Abstract
New enzyme functions often evolve through the recruitment and optimization of latent promiscuous activities. How do mutations alter the molecular architecture of enzymes to enhance their activities? Can we infer general mechanisms that are common to most enzymes, or does each enzyme require a unique optimization process? The ability to predict the location and type of mutations necessary to enhance an enzyme's activity is critical to protein engineering and rational design. In this review, via the detailed examination of recent studies that have shed new light on the molecular changes underlying the optimization of enzyme function, we provide a mechanistic perspective of enzyme evolution. We first present a global survey of the prevalence of activity‐enhancing mutations and their distribution within protein structures. We then delve into the molecular solutions that mediate functional optimization, specifically highlighting several common mechanisms that have been observed across multiple examples. As distinct protein sequences encounter different evolutionary bottlenecks, different mechanisms are likely to emerge along evolutionary trajectories toward improved function. Identifying the specific mechanism(s) that need to be improved upon, and tailoring our engineering efforts to each sequence, may considerably improve our chances to succeed in generating highly efficient catalysts in the future.
Keywords: activity‐enhancing mutations, ancestral sequence reconstruction, conformational dynamics, conformational tinkering, directed evolution, enzyme evolution, enzyme promiscuity, enzyme–substrate complementarity, molecular mechanisms, substrate repositioning
1. INTRODUCTION
How are catalytic functions optimized by evolution? This is a key question for protein scientists and evolutionary biochemists who are attempting to decipher the molecular principles of enzyme evolution. The answers to this question are critical for protein engineering and design to improve or modify enzyme activity, or to create enzymes endowed with novel functions that do not exist in nature. 1
The current model of enzyme evolution entails the recruitment and optimization of a preexisting activity in a protein scaffold. In addition to possessing high catalytic efficiency for their native substrate, many enzymes also exhibit promiscuous activities toward secondary substrates. 2 , 3 , 4 It has been extensively discussed that these latent promiscuous activities constitute a reservoir from which new chemistry can gradually emerge during functional optimization. 2 , 3 , 5 , 6 , 7 , 8 Many questions remain regarding the origin of promiscuity itself, and this topic has been debated in several articles over the last decade. 9 , 10 , 11 , 12 In this review, we focus on discussing another important aspect of enzyme evolution: optimization and its underlying molecular mechanisms. When an enzyme possessing a weak promiscuous catalytic activity is recruited or selected, how do mutations alter its molecular architecture to rapidly adapt to a new substrate upon environmental change?
To address this question, it is first necessary to reflect upon the mechanisms by which enzymes recognize their native cognate substrate, and their promiscuous adventitious substrates, respectively. Natural enzymes achieve high rate‐acceleration through the precise preorganization of active site residues that provide tailored electrostatic‐ and geometric‐complementarities with their native substrate, enabling specific transition state (TS) recognition and stabilization. 13 , 14 In addition, their structural motions have often been tuned by evolution to facilitate unique catalytic cycles, that is, the capture and release of chemical intermediates along the reaction coordinate. 15 , 16 In this context, low‐level promiscuous activities are likely to be the result of a suboptimal fit between the active site and the promiscuous TS, originating from poor geometric and electrostatic complementarities and/or unsynchronized dynamics. 8 , 10 Thus, the optimization of a promiscuous activity is likely to involve the tinkering of enzyme–substrate interactions to form a more optimal Michaelis complex and TS, 7 and the alteration of structural motions to adapt to new catalytic cycles. 17
In recent years, a number of studies have documented the molecular changes associated with an increase in activity during evolutionary cycles. Starting from the promiscuous activity of either a natural or computationally‐designed enzyme, these studies found that the optimization of enzyme function could be achieved by directed evolution. Other natural examples were identified via bioinformatic‐based approaches, such as ancestor sequence reconstruction (ASR) (Table 1). In the following sections, we present a collection of works that have shed new light on the mechanistic basis of enzyme evolution and outline the critical molecular determinants that govern functional transitions. First, we provide a global view of the distribution of activity‐enhancing mutations derived from several evolutionary trajectories, and quantify their role and contribution to the enhancement of novel catalytic activities. Second, we highlight studies that obtained atomic‐resolution snapshots along evolutionary trajectories and inferred the mechanisms by which mutations improve promiscuous functions. Finally, we provide some perspectives on, and directions for this matter, which we believe will further enhance our understanding of enzyme evolution and improve our ability to engineer novel catalysts.
TABLE 1.
A survey of the molecular mechanisms identified in natural and laboratory evolution
| Identified molecular mechanism of evolution | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Starting enzyme | Evolved function | New catalytic interaction | Active site reshaping | Conformational tinkering | Alteration of dynamics | Substrate repositioning | Repositioning cofactors | Description | Method | Refs |
| Atrazine chlorohydrolase (AtzA) | Melamine deaminase | + |
|
N | 18 | |||||
| Tyrocidine synthetase 1, A domain (TycA) | l‐alanine/l‐threonine adenylation | + |
|
RD | 19 | |||||
| (S)‐β‐Phe/l‐Phe specific adenylation | + |
|
RD | 20 | ||||||
| Carbonic anhydrase II (hCAII) | Esterase | + |
|
DE | 21 | |||||
| Cytochrome P450 | Propane hydroxylation | + |
|
RD/DE | 22, 23 | |||||
| ancDHCH1 | Methyl‐parathion hydrolase | + |
|
ASR | 24 | |||||
| ancMALS | Maltase | + |
|
ASR | 25 | |||||
| ancIMA1‐4 | Isomaltase | + | + |
|
||||||
| ancM/L‐malate/lactate dehydrogenase | Lactate dehydrogenase | + | + |
|
ASR | 26 | ||||
| Haloalkane dehalogenase (DhaA) | Enantioselectivity for β‐bromoalkanes | + | + |
|
RD | 27 | ||||
| β‐Lactamase (NDM1) | Phosphonate monoester hydrolase | + | + |
|
DE | 28 | ||||
| Arylsulfatase (PAS) | Phosphonate monoester hydrolase | + | + |
|
DE | 29 | ||||
| β subunit of tryptophan synthase (TrpB) | Non‐canonical AA synthesis (standalone Trp) | + | + |
|
DE | 30, 31 | ||||
| CypA proline isomerase (+ S99T) | Restoration of pro‐isomerization | + | + |
|
DE | 32 | ||||
| Lactonase (PON1) | Paraoxonase | + | + |
|
DE | 33, 34 | ||||
| Kemp Eliminase* (KE07) | Kemp eliminase | + | + |
|
RD/DE | 35 | ||||
| + | + | + |
|
DE | 36 | |||||
| Kemp Eliminase* (KE70) | Kemp eliminase | + | + | + |
|
RD/DE | 37 | |||
| TEM‐1 | Cefotaxime | + | + | + |
|
N | 38 | |||
| Diels‐alderase* (DA_20_00) | Diels‐alderase | + | + | + |
|
RD/DE | 39, 40 | |||
| Acyltransferase (LovD) | Acyltransferase (simvastatin synthesis) | + | + | + |
|
DE | 41 | |||
| N‐acyl homoserine lactonase (AiiA) | Paraoxonase | + | + | + |
|
DE | 42 | |||
| Metallo‐β‐lactamase (BcII) | Cephalexinase | + | + | + |
|
DE | 43, 44 | |||
| Phosphotriesterase (PTE) | Arylesterase | + | + | + | + |
|
DE | 45, 46 | ||
| AA–binding protein (AABP, AncCDT) | Cyclohexadienyl dehydratase | + | + | + | + |
|
ASR/DE | 47, 48 | ||
| Bifunctional PROFAR/PRA isomerase (HisA dup13‐15, D10G) | PRA isomerase (TrpF function) | + | + | + | + |
|
DE | 49, 50 | ||
| PROFAR isomerase (HisA function) | + | + | + | + |
|
|||||
| Kemp Eliminase* (HG3) | Kemp eliminase | + | + | + | + |
|
RD/DE | 51 | ||
| Anc chalcone isomerase (AncCHI) | Michael addition of chalconaringenin | + | + | + | + |
|
ASR/DE | 52 | ||
| Kemp Eliminase* (KE59) | Kemp eliminase | + | + | + | + |
|
RD/DE | 53 | ||
| PTE arylesterase (AE) | Paraoxonase | + | + | + | + |
|
DE | 54 | ||
| Retro‐aldolase* (RA95 and RA95.5) | (±)‐Methodol cleavage | + | + | + | + | + |
|
RD/DE | 55, 56 | |
|
RD/DE | 57, 58 | ||||||||
Note: The symbol + indicates that the mechanism has been specifically identified in the study. A blank indicates that the mechanism has not been identified in this model; note that it does not rule out the existence of other mechanisms at play. Asterisk symbol (*) denotes starting enzymes that were computationally designed.
Abbreviations: AA, amino acids; Anc, ancestor; AS, active site; ASR, ancestral sequence reconstruction; DE, directed evolution; E–S, enzyme−substrate complex; LFER, linear free‐energy relationships; LG, leaving group; MDs, molecular dynamics simulations; N, natural trajectory; TS, transition state; VDW, Van der Waals; RD, rational design.
2. A GLOBAL VIEW OF THE DISTRIBUTION OF ACTIVITY‐ENHANCING MUTATIONS
First, we would like to provide a bird's eye view on the mutational effects necessary to promote a novel promiscuous function by discussing the following questions: (a) What fraction of mutations is beneficial to the new enzymatic function? (b) How are these beneficial mutations distributed across an enzyme's structure? and (c) How many beneficial mutations are required to reach sufficient catalytic efficiency? While each model constitutes a unique case in terms of attributes and response to mutations, when observations are drawn from diverse enzymes they can reveal general trends and patterns that underlie the optimization of a novel catalytic activity.
2.1. What fraction of mutations improves catalytic activity?
Derived from numerous enzyme engineering efforts, the fraction of beneficial mutations is estimated to be very low: the screening of mutagenized libraries typically results in the identification of less than 10–20 activity‐enhancing mutations. Recent developments in massive‐scale mutational analysis platforms, that is, deep mutational scanning (DMS), are now providing a systematic and more statistical picture of the distribution of fitness effects (DFE) in proteins. 59 , 60 The overall DFE measured during several DMS experiments across different target enzymes is consistent with the observations inferred from enzyme evolution campaigns: ~60–70% of mutations are deleterious, 30–40% are neutral, and less than 5% of mutations confer improvements in function. 61 , 62 , 63 , 64 , 65 It should be noted that the selection pressure applied during DMS is often coupled to cell growth, such that the DFE not only reflects on changes in catalytic activity but also in protein expression, stability, and solubility. However, these observations must be put into perspective: assuming a protein sequence encompassing hundreds of amino acids, even a very small percentage of all available substitutions could still yield a substantial diversity of beneficial mutations. For example, nearly 2% of all possible single point substitutions in an enzyme of 300 amino acids (5,700 variants) would still correspond to 114 available beneficial mutations. Indeed, in a DMS study with VIM‐2 β‐lactamase, more than a hundred beneficial and specificity‐altering mutations were identified across 25 different positions (Figure 1a). 65 This represents a significant reservoir of accessible beneficial mutations within the local sequence space that can be harnessed.
FIGURE 1.
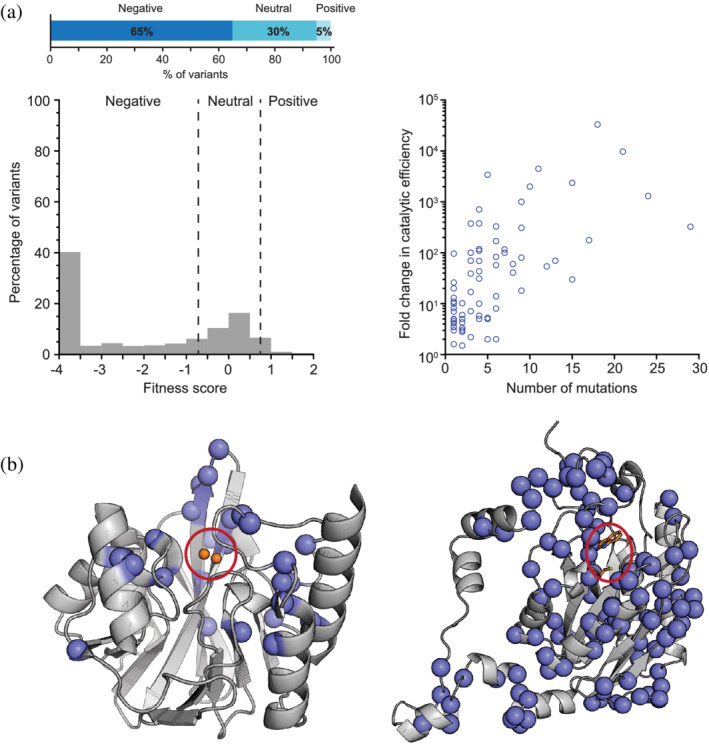
A global view of the distribution of activity‐enhancing mutations. (a) Histogram of the distribution of fitness effects for all missense mutations in the enzyme VIM2 under selection for growth at 128 μg/ml of ampicillin. The dashed vertical lines indicate fitness score cutoffs used to classify fitness effects as positive, neutral, or negative. The bar graph at the top indicates the total percentage of positive, neutral, or negative variants. (b) Cartoon representations of the crystal structures of VIM2 65 (left, PDB ID: 5yd7) and amiE 62 (right, PDB ID: 2uxy) with the positions of all activity‐enhancing mutations highlighted as blue spheres. Red circles indicate the location of the active site. The active site Zn2+ ions of VIM2 are depicted as orange spheres, whereas two active site residues of amiE, Trp138 and Cys166, are depicted as sticks. (c) Fold‐change in k cat/K M of several evolved enzymes against the number of missense mutations acquired during directed evolution. Panel A is adapted from Ref. 65, Panel C from Ref. 62
2.2. Where are activity‐enhancing mutations located?
While the fraction of beneficial mutations is largely consistent regardless of the enzyme model, the distribution of beneficial mutations on the tertiary structures seems to vary considerably among enzymes. In some cases, beneficial mutations cluster around the active site, for example, in the DMS study of VIM‐2 β‐lactamase, 23 out of 25 positions that contain at least 1 specificity altering mutation were located within 15 Å of the catalytic zinc ions (Figure 1b). 65 4‐Oxalocrotonate tautomerase and Tn5 transposon‐derived kinase have yielded similar patterns, where the majority of activity‐ and specificity‐altering mutations tended to cluster in the first or second shell. 63 , 64 In contrast, the majority of beneficial and specificity‐determining mutations occurring in amiE appear located far from the active site: most of the 395 mutations that specifically enhanced growth on isobutyramide were 9–21 Å away from the active site (Figure 1b). 62 Similarly, 53/106 (50%) of mutations in TEM‐1 β‐lactamase that were found to increase fitness toward cefotaxime are localized on the enzyme surface and far from the active site. 61 While the mutations observed in directed evolution and ASR studies are more biased, that is, they only represent mutations that were selected during adaptation, these studies have also unveiled the contributions of both proximal and distal mutations. In general, mutations closer to the active site (<10 Å) tend to have larger effects, however, a considerable number of activity‐enhancing mutations remain >10 Å away from the catalytic center. 66 , 67 , 68 Moreover, distal mutations can cause large improvements in activity. For example, a mutation located 13 Å away from the active site improved ceftazidime hydrolysis by a β‐lactamase by more than 600‐fold 69 and a mutation 19 Å away from the active site improved the activity of a fatty acid desaturase by ~30‐fold. 70
Overall, the wide distribution of activity‐enhancing mutations on protein structures is likely a reflection of the existence of multiple solutions for improving interactions between an enzyme's active site and its substrate. Mutations in the active site may generate critical residues that interact with the substrate and stabilize the TS; however, second‐shell mutations may also help to fine‐tune the key residues in the active site and/or binding pocket to be more complementary to the target substrate. In addition, surface mutations may function by altering conformational dynamics to more catalytically active conformations. Thus, the effects of proximal versus distal mutations will vary between different sequences and are dependent upon the underlying molecular bottleneck that needs to be improved upon (examples are discussed in greater detail in Section 3).
2.3. How many mutations are required to elicit a new function?
If the optimization of enzyme function cannot be achieved via single‐point mutations and requires multiple substitutions, how many are necessary to achieve a complete functional transition? A recent systematic analysis of directed evolution studies demonstrated that the overall improvement in activity compared to the total number of mutations varies substantially with the enzyme model (Figure 1c). 71 However, a prominent trend is that improvements greater than 1,000‐fold typically require the accumulation of at least 10 mutations. Moreover, the catalytic improvements along an evolutionary trajectory often exhibit “diminishing returns,” a hallmark of evolutionary optimization processes whereby the fitness improvement per mutation is large in early rounds of evolution but later becomes more incremental. 72 , 73 For example, during the evolution of phosphotriesterase (PTE) toward arylesterase activity, the first four rounds resulted in a ~1,100‐fold increase in activity, whereas the final 14 rounds only saw a ~30‐fold increase. 45 Thus, the accumulation of a large number of mutations (>10–20 mutations) appears essential to fully achieve the optimization of a novel catalytic activity, even though late‐occurring mutations may be comparatively less fruitful (Figure 1c). 74
2.4. The effect of mutational epistasis
An intriguing question underlying the distribution of beneficial mutations is the extent to which epistasis influences the effects of mutations. 75 , 76 For example, the effect of a mutation can switch from beneficial to deleterious (or the reciprocal), depending on the presence or absence of other mutations. Thus, as epistasis alters the nature of mutations, it restricts the accessibility of available substitutions and hence impacts evolutionary outcomes. On the scale of an entire protein sequence, epistasis appears relatively scarce, for example, DMS studies conducted on the RNA recognition motif of poly(A)‐binding protein and the IgG‐binding domain of protein G found that pairwise epistasis occurred in only 4–5% of double mutants. 77 , 78 By contrast, among activity‐enhancing mutations, epistasis appears highly prevalent and can drastically alter the effect of mutations. 18 , 24 , 79 , 80 , 81 A systematic analysis of nine examples of enzyme evolution revealed that 82% of functional mutations exhibit epistasis, with nearly half appearing either neutral or deleterious in the wild‐type background, only to become beneficial following the fixation of other mutations along the trajectory. 68 Thus, the distribution of activity‐enhancing mutations may be altered progressively as adaptation proceeds. Predicting epistasis and identifying mutations that can collectively improve the enzyme activity by several orders of magnitude is a key challenge for the field of protein evolution and engineering; it requires an in‐depth molecular understanding of the epistatic networks embedded within enzymes structures. A detailed discussion of the epistatic constraints on evolution and the molecular mechanisms underlying epistasis is beyond the scope of this review; thus, we refer readers to the excellent references therein for further insights into this matter. 68 , 75 , 76 , 82 , 83 , 84 , 85
3. MOLECULAR MECHANISMS OF ACTIVITY‐ENHANCING MUTATIONS
Understanding the mechanisms by which mutations alter the molecular architecture of enzymes to increase catalytic activity remains a great challenge in the field. Over the years, an increasing number of studies described the molecular changes occurring in enzymes, evolving in nature or the laboratory. Importantly, these studies also demonstrate how enzyme–substrate interactions are altered from promiscuous, suboptimal interactions to highly organized, optimal ones. In this section, we highlight and discuss several major mechanisms that have emerged from these recent examples, providing an atomic‐level view of the evolution of novel enzyme functions (Table 1).
3.1. Creation of new interactions with the substrate
One accessible solution for enzyme evolution is the creation of new interactions with a novel substrate. The introduction of a new residue in the active site may alter electrostatic interactions with the substrate and improve its catalysis. In some cases, the establishment of essential catalytic groups is needed to achieve radical functional transitions. A notable example was described during the natural evolution of cyclohexadienyl dehydratase (CDT) activity from an ancestral solute binding protein. 47 In this case of de novo enzyme evolution, the ancestral scaffold possessed an incomplete catalytic machinery; the introduction of a glutamate residue within the binding pocket early in CDT evolution, established general‐acid catalysis that was essential to the new function. Another remarkable example is the mechanistic transition from atrazine chlorohydrolase (AtzA) to melamine deaminase (TriA), two homologs performing herbicide degradation in nature (Figure 2a). The evolutionary transition requires the Ser–Asn catalytic dyad assisting atrazine dechlorination in AtzA to be substituted to a Cys–Asp for melamine deamination to occur. 18 A similar mechanism was reported during the directed evolution of a computationally designed retro‐aldolase, RA95.0 which initially contained a catalytic Lys210 designed to facilitate C─C bond cleavage of (±) methodol. 55 , 56 During the directed evolution of RA95.0, the function of Lys210 was taken over by a new mutation, T83K, which was inserted across the active site in an environment that better promotes catalysis (Figure 2b). 55 Even more surprisingly, further directed evolution resulted in the introduction of three additional mutations in the active site (V51Y, S110N, F180Y), which formed a hydrogen‐bonding network with Lys83. 57 This new catalytic tetrad catalyzes retro‐aldolization of (±) methodol via a completely retuned catalytic mechanism. 57 , 58
FIGURE 2.
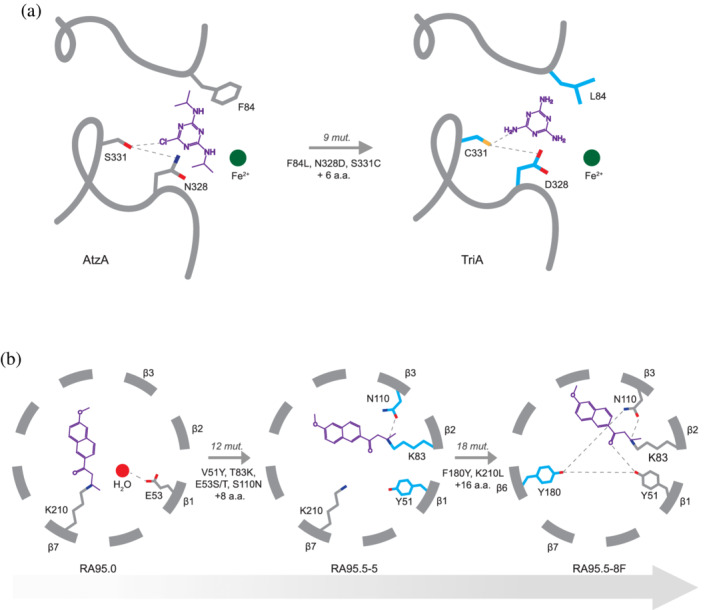
Creation of new enzyme–substrate interactions by evolution. (a) Schematic representation of key active site residues in AtzA and TriA. The Ser–Asn dyad, involved in atrazine dechlorination, is substituted to Cys–Asp during the evolution towards melamine deamination. 18 An additional active site residue, Phe84, is also mutated to leucine. The active site Fe2+ is depicted as a green sphere. (b) Schematic representation of the stepwise evolution of a designed retro‐aldolase, RA95.0. 55 , 57 (left) The catalytic dyad (K210‐E53 and a water molecule) from the initial RA95.0 design, (centre) was mutated to a distinct triad (K83‐N110‐Y51) in RA95.5–5 during directed evolution. (right) Further evolution resulted in the emergence of a catalytic tetrad (K83‐N110‐Y51‐Y180) in RA95.5‐8F. Key active site residues for Panels A and B are depicted as sticks; new residues installed at each evolutionary stage are highlighted in blue. The mutations fixed between each step of the trajectory are indicated on the gray arrows. The chemical structures of atrazine (left) and melamine (right) in Panel A, and the 1,3‐diketone mechanism‐based inhibitor of retro‐aldolases in Panel B are shown in purple. Panel B is adapted from Ref. 55
Beyond these extreme cases, new active site interactions appear to reinforce or complement preexisting ones by providing residue(s) that coordinate unique moieties(s) of the new substrate nonexistent in the native substrate. For instance, a six‐residue insertion within an active site loop was observed during the neofunctionalization of apicomplexan lactate dehydrogenase from an ancestral malate dehydrogenase (AncM/L). 26 This insertion displaces an active site arginine in favor of a new Trp107, forming hydrophobic interactions with the methyl substituent of the novel substrate, pyruvate. Several other studies, including the natural divergence of maltases and isomaltases 25 and HisA/TrpF isomerases, 49 , 50 or the laboratory evolution of arylester hydrolysis in PTE, 45 also described a similar phenomenon.
While the creation of a novel interaction with the substrate may be intuitive, this mechanism is not common in the literature. One explanation may be that promiscuous substrates typically interact with preexisting catalytic machinery, taking advantage of its inherent reactivity albeit in a suboptimal fashion. 86 , 87 Thus, the creation of new interactions may not be necessary as functional optimization can be achieved without drastic alterations of enzyme–substrate interactions, particularly when the target substrate closely resembles the native one.
3.2. Active site reshaping
A more prevalent evolutionary mechanism to enhance catalysis is the reshaping of the active site. This mechanism typically involves the tailoring of enzyme–substrate interactions to promote geometric complementarity. In this case, mutations typically occur around the active site to alter its overall shape and size, without necessarily affecting the electrostatic environment or catalytic machinery. These active site modifications are, of course, case‐specific: some enzymes require a narrowing of the active site cavity to promote a snug fit with smaller substrates. 22 , 23 , 37 , 41 For example, during the laboratory optimization of a computationally designed diels–alderase (DA_20_00), a ~9,700‐fold increase in chemical proficiency was achieved by the insertion of a 24‐residue helix‐turn‐helix motif, and the stepwise contraction of the cavity by multiple mutations to constrain the substrate in a productive orientation (Figure 3). This substantial remodeling of the binding pocket improved complementarity to the substrate without affecting the position of the designed catalytic residues. 39 , 40 Similar observations were reported during the evolution of cytochrome P450BM3 towards propane hydroxylation 22 , 23 and the evolution of the adenylation domain of tyrocidine synthetase (TycA) toward smaller, 19 or backbone‐modified amino acids. 20
FIGURE 3.
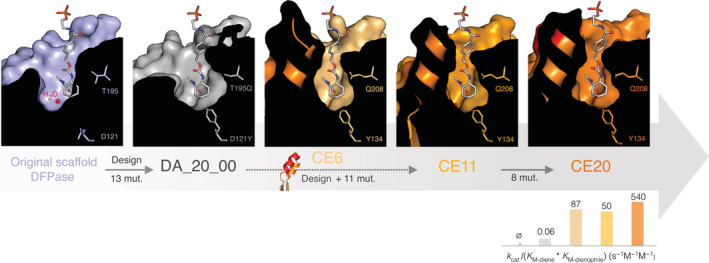
Active site reshaping by evolution. (top) Cutaway views of the substrate binding pocket of designed and evolved diels–alderases, in the same orientation, and overlaid with the phosphorylated product analog bound in CE20 (PDB ID: 4o5t) depicted as sticks. Two key catalytic residues, T195Q and D121Y, introduced by design in the scaffold of a diisopropylfluorophosphatase (DFPase, PDB ID: 1e1a) to generate diels–alderase activity in mutants DA_20_00 (PDB ID: 3ilc), CE6 (PDB ID: 3u0s), CE11 (PDB ID: 4o5s) and CE20 are highlighted as sticks. A designed 24‐residue helix‐turn‐helix motif that was incorporated into the structure is represented as cartoon, while a buried water molecule, present in all structures, is depicted as a red sphere. The number of mutations fixed between each step is indicated on the grey arrows. (bottom) Catalytic efficiency for the diels‐alderase reaction at each step of the trajectory, on a log scale. Adapted from Refs. 63 and 64
Conversely, an enlargement of the cleft may be required to remove steric hindrance and improve the access of larger substrates to the catalytic machinery. 21 , 28 , 29 , 47 , 52 For instance, the directed evolution of pseudomonas aeruginosa arylsulfatase (PAS) toward phosphonate hydrolysis resulted in the enlargement of its active site to better accommodate the bulkier new substrate. 29 Similarly, in the evolution of the metallo‐β‐lactamase NDM1 toward phosphonate hydrolysis, mutating a tryptophan to glycine removed a steric clash and enhanced enzyme–substrate complementarity. 28
Active site reshaping is the most frequently observed mechanism in the literature (24 out of the 32 examples herein, Table 1) and the consequences of active site reshaping can be substantial; several studies have observed more than 1,000‐fold increases in activity from this molecular strategy. The prevalence of this mechanism suggests, as previously discussed, that enzymes often possess preexisting residues that are able to form key interactions with promiscuous substrates, such that a modest active site reshaping may ensure rapid functional adaptation.
3.3. Conformational tinkering of active site residues
As previously discussed, enzyme evolution involves a significant number of remote mutations that do not directly interact with the substrate. While the effect of distal mutations is often hard to decipher, a significant number appear to contribute to fine‐tuning the position and dynamic motion of active‐site residues; a phenomenon known as “conformational tinkering.” 7 , 42 For example, during the laboratory evolution of an N‐acyl‐homoserine lactonase toward paraoxon, two distal mutations contributed to the shift of an active site residue, Phe68, by 3 Å, which promoted interactions with the leaving group of the new substrate. 42 Moreover, a fascinating sequence of mutational events has been reported by several other studies. (a) An initial mutation introduces a key residue that can potentially interact with the substrate but fails to efficiently do so due to mispositioning or the presence of conformational heterogeneity, that is, the sampling of multiple discrete rotamers of a residue. (b) Distal mutations subsequently alter the conformation of this key residue to become more catalytically competent. For example, during the directed evolution of PTE toward arylesterase activity, the first mutation that directly interacts with the new substrate, H254R, exhibited conformational heterogeneity. The subsequent fixation of multiple distal mutations contributed to the gradual shift of Arg254 to occupy a bent conformation that provides π‐cation interactions with the leaving group of the substrate (Figure 4a). 45 , 46 In addition, the directed evolution of an acyltransferase (LovD) for higher simvastatin synthesis also saw the accumulation of distal mutations that progressively restricted the conformational sampling of Tyr188 within the catalytic triad, stabilizing a conformation that restored the catalytically active geometry (Figure 4b). 41 Similar sequential events were observed during the directed evolution of a designed retro‐aldolase 55 and the natural evolution of chalcone isomerase (CHI). 52 Finally, during the evolution of a designed Kemp Eliminase (KE07), 35 the sampling of three distinct active site configurations, each with different catalytic efficiency, was observed along the evolutionary trajectory. 36 Over time, distal mutations resulted in the stabilization of the most catalytically efficient configuration, exhibiting improved positioning and orientation of a key Trp50 with respect to the substrate.
FIGURE 4.
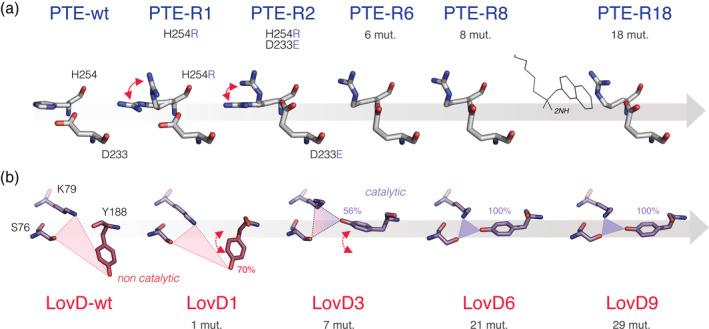
Conformational tinkering of active site residues by evolution. (a) Repositioning of the mutated active site residues, H254R and D233E (depicted as sticks), over the course of PTE evolution toward 2‐naphthyl hexanoate (2NH) hydrolysis (from left to right, PDB ID: 4pcp, 4xaf, 4xd5, 4xag, 4xay, and 4e3t). 46 (b) Representative snapshots of the reorganization of the K79‐S76‐Y188 catalytic triad during LovD evolution, observed by molecular dynamics simulations (MDs). Catalytic and noncatalytic regimes are depicted in purple and red, respectively. Tyr188 is gradually shifted by distal mutations from a noncatalytic to catalytic orientation. The percentages indicate the occupation of the representative side‐chain conformation in simulation time. Red (dotted) arrows indicate the sampling of multiple conformational rotamers. The total number of mutations acquired at each evolutionary stage is indicated beneath the names of the variants. Panel B was adapted from Ref. 41
The conformational tinkering of active site residues might be an important mechanism in enzyme evolution. While the introduction of a key residue in the active site may provide a stepping stone for further evolution, its configuration may be inadequate within its novel surrounding environment. Thus, the alteration of the residue's position and dynamics by other mutations, including distal ones, is essential to achieve efficient catalysis. The most extreme manifestation of this phenomenon is “conformational selection,” as described by Hong et al., which may be closely linked to the evolvability of promiscuous functions. 36 , 88 , 89 , 90 , 91
3.4. Repositioning or switching of metal cofactors
Analogous to the conformational tinkering of a key residue, the repositioning of active site cofactors, such as metal ions, has been described as a recurring evolutionary mechanism. 92 The laboratory evolution of serum paraoxonase PON1 toward phosphotriester hydrolysis identified an active site mutation, H115W, which altered the metal coordination of the catalytic Ca2+, following a 1.8 Å upward displacement of the cofactor. 33 , 34 , 93 Interestingly, molecular dynamics simulations (MDs) suggest that the Ca2+ position is plastic: the upward metal position appears to have preexisted in PON1 wild‐type; several mutations shifted the equilibrium toward the upward, more catalytically competent, metal coordination for the PTE activity. A similar phenomenon was also observed during PTE evolution toward arylesterase activity, where the distance between two active site Zn2+ ions decreased in the most evolved variant. 45 , 54 Intriguingly, the subsequent reverse evolution of PTE (back toward its native PTE activity) resulted in the precise restoration of the Zn2+ distances to the wild‐type configuration. 54 Finally, in the directed evolution of serine β‐lactamase BcII toward broader antibiotic specificity, a decrease in the distance between two‐active site Zn2+ appeared to be critical to stabilizing the new reaction intermediate and improving the rate‐limiting step of the reaction. 43 , 44
Furthermore, several studies have highlighted the importance of promiscuous metal binding in modulating metalloenzymes' specificity. 94 For instance, examination of the specificity of EcoRV restriction endonuclease revealed a single leucine to isoleucine mutation that was able to invert the enzyme's affinity from Mg2+ to Mn2+ and, in turn, alter specificity. 95 In natural or laboratory evolution, the switching of cofactor(s) is thus another avenue through which novel functions may emerge. 96
3.5. Alteration of enzyme conformational dynamics
Beyond local conformational tinkering, distal mutations can modulate the dynamics of larger structural elements, from loops to whole protein domains, which may be essential to complete catalytic cycles. 17 , 97 , 98 , 99 However, it may be difficult to capture the extent of, and the precision with which, conformational dynamics are fine‐tuned during enzyme evolution. A case in point was made by a study that attempted to enhance the enantioselectivity of a haloalkane dehalogenase, DhaA, for β‐bromoalkanes by transplanting the active site features (eight amino acid substitutions and an 11‐amino acid insertion) of a closely related enantioselective homolog, DbjA, into the DhaA scaffold. 27 While the active site geometry of the chimeric enzyme (DhaA12) was virtually identical to DbjA, the hybrid enzyme failed to become enantioselective due to inadequate amplitudes of motions and hydration levels (Figure 5a).
FIGURE 5.
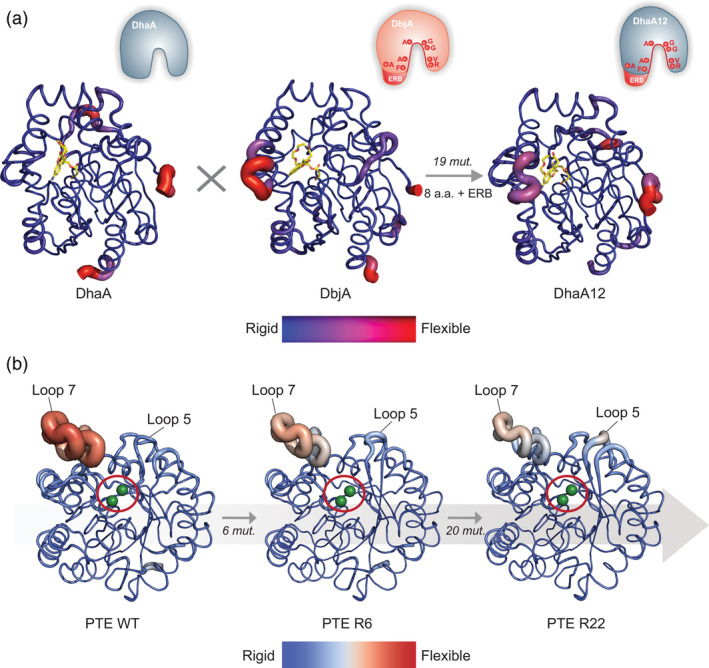
Alteration of enzyme conformational dynamics by evolution. Backbone B‐factors of (a) DhaA, DbjA, and DhaA12 haloalkane dehydrogenases, obtained by molecular dynamics simulations (MDs), 27 and (b) phosphotriesterase (PTE) WT, and variants R6, and R22 (PDB ID: 4pcp, 4xag, 4pcn) crystal structures, shown as cartoon putty representation. 46 The B‐factors are visualized by a color scale mapped onto the structure, and ribbon thickness. (a) The average structures of covalently bound fluorescent probes are shown as yellow sticks. A schematic of the active site transplantation events (mutations and ERB) is represented in the upper right corner of each enzyme. (b) The active site location of PTE is indicated by a red circle, and two Zn2+ ions shown as green spheres. The number of mutations fixed between each step of the trajectory is indicated on the gray arrow. Panel A is adapted with permission from Ref. 27
In enzymes for which catalytic cycles proceed via significant conformational transitions to accommodate distinct reaction intermediates, for example, via the sampling of open/closed states, an evolutionary reshaping of such transitions may be crucial to adapt to a new catalytic cycle. 30 , 31 , 32 , 100 , 101 Two aforementioned evolutionary studies illustrate this scenario: PTE and CDT. 46 , 47 , 48 In the former case, a series of mutations along the evolutionary trajectory anchored the active site Loop 7 in a productive (closed) conformation, while eliminating the sampling of the nonproductive (open) conformation (Figure 5b). 46 By contrast, other mutations increased the flexibility of another active site loop (Loop 5), thus resulting in markedly different motions when compared to the wild type enzyme. In the latter example, inactive or weakly active CDT ancestors were found to mainly adopt an open, noncatalytic conformation. 47 , 48 The conformational landscape of CDTs was gradually altered by evolution, such that the closed active conformation became more stabilized for greater improvements in catalytic efficiency.
While we have emphasized the importance of tailoring conformational dynamics, the extent to which such optimization becomes essential for enzyme evolution remains under debate. For instance, contrasting with previous examples, a study of two natural homologs of β‐lactamases and their chimeric variants demonstrated that the alteration of protein dynamics is not necessarily associated with changes in catalytic activity, nor required. 102 , 103 On the other hand, an extensive structural study of several variants of TEM‐1 β‐lactamase unveiled that two separate mutations caused opposite and incompatible dynamic changes to the structure of TEM‐1, despite both enhancing the hydrolysis of cefotaxime, a novel antibiotic. 38 Thus, we urgently need to advance our understanding of the role of enzyme dynamics in function and evolution in order to design better enzyme engineering strategies. 104 , 105
3.6. Substrate (re)positioning
In this final section, we highlight studies that provide an atomic‐depiction of the molecular changes directly impacting enzyme–substrate interactions during enzyme evolution. In a simple model, the optimization of enzyme function can be thought of as the process leading to a higher frequency of productive binding events between an enzyme and its substrate. 106 , 107 While it is challenging to capture molecular evidence of an increase in productive complexes per se, some studies have been able to observe a transition from predominantly nonproductive substrate binding modes to more productive ones. For instance, an early evolved mutant of a designed Kemp eliminases, HG2, was found to bind a TS analog (TSA), 6‐nitrobenzotriazole, in both productive and non‐productive or flipped, orientations (Figure 6a). 108 Further evolution fixed the mutation K50Q in mutant HG3 and introduced a new H‐bond donor with the TSA. This stabilized the productive complex and caused a >700‐fold increase in k cat/K M in subsequent mutants, HG3.17 and HG4. 51 , 105 The elimination of non‐productive binding modes was also described during the ancestor reconstruction of CHI 52 and the directed evolution of alcohol dehydrogenase A. 109 Counterintuitively, enhancing nonproductive binding may also be a molecular strategy, as disrupting the frequency of productive binding modes can alter catalysis: for example, the directed evolution of an α‐transglucosylase produced shorter sugar polymers as a result of enhanced nonproductive binding 110 .
FIGURE 6.
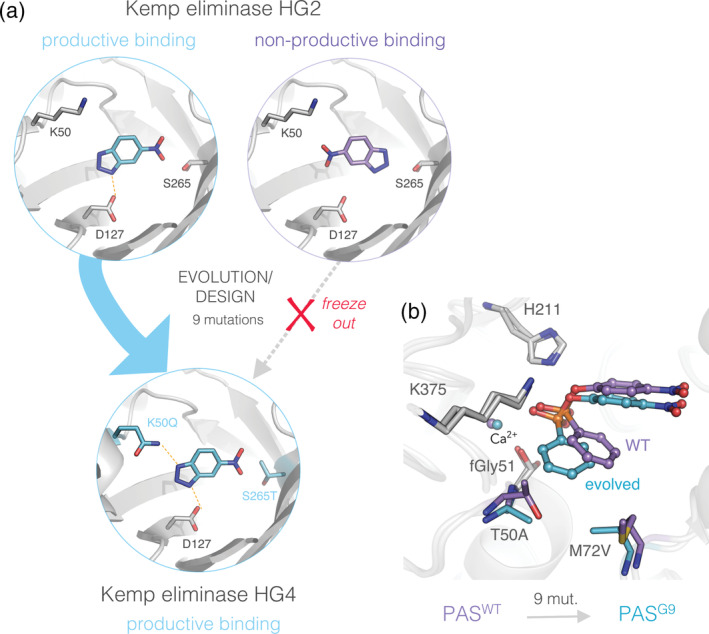
Substrate repositioning leads to novel enzyme‐substrate complexes in evolution. (a) The evolution of a designed kemp eliminase produced a weakly active mutant, HG2 (PDB ID: 3nyd), exhibiting two binding modes for the TS analog, 6‐nitrobenzotriazole. 108 (left) Asp127 binds the substrate in a productive orientation (teal sticks), as designed. (right) However, a nonproductive, flipped orientation (purple sticks), is also observed in the crystal structure. Further evolution stabilized the productive complex upon mutations K50Q and S265T, which eliminated the nonproductive orientation in subsequent mutants, for example, in the most evolved HG4 variant (PDB ID: 5rgf). 51 , 105 (b) Substrate repositioning induced by mutations T50A and M72V, from PASWT (PDB ID: 1hdh) to PASG9 (PDB ID: 4cxk) during arylsulfatase (PAS) evolution toward phosphonate monoester hydrolysis. 29 Evolution shifted the substrate closer to the catalytic machinery (Fgly nucleophile, K375 and H211) without altering the position of the catalytic residues, or creating new enzyme‐substrate interactions. Snapshots are representative stationary points from molecular dynamics simulations (MDs)
Beyond the dichotomy described above, it may be more realistic to consider the existence of a continuum of binding states (or E.S complexes), leading to diverse catalytic efficiencies that depend on the optimality of the enzyme–substrate interactions, for example, the orientation and proximity between the reactants. Indeed, promiscuous substrates may generally form less productive or inadequate interactions with noncognate enzymes 9 , 106 . Consequently, the optimization of an enzymatic activity could require the subtle tinkering of enzyme–substrate interactions toward more optimal, productive binding without major alterations of the catalytic machinery or overall structure. Such tailoring was demonstrated by the directed evolution of PAS toward phosphonate hydrolysis, where a modest enlargement of the active site cleft by two proximal mutations unlocked the access to the catalytic machinery for the bulkier substrate. 29 Interestingly, this active site reshaping was accompanied by the formation of a new Michaelis complex upon substrate repositioning (Figure 6b). As E.S interactions were more optimally oriented with respect to the catalytic center, in terms of distance and electrostatic complementation, evolution resulted in a large increase in TS stabilization. Similar observations were made during the evolution of a designed Kemp eliminase, KE59. 53
It remains difficult to establish which type of mechanism prevails in enzyme evolution, as only a handful of studies have extensively characterized the gradual changes of enzyme–substrate interactions during enzyme evolution, leaving this phenomenon largely undocumented. These limitations result from the challenge of gathering experimental evidence of subtle changes in enzyme–substrate interactions, in particular for promiscuous substrates that exhibit low catalytic efficiency in the initial stages of evolution. Yet, resolving atomic‐level views of such interactions is essential to advance our understanding of enzyme function and evolution. 111
4. PERSPECTIVES AND FUTURE DIRECTIONS
A growing number of studies have reported the evolution of enzymes performed in the laboratory or in nature. Here, we highlighted over 30 examples that provide detailed molecular insights into the mechanisms of enzyme evolution (Table 1). Needless to say, each of these cases present unique and distinct genetic and molecular solutions that result from the incredible diversity of enzyme attributes, for example, scaffold and active site architecture, type and level of native activity, and/or the acceptance of latent promiscuous substrates. Nonetheless, we observe several prominent mechanisms common amongst these examples of enzyme evolution. Importantly, we found that most of the examples herein do not rely on a single mechanism; rather, multiple strategies seem to be required to generate an efficient enzyme. In some cases, a defined sequence of events along a trajectory appears to be essential: initial mutations occur in the active site to generate a new interaction with the substrate, while later in the evolutionary process, distal mutations accumulate in the second‐shell to tinker the position of the initial key residue in order to fine‐tune its interaction with the novel substrate. 45 , 66 , 67 , 68 , 112 Such a sequence of mutational events is strongly associated with epistasis; the beneficial effects of later mutations only become realized after early mutations are fixed, which can severely restrict the accessibility of evolutionary trajectories. 68 , 113 To engineer and rationally design new and efficient enzymes, it is crucial to be able to identify the specific bottleneck that is being encountered, and hence, tailor our needs to overcome the issue.
Other molecular solutions that are not described in this review are likely to exist. Therefore, it is important to keep exploring enzyme systems, using both directed evolution and ASR, to expand our knowledge of the molecular mechanisms of enzyme evolution. In particular, the study of extensive functional transitions, such as transitions that bridge distinct catalytic mechanisms or the retracing of the evolution of catalysis from noncatalytic functions, should prove insightful. It is also intriguing to comprehensively explore multiple evolutionary transitions within an enzyme superfamily to study how diverse functions have emerged from the same scaffold. 114 , 115 , 116 To date, most studies have focused on single‐enzyme systems, however, understanding the dynamics of coevolution among multimeric enzyme systems remains challenging. Furthermore, continuous development of new technologies for directed evolution, such as new library generation and synthesis strategies, will allow us to more efficiently generate longer evolutionary trajectories under various conditions, and hence, to explore larger areas of the sequence space. These strategies include the effective incorporation of insertions and deletions, 117 high‐throughput screening methods 118 , 119 and continuous evolution systems 120 , 121 . Finally, a continuous expansion of available sequence information, including metagenomic DNA samples, is progressively filling in the critical gaps between functional transitions that are observed in nature, which until recently, were virtually impossible to retrace.
Importantly, more in‐depth analyses are urgently needed to improve our understanding of the underlying mechanisms of functional transitions. Traditional enzymology and structural techniques such as Brønsted plots, Arrhenius plots, stopped‐flow kinetics, pH profiles, kinetic isotope effects, crystallography, and nuclear magnetic resonance can all be employed to generate a chemically informed, atomic‐resolution view of how mutations directly affect protein function. On the other hand, cutting‐edge biochemical and biophysical techniques, such as recent advances in MDs, 97 , 122 femtosecond, time‐resolved and multitemperature crystallography 123 , 124 and single enzyme kinetics 125 , 126 should provide atomic‐resolution depictions of conformational and catalytic heterogeneity in enzymes, and inform on the extent of dynamic sampling. By observing catalysis in action, we may be able to gain incredible insights into the molecular basis of enzyme evolution and challenge the classical views of enzyme catalysis. 111 The combination of larger sequence space exploration, increased rounds of directed evolution, and detailed molecular characterizations of variants obtained along a trajectory, will provide a comprehensive view of the evolutionary bottlenecks that need to be overcome during the optimization of an enzyme function. To concur with Newton et al., “there has never been a better time to study enzyme evolution!” 127
AUTHOR CONTRIBUTIONS
Gloria Yang: Writing‐original draft; writing‐review and editing. Charlotte Miton: Writing‐original draft; writing‐review and editing. Nobuhiko Tokuriki: Funding acquisition; supervision; writing‐original draft; writing‐review and editing.
ACKNOWLEDGMENTS
We thank Moshe Goldsmith, Sílvia Osuna, Colin Jackson, Jiří Damborský, Rojo Rakotoharisoa, and Roberto Chica for providing material to build the figures of this review. We thank Janine N. Copp and John Z. Chen for critically reading the manuscript. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN 2017‐04909).
Yang G, Miton CM, Tokuriki N. A mechanistic view of enzyme evolution. Protein Science. 2020;29:1724–1747. 10.1002/pro.3901
Gloria Yang and Charlotte Miton contributed equally to this study.
Funding information Natural Sciences and Engineering Research Council of Canada
REFERENCES
- 1. Renata H, Wang ZJ, Arnold FH. Expanding the enzyme universe: Accessing non‐natural reactions by mechanism‐guided directed evolution. Angew Chem Int Ed Engl. 2015;54:3351–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Copley SD. Enzymes with extra talents: Moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol. 2003;7:265–272. [DOI] [PubMed] [Google Scholar]
- 3. Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. [DOI] [PubMed] [Google Scholar]
- 4. Mohamed MF, Hollfelder F. Efficient, crosswise catalytic promiscuity among enzymes that catalyze phosphoryl transfer. Biochim Biophys Acta. 2013;1834:417–424. [DOI] [PubMed] [Google Scholar]
- 5. O'Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6:R91–R105. [DOI] [PubMed] [Google Scholar]
- 6. Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. [DOI] [PubMed] [Google Scholar]
- 7. Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. [DOI] [PubMed] [Google Scholar]
- 8. Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27:157–167. [DOI] [PubMed] [Google Scholar]
- 9. Babtie A, Tokuriki N, Hollfelder F. What makes an enzyme promiscuous? Curr Opin Chem Biol. 2010;14:200–207. [DOI] [PubMed] [Google Scholar]
- 10. Copley, SD (2020). The physical basis and practical consequences of biological promiscuity. Physical Biology, 10.1088/1478-3975/ab8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pandya C, Farelli JD, Dunaway‐Mariano D, Allen KN. Enzyme promiscuity: Engine of evolutionary innovation. J Biol Chem. 2014;289:30229–30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Copley SD. An evolutionary biochemist's perspective on promiscuity. Trends Biochem Sci. 2015;40:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fersht AR. Catalysis, binding and enzyme‐substrate complementarity. Proc R Soc Lond B Biol Sci. 1974;187:397–407. [DOI] [PubMed] [Google Scholar]
- 14. Kirby AJ, Hollfelder F. From enzyme models to model enzymes. Cambridge: Royal Society of Chemistry, 2009. [Google Scholar]
- 15. Gatti‐Lafranconi P, Hollfelder F. Flexibility and reactivity in promiscuous enzymes. Chembiochem. 2013;14:285–292. [DOI] [PubMed] [Google Scholar]
- 16. Benkovic SJ, Hammes‐Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. [DOI] [PubMed] [Google Scholar]
- 17. Bhabha G, Biel JT, Fraser JS. Keep on moving: Discovering and perturbing the conformational dynamics of enzymes. Acc Chem Res. 2015;48:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noor S, Taylor MC, Russell RJ, et al. Intramolecular epistasis and the evolution of a new enzymatic function. PLoS One. 2012;7:e39822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Villiers B, Hollfelder F. Directed evolution of a gatekeeper domain in nonribosomal peptide synthesis. Chem Biol. 2011;18:1290–1299. [DOI] [PubMed] [Google Scholar]
- 20. Niquille DL, Hansen DA, Mori T, Fercher D, Kries H, Hilvert D. Nonribosomal biosynthesis of backbone‐modified peptides. Nat Chem. 2018;10:282–287. [DOI] [PubMed] [Google Scholar]
- 21. Gould SM, Tawfik DS. Directed evolution of the promiscuous esterase activity of carbonic anhydrase II. Biochemistry. 2005;44:5444–5452. [DOI] [PubMed] [Google Scholar]
- 22. Fasan R, Chen MM, Crook NC, Arnold FH. Engineered alkane‐hydroxylating cytochrome P450(BM3) exhibiting nativelike catalytic properties. Angew Chem Int Ed Engl. 2007;46:8414–8418. [DOI] [PubMed] [Google Scholar]
- 23. Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH. Evolutionary history of a specialized p450 propane monooxygenase. J Mol Biol. 2008;383:1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang G, Anderson DW, Baier F, et al. Higher‐order epistasis shapes the fitness landscape of a xenobiotic‐degrading enzyme. Nat Chem Biol. 2019;15:1120–1128. [DOI] [PubMed] [Google Scholar]
- 25. Voordeckers K, Brown CA, Vanneste K, et al. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012;10:e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boucher JI, Jacobowitz JR, Beckett BC, Classen S, Theobald DL. An atomic‐resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases. Elife. 2014;3:e02304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sykora J, Brezovsky J, Koudelakova T, et al. Dynamics and hydration explain failed functional transformation in dehalogenase design. Nat Chem Biol. 2014;10:428–430. [DOI] [PubMed] [Google Scholar]
- 28. Baier F, Hong N, Yang G, et al. Cryptic genetic variation shapes the adaptive evolutionary potential of enzymes. Elife. 2019;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miton CM, Jonas S, Fischer G, et al. Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset. Proc Natl Acad Sci USA. 2018;115:E7293–E7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maria‐Solano MA, Iglesias‐Fernández J, Osuna S. Deciphering the allosterically driven conformational ensemble in tryptophan synthase evolution. J Am Chem Soc. 2019;141:13049–13056. [DOI] [PubMed] [Google Scholar]
- 31. Buller AR, Brinkmann‐Chen S, Romney DK, Herger M, Murciano‐Calles J, Arnold FH. Directed evolution of the tryptophan synthase β‐subunit for stand‐alone function recapitulates allosteric activation. Proc Natl Acad Sci USA. 2015;112:14599–14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otten R, Liu L, Kenner LR, et al. Rescue of conformational dynamics in enzyme catalysis by directed evolution. Nat Commun. 2018;9:1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ben‐David M, Elias M, Filippi J‐J, et al. Catalytic versatility and backups in enzyme active sites: The case of serum paraoxonase 1. J Mol Biol. 2012;418:181–196. [DOI] [PubMed] [Google Scholar]
- 34. Aharoni A, Gaidukov L, Yagur S, Toker L, Silman I, Tawfik DS. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc Natl Acad Sci U S A. 2004;101:482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khersonsky O, Röthlisberger D, Dym O, et al. Evolutionary optimization of computationally designed enzymes: Kemp eliminases of the KE07 series. J Mol Biol. 2010;396:1025–1042. [DOI] [PubMed] [Google Scholar]
- 36. Hong N‐S, Petrović D, Lee R, et al. The evolution of multiple active site configurations in a designed enzyme. Nat Commun. 2018;9:3900–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khersonsky O, Röthlisberger D, Wollacott AM, et al. Optimization of the in‐silico‐designed kemp eliminase KE70 by computational design and directed evolution. J Mol Biol. 2011;407:391–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dellus‐Gur E, Elias M, Caselli E, et al. Negative epistasis and evolvability in TEM‐1 β‐lactamase – The thin line between an enzyme's conformational freedom and disorder. J Mol Biol. 2015;427:2396–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Preiswerk N, Beck T, Schulz JD, et al. Impact of scaffold rigidity on the design and evolution of an artificial diels–alderase. Proc Natl Acad Sci USA. 2014;111:8013–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siegel JB, Zanghellini A, Lovick HM, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular diels–alder reaction. Science. 2010;329:309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiménez‐Osés G, Osuna S, Gao X, et al. The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat Chem Biol. 2014;10:431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang G, Hong N, Baier F, Jackson CJ, Tokuriki N. Conformational tinkering drives evolution of a promiscuous activity through indirect mutational effects. Biochemistry. 2016;55:4583–4593. [DOI] [PubMed] [Google Scholar]
- 43. Tomatis PE, Fabiane SM, Simona F, Carloni P, Sutton BJ, Vila AJ. Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc Natl Acad Sci USA. 2008;105:20605–20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. González MM, Abriata LA, Tomatis PE, Vila AJ. Optimization of conformational dynamics in an epistatic evolutionary trajectory. Mol Biol Evol. 2016;33:1768–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tokuriki N, Jackson CJ, Afriat‐Jurnou L, Wyganowski KT, Tang R, Tawfik DS. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun. 2012;3:1257. [DOI] [PubMed] [Google Scholar]
- 46. Campbell E, Kaltenbach M, Correy GJ, et al. The role of protein dynamics in the evolution of new enzyme function. Nat Chem Biol. 2016;12:944–950. [DOI] [PubMed] [Google Scholar]
- 47. Clifton BE, Kaczmarski JA, Carr PD, Gerth ML, Tokuriki N, Jackson CJ. Evolution of cyclohexadienyl dehydratase from an ancestral solute‐binding protein. Nat Chem Biol. 2018;14:542–547. [DOI] [PubMed] [Google Scholar]
- 48. Kaczmarski JA, Mahawaththa MC, Feintuch A, et al. Altered conformational sampling along an evolutionary trajectory changes the catalytic activity of an enzyme. bioRxiv. 2020;57:320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Newton MS, Guo X, Söderholm A, et al. Structural and functional innovations in the real‐time evolution of new (βα)8 barrel enzymes. Proc Natl Acad Sci USA. 2017;114:4727–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Näsvall J, Sun L, Roth JR, Andersson DI. Real‐time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338:384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blomberg R, Kries H, Pinkas DM, et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature. 2013;503:418–421. [DOI] [PubMed] [Google Scholar]
- 52. Kaltenbach M, Burke JR, Dindo M, et al. Evolution of chalcone isomerase from a noncatalytic ancestor. Nat Chem Biol. 2018;14:548–555. [DOI] [PubMed] [Google Scholar]
- 53. Khersonsky O, Kiss G, Röthlisberger D, et al. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc Natl Acad Sci USA. 2012;109:10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaltenbach M, Jackson CJ, Campbell EC, Hollfelder F, Tokuriki N. Reverse evolution leads to genotypic incompatibility despite functional and active site convergence. Elife. 2015;4:e06492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Giger L, Caner S, Obexer R, et al. Evolution of a designed retro‐aldolase leads to complete active site remodeling. Nat Chem Biol. 2013;9:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Althoff EA, Wang L, Jiang L, et al. Robust design and optimization of retroaldol enzymes. Protein Sci. 2012;21:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Obexer R, Godina A, Garrabou X, et al. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat Chem. 2017;9:50–56. [DOI] [PubMed] [Google Scholar]
- 58. Zeymer C, Zschoche R, Hilvert D. Optimization of enzyme mechanism along the evolutionary trajectory of a computationally designed (retro‐)aldolase. J Am Chem Soc. 2017;139:12541–12549. [DOI] [PubMed] [Google Scholar]
- 59. Fowler DM, Fields S. Deep mutational scanning: A new style of protein science. Nat Methods. 2014;11:801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Araya CL, Fowler DM. Deep mutational scanning: Assessing protein function on a massive scale. Trends Biotechnol. 2011;29:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM‐1 β‐lactamase. Cell. 2015;160:882–892. [DOI] [PubMed] [Google Scholar]
- 62. Wrenbeck EE, Azouz LR, Whitehead TA. Single‐mutation fitness landscapes for an enzyme on multiple substrates reveal specificity is globally encoded. Nat Commun. 2017;8:15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. Comprehensive mutational scanning of a kinase in vivo reveals substrate‐dependent fitness landscapes. Nucleic Acids Res. 2014;42:e112–e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van der Meer J‐Y, Poddar H, Baas B‐J, et al. Using mutability landscapes of a promiscuous tautomerase to guide the engineering of enantioselective Michaelases. Nat Commun. 2016;7:10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen JZ, Fowler DM, Tokuriki N. Comprehensive exploration of the translocation, stability and substrate recognition requirements in VIM‐2 lactamase. Elife. 2020;9:e56707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wilding M, Hong N, Spence M, Buckle AM, Jackson CJ. Protein engineering: The potential of remote mutations. Biochem Soc Trans. 2019;47:701–711. [DOI] [PubMed] [Google Scholar]
- 67. Morley KL, Kazlauskas RJ. Improving enzyme properties: When are closer mutations better? Trends Biotechnol. 2005;23:231–237. [DOI] [PubMed] [Google Scholar]
- 68. Miton CM, Tokuriki N. How mutational epistasis impairs predictability in protein evolution and design. Protein Sci. 2016;25:1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vakulenko SB, Taibi‐Tronche P, Tóth M, Massova I, Lerner SA, Mobashery S. Effects on substrate profile by mutational substitutions at positions 164 and 179 of the class A TEM(pUC19) beta‐lactamase from Escherichia coli . J Biol Chem. 1999;274:23052–23060. [DOI] [PubMed] [Google Scholar]
- 70. Whittle E, Shanklin J. Engineering delta 9‐16:0‐acyl carrier protein (ACP) desaturase specificity based on combinatorial saturation mutagenesis and logical redesign of the castor delta 9‐18:0‐ACP desaturase. J Biol Chem. 2001;276:21500–21505. [DOI] [PubMed] [Google Scholar]
- 71. Goldsmith M, Tawfik DS. Enzyme engineering: Reaching the maximal catalytic efficiency peak. Curr Opin Struct Biol. 2017;47:140–150. [DOI] [PubMed] [Google Scholar]
- 72. Hartl DL, Dykhuizen DE, Dean AM. Limits of adaptation: The evolution of selective neutrality. Genetics. 1985;111:655–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stebbins J. The law of diminishing returns. Science. 1944;99:267–271. [DOI] [PubMed] [Google Scholar]
- 74. Goldsmith M, Aggarwal N, Ashani Y, et al. Overcoming an optimization plateau in the directed evolution of highly efficient nerve agent bioscavengers. Protein Eng Des Sel. 2017;30:333–345. [DOI] [PubMed] [Google Scholar]
- 75. Domingo J, Baeza‐Centurion P, Lehner B. The causes and consequences of genetic interactions (epistasis). Annu Rev Genomics Hum Genet. 2019;20:433–460. [DOI] [PubMed] [Google Scholar]
- 76. Starr TN, Thornton JW. Epistasis in protein evolution. Protein Sci. 2016;25:1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Melamed D, Young DL, Gamble CE, Miller CR, Fields S. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)‐binding protein. RNA. 2013;19:1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Olson CA, Wu NC, Sun R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr Biol. 2014;24:2643–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meini M‐R, Tomatis PE, Weinreich DM, Vila AJ. Quantitative description of a protein fitness landscape based on molecular features. Mol Biol Evol. 2015;32:1774–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lozovsky ER, Chookajorn T, Brown KM, et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci USA. 2009;106:12025–12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Moriuchi R, Tanaka H, Nikawadori Y, et al. Stepwise enhancement of catalytic performance of haloalkane dehalogenase LinB towards β‐hexachlorocyclohexane. AMB Express. 2014;4:10–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Storz JF. Compensatory mutations and epistasis for protein function. Curr Opin Struct Biol. 2018;50:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shah P, McCandlish DM, Plotkin JB. Contingency and entrenchment in protein evolution under purifying selection. Proc Natl Acad Sci USA. 2015;112:E3226–E3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lobkovsky AE, Koonin EV. Replaying the tape of life: Quantification of the predictability of evolution. Front Genet. 2012;3:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lunzer M, Golding GB, Dean AM. Pervasive cryptic epistasis in molecular evolution. PLoS Genet. 2010;6:e1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sunden F, AlSadhan I, Lyubimov A, Doukov T, Swan J, Herschlag D. Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution. J Biol Chem. 2017;292:20960–20974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bayer CD, van Loo B, Hollfelder F. Specificity effects of amino acid substitutions in promiscuous hydrolases: Context‐dependence of catalytic residue contributions to local fitness landscapes in nearby sequence space. Chembiochem. 2017;18:1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Aharoni A, Gaidukov L, Khersonsky O, McQ Gould S, Roodveldt C, Tawfik DS. The “evolvability” of promiscuous protein functions. Nat Genet. 2005;37:73–76. [DOI] [PubMed] [Google Scholar]
- 89. Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. [DOI] [PubMed] [Google Scholar]
- 90. Maria‐Solano MA, Serrano‐Hervás E, Romero‐Rivera A, Iglesias‐Fernández J, Osuna S. Role of conformational dynamics in the evolution of novel enzyme function. Chem Commun (Camb). 2018;54:6622–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ma B, Nussinov R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr Opin Chem Biol. 2010;14:652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sugrue E, Hartley CJ, Scott C, Jackson CJ. The evolution of new catalytic mechanisms for xenobiotic hydrolysis in bacterial metalloenzymes. Aust J Chem. 2016;69:1383–1395. [Google Scholar]
- 93. Ben‐David M, Soskine M, Dubovetskyi A, et al. Enzyme evolution: An epistatic ratchet versus a smooth reversible transition. Mol Biol Evol. 2020;37:1133–1147. [DOI] [PubMed] [Google Scholar]
- 94. Baier F, Chen J, Solomonson M, Strynadka NCJ, Tokuriki N. Distinct metal isoforms underlie promiscuous activity profiles of metalloenzymes. ACS Chem Biol. 2015;10:1684–1693. [DOI] [PubMed] [Google Scholar]
- 95. Vipond IB, Moon BJ, Halford SE. An isoleucine to leucine mutation that switches the cofactor requirement of the EcoRV restriction endonuclease from magnesium to manganese. Biochemistry. 1996;35:1712–1721. [DOI] [PubMed] [Google Scholar]
- 96. Anderson DW, Baier F, Yang G, Tokuriki N. Secondary environmental variation creates a shifting evolutionary watershed for the methyl‐parathion hydrolase enzyme. bioRxiv. 2019;65:1528–1544. [Google Scholar]
- 97. Petrović D, Risso VA, Kamerlin SCL, Sanchez‐Ruiz JM. Conformational dynamics and enzyme evolution. J R Soc Interface. 2018;15:20180330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Henzler‐Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. [DOI] [PubMed] [Google Scholar]
- 99. Fraser JS, Jackson CJ. Mining electron density for functionally relevant protein polysterism in crystal structures. Cell Mol Life Sci. 2011;68:1829–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Murciano‐Calles J, Romney DK, Brinkmann‐Chen S, Buller AR, Arnold FH. A panel of TrpB biocatalysts derived from tryptophan synthase through the transfer of mutations that mimic allosteric activation. Angew Chem Int Ed Engl. 2016;55:11577–11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gobeil SMC, Clouthier CM, Park J, et al. Maintenance of native‐like protein dynamics may not be required for engineering functional proteins. Chem Biol. 2014;21:1330–1340. [DOI] [PubMed] [Google Scholar]
- 103. Gobeil SMC, Ebert MCCJC, Park J, et al. The structural dynamics of engineered β‐lactamases vary broadly on three timescales yet sustain native function. Sci Rep. 2019;9:6612–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Davey JA, Damry AM, Goto NK, Chica RA. Rational design of proteins that exchange on functional timescales. Nat Chem Biol. 2017;13:1280–1285. [DOI] [PubMed] [Google Scholar]
- 105. Broom A, Rakotoharisoa RV, Thompson MC, et al. Evolution of an enzyme conformational ensemble guides design of an efficient biocatalyst. bioRxiv. 2020;8:729–749. [Google Scholar]
- 106. Bar‐Even A, Milo R, Noor E, Tawfik DS. The moderately efficient enzyme: Futile encounters and enzyme floppiness. Biochemistry. 2015;54:4969–4977. [DOI] [PubMed] [Google Scholar]
- 107. Gamage NU, Tsvetanov S, Duggleby RG, McManus ME, Martin JL. The structure of human SULT1A1 crystallized with estradiol. An insight into active site plasticity and substrate inhibition with multi‐ring substrates. J Biol Chem. 2005;280:41482–41486. [DOI] [PubMed] [Google Scholar]
- 108. Privett HK, Kiss G, Lee TM, et al. Iterative approach to computational enzyme design. Proc Natl Acad Sci USA. 2012;109:3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hamnevik E, Enugala TR, Maurer D, et al. Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer. FEBS J. 2017;284:3895–3914. [DOI] [PubMed] [Google Scholar]
- 110. Claverie M, Cioci G, Guionnet M, et al. Futile encounter engineering of the DSR‐M dextransucrase modifies the resulting polymer length. Biochemistry. 2019;58:2853–2859. [DOI] [PubMed] [Google Scholar]
- 111. Yabukarski F, Biel JT, Pinney MM, et al. Assessing positioning in enzymatic catalysis via ketosteroid isomerase conformational ensembles. bioRxiv. 2019;56:446–449. [Google Scholar]
- 112. Socha RD, Tokuriki N. Modulating protein stability – directed evolution strategies for improved protein function. FEBS J. 2013;280:5582–5595. [DOI] [PubMed] [Google Scholar]
- 113. Starr TN, Picton LK, Thornton JW. Alternative evolutionary histories in the sequence space of an ancient protein. Nature. 2017;549:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Akiva E, Copp JN, Tokuriki N, Babbitt PC. Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily. Proc Natl Acad Sci USA. 2017;114:E9549–E9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Baier F, Copp JN, Tokuriki N. Evolution of enzyme superfamilies: Comprehensive exploration of sequence‐function relationships. Biochemistry. 2016;55:6375–6388. [DOI] [PubMed] [Google Scholar]
- 116. Toth‐Petroczy A, Tawfik DS. The robustness and innovability of protein folds. Curr Opin Struct Biol. 2014;26:131–138. [DOI] [PubMed] [Google Scholar]
- 117. Emond S, Petek M, Kay E, et al. Access to unexplored regions of sequence space in directed enzyme evolution via insertion/deletion mutagenesis. bioRxiv. 2019;1179:136–151. [Google Scholar]
- 118. Markel U, Essani KD, Besirlioglu V, Schiffels J, Streit WR, Schwaneberg U. Advances in ultrahigh‐throughput screening for directed enzyme evolution. Chem Soc Rev. 2020;49:233–262. [DOI] [PubMed] [Google Scholar]
- 119. Zeymer C, Hilvert D. Directed evolution of protein catalysts. Annu Rev Biochem. 2018;87:131–157. [DOI] [PubMed] [Google Scholar]
- 120. Ravikumar A, Arzumanyan GA, Obadi MKA, Javanpour AA, Liu CC. Scalable, continuous evolution of genes at mutation rates above genomic error thresholds. Cell. 2018;175:1946–1957. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Badran AH, Liu DR. In vivo continuous directed evolution. Curr Opin Chem Biol. 2015;24:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Romero‐Rivera A, Garcia‐Borràs M, Osuna S. Computational tools for the evaluation of laboratory‐engineered biocatalysts. Chem Commun (Camb). 2016;53:284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. van den Bedem H, Fraser JS. Integrative, dynamic structural biology at atomic resolution – It's about time. Nat Methods. 2015;12:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Thompson MC, Barad BA, Wolff AM, et al. Temperature‐jump solution X‐ray scattering reveals distinct motions in a dynamic enzyme. Nat Chem. 2019;11:1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sakakihara S, Araki S, Iino R, Noji H. A single‐molecule enzymatic assay in a directly accessible femtoliter droplet array. Lab Chip. 2010;10:3355–3362. [DOI] [PubMed] [Google Scholar]
- 126. Sanabria H, Rodnin D, Hemmen K, et al. Resolving dynamics and function of transient states in single enzyme molecules. Nat Commun. 2020;11:1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Newton MS, Arcus VL, Gerth ML, Patrick WM. Enzyme evolution: Innovation is easy, optimization is complicated. Curr Opin Struct Biol. 2018;48:110–116. [DOI] [PubMed] [Google Scholar]


