Abstract
Kinesins are a diverse group of adenosine triphosphate (ATP)‐dependent motor proteins that transport cargos along microtubules (MTs) and change the organization of MT networks. Shared among all kinesins is a ~40 kDa motor domain that has evolved an impressive assortment of motility and MT remodeling mechanisms as a result of subtle tweaks and edits within its sequence. Several elegant studies of different kinesin isoforms have exposed the purpose of structural changes in the motor domain as it engages and leaves the MT. However, few studies have compared the sequences and MT contacts of these kinesins systematically. Along with clever strategies to trap kinesin–tubulin complexes for X‐ray crystallography, new advancements in cryo‐electron microscopy have produced a burst of high‐resolution structures that show kinesin–MT interfaces more precisely than ever. This review considers the MT interactions of kinesin subfamilies that exhibit significant differences in speed, processivity, and MT remodeling activity. We show how their sequence variations relate to their tubulin footprint and, in turn, how this explains the molecular activities of previously characterized mutants. As more high‐resolution structures become available, this type of assessment will quicken the pace toward establishing each kinesin's design–function relationship.
Keywords: cryo‐EM structure, crystal structure, kinesin, microtubules, motor protein, tubulin
1. INTRODUCTION
Microtubules (MTs) are straw‐shaped polymers of αβ‐tubulin heterodimers that string together in a head‐to‐tail fashion to form long protofilaments, which associate laterally. Kinesins are cytoskeletal motor proteins that can move cargos along MTs, manipulate the architecture of MT networks, and attach or detach tubulin subunits from MT ends. All of these activities result from their ability to convert ATP hydrolysis energy into nanoscale mechanical forces on tubulin lattices. Like other cytoskeletal motors (myosin and dynein), kinesins use the intrinsic polarity of MT filaments to dictate their direction of travel. Kinesins also take advantage of the dynamically different ends of MTs (plus and minus ends) to affect MT growth and shortening in an end‐specific manner. These activities help MTs to have wide‐ranging and versatile cellular roles. For example, in preparation for mitosis, kinesins help with the rapid disassembly of cytosolic MT networks of interphase cells, allowing the free tubulin to be repurposed for the mitotic spindle that partitions chromosomes to nascent daughter cells. During this process, certain teams of kinesins cooperate to establish the bipolar spindle arrangement, and then orchestrate spindle elongation when the cell is ready to separate sister chromosomes. 1 , 2 , 3 When mitosis is complete, other kinesins disassemble spindle MTs and guide reformation of the interphase MT network. 4 With these powers to form, manipulate and traverse massive structural arrays of MTs, kinesins provide a vital means with which to connect major cellular components and dynamically control their positions throughout the cell cycle. 5 , 6 , 7
The kinesin superfamily is composed of 14 unique members. Most kinesins move unidirectionally toward MT plus ends by coupling the difference in the free energy of adenosine triphosphate (ATP) binding to rotation and docking of a flexible region, named the neck linker, against their conserved motor domain (the MT‐binding part of the motor). The first glimpses of this nucleotide‐dependent conformational change mechanism were obtained from high‐resolution crystallographic structures of isolated kinesin‐1 and kinesin‐14 motor domains. 8 , 9 , 10 , 11 , 12 These models showed that the kinesin motor domain shares structural homology with the actin‐binding motor myosin, and G‐proteins, which transmit signals from stimuli outside cells to the inside. At the heart of the motor domain is a central β‐sheet that is flanked by three α‐helices on each side (α1 to α6) (Figure 1a). The nucleotide‐binding pocket resides on one side of the motor domain with α1, α2, and α3, while the MT‐binding interface is on the other with α4, α5, and α6. The structural elements surrounding the latter of these helices establish the remainder of the MT‐binding interface (loop‐2, ‐8, ‐11, and ‐12), and these regions have distinct signatures for each kinesin family.
FIGURE 1.
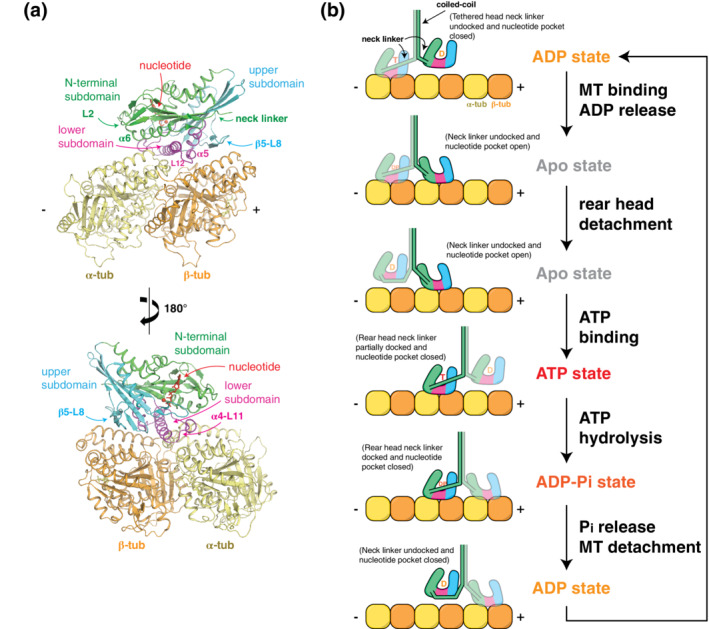
Kinesin–tubulin interaction and mechanochemical cycle. (a) The kinesin motor domain can be divided into three subdomains that move rigidly during the nucleotide/microtubule binding cycle. Each subdomain forms an interface with tubulin via: (1) loop‐2 and α6 in the N‐terminal subdomain; (2) the loop‐11‐α4‐loop‐12‐α5 (L11‐α4‐L12‐α5) cluster in the lower subdomain; and (3) the β5‐loop‐8 (β5‐L8) lobe in the upper subdomain. (b) A model for the kinesin stepping cycle is depicted for a processive kinesin dimer, whose two heads (motor domains) take turns in stepping toward the MT plus end. First, the tethered, ADP‐bound (D) head is swung around the coiled‐coil junction via docking of the neck‐linker against the rear motor domain. Once the tethered head finds a new tubulin binding site and becomes the lead head, adenosine diphosphate (ADP) is released and this “apo” form of the lead head binds strongly. Adenosine triphosphate (ATP) cannot enter the lead head until the trailing head finishes ATP hydrolysis, release phosphate, and detaches from the MT. Once ATP binding (T) occurs in the lead head, the neck‐linker is able to dock against its motor core, directing the tethered trailing head forward, in the plus end direction. Once this new lead head is firmly bound to tubulin, ATP hydrolysis in the new rear head makes detachment possible
The motor domain structure is specifically designed to couple MT‐binding interactions with its nucleotide state. When unbound from the MT, the motor domain (commonly referred to as the “head”) resides in the adenosine diphosphate (ADP)‐bound state, which has weak affinity for the MT (Figure 1b). MT contact encourages an increased twist in the central β‐sheet of the motor domain, opening the nucleotide pocket, and evicting the resident ADP. 13 , 14 In this “apo state,” the motor domain has high MT affinity, the nucleotide pocket remains open, and the Switch I and Switch II (SwI and SwII) regions remain in the closed conformation. This includes formation of a stringently conserved salt bridge between SwI and SwII necessary for catalysis. Once ATP enters and engages the P‐loop, the N‐terminal, and upper subdomains rotate relative to one another, closing the nucleotide pocket. These actions put SwI residues in position to interact with the γ phosphate of ATP and the active site Mg2+. SwII approaches ATP from the opposite side to bond with γ phosphate and to help SwI corral water molecules into position for nucleophilic attack and β‐γ‐phosphoanhydride bond hydrolysis. 13 , 15 , 16 These conformational changes expose a hydrophobic patch along the side of the motor domain that allows the neck‐linker element to partially dock. 17 , 18 In dimeric transport kinesins (e.g., kinesin‐1 and ‐3), neck‐linker docking provides the small, but not insignificant, amount of force needed to push the tethered head past the MT‐bound head, toward the MT plus‐end. 17 , 19 , 20 , 21 , 22 , 23 , 24 ATP hydrolysis completes the docking step and enables the tethered head to bind the MT. 25 Tethered head binding generates inter‐head strain that encourages departure of Pi from the rear head, followed by rear head release from the MT. 26 , 27 , 28 The result of one cycle of this tightly coordinated series of events is a single, 8 nm‐sized, step along the axis of the MT. 29 , 30 , 31 Depending on the processivity of the kinesin in question, hundreds more steps are possible before the motor detaches from its track.
Although all kinesins share the same nucleotide‐binding motifs and movements of these flexible elements, we are just beginning to learn how kinesin‐family‐specific variations have established dramatic divergence in properties like processivity, velocity, and MT‐remodeling activity. Some kinesins move very fast, whereas others are glacially slow and inefficient, hydrolyzing as many as seven ATP molecules per step. 32 What we know is that each kinesin family member has regions of unique sequence and structure that differentiate their enzymatic and mechanical properties (Figure 2). 80 Not surprisingly, this translates into differences in the mechanisms they use to accomplish their cellular roles. However, many important aspects of these structure–function relationships are not understood. Until we uncover this information, we cannot fully grasp how one family of kinesins mediates axonal transport of vesicles, while another regulates the structural dynamics of the mitotic spindle. This review examines the regions of kinesins that form major MT interactions. To avoid over‐interpreting kinesin‐MT interactions in cryo‐electron microscopy (EM) structures, we have chosen to compare structures with sub‐6 Å overall resolution. Our rationale is based on the fact that the resolution of the kinesin component is often several angstroms lower than the reported cryo‐EM data resolution. Where possible, we also compared the interfaces of each kinesin in their ATP‐like state to highlight similarities and differences in their tubulin footprints at the same stage in their catalytic cycle. We show how their sequence variations relate to their footprint on each tubulin subunit and highlight recent studies that have helped explain the functional significance of these interactions.
FIGURE 2.
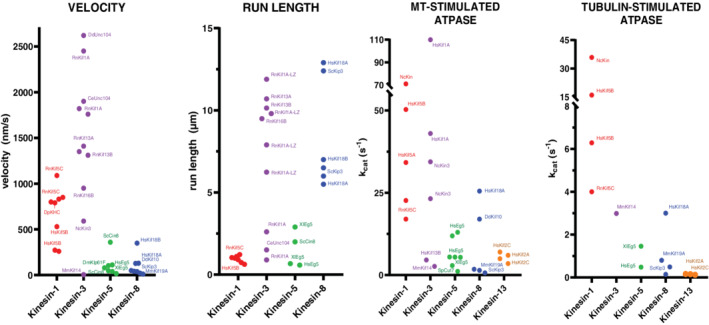
Motor specifications of kinesin families. Each data point corresponds to a reported value from an independent published experiment. Select motors are labeled. Values were obtained from: kinesin‐1, 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 kinesin‐3, 32 , 33 , 34 , 37 , 40 , 42 , 43 , 45 , 46 , 47 , 48 , 49 , 50 , 51 kinesin‐5, 41 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 kinesin‐8, 35 , 38 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 and kinesin‐13. 77 , 78 , 79 Note that several run length values for kinesin‐3 and ‐8 were limited by the length of MTs assembled in vitro
2. KINESIN MOTILITY MECHANISMS
Kinesins demonstrate three different forms of MT‐based motility: (a) processive stepping, in which a dimeric complex of two kinesin polypeptides advance in a hand‐over‐hand fashion along a single MT by alternating each motor domain's interaction with tubulin binding sites that are 8 nm apart; (b) nonprocessive motility, in which one motor domain contacts the MT and produces a powerstroke that translates the bound cargo before disengaging from the MT and resetting the motor domain for another binding event; and (c) one‐dimensional diffusion, in which a kinesin “surfs” along a MT by transient, electrostatic‐charge‐based interactions with the outer surface of the MT lattice.
Conventional kinesins (kinesin‐1) are plus‐end‐directed motors that rapidly transport molecules like mitochondria and synaptic vesicles along axons. These motors have been the prototype for understanding processive motility. Kinesin‐1 dimerizes via a parallel coiled‐coil that forms an extended stalk, placing two catalytic motor domains at one end of the complex (Figure 1b) and two cargo‐binding domains at the opposite end. Each motor domain cycles between conformations that are strongly attached to the MT (no‐nucleotide and ATP‐bound) and ones that are weakly attached (ADP‐bound), as described earlier. 81 Using this motility mechanism, kinesin‐1 can walk for distances as long as one micrometer, and at a speed of up to 1,000 nm/s (Figure 2). Astoundingly, dimeric kinesin‐3 motors can walk 10‐times as far, and over twice as fast, before detaching. 33 Some kinesin‐3 monomers can also move for long distances by 1D‐diffusion in their ADP state. 82 This “superprocessivity” is linked to their high affinity for the MT surface, making kinesin‐3s well‐suited for functions like long‐distance transport in neuronal cells. 34 , 83 , 84
Kinesin‐5 family members are also able to move processively, but the ATPase cycle and MT engagement of their motor domain is tuned for slow MT motility, rather than high‐speed/long‐distance cellular transport (Figure 2). 52 , 80 , 85 These motors form bipolar homotetrameric complexes, consisting of two motor dimers at opposite ends of a central helical rod. 86 This arrangement allows kinesin‐5s to crosslink spindle MTs into bundles to help establish mitotic spindle bipolarity. 86 , 87 , 88 , 89 , 90 Kinesin‐5 homotetramers also constrain and drive spindle elongation by sliding these MTs in opposite directions. 91 , 92 , 93 , 94 , 95 , 96
3. MICROTUBULE STABILIZERS AND DEPOLYMERIZERS
Kinesin‐8 and kinesin‐13 can use their motor domain for functions other than motility. 97 , 98 These motors actively shorten MTs by catalyzing tubulin subunit dissociation from MT ends using unique mechanical elements that recognize and/or promote tubulin protofilament bending. With this ability, kinesin‐8s and ‐13s can play critical roles in mitosis, such as pruning the length and number of dynamic astral MTs to help jiggle the spindle into the correct position across the plane cell division. 35 , 99 They also shorten kinetochore MTs to segregate chromosomes to the centers of daughter cells. 65 , 100 , 101 Failure in one or more of these activities can result in genomic instability, aneuploidy, and cell death.
Kinesin‐13 proteins are the major catalytic MT‐depolymerizing factors in higher eukaryotes. 102 , 103 Rather than stepping directionally along MTs, kinesin‐13 dimers move by rapid 1D diffusion across the MT lattice. 98 , 104 This activity is mediated by their unique motor core and an elongated helical neck domain. 102 , 105 Kinesin‐8 differs from kinesin‐13 on many levels. Their motor domain resides at the N terminus of the protein and they lack the long helical neck. Kinesin‐8 also combines the ability to destabilize MTs with highly processive, ATP‐driven movement along MTs. They also use length‐dependent accumulation at the plus end to preferentially disassemble long MTs. 65
4. SPECIFICATION OF KINESIN ACTIVITY BY DIVERGENT REGIONS OF THE MOTOR DOMAIN
All kinesin motor domains bind to the outside surface of MTs, along the backbone of the protofilament. In most cases, their tubulin interface can be separated into three distinct regions, stretching from the minus‐end to the plus‐end of one αβ‐tubulin heterodimer (Figure 1a): (a) loop‐2 (L2) and helix α6 in the N‐terminal subdomain; (b) the loop‐11, helix α4, loop‐12, helix α5 (L11‐α4‐L12‐α5) cluster in the lower subdomain; and (c) the β5 sheet—loop‐8 (β5‐L8) lobe in the upper subdomain. These regions are analogous to upper 50kD, lower 50kD, and N‐terminal subdomains in myosin, which undergo actin‐induced structural rearrangements that are coupled to release of ATP hydrolysis products and force generation. 106 Accordingly, these three regions move relative to one another in kinesins too. They can also bury nearly 2,000 Å2 of the solvent‐accessible surface on one tubulin dimer and can form upward of 25 H‐bonds and 13 salt bridges with tubulin residues (Figure 3). Kinesin‐1 family members have the smallest tubulin footprints, while kinesin‐13s and certain kinesin‐5s form the largest. In general, contacts with α‐tubulin predominate over β‐tubulin for all kinesins, but kinesin‐8 and ‐13 make many more interactions with α‐tubulin compared to the other motors. The ostensible purposes of these kinesin family‐specific interactions are described below.
FIGURE 3.
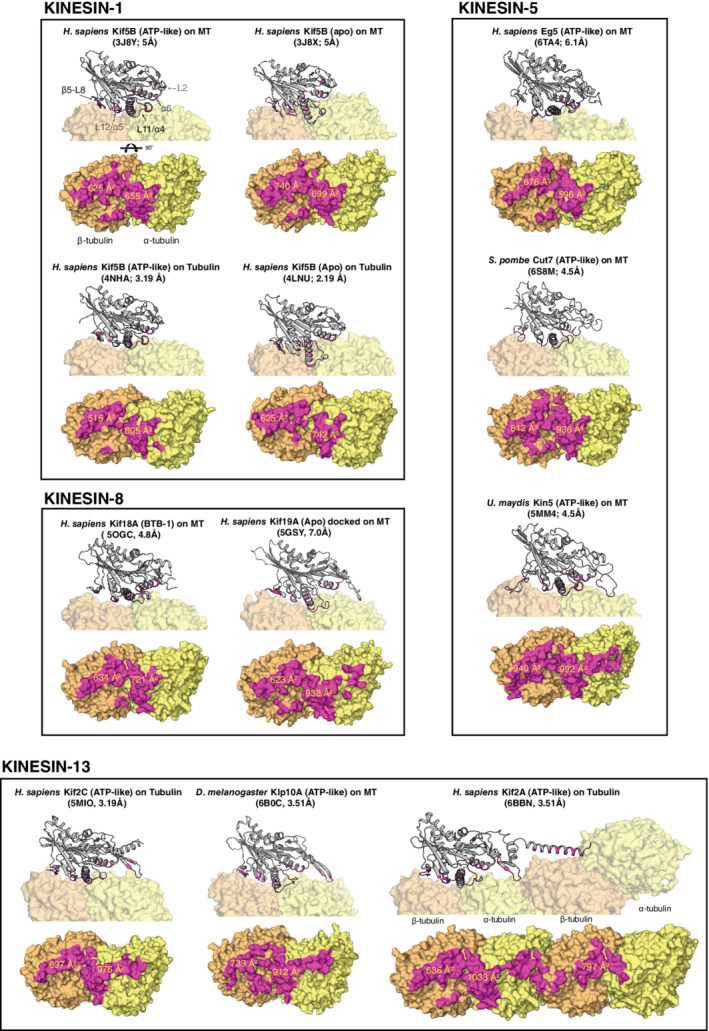
Microtubule‐binding surfaces for select families of kinesins. Kinesin structures are displayed as cartoons and tubulin is shown as surface representations. Interfaces were identified with ePISA 107 and are colored magenta. Values for buried surface are were calculated by ePISA using coordinate files retrieved from the Protein Data Bank (https://www.rcsb.org/). Nucleotide state, PDB IDs, and the overall map resolution for each structure are shown in parentheses
4.1. The β5‐L8 lobe
The β5‐L8 lobe is one of several flexible regions of the motor domain. It consists of two antiparallel β‐strands (β5a and β5b) separated by a short loop (L8b) and is connected to helix α3, which forms part of SwI (Figure 4). In all kinesin families, the β5‐L8 lobe faces toward the MT plus end and interacts with H12 of β‐tubulin (Figure 4b). 108 However, its sequence is not well conserved between the kinesin families, and varying levels of conservation exist within each family. As a result, there are several family‐specific differences in the configuration of the β5‐L8 lobe. These differences dramatically impact motor performance because β5‐L8 acts as structural communication pathway with α3‐SwI to regulate ATPase activity in accord with MT binding. 109 , 110
FIGURE 4.
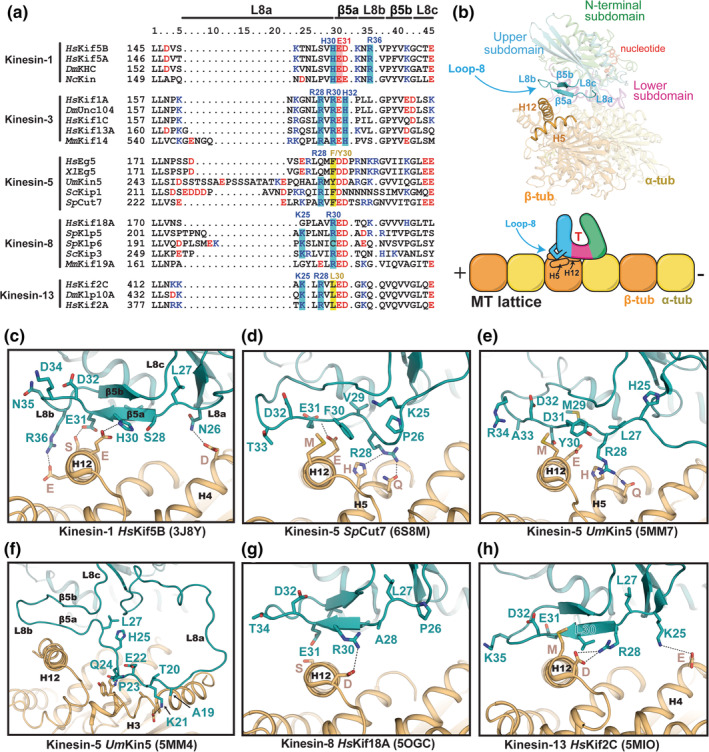
Comparison of β5‐L8 lobe structures and MT interactions. (a) Sequence alignment of β5‐L8 segments for selected kinesin family members. Positive and negative‐charged residues are colored blue and red, respectively. (b) Structural and cartoon representation highlighting the location of the β5‐L8 lobe and its interface with tubulin. (c–h) The conformation of the β5‐L8 lobe in each kinesin structure (cyan), and its contact with β‐tubulin (orange), are shown. Residue numbers in the kinesins have been changed to correspond to the columns in the sequence alignment for simplicity. PDB IDs are shown in parentheses. (e, f) Two different conformations of the β5‐L8 lobe that are observed in UmKin5 models (5MM4 and 5MM7)
All kinesin families contain a cluster of charged residues in β5a (Figure 4a). In most kinesins, these residues form electrostatic interactions with oppositely charged residues in β‐tubulin (Figure 4a). 108 Biophysical and computational studies of kinesin‐3 (not shown) suggest that β5‐L8 provides a large contribution to its MT affinity due to strong electrostatic contributions from an arginine (R28) at the end of L8a, and the R30 and H32 residues within β5a, which interact with helix H12 of β‐tubulin. 34 Some of these residues are also important for the enhanced processivity of kinesin‐3, whereas the less processive kinesin‐1 motors lack direct equivalents. Instead, kinesin‐1 makes sequence‐specific bonds with tubulin via other residues in β5a and L8b (H30, E31, R36) (Figure 4c). Kinesin‐1 uses these residues to differentiate between the tyrosinated and detyrosinated state of the carboxy‐terminus of α‐tubulin, and for polarized transport in neurons. 111
Kinesin‐5s show considerable sequence and structural variability in the β5‐L8 lobe. In the recent cryo‐EM structure of Eg5‐decorated MTs (not shown), no sequence‐specific contacts with tubulin are formed. 112 In yeast kinesin‐5s, insertions in loop L8a enable binding to noncanonical sites on the MT, in addition to H12 of β‐tubulin. 53 , 113 Another outcome of these L8a insertions is the formation of a small loop/kink that allows an arginine (R28) to H‐bond with polar residues (H and Q) on helix H5 of β‐tubulin (Figure 4d,e). Kinesin‐1, kinesin‐8, and kinesin‐13 do not form these contacts. In UmKin5, loop L8a makes a series of additional contacts with H3 of β‐tubulin (Figure 4f) and is large enough to contact β‐tubulin in the adjacent protofilament. 53 When the long L8 insert was deleted from the bidirectional kinesin‐5 in Saccharomyces cerevisiae, Cin8, its motility showed a directionality bias toward the minus end. 54 This suggests that the directionality switching ability of certain kinesin‐5s involves the β5‐L8 lobe.
Kinesin‐8s and ‐13s make multiple β5‐L8 contacts with β‐tubulin that are unique from other kinesins. 66 , 114 Both motors form a salt bridge between an arginine (R28 or R30) in β5a and an aspartate at the N‐term of H12 in β‐tubulin (Figure 4g,h). Kinesin‐13s, and some kinesin‐8s, also make a weak salt bridge with H4 via a lysine (K25) in L8a. Another contributor to the interaction of β5‐L8 with H12 in kinesin‐13s is a conserved leucine (L30), whose closest counterpart in in kinesin‐5s appears to be a bulky hydrophobic residue (F/Y30), which makes a similar interaction with a methionine in helix H12 of β‐tubulin (Figure 4a,d,e).
Charge neutralizing mutations of the conserved arginine (R30) in S. cerevisiae kinesin‐8 Kip3 had minimal effects on motility but reduced the depolymerization activity by lowering the plus‐end dwell time. 115 This suggests that β5‐L8 residues have an essential role in either binding or stabilizing curved tubulin structures at dynamic MT ends. In support of this, recent cryo‐EM structures of MT‐bound human Kif18A and Kif19A indicate that the β5‐L8 lobe of kinesin‐8s can either extend to adjust to the straight tubulin lattice in MTs, or retract toward the catalytic core on curved protofilaments. 66 How these movements in the β5‐L8 lobe specifically inhibit or activate MT depolymerization activity in kinesin‐8 is unclear and will likely require high‐resolution structures of kinesin‐8 bound to curved tubulin to understand fully.
The specific function of the β5‐L8 lobe in the unique mechanochemical cycle of kinesin‐13s is somewhat clearer. Here, β5‐L8 serves as the sensor to regulate ATP hydrolysis. 116 Its apex angles toward H12 of β‐tubulin, and there are no interactions with H5, perhaps owing to the slightly shorter and more rigid L8a sequence (Figure 4h). In MT‐bound Klp10A, the salt bridge formed between its conserved arginine (R28) and the initial aspartate (D) in H12 of β‐tubulin helps pull β5‐L8 away from the catalytic core, drawing α3 helix with it, away from SwII. 117 This holds the nucleotide pocket open, trapping kinesin‐13 in the ATP‐bound state. However, on curved tubulin subunits, β5‐L8 is pushed/pulled up toward the motor domain. 77 , 118 , 119 Helix α3 also changes conformation to allow the SwI loop‐9 to move closer to SwII, configuring the nucleotide‐binding pocket similarly to the hydrolysis competent ATP‐bound form of motile kinesins. 117
4.2. The L11‐α4‐L12‐α5 cluster
L11‐α4‐L12‐α5 form the SwII region (Figure 5a), which contacts tubulin at the intradimer interface and acts in combination with SwI (loop‐9) to couple MT binding to ATP turnover. The L11‐α4 portion contacts α‐tubulin on a small section of H3′ and H11′, and at the ends of H11 and H12. The L12‐α5 region binds H5 and S7 of β‐tubulin (Figure 5b). Despite high sequence conservation in SwII, select families show unique hallmarks that can have immense implications on motor activity (Figure 5a). Here, we summarize how the L11‐α4 junction of SwII couples MT‐binding to ATPase activity and motility, and how subtle tuning of its sequence and structure can allow kinesins to act as MT‐length regulatory factors.
FIGURE 5.
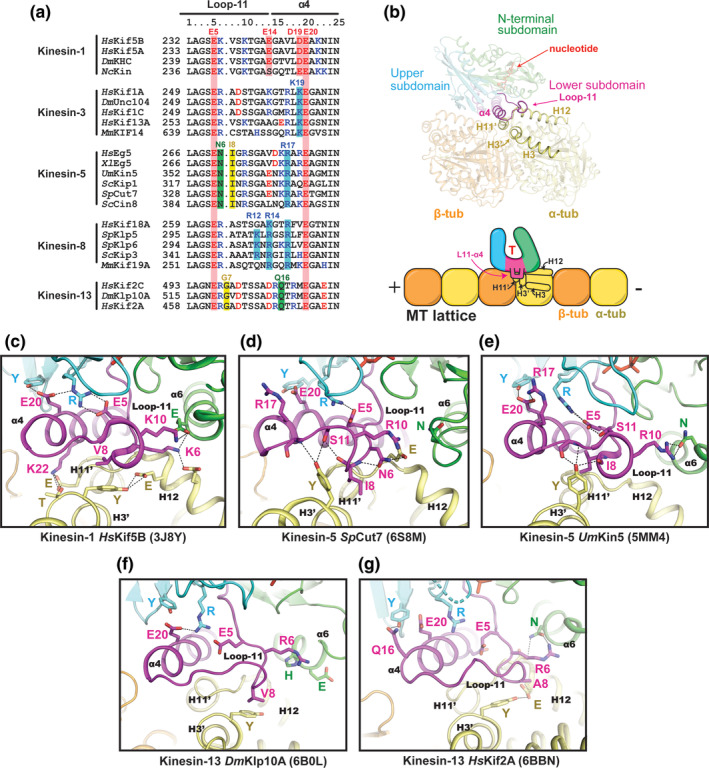
Comparison of the L11‐α4 region. (a) Sequence alignment of the L11‐α4 segments for selected kinesin family members. (b) Location of L11‐α4 within the motor domain and its interface with tubulin. (c–g) The conformation of L11‐α4 in each kinesin structure (magenta) and its contacts with α‐tubulin (yellow) are shown. Residue numbers in the kinesins have been changed to correspond to the columns in the sequence alignment for simplicity. PDB IDs are shown in parentheses
The large repository of kinesin‐1 structures bound to MTs and tubulin has so far presented the best explanation for how SwII couples MT‐binding to ATP turnover. 14 , 36 , 120 Upon MT binding, helix α4 elongates by 2.5 turns and loop‐11 forms a fully ordered structure. 10 This allows a glutamate (E20) within helix α4 to hydrogen bond to a tyrosine and a conserved arginine in loop‐7 and SwI, respectively (Figure 5c). These bonds move the arginine of SwI into position to form a salt bridge with the conserved glutamate (E5) of SwII. Glutamate E5 is responsible for initiating base‐catalyzed hydrolysis of ATP by indirectly activating a water nucleophile that attacks the γ‐phosphate. 15 , 36 Thus, MT‐binding greatly accelerates ATP hydrolysis by repositioning loop‐11 so that E5 is correctly situated for catalysis. This network of interactions remains in place in both apo and ATP‐bound conformations of MT‐bound kinesin‐1, anchoring the upper and lower subdomains together and conformationally priming the switch loops for ATP binding. 14 , 37
Kinesin‐3 experiences the same conformational changes in L11‐α4 upon MT binding as kinesin‐1. 37 Loop‐11 becomes ordered and SwII closes upon MT‐binding, and the cleft formed by the L11‐α4 junction widens after Pi departure. 14 , 37 , 121 However, unlike in kinesin‐1s, the unique L11‐α4 junction of kinesin‐3s appears to contribute to its super‐processivity (Figure 2). When the kinesin‐3‐specific lysine (K19) of RnKif1A was mutated to an aspartate, matching its kinesin‐1 equivalent, the velocity of motor movement slowed and its processivity was significantly reduced. 34 Unfortunately, without high‐resolution structures of MT‐bound kinesin‐3s, it is unclear exactly how L11‐α4 fosters superprocessivity. One prediction is that the K19 residue interacts with H3′ or H11′ of α‐tubulin to help strengthen MT binding. Another contributing factor appears to be the positively charged loop‐12 of kinesin‐3, named the “K‐loop.” When this loop was swapped with that of kinesin‐1, the MT attachment rates of these mutant kinesin‐3 motors were reduced to kinesin‐1 levels, but their velocity remained unchanged. 122 In kinesin‐1, family‐specific L11‐α4 residues afford the ability to discriminate between an expanded and compact MT lattice, whereas kinesin‐3s do not preferentially bind one or the other. 121 Lattice compaction sensing is mediated by E14 and D19 (Figure 5a), which interact with a conserved lysine and arginine in H3 and H3′ of tubulin, respectively. This tuning of the L11‐α4 junction provides a mechanism for kinesin‐1 motors to identify subpopulations of MTs within the cell.
Several new cryo‐EM structures have given insight into how kinesin‐5‐specific residues in L11‐α4 form distinct interactions with the intradimer interface of αβ‐tubulin 53 , 123 (Figure 5d). In SpCut7, an asparagine (N6) on loop‐11 (an R or K in other families) hydrogen bonds with the backbone of loop‐11 and a glutamate (E418) on helix H12 of α‐tubulin, causing loop‐11 to project downward instead of forming a loop. This places the kinesin‐5‐specific isoleucine (I8) of loop‐11 in position to form a non‐polar interaction with a tyrosine (Y112) on H3′ of α‐tubulin. This tyrosine also hydrogen bonds with the backbone of loop‐11. In all other structures of MT‐bound kinesins in the ATP‐like state, this tyrosine is rotated downward, often hydrogen bonding to a glutamate residue on H12 of α‐tubulin. Notably, H3′ is directly C‐terminal to turn T3 of α‐tubulin, which is a major structural component of the intradimer interface. Thus, in addition to tuning the mechanochemistry of kinesin‐5, loop‐11 could directly modulate the intradimer interface in a manner that affects MT structure. Recent studies in the Eg5 kinesin support this idea, showing that the L11‐α4 region is necessary for the MT polymerase activity of Eg5, as well as its slow motility and ATP turnover rate (Figure 2). 55 Based on this finding, it has been proposed that L11‐α4 of some kinesin‐5 motors induces straightening of αβ‐tubulin that promotes MT polymerization. 55 While it remains to be seen whether this activity translates across the entire kinesin‐5 family, the L11‐α4 junction remains highly conserved for all kinesin‐5s (Figure 5a).
In kinesin‐8s, loop‐11 may mediate MT‐depolymerization by suppressing ATPase activity once the motor reaches curved protofilaments at the MT plus end, forcing the kinesin to pause there and promote MT catastrophe. 38 The yeast kinesin‐8, ScKip3, appears to trigger this tight plus‐end binding by using a patch of basic residues within loop‐11 (specifically R12 and R14). These residues are positioned to interact with an aspartate residue (D118) on helix H3 of yeast α‐tubulin when tubulin is curved (not shown), but not when incorporated into the straight MT lattice. 38 , 115 This putative interaction led to the proposal that loop‐11 of kinesin‐8 forms a “tubulin‐binding switch” that enables the motor's MT depolymerization activity. However, it is not yet clear how this interaction stalls ATPase activity. Moreover, one of the two basic residues (R12) in loop‐11 is unique to fungal kinesin‐8s, and therefore this switch model may not apply to all kinesin‐8 family members (Figure 5a).
Loop‐11 of kinesin‐13 contains a single glycine insertion (G7) that is necessary for MT‐depolymerase activity (Figure 5a). 118 Mutational studies suggest that the added length of loop‐11 allows G7 to anchor kinesin‐13 to α‐tubulin, likely through an interaction with a glutamate on helix H12. Loop‐11 also undergoes conformational changes throughout the catalytic depolymerization cycle of kinesin‐13s (Figure 5f,g). On the straight MT lattice, kinesin‐13 remains in a catalytically inactive conformation, where the nucleotide pocket remains open. In this conformation, loop‐11 makes few interactions with other kinesin subdomains or the MT. 117 Upon binding the MT end, kinesin‐13 enters the catalytically active state, in which the salt bridge forms between E5 of SwII and R of SwI, closing the nucleotide pocket. These structural observations agree with evidence that kinesin‐13s have low levels of lattice stimulated ATPase activity, and high levels of MT‐end stimulated ATPase activity. 78 Lastly, when kinesin‐13 is complexed to a dramatically curved protofilament, a hydrogen bond forms between the conserved glutamine (Q16) at the start of helix α4 and a tyrosine residue in the upper subdomain (Figure 5g). 77 In transport kinesins, this tyrosine is usually hydrogen‐bonded to E20 of helix α4 (Figure 5c). This unique interaction may help to stabilize the catalytic intermediate of kinesin‐13 bound to a bent protofilament.
4.3. Loop‐2 and helix α6
In all kinesins, helix α6 of the N‐terminal subdomain interacts with helix H12 of α‐tubulin. However, certain kinesins contain family‐specific sequence modifications to loop‐2 that expand their α‐tubulin contacts to include the H8‐S7 motif (Figure 6a). Loop‐2 extends out of the small β1b/β1c sheet that points toward the minus end of the MT (Figure 6b). In kinesin‐13s, loop‐2 is an essential element for MT depolymerization activity. 124 , 125 It forms an elongated β‐hairpin, tipped with a conserved “KVD” motif that protrudes into the longitudinal interdimer interface between the β‐tubulin and α‐tubulin subunits (Figure 6c). 77 , 117 , 125 There, the valine residue inserts into a hydrophobic pocket bordered by H8‐S7 and H12 of α‐tubulin, and H11′ of the next β‐tubulin monomer. This penetrative interaction displaces the T7 loop—H8 helix motif of α‐tubulin, which forms one of the main longitudinal interfaces in protofilaments. 77 By shifting this interface, kinesin‐13 can incite protofilament bending and thus impart strain within the MT lattice. Loop‐2 may also operate in combination with loop‐8, but from the opposite end of the motor domain. Together, loop‐2 and loop‐8 could pull on the minus and plus ends of each tubulin heterodimer, respectively, stabilizing the curved configuration of individual tubulin dimers by a crossbow‐like mechanism (Asenjo, 2013).
FIGURE 6.
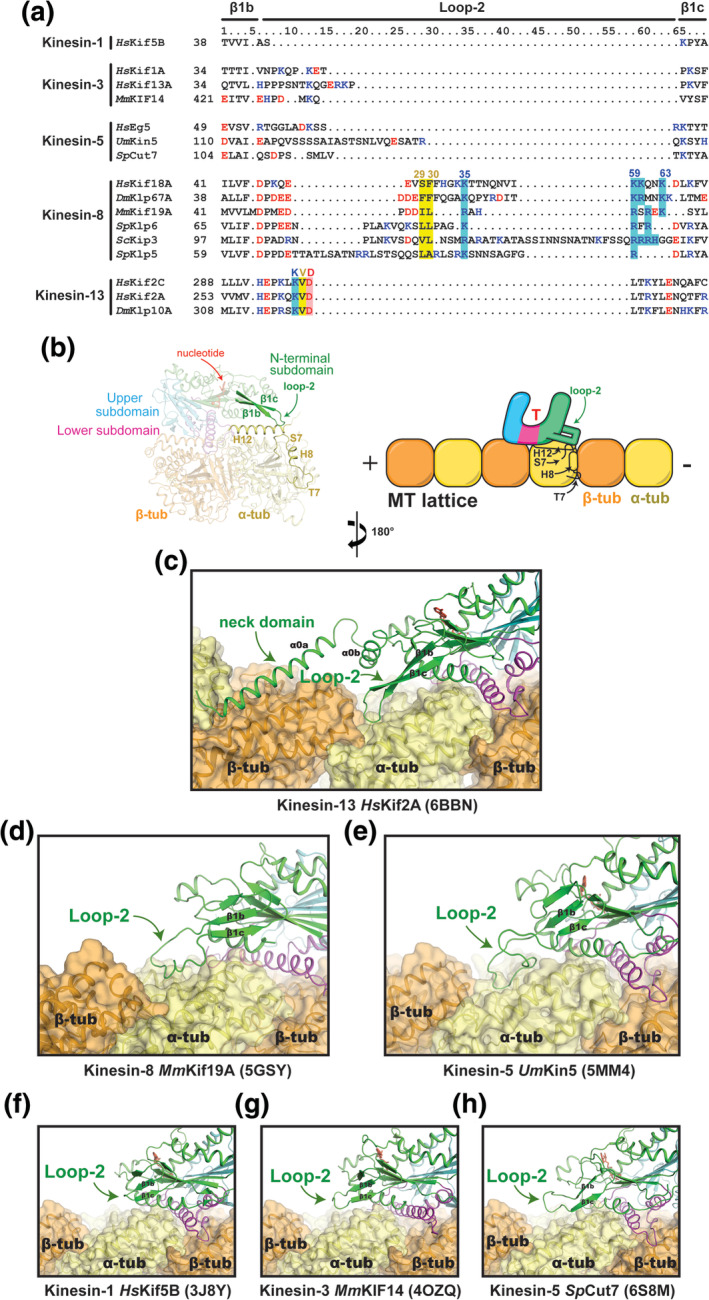
Comparison of loop‐2 structures and MT interactions. (a) Sequence alignment of the loop‐2 region for selected kinesin family members. (b) Location of loop‐2 within the motor domain and its interface with tubulin. (c–h) The conformation of loop‐2 in each kinesin structure (green) and its contacts with tubulin (orange and yellow) are shown. PDB IDs are shown in parentheses. (g) The coordinates for the MmKIF14 motor domain (4OZQ) were docked onto the MT‐bound structure of apo kinesin‐1 (3J8X)
Mutational studies emphasize the importance of loop‐2 in kinesin‐13‐mediated MT‐depolymerization. Shortening the β‐hairpin structure, or interchanging the charged residues of the KVD motif, debilitates MT depolymerization activity. 79 These mutations also diminish the MT‐stimulated ATPase activity of kinesin‐13. Therefore, lysine and aspartate of the KVD motif likely make complementary interactions with tubulin throughout the depolymerization cycle. This supports the functional relevance of the interaction between loop‐2 and interdimer interface in both MTs and curved protofilaments. 77 , 117 However, for robust MT depolymerization activity, kinesin‐13 requires the added leverage from the conserved 70‐residue N‐terminal extension of its motor domain, named the “neck domain” (Figure 6c). This region forms two α‐helices, α0a and α0b, the former of which makes extensive ionic interactions with helix H12 of β‐tubulin in the next tubulin dimer, toward the minus end of the protofilament (Figure 3). This interaction helps to guide loop‐2 into the interdimer interface. It also provides an additional anchor point to aid protofilament bending. Removal of the neck domain eliminates depolymerization activity from kinesin‐13s, 77 , 126 , 127 while point mutations that preserve the basic character of the neck domain retain depolymerization function. 128
More so than any other kinesin family, loop‐2 is extremely variable in length and sequence in kinesin‐8s (Figure 6a). The only consistent features are two hydrophobic residues (29 and 30) near the center of loop‐2, and a basic stretch of residues that leads back to β1c (Residues 59–63) (Figure 6a). Fungal kinesin‐8s such as SpKlp6, SpKlp5, and ScKip3 also contain additional residues before and/or after the tip of loop‐2. Unlike kinesin‐13, the extended loop‐2 of kinesin‐8 appears to be highly flexible. As such, loop‐2 is not resolved in the crystal structure of HsKif18A, 67 but it is visible in the structure of MmKif19A, 66 likely because it is 9–10 residues shorter than in other kinesin‐8s (Figure 6a,d). Upon binding the MT, loop‐2 of HsKif18A and MmKif19A adopts an ordered conformation that contacts the MT lattice. 66 , 68 This allows the hydrophobic tip (Residues 29 and 30) to enter the same hydrophobic pocket that is accessed by loop‐2 of kinesin‐13, near H8‐S7. It also allows, the basic cluster of residues of the kinesin‐8 loop‐2 to contact the negatively charged H12 helix of α‐tubulin.
Several studies have shown that loop‐2 is necessary for the high processivity and plus‐end accumulation of kinesin‐8s (Figure 2). Alanine substitutions at the basic residues of loop‐2 lowered MmKif19A motor affinity for MTs in vitro, 66 and dampened HsKif18A accumulation at microtubule ends in vivo. 129 Deletion of loop‐2 altogether prevented HsKif18 from stably associating with K‐fiber ends, and swapping loop‐2 of ScKip3 with the loop‐2 of kinesin‐1 significantly diminished run length and plus‐end dwell time. 38 What remains unclear is the role of loop‐2 in MT depolymerization by kinesin‐8s. As the hydrophobic tip of kinesin‐8 loop‐2 makes analogous interactions to H8‐S7 of the interdimer interface as the valine in kinesin‐13's KVD motif, the two families could operate through a similar mechanism. In support of this idea, mutations to hydrophobic and basic residues of the MmKif19A loop‐2 hinder the MT‐depolymerization activity of motor domain constructs. 66 However, a subsequent biochemical study on full‐length constructs of ScKip3 found no distinguishable difference between the depolymerization rate of wild‐type and a ScKip3 mutant that contained the loop‐2 sequence of kinesin‐1. 38 These differences in activity of fungal and human kinesin‐8 loop‐2 mutants may reflect alternate mechanisms of regulating MT dynamics by different kinesin‐8s. 69 , 130 , 131 Nonetheless, a true understanding of the role of loop‐2 will require functional studies of other kinesin‐8 isoforms, along with high‐resolution structures of kinesin‐8‐tubulin complexes that mimic MT depolymerization intermediates.
In transport motors like kinesin‐1 and kinesin‐3, loop‐2 appears to make a negligible contribution to tubulin binding and motility. It is too short to contact the MT in the ATP‐like state (Figure 6f,g), but can reach the MT lattice in the apo state to make one or more electrostatic interactions with helix H12 α‐tubulin due to a rotation of the N‐terminal subdomain. 14 The relevance of this interaction is unknown because mutations in loop‐2 do not impair the superprocessivity or velocity of kinesin‐3. 34 The function of loop‐2 in kinesin‐5 family members is also enigmatic. Its scant sequence conservation and varying length implies that each version is a unique adaptation that is unlikely to have functional relevance for the kinesin‐5 family as a whole (Figure 6a). The elongated loop‐2 of UmKin5 is an interesting example of this (Figure 6e). In the ATP‐like state, UmKin5 loop‐2 buries a hefty 290 Å2 of solvent accessible surface of α‐tubulin; at almost the same site as loop‐2 of kinesin‐8 and kinesin‐13 (Figure 3). 53 An obvious assumption would be that this interaction imparts UmKin5 with MT depolymerization abilities, but no evidence of this activity has been found. Alternatively, other kinesin‐5s of lower eukaryotes have shown the ability to regulate MT dynamics by acting as MT length‐dependent depolymerases, however, the involvement of loop‐2 has not been assessed. 132 , 133 , 134 , 135 Given that human kinesin‐5 Eg5 promotes MT polymerization activity, 55 , 136 rather than depolymerization, MT‐ and curved tubulin‐bound structures of each type of kinesin‐5 motor domain will be required to understand how their unique loop‐2 contacts may operate in these contrasting activities.
5. PERSPECTIVES
Figure 7 summarizes how the peripheral elements of the kinesin motor domain have evolved to impart specific families of motors with distinct enzymatic and MT‐remodeling capabilities. While kinesins serve as major MT binders, movers, crosslinkers, and length regulatory factors in all eukaryotes, they often need to work cooperatively or antagonistically with other microtubule‐associated proteins (MAPs). As with kinesins, high‐resolution cryo‐EM structures of MT‐bound MAPs are beginning to answer questions about how these proteins are specifically tuned to interact with MTs and regulate MT dynamics. Recent examples include the tubulin conformation‐sensing function of the Ndc80 complex, 137 the end‐tracking behavior of end‐binding proteins, 138 and the tubulin‐tethering activity of the axonal protein Tau. 139 Structural studies of MAPs are also revealing overlap in the MT binding sites of kinesins and MAPs, rationalizing how these proteins can impact each other's function. 139 , 140 , 141 Going forward, more of these high‐resolution structural characterizations will paint a detailed picture of the molecular mechanisms of other kinesins and MAPs, and will shows how suites of these proteins operate collectively to navigate and re‐shape MT networks.
FIGURE 7.
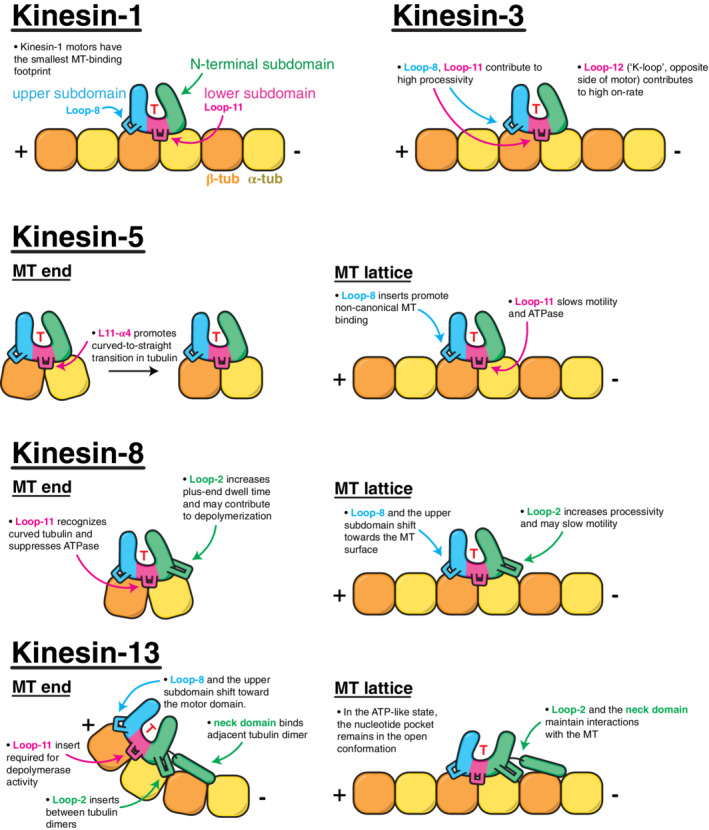
Summary of kinesin‐family‐specific adaptations and their role in motor activity
AUTHOR CONTRIBUTIONS
Byron Hunter: Conceptualization; writing‐original draft; writing‐review and editing. John Allingham: Conceptualization; funding acquisition; supervision; writing‐original draft; writing‐review and editing.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
ACKNOWLEDGMENTS
We thank our many colleagues in the kinesin field whose work contributed directly and indirectly to this effort, and we apologize for work that could not be cited due to space constraints. This work was supported by grants from CIHR and NSERC. B.H. is supported by an NSERC Postgraduate Scholarship.
Hunter B, Allingham JS. These motors were made for walking. Protein Science. 2020;29:1707–1723. 10.1002/pro.3895
Funding information Institute of Genetics, Grant/Award Number: MOP‐97832; Natural Sciences and Engineering Research Council of Canada, Grant/Award Number: RGPIN/356025‐2013; Canadian Institutes of Health Research (CIHR), Grant/Award Number: MOP‐97832
REFERENCES
- 1. Saunders WS, Hoyt MA. Kinesin‐related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. [DOI] [PubMed] [Google Scholar]
- 2. Roof DM, Meluh PB, Rose MD. Multiple kinesin‐related proteins in yeast mitosis. Cold Spring Harb Symp Quant Biol. 1991;56:693–703. [DOI] [PubMed] [Google Scholar]
- 3. Roof DM, Meluh PB, Rose MD. Kinesin‐related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woodruff JB, Drubin DG, Barnes G. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase‐promoting complex, aurora B kinase, and kinesin‐8. J Cell Biol. 2010;191:795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. [DOI] [PubMed] [Google Scholar]
- 6. Heald R, Nogales E. Microtubule dynamics. J Cell Sci. 2002;115:3–4. [DOI] [PubMed] [Google Scholar]
- 7. Egelman EH, Orlova A. New insights into actin filament dynamics. Curr Opin Struct Biol. 1995;5:172–180. [DOI] [PubMed] [Google Scholar]
- 8. Sack S, Kull FJ, Mandelkow E. Motor proteins of the kinesin family. Structures, variations, and nucleotide binding sites. Eur J Biochem. 1999;262:1–11. [DOI] [PubMed] [Google Scholar]
- 9. Sack S, Müller J, Marx A, et al. X‐ray structure of motor and neck domains from rat brain kinesin. Biochemistry. 1997;36:16155–16165. [DOI] [PubMed] [Google Scholar]
- 10. Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kull FJ, Endow SA. Kinesin: Switch I & II and the motor mechanism. J Cell Sci. 2002;115:15–23. [DOI] [PubMed] [Google Scholar]
- 12. Gulick AM, Song H, Endow SA, Rayment I. X‐ray crystal structure of the yeast Kar3 motor domain complexed with mg.ADP to 2.3 a resolution. Biochemistry. 1998;37:1769–1776. [DOI] [PubMed] [Google Scholar]
- 13. Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shang Z, Zhou K, Xu C, Csencsits R, Cochran JC, Sindelar CV. High‐resolution structures of kinesin on microtubules provide a basis for nucleotide‐gated force‐generation. eLife. 2014;3:e04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parke CL, Wojcik EJ, Kim S, Worthylake DK. ATP hydrolysis in Eg5 kinesin involves a catalytic two‐water mechanism. J Biol Chem. 2010;285:5859–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cross RA. Mechanochemistry of the kinesin‐1 ATPase. Biopolymers. 2016;105:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rice S, Lin AW, Safer D, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. [DOI] [PubMed] [Google Scholar]
- 18. Rosenfeld SS, Jefferson GM, King PH. ATP reorients the neck linker of kinesin in two sequential steps. J Biol Chem. 2001;276:40167–40174. [DOI] [PubMed] [Google Scholar]
- 19. Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nat Cell Biol. 2000;2:718–723. [DOI] [PubMed] [Google Scholar]
- 20. Asenjo AB, Weinberg Y, Sosa H. Nucleotide binding and hydrolysis induces a disorder‐order transition in the kinesin neck‐linker region. Nat Struct Mol Biol. 2006;13:648–654. [DOI] [PubMed] [Google Scholar]
- 21. Tomishige M, Stuurman N, Vale RD. Single‐molecule observations of neck linker conformational changes in the kinesin motor protein. Nat Struct Mol Biol. 2006;13:887–894. [DOI] [PubMed] [Google Scholar]
- 22. Khalil AS, Appleyard DC, Labno AK, et al. Kinesin's cover‐neck bundle folds forward to generate force. Proc Natl Acad Sci U S A. 2008;105:19247–19252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sindelar CV, Downing KH. An atomic‐level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci U S A. 2010;107:4111–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clancy BE, Behnke‐Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Milic B, Andreasson JO, Hancock WO, Block SM. Kinesin processivity is gated by phosphate release. Proc Natl Acad Sci U S A. 2014;111:14136–14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klumpp LM, Hoenger A, Gilbert SP. Kinesin's second step. Proc Natl Acad Sci U S A. 2004;101:3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crevel IM, Nyitrai M, Alonso MC, Weiss S, Geeves MA, Cross RA. What kinesin does at roadblocks: The coordination mechanism for molecular walking. EMBO J. 2004;23:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Block SM. Kinesin motor mechanics: Binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Atherton J, Yu IM, Cook A, et al. The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry. eLife. 2017;6:e27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ren J, Zhang Y, Wang S, Huo L, Lou J, Feng W. Structural delineation of the neck linker of kinesin‐3 for processive movement. J Mol Biol. 2018;430:2030–2041. [DOI] [PubMed] [Google Scholar]
- 31. Hesse WR, Steiner M, Wohlever ML, Kamm RD, Hwang W, Lang MJ. Modular aspects of kinesin force generation machinery. Biophys J. 2013;104:1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arora K, Talje L, Asenjo AB, et al. KIF14 binds tightly to microtubules and adopts a rigor‐like conformation. J Mol Biol. 2014;426:2997–3015. [DOI] [PubMed] [Google Scholar]
- 33. Soppina V, Norris SR, Dizaji AS, et al. Dimerization of mammalian kinesin‐3 motors results in superprocessive motion. Proc Natl Acad Sci U S A. 2014;111:5562–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scarabelli G, Soppina V, Yao XQ, et al. Mapping the processivity determinants of the kinesin‐3 motor domain. Biophys J. 2015;109:1537–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta ML Jr, Carvalho P, Roof DM, Pellman D. Plus end‐specific depolymerase activity of Kip3, a kinesin‐8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. [DOI] [PubMed] [Google Scholar]
- 36. Gigant B, Wang W, Dreier B, et al. Structure of a kinesin‐tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20:1001–1007. [DOI] [PubMed] [Google Scholar]
- 37. Atherton J, Farabella I, Yu IM, et al. Conserved mechanisms of microtubule‐stimulated ADP release, ATP binding, and force generation in transport kinesins. eLife. 2014;3:e03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arellano‐Santoyo H, Geyer EA, Stokasimov E, et al. A tubulin binding switch underlies Kip3/Kinesin‐8 depolymerase activity. Dev Cell. 2017;42:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alonso MC, Drummond DR, Kain S, Hoeng J, Amos L, Cross RA. An ATP gate controls tubulin binding by the tethered head of kinesin‐1. Science. 2007;316:120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS Biol. 2009;7:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krzysiak TC, Wendt T, Sproul LR, et al. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Norris SR, Soppina V, Dizaji AS, et al. A method for multiprotein assembly in cells reveals independent action of kinesins in complex. J Cell Biol. 2014;207:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. [DOI] [PubMed] [Google Scholar]
- 44. Walter WJ, Koonce MP, Brenner B, Steffen W. Two independent switches regulate cytoplasmic dynein's processivity and directionality. Proc Natl Acad Sci U S A. 2012;109:5289–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adio S, Bloemink M, Hartel M, Leier S, Geeves MA, Woehlke G. Kinetic and mechanistic basis of the nonprocessive Kinesin‐3 motor NcKin3. J Biol Chem. 2006;281:37782–37793. [DOI] [PubMed] [Google Scholar]
- 46. Adio S, Jaud J, Ebbing B, Rief M, Woehlke G. Dissection of kinesin's processivity. PLoS One. 2009;4:e4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hammond JW, Cai D, Blasius TL, et al. Mammalian Kinesin‐3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lessard DV, Zinder OJ, Hotta T, Verhey KJ, Ohi R, Berger CL. Polyglutamylation of tubulin's C‐terminal tail controls pausing and motility of kinesin‐3 family member KIF1A. J Biol Chem. 2019;294:6353–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okada Y, Hirokawa N. A processive single‐headed motor: Kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. [DOI] [PubMed] [Google Scholar]
- 50. Pollock N, de Hostos EL, Turck CW, Vale RD. Reconstitution of membrane transport powered by a novel dimeric kinesin motor of the Unc104/Kif1a family purified from Dictyostelium. J Cell Biol. 1999;147:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin‐3 motor GAKIN/KIF13B. Biochemistry. 2007;46:10039–10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cochran JC, Sontag CA, Maliga Z, Kapoor TM, Correia JJ, Gilbert SP. Mechanistic analysis of the mitotic kinesin Eg5. J Biol Chem. 2004;279:38861–38870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. von Loeffelholz O, Pena A, Drummond DR, Cross R, Moores CA. Cryo‐EM structure (4.5‐a) of yeast kinesin‐5‐microtubule complex reveals a distinct binding footprint and mechanism of drug resistance. J Mol Biol. 2019;431:864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gerson‐Gurwitz A, Thiede C, Movshovich N, et al. Directionality of individual kinesin‐5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011;30:4942–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen GY, Cleary JM, Asenjo AB, et al. Kinesin‐5 promotes microtubule nucleation and assembly by stabilizing a lattice‐competent conformation of tubulin. Curr Biol. 2019;29:2259–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cochran JC, Gilbert SP. ATPase mechanism of Eg5 in the absence of microtubules: Insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry. 2005;44:16633–16648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cochran JC, Krzysiak TC, Gilbert SP. Pathway of ATP hydrolysis by monomeric kinesin Eg5†. Biochemistry. 2006;45:12334–12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Duselder A, Thiede C, Schmidt CF, Lakamper S. Neck‐linker length dependence of processive Kinesin‐5 motility. J Mol Biol. 2012;423:159–168. [DOI] [PubMed] [Google Scholar]
- 59. Kaseda K, Crevel I, Hirose K, Cross RA. Single‐headed mode of kinesin‐5. EMBO Rep. 2008;9:761–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric inhibition of kinesin‐5 modulates its processive directional motility. Nat Chem Biol. 2006;2:480–485. [DOI] [PubMed] [Google Scholar]
- 61. Maliga Z, Kapoor TM, Mitchison TJ. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem Biol. 2002;9:989–996. [DOI] [PubMed] [Google Scholar]
- 62. Tao L, Mogilner A, Civelekoglu‐Scholey G, et al. A homotetrameric kinesin‐5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. [DOI] [PubMed] [Google Scholar]
- 63. Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weinger JS, Qiu M, Yang G, Kapoor TM. A non‐motor microtubule binding site in kinesin‐5 is required for filament crosslinking and sliding. Curr Biol. 2011;21:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin‐8 depolymerizes microtubules in a length‐dependent manner. Nat Cell Biol. 2006;8:957–962. [DOI] [PubMed] [Google Scholar]
- 66. Wang D, Nitta R, Morikawa M, Yajima H, Inoue S, Shigematsu H, Kikkawa M, Hirokawa (2016) Motility and microtubule depolymerization mechanisms of the kinesin‐8 motor, KIF19A. eLife 5:e18101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Peters C, Brejc K, Belmont L, et al. Insight into the molecular mechanism of the multitasking kinesin‐8 motor. EMBO J. 2010;29:3437–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Locke J, Joseph AP, Pena A, et al. Structural basis of human kinesin‐8 function and inhibition. Proc Natl Acad Sci U S A. 2017;114:E9539–E9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N. KIF19A is a microtubule‐depolymerizing kinesin for ciliary length control. Dev Cell. 2012;23:1167–1175. [DOI] [PubMed] [Google Scholar]
- 70. Jannasch A, Bormuth V, Storch M, Howard J, Schäffer E. Kinesin‐8 is a low‐force motor protein with a weakly bound slip state. Biophys J. 2013;104:2456–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Koonce MP, Tikhonenko I. Bioenergetics of the Dictyostelium kinesin‐8 motor isoform. Biomolecules. 2020;10:E563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Leong SY, Edzuka T, Goshima G, Yamada M. Kinesin‐13 and kinesin‐8 function during cell growth and division in the moss Physcomitrella patens . Plant Cell. 2020;32:683–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mayr MI, Storch M, Howard J, Mayer TU. A non‐motor microtubule binding site is essential for the high processivity and mitotic function of kinesin‐8 Kif18A. PLoS One. 2011;6:e27471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McHugh T, Gluszek AA, Welburn JP. Microtubule end tethering of a processive kinesin‐8 motor Kif18b is required for spindle positioning. J Cell Biol. 2018;217:2403–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stumpff J, Du Y, English CA, et al. A tethering mechanism controls the processivity and kinetochore‐microtubule plus‐end enrichment of the kinesin‐8 Kif18A. Mol Cell. 2011;43:764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Su X, Qiu W, Gupta ML Jr, Pereira‐Leal JB, Reck‐Peterson SL, Pellman D. Mechanisms underlying the dual‐mode regulation of microtubule dynamics by Kip3/kinesin‐8. Mol Cell. 2011;43:751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Trofimova D, Paydar M, Zara A, Talje L, Kwok BH, Allingham JS. Ternary complex of Kif2A‐bound tandem tubulin heterodimers represents a kinesin‐13‐mediated microtubule depolymerization reaction intermediate. Nat Commun. 2018;9:2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Friel CT, Howard J. The kinesin‐13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J. 2011;30:3928–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang W, Shen T, Guerois R, et al. New insights into the coupling between microtubule depolymerization and ATP hydrolysis by kinesin‐13 protein Kif2C. J Biol Chem. 2015;290:18721–18731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goulet A, Behnke‐Parks WM, Sindelar CV, Major J, Rosenfeld SS, Moores CA. The structural basis of force generation by the mitotic motor kinesin‐5. J Biol Chem. 2012;287:44654–44666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gennerich A, Vale RD. Walking the walk: How kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Okada Y, Hirokawa N. Mechanism of the single‐headed processivity: Diffusional anchoring between the K‐loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci U S A. 2000;97:640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Okada Y, Yamazaki H, Sekine‐Aizawa Y, Hirokawa N. The neuron‐specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. [DOI] [PubMed] [Google Scholar]
- 84. Siddiqui N, Straube A. Intracellular cargo transport by kinesin‐3 motors. Biochemistry. 2017;82:803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rosenfeld SS, Xing J, Jefferson GM, King PH. Docking and rolling, a model of how the mitotic motor Eg5 works. J Biol Chem. 2005;280:35684–35695. [DOI] [PubMed] [Google Scholar]
- 86. Cole DG, Saxton WM, Sheehan KB, Scholey JM. A "slow" homotetrameric kinesin‐related motor protein purified from drosophila embryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- 87. Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kashina AS, Scholey JM, Leszyk JD, Saxton WM. An essential bipolar mitotic motor. Nature. 1996;384:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gordon DM, Roof DM. The kinesin‐related protein Kip1p of Saccharomyces cerevisiae is bipolar. J Biol Chem. 1999;274:28779–28786. [DOI] [PubMed] [Google Scholar]
- 90. Acar S, Carlson DB, Budamagunta MS, et al. The bipolar assembly domain of the mitotic motor kinesin‐5. Nat Commun. 2013;4:1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. [DOI] [PubMed] [Google Scholar]
- 92. van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin‐5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Enos AP, Morris NR. Mutation of a gene that encodes a kinesin‐like protein blocks nuclear division in A. nidulans . Cell. 1990;60:1019–1027. [DOI] [PubMed] [Google Scholar]
- 94. Saunders AM, Powers J, Strome S, Saxton WM. Kinesin‐5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–R454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus‐end‐directed microtubule motor. Nature. 1992;359:540–543. [DOI] [PubMed] [Google Scholar]
- 96. Brust‐Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin‐5‐dependent poleward flux and spindle length control in drosophila embryo mitosis. Mol Biol Cell. 2009;20:1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol. 2011;21:1356–1365. [DOI] [PubMed] [Google Scholar]
- 98. Hunter AW, Caplow M, Coy DL, et al. The kinesin‐related protein MCAK is a microtubule depolymerase that forms an ATP‐hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Stout JR, Yount AL, Powers JA, Leblanc C, Ems‐McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22:3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin‐8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. West RR, Malmstrom T, Troxell CL, McIntosh JR. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol Biol Cell. 2001;12:3919–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule‐destabilizing enzymes. Cell. 1999;96:69–78. [DOI] [PubMed] [Google Scholar]
- 103. Walczak CE, Gayek S, Ohi R. Microtubule‐depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–441. [DOI] [PubMed] [Google Scholar]
- 104. Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. [DOI] [PubMed] [Google Scholar]
- 105. Lawrence CJ, Dawe RK, Christie KR, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sweeney HL, Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. [DOI] [PubMed] [Google Scholar]
- 107. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- 108. Woehlke G, Ruby AK, Hart CL, Ly B, Hom‐Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. [DOI] [PubMed] [Google Scholar]
- 109. Kull FJ, Endow SA. A new structural state of myosin. Trends Biochem Sci. 2004;29:103–106. [DOI] [PubMed] [Google Scholar]
- 110. Cochran JC, Sindelar CV, Mulko NK, et al. ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell. 2009;136:110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin‐1 motor domain to axons. Nat Neurosci. 2009;12:559–567. [DOI] [PubMed] [Google Scholar]
- 112. Pena A, Sweeney A, Cook AD, Topf M, Moores CA. Structure of microtubule‐trapped human kinesin‐5 and its mechanism of inhibition revealed using cryoelectron microscopy. Structure. 2020;28:450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bell KM, Cha HK, Sindelar CV, Cochran JC. The yeast kinesin‐5 Cin8 interacts with the microtubule in a noncanonical manner. J Biol Chem. 2017;292:14680–14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Asenjo AB, Chatterjee C, Tan D, et al. Structural model for tubulin recognition and deformation by kinesin‐13 microtubule depolymerases. Cell Rep. 2013;3:759–768. [DOI] [PubMed] [Google Scholar]
- 115. Arellano‐Santoyo H (2017) Regulation of microtubule plus‐end dynamics by molecular motors. Thesis or Dissertation, Harvard Library: Medical Sciences, Harvard University.
- 116. Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers‐M type kinesins stabilize curling of the protofilament using the class‐specific neck and loops. Cell. 2004;116:591–602. [DOI] [PubMed] [Google Scholar]
- 117. Benoit M, Asenjo AB, Sosa H. Cryo‐EM reveals the structural basis of microtubule depolymerization by kinesin‐13s. Nat Commun. 2018;9:1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wang W, Cantos‐Fernandes S, Lv Y, et al. Insight into microtubule disassembly by kinesin‐13s from the structure of Kif2C bound to tubulin. Nat Commun. 2017;8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Friel CT, Welburn JP. Parts list for a microtubule depolymerising kinesin. Biochem Soc Trans. 2018;46:1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Cao L, Wang W, Jiang Q, Wang C, Knossow M, Gigant B. The structure of apo‐kinesin bound to tubulin links the nucleotide cycle to movement. Nat Commun. 2014;5:5364. [DOI] [PubMed] [Google Scholar]
- 121. Shima T, Morikawa M, Kaneshiro J, et al. Kinesin‐binding‐triggered conformation switching of microtubules contributes to polarized transport. J Cell Biol. 2018;217:4164–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Arpag G, Norris SR, Mousavi SI, et al. Motor dynamics underlying cargo transport by pairs of kinesin‐1 and kinesin‐3 motors. Biophys J. 2019;116:1115–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. von Loeffelholz O, Moores CA. Cryo‐EM structure of the Ustilago maydis kinesin‐5 motor domain bound to microtubules. J Struct Biol. 2019;207:312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Shipley K, Hekmat‐Nejad M, Turner J, et al. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ogawa T, Saijo S, Shimizu N, Jiang X, Hirokawa N. Mechanism of catalytic microtubule depolymerization via KIF2‐tubulin transitional conformation. Cell Rep. 2017;20:2626–2638. [DOI] [PubMed] [Google Scholar]
- 126. Wang W, Jiang Q, Argentini M, et al. Kif2C minimal functional domain has unusual nucleotide binding properties that are adapted to microtubule depolymerization. J Biol Chem. 2012;287:15143–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Maney T, Wagenbach M, Wordeman L. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere‐associated kinesin. J Biol Chem. 2001;276:34753–34758. [DOI] [PubMed] [Google Scholar]
- 128. Ovechkina Y, Wagenbach M, Wordeman L. K‐loop insertion restores microtubule depolymerizing activity of a "neckless" MCAK mutant. J Cell Biol. 2002;159:557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kim H, Fonseca C, Stumpff J. A unique kinesin‐8 surface loop provides specificity for chromosome alignment. Mol Biol Cell. 2014;25:3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin‐8 motors act cooperatively to mediate length‐dependent microtubule depolymerization. Cell. 2009;138:1174–1183. [DOI] [PubMed] [Google Scholar]
- 131. Du Y, English CA, Ohi R. The kinesin‐8 Kif18A dampens microtubule plus‐end dynamics. Curr Biol. 2010;20:374–380. [DOI] [PubMed] [Google Scholar]
- 132. Gardner MK, Bouck DC, Paliulis LV, et al. Chromosome congression by Kinesin‐5 motor‐mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wang G, Gao X, Huang Y, Yao Z, Shi Q, Wu M. Nucleophosmin/B23 inhibits Eg5‐mediated microtubule depolymerization by inactivating its ATPase activity. J Biol Chem. 2010;285:19060–19067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. McCoy KM, Tubman ES, Claas A, et al. Physical limits on kinesin‐5‐mediated chromosome congression in the smallest mitotic spindles. Mol Biol Cell. 2015;26:3999–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Tubman E, He Y, Hays TS, Odde DJ. Kinesin‐5 mediated chromosome congression in insect spindles. Cell Mol Bioeng. 2018;11:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chen Y, Hancock WO. Kinesin‐5 is a microtubule polymerase. Nat Commun. 2015;6:8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Alushin GM, Ramey VH, Pasqualato S, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162:849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, Nogales E. Near‐atomic model of microtubule‐tau interactions. Science. 2018;360:1242–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kellogg EH, Howes S, Ti SC, et al. Near‐atomic cryo‐EM structure of PRC1 bound to the microtubule. Proc Natl Acad Sci U S A. 2016;113:9430–9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Atherton J, Jiang K, Stangier MM, et al. A structural model for microtubule minus‐end recognition and protection by CAMSAP proteins. Nat Struct Mol Biol. 2017;24:931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]


