Abstract
OBJECTIVE
Childhood socioeconomic disadvantage is associated with adulthood obesity risk, however epigenetic mechanisms are poorly understood. Objectives were to evaluate whether associations of childhood socioeconomic disadvantage with adulthood body mass index (BMI) are mediated by DNA methylation.
METHODS
Participants were 141 men and women from the New England Family Study, prospectively followed prenatally through mean age 47 years. Epigenome-wide DNA methylation was evaluated in peripheral blood and adipose tissue obtained at adulthood, using the Infinium HumanMethylation450K BeadChip. Childhood socioeconomic status (SES) at age 7 years was assessed directly from parent reports. Offspring adiposity was directly assessed using BMI at mean age 47 years. Associations of SES, DNA methylation, and BMI were estimated using least square estimators. Statistical mediation analyses were performed utilizing joint significance test and bootstrapping.
RESULTS
Of CpG sites significant at the 25% false discovery rate level in epigenome-wide methylation-BMI analyses, 91 sites in males and 71 sites in females were additionally significant for SES-methylation associations (P<0.001) in adipose tissue. Many involved genes biologically relevant for development of obesity, including fatty acid synthase (FASN), transmembrane protein 88 (TMEM88), signal transducer and activator of transcription 3 (STAT3), and neuritin 1 (NRN1). There was no evidence of epigenetic mediation in peripheral blood leukocytes.
CONCLUSIONS
DNA methylation at specific genes may be mediators of associations between childhood socioeconomic disadvantage and mid-life BMI in adipose tissue. Findings motivate continued efforts to study if and how childhood socioeconomic disadvantage is biologically embedded at the level of the epigenome in regions etiologically relevant for adiposity.
Keywords: DNA methylation, epigenetics, adiposity, body mass index, socioeconomic disadvantage, socioeconomic status
INTRODUCTION
Adiposity is an important risk factor for many aging-related conditions such as type 2 diabetes, cardiovascular disease, cancer, injuries, physical function disabilities, and premature mortality (1,2). With current trends, total healthcare costs attributable to obesity could reach $957 billion by 2030, accounting for approximately 18% of US health expenditures (3). The etiology of adiposity is multifactorial, and known to have genetic (4,5) and environmental determinants (6,7). What is becoming increasingly understood is that the origins of obesity are detectable early in the life course (8–10). While obesity disproportionately affects lower socioeconomic groups in adulthood, evidence suggests socioeconomic inequalities in obesity may also originate early in the life course, particularly among women (11). For example, a systematic review found inverse associations between childhood SES and adulthood obesity in 70% (14 of 20) of studies in females and 27% (four of fifteen) in males, indicating possible sex differences in the relation between childhood SES and adulthood adiposity (11).
Focus has turned to understanding the biological mechanisms by which genetic and environmental factors operate. Epigenetic mechanisms have been demonstrated to link the internal genetic landscape and external environmental influences, and it is recognized that adiposity and related cardiometabolic risk may arise as results of dysregulated cellular and metabolic programming via epigenetic mechanisms (12,13). Better knowledge of the precise nature of the alterations that give rise to this phenotype will improve the etiologic understanding of adiposity, and could potentially provide treatment targets. Several prior studies investigated epigenetic underpinnings of adiposity, as assessed by DNA methylation (14–23), however almost all studies used only blood (14–21,23). Blood may not be the most relevant tissue for research on the epigenetics of adiposity. Investigating DNA methylation in more targeted tissues, such as adipose tissue, may better elucidate the epigenetic mechanisms involved (24). Using adipose tissue, we recently showed that adiposity phenotypes (e.g. body mass index, android fat mass) were associated with DNA methylation in several genes that are biologically relevant to the development of adiposity, such as lipase E (LIPE), amine oxidase copper containing 3 (AOC3), extracellular superoxide dismutase 3 (SOD3), aquaporin 7 (AQP7), hypoxia-inducible factor 3 alpha (HIF3A), and cholesteryl ester transfer protein (CETP) (25). In contrast, blood DNA methylation profiles were not associated with adiposity, either before or after adjustment for leukocyte cell mixture effects (25). Research on epigenetic regulation of adiposity is still in the early phase, consequently many relevant genomic sites under epigenetic control likely remain to be discovered.
There is increasing interest in elucidating biological pathways by which adverse early life exposures affect adiposity, with strong plausibility for epigenetic mechanisms. Epigenetics play a central role during fetal development, where methylation is associated with somatic lineage differentiation. Early life is thus a sensitive period during which external environmental stimuli may have considerable influence on the establishment of epigenetic patterning. However, few studies have investigated whether socioeconomic status during childhood is related to DNA methylation profiles (26–31). Beyond this, none have had the opportunity to investigate DNA methylation profiles in adipose tissue (in contrast to blood), which may be the optimal strategy to investigate these hypotheses. Furthermore, to our knowledge, no studies to date have performed formal mediation analyses evaluating whether epigenetic methylation patterns may be mediators of observed associations between early life socioeconomic disadvantage and later life adiposity. Consequently, objectives of this study were to evaluate whether associations of childhood socioeconomic disadvantage with adulthood body mass index are mediated by alterations in DNA methylation in CpG (cytosine-phosphate-guanine) sites, assessed in peripheral blood leukocytes and subcutaneous adipose tissue samples obtained in adulthood. If DNA methylation explains part of the association between childhood disadvantage and adult adiposity, this should be detected by statistical mediation analyses. The mediation analyses themselves are not dispositive of a causal effect of disadvantage on methylation and a resulting causal effect of methylation on adiposity; such inferences require further assumptions regarding confounding, including no unmeasured SES-methylation confounders, and no unmeasured methylation-adiposity confounders.
METHODS
Sample
Study participants were from the New England Family Study (NEFS), which comprises a series of follow-up studies of subsamples of the 17,921 offspring of pregnant women enrolled in the Collaborative Perinatal Project (CPP) in the study’s Providence, Rhode Island, and Boston, Massachusetts, sites (United States) between 1959 and 1966 (32). The current NEFS sub-study, named the Longitudinal Effects on Aging Perinatal (LEAP) Project, is comprised of Providence-born participants assessed between 2010–2011. Of 796 participants eligible for assessment (i.e. not deceased, not incarcerated, had assessments taken at age 7 years, were located, and lived within 100 miles of the clinical assessment site), we were able to establish contact with 522 (76%) of the participants within the relatively brief 13-month data collection period, and invited them to participate in the study. Of these 522 participants, 19% (n=95) refused to participate, and a further 5% (n=27) agreed to participate but were unable to schedule assessments within the data collection period. This left 400 participants on whom assessments were made. Of these, 316 had adequate adipose tissue biopsy performed, 68 refused, and 16 had inadequate biopsy specimens. Blood and adipose tissue DNA methylation analyses were performed on a representative sample of 144 of these 316 participants. With the exception of adipose tissue for one participant, samples yielded adequate DNA methylation data for blood and adipose tissue analyses for all 144 participants. Of the 143 participants with both adipose and blood methylation data, 2 were missing age 7 SES. Consequently, analyses were performed on 141 participants for adipose tissue analyses, and 142 participants for blood analyses. Characteristics of participants with DNA methylation data did not differ significantly from LEAP participants without methylation data, except that mean BMI was greater in participants with vs. without DNA methylation data (31.5 vs. 29.7 kg/m2; p=0.02). This was likely because adipose tissue samples of adequate amount for methylation assays were more readily obtained when there were larger amounts of adipose tissue to harvest from. The study protocol was approved by the institutional review boards at Brown University and Memorial Hospital of Rhode Island. All participants provided informed consent.
Tissue Sample Collection, Methylation Profiling, and Data Processing
Subcutaneous adipose tissue samples were collected from the upper outer quadrant of the buttock using a 16-gauge needle and disposable syringe. DNA was extracted from adipose tissue samples using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) and the Zymo Genomic DNA Clean & Concentrator Kit, according to the manufacturers’ protocol. Whole blood samples were centrifuged to obtain buffy coat, and peripheral blood leukocyte DNA was extracted using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA), according to the manufacturers’ protocol. DNA was sodium-bisulfite converted using the EZ-96 DNA Methylation-Direct and EZ DNA Methylation-Direct kits (Zymo Research, Orange, CA; as per manufacturer’s instructions). Blood and adipose tissue samples were randomized across plates, and analyzed using the Infinium HumanMethylation450 BeadChip array (Illumina, San Diego, CA) at the Genomics Core Facility at the UCSF Institute for Human Genetics (San Francisco, CA), following standard Illlumina protocols. Control probe data was used to assess the quality of the array data (total signal, distribution of detection p-values, fraction of missing values). One adipose tissue sample was excluded due to having a detection p-value >0.05 for >1% of probes.
The DNA methylation data were out-of-band background corrected, dye bias-corrected (33), and further normalized using the Beta-Mixture Quantile Dilation (BMIQ) approach (34) (in order to obtain similar ranges for type I vs. type II probes on the infinium array). Methylation beta values for each CpG was calculated as M/(M+U+ε), where M and U refer to the signal from the probe corresponding to the methylated and unmethylated target CpG, respectively, and ε=100 in order to protect against division-by-zero. We also removed non-CpG sites (rs and ch probes), CpG sites located on sex chromosomes, CpG sites with SNPs in probe (35) (at the flanking G, within 10 nt of the target CpG, and 11–50 nt away from the target CpG), CpG sites that are considered cross-reactive (36) and CpG sites that had a detection p-value >0.05 for more than 1% of samples. The ‘rs’ (representSNPassays) probes are normal SNP probes; they do not measure methylation. The ‘ch’ probes represent non-CpG methylation sites.
Independent Variable
Socioeconomic status (SES) was assessed prospectively at age 7 by averaging the percentile of both parents’ educational attainment, occupation, and income relative to the US population (37).
Dependent Variables
Weight and height measures were obtained from participants wearing light clothing without shoes, using a calibrated stadiometer and weighing scale operated by trained nurse researchers. Heads were positioned in the Frankfurt plane. Body mass index (BMI) was calculated as weight per height squared (i.e. kg/m2).
Covariates
Age was directly assessed via date of birth (recorded directly in this prenatal cohort), subtracted from clinic visit date. Sex, race/ethnicity, and smoking status (current number of cigarettes smoked daily) were self-reported in adulthood. Childhood weight and height and resulting BMI (defined above) were directly assessed at age 7 years by medical staff according to a standard protocol (38).
Epigenome-wide associations between blood DNA methylation and adiposity could be confounded by differential prevalence of leukocytes from varying developmental lineages (24,39). Consequently, all epigenome-wide association studies (EWAS) analyses in blood were performed with adjustment for blood leukocyte distribution using the method developed by Houseman et al. (40). Estimates of cell type proportion for each of major 6 cell types (CD8T, CD4T, NK, B-cell, Monocytes, Granulocytes) were adjusted in the EWAS analyses as covariates.
Statistical Analyses
Variables including age, race, BMI at age 7 and cigarette smoking were compared between high and low SES groups in men and women, separately, using t-tests or chi-square tests.
We conducted the joint significance test (41) by two-stage EWAS analyses to identify CpG sites where methylation levels statistically mediate the association of SES with adult BMI. Specifically, we performed the first stage EWAS analyses to detect associations between CpG methylation and adult BMI. In stage 1 EWAS, with false discovery rate (FDR) (42) of 25%, we selected the candidate CpG sites with significant CpG-BMI associations. Among these we then conducted second stage EWAS analyses for the association of CpG methylation with SES. The candidate CpG sites with FDR<25% from the first stage EWAS, and p<0.001 in the second stage analyses, were reported as candidate mediating CpG sites. In the formal mediation analyses, suggestive evidence for mediation was inferred if the indirect effect p was <0.05. Note that in the light of the large number of tests performed in this study, these alpha thresholds are liberal. It should be noted that a more lenient FDR threshold of 25% was implemented due to our goal of performing mediation analyses, where data are subsetted twice; once for subsetting significant associations of BMI with methylation patterns (i.e. stage 1), and then from those significant associations, a second subset is identified that is also significantly associated with SES (i.e stage 2). To preserve the most signals from the first stage, we allowed for a 25% false discovery that would be further decreased in the second stage analyses. The FDR cutoff of 0.25 is arbitrary but not uncommon in practice (43–46). Similarly, the second stage cutoff of p<0.001 is arbitrary, but also within typical range of other studies that vary from 0.05 to 0.0005 (47–49). While we followed the convention of statistical significance cutoffs, we note that the optimal theoretical justification is still limited in literature. The candidate loci were further scrutinized with their biological plausibility to avoid false positive findings. Given the moderate sample size, these reasonably liberal cut-offs minimized the likelihood of false negative findings. In addition, among the CpG sites selected as candidate mediating sites, we emphasized particular attention on instances where several of the CpG sites were annotated to the same gene. These multiple hits occurring within the same gene could suggest that particular gene, if important for gene regulation and as a mediator between early life SES and later life body mass index, may have altered methylation patterns at multiple sites. As a technical note, mediation analyses make the genome-wide adjustment even more challenging since the second stage is a mixture of validation and additional investigation. The two-stage joint significance test was proposed in 2002 for low-dimensional analyses (41). To our knowledge, this study is the first to utilize the approach in a high-dimensional setting. To provide better statistical justification in the two-stage high-dimensional mediation analyses, we have another ongoing theoretical study, currently under review (Huang Y-T, Under Review), focusing on this issue where we aim to develop a testing procedure with well-justified theoretical property.
The first stage analyses were performed for associations of CpG methylation with adult BMI due to prior discoveries in this and other datasets showing significant associations of methylation patterns with BMI (25). Since associations of SES with CpG are less understood, we only conducted the analyses for this association focusing on CpG sites significantly associated with BMI in the first stage analyses. The sequence of the strategy better used the existing knowledge and may minimize the agnostic analyses for the CpG-SES association. However sensitivity analyses were performed using association of SES with CpG associations in stage 1 instead of 2. The joint significance test-based mediation analyses were conducted for DNA methylation in blood and adipose tissue separately.
All analyses were adjusted for age, race, BMI at age 7, smoking status in adulthood. Age, race and smoking status are related to methylation patterns and were adjusted for as potential confounders. BMI at age 7 years was adjusted for so that the estimated indirect effects in the mediation analyses were in reference to the change in BMI from childhood to adulthood; this is important in the CPP sample because the socioeconomic gradient in childhood BMI was in the direction of lighter children among lower SES families (Gilman et al., Under Review) during the era in the United States when the children were born (1959–1966). In all multivariate adjusted analyses, age 7 SES and BMI were included as continuous variables.
While the genome-wide joint significance test strategy only identified CpG sites with significant p-values, it did not provide effect estimates. Therefore, we utilized product-based mediation analyses to quantify the direct and indirect effects for the candidate mediating CpG sites that were identified through the two stage process. For each of these CpG sites, we fit two regression models using ordinary least square estimators. The first model regressed the methylation value on SES with adjustment of covariates: , and the second model regressed BMI on SES and methylation with adjustment of covariates: , where S, M, Y and X are SES, methylation M-value, adult BMI and demographic covariates, respectively, and α’s and β’s are the regression coefficients for the methylation M-value and the adiposity outcome (i.e., adult BMI), respectively. The M-value is where B is the beta-value of DNA methylation. T is transpose operator in matrix algebra and E is the expectation operator for the random variable. Under the framework of causal inference (50) using potential outcomes (51), the point estimates of direct effect (DE) and indirect effect (IE, or mediation effect) can be expressed by the combinations of regression parameters of the above two models: and (52). Their corresponding variance and confidence intervals were obtained using bootstrap procedures. Note that we conducted two different tests to identify mediation effects: we performed joint significance tests in epigenome-wide scan and directly testing IE from the above formula, termed as product significance tests for the candidate sites. It has been shown in the literature that the joint significance test is more powerful than the product significance test (41), and therefore, we only calculated the p-value from the product significance test for the candidate CpG sites. Proportion of mediation was calculated as: IE/(DE + IE) to indicate the proportion of total effect of SES on BMI mediated through CpG methylation (53); note that the proportion of mediation has plausible interpretation only if both DE and IE are in the same direction.
RESULT
Unadjusted analyses demonstrated significant associations of age 7 SES with adulthood BMI in females (p=0.004) and not males (p=0.29) (Table 1). There were no significant associations of SES with other covariates, such as age, race/ethnicity, and smoking (Table 1).
Table 1.
Demographics showing associations of age 7 socioeconomic status (SES) with covariates, stratified by sex.
| Male | Female | |||||
|---|---|---|---|---|---|---|
| Low SES | High SES | p | Low SES | High SES | p | |
| n | 34 | 33 | 38 | 36 | ||
| Age, mean years (SD) | 47.1 (1.4) | 47.0 (1.8) | 0.94 | 46.9 (1.7) | 46.7 (1.8) | 0.72 |
| Race/ethnicity, % white | 55.9 | 69.7 | 0.36 | 57.9 | 80.6 | 0.064 |
| Age 7 BMI, mean kg/m2 (SD) | 16.8 (3.7) | 16.2 (1.5) | 0.39 | 16.1 (2.4) | 16.2 (1.7) | 0.84 |
| Adult BMI, mean kg/m2 (SD) | 31.2 (6.1) | 32.7 (5.2) | 0.29 | 34.1 (8.2) | 28.3 (8.4) | 0.0036 |
| Current smoking, mean no. (SD) | 10.8 (13.4) | 9.7 (9.7) | 0.71 | 13.4 (14.5) | 8.3 (9.5) | 0.078 |
No., number of cigarettes smoked per day; SD, standard deviation.
High vs. low for SES was calculated using a median split.
In the first stage EWAS, adipose tissue DNA methylation at 110,452 CpG sites in males and 87,772 in females was associated with adult BMI at FDR<25%. Interestingly, no associations were observed in peripheral blood leukocytes at FDR <25%. Initial analyses showed an over-representation of DNA methylation-BMI associations at CpG sites in adipose tissue but not blood, for both males and females (Supplementary Figure 1). This suggests in initial analyses that methylation patterns are associated with body mass index in adipose tissue.
Of the 110,452 sites in males and 87,772 sites in females that were identified in the first stage EWAS of DNA methylation and adult BMI (FDR<25%), 91 sites in males and 71 sites in females showed suggestive p-values for SES-methylation associations (P<0.001), after adjusting for age, race, sex, age 7 BMI, and smoking in the adipose tissue samples. There was no evidence of epigenetic mediation in peripheral blood leukocytes, as no CpG sites for either males or females passed the first stage of statistical testing (FDR<25%). Furthermore, there was minimal evidence of differential leukocyte admixture in relation to SES or BMI, where proportions of B cells, CD4+ T cells, CD8+ T cells, granulocytes, monocytes, and Natural Killer cells were similar across tertiles of SES and BMI (Supplementary Figure 2).
The 20 CpG sites with the lowest FDR for the association of methylation with BMI (in adipose tissue) are shown in Table 2 (females) and Table 3 (males). The full lists of 71 and 91 CpG sites are displayed in Supplementary Table 1 (females) and Supplementary Table 2 (males). The candidate mediation CpG sites were different in analyses of females vs. males. R2 values ranged from 0.000 to 0.297 (maximum possible: 1.000) in the association of the methylation sites with BMI (Tables 2 and 3, Supplementary Tables 1 and 2).
Table 2.
Female adipose tissue mediation effects, for methylation sites in the 20 CpG sites having the lowest false discovery rate (FDR) values.
| CpG ID | Chrom | Gen Loc | Gene | pval(S->Mj) | pval(Mj->BMI) | FDR(Mj->BMI) | Indirect | 95% CI | P.indirect | Direct | 95% CI | P.direct | R2 | Prop. of Mediation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cg21120176 | 6 | 6534797 | LOC285780 | <0.001 | <0.001 | 0.036 | −0.17 | (−0.33,−0.06) | <0.001 | −0.13 | (−0.33,0.12) | 0.350 | 0.264 | 0.57 |
| cg22462726 | 3 | 184209261 | 0.001 | 0.001 | 0.046 | −0.05 | (−0.15,0.02) | 0.136 | −0.09 | (−0.21,0.06) | 0.220 | 0.276 | 0.38 | |
| cg11950105 | 17 | 80050646 | FASN | <0.001 | 0.001 | 0.050 | −0.29 | (−0.58,−0.13) | <0.001 | −0.21 | (−0.55,0.20) | 0.284 | 0.293 | 0.57 |
| cg25490145 | 17 | 80358850 | C17orf101 | 0.001 | 0.001 | 0.050 | 0.09 | (0.02,0.19) | 0.012 | 0.10 | (−0.04,0.25) | 0.164 | 0.295 | 0.47 |
| cg26750548 | 10 | 88441152 | LDB3 | <0.001 | 0.001 | 0.052 | −0.32 | (−0.64,−0.10) | 0.000 | −0.28 | (−0.73,0.21) | 0.230 | 0.257 | 0.54 |
| cg12930882 | 5 | 37825585 | GDNF | 0.001 | 0.003 | 0.060 | −0.08 | (−0.20,0.03) | 0.224 | −0.15 | (−0.33,0.06) | 0.168 | 0.210 | 0.33 |
| cg17918937 | 15 | 64992780 | OAZ2 | <0.001 | 0.003 | 0.062 | −0.11 | (−0.27,0.00) | 0.054 | −0.24 | (−0.49,0.03) | 0.098 | 0.168 | 0.32 |
| cg25840926 | 2 | 20647987 | RHOB | 0.001 | 0.003 | 0.062 | −0.11 | (−0.24,−0.02) | 0.010 | −0.16 | (−0.33,0.04) | 0.124 | 0.257 | 0.40 |
| cg21170029 | 1 | 209799330 | LAMB3 | 0.001 | 0.004 | 0.064 | −0.02 | (−0.13,0.05) | 0.540 | −0.13 | (−0.28,0.03) | 0.160 | 0.125 | 0.14 |
| cg02422603 | 13 | 114890566 | RASA3 | <0.001 | 0.004 | 0.064 | 0.30 | (0.05,0.64) | 0.022 | 0.30 | (−0.24,0.80) | 0.268 | 0.229 | 0.50 |
| cg18410680 | 17 | 7758397 | TMEM88 | 0.001 | 0.004 | 0.066 | −0.07 | (−0.18,0.00) | 0.060 | −0.13 | (−0.28,0.05) | 0.202 | 0.272 | 0.37 |
| cg27630153 | 16 | 88845038 | FAM38A | 0.001 | 0.005 | 0.071 | −0.23 | (−0.42,−0.08) | 0.002 | −0.21 | (−0.56,0.14) | 0.214 | 0.297 | 0.52 |
| cg14917100 | 2 | 127528627 | 0.001 | 0.006 | 0.072 | −0.14 | (−0.29,−0.04) | 0.006 | −0.20 | (−0.44,0.04) | 0.100 | 0.132 | 0.40 | |
| cg07568841 | 7 | 30362781 | ZNRF2 | <0.001 | 0.006 | 0.073 | 0.21 | (0.04,0.45) | 0.020 | 0.28 | (−0.10,0.63) | 0.138 | 0.218 | 0.42 |
| cg05522011 | 4 | 81122726 | PRDM8 | <0.001 | 0.006 | 0.074 | −0.07 | (−0.24,0.02) | 0.182 | −0.20 | (−0.45,0.10) | 0.246 | 0.215 | 0.26 |
| cg16850945 | 3 | 52488229 | TNNC1; NISCH | 0.001 | 0.007 | 0.077 | −0.10 | (−0.21, −0.03) | 0.008 | −0.05 | (−0.19,0.12) | 0.614 | 0.246 | 0.69 |
| cg06468133 | 11 | 3244441 | 0.001 | 0.008 | 0.079 | 0.26 | (0.04,0.54) | 0.020 | 0.26 | (−0.19,0.70) | 0.228 | 0.211 | 0.50 | |
| cg27296413 | 1 | 159037843 | AIM2 | 0.001 | 0.008 | 0.079 | 0.15 | (0.03,0.26) | 0.026 | 0.07 | (−0.11,0.33) | 0.440 | 0.133 | 0.67 |
| cg00278517 | 6 | 167070616 | RPS6KA2 | 0.001 | 0.012 | 0.092 | 0.09 | (−0.02,0.27) | 0.124 | 0.18 | (−0.10,0.41) | 0.254 | 0.194 | 0.34 |
| cg06608378 | 15 | 76016129 | ODF3L1 | <0.001 | 0.013 | 0.093 | −0.08 | (−0.22,0.00) | 0.062 | −0.15 | (−0.34,0.11) | 0.250 | 0.221 | 0.33 |
BMI, body mass index; Chrom, chromosome number; Direct, direct effect of methylation mediator in association of SEI with BMI; Gen Loc, gene location;FDR(Mj->BMI), false discovery rate for association of methylation mediator with BMI; Indirect, indirect effect of methylation mediator in association of SEI with BMI; P.direct, p value for direct effect; P.indirect, p value for indirect effect; Prop. of Mediation, proportion of total association between SES and BMI explained by DNA methylation (calculated as indirect effect/[direct effect+indirect effect]; proportion of mediation has plausible interpretation only if both direct and indirects effects are in the same direction; "N/A" represents effects in opposite directions); R2, percentage of the variance in BMI explaned by DNA methylation, without adjusting for covariates (range 0.000–1.000); SEI, age 7 y socioeconomic index; pval(S->Mj), p value for association of SEI with methylation mediator; pval(Mj->BMI), p value for assocation of methylation mediator with BMI; 95% CI, 95% confidence intervals.
Table 3.
Male adipose tissue mediation effects, for methylation sites in the 20 CpG sites having the lowest false discovery rate (FDR) values.
| CpG ID | Chrom | Gen Loc | Gene | pval(S->Mj) | pval(Mj->BMI) | FDR(Mj->BMI) | Indirect | 95% CI | P.indirect | Direct | 95% CI | P.direct | R2 | Prop. of Mediation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cg21120176 | 6 | 6534797 | LOC285780 | <0.001 | <0.001 | 0.036 | −0.17 | (−0.33,−0.06) | <0.001 | −0.13 | (−0.33,0.12) | 0.350 | 0.264 | 0.57 |
| cg22462726 | 3 | 184209261 | 0.001 | 0.001 | 0.046 | −0.05 | (−0.15,0.02) | 0.136 | −0.09 | (−0.21,0.06) | 0.220 | 0.276 | 0.38 | |
| cg11950105 | 17 | 80050646 | FASN | <0.001 | 0.001 | 0.050 | −0.29 | (−0.58,−0.13) | <0.001 | −0.21 | (−0.55,0.20) | 0.284 | 0.293 | 0.57 |
| cg25490145 | 17 | 80358850 | C17orf101 | 0.001 | 0.001 | 0.050 | 0.09 | (0.02,0.19) | 0.012 | 0.10 | (−0.04,0.25) | 0.164 | 0.295 | 0.47 |
| cg26750548 | 10 | 88441152 | LDB3 | <0.001 | 0.001 | 0.052 | −0.32 | (−0.64,−0.10) | 0.000 | −0.28 | (−0.73,0.21) | 0.230 | 0.257 | 0.54 |
| cg12930882 | 5 | 37825585 | GDNF | 0.001 | 0.003 | 0.060 | −0.08 | (−0.20,0.03) | 0.224 | −0.15 | (−0.33,0.06) | 0.168 | 0.210 | 0.33 |
| cg17918937 | 15 | 64992780 | OAZ2 | <0.001 | 0.003 | 0.062 | −0.11 | (−0.27,0.00) | 0.054 | −0.24 | (−0.49,0.03) | 0.098 | 0.168 | 0.32 |
| cg25840926 | 2 | 20647987 | RHOB | 0.001 | 0.003 | 0.062 | −0.11 | (−0.24,−0.02) | 0.010 | −0.16 | (−0.33,0.04) | 0.124 | 0.257 | 0.40 |
| cg21170029 | 1 | 209799330 | LAMB3 | 0.001 | 0.004 | 0.064 | −0.02 | (−0.13,0.05) | 0.540 | −0.13 | (−0.28,0.03) | 0.160 | 0.125 | 0.14 |
| cg02422603 | 13 | 114890566 | RASA3 | <0.001 | 0.004 | 0.064 | 0.30 | (0.05,0.64) | 0.022 | 0.30 | (−0.24,0.80) | 0.268 | 0.229 | 0.50 |
| cg18410680 | 17 | 7758397 | TMEM88 | 0.001 | 0.004 | 0.066 | −0.07 | (−0.18,0.00) | 0.060 | −0.13 | (−0.28,0.05) | 0.202 | 0.272 | 0.37 |
| cg27630153 | 16 | 88845038 | FAM38A | 0.001 | 0.005 | 0.071 | −0.23 | (−0.42,−0.08) | 0.002 | −0.21 | (−0.56,0.14) | 0.214 | 0.297 | 0.52 |
| cg14917100 | 2 | 127528627 | 0.001 | 0.006 | 0.072 | −0.14 | (−0.29,−0.04) | 0.006 | −0.20 | (−0.44,0.04) | 0.100 | 0.132 | 0.40 | |
| cg07568841 | 7 | 30362781 | ZNRF2 | <0.001 | 0.006 | 0.073 | 0.21 | (0.04,0.45) | 0.020 | 0.28 | (−0.10,0.63) | 0.138 | 0.218 | 0.42 |
| cg05522011 | 4 | 81122726 | PRDM8 | <0.001 | 0.006 | 0.074 | −0.07 | (−0.24,0.02) | 0.182 | −0.20 | (−0.45,0.10) | 0.246 | 0.215 | 0.26 |
| cg16850945 | 3 | 52488229 | TNNC1; NISCH | 0.001 | 0.007 | 0.077 | −0.10 | (−0.21, −0.03) | 0.008 | −0.05 | (−0.19,0.12) | 0.614 | 0.246 | 0.69 |
| cg06468133 | 11 | 3244441 | 0.001 | 0.008 | 0.079 | 0.26 | (0.04,0.54) | 0.020 | 0.26 | (−0.19,0.70) | 0.228 | 0.211 | 0.50 | |
| cg27296413 | 1 | 159037843 | AIM2 | 0.001 | 0.008 | 0.079 | 0.15 | (0.03,0.26) | 0.026 | 0.07 | (−0.11,0.33) | 0.440 | 0.133 | 0.67 |
| cg00278517 | 6 | 167070616 | RPS6KA2 | 0.001 | 0.012 | 0.092 | 0.09 | (−0.02,0.27) | 0.124 | 0.18 | (−0.10,0.41) | 0.254 | 0.194 | 0.34 |
| cg06608378 | 15 | 76016129 | ODF3L1 | <0.001 | 0.013 | 0.093 | −0.08 | (−0.22,0.00) | 0.062 | −0.15 | (−0.34,0.11) | 0.250 | 0.221 | 0.33 |
BMI, body mass index; Chrom, chromosome number; Direct, direct effect of methylation mediator in association of SEI with BMI; Gen Loc, gene location;FDR(Mj->BMI), false discovery rate for association of methylation mediator with BMI; Indirect, indirect effect of methylation mediator in association of SEI with BMI; P.direct, p value for direct effect; P.indirect, p value for indirect effect; Prop. of Mediation, proportion of total association between SES and BMI explained by DNA methylation (calculated as indirect effect/[direct effect+indirect effect]; proportion of mediation has plausible interpretation only if both direct and indirects effects are in the same direction; "N/A" represents effects in opposite directions); R2, percentage of the variance in BMI explaned by DNA methylation, without adjusting for covariates (range 0.000–1.000); SEI, age 7 y socioeconomic index; pval(S->Mj), p value for association of SEI with methylation mediator; pval(Mj->BMI), p value for assocation of methylation mediator with BMI; 95% CI, 95% confidence intervals.
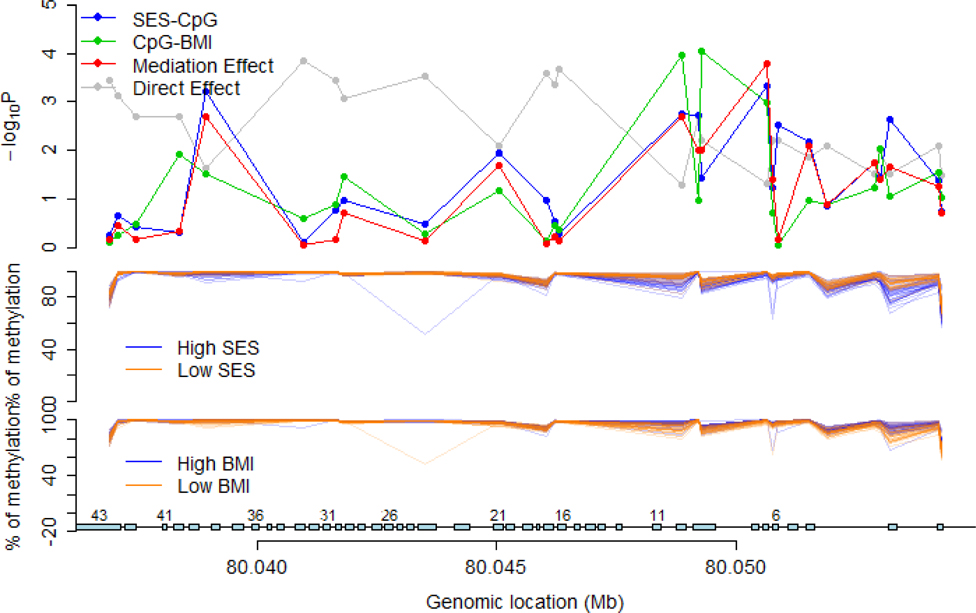
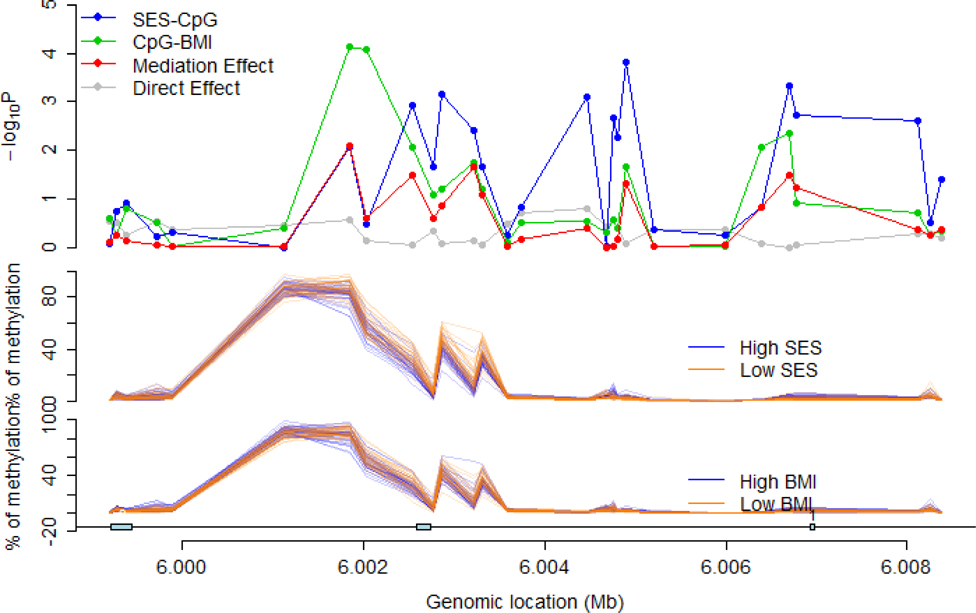
Among the 91 sites identified in males and 71 sites identified in females, there were several instances where multiple CpGs mapped to the same gene, including repeat hits in several genes that are biologically relevant for the development of obesity. These were fatty acid synthase (FASN – 2 sites), transmembrane protein 88 (TMEM88 – 2 sites), signal transducer and activator of transcription 3 (STAT3 – 2 sites) in females, and neuritin 1 (NRN1 – 3 sites) in males (Tables 2 and 3; Supplementary Tables 1 and 2). These tables showed suggestive evidence of statistical mediation where the indirect effect p value was <0.05 for several CpG methylation sites. While the main effect between SES and body mass index is significant in females (Table 1), the direct effect became non-significant in mediation analyses, possibly due to indirect effects through the methylation patterns identified in Table 2 and Supplementary Table 1. Figure 1 shows a detailed examination of the methylation profile for the FASN gene in females, where 11 of 26 methylation sites showed mediation p-values <0.05 (indirect effect; shown by red line in Figure 1). In this figure, it can be seen that there are robust associations between SES and methylation at several CpG sites (blue line), as well as between methylation and BMI at several CpG sites (green line). Similar analyses in other genes showed mediation p-values <0.05 in 5 of 28 sites for NRN1 in males (Figure 2), 3 of 11 sites for TMEM88 in females (Supplementary Figure 3), and 1 of 17 sites for STAT3 in females (Supplementary Figure 4).
Figure 1.
Formal mediation analyses of methylation patterns at 26 fatty acid synthase (FASN) CpG sites in female adipose tissue. Analyses evaluated whether FASN methylation patterns mediate the relation of perinatal socioeconomic status (SES) with age 47 body mass index (BMI). High vs. low for SEI and BMI was calculated using a median split.
Figure 2.
Formal mediation analyses of methylation patterns at 28 neuritin 1 (NRN1) CpG sites in male adipose tissue. Analyses evaluated whether NRN1 methylation patterns mediate the relation of perinatal socioeconomic status (SES) with age 47 body mass index (BMI). High vs. low for SES and BMI was calculated using a median split.
Sensitivity analyses for associations of CpG site methylation with BMI in adipose tissue using a more restrictive FDR<10% (instead of <25%) showed there were 21 sites in women and 35 sites in men identified as suggestive. Using FDR<5%, there were 2 sites in women and 16 sites in men identified. Further sensitivity analyses for associations of CpG site methylation with age 7 SES (instead of with adulthood BMI) at FDR<25%, showed 1 site in women and 0 sites in men as suggestive in adipose tissue. No sites in blood were identified as significant using this latter approach.
DISCUSSION
This study investigated the role of DNA methylation in the association between childhood SES and adult adiposity. Findings suggest that methylation of specific CpG sites in adipose tissue but not blood mediate the association between childhood SES and adulthood BMI. Specific gene sites identified were FASN, STAT3 and TMEM88 in females, and NRN1 in males. These epigenetic mediation findings should be interpreted with caution, as this is the first study to report them, and reasonably liberal thresholds for detection were implemented. As they are novel, these epigenetic mediation findings require replication to confirm the specific sites involved; more broadly, this study finds that early life conditions may be related to adult health partly through their influences on epigenetic mechanisms.
Prior Literature
Childhood socioeconomic disadvantage and adulthood adiposity
A systematic review demonstrated that childhood socioeconomic disadvantage is inversely associated with adulthood adiposity, with studies showing stronger associations in women than men. Specifically, inverse associations were found in 70% (14 of 20) of studies in women and 27% (4 of 15) of studies in men, where typical effect sizes for the differences in body mass index (BMI) between the highest and lowest SES were 1.0–2.0 kg/m2 in women and 0.2–0.5 kg/m2 in men (11). These findings are consistent with the current study, where associations of childhood socioeconomic disadvantage were statistically significantly associated with adulthood BMI in females and not males. Senese et al. have offered plausible explanations for these sex differences including parity and socioeconomic-patterned pressure for females to be slim (11).
Childhood socioeconomic disadvantage and methylation
Several previous studies have investigated associations of childhood disadvantage with DNA methylation patterns. To our knowledge, all studies to date used peripheral blood leukocytes or placental tissue. Our search of the literature revealed no studies of this issue that interrogated adipose tissue. With regard to studies in peripheral blood leukocytes, a genome-wide methylation study in 40 adult men from the 1958 British Birth Cohort showed associations of childhood SES with differential methylation of 1,252 gene promoters in whole blood DNA (26). Furthermore, a study in approximately 85 participants of the New York site of the Collaborative Perinatal Project showed associations of childhood SES with methylation of Sat2 repetitive elements in leukocyte DNA during adulthood, and no association with Alu and LINE-1 (27). Another study of 120 mother-offspring dyads showed that maternal education was significantly associated with methylation of INSIGF in blood at offspring age 17 months (28). A study, using purified monocytes, specifically examined 18 candidate genes related to stress reactivity and inflammation for 1,264 participants in the Multi-Ethnic Study of Atherosclerosis (MESA), demonstrating that low childhood SES was associated with adulthood DNA methylation in three stress-related genes (AVP, FKBP5, OXTR) and two inflammation-related genes (CCL1, CD1D) (29). A study of 92 adults showed substantial decreases in P-value distribution patterns for the relation of early life SES with methylation in peripheral blood mononuclear cells in samples that were corrected vs. not corrected for differential blood cells count (30). Most studies to date using leukocytes have not adjusted for cell admixture, consequently it is unclear if evidence of associations is due to DNA methylation or differential cell admixture. With regard to findings in placental tissue, one study showed significant associations of maternal education and maternal poverty status with HSD11B2 methylation in males (n=215) and not females (n=230) (31). Overall, evidence suggests some indication of methylation patterns associated with childhood socioeconomic disadvantage, however replication efforts are needed to determine consistency between studies and tissue types.
Methylation and adiposity
Exploration of epigenetic underpinnings of adiposity is receiving substantial research attention. In the New England Family Study, we recently showed that adiposity phenotypes (e.g. body mass index, android fat mass) were associated with DNA methylation in several genes that are biologically relevant to the development of adiposity, such as AOC3, LIPE, SOD3, AQP7, HIF3A, and CETP (25). Blood DNA methylation profiles were not associated with adiposity, before or after adjustment for leukocyte cell mixture effects. Two studies in adipose tissue showed blood DNA methylation at 3 CpG sites located in the HIF3A gene to be associated with BMI (25,54). Several other studies investigated associations of DNA methylation with adiposity (14–23,54), primarily in leukocytes (14–21,23). Research on epigenetic determinants of adiposity is still in the fairly early discovery phase, and consequently, many CpG sites that are potentially effectors of this phenotype likely remain to be discovered and replicated.
Mediation analyses of methylation as a pathway of how childhood SES could influence adiposity
There has been little exploration of the possible nature of the relationship of alterations in methylation as it pertains to effecting the childhood SES/adult adiposity association. Here, we applied a formal mediation analyses to investigate potential mediating roles of epigenetic changes in translating early life socioeconomic disadvantage into adiposity later in life. However in a study of early life stress, a mediation analysis in 31 offspring of mothers who went through the 1998 Quebec ice storm showed that methylation levels of genes implicated in diabetes (e.g. LTA, NFKBIA, PIK3CD) showed significant mediation of the relation between prenatal maternal stress and offspring’s central adiposity and BMI, where methylation was assessed in peripheral blood leukocytes (55). The current study’s findings of 91 CpG sites in males and 71 CpG sites in females having evidence of mediation between early life socioeconomic disadvantage and adulthood adiposity appear to be very novel. Particular enrichment in the FASN, TMEM88, STAT3, and NRN1 genes is biologically plausible. We reasoned that genes with multiple CpG sites showing significantly altered DNA methylation were most likely to have an important phenotypic change that significantly contributed to any SES-mediated epigenetic differences in adulthood. Each of the STAT3, FASN, TMEM88 and NRN1 genes showed alterations in DNA methylation in multiple CpG sites within each gene. Interestingly, these genes are all members of pathways that are known to be crucial in development. Members of the Nanog pathway that establishes and maintains pluripotency in embryonic stem cells include the JAK/STAT3 transcription factor axis (56). This pathway has been reported to be epigenetically modified in numerous chronic diseases, with evidence that STAT3 itself can be directly targeted by environmental exposures (57). The STAT3 pathway is also activated by leptin, indicating that it plays a role in the metabolic pathways central to obesity (58). The FASN protein has long been associated with cellular and organismal metabolism. Dysregulated metabolism is a hallmark of inflammation and the gene itself has been identified as a potential therapeutic target in numerous cancers (59). It is also well established that fatty acids are critically important in the development of cardiovascular diseases (60). In mice, high dietary fat alter FASN expression and this change is more pronounced in female mice than males (61). In addition, TMEM88 is a member of the WNT signaling pathway, which is now known to be critical in normal developmental patterning (62). Alterations in the pathway is associated with numerous adult degenerative diseases and cancers (63) and proteins with a similar structure are well known to be important for adipocyte differentiation (64). Finally, NRN1 has been reported to be expressed in high levels in adipose progenitor cells that give rise to white adipocytes (65) and, interestingly, it is selectively regulated by androgens in experiments conducted in-vitro (66). Hence, the sites that we identified as most likely to be phenotypically altered are all developmental mediators of pathways that play roles in chronic disease.
Mechanisms
With regard to how early life socioeconomic disadvantage could influence DNA methylation, there are a number of plausible mechanisms. One pathway may be through maternal and offspring diet related to socioeconomic disadvantage. It has been demonstrated that adults and children with lower socioeconomic status in the United States and other industrialized nations consume less of a number of methyl donor foods such as fruits and vegetables (67,68), as well as lower consumption of folate (68), compared to those with high SES. Socioeconomic disadvantage influences abilities to afford many of the sources of naturally occurring methylation donor such as citrus fruits and leafy greens (67,68). Furthermore, systematic reviews demonstrated that low childhood SES is predictive of adulthood obesity (11). Methylation patterns may be indicative of greater propagation of adipose cell number and size in the body, if the methylation influences genes involved with proliferation or enlargement of adipose cells. This could potentially be due in part to early life socioeconomic disadvantage. To the degree that socioeconomic disadvantage causes stress, stress has been shown to alter feeding behavior in animals and humans, where some respond with increased food consumption and others respond with decreased food consumption (69). It is important to note that stress can be dependent on the larger socioeconomic context in which participants are living. For example, parents and children living in poverty, with few employment opportunities, low education and inadequate support systems may be inherently more stressed (70), and more likely to express more adverse parenting styles, such as neglect, abuse and lower parental monitoring (71). There is evidence that obesity-related eating behaviors are enhanced when there is access to palatable food during periods of stress (69). Non-homeostatic eating has been suggested to activate the brain reward system (69). Early life environmental stressors such as disruption of mother-child relationships or abuse or neglect, have been shown to alter stress neurobiological outcomes and feeding behavior during adulthood (69). A prospective study by our group showed that elevated maternal cortisol levels during the gestational period were associated with increased coronary heart disease risk in female offspring 47 years later (72). These pathways, such as socioeconomic disadvantage-determined consumption of methyl-donor foods, likelihood of obesity, and stress-related consumption of palatable foods may be pathways through which early life socioeconomic disadvantage could influence methylation patterns in fat tissue, and eventually lead to greater adiposity.
Limitations and Strengths
Findings in this study should be interpreted with caution, as they have not been replicated in other studies. However, to our knowledge, there are no other studies that have sampled adipose tissue and have data on both individual estimates of early life SES as well as adulthood adiposity. Furthermore, in epigenome-wide methylation-BMI analyses, with 87,772 sites significant in females and 110,453 sites significant in males with FDR<25%, and p-value <0.001 for SES-methylation associations, we would expect 88 CpG sites in females, and 110 CpG sites in males, to be significant mediators simply due to chance. This is greater than the observed number of significant methylation sites (91 sites in males and 71 sites in females), suggesting the mediation findings may be due to chance. However, examination of each of the CpG sites interrogated by the array in the FASN and TMEM88 in females, and NRN1 in males, showed substantial enrichment of significant methylation in these regions, suggesting they may be biologically important. Secondly, it was not possible to adjust for cell admixture in the adipose tissue samples due to the large number of degrees of freedom needed in current statistical approaches for adipose tissue (40,73), and the relatively modest sample size. Consequently, it is uncertain if observed methylation patterns are due to alterations in methylation patterns, or due to alterations in adipose tissue cellular composition, between individuals with low vs. high socioeconomic disadvantage. Either of these pathways are of interest and biologically relevant. Future studies with larger sample sizes should clarify the underlying biology. In addition, race/ethnicity-specific differences in epigenetic mechanisms could be present. In the current study, only 25 male and 23 female participants were non-white (including African Americans, Native Americans and others). With analyses in this study being mediation analyses, and performed sex-specific, power was insufficient to implement effect modification analyses for race/ethnicity. However, a recent paper by our group showed race-specific epigenetic methylation patterns in adipose vs. blood tissue (74), suggesting some mechanisms may be unique by race/ethnicity. Finally, this study, although prospective from early childhood through mean age 47 y, is observational in design, leading to some limitations in causal inference if there are underlying unmeasured confounders. Inferring that the mediation analyses reflect a chain of causal effects requires two sets of assumptions regarding confounding: no unmeasured SES-methylation confounders, and no unmeasured methylation-adiposity confounders.
Strengths of the study include that we were able to utilize prospective assessments of childhood SES with both parent’s socioeconomic information, as well as direct prospective assessments of BMI at mean ages 7 years and 47 years. Furthermore, analyses were able to assess associations in both adipose tissue and blood instead of just blood as is usually performed. When exploring adiposity as an outcome, it is beneficial to assess the target tissue (i.e. adipose tissue), as shown in this study where all significant findings were in adipose tissue and not blood.
Conclusions
These findings provide evidence that the DNA methylation profile in adipose tissue at specific sites, including FASN and TMEM88 in females, and NRN1 in males, may explain part of the observed associations between childhood socioeconomic disadvantage and adulthood adiposity, particularly found in females (11). This adds to the small but growing literature on epigenetic etiologic explanations of social disparities in health. Plausible biological markers could one day serve as early biological indicators of socioeconomic intervention effectiveness, or as therapeutic targets. Findings are very early on in the discovery process, and it remains uncertain if DNA methylation could serve in this role.
Supplementary Material
Supplementary Table 1. Female adipose tissue mediation effects in all 71 CpG sites.
Supplementary Table 2. Male adipose tissue mediation effects in all 91 CpG sites.
ACKNOWLEDGEMENTS
Funding for this study was provided by NIH/NIA grants RC2AG036666 and R01AG048825 and by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Conflicts of Interest and Source of Funding: This work was supported by NIH/NIA grants RC2AG036666 and R01AG048825, and by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
REFERENCES
- 1.Pi-Sunyer FX. Medical hazards of obesity. Ann Intern Med. 1993;119:655–60. [DOI] [PubMed] [Google Scholar]
- 2.Jensen MD, Ryan DH, Apovian CM, Ard JD, Comuzzie AG, Donato KA, Hu FB, Hubbard VS, Jakicic JM, Kushner RF, Loria CM, Millen BE, Nonas CA, Pi-Sunyer FX, Stevens J, Stevens VJ, Wadden TA, Wolfe BM, Yanovski SZ. 2013 AHA/ACC/TOS Guideline for the Management of Overweight and Obesity in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and The Obesity Society. Circulation. 2013. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Beydoun MA, Liang L, Caballero B, Kumanyika SK. Will all Americans become overweight or obese? estimating the progression and cost of the US obesity epidemic. Obesity. 2008;16:2323–30. [DOI] [PubMed] [Google Scholar]
- 4.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256:51–4. [PubMed] [Google Scholar]
- 6.Lajunen HR, Kaprio J, Rose RJ, Pulkkinen L, Silventoinen K. Genetic and environmental influences on BMI from late childhood to adolescence are modified by parental education. Obesity. 2012;20:583–9. [DOI] [PubMed] [Google Scholar]
- 7.McCaffery JM, Papandonatos GD, Bond DS, Lyons MJ, Wing RR. Gene X environment interaction of vigorous exercise and body mass index among male Vietnam-era twins. Am J Clin Nutr. 2009;89:1011–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benyshek DC. The “early life” origins of obesity-related health disorders: new discoveries regarding the intergenerational transmission of developmentally programmed traits in the global cardiometabolic health crisis. Am J Phys Anthropol. 2013;152 Suppl 57:79–93. [DOI] [PubMed] [Google Scholar]
- 9.Bouret SG. Early life origins of obesity: role of hypothalamic programming. J Pediatr Gastroenterol Nutr. [Review]. 2009;48 Suppl 1:S31–8. [DOI] [PubMed] [Google Scholar]
- 10.Newnham JP, Pennell CE, Lye SJ, Rampono J, Challis JR. Early life origins of obesity. Obstet Gynecol Clin North Am. 2009;36:227–44. [DOI] [PubMed] [Google Scholar]
- 11.Senese LC, Almeida ND, Fath AK, Smith BT, Loucks EB. Associations between childhood socioeconomic position and adulthood obesity. Epidemiol Rev. 2009;31:21–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gluckman PD, Hanson MA. Developmental and epigenetic pathways to obesity: an evolutionary-developmental perspective. Int J Obes. 2008;32 Suppl 7:S62–71. [DOI] [PubMed] [Google Scholar]
- 13.Lavebratt C, Almgren M, Ekstrom TJ. Epigenetic regulation in obesity. Int J Obes. 2012;36:757–65. [DOI] [PubMed] [Google Scholar]
- 14.Almen MS, Jacobsson JA, Moschonis G, Benedict C, Chrousos GP, Fredriksson R, Schioth HB. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–7. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg AP, Irizarry RA, Fradin D, Aryee MJ, Murakami P, Aspelund T, Eiriksdottir G, Harris TB, Launer L, Gudnason V, Fallin MD. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med. 2010;2:49ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godfrey KM, Sheppard A, Gluckman PD, Lillycrop KA, Burdge GC, McLean C, Rodford J, Slater-Jefferies JL, Garratt E, Crozier SR, Emerald BS, Gale CR, Inskip HM, Cooper C, Hanson MA. Epigenetic Gene Promoter Methylation at Birth Is Associated With Child’s Later Adiposity. Diabetes. 2011;60:1528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groom A, Potter C, Swan DC, Fatemifar G, Evans DM, Ring SM, Turcot V, Pearce MS, Embleton ND, Smith GD, Mathers JC, Relton CL. Postnatal growth and DNA methylation are associated with differential gene expression of the TACSTD2 gene and childhood fat mass. Diabetes. 2012;61:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuehnen P, Mischke M, Wiegand S, Sers C, Horsthemke B, Lau S, Keil T, Lee YA, Grueters A, Krude H. An Alu element-associated hypermethylation variant of the POMC gene is associated with childhood obesity. PLoS Genet. 2012;8:e1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Relton CL, Groom A, St Pourcain B, Sayers AE, Swan DC, Embleton ND, Pearce MS, Ring SM, Northstone K, Tobias JH, Trakalo J, Ness AR, Shaheen SO, Davey Smith G. DNA methylation patterns in cord blood DNA and body size in childhood. PLoS ONE. 2012;7:e31821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Zhu H, Snieder H, Su S, Munn D, Harshfield G, Maria BL, Dong Y, Treiber F, Gutin B, Shi H. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med. 2010;8:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Goldberg J, Vaccarino V. Promoter methylation of serotonin transporter gene is associated with obesity measures: a monozygotic twin study. Int J Obes. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, Tornberg A, Dekker Nitert M, Eriksson KF, Jones HA, Groop L, Ling C. A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue. PLoS Genet. 2013;9:e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, Terry MB. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol. 2015;44:1277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, Hertzman C, Power C, Szyf M. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tehranifar P, Wu HC, Fan X, Flom JD, Ferris JS, Cho YH, Gonzalez K, Santella RM, Terry MB. Early life socioeconomic factors and genomic DNA methylation in mid-life. Epigenetics. 2013;8:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obermann-Borst SA, Heijmans BT, Eilers PH, Tobi EW, Steegers EA, Slagboom PE, Steegers-Theunissen RP. Periconception maternal smoking and low education are associated with methylation of INSIGF in children at the age of 17 months. J Dev Orig Hlth Dis. 2012;3:315–20. [DOI] [PubMed] [Google Scholar]
- 29.Needham BL, Smith JA, Zhao W, Wang X, Mukherjee B, Kardia SL, Shively CA, Seeman TE, Liu Y, Diez Roux AV. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: The multi-ethnic study of atherosclerosis. Epigenetics. 2015;10:958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, Kobor MS. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U S A. 2012;109 Suppl 2:17253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appleton AA, Armstrong DA, Lesseur C, Lee J, Padbury JF, Lester BM, Marsit CJ. Patterning in placental 11-B hydroxysteroid dehydrogenase methylation according to prenatal socioeconomic adversity. PLoS ONE. 2013;8:e74691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niswander K, Gordon M. The Women and their Pregnancies: The Collaborative Perinatal Study of the National Institute of Neurological Diseases and Stroke. Washington D.C.: National Institutes of Health, 1972. [Google Scholar]
- 33.Davis S DP, Bilke S, Triche T Jr. and Bootwalla M methylumi: Handle Illumina methylation data. R package version 2.10.0. 2014. [Google Scholar]
- 34.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price ME, Cotton AM, Lam LL, Farre P, Emberly E, Brown CJ, Robinson WP, Kobor MS. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myrianthopoulos NC, French KS. An application of the U.S. Bureau of the Census socioeconomic index to a large, diversified patient population. Soc Sci Med. 1968;2:283–99. [DOI] [PubMed] [Google Scholar]
- 38.Falkner F Office measurement of physical growth. Pediatr Clin NA. 1961;8:13–8. [DOI] [PubMed] [Google Scholar]
- 39.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacKinnon DP, Lockwood CM, Hoffman JM, West SG, Sheets V. A comparison of methods to test mediation and other intervening variable effects. Psychol Methods. 2002;7:83–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300. [Google Scholar]
- 43.Rubicz R, Zhao S, Geybels M, Wright JL, Kolb S, Klotzle B, Bibikova M, Troyer D, Lance R, Ostrander EA, Feng Z, Fan JB, Stanford JL. DNA methylation profiles in African American prostate cancer patients in relation to disease progression. Genomics. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dam PA, van Dam PJ, Rolfo C, Giallombardo M, van Berckelaer C, Trinh XB, Altintas S, Huizing M, Papadimitriou K, Tjalma WA, van Laere S. In silico pathway analysis in cervical carcinoma reveals potential new targets for treatment. Oncotarget. 2016;7:2780–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Z, Bolick SC, DeRoo LA, Weinberg CR, Sandler DP, Taylor JA. Epigenome-wide association study of breast cancer using prospectively collected sister study samples. J Nat Cancer Inst. 2013;105:694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordoba R, Sanchez-Beato M, Herreros B, Domenech E, Garcia-Marco J, Garcia JF, Martinez-Lopez J, Rodriguez A, Garcia-Raso A, Llamas P, Piris MA. Two distinct molecular subtypes of chronic lymphocytic leukemia give new insights on the pathogenesis of the disease and identify novel therapeutic targets. Leuk Lymphoma. 2016;57:134–42. [DOI] [PubMed] [Google Scholar]
- 47.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. [DOI] [PubMed] [Google Scholar]
- 49.Houtepen LC, Vinkers CH, Carrillo-Roa T, Hiemstra M, van Lier PA, Meeus W, Branje S, Heim CM, Nemeroff CB, Mill J, Schalkwyk LC, Creyghton MP, Kahn RS, Joels M, Binder EB, Boks MP. Genome-wide DNA methylation levels and altered cortisol stress reactivity following childhood trauma in humans. Nat Commun. 2016;7:10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robins JM, Greenland S. Identifiability and exchangeability for direct and indirect effects. Epidemiology. 1992;3:143–55. [DOI] [PubMed] [Google Scholar]
- 51.Rubin DB. Bayesian-Inference for Causal Effects - Role of Randomization. Ann Stat. 1978;6:34–58. [Google Scholar]
- 52.VanderWeele TJ, Vansteelandt S. Conceptual issues concerning mediation, interventions and composition. Stat Interface. 2009;2:457–68. [Google Scholar]
- 53.VanderWeele TJ. Bias formulas for sensitivity analysis for direct and indirect effects. Epidemiology. 2010;21:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, Waldenberger M, Peters A, Erdmann J, Hengstenberg C, Cambien F, Goodall AH, Ouwehand WH, Schunkert H, Thompson JR, Spector TD, Gieger C, Tregouet DA, Deloukas P, Samani NJ. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014. [DOI] [PubMed] [Google Scholar]
- 55.Cao-Lei L, Dancause KN, Elgbeili G, Massart R, Szyf M, Liu A, Laplante DP, King S. DNA methylation mediates the impact of exposure to prenatal maternal stress on BMI and central adiposity in children at age 13(1/2) years: Project Ice Storm. Epigenetics. 2015;10:749–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hawkins K, Joy S, McKay T. Cell signalling pathways underlying induced pluripotent stem cell reprogramming. World J Stem Cells. 2014;6:620–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J, Bai W, Niu P, Tian L, Gao A. Aberrant hypomethylated STAT3 was identified as a biomarker of chronic benzene poisoning through integrating DNA methylation and mRNA expression data. Exp Mol Pathol. 2014;96:346–53. [DOI] [PubMed] [Google Scholar]
- 58.Niswender KD, Schwartz MW. Insulin and leptin revisited: adiposity signals with overlapping physiological and intracellular signaling capabilities. Frontier Neuroendocrinol. 2003;24:1–10. [DOI] [PubMed] [Google Scholar]
- 59.Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calder PC. Functional Roles of Fatty Acids and Their Effects on Human Health. JPEN J Parenter Enteral Nutr. 2015;39:18S–32S. [DOI] [PubMed] [Google Scholar]
- 61.Kadota Y, Kawakami T, Takasaki S, Sato M, Suzuki S. Gene expression related to lipid and glucose metabolism in white adipose tissue. Obes Res Clin Pract. 2015. [DOI] [PubMed] [Google Scholar]
- 62.Kuhl M, Sheldahl LC, Park M, Miller JR, Moon RT. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16:279–83. [DOI] [PubMed] [Google Scholar]
- 63.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. [DOI] [PubMed] [Google Scholar]
- 64.Batrakou DG, de Las Heras JI, Czapiewski R, Mouras R, Schirmer EC. TMEM120A and B: Nuclear Envelope Transmembrane Proteins Important for Adipocyte Differentiation. PLoS ONE. 2015;10:e0127712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tews D, Schwar V, Scheithauer M, Weber T, Fromme T, Klingenspor M, Barth TF, Moller P, Holzmann K, Debatin KM, Fischer-Posovszky P, Wabitsch M. Comparative gene array analysis of progenitor cells from human paired deep neck and subcutaneous adipose tissue. Mol Cell Endocrinol. 2014;395:41–50. [DOI] [PubMed] [Google Scholar]
- 66.Marron TU, Guerini V, Rusmini P, Sau D, Brevini TA, Martini L, Poletti A. Androgen-induced neurite outgrowth is mediated by neuritin in motor neurones. J Neurochem. 2005;92:10–20. [DOI] [PubMed] [Google Scholar]
- 67.Giskes K, Avendano M, Brug J, Kunst AE. A systematic review of studies on socioeconomic inequalities in dietary intakes associated with weight gain and overweight/obesity conducted among European adults. Obes Rev. 2010;11:413–29. [DOI] [PubMed] [Google Scholar]
- 68.Darmon N, Drewnowski A. Does social class predict diet quality? Am J Clin Nutr. 2008;87:1107–17. [DOI] [PubMed] [Google Scholar]
- 69.Maniam J, Morris MJ. The link between stress and feeding behaviour. Neuropharmacol. 2012;63:97–110. [DOI] [PubMed] [Google Scholar]
- 70.Dowd JB, Simanek AM, Aiello AE. Socio-economic status, cortisol and allostatic load: a review of the literature. Int J Epidemiol. 2009;38:1297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Repetti RL, Taylor SE, Seeman TE. Risky families: family social environments and the mental and physical health of offspring. Psychol Bull. 2002;128:330–66. [PubMed] [Google Scholar]
- 72.Stinson LJ, Stroud LR, Buka SL, Eaton CB, Lu B, Niaura R, Loucks EB. Prospective evaluation of associations between prenatal cortisol and adulthood coronary heart disease risk: the New England family study. Psychosom Med. 2015;77:237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30:1431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang YT, Chu S, Loucks EB, Lin CL, Eaton CB, Buka SL, Kelsey KT. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigenetics. 2016:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Female adipose tissue mediation effects in all 71 CpG sites.
Supplementary Table 2. Male adipose tissue mediation effects in all 91 CpG sites.