The development of more than one primary malignant melanoma (MM) in a patient has been reported to range from 1.3 to 8.2%.1 The multiple primary melanomas (MPM) represent a valid study model for clinical and biomolecular characterization of Familial Melanoma Syndrome. Hereditary MM is the presence of MM in at least 2 first-degree relatives or in three relatives independent of degree.2 The pathogenesis of these inherited conditions is linked to germline cyclin-dependent kinase inhibitor 2A (CDKN2A) mutations.3–5 Among preliminary molecular screening tests, the role of immunohistochemical (IHC) analysis, particularly for p16 protein, is debated. Further studies are needed to confirm that IHC analysis is a suitable modality to screen for CDKN2A abnormalities in paraffin-embedded human MM tissue.
In this study, we evaluated the efficacy of p16 IHC analysis of MPM as preliminary screening to ascertain hereditary setting. We studied all patients with a diagnosis of at least two synchronous or metachronous primary MMs identified by a melanoma registry. Detailed family histories were collected for each MPM patient. Immunohistochemistry of p16, MLH1, MSH2, beta-catenin, E-cadherin and p53 proteins was performed on paraffin-embedded tumor samples. Altered expression of p16 was considered when loss of nuclear expression was observed.6,7 Analysis of germline CDK2NA gene mutations was performed on peripheral blood mononuclear cells of patients.8 Deletions of 9p21 locus were carried out on paraffin-embedded tumor samples using the fluorescence in situ hybridization method.
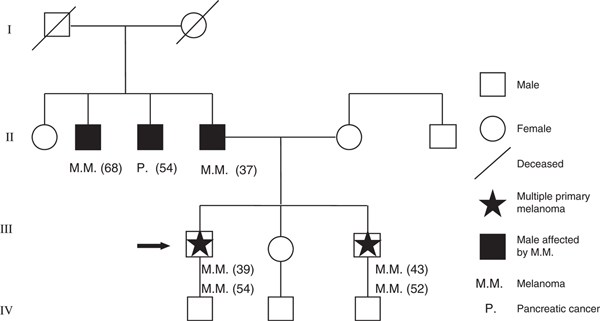
A total of 27 unrelated patients were identified who had been diagnosed with at least two primary MMs. Of these, 25 presented two lesions and two patients presented three lesions. The first two MMs were synchronous in nine patients; in those that were not synchronous, the median interval between the first two MMs was 18 months (range: 0–36 months). A careful evaluation of family history identified a positive MM history in nine of 27 (36.0%) patients and seven had at least 1 first-degree relative affected by MM, corresponding to the clinical criteria for definition of Familial MM. Among the MPM familial cases, a family with four patients having MMs (two of whom had two primary MMs) and one patient with pancreatic cancer was identified (Fig. 1).
Figure 1.
Genealogic tree of familial multiple primary melanomas patients. Numbers in parentheses indicate each patient’s age at diagnosis
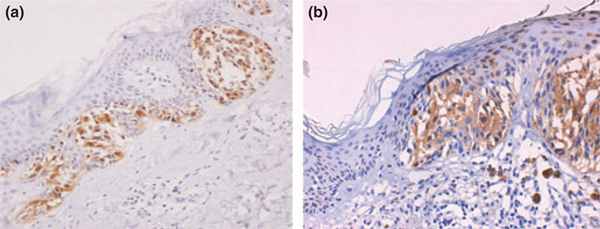
The main IHC result was that, in the MMs of three MPM patients, lack of p16 nuclear expression – with weak cytoplasmic expression – was observed (Fig. 2b). No significant association was found between p53 expression and other clinicopathologic variables as well as for β-catenin and E-cadherin expression. p53 protein was overexpressed in four cases and in two of these the overexpression was seen in both MMs of the same patient. Lack of expression of E-cadherin was shown in three patients. Nuclear expression of β-catenin was present in two cases in addition to a membranous pattern and in one case with prevalent nuclear localization. In three cases, we found a coexistence of several protein expression alterations, in particular, adhesion molecules, β-catenin and E-cadherin, with p53 and p16 proteins (Table 1).
Figure 2.
(a) Nuclear p16 expression in malignant melanoma; (b) Lack of nuclear p16 expression with weak cytoplasmic expression in hereditary multiple primary melanomas: (a) ×200; (b) ×200
Table 1.
Clinical and molecular features of MPM patients
| No. | Sex | Age (years) | MM thickness (mm) | Clark’s Level | Site | IHC | Other tumors | Tumors in family | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p16 | p53 | β-catenin | E-cadherin | MLH1 | MSH2 | CDKN2A/Del 9p21 | ||||||||
| 1 | M | 93 | 5 | IV | Nose | N | + | Focal | M | N | N | Basocellular epithelioma | ||
| 94 | 0.92 | II | Nose tip* | N | + | Focal | M | N | N | |||||
| 2 | F | 39 | 0.5 | III | Abdomen | N | − | M | M | N | N | Colonic adenomas | ||
| 39 | 1.26 | III | Left leg | N | + | M | Absent | N | N | |||||
| 52 | 0.49 | II | Left shoulder-blame | N | + | M | M | |||||||
| 3 | F | 65 | 0.59 | III | Right leg | N | − | M | M | N | N | |||
| 65 | 0.36 | II | Left leg (ex nevi) | N | − | M | M | N | N | |||||
| 4 | F | 30 | 1.8 | II | Left buttock | N | N | |||||||
| 33 | 0.3 | II | Left upper limb | N | − | M | M | N | N | |||||
| 5 | M | 67 | 0.3 | II | Right cheek | N | − | M | M | N | N | MM Brother | ||
| 67 | 1.44 | III | Right temple* | N | − | M | M | N | N | |||||
| 6 | F | 63 | Hypodermic | V | Left leg | N | − | M | M | N | N | 9p21 deletion | MM Daughter | |
| 67 | 1.65 | III | Left leg | Lack of nuclear p16 | − | M | M | N | N | |||||
| 7 | F | 32 | 0.44 | II | Lumbar region | N | − | M | M | N | N | MM Aunt | ||
| 34 | In situ | Right shoulder | N | − | M | M | N | N | ||||||
| 8 | M | 65 | 1.15 | IV | Abdomen | N | − | M | M | N | N | |||
| 66 | 0.35 | II | Left shoulder | N | − | M | M | N | N | |||||
| 9 | M | 35 | 0.6 | II | Back | N | − | N | M | N | N | MM Mother | ||
| 36 | 0.65 | II | Sacral region | N | − | N | M | N | N | |||||
| 10 | M | 70 | 0.28 | II | Back (ex nevi) | N | − | M | M | N | N | |||
| 70 | 0.47 | II | Left cervical region | N | − | M | M | N | N | |||||
| 11 | F | 59 | 1.2 | III | Right ankle | N | − | M | M | N | N | |||
| 61 | 1.73 | IV | Right leg | N | − | M | Absent | N | N | |||||
| 12 | M | 59 | 0.75 | II | Back | N | − | M | M | N | N | |||
| 59 | 1.15 | III | Hip | N | − | M | M | N | N | |||||
| 60 | 0.77 | Scalp | ||||||||||||
| 13 | M | 68 | 1 | III | Left forearm | N | − | M | M | N | N | Gist | MM Brother | |
| 68 | In situ | Left leg (ex nevi) | N | − | M | M | N | N | ||||||
| 14 | F | 24 | 0.65 | II | Back (ex nevi) | N | − | M | M | N | N | |||
| 25 | 0.21 | II | Back (ex nevi) | N | − | M | M | N | N | |||||
| 15 | M | 74 | 2.51 | IV | Left side of back | N | + | M | M | N | N | |||
| 74 | 0.44 | II | Chest | N | − | M | M | N | N | |||||
| 16 | F | 46 | 1.21 | IV | Right shoulder-blame | N | − | M | M | N | N | |||
| 46 | 0.46 | II | Right paravertebral region | N | − | M | M | N | N | |||||
| 17 | M | 53 | 0.25 | II | Back (ex nevi) | N | − | M | M | N | N | |||
| 54 | 1.07 | III | Right axilla | N | − | M | M | N | N | |||||
| 18 | M | 68 | 0.44 | II | Left cheek | N | − | M | M | N | N | Urothelial carcinoma | ||
| 69 | 0.14 | II | Back | N | − | M | M | N | N | |||||
| 19 | F | 50 | In situ | Forehead | N | − | M | M | N | N | Bowen Syndrome | MM Son | ||
| 60 | 0.22 | II | Forehead left upper side | N | − | M | M | N | N | |||||
| 20 | M | 39 | 2.77 | IV | Right lumbar region | Lack of nuclear p16 | − | M | M | N | N | Mut. L65P | Pancreas | MM Mother |
| 39 | In situ | Right shoulder | − | M | M | N | N | MM Brother | ||||||
| In situ | Nape (ex nevi) | MM Uncle | ||||||||||||
| 21 | M | 54 | 0.72 | IV | Left thigh | N | − | M | M | N | N | |||
| 55 | 0.39 | II | Back | N | − | M | M | N | N | |||||
| 22 | F | 21 | 0.3 | II | Thigh | N | − | M | M | N | N | |||
| 32 | In situ | Right hip | N | − | M | M | N | N | ||||||
| 23 | M | 37 | 0.7 | III | Abdomen | N | − | Focal | N | N | ||||
| 40 | 0.39 | II | Left shoulder | N | − | M | M | N | N | |||||
| 24 | F | 36 | 2.82 | IV | Abdomen (ex nevi) | Lack of nuclear p16 | + | M | Absent | N | N | 9p21 | MM Uncle | |
| 36 | In situ | Right shoulder | − | M | M | N | N | deletion | ||||||
| 25 | M | 31 | 1.88 | IV | Ear | N | − | M | M | N | N | |||
| 33 | 0.2 | II | Back | N | − | M | M | N | N | |||||
| 26 | M | 26 | 0.2 | II | Left thigh | N | − | M | ||||||
| 36 | In situ | Right side of chest (ex nevi) | N | − | M | |||||||||
| 36 | 0.23 | II | Right thigh | N | − | M | ||||||||
| 27 | M | 67 | 0.22 | II | Right shoulder-blame | N | − | M | M | N | N | MM Son | ||
| 69 | − | − | Choroid | N | − | M | M | N | N | Ocular MM | ||||
IHC: Immunohistochemistry analysis; N: nuclear expression; M: membranous expression; MM: malignant melanoma.
Germline CDKN2A mutation (T to C transition at bp 194 of exon 2, which results in a missense mutation L65p in p16 and in a silent mutation A79H in p14ARF) was identified in a MPM proband of a patient with Familial Melanoma Syndrome with lack of p16 nuclear expression (Fig. 1). 9p21 large deletion was identified in the two other MPM patients with positive family history for whom lack of p16 nuclear expression was detected at IHC analysis of MM.
Our clinical results indicate that more than 30% of MPM patients have a positive family history of MMs. We believe that our higher rate of familial cases among MPM patients is accurate as it was determined through a very detailed family history reconstruction including first-, second- and third-degree relatives.9 Increased MM risk is present in relatives of MPM patients so that screening programs and counseling may be offered to them. The collection of detailed family histories for the reconstruction of an accurate genealogic tree is crucial for familial lifetime risk evaluation.
The role of IHC evaluation enables us to suggest an approach such as preliminary molecular screening for all MPM patients. Hereditary MMs from patients with p16 germline mutations show disappearance of staining from the nucleus, and weak immunoreactivity in the cytoplasm (Fig. 2). Different mechanisms of p16 inactivation (promoter methylation, 9p21 large deletion, etc.) could be responsible for this staining pattern.10–12 Our IHC evidence revealed a concordance between lack of p16 nuclear expression and biomolecular evidence of CDKN2A aberrations (both L65P germline mutations and 9p21 large deletions).
We have shown that, in all MPM patients, p16 IHC analysis should be adopted for the selection of cases that may benefit from direct sequencing analysis of the CDKN2A gene. The combined use of clinical features and p16 IHC analysis can be useful for characterization of these multiple primary MM patients and recognition of the hereditary setting.
Acknowledgments
The authors thank Stefania Bettelli, Luca Fabbiani and Paola Manni for their technical assistance.
Contributor Information
Giovanni Ponti, Department of Oncology and Haematology, University of Modena and Reggio Emilia, Modena, Italy.
Gabriele Luppi, Department of Oncology and Haematology, University of Modena and Reggio Emilia, Modena, Italy.
Lorena Losi, Department of Pathology, University of Modena and Reggio Emilia, Modena, Ita.
Anna Maria Cesinaro, Department of Pathology, University of Modena and Reggio Emilia, Modena, Ita.
Giuliana Sartori, Department of Pathology, University of Modena and Reggio Emilia, Modena, Ita.
Antonio Maiorana, Department of Pathology, University of Modena and Reggio Emilia, Modena, Ita.
Giovanni Pellacani, Division of Dermatology, Department of Internal Medicine, University of Modena and Reggio Emilia, Modena, Italy.
Caterina Longo, Division of Dermatology, Department of Internal Medicine, University of Modena and Reggio Emilia, Modena, Italy.
Elisa Boni, Division of Dermatology, Department of Internal Medicine, University of Modena and Reggio Emilia, Modena, Italy.
Patrizia Pepe, Division of Dermatology, Department of Internal Medicine, University of Modena and Reggio Emilia, Modena, Italy.
Alberto Giannetti, Division of Dermatology, Department of Internal Medicine, University of Modena and Reggio Emilia, Modena, Italy.
Stefania Seidenari, Division of Dermatology, Department of Internal Medicine, University of Modena and Reggio Emilia, Modena, Italy.
Maria Teresa Landi, Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, DHHS, Bethesda, MD, USA.
References
- 1.Bhatia S, Estrada-Batres L, Maryon T, et al. Second primary tumors in patients with malignant melanoma. Cancer 1999; 86: 2014–2020. [DOI] [PubMed] [Google Scholar]
- 2.Lynch HT, Fusaro RM, Pester J, et al. Familial atypical multiple mole melanoma (FAMMM) syndrome: genetic heterogeneity and malignant melanoma. Br J Cancer 1980; 42: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borg A, Sandberg T, Nilsson K, et al. High frequency of multiple melanomas and breast and pancreatic carcinomas in CDKN2A mutation positive melanoma families. J Natl Cancer Inst 2000; 92: 1260–1266. [DOI] [PubMed] [Google Scholar]
- 4.Helsing P, Nymonen DA, Ariansen S, et al. Population-based prevalence of CDKN2A and CDK4 mutations in patients with multiple primary melanoma. Genes Chromosomes Cancer 2008; 47: 175–184. [DOI] [PubMed] [Google Scholar]
- 5.Hashemi J, Platz A, Ueno T, et al. CDKN2A germ-line mutations in individuals with multiple cutaneous melanomas. Cancer Res 2000; 60: 6864–6867. [PubMed] [Google Scholar]
- 6.Geradts J, Kratzke RA, Niehans GA, et al. Immunohistochemical detection of the cyclin-dependent kinase inhibitor 2/multiple tumor suppressor gene 1(CDKN2/MTS1) product p16INK4A in archival human solid tumors: correlation with retinoblastoma protein expression. Cancer Res 1995; 55: 6006–6011. [PubMed] [Google Scholar]
- 7.Sparrow LE, Eldon MJ, English DR, et al. p16 and p21WAF1 protein expression in melanocytic tumors by immunohistochemistry. Am J Dermatopathol 1998; 20: 255–261. [DOI] [PubMed] [Google Scholar]
- 8.Landi MT, Goldstein AM, Tsang S, et al. Genetic susceptibility in familial melanoma from northeastern Italy. J Med Genet 2004; 41: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries E, Bray FI, Eggermont AM, et al. European Network of Cancer Registries. Monitoring stage-specific trends in melanoma incidence across Europe reveals the need for more complete information on diagnostic characteristics. Eur J Cancer Prev 2004; 13: 387–395. [DOI] [PubMed] [Google Scholar]
- 10.Alonso SR, Ortiz P, Pollán M, et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles. A tissue microarray-based study. Am J Pathol 2004; 164: 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evangelou K, Bramis J, Peros I, et al. Electron microscopy evidence that cytoplasmic localization of the p16INK4a “nuclear” cyclin-dependent kinase inhibitor (CKI) in tumor cells is specific and not an artifact. A study in non-small cell lung carcinomas. Biotech Histochem 2004; 79: 5–10. [DOI] [PubMed] [Google Scholar]
- 12.Ghiorzo P, Villaggio B, Sementa AR, et al. Expression and localization of mutant p16 proteins in melanocytic lesions from familial melanoma patients. Hum Pathol 2004; 35: 25–33. [DOI] [PubMed] [Google Scholar]