Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has evolved into a major pandemic called coronavirus disease 2019 (COVID-19) that has created unprecedented global health emergencies, and emerged as a serious threat due to its strong ability for human-to-human transmission. The reports indicate the ability of SARS-CoV-2 to affect almost any organ due to the presence of a receptor known as angiotensin converting enzyme 2 (ACE2) across the body. ACE2 receptor is majorly expressed in the brush border of gut enterocytes along with the ciliated cells and alveolar epithelial type II cells in the lungs. The amino acid transport function of ACE2 has been linked to gut microbial ecology in gastrointestinal (GI) tract, thereby suggesting that COVID-19 may, to some level, be linked to the enteric microbiota. The significant number of COVID-19 patients shows extra-pulmonary symptoms in the GI tract. Many subsequent studies revealed viral RNA of SARS-CoV-2 in fecal samples of COVID-19 patients. This presents a new challenge in the diagnosis and control of COVID-19 infection with a caution for proper sanitation and hygiene. Here, we aim to discuss the immunological co-ordination between gut and lungs that facilitates SARS-CoV-2 to infect and multiply in the inflammatory bowel disease (IBD) and non-IBD patients.
Keywords: SARS-CoV-2, COVID-19, ACE2, Gut–lung axis, Inflammatory bowel disease, Gastrointestinal tract
1. Introduction
The human gut is an ecological niche for a huge population of enteric microbiota, majorly dominated by Bacteroidetes and Firmicutes (Foster and Neufeld, 2013) that produces several metabolites to maintain the gut homeostasis (Carabotti et al., 2015; Ahlawat et al., 2020). The gut microbiota plays important roles such as vitamin synthesis (Rowland et al., 2017), protection against pathogens (Hillman et al., 2017), development and maturation of host immune system (Proctor, 2019), intestinal angiogenesis (Baumgart and Carding, 2007), and differentiation and proliferation of intestinal epithelium (O’Hara and Shanahan, 2006). The gut microbial profile of each individual differs from another with alike relative abundance and dispersal among the healthy individuals. Also, the gut microbiota of a person keeps on changing throughout the life (Carabotti et al., 2015) and is most stable during adulthood (Nicholson et al., 2012). Such that, any deviation from normal gut microbial composition is defined as “microbial dysbiosis” that is characterized by the bloom in pathobionts and instability or reduction in the populations of ‘keystone’ taxa like Bacteroidetes and Firmicutes (Ahlawat et al., 2020; Duboc et al., 2013). Purposefully, the concept of gut microbiota dysbiosis has been discussed in a mini-review published by (Brüssow, 2020).
Besides, the lungs of healthy people harbour Fusobacterium, Haemophilus, Prevotella, Streptococcus, and Veillonella as main genera, which are relatively small in size when compared to the enteric microbiota (He et al., 2017). The emergence and maintenance of lung microbiota is governed by the equilibrium between microbial migration from the upper respiratory tract and microbial removal by the host defense systems, with small contribution from the multiplication of native microbes. Even in small concentrations, the airway microbiome is crucial to the host immunity such that an imbalance between the microbial immigration and removal predisposes its host towards the progression and exacerbations of respiratory diseases (He et al., 2017; Wypych et al., 2019). For instance, the patients with cystic fibrosis have heightened bacterial burden in their lower airways with species like Burkholderia spp., Pseudomonas aeruginosa, and Staphylococcus aureus. Asthma is another example of a multifactorial respiratory disease where the patients have greater diversity of Actinobacteria, Firmicutes, and Proteobacteria (Marri et al., 2013). There is a recent report of a possible link of gut microbiota with coronavirus disease (COVID-19) (Dhar and Mohanty, 2020); however, an updated appraisal is still needed due to the rapid generation of clinical data on gastrointestinal (GI) symptoms. Therefore, the present review summarizes the most updated evidences that suggest the existence of an immunological co-ordination between two vital organs, i.e., the gut and lungs, throughout the course of COVID-19 infection.
2. SARS-CoV-2 and COVID-19 outbreak
The coronavirus disease (COVID-19) is an ongoing pandemic with number of confirmed cases approaches 10.5 million and deaths surpasses 600,000 globally. The disease is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a positive-sense, single-stranded RNA virus of family Coronaviridae and genus Betacoronavirus (Wong et al., 2020). The proteome of SARS-CoV-2 consists of 4 structural proteins (membrane (M), envelope (E), nucleocapsid (N), and spike (S)) (He et al., 2020), 15 mature non-structural proteins (nsp1−10 and nsp12−16), and 9 accessory proteins (Prates et al., 2020). In general, coronaviruses are enveloped, positive-sense, non-segmented, and single-strand RNA viruses with six known species to cause human disease. SARS-CoV-2 has emerged as seventh species known to infect humans. Majority of them mostly cause mild respiratory disease. However, fatal coronaviruses have appeared sporadically in the past decades, such as the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, which also belongs to genus Betacoronavirus (Zaki et al., 2012). Very recently in December 2019, the cases of pneumonia with unfamiliar etiology were diagnosed in Wuhan, Hubei province of China. Later, a new coronavirus, that is, SARS-CoV-2 was obtained from the samples of lower respiratory tract of various patients (Repici et al., 2020). The disease was noted to be like influenza, with symptoms ranging from mild respiratory to drastic lung injury, multiple organ failure driven by hyper-inflammation and “cytokine storm” syndrome (Neurath, 2020), and death (Lamers et al., 2020). Fortunately, SARS-CoV-2 has lower (∼4%) mortality rate compared to other zoonotics like Ebola, SARS, and MARS, which have higher mortality rate ranging from 15 to 90%. However, it’s unfortunate that SARS-CoV-2 could not been contained like other coronaviruses, may be due to its higher rates of asymptomatic transmission. Further, comparative genome studies have found variations in the small fragment composed of 380 amino acids across various SARS-like coronaviruses and SARS-CoV-2. The reported variations have been contemplated to be important for determining the pathogenic divergence of COVID-19 (Prates et al., 2020).
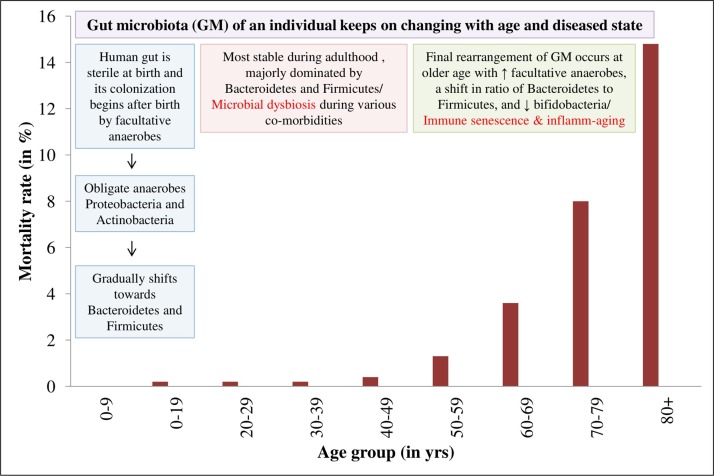
Additionally, co-morbidities like respiratory diseases, cardiovascular diseases, hypertension, diabetes, and patient age may worsen the COVID-19 manifestations. Aging is linked to the impairment of acquired immune system, characterized by immune senescence and inflamm-aging or the slow occurrence of the chronic sub-clinical inflammation. Thus, it is proposed that SARS-CoV-2 infection in older men with unregulated hyper-inflammation, drastically reduced B lymphocyte-driven acquired immunity, impaired plasmacytoid dendritic cells (DCs) type I interferon (IFN) pathway, and reduced ACE2 expression, induces high mortality (Gubernatorova et al., 2020) (Fig. 1 ) and also opens a Pandora’s box of disease etiology.
Fig. 1.
Graphical representation of gut microbiota alterations and COVID-19-associated mortality rate among various age groups. Source: Ahlawat et al., 2020; Novel, 2020.
3. How SARS-CoV-2 affects the human body?
Currently, the pathologists and clinicians are trying really hard to figure out the damage made by SARS-CoV-2 as it spread through the human body. They have discovered that even if our lungs are at prime risk, the virus can surprisingly move to other organs like the kidneys, heart and blood vessels, brain, and gut with destructive motives (Wadman et al., 2020).
When novel coronavirus enters the nose and throat through the inhalation of virus-laden-droplets expelled from an infected person, it gets unprecedented welcome by the lining of the nose due to the presence of a receptor known as angiotensin converting enzyme 2 (ACE2). The receptor is present all over the body to assist in regulating the host blood pressure (Fig. 2 ). However, in the case of COVID-19 infection the host tissue becomes potentially accessible to the infection as the virus needs the receptors to enter the cell. Thereafter, the virus takes over the cell’s machinery to replicate and invade the new cells. During this early stage, if the immune system doesn’t resist SARS-CoV-2, the virus then moves down to invade the lungs, where it can grow to fatal level. The battling immune system with the invader disrupts the oxygen transfer from the air sacs to rest of the body parts. Further, the fore-front warriors, that is, white blood cells (WBCs) release chemokines that consequently directs more immune cells to target and kill the virus-infected cells, thereby leaving behind the basic pathology of pneumonia, i.e., the fluid-filled sacs with the dead cells and pus. Autopsies showed the fluid-stuffed alveoli with WBCs, mucus, and detritus of damaged lung cells (Wadman et al., 2020). Some patients also contract secondary bacterial and fungal airway infections (Tay et al., 2020). However, the suspected driving force in severely-ill patients is a devastating over-reaction of the human immune system called “cytokine storm”, where the levels of specific cytokines rise far beyond what’s generally required by the body, such that the immune cells start attacking the host’s healthy tissues.
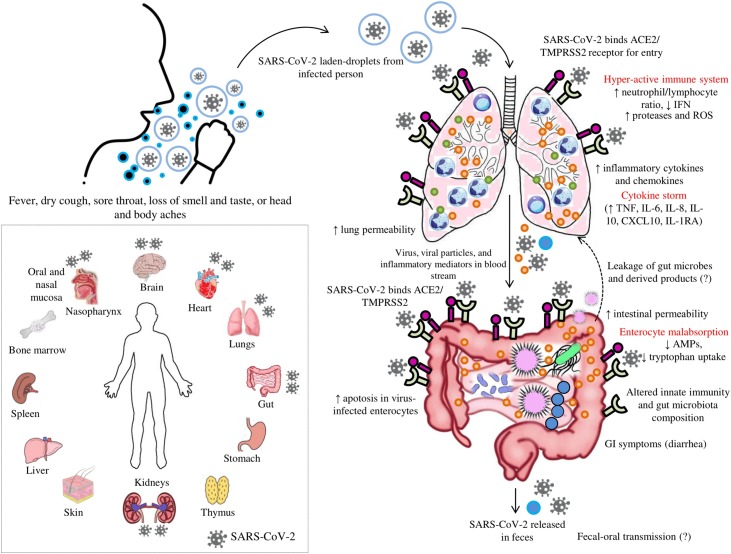
Fig. 2.
Representation of a probable “gut-lung axis” in SARS-CoV-2 caused COVID-19 disease. Inhalation of SARS-CoV-2-laden-droplets expelled from an infected person leads to binding of SARS-CoV-2 to angiotensin converting enzyme 2 (ACE2) and other receptors for entry to host cells. The hyperactive host immune system releases inflammatory mediators leading to “cytokine storm”. Increased inflammatory mediators lead to lung hyper-permeability such that the virus along with the inflammatory mediators via circulation migrates to intestine and binds highly expressed ACE2 receptors on the enterocytes. SARS-CoV-2 reduces the expression of ACE2 receptors and affects the microbial composition and host immune system. The inflammatory mediators disrupt the intestinal permeability leading to the leakage of gut microbes and associated metabolites into circulation. The leaked microbes and products via circulation migrate to organs including lungs and produce abnormalities. “Microbial dysbiosis” is also suspected due to the observation of diarrhea as a main GI symptom in patients with the COVID-19 disease. Inset: Human organs that express ACE2 receptor with the representation of the organs where SARS-CoV-2 reach have been reported previously.  : ACE2 receptor,
: ACE2 receptor,  : TMPRSS2 receptor,
: TMPRSS2 receptor,  : neutrophils,
: neutrophils,  : lymphocytes,
: lymphocytes,  : mucus, and
: mucus, and  : inflammatory mediators. *ACE2 and TMPRSS2 are expressed in the brush border of host cells. In figure, the localisation of ACE2 and TMPRSS2 outside of the gut or lungs instead of in brush border contact is just for easy representation to the readers.
: inflammatory mediators. *ACE2 and TMPRSS2 are expressed in the brush border of host cells. In figure, the localisation of ACE2 and TMPRSS2 outside of the gut or lungs instead of in brush border contact is just for easy representation to the readers.
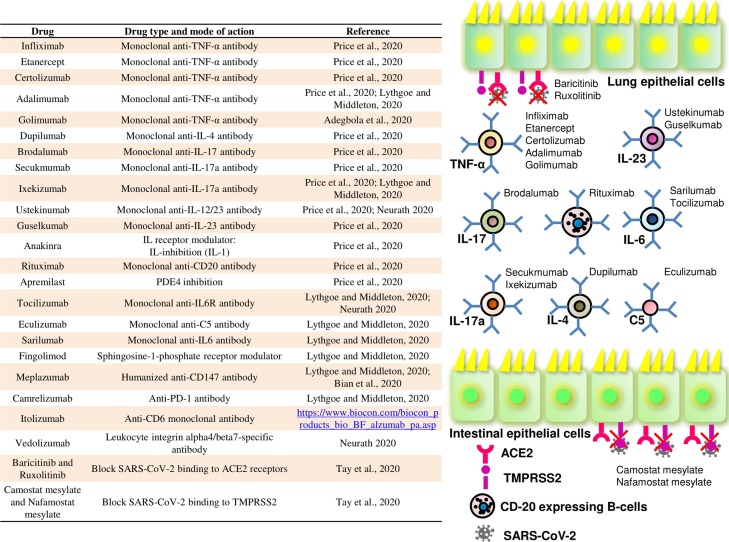
Studies have reported the increased concentrations of these cytokines in blood of hospitalized COVID-19 patients (Wadman et al., 2020; Lin et al., 2020). The majority of serious COVID-19 cases were linked to high systemic levels of TNF, IL-1b, and IL-6 (Gubernatorova et al., 2020), where C-reactive protein (CRP), d-dimers, ferritin (Merad and Martin, 2020), and IL-6 were found as the most significant clinical predictors of the COVID-19-associated mortality. IL-6 is a main pro-inflammatory cytokine at mucosal site during infection onset that performs various functions such as hematopoiesis, regulation to inflammation, auto-immunity, acute-phase response, and modulates host defense via several immune-stimulating mechanisms (Gubernatorova et al., 2020). Its up-regulation is a typical feature of aging (especially in men) and continuous elevation of IL-6 is known to foster the viral replication and promote inflammation and injury of the lung tissues. Thus, the chronic elevation of IL-6, especially in older men, pre-disposes them towards high SARS-CoV-2-associated mortality (Bonafè et al., 2020). Further, a negative correlation between cytokines concentrations and T-cell (CD4+ and CD8+) counts suggests that the “cytokine storm” actually dampens the host adaptive immunity against infection (Gubernatorova et al., 2020). In support, the recent analysis of multi-omics data suggests a functional immune deficiency syndrome due to the repression or destruction of specific cells of the host immune system in lungs (Prates et al., 2020). In addition, reduced IFN-γ production by CD4 + T cells (Chen et al., 2020b), increased neutrophils, elevated naïve/memory T-cells ratio (Neurath, 2020), enhanced inflammatory monocyte-derived macrophages, and depleted tissue-resident alveolar macrophages were also observed in severe COVID-19 patients (Merad and Martin, 2020). Altogether, many studies have indicated elevated neutrophil/lymphocyte ratio as an independent major risk factor for severe COVID-19 cases (Liu et al., 2020a; Kuri-Cervantes et al., 2020; Zhang et al., 2020). In this regard, clinical trials integrating IL-6 receptor and IL-1b blockade in COVID-19 patients have been initiated with the early encouraging results (Fig. 3 ). Further, determining the prevalence and severity of COVID-19 in patients with immune-modulatory biologics due to other co-morbidities, will provide additional insights on COVID-19 pathophysiology. It may be explored and used as a potential candidate to block a specific immune pathway to control the disease severity (Merad and Martin, 2020). Very recently, the Drugs Controller General of India (DGCI) has granted an emergency approval to Itolizumab for treating COVID-19 patients with moderate-to-severe acute respiratory distress (https://www.hindustantimes.com/india-news/dcgi-approves-limited-use-of-psoriasis-injection-for-covid/story-bkVPzdJ7Y9oaCiX2NJkypO.html). Itolizumab is a humanized recombinant anti-CD6 monoclonal antibody, generally used in treatment of patients with chronic plaque psoriasis by inhibiting T-cell proliferation and pro-inflammatory cytokines production (https://www.biocon.com/biocon_products_bio_BF_alzumab_pa.asp).
Fig. 3.
Representation of various immunomodulatory drugs that are proposed to treat the SARS-CoV-2 caused COVID-19 disease, and their action mechanism.
4. The bi-directional gut–lung axis: a pre-COVID-19 appraisal
An alteration in gut microbiota is associated to a bi-directional deviation in the relationship between the gut and several vital human organs that eventually cause severe diseases symptoms. Recently, our gut microbiome group has reviewed the bi-directional communication network between the gut microbes and vital human organs, except the lungs (Ahlawat et al., 2020). Interestingly, the alterations in the lungs microbial community including airways also affect the composition of intestinal microbiota. Other way, several GI disorders have manifestations in respiratory tract, for example, about half of the inflammatory bowel disease (IBD) patients with known alterations in their intestinal microbiota composition have reduced lung function. Thus, suggesting the “gut–lung axis” as a bi-directional communication network where many respiratory infections are often accompanied by GI symptoms (Wypych et al., 2019) or gut dysfunction or secondary gut dysfunction complications (Gao et al., 2020). For instance, the respiratory infection with Pneumocystis murina or influenza virus or intra-tracheal instillation of lipo-polysaccharide (LPS) in animal models drives changes in their gut microbiota (Wypych et al., 2019). The respiratory infection with influenza virus in mice models elevates Enterobacteriaceae and decreases Lactococci and Lactobacilli in their gut microbiota. Moreover, the dysbiosis in airway microbiota upon LPS-administration in mice disturbs their gut microbiota via bacterial movement from lungs into blood. Manifestations of pneumonia due to P. aeruginosa or multi-drug resistant S. aureus in lungs are believed to trigger gut injury, as P. aeruginosa-induced pneumonia leads to decreased intestinal epithelial proliferation (Anand and Mande, 2018). Further, acute lung injury disturbs airway microbiota, triggers transitory translocation of bacteria into bloodstream, and causes an acute heightened bacterial load in cecum. The chronic obstructive pulmonary disease (COPD) patients show the intestinal hyper-permeability with a high prevalence of IBD. On contrary, the healthy microbiota maintains tolerogenic immune-modulatory effects in gut and safeguards against the systemic inflammatory diseases (He et al., 2017).
In other way, dysbiosis in intestinal microbiota is linked to respiratory disorders and infections. For instance, increase in Clostridia and reduction in Bifidobacteria in gut is linked to asthma in early life (Anand and Mande, 2018). Moreover, the “gut–lung axis” also involves the migration of immune cells from gut to respiratory tract through circulation, where it encourages the host’s ability to fight infections. However, similar interplay also occurs in a disease. Further, the gut regulates the responses in lungs via host-acquired inflammatory mediators in the circulation. The elevated levels of these inflammatory mediators detected in serum of patients with gut disorders influences immune responses, thereby suggesting an another way of determining the local micro-environment in lung (Wypych et al., 2019). Also, the respiratory viral infections can alter the intestinal microbiome, where the intestinal microbiome determines the adaptive immune responses against the respiratory pathogens and is necessary for priming the innate immune responses against the pulmonary infections. During respiratory viral infections, the level of macrophage response to the respiratory viruses depends on the presence of intestinal microbes (Hanada et al., 2018). This suggests that the lung and the gut are closely linked organs that affect each other’s homeostasis via an immunological co-ordination between them (Fig. 2). Indispensably, microbes (among environmental factors) have central role in shaping the normal and pathologic immune responses in both the lung and gut (He et al., 2017).
Similar cross-talk between gut and lungs occurs in COVID-19 cases. A study with small sample size of eight patients found that lung microbial composition in broncho-alveolar lavage fluid samples of COVID-19 patients is dominated by bacteria generally found in oral and upper respiratory tract. This was similar to patients with the community-acquired pneumonia. The microbial signatures in lung may predict the acute respiratory distress syndrome (ARDS), most common complication from COVID-19, and the long-term outcomes of the outbreak. Emerging data identifies the role of gut microbiota in improving the antiviral immunity (He et al., 2020). Such that, various reports suggest the importance of gut microbiota modulation in reducing enteritis, ventilator-associated pneumonia, and reversing the side effects of antibiotics in order to avoid the replication of influenza virus in lung epithelium. However, at current there is no clinical evidence of gut microbiota modulation as therapeutic for treating COVID-19, but few emerging reports speculate the role of targeting the gut microbiota as a new therapeutic choice or adjuvant therapeutic option. In this way, probiotics can be used to alter the GI symptoms favourably by sustaining the balance of gut microecology and protect the respiratory system by preventing the secondary bacterial infections; thus, suggesting the crucial role of gut microbiota in ongoing COVID-19 disease (Gao et al., 2020; Tiwari et al., 2020).
5. The gut–lung axis in post-COVID-19 disease outbreak
The genomic characterization of this novel virus showed a high homology [72 %] (in regard to the structure of receptor-binding domain) with the SARS-CoV, thereby indicating that the new virus via spike protein S2 could bind ACE2 and infect human cells (D’Amico et al., 2020; Gao et al., 2020). Out of spike proteins S1 and S2, the S1 mediates the attachment of virus to the host cell membrane and S2 favours the fusion of the cell membranes. This process needs priming by cellular serine proteases (TMPRSS2) that enable S-protein cleavage, thereby controlling the entire mechanism (D’Amico et al., 2020). Evidence suggests that ACE2 receptor has protective anti-inflammatory effects and S-protein of the virus down-regulates its expression via increased release of pro-inflammatory chemokines and cytokines, thereby eliciting inflammation, vascular permeability, and neutrophil recruitment to the lungs (Bonafè et al., 2020). Consistent with this, many other studies supported the declined ACE2 expression in pulmonary viral infections such as SARS-CoV and influenza viruses like H1N1, H5N1, and H7N9 (Cole-Jeffrey et al., 2015). Thus, coronaviruses including SARS-CoV-2 have evolved to use proteases in Renin-Angiotensin system (RAS) for cell entry. RAS proteases other than ACE2 such as ACE and ANPEP may also be crucial in COVID-19 disease due to their ability to activate pro-inflammatory genes, which are the components of innate anti-viral response. By integrating the proteome structural analyses with the multi-omics data, a recent study, suggested that COVID-19 not only involves a direct binding of SARS-CoV-2 to ACE2 for cell entry but it also engages an imbalance of various other components of RAS. The expression profile of RAS genes from cells of broncho-alveolar lavage samples of COVID-19 patients suggested significantly up-regulated angiotensin, renin, MAS, ACE, and ACE2. In addition, the expression data from several sources and tissues suggest that the SARS-CoV-2 has evolved to target the host GI system (Prates et al., 2020).
A recent study identified ACE2 receptor expressing in the GI epithelial cells. This suggests that SARS-CoV-2 just like its 2003 predecessor can actively infect and multiply in GI tract (Wong et al., 2020), especially in ACE2 receptor-rich lining of the lower digestive tract (Wadman et al., 2020; Li et al., 2020). Studies showed that the new virus binds 10–20 times strongly to human ACE2 than its cousin SARS. Many studies provided additional evidence that the coronaviruses may infect GI tract due to the high level co-expression of TMPRSS2 and ACE2 in enterocytes along with lungs and esophagus. The proposed mechanism involves the alteration of intestinal permeability due to the novel virus infection that results in enterocyte malabsorption (Fig. 2). The intestinal ACE2 via its involvement in dietary amino acids uptake regulates the expression of anti-microbial peptides (AMPs) and promotes intestinal homeostasis (D’Amico et al., 2020; Hashimoto et al., 2012). Evidences suggest that ACE2 plays a crucial non-catalytic role in gut biology and modulation of intestinal microbiota composition, thereby leading to a speculation that the beneficial effects of ACE2 are partially mediated through the alteration in intestinal microbiome. For instance, ACE2-KO animals display altered gut microbial composition, decreased expression of AMPs, and declined levels of neutral amino acids in their serum with specific impairment of tryptophan (Trp) uptake, which can be restored by the Trp administration. In agreement to this, the probiotics were shown to reduce the oxidative stress, positively alter the cholesterol levels, release vaso-deleterious ACE-inhibiting peptides, and attenuate stress-induced hyper-permeability (Cole-Jeffrey et al., 2015). In addition, ACE2 regulates innate immune responses and influences the composition of host intestinal microbiota (Perlot and Penninger, 2013). Host Trp metabolites such as the melatonin, serotonin, and kynurenines and the bacterial Trp metabolites like indole, indolic acid, tryptamine, and skatole have effects on intestinal microbial composition, microbial metabolism, host immune system, host-microbiome interface, and host immune system-intestinal microbiota interactions. Thus, ACE2 has role in regulating intestinal amino acid homeostasis, expression of AMPs, innate immunity, gut microbial ecology, and transmissible susceptibility to colitis as the transplantation of altered gut microbiota from ACE2 mutant mice into germ-free (GF) wild-type hosts increases the propensity to develop severe colitis. Additionally, the deficiency in ACE2 results in a highly enhanced susceptibility to intestinal inflammation caused by epithelial damage (Hashimoto et al., 2012).
According to a preliminary data, the S-protein of SARS-CoV-2 can bind another surface molecule, i.e., CD147, and is considered as an alternative pathway for virus entry inside host cells. CD147 is mainly found on hematopoietic cells including red blood cells (RBCs), neuronal, and epithelial cells. It is also linked to SARS-CoV infection, where SARS-CoV protein binds to CD147 in association with cyclophilin A and the N-protein of SARS-CoV binds to cyclophilin A in ACE2-expressing infected-host cells. Further, they suggested that the in vitro inoculation of human lung epithelial cells with SARS-CoV-2 produces cytopathic effects and ceases the cilium beating of the epithelial cells (Gubernatorova et al., 2020). In this regard, one strategy for developing the therapeutics against the SARS-CoV-2 is by blocking the ACE2 or TMPRSS2 using compounds like baricitinib and ruxolitinib for ACE2 and camostat mesylate and nafamostat mesylate for TMPRSS2 that have been clinically approved for other co-morbidities (Fig. 3) or by delivering the high concentrations of a soluble recombinant form of ACE2, i.e., APN01 that can probably reduce the virus entry into the host cells. Further, monoclonal antibodies targeting the S-protein of SARS-CoV-2 may also inhibit the virus entry or membrane fusion (Tay et al., 2020).
Another very recent study performed to determine the target cell-type reveals that both SARS-CoV and SARS-CoV-2 can infect the ciliated cells in differentiated airway cultures and the enterocyte progenitors in human small intestinal organoids (hSIOs) with apical secretion of the virus (Lamers et al., 2020). SARS-CoV-2 infects both the post-mitotic enterocytes labelled by apolipoprotein A1 (APOA1) and K167-positive dividing cells, thereby displaying the ApoA1+ enterocyte phenotype and proliferative enterocyte progenitor-phenotype. Initially, single virus-infected cells were observed after 24 h of infection, suggesting small infection clusters, which later spread all over the organoid such that at 60 h, the number of virus-infected cells as well as apoptosis in virus-infected enterocytes became prominent. The transmission electron microscopy (TEM) analysis of SARS-CoV-2-infected hSIOs showed the onset of viral infection in the intact organoid at various stages of lifecycle, i.e., early double membrane vesicles (DMVs) as areas of viral replication, initial production of virus in golgi apparatus, and complete occupation of virus particles in the endomembrane system. This is in support with the fact that coronavirus (CoV) virions to efficiently release from the host cells must bud into the lumen of secretory pathway at endoplasmic reticulum-Golgi intermediate compartment (ERGIC), navigate through the Golgi, and anterograde the endomembrane system (Westerbeck and Machamer, 2019). Further, the changes in the gene expression studied via mRNA sequencing suggest that the SARS-CoV-2 infection elicits specific cytokines and interferon-stimulated genes (ISGs) of type I and III interferon responses, which was stronger than SARS-CoV (Lamers et al., 2020).
Interestingly, a study revealed the probable role of gut microbiota in pre-disposition of normal persons to abnormal inflammatory states, which accounts for severe COVID-19. Among the identified OTUs: Bacteroides, Streptococcus, and Clostridiales were negatively correlated with most of the tested inflammatory cytokines, whereas Blautia, Ruminococcus, and Lactobacillus showed positive associations. In addition, 45 fecal metabolites categorized as amino acids, bile acids, and fatty acids from three pathways namely arginine biosynthesis, aminoacyl-tRNA biosynthesis, and branched-chain amino acids (BCAAs) biosynthesis showed significant correlations with greater than half of the selected OTUs. These metabolites might play an important role in mediating the effect of intestinal microbiota on host metabolism and inflammation. Therefore, this suggests a probable role of intestinal microbiota in the progression and severity of COVID-19 (Gou et al., 2020).
Emerging data have identified the viral RNA of SARS-CoV-2 in fecal specimens (Zhou et al., 2020; Li et al., 2020) and anal/rectal swabs of the COVID-19 patients (Gao et al., 2020) even after the clearance of virus from the upper respiratory tract (Wong et al., 2020). About 53 % of sampled patient’s reported viral RNA in their stool samples and up to half of patients suffer diarrhea. A recent study identified the SARS-CoV-2 protein localized in duodenal, rectal, and gastric cells in biopsies from a COVID-19 patient. Moreover, the accumulating data with the evolution and expansion of the pandemic suggests that the digestive symptoms are significantly common in COVID-19 patients (Pan et al., 2020). Numerous reports across the globe suggest the occurrence of GI symptoms in COVID-19 patients (Table 1 ) and surprisingly, a subset of patients initially presented with only GI symptoms were also reported (Song et al., 2020). However, reason for variations across various studies is unclear, possible reasons includes differences in reporting and different patient populations and viral strains. Additionally, COVID-19 patients with GI symptoms appear to have significantly longer illness duration and viral clearance time than patients without GI symptoms. Although, GI symptoms may not be dominant but they would be helpful in identifying and preventing the spread of COVID-19 at an early stages with increased requirements to protect and treat the patients with underlying digestive diseases like IBD (Li et al., 2020). Thus, suggesting the fact that GI symptoms can’t be under-represented against the typical respiratory symptoms even though they dominate the clinical representation of the COVID-19 disease. Furthermore, the SARS-CoV-2 presence in GI tract intensifies the alarming danger of its probable transmittance through the feces of infected persons. Therefore, it alerts and unease as well due to the possibilities of additional mode of transmission through open defecation in most of the under-developed nations. However, till date there are no reported evidences of fecal transmission and it’s still not evident that whether feces contain intact or infectious virus or just the viral RNA and proteins (Wadman et al., 2020). Also, it’s unknown whether the virus in feces is acquired from the cellular fragments of the respiratory tract or consists of the replicates from the GI tract. Anyway, it’s logical to take precautionary steps to prevent the fecal-oral transmission (Li et al., 2020).
Table 1.
Summary of various studies reporting the gastrointestinal (GI) symptoms in SARS-CoV-2 infected patients.
| Sr. No. | No. of COVID-19 patients analyzed | No. of COVID-19 patients with GI symptoms | Average age (in years) |
Digestive symptoms | Country | Comments | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 204: 107 male and 97 female | 38 (18.6 %) | 52.9 ± 16 | Diarrhea (34 %), vomiting (3.9 %), and abdominal pain (1.9 %) | Hubei, China | 6 patients had digestive symptoms without respiratory symptoms; digestive symptoms intensify with the severity of disease | (Pan et al., 2020) |
| 2 | 651 | 74 (11.4 %) with at least one (nausea, vomiting or diarrhea) symptom | 46.14 ± 14.19 | Diarrhea (53), vomiting (11), nausea (10), nausea/vomiting (4), and nausea/ vomiting/diarrhea (3) | Zhejiang, China | COVID-19 patients with GI symptoms showed family clustering, severe/critical tendency, and fever >38.5 °C | (Jin et al., 2020) |
| 3 | 138: 75 male and 63 female | _ | 56 | Nausea (10.1 %), diarrhea (10.1 %), vomiting (3.6 %), and abdominal pain (2.2 %) | Wuhan, China | GI symptoms were more frequent in 36 ICU patients (diarrhea (16.7 %), nausea (11.1 %), vomiting (8.3 %), and abdominal pain (8.3 %)) | (Wang et al., 2020) |
| 4 | 73 | 39 (53.42 %): 25 male and 14 female had RNA of SARS-CoV-2 in stool | 43 | Diarrhea (26) and GI bleeding (10) | China | Infiltration of lymphocytes and plasma cells with interstitial edema was observed on endoscopy; 23.29 % patients had positive results in stool samples even after negative results in respiratory samples | (Xiao et al., 2020) |
| 5 | 10: 6 male, 4 female | 50 ± 18 | Diarrhoea (60 %), abdominal pain (30 %), and vomiting (10 %) | Piacenza, Italy | _ | (Poggiali et al., 2020) | |
| 6 | 95: 45 male, 50 female |
58 (61.1 %): 27 male, 31 female | 45.3 ± 18.3 | Diarrhoea (24.2 %), nausea (17.9 %), vomiting (4.2 %), and upper GI haemorrhage (2.1 %) | Zhuhai, China | Out of 65 hospitalised patients including 42 with and 23 without GI symptoms, 22 (52.4 %) and 9 (39.1 %) had SARS-CoV-2 positive faeces, respectively |
(Lin et al., 2020) |
| 7 | 206 | 117 (48: digestive symptoms only; 69: digestive & respiratory symptoms) | 62.5 | Diarrhea (32.5 %), vomiting (11.7 %), and abdominal pain (4.4 %) | Wuhan, China | Patients with digestive symptoms had a longer duration between symptom onset and viral clearance | (Han et al., 2020) |
| 8 | 42: 27 female, 15 male | 8 (19.05 %) patients had at least one GI symptom |
51 | Diarrhea (16.67 %), abdominal pain (11.95 %), nausea (9.52 %), and vomiting (7.14 %) | Wuhan, China | 18 (64.29 %) patients remained positive for viral RNA in feces even after pharyngeal swabs turned negative | (Chen et al., 2020a) |
| 9 | 254: 139 females, 115 males | 66 (26 %) | 50.6 | Diarrhea (18.1 %), nausea (8.3 %), vomiting (5.9 %), and abdominal pain (1.2 %) | GI symptoms in females were higher than in males (62.8 % v/s 37.2 %); ↓ Hemoglobin, ↑ CRP, ↑ alanine aminotransferase in GI symptom group than in non-GI symptom group |
(Zhou et al., 2020) | |
| 10 | 412: 241 male, 171 female | 3.2 % | 57 | Bowel wall abnormalities, unusual yellow discoloration of bowel (3), and bowel infarction (2) |
Boston, USA | Bowel abnormalities were more frequent in ICU patients | (Bhayana et al., 2020) |
| 11 | 116: 62 male, 54 female | 31.9 % | 50 | Nausea/vomiting (12 %), diarrhea (12 %), and abdominal pain (8.8 %) | California, USA | – | (Cholankeril et al., 2020) |
| 12 | 232 | 21 % | 47.5 | Diarrhea (21 %) | Hubei, China | Patients with diarrhoea showed severe symptoms of pneumonia than those without diarrhoea | (Wan et al., 2020) |
| 13 | 1099: 640 male, 459 female | – | 47 | Nausea or vomiting (5%), diarrhea (3.8 %) | China | – | (Guan et al., 2020) |
| 14 | 41: 30 male, 11 female | 3% | 49 | Diarrhea (3%) | Wuhan, China | – | (Huang et al., 2020) |
| 15 | 1141 | 183 (16 %): 102 male, 81 female | – | Anorexia (98 %), nausea (73 %), diarrhea (37 %), diffuse abdominal pain (25 %), nausea or vomiting (20 %), abdominal pain or diarrhea (9%), and all of these symptoms (7%) | Wuhan, China | – | (Luo et al., 2020) |
| 16 | 52: 35 male, 17 female | – | 59.7 ± 13.3 | GI haemorrhage (4%) and vomiting (4%) | Wuhan, China | GI haemorrhage occurred in critically ill patients | (Yang et al., 2020) |
| 17 | 305 | – | 57 | Diarrhea (49.5 %), loss of appetite (50.2 %), nausea (29.4 %), vomiting (15.9 %), and abdominal pain (6%) | Wuhan, China | 22.2 % patients had nondrug-related diarrhoea |
(Fang et al., 2020) |
| 18 | 99: 67 male, 32 female | – | 55.5 | Diarrhoea (2%) and nausea and vomiting (1%) | Wuhan, China | ↓ Lymphocytes and haemoglobin in many patients; Acinetobacter baumannii, Klebsiella pneumoniae, and Aspergillus flavus were cultured in one patient; Candida glabrata and Candida albicans was diagnosed in one and three cases, respectively | (Chen et al., 2020c) |
| 19 | 142 | 7 (4.9 %): 4 male, 3 female | 35−75 | Anorexia (7), diarrhea (6), upper abdominal discomfort (6), nausea (4), vomiting (2), and heartburn (1) | China | All the patients with GI symptoms had fever and showed no respiratory symptoms or developed during later stages | (Ai et al., 2020) |
| 20 | 157: 74 male, 83 females | 63 (40.1 %): 24 male, 39 female | 49.3 ± 14.5 | Anorexia (74.6 %), diarrhea (39.7 %), and nausea (33.3 %) | China | GI symptoms are frequent in COVID-19 but are not associated with the severity of diseases or worse outcomes | (Cao et al., 2020) |
| 21 | 1320: 579 male, 741 females | 192 (14.5 %): 90 male, 102 female | 50 | Diarrhea (55.7 %), anorexia (32.3 %), nausea and vomiting (29.7 %), and abdominal pain (5.7 %) | Wuhan, China | Patients with GI symptoms are at the higher risk of clinical deterioration | (Zheng et al., 2020) |
| 22 | 17: 8 male, 9 female | 14 | 8 | Abdominal pain, vomiting, and/or diarrhea; one showed acute ileocolitis | New York, USA | Levels of inflammatory markers were elevated in all patients | (Cheung et al., 2020) |
| 23 | 58: 25 male, 33 female | All patients represent one or other GI symptom | 9 | Abdominal pain (53 %), diarrhea (52 %), and vomiting (45 %) | England | – | (Whittaker et al., 2020) |
| 24 | 318 | 195 (61.3 %) had at least one GI symptom | Adults ≥18 years | Anorexia (34.8 %), diarrhea (33.7 %), nausea (26.4 %), vomiting (15.4 %), and abdominal pain (14.5 %) | USA | – | (Redd et al., 2020) |
| 25 | 278: 145 male, 133 female | 97 (35 %) | Adults ≥18 years | Diarrhea (56) and nausea and vomiting (63) | USA | Patients with GI symptoms were significantly more likely to test positive for COVID-19 | (Nobel et al., 2020) |
| 26 | 1992: 1128 male, 864 female | 1052 (53 %) had at least one GI symptom | 60.1 ± 16.3 | Diarrhea (34 %), nausea (27 %), vomiting (16 %), and abdominal pain (11 %) | USA and Canada | Among hospitalized patients, GI symptoms were common. Majority were mild and not associated with a severe clinical course |
(Elmunzer et al., 2020) |
| 27 | 29,393 | 2289 (7.8 %) | 47 | 1785 had GI symptoms with fever | China | Patients with both fever and GI symptoms had a 85 % higher risk of severe disease | (Liu et al., 2020b) |
| 28 | 150: 83 male, 67 female | 31 (20.6 %) had at least one GI symptom | 57.6 ± 17.2 | Diarrhea (14.7 %), nausea or vomiting (10.7 %), and abdominal pain (2%) | USA | Presence of GI manifestations at the time of admission was not associated with increased secondary outcomes |
(Ramachandran et al., 2020) |
*Studies having small sample size of (<5) COVID-19 patients with GI symptoms were not considered in the table.
6. Recommendatory note on IBD during COVID-19
There is a further need to raise awareness regarding the management of patients with pre-existing digestive diseases such as IBD (Queiroz et al., 2020), which is characterized by the periods of remission and relapse that are triggered by an inappropriate chronic immune response, whose therapeutics involves immunosuppressive medications. Some COVID-19 manifestations may mimic an IBD exacerbation; therefore, it is suggested to test IBD patients for SARS-CoV-2 before assuming flare diagnosis as currently there is no evidence in support of SARS-CoV-2 infection as a cause of IBD flare. The immunosuppressed patients often represent atypical presentations of viral diseases (Estevinho and Magro, 2020). However, data reported so far did not recognize the use of immunomodulator as a risk factor for COVID-19 severity (Queiroz et al., 2020) despite the theoretical fact that decreased host immune surveillance increases virus burden (Estevinho and Magro, 2020). It was assumed that these patients are at higher risk of developing COVID-19 complications (Queiroz et al., 2020) as the ACE2 expression and activity of host cell trypsin-like proteases are increased in inflamed gut of IBD patients, thereby suggesting inflamed gut as a doorway for viral entry into host tissues (Monteleone and Ardizzone, 2020). But reports suggest that having IBD does not increase the risk of developing COVID-19 (Grossberg et al., 2020; Monteleone and Ardizzone, 2020; Estevinho and Magro, 2020) as neither inflammation nor IBD medication increases the gut expression of ACE2 and TMPRSS2 (Burgueño et al., 2020). Further, ACE2 has two forms, full-length ACE2 and soluble form, and notably the latter form is up-regulated in IBD patients. The soluble ACE2, by acting as competitive interceptor of virus prevents its binding to the full-length ACE2, thereby raises the possibility towards limiting COVID-19 infection (Monteleone and Ardizzone, 2020). On other hand, considering the systemic hyper-inflammation as a driver of COVID-19 severity, it could be hypothesized that patients on immunosuppressant could have a milder disease (Fig. 3). The protection against COVID-19 might be due to the ongoing therapy of IBD and it should not be stopped (Aziz et al., 2020). Therefore, priority must be given to the maintenance of IBD remission, as risk of flares outweighs the probability of contracting SARS-CoV-2 infection. Additionally, considering the possibility of transmission via fecal-oral route, all invasive procedures like surgeries and elective endoscopies must be delayed; however, in emergent procedures the patients should be tested for SARS-CoV-2 (Estevinho and Magro, 2020).
Conclusively, the probable “gut-lung axis” in SARS-CoV-2 caused COVID-19 disease involves the inhalation of SARS-CoV-2-laden-droplets expelled from an infected person. This leads to the binding of spike protein of SARS-CoV-2 to ACE2 and other receptors for the entry inside the host cells. As a result, the hyperactive immune system releases inflammatory mediators that cause lung hyper-permeability such that the virus along with inflammatory mediators via circulation migrates to intestine and binds to highly expressed ACE2 receptors on enterocytes. This disrupts the intestinal permeability leading to the leakage of gut microbes and associated metabolites into circulation. The leaked microbes and products via circulation migrate to organs including lungs, and cause damage. Therefore, “microbial dysbiosis” is also suspected due to the observation of diarrhea as a main GI symptom in patients with the COVID-19 disease (Fig. 2).
7. Conclusion
In the current pandemic situation it is necessary to highlight the fact that the cases of COVID-19 patients suffering with GI symptoms of the disease are significant and can’t be ignored. Some patients have shown only GI symptoms, which lead to the under-estimation of COVID-19 cases, as the correct evaluation may not be done in patients having the mild symptoms. The viral detection and long-term persistence in fecal samples of COVID-19 patient’s suggest the possible oral-fecal route of transmission, thereby requiring additional precautions to avoid the exposure with possible sources of contamination like feces, vomitus, and other body fluids. There are possibilities of the transmission of virus via fecal-oral route, therefore it has been emphasized on proper hand washing, taking additional precautions while using public toilets, and control on open defecation in the developing and poor countries. Maximum mortality of COVID-19 disease was reported in elderly or persons with serious illness and the one with altered gut microbiota (Fig. 1). Therefore, we propose a possible link among dysbiosis, altered immune system, and reduced recovery from SARS-CoV-2 infection. Further, it is a well-known fact that the flora associated with both the organs drives the functioning of immune system to combat severe infections. It is possible that the virus induces microbial dysbiosis that causes diarrhea; therefore, there is a need of an exhaustive multi-omics study to explore the alterations in gut microbiome, mycobiome, and virome. Here, gut microbiome can play a crucial role in modulating the immune responses of COVID-19 infected individual, and prevent the damage of vital organs, including lungs. Therefore, re-formulating the gut microbiota may emerge as a new therapeutic target in the disease management of COVID-19 patients employing nutritional therapy, probiotics or fecal microbiota transplantation (using standard guidelines).
Funding information
This research was supported by the grant from the Indian Council of Medical Research (ICMR)-New Delhi [grant no. 5/4/8-1/2019-NCD-II].
Declaration of Competing Interest
The authors declare they have no conflict of interest.
References
- Ahlawat S., Asha, Sharma K.K. Gut–organ axis: a microbial outreach and networking. Lett. Appl. Microbiol. 2020 doi: 10.1111/lam.13333. [DOI] [PubMed] [Google Scholar]
- Ai J.W., Zi H., Wang Y., Huang Q., Wang N., Li L.Y., Pei B., Ji J., Zeng X.T. Clinical characteristics of COVID-19 patients with gastrointestinal symptoms: an analysis of seven patients in China. Front. Med. 2020;7:308. doi: 10.3389/fmed.2020.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S., Mande S.S. Diet, microbiota and gut-lung connection. Front. Microbiol. 2018;9:2147. doi: 10.3389/fmicb.2018.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M., Fatima R., Haghbin H., Lee-Smith W., Nawras A. The incidence and outcomes of COVID-19 in IBD patients: a rapid review and meta-analysis. Inflamm. Bowel Dis. 2020 doi: 10.1093/ibd/izaa170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart D.C., Carding S.R. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- Bhayana R., Som A., Li M.D., Carey D.E., Anderson M.A., Blake M.A., Catalano O., Gee M.S., Hahn P.F., Harisinghani M., et al. Abdominal imaging findings in COVID-19: preliminary observations. Radiology. 2020 doi: 10.1148/radiol.2020201908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafè M., Prattichizzo F., Giuliani A., Storci G., Sabbatinelli J., Olivieri F. Inflamm-aging: why older men are the most susceptible to SARS-CoV-2 complicated outcomes. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H. Problems with the concept of gut microbiota dysbiosis. Microbial Biotech. 2020;13(2):423–434. doi: 10.1111/1751-7915.13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgueño J.F., Reich A., Hazime H., Quintero M.A., Fernandez I., Fritsch J., Santander A.M., et al. Expression of SARS-CoV-2 entry molecules ACE2 and TMPRSS2 in the gut of patients with IBD. Inflamm. Bowel Dis. 2020;26(6):797–808. doi: 10.1093/ibd/izaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Chen M., He L., Xie J., Chen X. Clinical features and outcomes of COVID-19 patients with gastrointestinal symptoms. Crit. Care. 2020;24(1):1–3. doi: 10.1186/s13054-020-03034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabotti M., Scirocco A., Maselli M.A., Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015;28(2):203–209. [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung E.W., Zachariah P., Gorelik M., Boneparth A., Kernie S.G., Orange J.S., Milner J.D. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York City. JAMA. 2020 doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholankeril G., Podboy A., Aivaliotis V.I., Tarlow B., Pham E.A., Spencer S., Kim D., Hsing A., Ahmed A. High prevalence of concurrent gastrointestinal manifestations in patients with SARS-CoV-2: early experience from California. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole-Jeffrey C.T., Liu M., Katovich M.J., Raizada M.K., Shenoy V. ACE2 and microbiota: emerging targets for cardiopulmonary disease therapy. J. Vardiovasc. Pharmacol. 2015;66(6):540–550. doi: 10.1097/FJC.0000000000000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico F., Baumgart D.C., Danese S., Peyrin-Biroulet L. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention and management. Clin. Gastroenterol. Hepatol. 2020 doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D., Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc H., Rajca S., Rainteau D., Benarous D., Maubert M.A., Quervain E., Thomas G., Barbu V., Humbert L., Despras G., et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62(4):531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- Elmunzer B.J., Spitzer R.L., Foster L.D., Merchant A.A., Howard E.F., Patel V.A., West M.K., et al. Digestive manifestations in patients hospitalized with COVID-19. medRxiv. 2020 doi: 10.1101/2020.07.07.20143024. [DOI] [Google Scholar]
- Estevinho M.M., Magro F. The impact of SARS-CoV-2 on inflammatory bowel disease. GE-Port. J. Gastroenterol. 2020;27(4):227–229. doi: 10.1159/000508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., Ma J., Guan J., et al. Manifestations of digestive system in hospitalized patients with novel coronavirus pneumonia in Wuhan, China: a single-center, descriptive study. Chin. J. Dig. Dis. 2020 doi: 10.3760/cma.j.issn.0254-1432.2020.0005. [DOI] [Google Scholar]
- Foster J.A., Neufeld K.A.M. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36(5):305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Gao Q.Y., Chen Y.X., Fang J.Y. 2019 novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020;21(3):125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou W., Fu Y., Yue L., Chen G.D., Cai X., Shuai M., Xu F., Yi X., Chen H., Zhu Y.J., et al. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv. 2020 doi: 10.1101/2020.04.22.20076091. [DOI] [Google Scholar]
- Grossberg L.B., Pellish R.S., Cheifetz A.S., Feuerstein J.D. Review of societal recommendations regarding management of patients with inflammatory bowel disease during the SARS-CoV-2 pandemic. Inflamm. Bowel Dis. 2020 doi: 10.1093/ibd/izaa174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubernatorova E.O., Gorshkova E.A., Polinova A.I., Drutskaya M.D. IL-6: relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Duan C., Zhang S., Spiegel B., Shi H., Wang W., Zhang L., Lin R., Liu J., Ding Z., et al. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am. J. Gastroenterol. 2020;115(6):916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada S., Pirzadeh M., Carver K.Y., Deng J.C. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front. Immunol. 2018;9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., Sigl V., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wen Q., Yao F., Xu D., Huang Y., Wang J. Gut–lung axis: the microbial contributions and clinical implications. Crit. Rev. Microbiol. 2017;43(1):81–95. doi: 10.1080/1040841X.2016.1176988. [DOI] [PubMed] [Google Scholar]
- He Y., Wang J., Li F., Shi Y. Microbiota might play a role in SARS-CoV-2 infection. Front. Microbiol. 2020;11:1302. doi: 10.3389/fmicb.2020.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman E.T., Lu H., Yao T., Nakatsu C.H. Microbial ecology along the gastrointestinal tract. Microbes Environ. 2017;32(4):300–313. doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Lian J.S., Hu J.H., Gao J., Zheng L., Zhang Y.M., Hao S.R., Jia H.Y., Cai H., Zhang X.L., et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69(6):1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A., Weisman A.R., Agyekum R., Mathew D., Baxter A.E., Vella L., et al. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.05.18.101717. [DOI] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Schayck J.P., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.Y., Wu W., Chen S., Gu J.W., Li X.L., Song H.J., Du F., Wang G., Zhong C.Q., Wang X.Y., Chen Y., et al. Digestive system involvement of novel coronavirus infection: prevention and control infection from a gastroenterology perspective. J. Dig. Dis. 2020;21(4):199–204. doi: 10.1111/1751-2980.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., Gu Z., Gao L., Shi H., Mai L., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69(6):997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., et al. Neutrophil-to-Lymphocyte ratio predicts severe illness patients with 2019 novel coronavirus in the early stage. medRxiv. 2020 doi: 10.1101/2020.02.10.20021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Tao L., Liu X., Yao H., Yu S., Wang Q., et al. GI symptoms and fever increase the risk of severe illness and death in patients with COVID-19. Gut. 2020 doi: 10.1136/gutjnl-2020-321751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Zhang X., Xu H. Don’t overlook digestive symptoms in patients with 2019 novel coronavirus disease (COVID-19) Clin. Gastroenterol. Hepatol. 2020;18(7):1636–1637. doi: 10.1016/j.cgh.2020.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri P.R., Stern D.A., Wright A.L., Billheimer D., Martinez F.D. Asthma-associated differences in microbial composition of induced sputum. J. Allergy Clin. Immunol. 2013;131:346–352. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G., Ardizzone S. Are patients with inflammatory bowel disease at increased risk for Covid-19 infection? J. Crohns Colitis. 2020 doi: 10.1093/ecco-jcc/jjaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath M.F. Covid-19 and immunomodulation in IBD. Gut. 2020 doi: 10.1136/gutjnl-2020-321269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Nobel Y.R., Phipps M., Zucker J., Lebwohl B., Wang T.C., Sobieszczyk M.E., Freedberg D.E. Gastrointestinal symptoms and COVID-19: case-control study from the United States. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel C.P.E.R.E. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- O’Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020;115(5):766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T., Penninger J.M. ACE2–from the renin–angiotensin system to gut microbiota and malnutrition. Microb. Infect. 2013;15(13):866–873. doi: 10.1016/j.micinf.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggiali E., Ramos P.M., Bastoni D., Vercelli A., Magnacavallo A. Abdominal pain: a real challenge in novel COVID-19 infection. Eur. J. Case Rep. Intern. Med. 2020;7(4) doi: 10.12890/2020_001632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prates E.T., Garvin M.R., Pavicic M., Jones P., Shah M., Alvarez C., Kainer D., Demerdash O., Amos B.K., Geiger A., et al. Functional immune deficiency syndrome via intestinal infection in COVID-19. bioRxiv. 2020 doi: 10.1101/2020.04.06.028712. [DOI] [Google Scholar]
- Proctor L. What’s next for the human microbiome? Nature. 2019;569:623–625. doi: 10.1038/d41586-019-01654-0. [DOI] [PubMed] [Google Scholar]
- Queiroz N.S.F., Barros L.L., Azevedo M.F.C., Oba J., Sobrado C.W., Carlos A.S., Milani L.R., Sipahi A.M., Damião A.O.M.C. Management of inflammatory bowel disease patients in the COVID-19 pandemic era: a Brazilian tertiary referral center guidance. Clinics. 2020;75 doi: 10.6061/clinics/2020/e1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P., Onukogu I., Ghanta S., Gajendran M., Perisetti A., Goyal H., Aggarwal A. Gastrointestinal symptoms and outcomes in hospitalized COVID-19 patients. Dig. Dis. 2020 doi: 10.1159/000509774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd W.D., Zhou J.C., Hathorn K.E., McCarty T.R., Bazarbashi A.N., Thompson C.C., Shen L., Chan W.W. Prevalence and characteristics of gastrointestinal symptoms in patients with SARS-CoV-2 infection in the United States: a multicenter cohort study. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repici A., Maselli R., Colombo M., Gabbiadini R., Spadaccini M., Anderloni A., Carrara S., Fugazza A., Leo M.D., Galtieri P.A., et al. Coronavirus (COVID-19) outbreak: what the department of endoscopy should know. Gastrointest. Endosc. 2020 doi: 10.1016/j.gie.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland I., Gibson G., Heinken A., Scott K., Swann J., Thiele I., Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 2017;57(1):1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Liu P., Shi X.L., Chu Y.L., Zhang J., Xia J., Gao X.Z., Qu T., Wang M.Y. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69(6):1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.K., Dicks L.M., Popov I.V., Karaseva A., Ermakov A.M., Suvorov A., Tagg J.R., Chikindas M.L. Probiotics at war against viruses: what is missing from the picture? Front. Microbiol. 2020 doi: 10.3389/fmicb.2020.01877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman M., Couzin-Frankel J., Kaiser J., Matacic C. A rampage through the body. Science. 2020;368:356–360. doi: 10.1126/science.368.6489.356. [DOI] [PubMed] [Google Scholar]
- Wan Y., Li J., Shen L., Zou Y., Hou L., Zhu L., Faden H.S., Tang Z., Shi M., Jiao N., et al. Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. Lancet Gastroenterol. Hepatol. 2020;5(6):534–535. doi: 10.1016/S2468-1253(20)30118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J.W., Machamer C.E. The infectious bronchitis coronavirus envelope protein alters Golgi pH to protect the spike protein and promote the release of infectious virus. J. Virol. 2019;93(11):e00015–00019. doi: 10.1128/JVI.00015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker E., Bamford A., Kenny J., Kaforou M., Jones C.E., Shah P., Ramnarayan P., Fraisse A., Miller O., Davies P., Kucera F., et al. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Lui R.N., Sung J.J. Covid-19 and the digestive system. J. Gastroenterol. Hepatol. 2020;35(5):744–748. doi: 10.1111/jgh.15047. [DOI] [PubMed] [Google Scholar]
- Wypych T.P., Wickramasinghe L.C., Marsland B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019;20(10):1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Resp. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zhou X., Zhu C., Feng F., Qiu Y., Feng J., Jia Q., Song Q., Zhu B., Wang J. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.12.20035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Yang C., Wang H.Y., Chen X., Yu L., Wu Z.L., Sun H. Clinical characteristics and outcomes of COVID‐19 patients with gastrointestinal symptoms admitted to Jianghan Fangcang Shelter Hospital in Wuhan, China. J. Med. Virol. 2020 doi: 10.1002/jmv.26146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Zhao N., Shu Y., Han S., Chen B., Shu X. Effect of gastrointestinal symptoms on patients infected with coronavirus disease 2019. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]