Abstract
Background and Objective
Without a specific antiviral treatment or vaccine, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic, affecting over 200 countries worldwide. A better understanding of B- and T-cell immunity is critical to the diagnosis, treatment and prevention of coronavirus disease 2019 (COVID-19).
Methods
A cohort of 129 patients with COVID-19 and 20 suspected cases were enrolled in this study, and a lateral flow immunochromatographic assay (LFIA) and a magnetic chemiluminescence enzyme immunoassay (MCLIA) were evaluated for SARS-CoV-2 IgM/IgG detection. Additionally, 127 patients with COVID-19 were selected for the detection of IgM and IgG antibodies to SARS-CoV-2 to evaluate B-cell immunity, and peripheral blood lymphocyte subsets were quantified in 95 patients with COVID-19 to evaluate T-cell immunity.
Results
The sensitivity and specificity of LFIA-IgM/IgG and MCLIA-IgM/IgG assays for detecting SARS-CoV infection were > 90%, comparable with reverse transcription polymerase chain reaction detection. IgM antibody levels peaked on day 13 and began to fall on day 21, while IgG antibody levels peaked on day 17 and were maintained until tracking ended. Lymphocyte and subset enumeration suggested that lymphocytopenia occurred in patients with COVID-19.
Conclusions
LFIA-IgM/IgG and MCLIA-IgM/IgG assays can indicate SARS-CoV-2 infection, which elicits an antibody response. Lymphocytopenia occurs in patients with COVID-19, which possibly weakens the T-cell response.
Key Points
| Lateral flow immunochromatographic assay-IgM/IgG and magnetic chemiluminescence enzyme immunoassay-IgM/IgG assays can indicate SARS-CoV-2 infection, which elicits an antibody response. |
| Lymphocytopenia occurs in patients with COVID-19, which possibly weakens the T-cell response. |
Introduction
According to the World Health Organization report on 24 April, 2020, 2,591,015 coronavirus disease 2019 (COVID-19) cases were confirmed with 178,686 deaths globally. The novel severe acute respiratory coronavirus 2 (SARS-CoV-2) is the causative agent of the COVID-19 pandemic, and a lack of approved vaccines or specific treatments to SARS-CoV-2 leaves patients dependent on their own immune responses [1].
The SARS-CoV-2 single-stand RNA genome is ~ 29.8 kb in length and encodes ~ 28 proteins (four structural proteins, eight accessory proteins and 15 non-structural proteins) [1]. High infectivity, similar syndromes and considerable mortality make laboratory diagnosis particularly important. Furthermore, laboratory diagnosis is critical to SARS-CoV-2 patient management, contact tracing and epidemiological studies. Timely diagnosis of suspected COVID-19 cases could reduce and interrupt the transmission of COVID-19 from person to person. Nucleic acid amplification tests (NAT) and serological testing are two main laboratory testing methods [2–4].
IgM antibodies elicited immediately after coronavirus infection indicate a current or new infection, while IgG antibodies produced subsequently indicate recovery or a prior infection [5, 6]. Therefore, antibody detection could indicate the occurrence of infection and disease progress. Liu et al. [7] demonstrated that an IgM/IgG assay for antibody detection was superior to real-time reverse transcription polymerase chain reaction (RT-PCR) for NAT, which is the gold standard for SARS-CoV-2 detection [8], and it also exhibited a lower false-negative rate [9, 10]. One of the main reasons for the high false-positive rate of RT-PCR assays is the difference between the main virus colonisation site (lower respiratory tract) and typical sampling sites (upper respiratory tract) [9]. Unlike RT-PCR assays, antibody assays do not require sophisticated instruments, laboratory conditions or tedious experimental procedures. For example, the lateral flow immunochromatographic assay (LFIA) can be completed within 15 minutes without specialised equipment or professionals. IgM and IgG antibodies appear earlier in severe patients than in non-severe patients, and their titres are significantly higher, indicating that a strong antibody response is associated with delayed viral clearance and disease severity [11]. The antibody level can be measured by a magnetic chemiluminescence enzyme immunoassay (MCLIA) to guide treatment during disease progression. The humoural immunity of patients can be evaluated by serological detection, as exemplified by quantitative detection of IgG and IgM [12], which provide essential complementary methods to NAT for pathogenic diagnosis.
Cell-mediated immunity of patients can be evaluated by quantitative monitoring of lymphocytes and their subsets (e.g., T cells, CD4 + T cells and CD8 + T cells), which are the primary effector cells [13]. Lymophocyte enumeration represents immune status on a daily clinical routine [14]. T cells coordinate multiple aspects of adaptive immunity to pathogens throughout life [15]. Naive T cells that play a central role in cell-mediated immunity fall into two large classes, including CD4 + and CD8 + T cells [16]. CD4 + T cells differentiate into several subsets of effector T cells (T helper-1, T helper-2, T helper-17, regulatory T cells) that orchestrate different immune functions. CD8 + T cells are critical for mediating clearance following many acute viral infections in the lung [17]. Immunocytes also play a critical role in determining the outcomes of virus infection [18]. Therefore, changes in the relative abundance of immunocytes can serve as indicators for cell-mediated immunity. Large numbers of subsets of lymphocytes can be simultaneously measured by multi-parametric flow cytometry, which is a sensitive, specific and cost-effective method of counting lymphocyte subsets [13]. Peripheral blood lymphocyte populations and subpopulations enumerated by flow cytometry indicate the immunological and pathological status [19, 20].
B-cell responses mediate humoural immunity, while T-cell responses regulate immune response and directly lead to cellular immunity [15]. In the present study, we preliminarily evaluated humoural and cellular immunity in response to SARS-CoV-2 infection based on IgG/IgM antibody detection and lymphocyte subset enumeration, respectively.
Materials and Methods
Participants
A cohort of 129 patients with COVID-19 and 20 suspected cases were enrolled in this study. Patients with COVID-19 confirmed by an RT-PCR assay were hospitalised in Tianjin Haihe Hospital (Tianjin, China). Among them, 64 were men and 65 were women, aged 44.19 years (95% confidence interval [CI] 41.63–46.75; range 8–91 years). The suspected cases with a negative RT-PCR assay did not have previous virus contact but similar symptoms.
Additionally, 127 patients (63 men and 64 women) aged 48.51 years (95% CI 45.56–51.46; range 8–91 years) of this cohort were selected for detection of IgM and IgG antibodies against SARS-CoV-2 to evaluate B-cell immunity. Of 127 patients, 54 were sampled at 0–7 days from the onset of illness, 36 at 8–15 days, and 37 at 16–38 days. Furthermore, 95 patients (56 men and 39 women) aged 40.23 years (95% CI 36.14–44.32; range 8–91 years) of this cohort were included in a retrospective study to enumerate lymphocytes and their subsets to evaluate T-cell immunity. The study was approved by the Ethics Commission of Tianjin Haihe Hospital (2020HHQX-001-2020HHQX-004). Informed consent was obtained from all individual participants.
Detection of IgG and IgM Antibodies Against SARS-CoV-2
Two serological assays, LFIA and MCLIA, were supplied by Beier Bioengineering Co. Ltd (Beijing, China) to qualitatively and/or quantitatively measure IgG and IgM antibodies against SARS-CoV-2. Serum samples were inactivated at 56 °C for 30 min for antibody detection. Tandem antigenic determinants from nucleoprotein and spike proteins of SARS-CoV-2 were employed as diagnostic antigens in both assays.
The 2019-New Coronavirus IgM/IgG Rapid Test Cassette for LFIA (Beier, Beijing, China) was applied to qualitatively detect IgM and IgG following the manufacturer’s instructions. Briefly, serum/plasma diluted in a sample diluent was placed in a specimen well and the results were recorded within 15 min. With the control illuminated, the appearance of an M line indicated an IgM-positive result, a G line indicated an IgG-positive result, and both lines indicated positivity for both IgM and IgG.
The 2019-New Coronavirus IgM/IgG Test for MCLIA (Beier, Beijing, China) was used to detect quantitatively detect IgM and IgG antibodies according to the manual. Briefly, calibration and quality control were performed, diluted serum/plasma samples were loaded onto the machine and the results were auto-calculated by the machine. Samples whose results were below 5 AU/mL were considered negative, those above 8 AU/mL were positive and those in the range of 5–8 AU/mL were potentially positive.
Peripheral Blood Lymphocyte Subset Enumeration and Reference Range Determination
Flow cytometry (FCM) was applied to enumerate peripheral blood mononuclear cell subsets in 95 patients with COVID-19, and a re-examination of peripheral blood mononuclear cell subsets was performed in 11 patients after clinical therapy. Peripheral blood mononuclear cells prepared from anticoagulant venous blood [21] and related reagents were loaded onto a DxFLEX FCM instrument (Beckman Coulter, Fullerton, CA, USA). The following nine indicators were measured according to the manufacturer’s instructions: lymphocyte percentage (reference range [RR] 27.9–37.3%, lymphocytes [1752–2708]), T-cell percentage (62.6–76.8%), T cells (1185–1901), CD4 + T-cell percentage (30–46%), CD4 + T cells (561–1137), CD8 + T-cell percentage (19.2–33.6%), CD8 + T cells (404–754) and CD4 +/CD8 + (1.4–2.0).
Statistical Analysis
Measurement and enumeration results are expressed as mean ± standard deviation (SD) and compared with the Mann–Whitney U test. Categorical variables are expressed as percentages and compared by chi-squared test or Fisher’s exact test. The Shapiro–Wilkes test and Levene’s test were used to assess normality and variance homogeneity. The independent-group Student’s t test and the Bonferroni test were utilised to compare the mean value of every group. A p value < 0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS (version 26.0; Chicago, IL, USA) and GraphPad Prism (version 8.0; San Diego, CA, USA).
Results
Evaluation of Two Serodiagnostic Methods
Lateral Flow Immunochromatographic Assay-IgM/IgG Assay
Of 127 patients confirmed positive for COVID-19 by RT-PCR, 94.48% were positive by the LFIA-IgM assay, with a diagnostic consistency of 95.24% (95% CI 91.8–98.7; Table 1). Moreover, all the 20 suspected cases were negative by the LFIA-IgM assay (Table 1). The sensitivity and specificity of the LFIA-IgM assay were 94.48% (90.5–98.5%) and 100%, respectively. There was no statistically significant difference between RT-PCR and LFIA-IgM assays (p > 0.05), with a Kappa value of 0.823 (0.752–0.946), indicating excellent consistency between the two diagnostic methods.
Table 1.
Comparison between lateral flow immunochromatographic assay (LFIA)-IgM/IgG and reverse transcription polymerase chain reaction (RT-PCR) assays
| RT-PCR | Total | ||
|---|---|---|---|
| Positive | Negative | ||
| LFIA-IgM | |||
| Positive | 120 | 0 | 120 |
| Negative | 7 | 20 | 27 |
| LFIA-IgG | |||
| Positive | 121 | 0 | 121 |
| Negative | 6 | 20 | 26 |
| Total | 127 | 20 | 147 |
Of 127 patients confirmed positive for COVID-19 by RT-PCR, 121 (95.28%) were positive by the LFIA-IgG assay, with a diagnostic consistency of 95.92% (95% CI 92.7–99.2; Table 1). Moreover, all the 20 suspected cases were negative by the LFIA-IgG assay (Table 1). The sensitivity and specificity of the LFIA-IgG assay were 95.28% (95% CI 91.5–99.0) and 100%, respectively. There was no statistically significant difference between RT-PCR and LFIA-IgG assays (p > 0.05), with a Kappa value of 0.846 (0.715–0.960), suggesting excellent consistency between the two diagnostic methods.
Magnetic Chemiluminescence Enzyme Immunoassay-IgM/IgG Assay
Of 127 patients confirmed positive for COVID-19 by RT-PCR, 117 were confirmed positive by the MCLIA-IgM assay, with a diagnostic consistency of 93.20% (95% CI 89.1–97.3; Table 2). Moreover, all the 20 suspected cases were negative by the MCLIA-IgM assay (Table 2). The sensitivity and specificity of the MCLIA-IgM assay were 92.13% (87.4–96.9%) and 100%, respectively. There was no statistically significant difference between RT-PCR and MCLIA-IgM assays (p > 0.05), with a Kappa value of 0.761 (0.609–0.878), implying excellent consistency between the two diagnostic methods.
Table 2.
Comparison between magnetic chemiluminescence enzyme immunoassay (MCLIA)-IgM/IgG and reverse transcription polymerase chain reaction (RT-PCR) assays
| RT-PCR | Total | ||
|---|---|---|---|
| Positive | Negative | ||
| MCLIA-IgM | |||
| Positive | 117 | 0 | 117 |
| Negative | 10 | 20 | 30 |
| MCLIA-IgG | |||
| Positive | 115 | 0 | 115 |
| Negative | 12 | 20 | 36 |
| Total | 127 | 20 | 147 |
Of 127 patients confirmed positive for COVID-19 by RT-PCR, 90.55% were positive by the MCLIA-IgG assay, with a diagnostic consistency of 91.84% (87.4–96.3%; Table 2). Moreover, all the 20 suspected cases were negative by the MCLIA-IgG assay (Table 2). The sensitivity and specificity of the MCLIA-IgG assay were 90.55% (85.4–95.7%) and 100%, respectively. There was no statistically significant difference between RT-qPCR and MCLIA -IgG assays (p > 0.05), with a Kappa value of 0.723 (0.548–0.864), reflecting moderate consistency between the two diagnostic methods.
Antibody Response Profiles of 127 Patients with COVID-19
The concentration of IgM/IgG in patients with COVID-19 was measured by the MCLIA-IgM/IgG assay. The 290 tests included 175 replicated tests on 50 patients at different time points from the onset of illness. Among the replicated tests, 4.0% (2/50), 26.0% (13/50), 26.0% (13/50), 28.0% (14/50) and 16.0% (8/50) of patients were tested two, three, four, five and six times, respectively.
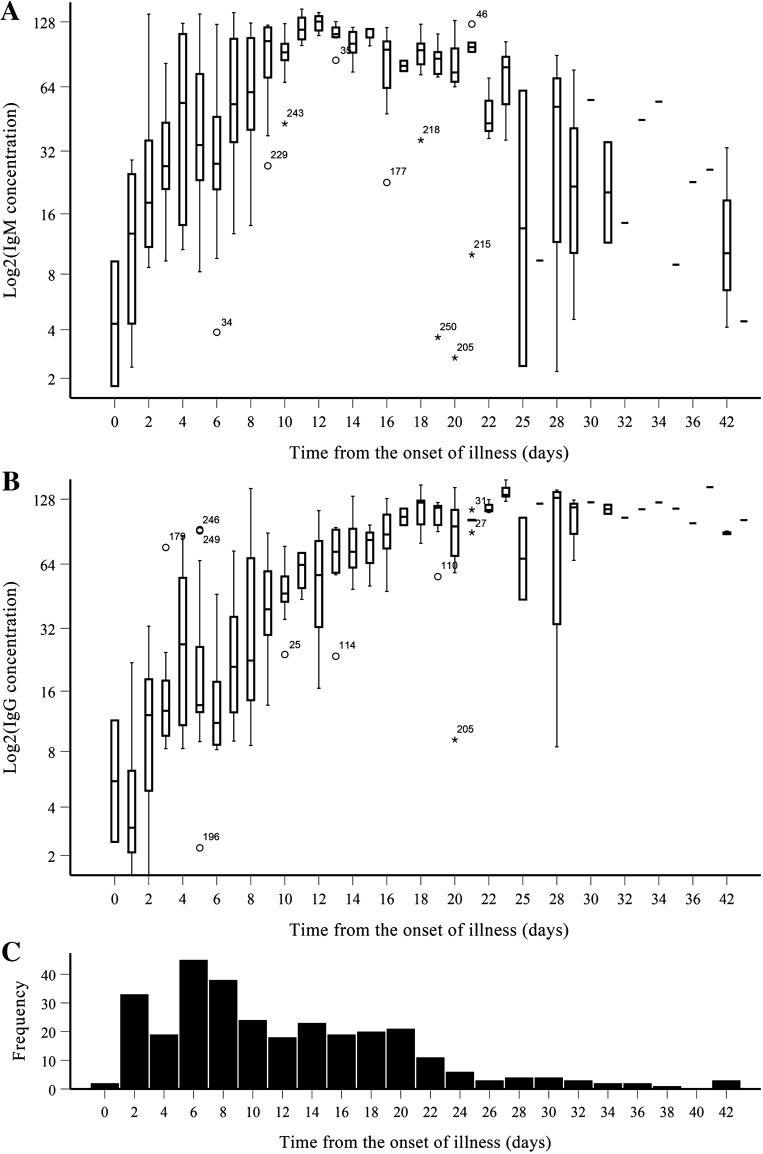
The concentration of IgM and IgG was 67.96 ± 42.82 AU/mL and 58.67 ± 44.90 AU/mL, respectively, without significant differences in age (< 18 years, 18–59 years and > 59 years; p > 0.05) and sex distribution (p > 0.05). According to the time from the onset of illness, all tests were divided into three groups (0–7 days, 8–15 days and > 15 days). For IgM, there were statistically significant differences among the three groups (0–7 days = 40.09 ± 36.28 AU/mL; 8–15 days = 108.45 ± 22.01 AU/mL; > 15 days = 72.84 ± 33.61 AU/mL; p < 0.05). There were also statistically significant differences among the three groups based solely on the repetition tests from 50 patients (0–7 days = 39.60 ± 36.34 AU/mL; 8–15 days = 106.61 ± 24.91 AU/mL; > 15 days = 73.25 ± 33.32 AU/mL; p < 0.05). Furthermore, IgM antibody levels peaked on day 13 and began to decline on day 21, indicating a rise and fall with disease progression (Fig. 1a).
Fig. 1.
Profiles of antibody responses in 127 patients with COVID-19. a IgM; b IgG; and c frequencies
For IgG, there were statistically significant differences among the three groups (0–7 days = 13.05 ± 10.53 AU/mL; 8–15 days = 60.32 ± 26.00 AU/mL; > 15 days = 111.30 ± 22.41 AU/mL). Again, there were also statistically significant differences among the three groups based solely on the repetition tests from 50 patients (0–7 days = 13.07 ± 10.61 AU/mL; 8–15 days = 61.24 ± 26.21 AU/mL; > 15 days = 111.76 ± 22.64 AU/mL; p < 0.05). Moreover, IgG antibody levels peaked on day 17 then plateaued thereafter (Fig. 1b).
Lymphocyte and Lymphocyte Subset Enumeration
Ninety-five patients with COVID-19 were subjected to T-cell response analysis, involving 106 tests with 12 replicated tests from ten patients (Table 3). Among the replicated tests, nine patients underwent two repetitions and one patient had four repetitions. In terms of disease severity, eight cases were mild (7.50%), 72 were common (67.9%), 12 were severe (11.3%) and three were critical (2.80%) among the 95 patients with COVID-19. Mantel–Haenszel chi-squared tests revealed a linear relationship between age and disease severity (χ2 = 10.770, p < 0.05), with a Pearson’s r value of 0.340 (p < 0.05), suggesting that disease severity increased with age.
Table 3.
Lymphocytes and subsets enumeration in 95 patients with COVID-19
| Indicators | Mean ± SD | Proportiona |
|---|---|---|
| Lymphocytes | ||
| Count (× 106/L) | 1848.66 ± 1024.83 | 56:31:19 |
| % | 21.98 ± 10.42 | 71:28:7 |
| T cells | ||
| Count (× 106/L) | 1319.71 ± 768.41 | 50:32:24 |
| % | 70.10 ± 9.04 | 22:57:27 |
| CD4 + T cells | ||
| Count (× 106/L) | 758.38 ± 433.85 | 39:46:21 |
| % | 40.83 ± 8.79 | 8:68:30 |
| CD8 + T cells | ||
| Count (× 106/L) | 476.04 ± 317.01 | 51:32:23 |
| % | 24.89 ± 5.31 | 16:83:7 |
| CD4 +/CD8 + | 1.74 ± 0.62 | 37:43:25 |
SD standard deviation
aIndicates the proportion of patients below, within and above the reference range
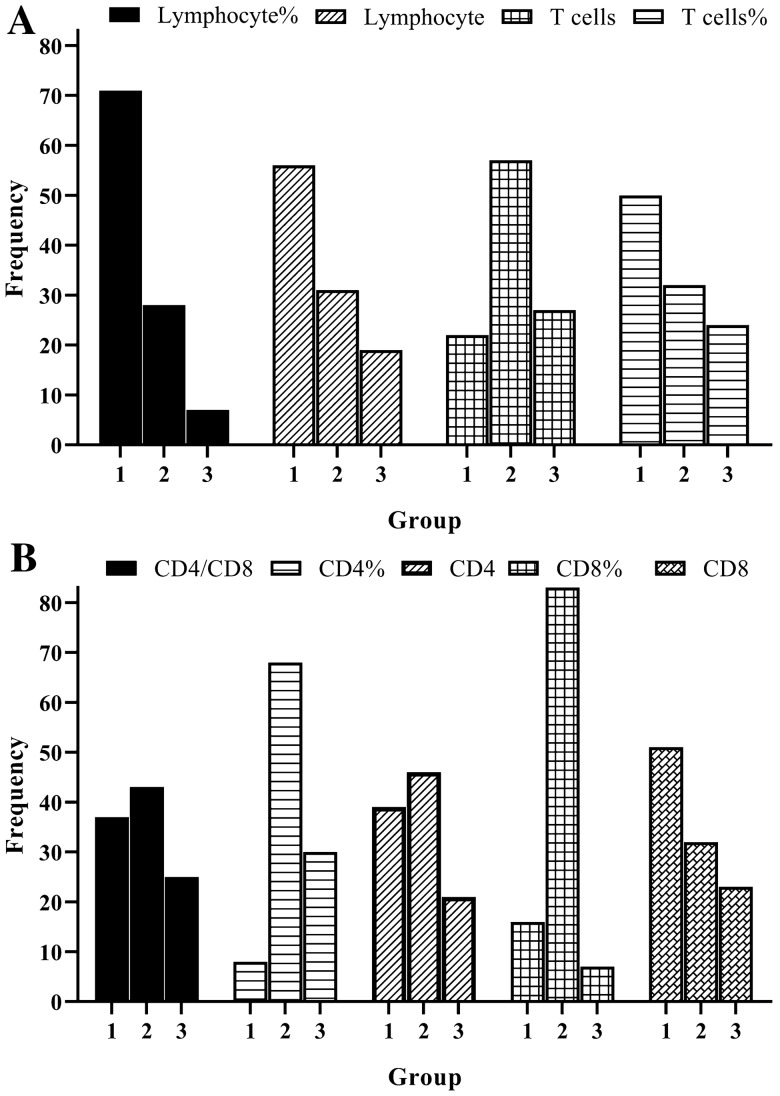
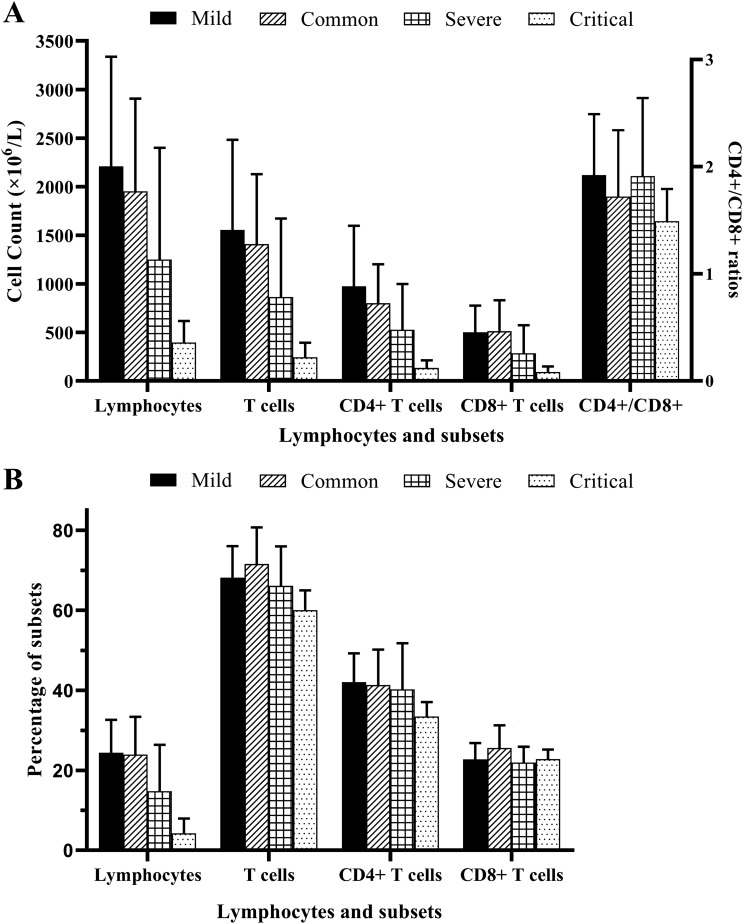
The lymphocyte percentage in 67.0% (71/106) of patients was below the RR (Fig. 2a). However, the more serious the condition, the greater the proportion of patients below the RR (p < 0.05), not considering mild cases. Lymphocyte enumeration was above the RR in only 17.9% of patients (Fig. 2a). Likewise, there were statistically significant differences between patients with mild (2208.84 ± 1130.17) and common (1952.28 ± 953.16) disease severity, and between patients with severe (1250.42 ± 1150.30) and critical (397.38 ± 219.90) disease severity (p < 0.05) [Fig. 3a].
Fig. 2.
Frequencies of lymphocytes and lymphocyte subset enumeration according to reference ranges. a Enumeration and percentage of lymphocytes and T cells and b CD4/CD8 ratio, enumeration and percentage of CD4 + and CD8 + T cells. 1 = below the reference range; 2 = within the reference range; 3 = above the reference range
Fig. 3.
Enumeration and percentage of lymphocytes and subsets in patients with COVID-19 of different disease statuses. a Enumeration of lymphocytes and subsets, and CD4/CD8 ratio and b percentage of lymphocytes and subsets
The T-cell percentage was normal in 53.8% of patients (Fig. 2a), and there were no statistically significant differences in the proportion of patients with differences in disease severity (mild = 68.16% ± 7.84%, common = 71.57% ± 9.15%, severe = 66.12% ± 9.88%, critical = 60.03% ± 4.96%; p > 0.05) (Fig. 3b). As the condition worsened, the percentage of patients within the normal reference range (NRR) decreased (p < 0.05). T-cell enumeration in 47.2% of patients was below the NRR, with 30.2% in the NRR (Fig. 2a). As the condition worsened, the percentage of patients below the NRR increased (p < 0.05). Similarly, there were statistically significant differences between patients with mild (1554.59 ± 928.43) and common (1408.38 ± 721.05) disease severity, and between patients with severe (864.11 ± 807.83) and critical (244.85 ± 149.35) disease severity (p < 0.01) (Fig. 3a).
The CD4 + T-cell percentage in 64.2% (68/106) of patients was normal, with 28.3% above the NRR (Fig. 2b). As the condition worsened, the percentage of patients below the NRR increased (p < 0.05). CD4 + T-cell enumeration in 43.3% of patients was normal, with 36.8% below the RR (Fig. 2b). As the condition worsened, the percentage of patients above the NRR decreased (p < 0.05). There were statistically significant differences between patients with mild (976.05 ± 622.08) and common (801.42 ± 400.83) disease severity, and between patients with severe (528.42 ± 470.17) and critical (135.28 ± 78.22) disease severity (p < 0.01) (Fig. 3a).
The CD8 + T-cell percentage in 78.3% of patients was normal, with 15.1% below the NRR (Fig. 2b). CD8 + T-cell enumeration in 48.1% of patients was below the NRR, and above the NRR in 21.7% (Fig. 2b). As the condition worsened, the percentage of patients above the NRR decreased (p < 0.01). Moreover, there were statistically significant differences between patients with common (514.03 ± 316.50) and critical (91.93 ± 57.52) disease severity (p < 0.01) (Fig. 3a).
The CD4 +/CD8 + ratio in 34.9% of patients was below the NRR, and above the NRR in 23.6% of patients (Fig. 2b). Regarding the 12 replicated tests from ten patients, every indicator was markedly increased with disease progression (p < 0.05).
Discussion
The ongoing COVID-19 pandemic response requires the development of diagnostics, therapeutics and vaccines. In the present study, we analysed antibody levels, lymphocytes, and lymphocyte subsets in patients with COVID-19 to preliminarily evaluate humoural and cellular immunity. The results could provide guidance for clinical diagnostics, specific treatments and vaccine development.
Simple and reliable diagnostic methods are being developed, but NAT based on RT-PCR remains the gold standard for laboratory diagnosis of SARS-CoV-2. Regardless of the advantages and disadvantages of RT-PCR, the sampling method alone greatly reduces the representativeness of samples, which are mostly upper respiratory tract samples (mainly throat swabs) [22]. Furthermore, NAT results from recovering patients can be negative. Serodiagnosis effectively avoids this problem. Herein, we demonstrated that the sensitivity and specificity of LFIA-IgM/IgG and MCLIA-IgM/IgG assays were > 90%, with excellent consistency with RT-PCR (Tables 1, 2), suggesting that these two serodiagnostic assays could rapidly indicate SARS-CoV-2 infection.
SARS-CoV-2 infection involves innate immune response and T- and B-cell immunity and an anti-viral neutralising antibody response [23]. An understanding of humoural and cellular immunity is helpful for clinical diagnosis and treatment. Fluctuation in antibody responses and levels could also provide reference values for clinical treatment, as it reflects the immune responses of patients to SARS-CoV-2. Herein, IgM antibody levels peaked on day 13 and began to decline on day 21, while IgG antibody levels peaked on day 17 and were maintained until the ending of tracking (Fig. 1). Most patients elicited an antibody response to SARS-CoV-2. Similar to the conclusion by Azkur et al. [23], an increase in virus-specific IgM in the acute phase followed by an increase in virus-specific IgG at later phases has been observed in the course of COVID-19 (Fig. 1). Profiling of anti-SARS-CoV-2 antibodies may be useful for diagnosis and in epidemiological analysis. Based on knowledge of antibody fluctuation, doctors could quickly judge the immune response to SARS-CoV-2, which is the main driver of virus elimination [23].
Lymphocyte enumeration could preliminarily reflect cellular immunity. Herein, we found that most patients displayed numbers below the RR (Fig. 2), suggesting that lymphocytopenia occurred during SARS-CoV-2 infection. SARS-CoV-2 can infect human T-cell lines [23], which is a reason for lymphocytopenia. Low lymphocyte count occurred frequently during systemic inflammatory response [24]. Lymphopenia as an early predictor of high disease severity in patients with COVID-19 was thought to cause a defect in antiviral and immune regulatory immunity [23]. The reduction in lymphocyte and subset enumeration was significant, especially in the critical patients of our study (Fig. 3).
Based on nine indicators, the infection provoked strong antibody responses but weakened the ability of cellular immunity to eliminate virus-infected cells. This is unfortunate because strong antibody responses and weak cellular immunity hamper the fight against SARS-CoV-2. Antibody-dependent enhancement could increase viral infection owing to the production of related antibodies [25], and weaker cellular immunity may be insufficient to kill all infected cells. Indeed, anti-SARS-CoV spike protein antibodies are responsible for infecting immune cells [26, 27]. Current clinical evidence suggests that antibody-dependent enhancement of SARS-CoV-2 may occur, which hinders the ability to manage inflammation in the lungs and elsewhere [25].
Because of the COVID-19 pandemic, we did not choose cases depending on certain control factors (e.g. age, sex) and sample at the exact time point but included all hospitalised cases and sampled near the exact time point. Additionally, the effect of heat inactivation on antibody detection has been a concern for a long time. Hu et al. suggested that heat inactivation interfered with the immunoanalysis of antibodies [28], while Xue et al. also demonstrated that there was no significant effect on the results of immunochromatography (LFIA) and chemiluminescence (MCLIA) after heat inactivation [29]. For the sake of biosafety, the sera were inactivated at 56 °C for 30 min before antibody detection in the study.
Conclusions
Understanding humoural and cellular immune responses is of great significance for the prevention and treatment of SARS-CoV-2 infection. Monitoring the immune response to SARS-CoV-2 may provide an early warning for potential risks associated with vaccine immunisation and reinfection. LFIA-IgM/IgG and MCLIA-IgM/IgG assays can indicate SARS-CoV-2 infection, which elicits an antibody response. Lymphocytopenia occurs in patients with COVID-19, which possibly weakens the T-cell response.
Acknowledgements
We thank International Science Editing (http://www.internationalscienceediting.com) for polishing this manuscript.
Author Contributions
JS: supervised the entire project; L-XZ and JS: designed the experiments; J-PW: screened the cases; H-BS: prepared the related reagents; L-XZ, S-YM, Y-QD and Z-HQ: performed the experiments; H-YC: carried out the statistical analysis; YX: collected the data; L-XZ: contributed to the data analysis and manuscript writing.
Compliance with Ethical Standards
Funding
This study was supported by the National Natural Science Foundation of China (No. 81773394). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of Interest
Li-Xia Zhang, Shu-Yan Miao, Zhong-Hua Qin, Jun-Pin Wu, Huai-Yong Chen, Hai-Bai Sun, Yi Xie, Yan-Qing Du, and Jun Shen have no known competing financial interests or personal relationships that could have influenced the work reported in this article.
Ethics Approval
Ethics approval for the study was obtained from the Ethics Commission of Tianjin Haihe Hospital (2020HHQX-001-2020HHQX-004).
Consent to Participate
Informed consent was obtained from all individual participants.
Consent for Publication
All participants consent to participate in a study and having their data published in a journal article.
Availability of Data and Material
The datasets used or analysed during the current study are available from the corresponding author on reasonable request.
Code Availability
All data, models, and code generated or used during the study appear in the submitted article.
Footnotes
Li-Xia Zhang, Shu-Yan Miao and Zhong-Hua Qin contributed equally to this article.
References
- 1.Jiang HW, Li Y, Zhang HN, Wang W, Men D, Yang X, et al. Global profiling of SARS-CoV-2 specific IgG/IgM responses of convalescents using a proteome microarray. medRxvi. 2020 doi: 10.1101/2020.03.20.20039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan Y, Chang L, Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev Med Virol. 2020;30(3):e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu F, Wang H, Zhang Z, Cao L, Wang C, Wu J, et al. Laboratory testing techniques for SARS-CoV-2. Nan Fang Yi Ke Da Xue Xue Bao. 2020;40(2):164–167. doi: 10.12122/j.issn.1673-4254.2020.02.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 5.Mo H, Zeng G, Ren X, Li H, Ke C, Tan Y, et al. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11(1):49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu LP, Wang NC, Chang YH, Tian XY, Na DY, Zhang LY, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13(10):1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu R, Liu X, Han H, Shereen MA, Niu Z, Li D, et al. The comparative superiority of IgM–IgG antibody test to real-time reverse transcriptase PCR detection for SARS-CoV-2 infection diagnosis. MedRxiv. 2020 doi: 10.1101/2020.03.28.20045765. [DOI] [Google Scholar]
- 8.Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020;81(1):e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6):e00461. doi: 10.1128/jcm.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lou B, Li TD, Zheng SF, Su YY, Li ZY, Liu W, et al. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J. 2020;19:2000763. doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z, et al. Viral kinetics and antibody responses in patients with COVID-19. MedRxiv. 2020 doi: 10.1101/2020.03.24.20042382. [DOI] [Google Scholar]
- 12.van Zelm MC, Szczepanski T, van der Burg M, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J Exp Med. 2007;204(3):645–655. doi: 10.1084/jem.20060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milioglou I, Kalaitzidou I, Ladomenou F. Interpretation of lymphocyte subset counts by the general pediatrician. Pediatr Int. 2019;61(1):16–22. doi: 10.1111/ped.13701. [DOI] [PubMed] [Google Scholar]
- 14.Blum KS, Pabst R. Lymphocyte numbers and subsets in the human blood. Do they mirror the situation in all organs? Immunol Lett. 2007;108(1):45–51. doi: 10.1016/j.imlet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Kumar BV, Connors TJ, Farber DL. Human T cell development, localization, and function throughout life. Immunity. 2018;48(2):202–213. doi: 10.1016/j.immuni.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, Jin F, Ma L. The detection and significance of T cells in nasopharyngeal carcinoma patients. J Cancer Res Ther. 2018;14:S331–S335. doi: 10.4103/0973-1482.235350. [DOI] [PubMed] [Google Scholar]
- 17.Retamal-Díaz A, Covián C, Pacheco GA, Castiglione-Matamala AT, Bueno SM, González PA, et al. Contribution of resident memory CD8(+) T cells to protective immunity against respiratory syncytial virus and their impact on vaccine design. Pathogens. 2019;8(3):147. doi: 10.3390/pathogens8030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao MQ, Wang LH, Lian GW, Lin ZF, Li YH, Guo M, et al. Characterization of lymphocyte subsets in peripheral blood cells of children with EV71 infection. J Microbiol Immunol Infect. 2019;S1684–182(18):30401–30408. doi: 10.1016/j.jmii.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Louati N, Rekik T, Menif H, Gargouri J. Blood lymphocyte T subsets reference values in blood donors by flow cytometry. Tunis Med. 2019;97(2):327–334. [PubMed] [Google Scholar]
- 20.Al Kahlout RA, Nasrallah GK, Farag EA, Wang L, Lattwein E, Muller MA, et al. Comparative serological study for the prevalence of anti-MERS coronavirus antibodies in high- and low-risk groups in Qatar. J Immunol Res. 2019;2019:1386740. doi: 10.1155/2019/1386740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Wang L, Feng Z, Geng D, Sun Y, Yuan G. Dynamic changes of laboratory parameters and peripheral blood lymphocyte subsets in severe fever with thrombocytopenia syndrome patients. Int J Infect Dis. 2017;58:45–51. doi: 10.1016/j.ijid.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen MC, et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020 doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Núñez J, Miñana G, Bodí V, Núñez E, Sanchis J, Husser O, et al. Low lymphocyte count and cardiovascular diseases. Curr Med Chem. 2011;18(21):3226–3233. doi: 10.2174/092986711796391633. [DOI] [PubMed] [Google Scholar]
- 25.Tetro JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22(2):72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yip MS, Leung NHL, Cheung CY, Li PH, Lee HHY, Daëron M, et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J. 2014;11(1):82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yip MS, Leung HL, Li PH, Cheung CY, Dutry I, Li D, et al. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J. 2016;22(3 Suppl. 4):25–31. [PubMed] [Google Scholar]
- 28.Hu X, An T, Situ B, Hu Y, Ou Z, Li Q, et al. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. 2020. 10.1101/2020.03.12.20034231. [DOI] [PMC free article] [PubMed]
- 29.Xue X, Zhu C, Huang S, Pan L, Xu J, Li W. Effect of heat inactivation of blood samples on the efficacy of three detection methods of SARS-CoV-2 antibodies. Nan Fang Yi Ke Da Xue Xue Bao. 2020;40(3):316–320. doi: 10.12122/j.issn.1673-4254.2020.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]