Abstract
Abstract - Growth Regulating Factors (GRFs) comprise a transcription factor family with important functions in plant growth and development. They are characterized by the presence of QLQ and WRC domains, responsible for interaction with proteins and DNA, respectively. The QLQ domain is named due to the similarity to a protein interaction domain found in the SWI2/SNF2 chromatin remodeling complex. Despite the occurrence of the QLQ domain in both families, the divergence between them had not been further explored. Here, we show evidence for GRF origin and determined its diversification in angiosperm species. Phylogenetic analysis revealed 11 well-supported groups of GRFs in flowering plants. These groups were supported by gene structure, synteny, and protein domain composition. Synteny and phylogenetic analyses allowed us to propose different sets of probable orthologs in the groups. Besides, our results, together with functional data previously published, allowed us to suggest candidate genes for engineering agronomic traits. In addition, we propose that the QLQ domain of GRF genes evolved from the eukaryotic SNF2 QLQ domain, most likely by a duplication event in the common ancestor of the Charophytes and land plants. Altogether, our results are important for advancing the origin and evolution of the GRF family in Streptophyta.
Keywords: GRF, molecular evolution, QLQ domain, Bayesian analysis
Introduction
Growth Regulating Factors (GRFs) compose an important transcription factor family that plays diverse roles in plant development. These transcription factors are characterized by the obligatory presence of 2 conserved domains named QLQ (Gln, Leu, Gln) and WRC (Trp, Arg, Cys) (van der Knaap et al., 2000). The QLQ domain is usually located at the protein N-terminus and contains the motif QX3LX2Q. This region is named QLQ due to the similarity to the protein-protein interaction domain of the yeast SWI2/SNF2 (Switch/Sucrose non-fermentable), which is a subunit of a chromatin-remodeling complex (van der Knaap et al., 2000). Located after the QLQ, the WRC domain contains a nuclear localization signal and a CX9CX10CX2H motif (van der Knaap et al., 2000), which is an atypical C3H Zinc-finger motif found in barley HRT (Hordeum repressor of transcription), a transcriptional repressor of the Gibberellin Response Element (GARE) (Raventós et al., 1998). Further studies have demonstrated that the WRC domain from GRFs acts as DNA binding domain in barley, Arabidopsis, and rice (Osnato et al., 2010; Kim et al., 2012; Kuijt et al., 2014), and that some GRFs possess more than one WRC, such as AtGRF9 (Kim et al., 2003) and BrGRF12 (Wang et al., 2014). Besides that, there are other conserved regions found in the C-termini of some but not all GRFs, such as FFD (Phe, Phe, Asp), TQL (Thr, Gln, Leu), and GGPL (Gly, Gly, Pro, Leu) (van der Knaap et al., 2000; Kim et al., 2003; Zhang et al., 2008); however, their roles were not yet unveiled (Kim and Tsukaya, 2015).
Most of the studies in recent years have focused on the understanding of the specific roles of GRFs in different plant species (Omidbakhshfard et al., 2015; Kim and Tsukaya, 2015). The first known functions described for these proteins were in stem and leaf growth, particularly in GA-induced stem elongation (van der Knaap et al., 2000), regulation of cell proliferation in leaf primordia (Horiguchi et al., 2005; Kim and Lee, 2006), cotyledons and shoot apical meristem (SAM) (Kim and Lee, 2006; Kuijt et al., 2014). Other functions related to plant development were also revealed, including participation in flower organogenesis (Liu et al., 2014), organ longevity (Debernardi et al., 2014; Vercruyssen et al., 2015), seed oil production (Liu et al., 2012), photosynthetic efficiency (Liu et al., 2012; Vercruyssen et al., 2015), control of grain size and yield (Che et al., 2015; Duan et al., 2015; Hu et al., 2015; Li et al., 2016; Sun et al., 2016). Importantly, GRF genes are known to be upstream regulators of class I KNOX (KNOTTED1-like homeobox) genes required to maintain an appropriate level of SAM activity, together with other regulators of KNOX I expression, and this function is conserved in monocot and eudicot species (Kuijt et al., 2014; Tsuda and Hake, 2015). Under adverse environmental conditions, GRFs also play important roles, such as coordination of growth in response to osmotic and ABA-induced stresses (Kim et al., 2012) and host transcriptional reprogramming during cyst nematode infection (Hewezi et al., 2012) and in response to fungal pathogens (Soto-Suárez et al., 2017).
GRFs can physically interact with GRF-Interacting Factors (GIFs), a small family of transcriptional co-activators. This interaction occurs between the QLQ domain of GRF and the SNH (SSXT N-terminal homolog) domain present in GIF proteins (Kim and Kende, 2004). However, this interaction does not seem to be mandatory for GRF function because GRFs are capable of acting as negative regulators (Kim et al., 2012; Kuijt et al., 2014). Recently, it was demonstrated that the functioning of the GRF-GIF duo may be associated with the auxin signaling network (Lee et al., 2018). Also, it is not clear whether distinctive heterodimers of GRF and GIF have different functions in the downstream pathways (Kim and Tsukaya, 2015).
GRFs are part of a complex regulatory module. Some GRF members are negatively regulated at the transcript level by miR396 (Rodriguez et al., 2010; Wang et al., 2011; Hewezi et al., 2012; Debernardi et al., 2014). The miRNA396 responds to different stress conditions such as drought, cold, high-salinity, UV-B light, and pathogens (Liu et al., 2008; Zhou et al., 2012; Casadevall et al., 2013; Soto-Suárez et al., 2017), and it is also regulated by the TCP family (TEOSINTE BRANCHED1, CYCLOIDEA, and PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR1) (Schommer et al., 2014), which also modulates the gene expression of GRFs and GIFs directly (Rodriguez et al., 2010). Moreover, GRFs affect miR396 transcript levels and then, the gene expression of other GRFs (Hewezi et al., 2012), in an intricate cascade of regulation.
In Arabidopsis, GIF1, also called ANGUSTIFOLIA3 (AN3), is a homolog to the human Synovial Translocation Protein (SYT) (Kim and Kende, 2004). Interestingly, SYT interacts with the human SNF2 proteins, BRM (Brahma), and BRG (Brahma-related gene 1) (Nagai et al., 2001). Also, in Arabidopsis, GIF1 can associate with 2 different SWI/SNF complexes through the interaction with BRM or SYD (Splayed), the SNF2 homologs in this species (Debernardi et al., 2014).
SNF2 protein is part of a homonymous subfamily of the SNF2 family. Whereas the SNF2 family is characterized by the presence of a conserved SNF2 domain, QLQ is found only in the SNF2 subfamily (Eisen et al., 1995; Ryan and Owen-Hughes, 2011). Although the SNF2 and GRF proteins are known to share a conserved QLQ domain located at the N-termini of both proteins, and have the same molecular partner GIF or its ortholog SYT, to date, there has been no study addressing the evolutionary aspects related to the origin of the GRFs or exploring the divergence between GRF and SNF2.
GRF-encoding genes are found in plant genomes, including the Charophyte Klesormidium nitens (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015; Cao et al., 2016; Catarino et al., 2016; Wilhelmsson et al., 2017), suggesting that the emergence of this transcription factor may precede the occurrence of the land plants. Based on phylogenetic analysis, previous studies proposed divisions of GRFs in six (Omidbakhshfard et al., 2015) or five (Cao et al., 2016) groups. The former study claims that the GRF genes evolved via an eudicot whole-genome triplication and other independent WGD events, followed by gene retention in the ancestors of soybean and poplar Among the 6 groups, the authors found two groups specific to eudicot species and no group exclusive to monocots (Omidbakhshfard et al., 2015). The latter study focused on Arabidopsis, rice, Chinese pear, poplar, and grape genes. Among the five groups, three contain genes from the five species, whereas the other two groups include genes from one, two, or three species. Also, they found one group specific to monocots and one exclusive to eudicot species (Cao et al., 2016).
Many aspects of the biological functions of GRFs are already well known. However, the evolutionary history and diversification of these proteins are not yet completely elucidated and need to be more deeply comprehended based on different methods and discussed in detail. In this work, we conducted a phylogenetic approach to understand the evolution and diversification of the GRF gene family.
Based on the divergence within the QLQ domain found in SNF2 and GRF and on the distribution of each family across distinct taxa, we hypothesize that GRFs evolved from SNF2 and were established as a new transcription factor in the common ancestor of the Charophytes and land plants. In addition, we suggest that SNF2 and GRFs’ QLQ domains diverged particularly early in the course of evolution, most likely as a result of a duplication event. Also, we found well-supported data for eleven groups of GRF genes in flowering plants, six groups exclusive to eudicots, and five groups exclusive to monocot species, suggesting that the GRF family evolved mostly independently in monocot and eudicot species.
Material and Methods
Sequence retrieval
The sequences were retrieved from the public databases Phytozome v12.0 (Goodstein et al., 2012) (www.phytozome.jgi.doe.gov/pz/portal.html), Metazome v3.2 (available at www.metazome.jgi.doe.gov/pz/portal.html), NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi), FernBase (https://www.fernbase.org/), Congenie (http://congenie.org/), MarpolBase (http://marchantia.info/), and Klebsormidium nitens NIES_2285 genome project v1.1 (Hori et al., 2014) (available at: www.plantmorphogenesis.bio.titech.ac.jp/~algae_genome_project/klebsormidium). A detailed list of all species and loci used in this work is provided in Tables S1 (122.9KB, pdf) , S2 (149.2KB, pdf) , and S3 (121.6KB, pdf) .
For GRF sequences, two previously identified sequences - OsGRF1 (van der Knaap et al., 2000) and AtGRF1 (Kim et al., 2003) - were used as queries in blastp, besides searches for QLQ and WRC annotated domains in the Phytozome database. The searches were conducted against 40 sequenced plant genomes (Table S2 (149.2KB, pdf) ) and four Chlorophytes (green macroalgae) genomes (Chlamydomonas reinhardtii, Volvox carteri, Micromonas sp. RCC299 and Ostreococcus lucimarinus), available at Phytozome. The charophytes are the extant group of green algae that are most closely related to modern land plants. We conducted a blast search against the Charophyte species Klebsormidium nitens NIES 2285 genome to check the presence of GRFs in this organism.
A tree of the 45 species was reconstructed with phyloT (available at http://phylot.biobyte.de) to facilitate the visualization of GRF expansion in different species (Figure 1). Because K. nitens is a unique Charophyta alga with a sequenced genome available, we performed blast searches using transcriptomic data from Spirogyra pratensis, Nitella mirabilis, Mesostigma viride, Closterium peracerosum-strigosum-littorale, and Klebsormidium crenulatum. The blast search was conducted using the GRF sequence from K. nitens as query. The retrieved sequences were analyzed in ScanProsite (Castro et al., 2006) to verify the presence of both QLQ and WRC domains. Complete protein sequences were subjected to domain analysis, and only sequences presenting both domains were considered to be GRFs. We found GRFs only in Charophyta and land plants. From 415 GRF sequences, three were discarded from the phylogenies due to low-score domains or bad-quality alignments (Table S2 (149.2KB, pdf) ).
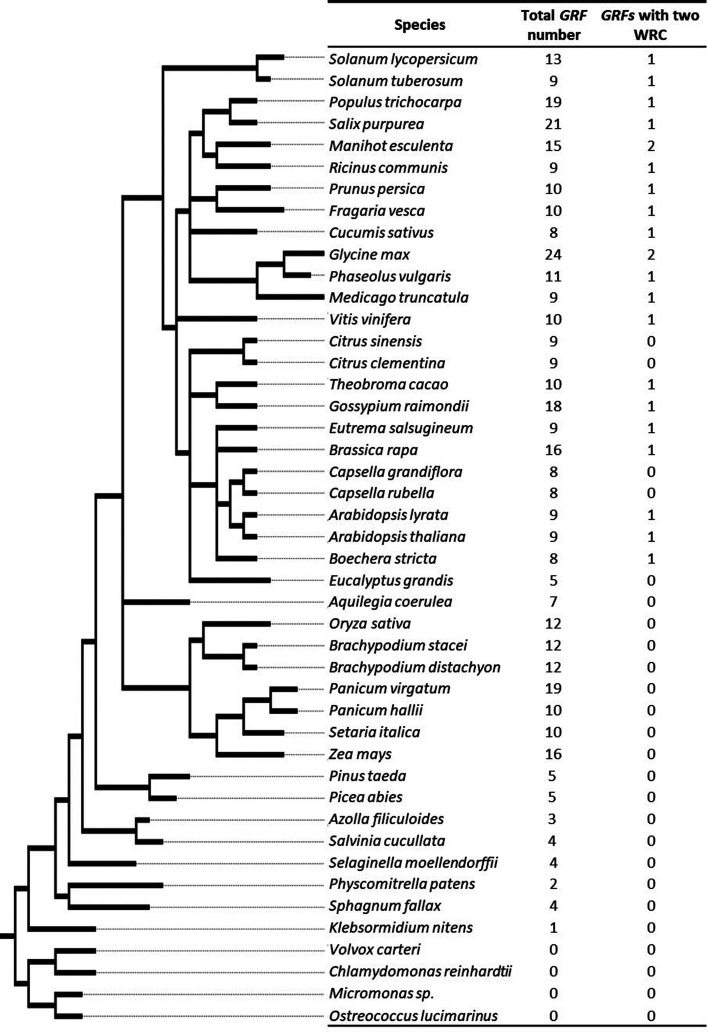
Figure 1. Tree of species. Species searched and the number of genes found in each species. The tree was based on NCBI Taxonomy and was constructed with phyloT, available at: http://phylot.biobyte.de .
For SNF2 analysis, SNF2 from Saccharomyces cerevisiae (NM_001183709) was used as the query for blastp in the NCBI database against nine fungi species and in the Metazome against 11 complete sequenced genomes. Plant and K. nitens sequences were searched using the SNF2-related BRAHMA (BRM) from Arabidopsis as the query in blastp against the genomes of seven plant and five algae species in Phytozome and K. nitens genomes (Table S1 (122.9KB, pdf) ).
Sequence alignments and evolutionary analyses
Sequence alignments were performed using CDS sequences from QLQ and WRC considering codon position, using the MUSCLE algorithm (Edgar, 2004), available at MEGA 7.0 (Molecular Evolutionary Genetics Analysis) (Kumar et al., 2016). The sequences were checked to find QLQ and WRC domains, which were used for phylogenetic analysis. Phylogenetic trees were reconstructed using nucleotide sequences of QLQ and WRC domains by Bayesian inference using BEAST2.4.5 (Bouckaert et al., 2014).
For GRF sequences, the best fit model for nucleotide evolution was GTR with invariable sites and gamma-distributed rates. A smaller tree containing sequences from Arabidopsis, rice, and the moss P. patens was reconstructed with the same parameters to allow a better understanding of the gene structure analysis of these species. For SNF2-GRF analysis, the best fit model for nucleotide evolution was TPM2 with invariable sites and gamma-distributed rates. Both models were selected with jModeltest v2.1.7 (http://jmodeltest.org/). The Birth and Death Model was selected as tree prior, and 100,000,000 generations were performed with Markov Chain Monte Carlo algorithm (MCMC) (Gilks, 2005) for evaluation of posterior distributions in all cases.
After manual inspection of the alignments, 415 sequences were used based on alignment quality and the presence of both QLQ and WRC domains for GRF analysis, totaling 243 DNA sites, 108 corresponding to QLQ, and 135 corresponding to WRC. For SNF2-GRF analysis, 131 sequences from QLQ domains with 108 DNA sites were used. In both cases, convergence was verified with Tracer v.1.6 (Rambaut et al., 2014) (http://beast.bio.ed.ac.uk/Tracer), and consensus trees were generated using TreeAnnotator, available at BEAST package. The resulting trees were viewed and edited using FigTree v.1.4.3.
Using GRF-SNF2 alignment and the respective phylogenetic tree as input, the rates of nonsynonymous to synonymous substitutions (dN/dS or ω) were computed, and homogeneity and positive selection were determined using maximum-likelihood models in the program CODEML in PAML (v.4.9) (Yang, 2007). For site model analysis, models M0 (basic), M1 (nearly neutral), M2 (selection), M3 (discrete), M7 (beta distribution, ω > 1 disallowed), and M8 (beta distribution, ω > 1 allowed) were considered (Goldman and Yang, 1994; Yang and Nielsen, 1998; Yang, 2000; Yang et al., 2005). The branch-site model was carried out comparing the alternative model (model = 2, Nsites = 2, fix_omega = 0, and omega = 0) with its null model (model = 2, Nsites = 2, fix_omega = 1, and omega = 1) (Zhang et al., 2005; Yang and Reis, 2011). The GRF branch was selected as the foreground, and statistical significance was addressed using LRT. CodeML was set to estimate branch lengths by using random starting points (fix_blenght = -1) and the F3x4 option for expected codon frequencies based on 3-codon positions. Naive empirical Bayes and Bayes empirical Bayes approaches were used to calculate the posterior probability of each site within the alternative model.
Domain architecture and gene structure analysis
Complete protein sequences of 392 GRFs were submitted to MEME Suite v4.12.0 (Bailey et al., 2009) (http://meme-suite.org/) to search for five different motifs in any number of repetitions, in order to find different combinations of QLQ, WRC, FFD, TQL, and GGPL in GRF proteins. We set a cut-off E-value of 10-6 to avoid false positives. The specific positions of the domains were used to construct a diagram presented in Figure S1 (537KB, pdf) . Protein sequences corresponding to the domains of all genes used in phylogeny analysis were used to construct the logos of the five domains on WebLogo3 (Crooks et al., 2004). For gene structure analysis, we used genomic sequences of three representative species, Arabidopsis, rice, and P. patens. The information about intron/exon organization was retrieved from Phytozome.
Synteny analysis and chromosomal locations
To better understand the pattern of expansion of GRFs, we conducted synteny analysis on PLAZA 4.0 (Van Bel et al., 2018). Synteny is based on the occurrence of collinear blocks between genomes, and these blocks are identified by the presence of homolog genes, also referred to as anchors, in both genomes or in different segments inside a genome.
The loci of GRFs from Arabidopsis, soybean, tomato, rice, maize, and purple false brome were searched in PLAZA 4.0 to find anchor points between different GRFs. The synteny relationships between the genomes were illustrated using CIRCOS (Krzywinski et al., 2009). The chromosomal positions and duplications of Arabidopsis and rice GRFs were drawn from information obtained from NCBI and PLAZA 4.0 databases, respectively.
Identification of OsGRF putative targets
To identify putative targets of the rice GRFs, we determined the location of the conserved motif “TGTCAG” or the reverse complement “CTGACA” in the rice genome using the fuzznuc tool from EMBOSS (Rice et al., 2000). All the hits were annotated back in the rice genome using the ChIPpeakAnno package (Zhu et al., 2010) for the R environment. Genes containing at least two motifs within 1500 bp upstream of ATG were selected using a customized R script. The functional annotation of Gene Ontology terms and a statistical overrepresentation test were performed using the PANTHER 11 (Mi et al., 2017) database with default settings, and only results with P<0.05 were considered.
Results
Identification of GRF genes and QLQ divergence from SNF2
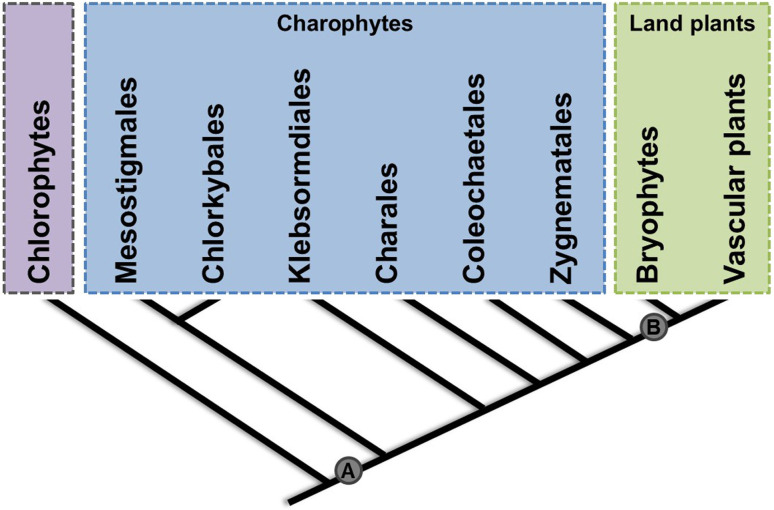
We analyzed 45 plant genomes and found GRF genes in 41 of them. Viridiplantae separated into Chlorophyta and Streptophyta approximately 629 to 890 million years ago (Morris et al., 2018). Streptophyta comprises Embryophyta, referred to as “land plants”, and six distinct groups of Charophyte algae: Mesostigmales, Chlorkybales, Klebsormdiales, Charales, Coleochaetales, and Zygnematales.
We also found a GRF gene in the genome of the Charophyte algae Klebsormidium nitens (formerly Klebsormidium flaccidum). As previously reported, we did not find GRFs in Chlorophytes. A total of 410 GRF-encoding genes were identified, of which 22 produce proteins containing 2 WRC domains (Figure 1).
In addition to the GRF genes previously described (Zhang et al., 2008; Filiz et al., 2014; Cao et al., 2016), we found four extra genes in the maize genome, ZmGRF15 to 18. We discarded ZmGRF8 and 12 from our analyses because the former contains only a partial WRC domain and does not have QLQ, and in the latter one, both domains are absent; however, we kept the nomenclature to avoid future confusion. We also found two additional genes in purple false brome (BdiGRF11 and 12) and two extra genes in grapevine (VviGRF9 and VviGRF10) (Tables S2 and S3).
We also analyzed the divergence between GRF and SNF2 because both share a conserved QLQ domain, which allows the interaction with SNH domains present in the homologous SYT and GIF families. Whereas GRFs are exclusive to Streptophyta, SNF2 genes are widely present in eukaryotes, such as fungi, metazoan, and plants, and compose a subfamily of the SNF2 family (Eisen et al., 1995; Ryan and Owen-Hughes, 2011). The SNF2 subfamily genes are the only representatives of the SNF2 family that have a QLQ domain. These genes have different names, such as SNF2 in fungi, BRM and BRG1 in metazoans, and BRM and SPLAYED (SYD) in plants. In this work, the general term “SNF2” was used for all of the SNF2-type genes. However, when referring to a particular gene, the specific gene name was used.
The binding region between BRM (the human SNF2) and SYT (the GIF homolog) was shown to be located between the amino acids 156 to 205 for BRM, and 1 to 181 for SYT (Nagai et al., 2001). Analyses in SMART (Simple Modular Architecture Tool) (Letunic et al., 2015) showed that these regions correspond to QLQ (172 to 208) and SNH (17 to 77) domains, respectively. The interaction between GRFs and GIFs also occurs via these domains (Kim and Kende, 2004). From blastp analyses, we were able to identify SNF2 homologs in fungi, metazoans, algae, and plants. Fifty-two encoding genes from 32 species were further selected for our analysis (Table S1).
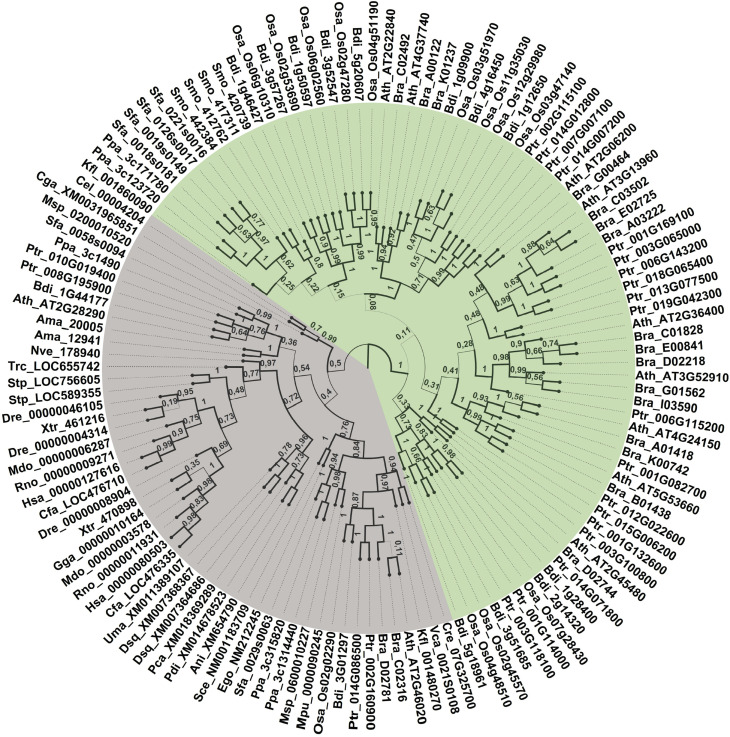
The phylogenetic relationships of SNF2 and GRF gene families revealed that GRFs and SNF2 grouped in distinct clades. While all GRFs are grouped in a well-supported cluster, SNF2 members are organized into smaller groups. Higher divergence within the QLQ domain found in SNF2 is consistent with its prevalence across distant taxa because it is present in diverse eukaryotic species. Also, the extent of conservation within the QLQ domain that comprises GRFs transcription factors stems from these proteins having evolved more recently (Figures 2 and 6).
Figure 2. Phylogenetic relationship among QLQ from SNF2 and GRFs. The tree with SNF2- and GRF-coding sequences of QLQ domains of algae, land plants, animals, and fungi was reconstructed by Bayesian inference. GRFs are colored in green, SNFs are colored in gray. The species and loci information are detailed in Table S1.
Figure 6. Analysis of GRF motif conservation. Logos of QLQ (36 aa), WRC with the C3H motif (45 aa), FFD (19 aa), TQL (15 aa), and GGPL (18 aa) domains. The height of the letters is based on the probability of the residue in the position, and the width was adjusted to fit. Domain colors: QLQ in green, WRC in pink, FFD in orange, TQL in blue, and GGPL in purple.
AtBRM and AtSYD are paralogous that grouped into distinct subclades in the SNF2 clade. In addition to Arabidopsis, purple false brome, turnip, and populus possess both genes, whereas rice and turnip have only BRM. Both BRM and SYD suffered specific duplications in populus, and BRM was duplicated in turnip, probably in a WGD event. The detailed information on species, loci, and taxa terminologies of SNF2 and GRFs are provided in Table S1 (122.9KB, pdf) and S2 (149.2KB, pdf) , respectively.
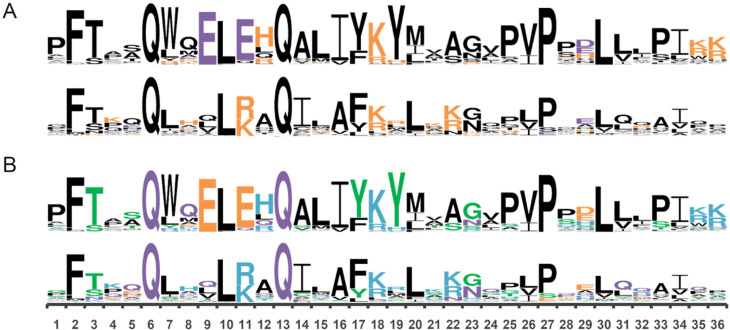
The early divergence between the QLQ domains of SNF2 and GRFs was accompanied by changes in the amino acid composition and therefore in the properties of QLQ domain. In GRF proteins, QLQ domain presents two conserved glutamic acid (E) residues in positions 9 and 11, conferring a negative charge and acidic property to the core. In the case of SNF2, the charge is neutral to positive, and the chemical property varies from neutral to basic in the same positions (Figure 3A and 3B). Other prominent differences are observed in positions 12, 22, 35, and 36. Besides the canonical QX3LX2Q, the most conserved residues are the phenylalanine (F) in position 2, the proline (P) in position 27, and the leucine (L) at position 30 (Figure 3A and 3B).
Figure 3. QLQ properties in SNF2 and GRF. (A) Logos representing the charge of the amino acids from the QLQ domain present in GRF (top) and SNF2 (bottom). Negative residues in purple and positive in orange. (B) Logos representing chemical properties of the amino acids from QLQ domain of GRF (top) and SNF2 (bottom). Basic residues in blue, acidic in orange, neutral in purple, polar in green, and hydrophobic in black. Protein sequences corresponding to the QLQ domain from SNF2 and GRFs were used separately to construct the diagrams in WebLogo3.
To analyze which protein sites might be under positive selection, we performed site model and branch-site model analyses. The site model assumes that some sites are under positive selection on all tree branches, whereas the branch-site model assumes that positive selection may be taking place on the foreground branch only. The site model analyses of the SNF2-GRF group revealed significant evolutionary constraints in the QLQ domain. The log-likelihood difference between models M0 and M3 was statistically different (Table S4 (94.9KB, pdf) ), suggesting that ω is heterogenous among the analyzed sites; however, positive selection was not detected through this approach. On the other hand, branch-site model analysis revealed positive selection for the branch leading to GRF (Table S4 (94.9KB, pdf) ). When comparing GRF, defined as the foreground, with the SNF2 representatives, it was possible to detect positive selection at positions 11, 12, and 22, which are associated with significant changes within the QLQ domain. Positive selection was also detected at QLQ positions 7, 16, 19, 24, 31, 32, and 34.
Diversification of GRF family in Streptophyta
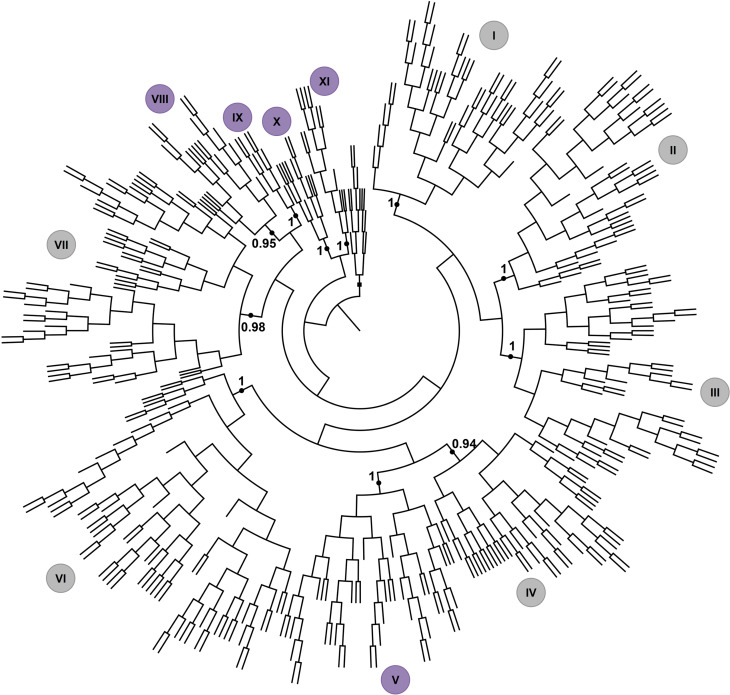
The GRF family has undergone a significant expansion in land plants. The phylogenetic tree, reconstructed from 392 sequences by the Bayesian method, allowed us to identify 11 well-supported groups of GRF proteins in flowering plants, as shown by the posterior probability (Figure 4 and Figure S1 (537KB, pdf) ). The composition of the groups is consistent with domain distribution in all GRF proteins (Figure S1 (537KB, pdf) ), and with gene structural organization (Figure 5) and synteny analysis (Figure 7A and 7B) from selected species.
Figure 4. Phylogenetic relationship of GRFs. Coding sequences of QLQ and WRC domains of K. nitens and land plants were used to generate a phylogenetic tree reconstructed by Bayesian inference. Groups I to XI correspond to flowering plants, and the others correspond to algae and moss sequences. The dicot groups are colored in gray, whereas the monocot groups are colored in purple. Species, loci, and taxa terminologies are available in Table S2. A detailed tree is available in Figure S1 (537KB, pdf) .
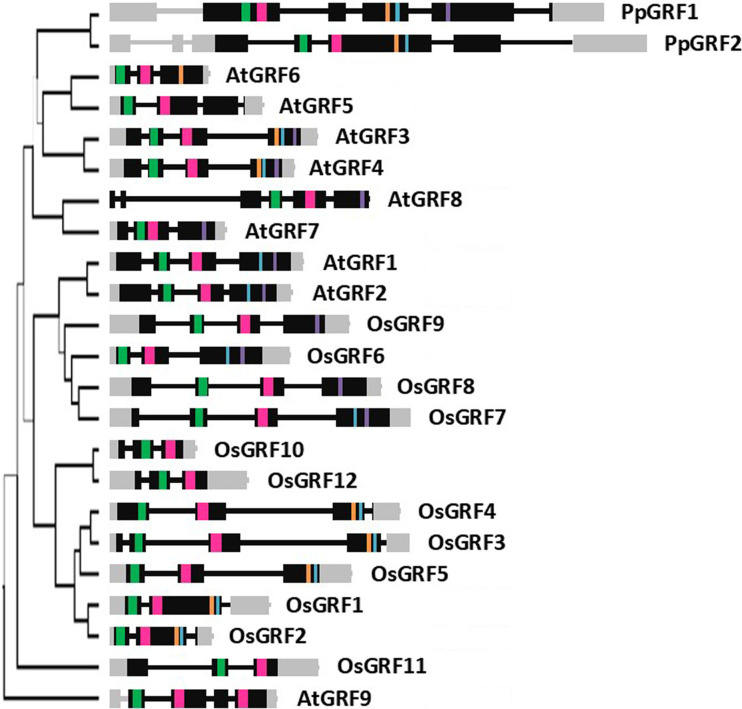
Figure 5. Structural organization of GRF genes. QLQ and WRC coding sequences of GRFs of P. patens (Pp), A. thaliana (At), and O. sativa (Os) were used to generate a phylogenetic tree by Bayesian inference (line width by posterior probability). The graphical representation of gene structures was based on genomic information available at Phytozome. Gray color corresponds to 5’ and 3’ untranslated regions. Black bars and lines indicate exons and introns, respectively. The different domains are colored in green (QLQ), pink (WRC), orange (TQL), blue (TQL), and purple (GGPL). For scale, QLQ domain (green) corresponds to 118bp.
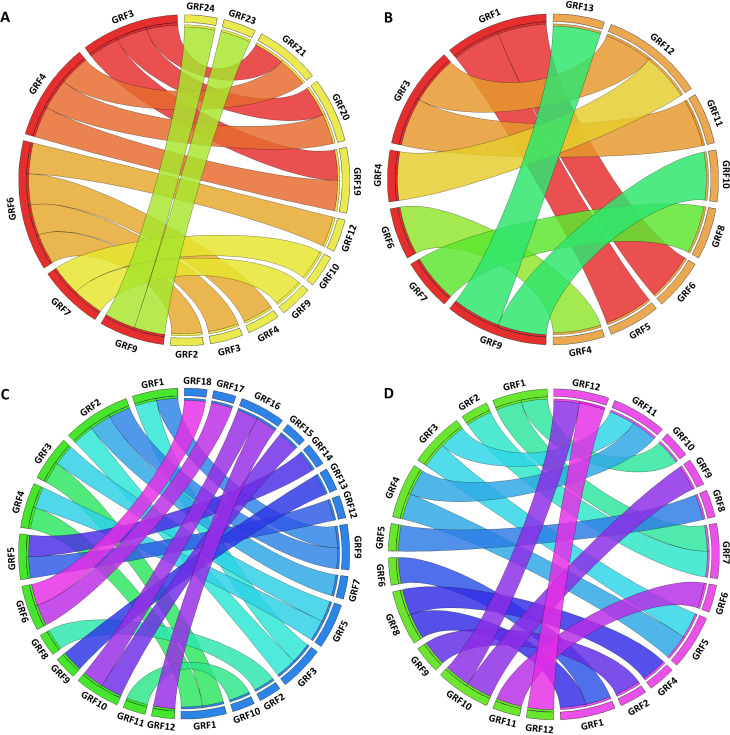
Figure 7. Syntenic analysis of GRFs in different species. Synteny between (A) Arabidopsis (red) and soybean (yellow), (B) Arabidopsis (red) and tomato (orange); (C) rice (green) and maize (blue), (D) rice (green) and purple false brome (purple). Probable orthologs are linked by ribbons.
The expansion of the GRF gene family occurred independently for the current monocot and eudicot species. There are six groups exclusive to eudicots: groups I to IV, VI and VII; and five groups exclusive to monocots: groups V and VIII to XI. For both monocots and eudicots, duplication events occurred mainly on the basis of each one of these groups of species because the genes from different species are almost ubiquitous throughout each group. A phylogenetic tree showing the separation of the 11 groups is provided in Figure 4, and the complete tree containing all taxa terminologies, group relationships, and domain characterization of all GRFs is found in Figure S1 (537KB, pdf) . Sequences derived from the Klebsormidium genome and from bryophytes and lycophytes representatives did not cluster in well-supported groups in our analysis (Figure 4).
To gain further insight into GRF diversification, sequences from fern (Azolla Filiculoides and Salvinia cucullata) and Gymnosperm (Picea abies and Pinus taeda) genomes, as well as from Spirogyra pratensis, Nitella mirabilis, Mesostigma viride, Closterium peracerosum-strigosum-littorale, and Klebsormidium crenulatum transcriptomes, were added to the analysis. This second analysis recovered the same 11 well-supported groups than the previous analysis (Figure S2 (381.5KB, pdf) ), for which reason we deepened our discussion within these groups.
In general, Group I is characterized by the presence of proteins containing five domains. A duplication in the basis of Brassicaceae originated AtGRF3 and AtGRF4 and their orthologs. Duplications in the basis of eudicots were responsible for the Group II expansion, giving rise to a subgroup of 15 genes encoding proteins containing one WRC domain and 22 GRFs presenting 2 WRC domains. In most cases, there is no additional motif, with some exceptions where TQL is present.
Group III is characterized by the presence of the GGPL, which corresponds to the only additional domain, and basal duplications gave rise to subgroups containing AtGRF7 and AtGRF8. Proteins from Groups IV and V have similar structures, with the presence of FFD and TQL additional domains. Group IV is exclusive to eudicots, whereas group V comprises GRFs from monocot species. The similarity between these groups may be explained by a common ancestor gene that evolved independently in monocot and dicot species. The expansion of group IV occurred on the basis of eudicots, and the Brassicaceae ancestor probably suffered gene loss because there are no members in this group. The expansion of the Group V occurred mainly via duplications in the origins of Poaceae, originating 5 subgroups. These duplications gave rise to the paralogous OsGRF1 and OsGRF2, OsGRF3 and OsGRF4, and the closely related gene OsGRF5.
Group VI is exclusive to eudicots and present subgroups containing different extra domains. The subset containing AtGRF5 and AtGRF6 is specific to Brassicaceae. The first possess only QLQ and WRC, and the second also contains the FFD domain. Although this group has a diversified protein structure, AtGRF5 and AtGRF6 are syntenic to other genes present in this group. Group VII arose in the basis of eudicots. In general, members of this group possess TQL and GGPL, with some exceptions. Duplication in the basis of Brassicaceae gave rise to the paralogous AtGRF1 and AtGRF2, presenting TQL and GGPL.
Groups VIII, IX, X, and XI evolved from an ancestor of Poaceae. Groups VIII and IX are more related to the eudicot Group VII and may have a common ancestor gene that diverged independently in monocot and dicot species. A basal duplication in Group VIII originated 2 subgroups, the first containing OsGRF6 and its orthologs, possessing TQL and GGPL, and the other subgroup containing OsGRF7, OsGRF8, and its orthologs, with the GGPL domain. Group IX, in which OsGRF9 is present, originated in the Poaceae ancestor and has only a GGPL extra domain.
Groups X and XI probably evolved with basal duplications. The structure of the members of these groups is formed by QLQ and WRC only, without the presence of additional domains. Group X is formed by OsGRF11, ZmGRF10, and other genes, whereas group XI comprises OsGRF10 and OsGRF12, among others. Also, the GRFs in these groups have an extremely short C-terminal region and the absence of additional domains. Despite the similarity between these groups, the low posterior probability in the consensus tree did not support a single clade between the groups X and XI, suggesting the existence of some level of divergence between them.
Structural organization of GRF genes from Arabidopsis, rice, and moss
We selected Arabidopsis, rice, and moss as representative species of eudicots, monocots and mosses to analyze the structural organization of GRF genes between these clades. Among these three species, the two GRF genes from the moss Physcomitrella patens are the largest, with 6200 and 6399 bp, respectively. OsGRFs ranged from 1126 to 3948 bp and AtGRFs from 1053 to 3416 bp. To facilitate comparison of gene structures, a tree was reconstructed with sequences of only these three species (Figure 5). Most of the genes have QLQ and WRC in separate exons, except for PpGRF1 and AtGRF7. The number of introns interrupting the coding region varied from 2 to 4, and domain position follows the order QLQ, WRC, FFD, TQL, and then GGPL.
In general, genes positioned in the same group have highly similar gene structures, besides domain composition. The two PpGRFs have 4 introns interrupting the coding region. AtGRF5 and 6, both from Group VI have a similar organization; however, AtGRF5 lost the FFD domain. From Group I, both AtGRF3 and AtGRF4 have 4 exons and 3 introns interrupting the coding region and possess all the 5 domains. AtGRF7 and 8, from Group III, have GGPL as the only extra domain but present different genetic structures. AtGRF1 and AtGRF2, from Group VII, both possess 4 exons and 3 introns, with TQL and GGPL in the last exon. OsGRF9 from Group IX has 3 introns, 4 exons, and a GGPL domain. From Group VIII, OsGRF6 have 2 introns and 3 exons, and the closely related OsGRF7 and OsGRF8 have 3 introns and 4 exons. OsGRF10 and OsGRF12, both from Group XI, have 2 introns and 3 exons, and no additional domain. OsGRF1 to 5, from Group V, have similar gene structures, with the presence of FFD and TQL domains. The subgroup including OsGRF1 and OsGRF2 contains 3 exons and 2 introns, whereas the subgroup of OsGRF3 to 5 has an additional intron separating WRC from the extra domains. OsGRF11, from Group X, have no additional domain, and AtGRF9, from Group II, possess an extra WRC motif.
Domain conservation of GRFs
We also analyzed the amino acid sequence conservation of the five domains in 392 GRF sequences to identify the pattern of conservation and the polymorphic sites (Figure 6). Among the five domains, WRC is the most conserved, except for the region between the positions 19 to 25 that is less conserved. We found an absolute conservation of the C3H motif, suggesting the importance of this motif for GRF function. QLQ domain has some sites with high conservation, importantly, the QX3LX2Q, the phenylalanine (F) in position 2, 2 glutamic acid (E) residues in positions 9 and 11, the proline (P) in position 27, and the leucine (L) in position 30, among others. FFD have a higher conservation in the core of the motif. Besides 2 phenylalanine (F) and the aspartic acid (D) residues in positions 8 to 10, this domain possesses tryptophan (W) in position 12, and proline (P) in 13. TQL has three sites even more conserved than the amino acids present in positions 3 to 5 that appoint the domain. Two serine (S) and one proline (P) residues, localized in the sites 6, 8, and 10 respectively, are almost absolutely conserved. The GGPL domain also has core conservations, with glutamic acid (E) and leucine (L) in positions 11 and 13, besides the two glycines (G), the proline (P), and the leucine (L) that names the motif, located at positions 6 to 9.
Synteny analysis and genomic organization of GRF genes
To find probable orthologs of AtGRFs and OsGRFs, we conducted searches in PLAZA 4.0 (Van Bel et al., 2018). Arabidopsis GRFs were searched against tomato and soybean genomes, and rice GRFs were searched against maize and purple false brome genomes. The pairs of probable orthologs found are summarized in Figure 7. In general, these pairs are consistent with the distribution of the genes in the groups of the phylogenetic tree.
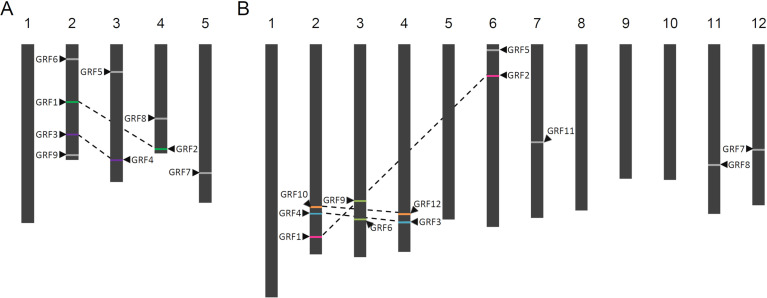
We also analyzed the intraspecific duplications of GRFs in Arabidopsis and rice using the same database. The relative positions of the genes and the duplicated blocks are graphically displayed in Figure 8. AtGRF1 and AtGRF2 are located on chromosomes 2 and 4, respectively. Both genes are members of Group VII. Group I members are AtGRF3, located on chromosome 2, and AtGRF4, located on chromosome 3. OsGRF1 and OsGRF2 are located on chromosomes 2 and 6. OsGRF3 and OsGRF4 are located on chromosomes 4 and 2. These latter 4 genes are members of Group V, and the syntenic genes form different subsets inside the main group. OsGRF6 and OsGRF9 are both located on chromosome 3.
Figure 8. Distribution of GRFs in nuclear chromosomes of Arabidopsis and rice. The positions of the GRFs from (A) Arabidopsis and (B) rice are indicated by arrows. Genes in duplicated regions are represented with the same colors. Genes not duplicated are represented in gray. Duplicated regions in different chromosomes are linked with dashed lines.
In silico prediction of GRF targets and biological processes
A target cis-element for AtGRF6 and 7 transcription factors were characterized by functional (Kim et al., 2012) and cistrome (O’Malley et al., 2016) analyses. The regulatory sequences described in these previous works present the core nucleotides “TGTCAG” that was first discovered in the DREB2A promoter, which is regulated by AtGRF7 (Kim et al., 2012). In rice, one study showed that GRF binding activity to the promoter of the KNOX gene Oskn2 was associated with the presence of CTG or CAG repeats (Kuijt et al., 2014). It is not known whether these target sequences are conserved among different species; however, one hypothesis for the maintenance of multiple binding sites is that it contributes to the regulation of a plethora of genes.
Initial studies from our group suggest the functionality of “TGTCAG” in the regulation of OsGRF11 targets in rice (Fonini, 2017). Here, we conducted an in silico analysis to find putative targets of GRFs by the cis-element core “TGTCAG” or the reverse complement “CTGACA” in this species, whereas the CTG or CAG repeats are not suitable for this type of analysis.
The identification of putative targets was conducted in the fuzznuc tool from EMBOSS (Rice et al., 2000). We identified genes containing at least two core motifs in a region of 1,500 bp upstream of ATG. A set list containing 1270 putative targets of GRFs was submitted to Gene Ontology analysis and an overrepresentation test in the PANTHER (Mi et al., 2017) database. From these, 83 were not annotated in the database, and seven had multiple mapping information. The complete list of enriched GO terms and the 1270 putative targets are available in Tables S5 (208.6KB, pdf) and S6 (241.1KB, pdf) , respectively. The statistic overrepresentation test demonstrates that the enriched targets are involved in several biological processes related to GRF functions. Among these processes are the regulation of leaf development (GO:2000024), regulation of endosperm development (GO:2000014), adaxial/abaxial pattern formation (GO:2000011), regulation of meristem structural organization (GO:0009934), reproductive process (GO:0022414), cell cycle (GO:0007049), cell division (GO:0051301), and regulation of cell proliferation (GO:0042127).
This result suggests that the cis-element is conserved (at least in rice) because several biological processes of the putative targets match with already-characterized GRF functions. Also, this target library may contribute to functional GRF studies in rice and in other species.
Discussion
In this work, we analyzed 45 plant and algal genomes and reconstructed the evolutionary history of the GRF family from algae to modern angiosperms. We also found GRF genes in the genome of Charophyte algae species (K. nitens) and in the transcriptomes of other Charophytes Spirogyra pratensis, Nitella mirabilis, Mesostigma viride, Closterium peracerosum-strigosum-littorale, and Klebsormidium crenulatum, showing that the GRF family arose earlier than previously thought during the evolution of Streptophyta, most likely by a duplication event in the common ancestor of Charophyte and land plants. This finding and the phylogenetic results allowed us to suggest that GRFs may arise after the division between Charophyta and Chlorophyta due to the fact that the GRF is not present in genomes of Chlorophyta (Figures 1 and 9).
We found evidence for the emergence of GRFs in Mesostigma viride, from the basal Charophyte Mesostigmales (Figure 9). We also conducted searches on available public transcriptome databases and found GRF encoding sequences in the Charophytes Spirogyra pratensis, Nitella mirabilis, Mesostigma viride, Closterium peracerosum-strigosum-littorale, and Klebsormidium crenulatum. Previous studies proposed that the GRF genes have originated after the emergence of Embryophyta (Omidbakhshfard et al., 2015; Kim and Tsukaya, 2015), mainly because of the absence of GRFs in Chlorophyta species. However, the availability of the Charophyta genome allowed to demonstrate that GRFs most likely originated earlier (Catarino et al., 2016; Wilhelmsson et al., 2017). Hence, this family existed even before the first multicellular green plants, which arose after the divergence between Mesostigmales and Chlorkybales (Jill Harrison, 2017).
Figure 9. Emergence and expansion of GRF family in Viridiplantae. (A) GRF emerged in a charophyta ancestor previous to Mesostigmales. (B) The expansion of the GRF family occurred in land plants. Diagram of the relationships between algae and plant lineages is based on data already published (Morris et al., 2018).
Previous works reported the presence of QLQ in both SNF2 and GRF genes (van der Knaap et al., 2000; Omidbakhshfard et al., 2015; Cao et al., 2016; Fina et al., 2017; Khatun et al., 2017), but, to date, no study had been dedicated to explore the divergence between these 2 families. GIFs are known to be molecular partners of GRFs in the regulation of cell proliferation (Horiguchi et al., 2005), ear development (Zhang et al., 2008), flower development (Liu et al., 2014), and plant longevity (Debernardi et al., 2014). Also, it is already known that the interaction of GRFs and GIFs occurs via QLQ and SNH domains, respectively. SNF2 proteins interact with SYT proteins, the GIF homologs. Because the region of the interaction of SNF2 with SYT was already described, we analyzed the protein sequences and found that the regions correspond to QLQ and SNH domains, respectively. Beyond that, AtGIF1 is shown to interact with SWI/SNF complexes through the interaction with BRM and SYD (Vercruyssen et al., 2014). These dimer formations prompted us to investigate the divergence between GRFs and SNF2 genes.
We also demonstrated that the QLQ domain from GRF and SNF2 diversified particularly early in the course of evolution, although both maintained the protein interaction function with the SNH domains present in the homologous GIFs or SYT, respectively. Whereas SNF2 remained as chromatin remodeling proteins, GRFs evolved as specific transcription factors.
We hypothesized that the QLQ present in GRFs arose from a duplication of an SNF2 QLQ in the common ancestor of the Charophytes and land plants, and the divergence between these genes appears to have occurred early in the evolution. Our phylogenetic analysis revealed that SNF2 and GRF genes are grouped into distinct clades with the presence of algae and moss sequences in both clades. This observation suggests that the divergence between both SNF2 and GRF QLQ domains occurred before the emergence of land plants and after the divergence between the Chlorophyta and Charophyta lineages. Interestingly, although sequences from Spirogyra pratensis, Closterium peracerosum-strigosum-littorale, and Klebsormidium crenulatum grouped within the GRF cluster, as well as the sequence derived from Klebsormidium nitens, GRF coding sequences from Nitella mirabilis and Mesostigma viride were kept out. A detailed analysis from these sequences revealed that QLQ positions 9 and 11 are not occupied by glutamic acids, as observed for almost every GRF encoding sequence analyzed (Figure 3). In fact, these positions are occupied either by an isoleucine or glutamine and by a glutamine or aspartate, respectively. Despite having a WRC domain and high similarity to other GRFs, both proteins harbor a QLQ that resembles SNF2 proteins, presenting at least one neutral residue within these positions. Also, our analyses from the rates of nonsynonymous to synonymous substitutions suggest a positive selection in QLQ from GRFs (Table S4 (94.9KB, pdf) ).
The expansion of the GRF family accompanied the rapid evolution of plants, since the basal Charophytes until the modern angiosperms (Figures 1 and 9). Remarkably, GRFs evolved with the expansion of gene number and remained as families. Whereas in the Charophyte K. nitens genome there is just one gene, the family encompasses 24 genes in soybean, the one with the highest number of genes among the species analyzed. Other species with a high number of genes, such as switchgrass, maize, turnip, cotton, and Salicaceae, underwent whole-genome duplication (WGD) events at some moment in the course of evolution (Renny-Byfield and Wendel, 2014). This finding supports the data obtained by the phylogenetic analyses, suggesting that besides ancestral duplications in basal monocot and eudicot, recent WGD events were crucial for the expansion of the family.
The conservation among the sequences of QLQ and WRC in different GRFs did not allow further characterization of the relations between the groups. However, through the analysis of the domain composition, we observed that the dicot Group IV and the monocot Group V are somewhat related. We also noticed a relationship between monocot groups X and XI, both with no additional domains and a short C-terminal region. ZmGRF10, a member of Group X, can interact with GIFs. However, it lacks the C-terminal domain and the transactivation activity (Wu et al., 2014). The other members of Groups X and XI share the same structure of ZmGRF10; hence, it is possible that the other GRFs in both groups also lack this transactivation ability. Our analyses also suggest that duplications on the basis of monocots and eudicots and species-specific WGD events were crucial for the expansion of the GRF family in Viridiplantae.
Functional studies on Arabidopsis and rice illustrated that GRFs play diverse roles in important agronomic traits such as plant growth, grain productivity, stress responses, and integration of defense with growth processes (see Kim and Tsukaya, 2015 and Omidbakhshfard et al., 2015 for reviews). In this work, we identified several paralogous and probable orthologs of genes related to these traits that could be manipulated in order to favor characteristics of interest. In this work, we identified putative targets of this transcription factor family in rice and orthologs of GRFs known to play important agronomic roles, findings that may be important in guiding future studies in diverse species.
OsGRF4 and OsGRF6 have been linked to yield-related traits, regulating grain size (Che et al., 2015; Duan et al., 2015) and panicle branching (Gao et al., 2015), respectively. The expression of both genes and their homologs could be explored to improve plant productivity, alone or in combination, mainly in cereal crops. We identified paralogous and ortholog versions of both genes. OsGRF3 is paralogous of OsGRF4, whereas BdGRF5 and 11, and ZmGRF1 and 5 are their putative orthologs. Also, OsGRF6 and OsGRF9 are contained in a syntenic block of duplication and seem to be paralogous, and the probable orthologs of OsGRF6 are BdGRF1, ZmGRF17, and ZmGRF18.
Regarding stress responses, AtGRF7 has been implicated in the regulation of osmotic stress-responsive genes to prevent growth inhibition under stress conditions (Kim et al., 2012). We found probable orthologs of AtGRF7 in the genomes of tomato (SlGRF8) and soybean (GmGRF9 and 10). The expression of AtGRF7 or its orthologs, in combination with osmotic defense genes, could be utilized to balance growth and defense processes during stress. Alterations in the expression of AtGRF1 and 2 in response to infection with cyst nematodes were already related to the development of the syncytium, a feeding structure that enables nematode establishment in roots (Hewezi et al., 2012). Modulation of the expression of both genes, or their putative orthologs SlGRF5 and 6, could be important for preventing the formation of the feeding structure, avoiding nematode infection. All these genes are promising candidates for genetic engineering of important agronomic traits and could be further investigated in future studies.
Data Access
The alignments are availabe at: https://data.mendeley.com/datasets/p25czj44sn/draft?a=801f6365-5d02-48f5-aef4-e6da1f3a510b.
Acknowledgments
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES - Finance code 001); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); Fundação de Apoio à Pesquisa do Rio Grande do Sul (FAPERGS); and Fundação para a Ciência e a Tecnologia de Portugal (FCT) through the R&D Unit, UIDB/04551/2020 (GREEN-IT - Bioresources for Sustainability).
Supplementary Material
The following online material is available for this article:
Species, loci and taxa terminologies of SNF2-type genes used in the tree.
Species, loci and taxa terminologies of GRF genes used in the trees.
GRF numbers and loci used in the syntenyc analysis.
Site and branch-site model analysis.
GO terms and enrichment analysis of putative targets of GRFs in rice.
Annotation of 1270 putative targets of GRFs in rice genome.
Phylogenetic tree of GRFs and protein domain composition.
Phylogenetic relationship of GRFs.
Footnotes
Associate Editor: Carlos F. M. Menck
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Han Y, Jin Q, Lin Y, Cai Y. GRF genes in Chinese pear (Pyrus bretschneideri Rehd), poplar (Populous), grape (Vitis vinifera), Arabidopsis and rice (Oryza sativa) Front Plant Sci. 2016;7:1750. doi: 10.3389/fpls.2016.01750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall R, Rodriguez RE, Debernardi JM, Palatnik JF, Casati P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell. 2013;25:3570–3583. doi: 10.1105/tpc.113.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino B, Hetherington AJ, Emms DM, Kelly S, Dolan L. The stepwise increase in the number of transcription factor families in the precambrian predated the diversification of plants on land. Mol Biol Evol. 2016;33:2815–2819. doi: 10.1093/molbev/msw155. [DOI] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2015;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, Rodriguez RE, Palatnik JF. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014;79:413–426. doi: 10.1111/tpj.12567. [DOI] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants. 2015;2:15203. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiz E, Koç I, Tombuloglu H. Genome-wide identification and analysis of growth regulating factor genes in Brachypodium distachyon: in silico approaches. Turk J Biol. 2014;38:296–306. [Google Scholar]
- Fina J, Casadevall R, AbdElgawad H, Prinsen E, Markakis MN, Beemster GTS, Casati P. UV-B inhibits leaf growth through changes in growth regulating factors and gibberellin levels. Plant Physiol. 2017;174:1110–1126. doi: 10.1104/pp.17.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonini LS. Caracterização do gene Osbhlh35 e dos fatores de transcrição envolvidos na regulação de sua expressão. 2017. D. Sc. Thesis. [Google Scholar]
- Gao F, Wang K, Liu Y, Chen Y, Chen P, Shi Z, Luo J, Jiang D, Fan F, Zhu Y, et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat Plants. 2015;2:15196. doi: 10.1038/nplants.2015.196. [DOI] [PubMed] [Google Scholar]
- Gilks WR. Markov Chain Monte Carlo. Encyclopedia of Biostatistics. 2005 doi: 10.1002/0470011815.b2a14021. DOI: [DOI] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012;159:321–335. doi: 10.1104/pp.112.193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana . Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Jill Harrison C. Development and genetics in the evolution of land plant body plans. Philos Trans R Soc Lond B, Biol Sci. 2017;372:20150490. doi: 10.1098/rstb.2015.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatun K, Robin AHK, Park JI, Nath UK, Kim CK, Lim KB, Nou IS, Chung MY. Molecular characterization and expression profiling of tomato GRF transcription factor family genes in response to abiotic stresses and phytohormones. Int J Mol Sci. 2017;18:E1056. doi: 10.3390/ijms18051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313x.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BH. GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol. 2006;49:463468. [Google Scholar]
- Kim JH, Tsukaya H. Regulation of plant growth and development by the growth-regulating factor and grf-interacting factor duo. J Exp Bot. 2015;66:6093–6107. doi: 10.1093/jxb/erv349. [DOI] [PubMed] [Google Scholar]
- Kim JS, Mizoi J, Kidokoro S, Maruyama K, Nakajima J, Nakashima K, Mitsuda N, Takiguchi Y, Ohme-Takagi M, Kondou Y, et al. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell. 2012;24:3393–3405. doi: 10.1105/tpc.112.100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt SJH, Greco R, Agalou A, Shao J, ‘t Hoen CC, Overnäs E, Osnato M, Curiale S, Meynard D, van Gulik R, et al. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 2014;164:1952–1966. doi: 10.1104/pp.113.222836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Lee BH, Jung JH, Park SK, Song JT, Kim JH. GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 2018;176:717–729. doi: 10.1104/pp.17.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–60. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, He Z, Ren Y, Li W, Deng Q, et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol J. 2016;14:2134–2146. doi: 10.1111/pbi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana . RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hua W, Yang HL, Zhan GM, Li RJ, Deng LB, Wang XF, Liu GH, Wang HZ. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J Exp Bot. 2012;63:3727–3740. doi: 10.1093/jxb/ers066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Guo S, Xu Y, Li C, Zhang Z, Zhang D, Xu S, Zhang C, Chong K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014;165:160–174. doi: 10.1104/pp.114.235564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, Wellman CH, Yang Z, Schneider H, Donoghue PCJ. The timescale of early land plant evolution. Proc Natl Acad Sci USA. 2018;115:E2274–E2283. doi: 10.1073/pnas.1719588115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Tanaka S, Tsuda M, Endo S, Kato H, Sonobe H, Minami A, Hiraga H, Nishihara H, Sawa H, et al. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci USA. 2001;98:3843–3848. doi: 10.1073/pnas.061036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SSC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B. Growth-Regulating Factors (GRFs): A Small transcription factor family with important functions in plant biology. Mol Plant. 2015;8:998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Osnato M, Stile MR, Wang Y, Meynard D, Curiale S, Guiderdoni E, Liu Y, Horner DS, Ouwerkerk PBF, Pozzi C, et al. Cross talk between the KNOX and ethylene pathways is mediated by intron-binding transcription factors in barley. Plant Physiol. 2010;154:1616–1632. doi: 10.1104/pp.110.161984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J. HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem. 1998;273:23313–23320. doi: 10.1074/jbc.273.36.23313. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Wendel JF. Doubling down on genomes: polyploidy and crop plants. Am J Bot. 2014;101:1711–1725. doi: 10.3732/ajb.1400119. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Owen-Hughes T. Snf2-family proteins: chromatin remodellers for any occasion. Curr Opin Chem Biol. 2011;15:649–656. doi: 10.1016/j.cbpa.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF. Repression of cell proliferation by miR319-regulated TCP4. Mol Plant. 2014;7:1533–1544. doi: 10.1093/mp/ssu084. [DOI] [PubMed] [Google Scholar]
- Soto-Suárez M, Baldrich P, Weigel D, Rubio-Somoza I, San Segundo B. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci Rep. 2017;7:44898. doi: 10.1038/srep44898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J Integr Plant Biol. 2016;58:836–847. doi: 10.1111/jipb.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Hake S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr Opin Plant Biol. 2015;27:91–96. doi: 10.1016/j.pbi.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018;46:D1190–D1196. doi: 10.1093/nar/gkx1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Kim JH, Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000;122:695–704. doi: 10.1104/pp.122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Tognetti VB, Gonzalez N, Van Dingenen J, De Milde L, Bielach A, De Rycke R, Van Breusegem F, Inzé D. GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 2015;167:817–832. doi: 10.1104/pp.114.256180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, Jégu T, Archacki R, Van Leene J, Andriankaja M, et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014;26:210–229. doi: 10.1105/tpc.113.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot. 2011;62:761–773. doi: 10.1093/jxb/erq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Qiu N, Ding Q, Li J, Zhang Y, Li H, Gao J. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis) BMC Genomics. 2014;15:807. doi: 10.1186/1471-2164-15-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson PKI, Mühlich C, Ullrich KK, Rensing SA. Comprehensive genome-wide classification reveals that many plant-specific transcription factors evolved in streptophyte algae. Genome Biol Evol. 2017;9:3384–3397. doi: 10.1093/gbe/evx258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang D, Xue M, Qian J, He Y, Wang S. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J Integr Plant Biol. 2014;56:1053–1063. doi: 10.1111/jipb.12220. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J Mol Evol. 2000;51:423–432. doi: 10.1007/s002390010105. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Reis M. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28:1217–1228. doi: 10.1093/molbev/msq303. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhang DF, Li B, Jia GQ, Zhang TF, Dai JR, Li JS, Wang SC. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.) Plant Sci. 2008;175:809–817. [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu M, Jiang J, Qiao G, Lin S, Li H, Xie L, Zhuo R. Expression profile of miRNAs in Populus cathayana L. and Salix matsudana Koidz under salt stress. Mol Biol Rep. 2012;39:8645–8654. doi: 10.1007/s11033-012-1719-4. [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014. http://beast.bio.ed.ac.uk/Tracer
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Species, loci and taxa terminologies of SNF2-type genes used in the tree.
Species, loci and taxa terminologies of GRF genes used in the trees.
GRF numbers and loci used in the syntenyc analysis.
Site and branch-site model analysis.
GO terms and enrichment analysis of putative targets of GRFs in rice.
Annotation of 1270 putative targets of GRFs in rice genome.
Phylogenetic tree of GRFs and protein domain composition.
Phylogenetic relationship of GRFs.