Genomic long reads of the interspecific grapevine rootstock cultivar ‘Börner’ (Vitis riparia GM183 × Vitis cinerea Arnold) were used to assemble its chloroplast and mitochondrion genome sequences. We annotated 133 chloroplast and 172 mitochondrial genes, including the RNA editing sites. The organelle genomes in ‘Börner’ were maternally inherited from Vitis riparia.
ABSTRACT
Genomic long reads of the interspecific grapevine rootstock cultivar ‘Börner’ (Vitis riparia GM183 × Vitis cinerea Arnold) were used to assemble its chloroplast and mitochondrion genome sequences. We annotated 133 chloroplast and 172 mitochondrial genes, including the RNA editing sites. The organelle genomes in ‘Börner’ were maternally inherited from Vitis riparia.
ANNOUNCEMENT
Long reads generated by single-molecule real-time (SMRT) DNA sequencing technology (Pacific Biosciences) are one starting point for high-quality chloroplast (1, 2) and mitochondrion genome sequence assemblies. The cultivated grapevine Vitis vinifera is highly susceptible to pathogens. Resistant cultivars like the interspecific hybrid ‘Börner’ (V. riparia GM183 [mother plant] × V. cinerea Arnold [pollen donor]) are used as rootstocks for growing elite grapevine varieties. We assembled and annotated the chloroplast (cp_Boe) and mitochondrion (mt_Boe) genome sequences of ‘Börner’ from SMRT reads. All bioinformatics tools were applied with default parameters unless otherwise noted.
Genomic DNA was extracted from young leaves of cultivar ‘Börner’ (3) and sequenced on a Sequel I sequencer (1Mv3 SMRT cells, binding kit v3.0, sequencing chemistry v3.0, all from PacBio). Potential plastid or mitochondrial reads were filtered by BLASTN (BLAST 2.7.1+) searches (4) against plastid or mitochondrial sequences (RefSeq release 91). The following criteria were used: read length, above 500 nucleotides (nt); identity, above 70%; query coverage, above 30%. The 292,574 potential plastid reads (2,715,983,671 nt in total; N50, 12,829 nt) and the 426,918 potential mitochondrial reads (3,928,350,102 nt; N50, 12,624 nt) were separately assembled with Canu v1.7 (5). Each longest contig displayed high similarity to the chloroplast (6) or mitochondrion (7) genome sequence of V. vinifera. Subsequently, Bandage (8) was used to confirm that the assembly was correct. Overlapping end sequences from the circular genomes were manually trimmed, and the start was aligned to that of the grapevine reference sequences. The assemblies were polished three times with Arrow (SMRT Link release 5.1.0.26412). The last round of polishing was carried out with the start shifted to the opposite position of the sequence.
To aid annotation, RNA was extracted from ‘Börner’ tissues using the peqGOLD plant RNA kit (Peqlab) according to the manufacturer’s instructions. Indexed Illumina sequencing libraries were prepared from 1,000 ng total RNA according to the TruSeq RNA Sample Preparation v2 Guide. The resulting transcriptome sequencing (RNA-Seq) libraries were pooled in equimolar amounts and sequenced in a 2 × 100-nt paired-end format on a HiSeq 1500 instrument.
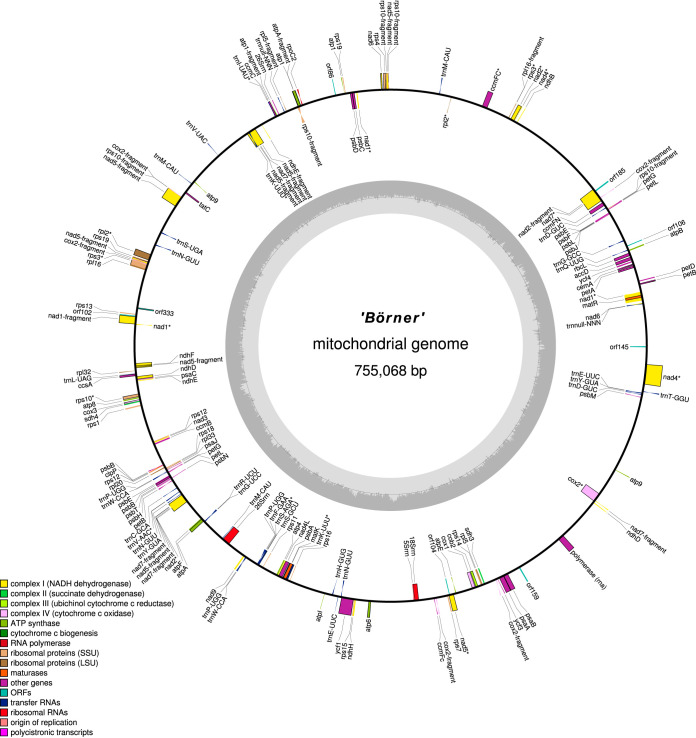
cp_Boe (161,008 bp; GC content, 37.4%) and mt_Boe (755,068 bp; GC content, 44.3%) were annotated with the Web service GeSeq v1.66 (specific settings for cp_Boe: annotate plastid IR enabled, HMMER profile search [9] enabled, reference sequence V. vinifera chloroplast annotation [6], and MPI-MP chloroplast references enabled; specific settings for mt_Boe: reference sequence V. vinifera mitochondrion annotation [7]; settings for both: tRNA annotators tRNAscan-SE v2.0 [10, 11], ARAGORN v1.2.38 [12] with “Allow overlaps” and “Fix introns” enabled) (13), which uses OGDRAW v1.3 (14, 15) to visualize the annotation (Fig. 1). RNA editing sites were determined (16) using RNA-Seq data from five different ‘Börner’ tissues. A total of 133 genes with 90 editing sites were identified for cp_Boe, encoding 85 mRNAs, 39 tRNAs, 8 rRNAs, and 1 pseudogene. For mt_Boe, 172 genes with 624 editing sites were identified that encode 67 mRNAs, 38 tRNAs, 4 rRNAs, and 63 pseudogenes/gene fragments. While cp_Boe confirms the maternal inheritance of the chloroplast from V. riparia due to its high similarity to the chloroplast sequence from V. riparia voucher Wen 12938 (17), mt_Boe is the first mitochondrion genome sequence from V. riparia and differs from the V. vinifera mitochondrion (7) at 141 positions in the coding regions.
FIG 1.
Annotation of the ‘Börner’ mitochondrial genome. The annotation was created with GeSeq and visualized with OGDRAW. Genes containing introns are marked with an asterisk (*).
Data availability.
‘Börner’ RNA-Seq reads (leaves, ENA accession no. ERR3894001; winter leaves, ERR3895010; inflorescences, ERR3894002; tendrils, ERR3894003; roots, ERR3895007), raw SMRT sequence reads (plastid, ERR3610907; mitochondrion, ERR3610837), and chloroplast and mitochondrion genome sequences, including annotation, have been deposited in GenBank/DDBJ/ENA (cp_Boe, ENA accession no. LR738917; mt_Boe, LR738918) under project no. PRJEB34983. The RNA editing tables, coding sequences, and protein sequences of genes subject to RNA editing in edited and unedited form are available as data publications (cpBoe_RNAedit, https://pub.uni-bielefeld.de/record/2941430; mtBoe_RNAedit, https://pub.uni-bielefeld.de/record/2941437).
ACKNOWLEDGMENTS
We thank the members of the Chair of Genetics and Genomics of Plants at Bielefeld University as well as the members of the Julius Kühn Institute for Grapevine Breeding Geilweilerhof for their support.
The project was supported by funds from the Federal Ministry of Food and Agriculture (BMEL), based on a decision by the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE) under the innovation support program (project acronym MureViU, 28-1-82.066-15), as well as by the EU COST Action INTEGRAPE (CA 17111). We acknowledge support for the article processing charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University.
REFERENCES
- 1.Ferrarini M, Moretto M, Ward JA, Surbanovski N, Stevanovic V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, Sargent DJ. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genomics 14:670. doi: 10.1186/1471-2164-14-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stadermann KB, Weisshaar B, Holtgräwe D. 2015. SMRT sequencing only de novo assembly of the sugar beet (Beta vulgaris) chloroplast genome. BMC Bioinformatics 16:295. doi: 10.1186/s12859-015-0726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pucker B, Holtgräwe D, Stadermann KB, Frey K, Huettel B, Reinhardt R, Weisshaar B. 2019. A chromosome-level sequence assembly reveals the structure of the Arabidopsis thaliana Nd-1 genome and its gene set. PLoS One 14:e0216233. doi: 10.1371/journal.pone.0216233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H. 2006. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol 6:32. doi: 10.1186/1471-2148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goremykin VV, Salamini F, Velasco R, Viola R. 2009. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol 26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 8.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler TJ, Eddy SR. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 16.Brenner WG, Mader M, Muller NA, Hoenicka H, Schroeder H, Zorn I, Fladung M, Kersten B. 2019. High level of conservation of mitochondrial RNA editing sites among four Populus species. G3 (Bethesda) 9:709–717. doi: 10.1534/g3.118.200763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen J, Harris AJ, Kalburgi Y, Zhang N, Xu Y, Zheng W, Ickert-Bond SM, Johnson G, Zimmer EA. 2018. Chloroplast phylogenomics of the New World grape species (Vitis, Vitaceae). J Syst Evol 56:297–308. doi: 10.1111/jse.12447. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
‘Börner’ RNA-Seq reads (leaves, ENA accession no. ERR3894001; winter leaves, ERR3895010; inflorescences, ERR3894002; tendrils, ERR3894003; roots, ERR3895007), raw SMRT sequence reads (plastid, ERR3610907; mitochondrion, ERR3610837), and chloroplast and mitochondrion genome sequences, including annotation, have been deposited in GenBank/DDBJ/ENA (cp_Boe, ENA accession no. LR738917; mt_Boe, LR738918) under project no. PRJEB34983. The RNA editing tables, coding sequences, and protein sequences of genes subject to RNA editing in edited and unedited form are available as data publications (cpBoe_RNAedit, https://pub.uni-bielefeld.de/record/2941430; mtBoe_RNAedit, https://pub.uni-bielefeld.de/record/2941437).