablab and commonly referred to as the hyacinth bean or lablab bean, is a legume in the Fabaceae family, mentioned in the Chinese traditional medicine text Compendium of Materia Medica as having properties of “strengthening the spleen and reducing dampness.” D. lablab lectin 1 (DLL-I) is a glucose/mannose lectin (Mo et al., 1999) isolated from the hyacinth bean. DLL-I has also been referred to as Flt3 Receptor Interacting Lectin (FRIL), after Colucci et al. (1999) discovered it having the unique property of sustaining hematopoietic progenitor cells in suspension culture by binding to cellular Flt3 receptors. FRIL has also been shown to preserve neural progenitor cells and evokes anti-tumor activity by reducing tumor neoangiogenesis through immunomodulation (Yao et al., 2008; Vigneshwaran et al., 2017). It is a typical legume lectin that has a 48% sequence identity to the well-known concanavalin A (ConA), with a similar β-prism type-II fold and one carbohydrate-binding domain (CBD) per monomer. Previous studies have suggested that FRIL is a glucose/mannose-specific lectin based on its affinity for the monosaccharides mannose, glucose, and N-acetylglucosamine, with a strong preference for the α-anomeric configuration (Mo et al., 1999). However, no study has been done on FRIL’s binding to higher-order sugars, such as the N-glycans commonly found on cell or viral glycoproteins, nor has FRIL been reported to have anti-viral activity.
Our current research stemmed from a screening of various ingredients used in Chinese traditional medicine for microneutralization (MN) activity against the influenza virus. From this screening, we discovered that the aqueous extract from Lablab purpureus has potent anti-influenza activity against a broad spectrum of influenza strains, and this activity was abrogated by heat or proteinase K treatment. The unexpected emergence of COVID-19 in the midst of our ongoing study compelled us to also explore its effects on SARS-CoV-2. Here, we elucidate that the protein FRIL isolated from this extract is responsible for our observed neutralization effect, and we characterize its neutralization breadth, potency, ligand binding, and stoichiometry, as well as its mechanism of action.
Results
Lablab Extract Neutralizes Influenza Virus
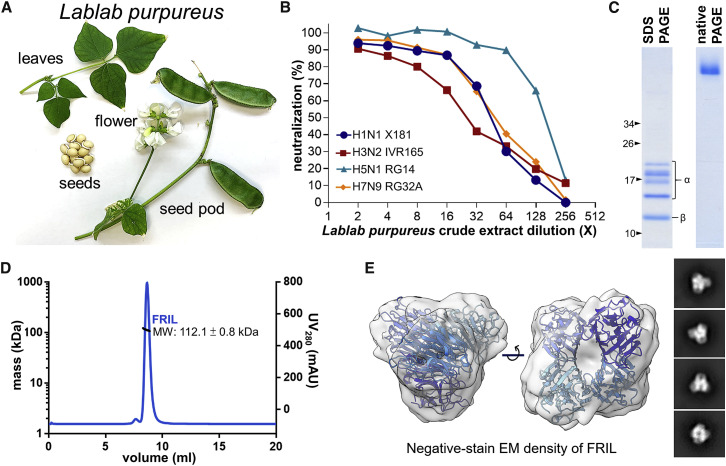
First, we assessed the MN ability of serially diluted crude aqueous extract of Lablab purpureus seeds (Figure 1 A) against four influenza vaccine strains spanning both group 1 and group 2 influenza A viruses, including A/California/7/2009-like (H1N1 X181), A/Vietnam/1194/2004-like (H5N1 RG14), A/Victoria/361/2011-like (H3N2 IVR-165), and A/Shanghai/2/2013-like (H7N9 RG32A). Results showed that Lablab crude extract exhibited neutralization ability against all four strains tested (Figure 1B).
Figure 1.
The Isolation and Characterization of FRIL from Lablab Extract
(A) Display of the plant Lablab purpureus.
(B) Microneutralization of Lablab purpureus seed crude aqueous extract against X181 (H1N1), RG14 (H5N1), IVR-165 (H3N2), and RG32A (H7N9) viruses. A single experiment was performed in this screening.
(C) Purified anti-viral reagent exhibits five bands on SDS-PAGE (left) that are confirmed as different truncations of the α and β subunits of FRIL by mass spectrometry. The same sample exhibits only one single band of higher molecular weight on native PAGE (right). Black arrows indicate molecular weights (kD) of the protein ladder for SDS-PAGE; no marker was used on native PAGE. Data are representative of 3 independent experiments.
(D) SEC-MALS of purified FRIL in solution (PBS) shows a single narrow peak. The MALS trace (black line) indicates a molecular mass of 112.1 ± 0.8 kDa. Data are representative of 2 independent experiments.
(E) Negative-stain EM density of purified FRIL (gray) fitted with its previously solved crystal structure (PDB: 1qmo) confirms its tetrameric state in solution, with different shades of blue for each monomer. Representative two-dimensional classes are shown on the right. See also Figure S1A.
FRIL Is Isolated and Characterized from Lablab Extract
To isolate the agent in the aqueous extract of Lablab purpureus seeds that is responsible for the observed influenza MN effect, we performed sequential protein fractionations, including ammonium sulfate precipitation, anion-exchange chromatography, size-exclusion, and affinity chromatography with Cibacron Blue beads. In each step, fractions with the highest MN titer against H5N1 RG14 were selected to proceed to the next step. Finally, five SDS-PAGE bands ranging from 10 to 20 kDa remain, resisting any further attempts at separation. The final fraction containing these five bands appeared as a single protein band when run on a native PAGE (Figure 1C). Additionally, this band was excised and eluted from the native PAGE, and the eluted protein retained neutralization activity against RG14. Mass spectrometry analysis revealed these five bands contained peptide sequences identical to the Lablab purpureus FRIL (Colucci et al., 1999) and Dolichos lablab lectin (DLL, later named DLL-I) peptide (Gowda et al., 1994), with the four higher kDa bands corresponding to FRIL’s differentially digested and N-glycosylated α subunit and the lowest band corresponding to the β subunit (B S et al., 2014).
FRIL has one CBD per monomer, but there have been conflicting reports on whether FRIL is a dimer or tetramer in solution (Mo et al., 1999; Gowda et al., 1994; Güran et al., 1983). To determine the oligomeric state of FRIL, we used size-exclusion chromatography with multiple angle light scattering (SEC-MALS), dynamic light scattering (DLS), and negative-stain electron microscopy (EM). SEC-MALS analysis suggests that our purified FRIL forms a 112.1-kDa tetramer in solution (Figure 1D). Using negative-staining EM, we saw a tetrameric electron density map with a ∼27-Å resolution (Figure 1E; Figure S1A), similar to its previously reported crystal structure (Hamelryck et al., 2000). DLS results also showed that FRIL’s size resembles known tetrameric legume lectin ConA, which has a similar tertiary structure as FRIL (Figure S1B).
FRIL Has Potent Anti-influenza Activity In Vitro and In Vivo
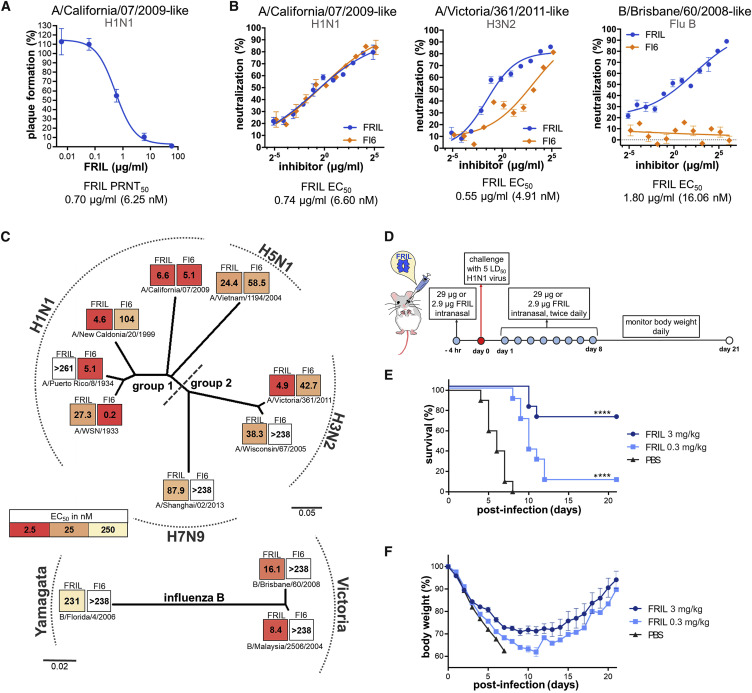
The anti-influenza activity of FRIL in vitro was assessed with plaque reduction assay and MN. FRIL exhibited a 50% plaque reduction neutralization test (PRNT50) value of 0.697 μg/mL against X181, similar to its MN half maximal effective concentration (EC50) of 0.74 μg/mL (Figures 2A and 2B; Figure S2A). Next, we explored the MN breadth of the purified FRIL with a representative panel of 11 influenza viruses spanning group 1 (H1N1), group 2 (H3N2), and influenza B (Yamagata and Victoria linages), along with avian strains of H7N9 and H5N1. The EC50 results were compared with the broadly neutralizing influenza antibody (bnAb) FI6v3 (Corti et al., 2011). With the exception of the laboratory strain A/Puerto Rico/8/1934 (PR8), FRIL was able to achieve low nanomolar levels of EC50 against group 1 viruses (H1N1 and H5N1) and H3N2. FRIL also had nanomolar neutralization titers against H7N9 and influenza B, ranging from an EC50 of 8.39 nM for a B/Malaysia/2506/2004-like virus to 231.4 nM for a B/Florida/4/2006-like virus (Figure 2C; Figure S2B). The EC50 values of FI6v3 were in a similar range to what has been previously reported (Corti et al., 2011). In addition, a FRIL MN experiment comparing either egg-produced or MDCK-cell-produced X181 viruses shows that the source of virus does not affect FRIL’s EC50 (Figure S2C).
Figure 2.
FRIL Exhibits Potent Broad-Spectrum Anti-influenza Activity In Vitro and In Vivo
(A) FRIL plaque reduction assay with H1N1 X181 virus. Data are representative of 3 independent experiments performed in triplicate (mean ± SEM). See also Figure S2A.
(B) FRIL (blue) and bnAb FI6v3 (FI6, orange) MN of H1N1 X181 (left), (middle) H3N2 IVR-165 (middle), and (right) an influenza B (B/Brisbane/60/2008-like) (right) vaccine strain. Data are representative of 2 independent experiments performed in triplicate (mean ± SEM) for each strain.
(C) HA phylogenetic tree created with MEGA-X shows microneutralization EC50 values (nM) of FRIL and bnAb FI6v3 (FI6) against 11 representative vaccine and laboratory strains of group 1, group 2, and influenza B viruses. Each block is colored by EC50 values (nM): the darker the color, the higher the neutralizing activity. Data are representative of 2 independent experiments performed in triplicate (mean ± SEM) for each strain. See also Figure S2B.
(D) Treatment schedule of X181 challenge and FRIL administration. A total of 29 or 2.9 μg of FRIL protein was given intranasally to BALB/c mice (n = 10) 4 h before challenge. Influenza virus intranasal challenge was conducted using 5 LD50 of X181 virus, and 29 or 2.9 μg of FRIL protein was then given intranasally twice per day for 8 days following challenge.
(E and F) Survival (E) and body weight (F) were tracked for 21 days following influenza virus challenge. Intranasal administration for the PBS group was halted after 4 days due to declining conditions of mice. Significance (compared to PBS group) was determined by log-rank (Mantel-Cox) test; ∗∗∗∗p < 0.0001.
Having demonstrated FRIL’s anti-influenza effects in vitro, we next evaluated FRIL’s in vivo activity by intranasal administration in mice. FRIL was first given intranasally to 10 BALB/c mice 4 h before intranasal infection with 5 LD50 (median lethal dose) of X181 virus. Afterward, FRIL was administered every 12 h for 8 days, and survival and body weight were monitored for 21 days post-infection (Figure 2D). We found that 58.6 μg/day (2.93 mg/kg/day) FRIL intranasal treatment offered protection in 7 out of 10 mice, with the 3 deaths delayed compared to PBS controls. At a 5.86-μg/day (0.29 mg/kg/day) treatment, the median time of death was also significantly delayed by 4 days (Figures 2E and 2F), although only 1/10 mice survived.
Taken together, these results demonstrate that the purified FRIL possesses broad-spectrum anti-influenza activity in vitro and has dose-dependent anti-viral activity when given intranasally in vivo.
FRIL Neutralizes Only Influenza Viruses with Complex-Type N-Glycans
Although previous reports have shown FRIL to be a mannose/glucose-specific lectin, its affinity to higher-order sugar structures found on influenza envelope proteins have not been studied. To explore what types of oligosaccharides FRIL binds to, we created differentially glycosylated egg-based influenza viruses of H1N1 X181 by subjecting them to treatment by the mannosidase I inhibitor kifunensine (KIF) during seeding into the allantoic cavity of eggs, followed by a high-mannose-cleaving endoglycosidase H (endo H) after harvest (Tseng et al., 2019). Four types of virus particles were thus created: no treatment (both complex and high-mannose-type glycans naturally exist on the virus surface), KIF-treated (high mannose only), endo-H-treated (complex glycans remain intact, high-mannose glycans digested down to single GlcNAc), and KIF/endo H-treated (single GlcNAc only) (Figure 3 A). These viruses were then purified by sucrose density gradient centrifugation, and its N-glycan constituency was confirmed by glycopeptide analysis with tandem mass spectrometry (Figure S3A).
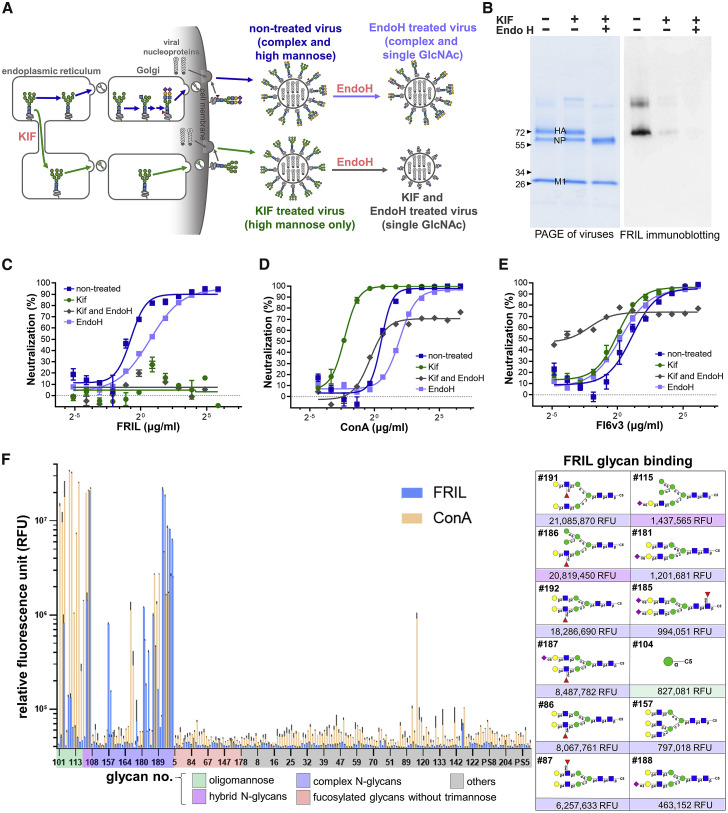
Figure 3.
FRIL Binds to and Neutralizes Only Influenza Viruses with Complex-Type N-Glycans
(A) Schematic diagram showing the generation of non-treated (KIF (–) Endo H (–); complex- and high-mannose-type glycans), KIF-treated (KIF (+) Endo H (–); high-mannose-type glycans only), endo-H-treated (KIF (–) Endo H (+); complex- and single-GlcNAc residues on former high-mannose sites), and KIF- and endo-H-treated (KIF (+) Endo H (+); single-GlcNAc residue on all N-glycosylation sites) influenza virus particles.
(B) SDS-PAGE (left) and FRIL immunoblotting (right) of non-treated, KIF-treated, and KIF- and endo-H-treated lysed virus particles. FRIL immunoblotting was done by incubating FRIL with viral proteins transferred onto the membrane, followed by detection with anti-FRIL antibodies. Data are representative of 3 individual experiments.
(C–E) Microneutralization assay of non-treated (blue square), endo-H-treated (light blue square), KIF-treated (green circle), and KIF- and endo-H-treated (gray triangle) viruses with FRIL (C), ConA (D), and bnAb FI6v3 (E). Mean ± SEM of three replicates.
(F) Left: glycan array with Cy3-labeled FRIL (blue) and ConA (orange). Right: list of the 12 glycans that bound to FRIL with the highest intensity. Symbol Nomenclature for Glycans (SNFG) is used to represent oligosaccharides on the graph (blue square for GlcNAc, green circle for mannose, yellow circle for galactose, red triangle for fucose, and purple diamond for NeuAc). Data are representative of 2 independent experiments performed in triplicate (mean ± SEM).
We tested the binding of FRIL to these three types of differentially glycosylated viruses by FRIL immunoblotting (where lysed whole viruses were blotted onto a PVDF membrane, then probed with FRIL) and live virus ELISA. Immunoblotting showed FRIL binds primarily to HA, the glycoprotein that mediates virus attachment and entry (Figure 3B). Surprisingly, FRIL bound to non-treated, mostly complex-type virus particles at a higher intensity than KIF-treated high-mannose viruses, which is different from the oligosaccharide binding affinity of most other known antiviral lectins (Figure 3B). In a live virus ELISA, (Figure S3B) FRIL also showed significantly higher binding to complex-type particles compared with high-mannose or single GlcNAc particles, in spite of possible interference by glycolipids and other non-specific components of the glycocalyx in this assay. This preference for complex-type glycans is also reflected in MN assays: FRIL shows neutralization only against non-treated and endo-H-treated viruses containing complex-type sugars, but not against high-mannose and single-GlcNAc viruses (Figure 3C). In contrast, the well-documented high-mannose-binding lectin ConA had the highest neutralization titers against KIF-treated viruses (Figure 3D), and bnAb FI6v3 (an HA stem-specific antibody possibly affected by steric hindrance of N-glycans near its epitope) (Magadán et al., 2014) showed the best neutralization activity against virus particles that contained only a single GlcNAc (Figure 3E).
To further investigate this interesting phenomenon, we used glycan array analysis with fluorescent dye Cy3-labeled FRIL (Figure 3F). FRIL demonstrated the best binding to complex-type N-glycans with α1-3 or α1-4 fucosylated sub-terminal GlcNAc, including Galβ1-4(Fucα1-3)GlcNacβ1-2Man-R (Lewis X/SSEA-1/CD15-carrying N-glycans) and Galβ1-4(Fucα1-4)GlcNacβ1-2Man-R, ranging from 21,085,870 to 6,257,633 relative fluorescent units (RFUs). Slightly weaker binding was seen on non-terminally fucosylated complex- and hybrid-type N-glycans (1,437,565 to 463,152 RFUs), as well as its previously documented binding to the singular mannose residue (827,081 RFUs) and GlcNAc-linked trimannose (142,360 RFUs). Signals from oligomannose residues Man9 (49,933 RFUs) and Man5 (64,093 RFUs) or Lewis X (39,490 RFUs) and Lewis A (42,606 RFUs) antigens that were not attached to the N-glycan trimannose core were no higher than other unrelated glycan structures. Terminal desialyation, as would be anticipated from influenza NA activity, did not appear to affect FRIL binding (Figure S3C). In contrast, Cy3-labeled ConA exhibited the strongest binding to oligomannose structures, such as Man3, Man5, and Man9. The result of direct Cy3 labeling was confirmed by using polyclonal anti-FRIL antibodies (Figure S3D).
In summary, our glycan array analysis supports our assays with differentially glycosylated influenza viruses, indicating that FRIL binds preferentially to complex type N-glycans and only neutralize viruses with complex-type glycans on their surface. Compared to other high-mannose-binding anti-viral lectins in previous studies, such as GRFT, CVN, and H84T Banlec, the complex-type-binding FRIL may have an advantage in targeting viral glycoproteins whose complex-type glycans are in majority, such as HA of H1N1 and S of SARS-CoV-2. The molecular mechanism underlying such a binding preference is, however, yet to be revealed.
FRIL Sequesters Influenza Virions in Late Endosome and Prevents Nuclear Entry
Due to the surprising finding that FRIL’s oligosaccharide preference is different from most known antiviral lectins, we were interested in its antiviral mechanism. First, a competitive MN inhibition assay using monosaccharides shows that the two sugars that can inhibit FRIL hemagglutination, namely, α-methylmannopyranoside and D-glucose, were also able to inhibit FRIL MN (Figure 4 A), suggesting that FRIL’s observed neutralization effect is dependent on its carbohydrate-binding function. Second, pre-treatment of cells with either FRIL or virus, followed by 18 h of incubation with the opposite agent (without either coming into direct contact with one another) both failed to neutralize the influenza virus (Figure 4B), confirming that FRIL is an inhibitor of virus entry and must bind directly to the virus particle.
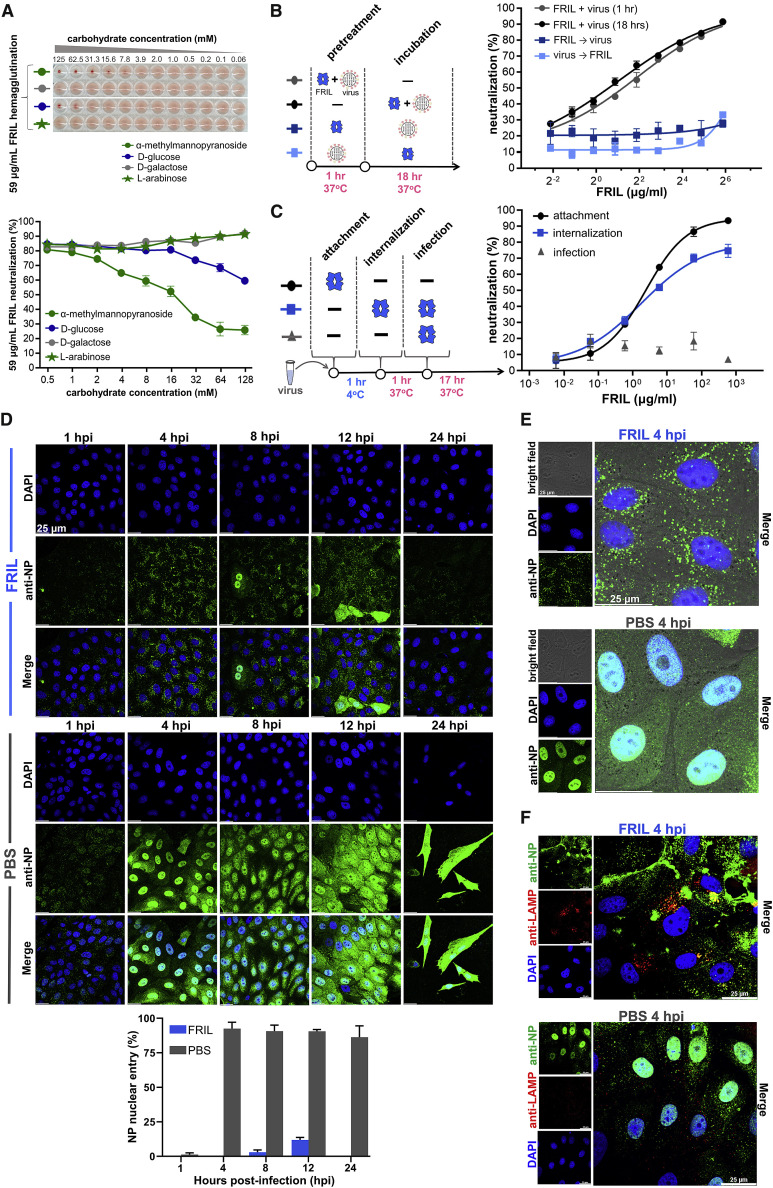
Figure 4.
FRIL Halts Influenza Virus Entry in the Late Endosome
(A) Top: hemagglutination inhibition of 59 μg/ml FRIL (4 HAU) by the monosaccharides α-methylmannopyranoside (green circle), D-galactose (gray circle), D-glucose (blue circle), and L-arabinose (green star). The panel is representative of three replicates. Bottom: competitive FRIL microneutralization inhibition with the same monosaccharides as the top panel. Mean ± SEM of three replicates.
(B) FRIL microneutralization where FRIL and virus were added, either together (circle) or sequentially (square), in a 1-h pre-treatment step, after which they were removed or subject to an ensuing 18-h incubation step. Mean ± SEM of three replicates.
(C) FRIL microneutralization with a 1-h low-temperature arrest of viral endocytosis, separating viral entry steps into attachment (black circle), internalization (blue square), and infection (gray triangle), with FRIL added during the different steps. Data are representative of 3 independent experiments performed in triplicate (mean ± SEM).
(D) Progression of influenza RNP after virion endocytosis with or without FRIL inhibition, visualized by immunofluorescence tracking of viral NP protein (anti-NP; green) and nuclei (DAPI; blue). Quantification of the top panel plus two others for the percentage of FRIL-treated (blue) and PBS-treated (gray) cells with nuclear NP signal is shown in the bottom panel. Error bars represent SEM. Data are representative of 3 independent experiments.
(E) Higher magnification showing NP signal (anti-NP; green) clustered in the perinuclear region at 4 hpi for FRIL (top panel), as opposed to RNP entry into cell nucleus (DAPI, blue) for PBS control (bottom panel).
(F) Co-localization (yellow) of influenza NP signal (green) with the late endosomal marker LAMP-1 (red) is observed in FRIL-treated cells at 4 hpi (top panel) but not in PBS control (bottom panel). Data are representative of 2 independent experiments.
Influenza virus cell entry includes the sequential steps of virus attachment, endocytosis, uncoating, and nuclear import. To test whether FRIL blocks attachment, we used a FRIL hemagglutination inhibition assay (HAI) against H1N1, H3N2, H5N1, and H7N9 viruses (Figure S4A). In all four strains, no HAI was observed up to 11.7 μg/mL FRIL. An ELISA done on influenza NP in MDCK cells given 1 h to bind with virus also showed no significant difference in virus attachment whether FRIL was added or not (Figure S4B). We therefore conclude FRIL does not inhibit influenza virus attachment. The next step after virus attachment is cell endocytosis, a process that can be delayed by lowering the temperature to 4°C (Matlin et al., 1981). To confirm that FRIL’s neutralization effect occurs after virus attachment, virus particles were added to the cell surface at low temperatures so that attachment takes place but endocytosis is inhibited, and FRIL was then added to the virus-attached cells (Figure 4C). Under this condition, FRIL exhibited a similar neutralization EC50 compared with when the lectin and virus were mixed and added to cells together (EC50s of 4.25 μg/mL and 2.35 μg/mL, respectively). In contrast, giving the virus 1 h at 37°C to be endocytosed before FRIL treatment results in a complete loss of neutralization, indicating that FRIL exerts its antiviral effect after virus attachment, and only after being in contact with the virus before its endocytosis and subsequent infection (Figure 4C).
Finally, we followed the progression of the influenza virus after its endocytosis into the host cell by tracking the viral NP protein with immunofluorescence staining by anti-NP antibody (Figures 4D and 4E). Multiple-round infections after the initial addition of virus to the slide were inhibited by a lack of L-(tosylamido-2-phenyl) ethyl chloromethyl ketone-treated trypsin (TPCK-trypsin) in cell culture medium. In non-treated controls (PBS), the virus is carried by endosomes to the perinuclear region at 1 h post-infection (hpi), where the virus would then undergo endosomal acidification and HA-mediated viral fusion. At 4 hpi, the viral ribonucleoprotein complex (vRNP; including NP) for the non-treated control was observed in the nucleus, where influenza transcription and replication take place. The production of new NP in the cytoplasm was seen at 8 and 12 hpi, and widespread cell death occurred at 24 hpi for the control group. In contrast, 33-μg/ml FRIL treatment caused a retention of NP signal in the perinuclear region for up to 12 hpi. Staining with the late endosomal/lysosomal marker LAMP-1 confirmed that the NP signal remained in late endosomes/lysosomes (Figure 4F). NP signals became weaker by 24 hpi (Figure 4D), presumably due to eventual breakdown in lysosomes. A ConA treatment of 33 μg/mL also exhibited a pattern of numerous NP vacuoles sequestered in the perinuclear region at 4 hpi (Figure S4C), while treatment of MDCK cells with FRIL alone in the absence of influenza infection found that FRIL binds mostly to the cell membrane (Figure S4D). FRIL immunoblotting under different pH conditions was used to confirm that FRIL is active at endosomal pH (Figure S4E). A trypsin susceptibility assay performed with recombinant HA showed that unlike the HA-stem-targeting antibody FI6, FRIL does not prevent pH-dependent HA conformational change (Figure S4F).
Overall, our results establish that FRIL exerts its anti-influenza effect by binding to the virus, halting its infection in the late endosome/lysosome, and preventing its nuclear import through ways other than affecting HA conformational change for virus fusion.
FRIL Displays Multiple Layers or Dimensions of Multivalency
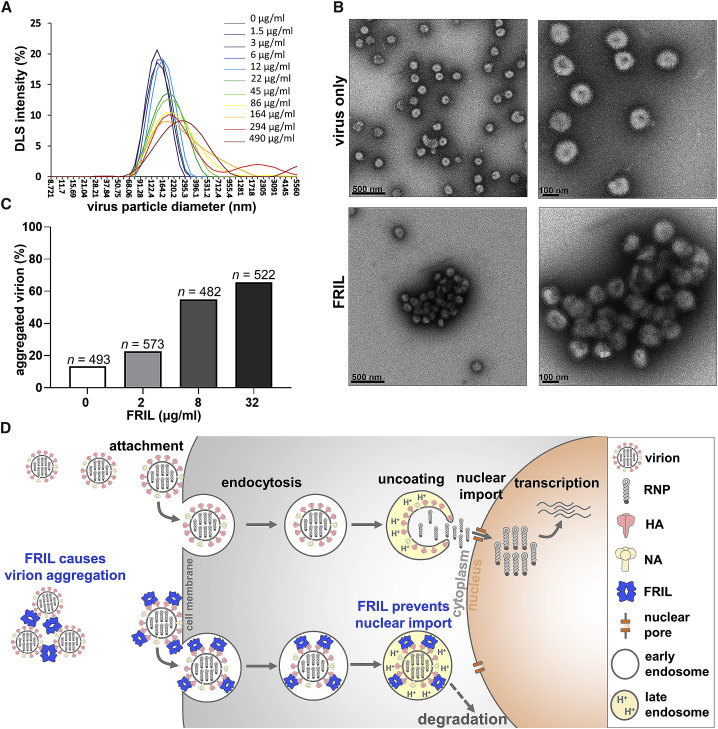
During our immunofluorescence staining of influenza NP, we noticed granules of large, intense anti-NP signal in our FRIL-treated group but not in the PBS-treated control. As FRIL is a tetramer (Figures 1D and 1E) capable of multivalent cross-linking (Hamelryck et al., 2000) and erythrocyte aggregation (Figure S4A), we suspect these granules are large aggregates of influenza virions agglutinated by FRIL displaying multiple dimensions of multivalency characteristic of potent anti-viral lectins (Lusvarghi et al., 2016). To explore this possibility, we first performed DLS analysis on purified influenza X181 virus particles that were then treated with 1.5 μg/mL to 490 μg/mL of FRIL. Starting at 22 μg/mL, an increase in particle diameter intensity distribution can be observed (147.6 nm ± 38.4 at 0 μg/mL increased to 202.7 nm ± 80.2 at 22 μg/mL of FRIL), indicating the formation of aggregates (Figure 5 C). In contrast, no such aggregation was observed for KIF- and endo-H-treated viruses (with only a single GlcNAc at each N-glycosylation site) even at the highest concentration used (145.2 nm ± 39.7 at 0 μg/mL compared with 151.2 nm ± 47.0 at 490 μg/mL of FRIL) (Figure S5).
Figure 5.
FRIL Aggregates Influenza Virus Particles
(A) Dynamic light scattering (DLS) analysis of virus particle aggregation under increasing concentrations of FRIL (from 1.5 μg/ml in purple line to 490 μg/ml in dark red line).
(B) Negative-stain EM images of purified X181 virions alone (top panels) and aggregated X181 virus particles after mixing with 150 μg/ml FRIL (bottom panels). Data are representative of 3 independent experiments.
(C) Quantitation of aggregated virions calculated from 20 images for each FRIL concentration. Virions that directly contact each other are considered aggregated. Concentrations above 32 μg/ml proved difficult to ascertain due to the formation of large overlapping aggregates.
(D) A proposed model for FRIL’s anti-influenza mechanism: large FRIL/virus aggregations may occur outside the cell to prevent virus entry, while FRIL endocytosed together with the virus may trap the virus in the late endosome and prevent its nuclear import.
Under negative-stain EM, large three-dimensional aggregations of overlapping influenza X181 particles were observed at a 150-μg/mL FRIL concentration, whereas little aggregation was seen in untreated virus particles (Figure 5B). Quantification of aggregation was performed by manually counting virus particles in close proximity to one another (Figure 5C). We observed a dose-dependent increase in the percentage of aggregates up to 32 μg/mL FRIL, although concentrations higher than that formed densely packed clumps of layered viruses that make it difficult to visually quantify individual particles.
These results indicate that FRIL’s outward projection of four CBDs is capable of bridging multiple virus particles and creating large, three-dimensional aggregations of influenza virions at 32–150 μg/mL. The clustering of virions may partially explain the FRIL-mediated inhibitory mechanism of viral entry and nuclear import and will serve as the first step toward a molecular understanding of these events. Furthermore, endogenous airway lectins SP-D and MBL are known to aggregate influenza virus particles and facilitate viral clearance by the immune system in vivo (Hartshorn et al., 1997), and it is possible that FRIL has a similar effect.
FRIL Exhibits Potent Neutralization of SARS-CoV-2 but Not HIV
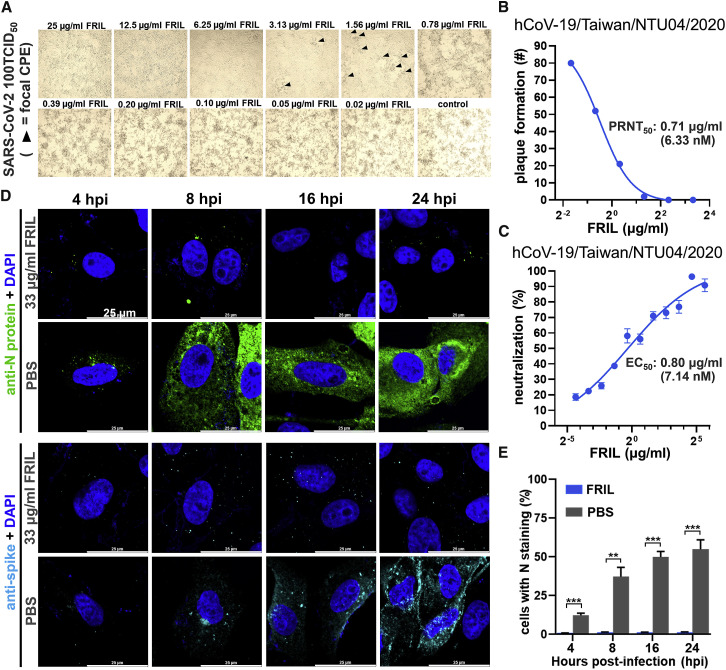
We assessed the antiviral activity of FRIL against the SARS-CoV-2 strain hCoV-19/Taiwan/NTU04/2020 with microscopic observation of cytopathic effect (CPE), a plaque reduction neutralization assay visualized by crystal violet staining, and MN assay with polyclonal anti-SARS-CoV-2 nucleoprotein (N). From direct observation of CPE, the Vero cell monolayer was morphologically unaffected by virus introduction down to 6.25 μg/mL of FRIL, with focal CPE appearing as the FRIL concentration dropped to 3.13–1.56 μg/mL, and widespread cell detachment below 0.39 μg/mL (Figure 6 A). In the plaque reduction neutralization assay, FRIL exhibited a PRNT50 value of 0.71 μg/mL (6.36 nM) against hCoV-19/Taiwan/NTU04/2020 (Figure 6B; Figure S6B) after 4 days of incubation, similar to its MN EC50 of 0.80 μg/mL (7.15 nM) (Figure 6C).
Figure 6.
FRIL Exhibits Potent Neutralization of SARS-CoV-2
(A) FRIL inhibition of Vero E6 CPE under 100 50% infective dose (TCID50) of SARS-CoV-2 (hCoV-19/Taiwan/NTU04/2020). Black triangles represent areas of focal CPE, whereas FRIL concentrations below 0.78 μg/ml (6.96 nM) were unable to prevent diffuse and widespread CPE. Data are representative of 2 experiments.
(B) FRIL plaque reduction neutralization assay with hCoV-19/Taiwan/NTU04/2020 virus. A single experiment was performed. See also Figure S6B.
(C) FRIL MN of hCoV-19/Taiwan/NTU04/2020. Data are representative of 2 experiments performed in quadruplicate (mean ± SEM).
(D) Immunofluorescence tracking of SARS-CoV-2 N (green) and S protein (cyan) production at 4 to 24 hpi, with or without 33 μg/ml (0.29 μM) FRIL inhibition. Nuclei are counterstained with DAPI (blue). Data are representative of 2 experiments.
(E) N-protein-positive cells at different time points after infection, with (blue bars) or without (gray bars) 33 μg/ml (0.29 μM) of FRIL. Three images with 519∼1,279 cells per image were quantified using ImageJ software. Significance versus PBS control was determined by multiple t tests with Prism 8 software (∗∗p < 0.01, ∗∗∗p < 0.001). See also Figure S6C.
Furthermore, we explored the effect of FRIL on viral protein production over time inside Vero E6 cells by using polyclonal anti-SARS-CoV-2 N protein and SARS-CoV-2 S protein antibodies at 1, 2, 4, 8, 16, and 24 hpi (Figures 6D and 6E; Figure S6C). In non-FRIL-treated controls (PBS), viral N protein signals were observed inside cell punctae starting at 4 hpi, and the percentage of cells that stained strongly with N protein signal in its cytosol (signifying new N protein production) steadily increased from 8 hpi onward. In contrast, very little N protein production was seen inside FRIL-treated cells up to 24 hpi. Viral S protein displayed a similar trend, although the patchy distribution of the membrane-bound S protein makes them harder to quantify.
In contrast to FRIL’s potent effects against SARS-CoV-2, MN using pseudotyped HIV-1 JR-FL virus only yielded a moderate EC50 of 8.41 μg/mL (74.96 nM; Figure S6A), and more importantly, FRIL was not able to achieve a >90% neutralization even at the highest concentration used (316 μg/mL).
FRIL Binds to SARS-CoV-2 S Protein
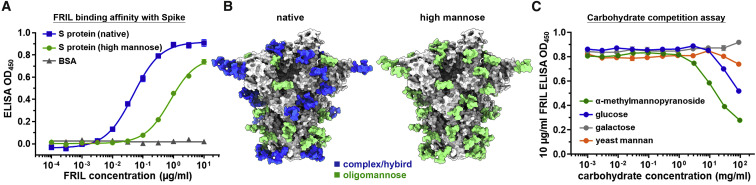
ELISA was used to determine FRIL’s binding affinity to the recombinant SARS-CoV-2 S protein produced in HEK293T (with native glycosylation) and HEK293S (GnTI−; high-mannose N-glycosylation only) cells. We found FRIL was able to bind to recombinant S protein at concentrations as low as 10 ng/mL (89.1 picomole), and the binding to S proteins with predominantly complex-type N-glycans (native glycosylation) is 30 times stronger than those with only high-mannose glycans (Figure 7 A). This result is consistent with the published glycan profile of SARS-CoV-2 S protein that shows most of its glycosylation sites are complex type or hybrid, except two sites (N234 and N709) which are >80% high mannose (Figure 7B).
Figure 7.
FRIL Binds to Spike Protein by Complex-Type Glycans
(A) ELISA of FRIL binding to SARS-CoV-2 S protein, with either native N-glycosylation (containing complex type, hybrid, and high mannose glycans; blue square) or high mannose only (green circle). The unglycosylated bovine serum albumin (BSA; gray triangle) was used as control. Mean ± SEM of three replicates.
(B) Cryo-EM structure of SARS-CoV-2 S protein (EMD-30419; PDB: 7CN9) modeled with glycans according to either the published glycan profile (left; native) or the high-mannose (Man5) type produced by HEK293S cell line (right). Glycans were colored blue (>50% complex or hybrid) or green (>50% oligomannose). See also Figure S7.
(C) ELISA values of 10 μg/ml FRIL under competitive inhibition from the monosaccharides α-methylmannopyranoside (green), D-glucose (blue), and D-galactose (gray) and yeast mannan from Saccharomyces cerevisiae (orange). Mean ± SEM of three replicates.
Competitive inhibition of FRIL’s S protein binding was performed with the monosaccharides α-methylmannopyranoside, glucose, galactose, and yeast mannan from Saccharomyces cerevisiae (Figure 7C). Results show that, as expected, glucose and the α-anomeric configuration of mannose were able to inhibit FRIL binding to S protein, whereas galactose, which is not a ligand of FRIL, had no such effect. Yeast mannan had only a slight inhibitory effect on FRIL binding, in contrast to the single α-anomeric mannose.
Discussion
The evolutionary dynamics of sustained influenza virus circulation in a population with pre-existing immunity favors the addition of N-glycans to surface glycoproteins, masking immunogenic epitopes that would otherwise be recognized by the host immune system (Tate et al., 2014). Encountering carbohydrate-binding agents that target this glycan shield puts the virus in a difficult position, in which mutations that delete surface glycans will render the virus more susceptible to antibody recognition and the addition of glycans increase their vulnerability to lectins. This carbohydrate-binding agent concept has been hypothesized as a novel strategy in the treatment of HIV (Balzarini, 2005).
A panel of 11 influenza viruses showed that FRIL has potent MN effects against group 1, group 2, and influenza B viruses and against strains without heavily N-glycosylated HAs, such as WSN (2 sites) and RG32A (3 sites). Only a weak correlation exists between the number of predicted glycans on the virus and its FRIL EC50, indicating that aside from the number of N-glycan sites, the type and position of N-glycans also play a role in FRIL neutralization. The laboratory strain PR8, which does not have any glycosylation on its HA head and has previously been found to be resistant to CVN (O’Keefe et al., 2003), was also resistant to FRIL.
The discovery that FRIL showed stronger binding to complex-type glycans than high mannose was unexpected, as most exogenous lectins that have antiviral properties interact predominantly with high-mannose structures (Mitchell et al., 2017). FRIL has previously been characterized as a mannose/glucose lectin that bound to both α1-3- and α1-6-linked mannose but did not precipitate yeast mannans (Mo et al., 1999). Our glycan array results confirm that FRIL binds to single mannose and branched trimannoside as Mo et al. (1999) reported, but it also had an equal or stronger affinity to various complex-type N-glycans, especially those with α1-3 or α1-4 fucosylated sub-terminal GlcNAc. It is worth noting that, as Figures 7A, 7C, and S3C suggest, FRIL does not entirely abhor attachment to oligomannose glycans. The binding is just considerably weaker.
This affinity for complex-type glycans may explain why FRIL was able to demonstrate good neutralization against X181 and SARS-CoV-2, but not HIV. Watanabe et al. (2020) have determined that complex- and hybrid-type glycans comprise 71% of all N-glycosylation on the SARS-CoV-2 S protein, with oligomannose and unoccupied taking up the remaining 28% (Watanabe et al., 2020). Our own mass spectrometry results revealed that 62%–67% of glycosylation on egg-origin influenza X181 HA were complex type, with 33%–38% oligomannose (Tseng et al., 2019; Figure S3A). In contrast, HIV-1 JR-FL Env protein N-glycosylation is ∼60% oligomannose (Struwe et al., 2018). This could explain why the high-mannose-binding lectin GRFT exhibited a lower EC50 in HIV than in coronavirus, whereas FRIL had the opposite effect. Nevertheless, all three viruses contain a mixture of oligomannose and complex-type glycans, so it might be worthwhile to explore the possibility of synergistic combinations of high-mannose lectins with FRIL to cover a broader spectrum of possible oligosaccharides.
However, this brings up the concern that because complex-type N-glycans are commonly expressed on host cell glycoproteins, FRIL given intranasally would bind to host cells, inducing adverse effects. This trepidation can be alleviated by the fact that our in vivo challenge experiment at the highest dosage of 2.9 mg/kg/day FRIL was well tolerated. In contrast, 2 mg/kg/day of CVN treatment was found to be lethal to mice (Smee et al., 2008). Furthermore, previous studies that used an intraperitoneal administration route for FRIL was well tolerated at dosages as high as 30 mg/kg, and no significant cytotoxicity was observed in A549 and cancer cell lines (Vigneshwaran et al., 2017).
FRIL’s strong binding to antennary-Lewis-X-carrying N-glycans may offer a possible explanation for previous reports on its ability to preserve hematopoietic or neural progenitor cells in culture (Colucci et al., 1999; Yao et al., 2008). Lewis X, otherwise known as SSEA-1 or CD15, is a known undifferentiation marker found on stem cells. Although its active role in embryonic development is still unknown, it has been reported that antennary Lewis X serves as an activator of Notch signaling and maintenance of neural stem cell stemness (Yagi et al., 2012). This may explain why Yao et al. (2008) found Notch upregulated after neuronal progenitor cells were treated with FRIL.
A recent report indicates that the molecularly engineered H84T Banlec inhibits influenza virus uncoating at the late endosome/lysosome stage (Covés-Datson et al., 2020). In our current study, we observed a similar phenomenon for both the high-mannose binding ConA and the complex-type binding FRIL (Figure 4E; Figure S4D), hinting that this might be a common mechanism of anti-influenza action for diverse categories of lectins. However, pre-treatment of cells with FRIL 1 h before virus infection did not neutralize the virus (Figure 4B), and most of FRIL remained extracellular when FRIL was applied to non-infected cells (Figure S4E), indicating that unlike H84T, FRIL must first bind to the virus particle before being endocytosed. These results, coupled with our finding that FRIL aggregates influenza virions, allows us to put forth a model for FRIL’s anti-influenza action (Figure 5D): FRIL first binds and extracellularly cross-links virions, which results in either large aggregates rapidly cleared by the host immune system, or the FRIL-virus complex is endocytosed into host cells. The FRIL-bound virus is subsequently retained in the late endosome/lysosome and prevented from nuclear entry, until its ultimate degradation 24 hpi. However, there is the possibility that the mechanism for FRIL inhibition of SARS-CoV-2 may be different from influenza virus, given that we did not observe strong N protein signal retained inside cell punctae in FRIL-treated samples, and further investigations will be needed for a more complete mechanistic understanding.
An issue that remains unresolved in our current study is generating recombinant FRIL. This may or may not cause problems in practical application of this agent, as other antiviral lectins such as H84T Banlec and Q-GRFT have used extensive molecular engineering to uncouple mitogenicity (Swanson et al., 2015) or increase oxidation resistance (Corman et al., 2020). We have attempted various approaches, including using prokaryotic and yeast cells, but so far production of bioactive recombinant FRIL remains elusive. It is possible that FRIL undergoes extensive post-translational processing comparable to the structurally similar lectin ConA (Chrispeels et al., 1986).
In conclusion, we found that FRIL is a tetrameric lectin with potent anti-influenza and anti-SARS-CoV-2 activity. It preferentially binds to complex-type N-glycans to halt influenza virus entry at the late endosomal stage, and we have demonstrated that FRIL is effective both in vitro and in vivo. Furthermore, FRIL’s neutralizing ability is at least on par with most known antiviral, neutralizing monoclonal antibodies. We believe its utility as a preventive or therapeutic agent in influenza and the current COVID-19 pandemic warrants further investigation: for example, to coat it on masks or be included in aerosol mists in a closed space, such as an airplane cabin for reducing transmission, or to be used in an inhaler (like Relenza for influenza), which will require vigilant clinical trials to evaluate its safety and efficacy.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-NP polyclonal antibody | This study | N/A |
| Rabbit anti-FRIL polyclonal antibody | This study | N/A |
| Mouse anti-SARS-CoV-2 N protein | This study | N/A |
| Mouse anti-SARS-CoV-2 S protein | This study | N/A |
| Rabbit anti-LAMP1 polyclonal antibody | abcam | Cat# ab24170; RRID: AB_775978 |
| Goat anti-Rabbit IgG Alexa Fluor 647 | abcam | Cat# ab150079; RRID: AB_2722623 |
| Goat anti-Mouse IgG Alexa Fluor 488 | Invitrogen | Cat# A32723; RRID: AB_2633275 |
| Goat anti-Rabbit IgG Alexa Fluor 488 | Invitrogen | Cat# A32731; RRID: AB_2633280 |
| Goat anti-Rabbit IgG HRP | Jackson ImmunoResearch | Cat# 111-035-144; RRID: AB_2307391 |
| FI6 | This study | N/A |
| Bacterial and Virus Strains | ||
| A/California/7/2009-like | Adimmune | N/A |
| A/Vietnam/1194/2004-like | Adimmune | N/A |
| A/Victoria/361/2011-like | Adimmune | N/A |
| A/Wisconsin/67/2005-like | Adimmune | N/A |
| A/Shanghai/2/2013-like | Adimmune | N/A |
| B/Brisbane/60/2008-like | Adimmune | N/A |
| B/Florida/04/2006-like | Adimmune | N/A |
| B/Malaysia/2506/2004-like | Adimmune | N/A |
| A/WSN/1933 | NIBSC | N/A |
| A/New Caledonia/20/1999 | NIBSC | N/A |
| A/Puerto Rico/8/1934 | NIBSC | N/A |
| hCoV-19/Taiwan/NTU04/2020 | NTU | N/A |
| Biological Samples | ||
| Lablab purpureus | Market in Hong Kong | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant A/California/09 (H1N1) hemagglutinin ectodomain | This study | N/A |
| Recombinant SARS-CoV-2 spike protein ectodomain | This study | N/A |
| Recombinant endoglycosidase H | This study | N/A |
| Kifunensine | Sigma-Aldrich | CAS 109944-15-2 |
| DAPI staining solution | abcam | ab228549 |
| Concanavalin A | Sigma-Aldrich | L7647 |
| Mannan from Saccharomyces cerevisiae | Sigma-Aldrich | M7504 |
| Deposited Data | ||
| Negative stain EM structure of FRIL tetramer | This study | EMD-30380 |
| Cryo-EM structure of SARS-CoV-2 spike protein | This study | EMD-30419, PDB: 7CN9 |
| Experimental Models: Cell Lines | ||
| Madin-Darby canine kidney | ATCC | CCL-34 |
| A549 | ATCC | CCL-185 |
| Vero E6 | ATCC | CRL-1586 |
| HEK293T | ATCC | CRL-3216 |
| HEK293S GnTI- | ATCC | CRL-3022 |
| Experimental Models: Organisms/Strains | ||
| BALB/c | LASCo | N/A |
| Embryonated chicken eggs | AHRI | N/A |
| Software and Algorithms | ||
| Prism 8.0 | Graphpad software | N/A |
| ImageJ | NIH | N/A |
| ChimeraX | UCSF | N/A |
| cisTEM | Tim Grant, Alexis Rohou, Nikolaus Grigorieff | N/A |
| Relion | MRC Laboratory, Cambridge, UK | N/A |
| Coot | Emsley et al. | NA |
| Phenix | Liebschner et al., 2019 | NA |
| Other | ||
| Turkey red blood cells | Jianrong | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Che Ma (cma@gate.sinica.edu.tw).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Negative stain EM density of FRIL and cryo-EM map/structure of SARS-CoV-2 spike protein generated during this study have been deposited in the Electron Microscopy Data Bank. Negative stain EM structure of FRIL tetramer (EMD-30380), cryo-EM structure of SARS-CoV-2 spike protein (EMD-30419, PDB: 7CN9).
Experimental Model and Subject Details
Virus strains
The vaccine strains of A/California/7/2009-like, A/Vietnam/1194/2004-like, A/Victoria/361/2011-like, A/Wisconsin/67/2005-like, A/Shanghai/2/2013-like, B/Brisbane/60/2008-like, B/Florida/04/2006-like, and B/Malaysia/2506/2004-like viruses were obtained from Adimmune Corporation, Taichung, Taiwan. A/WSN/1933, A/New Caledonia/20/1999, and A/Puerto Rico/8/1934 were obtained from National Institute for Biological Standards and Control, Hertfordshire, UK. hCoV-19/Taiwan/NTU04/2020 was obtained from National Taiwan University, Taipei, Taiwan.
Plants and cell lines
Madin-Darby canine kidney (CCL-34), A549 (CCL-185) and Vero E6 (CRL-1586) cell lines were obtained from American Type Culture Collection, Manassas, Virginia. The seeds of Lablab purpureus were purchased from a market in Hong Kong.
Animals
9 day old embryonated chicken eggs were obtained from Animal Health Research Institute, New Taipei City, Taiwan. 8 week old female BALB/c mice were obtained from LASCo, Taipei, Taiwan.
Method Details
FRIL purification
Lablab purpureus bean powder was first extracted by PBS, then dialyzed overnight with decreased salt concentration. The sediment was resolubilized in 20 mM phosphate buffer pH 8, and loaded onto an Unosphere Q column (BioRad, Hercules, California). Bound proteins were eluted with a 0∼0.5M NaCl gradient, and fractions exhibiting the highest MN titers (peak 2) against RG14 (H5N1) strain were pooled and concentrated. The concentrated sample was then loaded onto a Superdex s200 10/300GL size exclusion column (GE, Boston, Massachusetts), and fractions exhibiting the highest MN titers (peak 4) against RG14 were pooled and concentrated. Finally, the 5 bands representing FRIL were separated from 2 nonspecific bands at approximately 30 and 40 kDa by collecting cibaron blue affinity chromatography (Affi-Gel. BioRad) flow-through.
Microneutralization and plaque reduction assays
The 50% infective dose (TCID50) and immunoplaque assay (PFU/ml) of viruses in MDCK or Vero E6 cells were determined beforehand. A protocol for the serological diagnosis of influenza by MN assay was used, with FRIL in place of sera (World Health Organization, 2011). FRIL and viruses were incubated at 37°C for 1 hour in a 96 well tissue culture plate, then 1.5x104 cells/well were added to the mixture. The plate was then cultured in serum-free medium for 18∼20 hours, then washed and fixed with 50% methanol 50% acetone. Anti-NP (influenza virus) or anti-N (coronavirus) ELISA was then used to determine virus titer. Plates were blocked with 5% skim milk 0.5% BSA, and rabbit polyclonal anti-NP or mouse polyclonal anti-N primary antibody and HRP-conjugated secondary antibody were sequentially added. Peroxidase substrate solution (TMB) and 1M H2SO4 stop solution were used and the absorbance (OD 450 nm) read by a microplate reader (Victor3. Perkin Elmer, Waltham, Massachusetts).
For plaque reduction assay, MDCK or Vero E6 cells were plated onto a 6-well plate at 2x105 cells/well overnight for 90% confluence. FRIL and viruses were co-incubated at 37°C for 1 hour, before the mixture is added onto the monolayer for another hour. The virus/FRIL mixture is aspirated, the cells washed with PBS, and a 0.5% low-melting agarose in serum-free media is layered onto the cells. The plates are allowed to solidify at room temperature for 30 minutes, then incubated at 37°C for 4∼5 days or until cytopathic effects (CPE) are observed. Afterward, cells are fixed with 7.4% formalin 1% tween 20, and agarose plugs removed. For influenza virus, immunoplaque assay performed with rabbit polyclonal anti-nucleoprotein (NP) primary antibody and HRP-conjugated secondary antibody, and plaques are visualized by incubating with KPL TruBlue peroxidase substrate (Seracare, Milford, Massachusetts) overnight. For coronavirus, the plate was stained with 0.5% crystal violet.
Production and purification of differentially-glycosylated viruses
The method for egg-based production of differentially-glycosylated viruses has been described previously (Tseng et al., 2019). Briefly, 9 day old embryonated chicken eggs were inoculated with A/California/7/2009-like (X181) virus at 10,000-fold dilution of seed stock in allantoic cavity, with or without the presence of 0.2 mg/ml KIF. After 48 hr incubation (35°C), the allantoic fluid was harvested and concentrated, and half of the KIF-treated virus were subjected to endoglycosidase H (20 μg/ml final concentration) treatment overnight at 4°C. All three treatments (no treatment, KIF, KIF and EndoH) were loaded onto 25x89 centrifugal tubes with a 25∼55% sucrose gradient, and influenza virus particles purified using sucrose gradient centrifugation (Optima L-90K. Beckman Coulter, Brea, California) at 20000 rpm overnight. Finally, sucrose is removed from the purified viruses through dialysis (10k MWCO Snakeskin tubing. Thermo Fisher Scientific, Waltham, Massachusetts) with PBS.
Glycan array analysis
Glycan array analysis was performed as described previously (Shivatare et al., 2016). Briefly, FRIL was conjugated with Cy3 in a 1:1 ratio, and bioactivity checked with hemagglutination assay. Glycan microarray slides were blocked for 1 hour with blocking buffer (Superblock, Thermo Fisher Scientific) and then washed with PBST (PBS buffer, 0.05% Tween 20). 10 μg/ml conjugated FRIL-Cy3 was added to the array and incubated at room temperature for 1 hour, and washed to remove unbound FRIL. Microarray slides were spun dry prior to scanning with a GenePix 4300A reader (Molecular Devices, San Jose, California) at OD 532 nm and analyzed with GenePix Pro 7.0 software (Molecular Devices).
FRIL live-virus ELISA and Western Blotting
For FRIL ELISA, live sucrose-gradient purified X181 viruses were absorbed onto an empty ELISA plate at 4°C overnight. 10% BSA in PBS was used for blocking, then serially-diluted FRIL was added onto the plate. Polyclonal anti-FRIL primary antibody and HRP-conjugated secondary antibody were sequentially added. Peroxidase substrate solution (TMB) and 1M H2SO4 stop solution were used and the absorbance (OD 450 nm) read by a microplate reader (Victor3. Perkin Elmer). All steps up to the addition of stop solution were done in a biosafety cabinet.
For western blotting, purified X181 viruses were loaded onto a 4∼15% SDS-PAGE (Bio-Rad) with a non-reducing loading dye, the transferred onto PVDF membrane with semi-dry method (Trans-Blot SD, Bio-Rad). The membrane is blocked with 5% BSA in PBST for 1 hour at room temperature, then 1.2 μg/ml FRIL protein is added. The membrane was then sequentially treated with polyclonal anti-FRIL primary antibody and HRP-conjugated secondary antibody. Finally, Clarity ECL substrate (Bio-Rad) was added for chemiluminescence, and visualized with ImageQuant LAS4000 (GE).
Intranasal challenge in mice
Our mouse challenge experiments were performed by following an intranasal administration method described previously (Sidwell et al., 1998), with some modifications. Briefly, LD50 of the A/California/7/2009-like (X181) virus in BALB/c mice were determined before experiments. Ten 8 week old female BALB/c mice per group were sedated with tiletamine and zolazepam (Zoletil. Virbac, Carros, France) i.p. and given a pre-infection 50 μL intranasal dose of FRIL, at 29 or 2.9 μg/dose. Control group was given PBS. 4 hours after treatment, 5LD50 of X181 virus was given intranasally to all groups. Thereafter, 29 or 2.9 μg/dose of FRIL was given i.n. every 12 hours for 8 days, and mice body weight and survival were recorded for 21 days. Intranasal administration of the PBS group was halted after 4 days due to declining condition of mice.
All animal experiments were conducted in accordance with the guidelines established by the Institutional Animal Care and Use Committee of Academia Sinica (approval no. 18-12-1272), and all procedures were performed by a licensed veterinarian. Animals were humanely sacrificed by CO2 inhalation at the end of experiment.
Dynamic light scattering
Purified X181 viruses (no treatment, KIF and endo H-treated) were incubated with increasing concentrations of FRIL protein at 37°C for 30 minutes. The mixture was then transferred to a plastic cuvette and measured with Zetasizer Nano-ZS (Malvern Instruments, Malvern, UK).
SEC-MALS
FRIL size exclusion chromatography was performed by running the lectin through a silicon-based BioSEC-3 column (Agilent Technologies, Santa Clara, California), on an Akta FPLC (GE) connected to a three-angle light-scattering detector (mini-DAWN TREOS) and a refractive index detector (Optilab T-rEX. Wyatt Technology, Santa Barbara, California). Data analysis was done with ASTRA.
Hemagglutination inhibition assay
The hemagglutination titer of FRIL was determined beforehand. Methyl-α-D-mannopyranoside, D-galactose, D-glucose and L-arabinose were serially diluted in V-bottom 96-well plates with PBS, 25 μl/well. 25 μL of 59 μg/ml FRIL (4.1 HAU) were then added to each well, followed by 50 μL of 0.5% turkey red blood cells (Jianrong, New Taipei City, Taiwan). After 30 minutes of incubation, hemagglutination inhibition titers were assessed by RBC sedimentation at the bottom of the wells.
Immunofluorescence microscopy
MDCK or Vero E6 cells were seeded onto 35x12mm glass bottom dishes (αplus, Taoyuan City, Taiwan) at 2.75x105 cells/ml for 16 hours. Purified X181 or unpurified hCoV-19/Taiwan/NTU04/2020 virus (MOI: 1.2 and 1, respectively) were incubated with 100 μg/ml (final concentration: 33 μg/ml) of lectin for 1 hour before the mixture was added to dishes for 1∼24 hours of infection. Afterward, cells were fixed and permeabilized before being immunostained with either rabbit polyclonal anti-NP (1:1000 dilution), mouse polyclonal anti-N (1:1000 dilution), mouse polyclonal anti-S (1:100 dilution), or rabbit polyclonal anti-FRIL (1:10000 dilution) antibody and Alexa Fluor 488-labeled goat anti-rabbit secondary antibody. Counterstaining was performed with DAPI. Samples were analyzed by a Leica TCS SP8X confocal microscope with HC PL APO CS2 63x/1.40 oil immersion lens (Leica AG, Wetzlar, Germany).
Negative stain electron microscopy
For negative stain EM density of the oligomeric FRIL protein, FRIL (18 μg/mL) was applied for 60 s to a carbon-coated 400 mesh copper grid (Electron Microscopy Sciences, Hatfield, Pennsylvania), then negatively stained with 2% uranyl formate for 60 s. Data was collected under a FEI Tecnai G2 F20 S-TWIN electron microscope (Thermo Fisher Scientific) operating at 120 keV and a magnification of 470K that resulted in a pixel size of 2.06 Å at the specimen plane. Particles selection, 2D classification and 3D reconstruction were processed by cisTEM. 67,041 particles were chosen to reconstruct the 3D map using a D2 symmetry. Structural figures were generated by UCSF ChimeraX package.
For visualization and counting of virus particle aggregation, sucrose-gradient purified X181 viruses and FRIL were co-incubated at 37°C for 30 minutes. After incubation, samples were diluted with PBS and applied to a carbon-coated 400 mesh copper grid and negatively stained with Nano-W (Nanoprobes, Yaphank, New York). Viruses were observed under transmission electron microscope (JEM-1400. JEOL, Peabody, Massachusetts) operating at 120 kV coupled to a CCD camera (Gatan 895. Gatan, Pleasanton, California). Images were captured by Gatan Digital Micrographic software at the magnifications of 2.6K and 5K. Virus aggregation was quantified by calculating the percentage of virus aggregates captured on 20 images (four corners of 5 randomly-chosen intact grids) under 5K magnification.
Cryo-EM
HEK293E (Ebna) and HEK293S cells were used to overexpress the SARS-CoV-2 Spike protein (GenBank: YP_009724390.1) ectodomain (14-1209) by transfection with polyethylenimine (PEI) in FreeStyle 293 expression medium (Life technologies) at 37°C with 8% CO2 for 6 days. The supernatant was harvested and purified using Ni-NTA affinity resin (GE Healthcare) and further purified by size-exclusion chromatography Superose 6 10/300 GL (GE Healthcare) in a buffer containing 20 mM Tris pH 8.0 and 100 mM NaCl. 4 μL of fresh purified protein sample (0.48mg/ml) were loaded onto a fresh glow discharged (60 s) holey carbon grid (Quantifoil R1.2/1.3) and plunge freezing with a Vitrobot Mark IV (Thermo Fisher Scientific) at 3 s blot time in 4°C and 100% humidity. Automated data collection was performed by the EPU software (Thermo Fisher Scientific) in a Titan Krios G3 operating at 300 kV with Gatan BioQuantum energy filter and K2 camera. Total of 3,417 micrographs were recorded at magnification of 165,000 × (0.82 Å/pixel) with a defocus range between −1.0 μm to −2.5 μm. The total dose rate was approximately 57 e-/Å2 for 60 frames in a 4.5 s exposure time. Movie alignment, contrast-transfer function (CTF) estimation and particle extraction were carried out using cisTEM (Grant et al., 2018). The particle stacks were transferred to Relion 3.0 (Zivanov et al., 2018) for 2D and 3D classification followed by CTF refinement, 3D auto-refine and post-processing without symmetry. The overall resolution (4.7Å) of cryo-EM map was reported by the gold-standard FSC with 0.143 cut-off. UCSF ChimeraX (Goddard et al., 2018) and Coot (Emsley et al., 2010) were used to fit atomic models (PDB 6VSB) into the final map. The model was manually rebuilt using Coot (Emsley et al., 2010) and subsequently real-space refined by Phenix (Liebschner et al., 2019). N-linked glycans were hand-modeled using Coot Glyco extension based on the published glycan profile (Watanabe et al., 2020). Structure figures were generated using UCSF ChimeraX.
Trypsin susceptibility assay
Trypsin susceptibility assay was performed as described previously (Kadam et al., 2017). Briefly, recombinant A/California/7/2009 HA was diluted to 2 mg/ml in PBS (pH 7.4), and FRIL (FRIL:HA 10:1 or 2:1 molar ratio) or FI6 (2:1 molar ratio) were added and allowed to incubate for 30 minutes at room temperature. A 200 mM sodium acetate buffer (pH 5) was then added to the mixture to lower the pH down to 5, and incubated at 37°C for 20 minutes. A 200 mM Tris buffer (pH 8.5) was used to bring the pH back up to 7. Finally, TPCK-trypsin was added to the mixture at a 1:50 molar ratio and the mixture incubated at 37°C for 30 minutes before the digestion was stopped by the addition of SDS-PAGE loading dye (non-reducing), and denatured at 100°C for 10 mins. Samples were run on a 4∼15% SDS-PAGE (Bio-Rad) to assess trypsin susceptibility.
Quantification and Statistical Analysis
All data are presented as mean ± SEM except Figures 1B and 6B, where only a single sample was tested against each virus. Absolute EC50 values were calculated with Prism 8 software. For Figure 2E significance was determined by log-rank (Mantel-Cox) test. For Figure 6E significance versus PBS control was determined by multiple t tests. For Figure S3 significance was determined by 2-way ANOVA with Tukey’s multiple comparison. For all data, significance is presented as p < 0.05 (∗), with p < 0.01 (∗∗), p < 0.001 (∗∗∗) and p < 0.0001 (∗∗∗∗) indicated by an increasing number of asterisks. The n number of individual experiments are mentioned in figure legends.
Acknowledgments
We thank Sue-Jane Chen and Yung-Chieh Tseng for insightful discussions. We thank Academia Sinica Biological Electron Microscopy Core Facility for EM technical support. The core facility is funded by the Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-119). We thank Academia Sinica Cryo-EM Center (grant no. AS-CFII-108-110) and Taiwan Protein Project (grant no. AS-KPQ-109-TPP2). We thank Chien-Hung Chen for glycopeptide liquid chromatography MS/MS analysis and Tsung-Wei Su for SEC-MALS operation. Funding of this project was provided by Academia Sinica Genomics Research Center Summit Project AS-SUMMIT-109 and Taiwan Ministry of Science and Technology grant 108-2113-M-001-014 (to C.M.).
Author Contributions
C.M., J.-T.J., C.-H.W., C.-Y.W., and Y.-M.L. designed the experiments; C.M. and Y.-M.L. wrote the paper; X.C. and Y.-M.L. worked on figures and diagrams. J.-T.J., M.S.-A.-M., and Y.-M.L. carried out the initial antiviral screening and animal studies; K.-S.L. performed the glycan array; M.S.-A.-M. conducted immunofluorescent imaging; J.-T.J. and Y.-M.L. did the MNs and other in vitro work; J.M.L. created pseudotyped HIV; T.-H.C. performed FRIL EM and 3D reconstruction of FRIL EM images; J.M.L., X.C., Y.-M.W., and M.-C.H. performed SARS-CoV-2 spike EM and 3D reconstruction of spike EM images.
Declaration of Interests
A patent application has been submitted by Academia Sinica based on the results shown in this study, with C.M., J.-T.J., and Y.-M.L. as inventors.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108016.
Supplemental Information
References
- B S G.K., Pohlentz G., Schulte M., Mormann M., Nadimpalli S.K. N-glycan analysis of mannose/glucose specific lectin from Dolichos lablab seeds. Int. J. Biol. Macromol. 2014;69:400–407. doi: 10.1016/j.ijbiomac.2014.05.073. [DOI] [PubMed] [Google Scholar]
- Balzarini J. Targeting the glycans of gp120: a novel approach aimed at the Achilles heel of HIV. Lancet Infect. Dis. 2005;5:726–731. doi: 10.1016/S1473-3099(05)70271-1. [DOI] [PubMed] [Google Scholar]
- Balzarini J., Neyts J., Schols D., Hosoya M., Van Damme E., Peumans W., De Clercq E. The mannose-specific plant lectins from Cymbidium hybrid and Epipactis helleborine and the (N-acetylglucosamine)n-specific plant lectin from Urtica dioica are potent and selective inhibitors of human immunodeficiency virus and cytomegalovirus replication in vitro. Antiviral Res. 1992;18:191–207. doi: 10.1016/0166-3542(92)90038-7. [DOI] [PubMed] [Google Scholar]
- Chrispeels M.J., Hartl P.M., Sturm A., Faye L. Characterization of the endoplasmic reticulum-associated precursor of concanavalin A. Partial amino acid sequence and lectin activity. J. Biol. Chem. 1986;261:10021–10024. [PubMed] [Google Scholar]
- Colucci G., Moore J.G., Feldman M., Chrispeels M.J. cDNA cloning of FRIL, a lectin from Dolichos lablab, that preserves hematopoietic progenitors in suspension culture. Proc. Natl. Acad. Sci. USA. 1999;96:646–650. doi: 10.1073/pnas.96.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman J.M., Hamorsky K.T., Shepherd J.W., Hiatt E., Fuqua J.L., Palmer K.E. Stability of plasmid and viral banks supporting the cGMP manufacture of Q-Griffithsin from a TMV-based viral vector. J. Biotechnol. 2020;320:74–76. doi: 10.1016/j.jbiotec.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Covés-Datson E.M., King S.R., Legendre M., Gupta A., Chan S.M., Gitlin E., Kulkarni V.V., García J.P., Smee D.F., Lipka E. A molecularly engineered antiviral banana lectin inhibits fusion and is efficacious against influenza virus infection in vivo. Proc. Natl. Acad. Sci. USA. 2020;117:2122–2132. doi: 10.1073/pnas.1915152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianchecchi E., Torelli A., Montomoli E. The use of cell-mediated immunity for the evaluation of influenza vaccines: an upcoming necessity. Hum. Vaccin. Immunother. 2019;15:1021–1030. doi: 10.1080/21645515.2019.1565269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordts S.C., Renders M., Férir G., Huskens D., Van Damme E.J., Peumans W., Balzarini J., Schols D. NICTABA and UDA, two GlcNAc-binding lectins with unique antiviral activity profiles. J. Antimicrob. Chemother. 2015;70:1674–1685. doi: 10.1093/jac/dkv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda L.R., Savithri H.S., Rao D.R. The complete primary structure of a unique mannose/glucose-specific lectin from field bean (Dolichos lab lab) J. Biol. Chem. 1994;269:18789–18793. [PubMed] [Google Scholar]
- Grant T., Rohou A., Grigorieff N. cisTEM, user-friendly software for single-particle image processing. eLife. 2018;7:e35383. doi: 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig A.S., Bouillant A.M. Binding effects of concanavalin A on a coronavirus. Can. J. Comp. Med. 1977;41:122–126. [PMC free article] [PubMed] [Google Scholar]
- Güran A., Tichá M., Filka K., Kocourek J. Isolation and properties of a lectin from the seeds of the Indian bean or lablab (Dolichos lablab L.) Biochem. J. 1983;209:653–657. doi: 10.1042/bj2090653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelryck T.W., Moore J.G., Chrispeels M.J., Loris R., Wyns L. The role of weak protein-protein interactions in multivalent lectin-carbohydrate binding: crystal structure of cross-linked FRIL. J. Mol. Biol. 2000;299:875–883. doi: 10.1006/jmbi.2000.3785. [DOI] [PubMed] [Google Scholar]
- Hartshorn K.L., White M.R., Shepherd V., Reid K., Jensenius J.C., Crouch E.C. Mechanisms of anti-influenza activity of surfactant proteins A and D: comparison with serum collectins. Am. J. Physiol. 1997;273:L1156–L1166. doi: 10.1152/ajplung.1997.273.6.L1156. [DOI] [PubMed] [Google Scholar]
- Heo Y.A. Baloxavir: First Global Approval. Drugs. 2018;78:693–697. doi: 10.1007/s40265-018-0899-1. [DOI] [PubMed] [Google Scholar]
- Hsieh L.E., Lin C.N., Su B.L., Jan T.R., Chen C.M., Wang C.H., Lin D.S., Lin C.T., Chueh L.L. Synergistic antiviral effect of Galanthus nivalis agglutinin and nelfinavir against feline coronavirus. Antiviral Res. 2010;88:25–30. doi: 10.1016/j.antiviral.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Yamashita M., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Kiso M., Murakami J., Yasuhara A., Takada K., Ito M., Nakajima N. Influenza A variants with reduced susceptibility to baloxavir isolated from Japanese patients are fit and transmit through respiratory droplets. Nat. Microbiol. 2020;5:27–33. doi: 10.1038/s41564-019-0609-0. [DOI] [PubMed] [Google Scholar]
- Kadam R.U., Juraszek J., Brandenburg B., Buyck C., Schepens W.B.G., Kesteleyn B., Stoops B., Vreeken R.J., Vermond J., Goutier W. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Pannecouque C., Van Damme E., Peumans W., Egberink H., Balzarini J., Van Ranst M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 2007;75:179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszalka P., Tilmanis D., Hurt A.C. Influenza antivirals currently in late-phase clinical trial. Influenza Other Respir. Viruses. 2017;11:240–246. doi: 10.1111/irv.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y., Wandersee M.K., Smith A.J., Zhou Y., Simmons G., Nelson N.M., Bailey K.W., Vest Z.G., Li J.K.K., Chan P.K.-S. Inhibition of severe acute respiratory syndrome coronavirus replication in a lethal SARS-CoV BALB/c mouse model by stinging nettle lectin, Urtica dioica agglutinin. Antiviral Res. 2011;90:22–32. doi: 10.1016/j.antiviral.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V., Abraham J., Adair T., Aggarwal R., Ahn S.Y. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusvarghi S., Lohith K., Morin-Leisk J., Ghirlando R., Hinshaw J.E., Bewley C.A. Binding Site Geometry and Subdomain Valency Control Effects of Neutralizing Lectins on HIV-1 Viral Particles. ACS Infect. Dis. 2016;2:882–891. doi: 10.1021/acsinfecdis.6b00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadán J.G., Altman M.O., Ince W.L., Hickman H.D., Stevens J., Chevalier A., Baker D., Wilson P.C., Ahmed R., Bennink J.R., Yewdell J.W. Biogenesis of influenza a virus hemagglutinin cross-protective stem epitopes. PLoS Pathog. 2014;10:e1004204. doi: 10.1371/journal.ppat.1004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin K.S., Reggio H., Helenius A., Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol. 1981;91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Séron K., Labitt R.N., Danneels A., Palmer K.E., Whittaker G.R., Dubuisson J., Belouzard S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antiviral Res. 2016;133:1–8. doi: 10.1016/j.antiviral.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C.A., Ramessar K., O’Keefe B.R. Antiviral lectins: Selective inhibitors of viral entry. Antiviral Res. 2017;142:37–54. doi: 10.1016/j.antiviral.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo H., Meah Y., Moore J.G., Goldstein I.J. Purification and characterization of Dolichos lablab lectin. Glycobiology. 1999;9:173–179. doi: 10.1093/glycob/9.2.173. [DOI] [PubMed] [Google Scholar]
- O’Keefe B.R., Smee D.F., Turpin J.A., Saucedo C.J., Gustafson K.R., Mori T., Blakeslee D., Buckheit R., Boyd M.R. Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin. Antimicrob. Agents Chemother. 2003;47:2518–2525. doi: 10.1128/AAC.47.8.2518-2525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe B.R., Giomarelli B., Barnard D.L., Shenoy S.R., Chan P.K., McMahon J.B., Palmer K.E., Barnett B.W., Meyerholz D.K., Wohlford-Lenane C.L., McCray P.B., Jr. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010;84:2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivatare S.S., Chang S.H., Tsai T.I., Tseng S.Y., Shivatare V.S., Lin Y.S., Cheng Y.Y., Ren C.T., Lee C.C., Pawar S. Modular synthesis of N-glycans and arrays for the hetero-ligand binding analysis of HIV antibodies. Nat. Chem. 2016;8:338–346. doi: 10.1038/nchem.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell R.W., Huffman J.H., Barnard D.L., Bailey K.W., Wong M.H., Morrison A., Syndergaard T., Kim C.U. Inhibition of influenza virus infections in mice by GS4104, an orally effective influenza virus neuraminidase inhibitor. Antiviral Res. 1998;37:107–120. doi: 10.1016/s0166-3542(97)00065-x. [DOI] [PubMed] [Google Scholar]
- Smee D.F., Bailey K.W., Wong M.H., O’Keefe B.R., Gustafson K.R., Mishin V.P., Gubareva L.V. Treatment of influenza A (H1N1) virus infections in mice and ferrets with cyanovirin-N. Antiviral Res. 2008;80:266–271. doi: 10.1016/j.antiviral.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe W.B., Chertova E., Allen J.D., Seabright G.E., Watanabe Y., Harvey D.J., Medina-Ramirez M., Roser J.D., Smith R., Westcott D. Site-Specific Glycosylation of Virion-Derived HIV-1 Env Is Mimicked by a Soluble Trimeric Immunogen. Cell Rep. 2018;24:1958–1966.e5. doi: 10.1016/j.celrep.2018.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.D., Boudreaux D.M., Salmon L., Chugh J., Winter H.C., Meagher J.L., André S., Murphy P.V., Oscarson S., Roy R. Engineering a therapeutic lectin by uncoupling mitogenicity from antiviral activity. Cell. 2015;163:746–758. doi: 10.1016/j.cell.2015.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate M.D., Job E.R., Deng Y.M., Gunalan V., Maurer-Stroh S., Reading P.C. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses. 2014;6:1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.C., Wu C.Y., Liu M.L., Chen T.H., Chiang W.L., Yu Y.H., Jan J.T., Lin K.I., Wong C.H., Ma C. Egg-based influenza split virus vaccine with monoglycosylation induces cross-strain protection against influenza virus infections. Proc. Natl. Acad. Sci. USA. 2019;116:4200–4205. doi: 10.1073/pnas.1819197116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneshwaran V., Thirusangu P., Vijay Avin B.R., Krishna V., Pramod S.N., Prabhakar B.T. Immunomodulatory glc/man-directed Dolichos lablab lectin (DLL) evokes anti-tumour response in vivo by counteracting angiogenic gene expressions. Clin. Exp. Immunol. 2017;189:21–35. doi: 10.1111/cei.12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., Mclellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. [Google Scholar]
- Wu N.C., Young A.P., Al-Mawsawi L.Q., Olson C.A., Feng J., Qi H., Chen S.H., Lu I.H., Lin C.Y., Chin R.G. High-throughput profiling of influenza A virus hemagglutinin gene at single-nucleotide resolution. Sci. Rep. 2014;4:4942. doi: 10.1038/srep04942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Lin C.W., Tsai T.I., Lee C.D., Chuang H.Y., Chen J.B., Tsai M.H., Chen B.R., Lo P.W., Liu C.P. Influenza A surface glycosylation and vaccine design. Proc. Natl. Acad. Sci. USA. 2017;114:280–285. doi: 10.1073/pnas.1617174114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H., Saito T., Yanagisawa M., Yu R.K., Kato K. Lewis X-carrying N-glycans regulate the proliferation of mouse embryonic neural stem cells via the Notch signaling pathway. J. Biol. Chem. 2012;287:24356–24364. doi: 10.1074/jbc.M112.365643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.R., Lin Y.C., Huang Y.P., Su C.H., Lo J., Ho Y.L., Yao C.Y., Hsu L.C., Wu H.S., Liu M.T. Reassortment and mutations associated with emergence and spread of oseltamivir-resistant seasonal influenza A/H1N1 viruses in 2005-2009. PLoS One. 2011;6:e18177. doi: 10.1371/journal.pone.0018177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Xie X., Li Y., Wang D., Han S., Shi S., Nan X., Bai C., Wang Y., Pei X. Legume lectin FRIL preserves neural progenitor cells in suspension culture in vitro. Clin. Dev. Immunol. 2008;2008:531317. doi: 10.1155/2008/531317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Negative stain EM density of FRIL and cryo-EM map/structure of SARS-CoV-2 spike protein generated during this study have been deposited in the Electron Microscopy Data Bank. Negative stain EM structure of FRIL tetramer (EMD-30380), cryo-EM structure of SARS-CoV-2 spike protein (EMD-30419, PDB: 7CN9).