Abstract
The abnormal m6A modification caused by m6A modulators is a common feature of various tumors; however, little is known about which m6A modulator plays the most important role in triple-negative breast cancer (TNBC). In this study, when analyzing the influence of m6A modulators (METTL3, METTL14, WTAP, FTO, and ALKBH5) on the prognosis of breast cancer, especially in TNBC using several on-line databases, methyltransferase-like 3 (METTL3) was found to have low expression in breast cancer, and was closely associated with short-distance-metastasis-free survival in TNBC. Further investigation showed that knockdown of METTL3 could enhance the ability of migration, invasion, and adhesion by decreasing m6A level in TNBC cell lines. Collagen type III alpha 1 chain (COL3A1) was identified and verified as a target gene of METTL3. METTL3 could down-regulate the expression of COL3A1 by increasing its m6A methylation, ultimately inhibiting the metastasis of TNBC cells. Finally, with immunohistochemistry staining in breast cancer tissues, it was proved that METTL3 expression was negatively correlated with COL3A1 in TNBC, but not in non-TNBC. This study demonstrated the potential mechanism of m6A modification in metastasis and provided potential targets for treatment in TNBC.
Keywords: METTL3, m6A, triple-negative breast cancer, metastasis, COL3A1
Introduction
Breast cancer, the most common cancer in women, poses a serious threat to the health of women (1). Despite the improvement of treatment strategies, the prognosis of breast cancer, especially triple-negative breast cancer (TNBC), remains poor due to metastasis. In recent years, epigenetic regulation, such as DNA methylation, histone acetylation, and non-coding RNAs, has been reported to play a critical role in the development of breast cancer including TNBC. Especially, as a new emerging epigenetic modification, RNA methylation has attracted much attention due to its non-negligible function in cancer development; however, current studies of RNA methylation-related cancer development are just the tip of the iceberg in this cognate area. It is necessary to clarify the mechanisms underlying RNA methylation-involved metastasis in TNBC.
As the most prevalent RNA methylation modification, N6-methyladenosine (m6A) infers that the nitrogen-6 position of adenosine base in RNA is methylated by the regulation of m6A methyltransferases and m6A demethylases. Methyltransferase, including methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms tumor 1-associated protein (WTAP) can form into complexes and mediate the cellular deposition of m6A on mammalian mRNAs, whereas demethylases including FTO and its homolog AlkB family member 5 (ALKBH5) can selectively reverse m6A to adenosine (2–5). RNA m6A methylation is known to be involved in various biological processes, such as stem cell differentiation and pluripotency, circadian periods, embryogenesis, and DNA damage response (2, 3). Cumulative studies have proved that the change of RNA m6A modification caused by the aberrant expression of m6A modulators can also influence the development of cancers. The methyltransferase METTL14 can inhibit tumor metastasis in HCC by positively regulating the m6A level of DGCR8 and promoting the binding of DGCR8 to pri-miRNAs (6); similarly, the demethylase FTO can promote cell proliferation via down-regulating the m6A level of USP7 in advanced non-small cell lung cancer, indicating the repression role of m6A in cancer development (7), however, other studies obtained contradictory results wherein METTL3 could promote the proliferation of prostate cancer cell via enhancing the m6A level of GLI15 (8); similarly, ALKBH5 was found to be able to inhibit pancreatic cancer metastasis by down-regulating KCNK15-AS1, suggesting that m6A modification of RNA plays an oncogenic role in cancer (9). Therefore, it seems that an m6A modulator might play both promotional, and inhibitory roles in different types of cancers by regulating different specific target genes. To date, the role of RNA m6A methylation in the development of breast cancer remains unclear. The only studies of breast cancer have shown that METTL3-mediated enhancement of m6A level could promote the proliferation of breast cancer cells (10), while the high level of m6A caused by FTO knockdown could inhibit the proliferation and metastasis of breast cancer (11). Therefore, it seems that, although the changes in m6A level are consistent, the effects of different modulators on breast cancer differ because the specific target genes are different. The underlying role and epigenetic regulation of m6A modulators in breast cancer, especially in TNBC, still needs to be investigated.
In this study, we analyzed the prognostic role of m6A modulators in TNBC using several online databases and found that the low expression of METTL3 was associated with the poor prognosis of TNBC. Further molecular mechanism investigation indicated that silence of METTL3 could up-regulate the expression of Collagen type III alpha 1 chain (COL3A1) by increasing m6A-levels, ultimately promoting metastasis of TNBC cells. This study revealed the important role of m6A modification mediated by METTL3 in TNBC and suggests that METTL3 might act as a novel therapeutic target in TNBC metastasis.
Materials and Methods
Data Sources: On-Line Databases
KM plotter (http://kmplot.com/analysis/) is a website used for on-line analysis, which is capable of assessing 54k genes on the survival of 21 cancer types, including breast cancer. The association between the distance-metastasis-free survival (DMFS) and the expression of m6A modulators (METTL3, METTL14, WTAP, FTO, and ALKBH5), COL3A1 was analyzed using KM plotter, respectively. GEPIA (http://gepia.cancer-pku.cn) is an on-line database including the RNA sequencing expression data of 9,736 tumors and 8,587 normal tissue samples from the Cancer Genome Atlas (TCGA) and the Genotype Tissue Expression (GTEx). The transcriptional levels of five m6A modulators above in breast cancer tissues and normal breast tissues were obtained from GEPIA. The mRNA expression data of 91 patients with TNBC were downloaded from TCGA (https://www.cancer.gov/) for analysis of the correlation between the m6A modulators and target genes.
Cell Culture
The human breast cancer cell lines, MDA-MB-231, and MDA-MB-468, were acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All the cells were incubated in L15 culture medium (Gibco, NY, USA) supplemented with 10% FBS at 37°C under 5% CO2 and saturated humidity.
Antibodies
The primary antibodies for western blot, anti-METTL3 (#96391) were sourced from Cell Signaling Technology (MA, USA), anti-COL3A1 (sc-514601) was sourced from Santa Cruz (CA, USA), anti-α-tubulin (ab7291) was sourced from Abcam (CA, USA). HRP-conjugated goat anti-mouse/rabbit secondary antibodies (ZDR-5306/5307) were sourced from ZSBIO (Beijing, China). The antibodies for immunohistochemistry, anti-METTL3 (ab195352) were sourced from Abcam (CA, USA), and anti-COL3A1 (sc-166316) was sourced from Santa Cruz (CA, USA).
Trans-well Migration and Invasion Assays
For the migration assay, ~2 × 104 cells were suspended in 200 μl serum-free L15 medium and added into the upper chamber of a trans-well plate (Corning, USA) with an 8-μm pore size polycarbonate filter, and 500 μl L15 medium with 10% FBS were dispensed into the lower chambers, and incubated for 24 h. Then the upper chambers were fixed in 75% ethanol, and the cells on the upper surface of the filter were removed manually with a cotton swab. Then the migrating cells were stained with Wright–Giemsa stain.
The invasion assay was similar to the migration assay except that 3% matrigel was dispensed into the upper chamber before seeding 3 × 104 cells into the culture system. Migrating and invading cells were observed under an optical microscope. The cells from three fields were counted with Image J (https://imagej.nih.gov/ij/download.html).
Cell Adhesion Assay
The cells were seeded at 3 × 104 per well into 96-well-plates pre-coated with 10% matrigel overnight. After 30 min-incubation at 37 °C, the non-adherent cells were removed by PBS washing. Then the cells were fixed with 75% ethanol for 10 min and stained with Wright–Giemsa stain. Images were acquired by microscope and the quantities of cells were counted with Image J.
RNA Isolation and Quantitative Real-Time PCR Assay
RNA was isolated with TRIzol reagent (Invitrogen, CA, USA) and identified using a NanoDrop spectrophotometer (Thermo Scientific, Rockford, IL, USA). The Reverse Transcription Kit (Promega, WI, USA) was used for mRNA reverse transcription. Quantitative real-time PCR (qRT-PCR) assay was then performed using the SYBR Green kit (Promega, WI, USA) on the ABI7500 instrument (Thermofisher, IL, USA). All the reactions were conducted for triplicates. 18S RNA was used as the internal control. Primers of METTL3, COL3A1, MYH11, and 18S were used as follows:
METTL3 forward: 5′-TTGTCTCCAACCTTCCGTAGT-3′
METTL3 reverse: 5′-CCAGATCAGAGAGGTGGTGTAG-3′
COL3A1 forward: 5′-CCCACTATTATTTTGGCACAACAG-3′
COL3A1 reverse: 5′-AACGGATCCTGAGTCACAGACA-3′
MYH11 forward: 5′-AGTTCGAAAGGGATCTCCA-3′
MYH11 reverse: 5′-CATACTCGTGAAGCTGTCTC-3′
18S forward: 5′CCCGGGGAGGTAGTGACGAAAAAT-3′
18S reverse: 5′-CGCCCGCCCGCTCCCAAGAT-3′.
Methylated RNA Immunoprecipitation-qRT-PCR
The methylated RNA immunoprecipitation-qRT-PCR (MeRIP-qRT-PCR) assay was conducted according to the standard protocol of Magna MeRIP m6A Kit (Millipore, MA, USA, 17-10499) with a slight modification. The total RNA (300 μg) was isolated with TRIzol reagent and fragmented. Except for 3 μg of the total RNA as input, the remaining RNA was used for m6A-immunoprecipitation with m6A antibody. The Protein A/G Magnetic Beads were prepared by 30 min-incubation with m6A-specific antibody in immunoprecipitation buffer at room temperature, then incubated with the MeRIP reaction mixture and RNA for 2h at 4°C. Finally, the MeRIPed-RNA was cleaned up and concentrated with RNeasy MinElute Clean-up Kit (Qiagen, Hilden, Germany). The enriched RNA level of COL3A1 was analyzed by qRT-PCR. The primers were as same as the primers used in real-time PCR assay.
Transfection
For the knockdown of METTL3 and COL3A1, the cells were seeded at 105 cells/well in a 6-well-plate and the siRNAs targeted to METTL3 and COL3A1 (GENEWIZ, Beijing, China) at a final concentration of 50 nM were transfected using JetPRIME Transfection Reagent (Polyplus, Illkirch, France) according to the manufacturer's instructions. Negative control (NC) siRNA was used as a control. For the overexpression of METLL3, the cells were seeded at 105 per 6-well-plate and transfected with plasmids pcDNA3.1-FLAG and pcDNA3.1-METTL3 (Obio, Shanghai, China) at a final concentration of 1 μg/ml using JetPRIME Transfection Reagent. The siRNA sequences of NC, METTL3, and COL3A1 were used as follows:
si-METTL3-1126 (KD1): 5′-CCUGCAAGUAUGUUCACUATT-3′
si-METTL3-1400 (KD2): 5′-GCUCAACAUACCCGUACUATT-3′
si-METTL3-1604 (KD3): 5′-GGUUGGUGUCAAAGGAAAUTT-3′
si-COL3A1-1 (KD1): 5′-GGAUGCAAAUUGGAUGCUAtt-3′
si-COL3A1-2 (KD2): 5′-CCCUCCUAAUGGUCAAGGAtt-3′
si-NC: 5′-UUCUCCGAACGUGUCACGUtt-3′.
Western Blot
The harvested cells were lysed in a lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 10 mM EDTA, 50 mM NaF, 1 mM NaVanadate, 1% Triton X-100, 1 mM PMSF, and 0.5% aprotin) and the protein concentration was quantified according to the Coomassie blue G250 staining technique. Equivalent protein was electrophoresed on 8–10% SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Perkin-Elmer, Waltham, MA, USA). Then the membranes were blocked with 5% skimmed milk in TBST for 1 h and incubated with the primary antibodies overnight (more than 6 h). The concentration of anti-METTL3 (#96391) was 1:1,000, the concentration of anti-COL3A1 (sc-514601) was 1:200, the concentration of anti-α-tubulin (ab7291) was 1:1,000. After being immunoblotted with HRP-conjugated goat anti-mouse/rabbit antibody (1:2,000) for 40 min, the signal strength of revealed protein bands could be detected with enhanced chemiluminescence reagent (SuperSignal Western Pico Chemiluminescent Substrate; Pierce, Rockford, IL, USA) and visualized with the Electrophoresis Gel Imaging Analysis System (DNR Bio-Imaging Systems, Jerusalem, Israel). The blots were scanned and the abundance assessed quantitatively using ImageJ.
ELISA
Cells at a density of 1 × 105/well in a 6-well-plate were incubated for 72 h, and Collagen α1(III) levels in cell culture medium and cell lysate were measured, respectively, using a commercially available Collagen α1(III) ELISA kit at 450 nm by a microplate spectrophotometer (CSB-E13446h, Cusabio, Wuhan, China). The experiment process was carried out according to the instructions of the kit. The samples were added into wells and incubated 2 h in 37°C. After removing the liquid of each well, Biotin-antibody was added into each well. After incubated 1 h in 37°C, HRP-avidin and TMB Substrate were used for color rendering. The standard curve was constructed by the mean absorbance of each standard and the concentration. The concentration of each sample was determined according to standard curve. To acquire the total amount of Collagen α1(III) in each sample, the concentration was multiplied by the total volume. Then the amount of Collagen α1(III) in each sample was normalized to the amount of Collagen α1(III) in 107 cells. All samples and standards were detected in duplicate.
Immunohistochemistry
The tissue microarray (TMA) sections (HBreD140Su06) and the relevant clinical data were obtained from Shanghai Outdo Biotechnology Company. This study was approved by the Ethics Committee of Shanghai Outdo Biotechnology Company (YB M-05-02), and all patients have given their informed consent. The TMA sections were deparaffinized and rehydrated with an ethanol gradient. Then antigen retrieval was performed with citrate buffer (MXB, Fuzhou, China, MVS-0066) and the TMA sections were blocked with endogenous peroxidases in UltraSensitive™ SP (Mouse/Rabbit) IHC Kit (MXB, Fuzhou, China, KIT-9730-A&B). The concentration of anti-METTL3 (ab195352) was 1:500, the concentration of anti-COL3A1 (sc-166316) was 1:50. After overnight incubation with primary antibody, the TMA sections were incubated with biotinylated secondary antibody for 10 min in UltraSensitive™ SP (Mouse/Rabbit) IHC Kit (MXB, Fuzhou, China, KIT-9730-C&D) and developed with DAB Kit (MXB, China, DAB-0031). Finally, the TMA sections were counterstained with hematoxylin (Solarbio, Beijing, China), and dehydrated with an ethanol gradient and mounted with neutral balsam (Solarbio, Beijing, China). METTL3 expression was evaluated by two independent reviewers by calculating the average positively stained tumor cells at 400×magnification. The positive signal of COL3A1 was quantified as integrate optical density (IOD) value using ImageJ software.
Statistical Analysis
Statistical analyses were carried out using SPSS (version 16.0) and R (V.3.2.5). Limma package analysis was conducted to explore the correlations between METTL3 and other genes. One-way ANOVA and Student's t-test were used to determine statistical significance. Statistical significance was identified as P-values of <0.05.
Results
Low Expression of METTL3 Was Associated With Poor Prognosis in TNBC
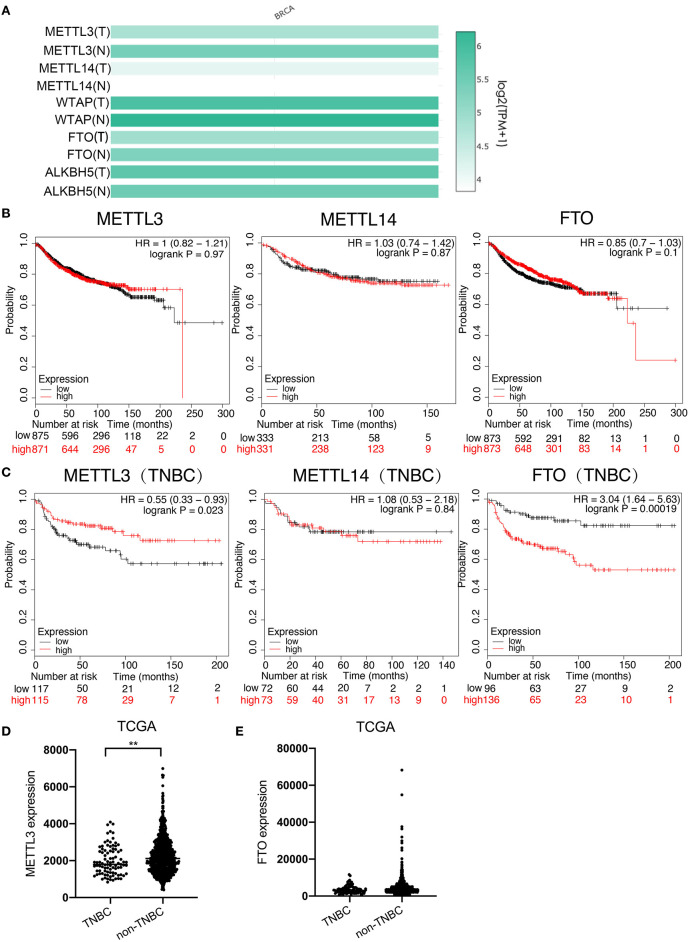
To investigate which m6A modulator plays an important role in breast cancer, especially in TNBC, the mRNA expressions of m6A methyltransferases (METTL3, METTL14, and WTAP) and demethylases (FTO and ALKBH5) were first compared between breast cancer tissues and normal breast tissues using the RNA sequencing expression data in GEPIA (http://gepia.cancer-pku.cn) on-line database. The result showed that the expression of METTL3 (T = 4.8, N = 5.4) and FTO (T = 4.9, N = 5.3) was significantly lower, whereas that of METTL14 (T = 4.2, N = 3.8) was higher in breast cancer tissues than that in normal tissues, but the difference in the expression of WTAP (T = 5.9, N = 6.2) and ALKBH5 (T = 5.7, N = 5.5) was not very significant (Figure 1A). Next, the effects of METTL3, METTL14, and FTO on distant metastasis free survival (DMFS) of total breast cancer patients and TNBC patients were analyzed using KM-plotter on-line database, respectively. For overall patients, the analysis result indicated that no significant difference of DMFS was obtained between the patients with all three modulators in high-expression groups and low-expression groups (Figure 1B); however, for the TNBC patients, although no significant difference was found between the DMFS of the METTL14 high-expression group and low-expression group, the DMFS of METTL3 high expression group was shown to be longer than that in the METTL3 low-expression group, whereas the DMFS of the FTO high-expression group was shorter than that of the FTO low-expression group, indicating that METTL3 is a protective factor, but FTO is a risk factor for DMFS of TNBC (Figure 1C). As the result, the low expression of FTO in TNBC tissues was contradictory to its role as a risk factor (Figures 1A,E) and only METTL3 was shown to play an important inhibitory role in the metastasis of TNBC (Figures 1A,D), indicating that METTL3 might contribute to the metastasis of TNBC. Therefore, the role of METTL3 in TNBC metastasis was focused on in the following investigation.
Figure 1.
Low expression of METTL3 was associated with poor prognosis in TNBC. (A) Expression analysis of METTL3, METTL14, WTAP, FTO, and ALKBH5 in BC tissue (1085) and normal tissue (291) using TCGA and GTEx online database. (B) Kaplan–Meier analysis for the DMFS of METTL3, METTL14, and FTO in overall BC patients using KM-plotter online database. (C) Kaplan–Meier analysis for the DMFS of METTL3, METTL14, and FTO in TNBC patients using KM-plotter online database. (D) METTL3 expression analysis in TNBC patients (n = 91) and non-TNBC patients (n = 1,005) using TCGA. (E) FTO expression analysis in TNBC patients (n = 1,005) and non-TNBC patients (n = 584) using TCGA. **P < 0.01.
METTL3 Suppressed Metastasis of TNBC Cells by Enhancing m6A Modification
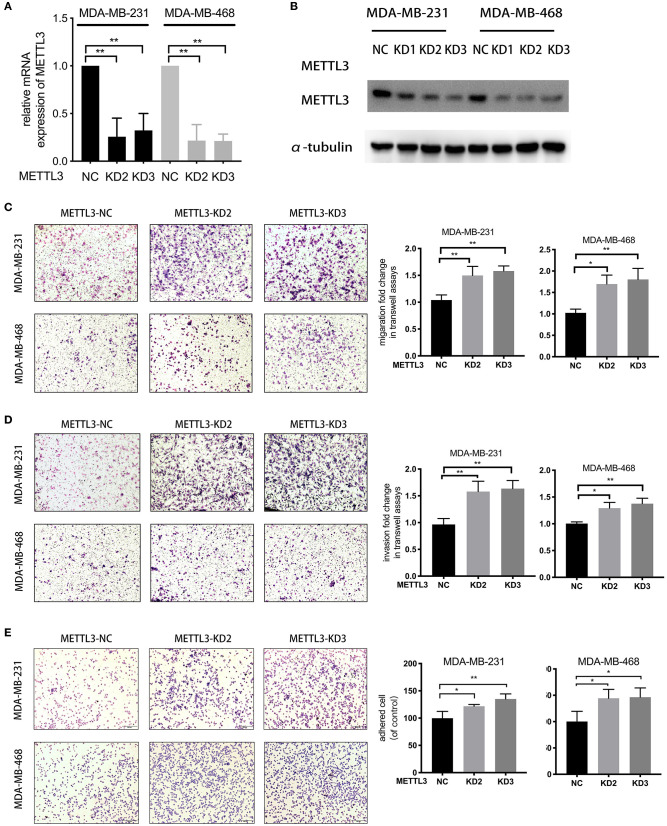
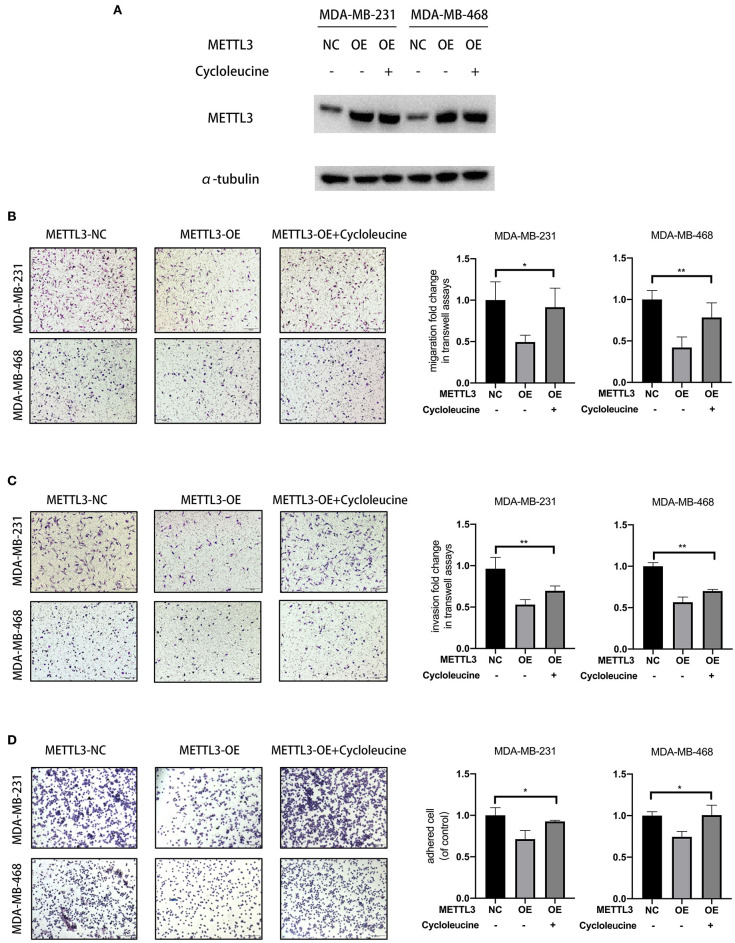
For the knockdown of METTL3, three siRNAs targeted to METTL3 were transfected according to the manufacturer's instructions (Figures 2A,B). Two sequences, si-METTL3-1604 (KD2) and si-METTL3-1126 (KD3), were selected for subsequent experiments. To investigate whether METTL3 could inhibit TNBC metastasis, the effect of METTL3 on migration, invasion, and adhesion to cell-extracellular matrix (ECM) was detected by trans-well assay or adhesion assay in TNBC cell lines, MDA-MB-231, and MDA-MB-468. The results showed that METTL3 knockdown (KD) significantly increased the ability of migration and invasion, as well as the adhesion capability to ECM in both MDA-MB-231 and MDA-MB-468 cells (Figures 2C–E), indicating that METTL3 could inhibit the potential of cell mobility of TNBC cells. Then, to determine whether METTL3-inhibited potential of cell mobility was related to m6A modification or not, METTL3 was transiently overexpressed in MDA-MB-231 and MDA-MB-468 cells followed by the treatment with cycloleucine, a small molecule inhibitor of m6A modification. The results of trans-well assays demonstrated that METTL3 overexpression (OE)-suppressed migration, invasion, and adhesion were significantly recovered by cycloleucine (Figures 3A–D). These results strongly suggested that METTL3 inhibited the potential of cell mobility of TNBC cells by enhancing m6A modification.
Figure 2.
METTL3 suppressed metastatic ability in TNBC cells. (A) qRT-PCR was used to detect METTL3 expression in MDA-MB-231 and MDA-MB-468 cells transfected with si-NC or si-METTL3. 18S was used as an internal control. (B) Western blot was used to detect METTL3 expression in MDA-MB-231 and MDA-MB-468 cells transfected with si-NC or si-METTL3. α-tubulin was used as a loading control. (C,D) Transwell assay was used to detect the migration and invasion ability in MDA-MB-231 and MDA-MB-468 cells with METTL3 transient knockdown (left panels). Relative fold change was shown as the proportion of the number of control cells transfected with si-NC (right panels). Original magnification, 100×. (E) Adhesion assay was used to detect the adhesion ability of MDA-MB-231 and MDA-MB-468 cells with METTL3 transient knockdown (left panels). Relative fold change was shown as the proportion of the number of control cells transfected with si-NC (right panels). Original magnification, 100×. *P < 0.05, **P < 0.01. Error bars represent the mean ± SD of three independent experiments.
Figure 3.
METTL3 overexpression-suppressed migration, invasion, and adhesion were significantly rescued by cycloleucine. (A) Western blot was used to detect METTL3 expression in MDA-MB-231 and MDA-MB-468 cells in rescued assay. α-tubulin was used as a loading control. (B,C) Transwell assay was used to detect the migration and invasion ability in MDA-MB-231 and MDA-MB-468 cells with METTL3 transient overexpression and METTL3 overexpression rescued with cycloleucine (left panels). Relative fold change was shown as the proportion of the number of control cells transfected with pcDNA3.1-FLAG (right panels). Original magnification, 100×. (D) Adhesion assay was used to detect the adhesion ability of MDA-MB-231 and MDA-MB-468 cells with METTL3 transient overexpression (left panels). Relative fold change was shown as the proportion of the number of control cells transfected with pcDNA3.1-FLAG (right panels). Original magnification, 100×. *P < 0.05, **P < 0.01. Error bars represent the mean ± SD of three independent experiments.
COL3A1 Was Identified as a Potential Target of METTL3 in TNBC
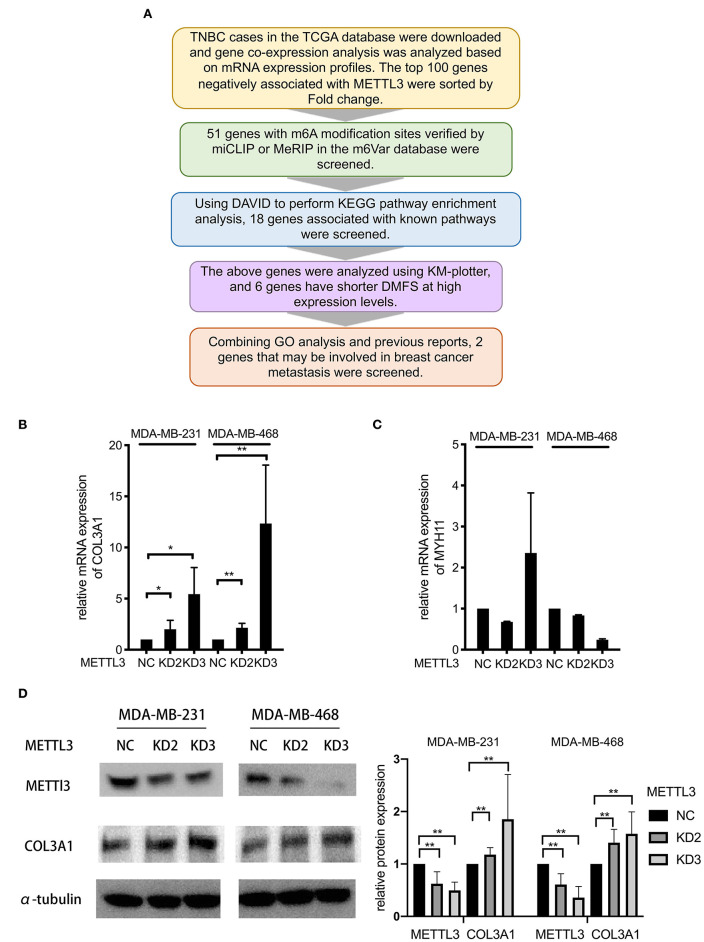
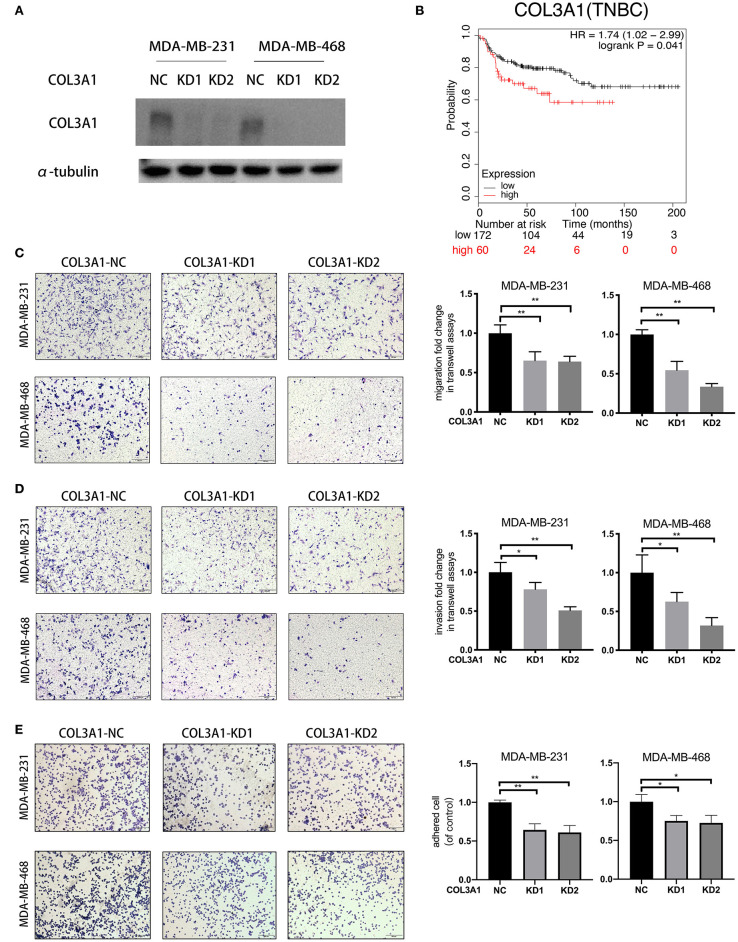
It was known that m6A modification could down-regulate gene expression by accelerating RNA degradation (12–14). Therefore, to identify the target gene of METTL3 involved in METTL3-inhibited metastasis, multi-step screening was performed as summarized in Figure 4A. Firstly, mRNA expression profiles of 91 TNBC patients in the TCGA dataset were downloaded, and the differentially expressed genes (DEG) were screened to identify those with P < 0.05 and log fold change (FC) using the “limma” package in R. The Log FC of DEG genes more than 0 was identified as representing positive related genes, whereas that <0 represented negative related genes. Among the genes for which mRNA expression was negatively related to METTL3, the top 100 genes according to the correlation coefficient were selected for further m6A methylation analysis using the m6Avar database (http://m6avar.renlab.org). All these genes and related information were listed in Table S1. As a result, 51 genes, which were verified to be able to be modified by m6A, were screened. Then, with the KEGG pathway enrichment analysis by DAVID (https://david.ncifcrf.gov), 18 genes were shown to be associated with the focal adhesion pathway, metabolism pathway, and so on, suggesting close involvement with metastasis in breast cancer. Subsequently, the association of 18 genes with the DMFS of TNBC patients was further analyzed using KM-plotter, and six alternative genes were found to have shorter DMFS at high levels in TNBC (Table 1). In particular, COL3A1 and MYH11 aroused our attention, because, according to the Gene Ontology (GO) analysis in DAVID, it was shown that COL3A1 was involved in the biological process of skeletal system development and cell-matrix adhesion, and MYH11 was involved in the biological process of elastic fiber assembly, which were similar to the findings of previous studies that they could promote metastasis in breast cancer. Therefore, the two genes were chosen as target gene candidates for METTL3. The further verification result of qRT-PCR detection showed that METTL3-KD only up-regulated the mRNA expression of COL3A1, but not MYH11 in MDA-MB-231 and MDA-MB-468 cells (Figures 4B,C). Similarly, the result of western blot assay also confirmed that METTL3-KD increased the protein level of COL3A1 in TNBC cells (Figure 4D). All of the above data strongly suggested that COL3A1 might be the target gene of METTL3.
Figure 4.
COL3A1 was identified as a potential target of METTL3 in TNBC. (A) Flowchart for screening potential target genes. (B,C) qRT-PCR was used to detect COL3A1 and MYH11 expression in MDA-MB-231 and MDA-MB-468 cells transfected with the si-NC or the si-METTL3. 18S was used as an internal control. (D) Western blot was used to detect COL3A1 expression in MDA-MB-231 and MDA-MB-468 cells transfected with the si-NC or the si-METTL3. α-tubulin was used as a loading control (left panel). The blots were scanned and the abundance assessed quantitatively using ImageJ (right panel). *P < 0.05, **P < 0.01. Error bars represent the mean ± SD of three independent experiments.
Table 1.
18 genes which were associated with pathways involved in metastasis and relative KEGG pathway, and DMFS analysis.
| Gene name | KEGG Pathway enrichment analysis |
DMFS analysis HR (P) |
|
|---|---|---|---|
| 1 | FZD4 | Wnt signaling pathway, Hippo signaling pathway, signaling pathways regulating pluripotency of stem cells, melanogenesis, HTLV-I infection, pathways in cancer, proteoglycans in cancer, basal cell carcinoma | 0.47 (0.034) |
| 2 | UTS2R | Neuroactive ligand-receptor interaction | 1.42 (0.350) |
| 3 | PTPRJ | Adherens junction | 1.66 (0.048) |
| 4 | CPSF3 | mRNA surveillance pathway | 0.50 (0.088) |
| 5 | MTHFD2L | One carbon pool by folate, Metabolic pathways | 1.77 (0.035) |
| 6 | MYH11 | Tight junction | 2.06 (0.033) |
| 7 | LPGAT1 | Glycerophospholipid metabolism | 1.57 (0.270) |
| 8 | LAPTM4B | Lysosome | 1.36 (0.230) |
| 9 | IL1R1 | MAPK signaling pathway, cytokine–cytokine receptor interaction, NF-kappa B signaling pathway, osteoclast differentiation, hematopoietic cell lineage, inflammatory mediator regulation of TRP channels, amoebiasis, HTLV-I infection | 1.59 (0.160) |
| 10 | COL3A1 | PI3K-Akt signaling pathway, focal adhesion, ECM-receptor interaction, platelet activation, protein digestion and absorption, amoebiasis | 1.74 (0.041) |
| 11 | ALG10B | N-Glycan biosynthesis, metabolic pathways | 2.14 (0.033) |
| 12 | GMPPB | Fructose and mannose metabolism, amino sugar and nucleotide sugar metabolism, metabolic pathways | 1.43 (0.190) |
| 13 | RNASEH1 | DNA replication | 1.53 (0.100) |
| 14 | UBE4B | Ubiquitin mediated proteolysis, protein processing in endoplasmic reticulum | 2.46 (0.007) |
| 15 | NCOA1 | Thyroid hormone signaling pathway | 1.47 (0.200) |
| 16 | SNRPE | Spliceosome | 1.45 (0.160) |
| 17 | TGM2 | Huntington's disease | 0.54 (0.043) |
| 18 | FDFT1 | Steroid biosynthesis, metabolic pathways, biosynthesis of antibiotics | 1.28 (0.340) |
METTL3 Down-Regulated the Expression of COL3A1 by Increasing m6A Level
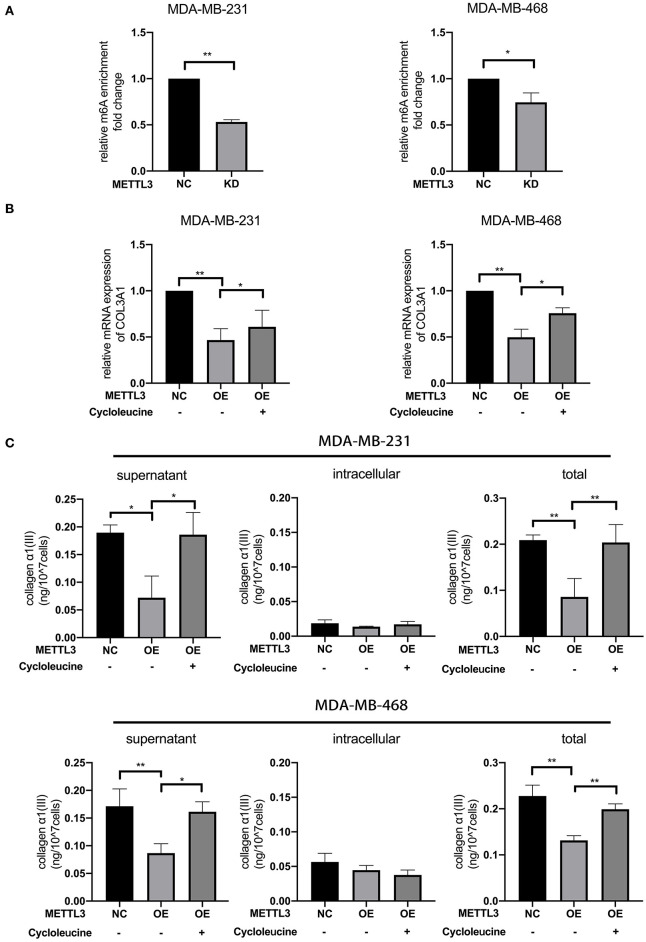
To investigate whether COL3A1 was regulated by METTL3-mediated m6A methylation or not, the relative m6A enrichment level change of COL3A1 before and after METTL3-KD was detected by MeRIP-qRT-PCR in MDA-MB-231 and MDA-MB-468 cells. As shown in Figure 5A, METTL3-KD significantly reduced m6A-methylated COL3A1 mRNA expression. Furthermore, with qRT-PCR detection, it was shown that METTL3- OE decreased the mRNA expression of COL3A1, while cycloleucine partially recovered METTL3-OE-down-regulated COL3A1 (Figure 5B). As shown in Figure 5C, the results of ELISA showed that the secretion level of Collagen α1(III) in supernatant decreased by METTL3-OE could be recovered by cycloleucine while the METTL3-OE decreased the intracellular level of Collagen α1(III), the cycloleucine could not recover the reduction of the intracellular level of Collagen α1(III). Considering the amount of Collagen α1(III) secreted into the supernatant is greater than the amount in the cell, the total levels of Collagen α1(III) which were decreased by METTL3-OE could be recovered by cycloleucine after normalizing (Figure 5C). These results proved that COL3A1 was down-regulated by METTL3-mediated m6A modification on COL3A1 (Figure 5C).
Figure 5.
METTL3 down-regulated the expression of COL3A1 by increasing m6A levels. (A) MeRIP-qRT-PCR was used to detect the m6A modification level of COL3A1 in MDA-MB-231 and MDA-MB-468 cells transfected with si-NC or si-METTL3. The relative enrichment fold changes were shown as proportions of control cells enrichment. (B) qRT-PCR was used to detect COL3A1 expression in MDA-MB-231 and MDA-MB-468 cells with METTL3 transient overexpression or the combination of METTL3 overexpression with cycloleucine. 18S was used as an internal control. (C) ELISA was used to detect the secretion level of collagen α1 (III) of the supernatant (left panels), intracellular (middle panels) and total secreted protein (right panels) in MDA-MB-231 and MDA-MB-468 cells with METTL3 transient overexpression or the combination of METTL3 overexpression with cycloleucine. *P < 0.05, **P < 0.01. Error bars represent the mean ± SD of three independent experiments.
COL3A1 Promoted the Metastatic Ability of TNBC Cells
The role of COL3A1 in TNBC metastasis was further investigated. The influence of COL3A1 on DMFS of TNBC patients analyzed by KM-plotter is shown in Figure 6B, the DMFS of TNBC patients with high-expression COL3A1 was shorter than in those with a low expression thereof. There was no significant difference in the expression level of COL3A1 and DMFS in the overall patients (Figure S1). In addition, when COL3A1 was knocked-down in MDA-MB-231 and MDA-MB-468 cells (Figure 6A), the migration, invasion, and adhesion to ECM were significantly suppressed (Figures 6C–E). These data indicated that COL3A1 played an important role in promoting metastasis in TNBC.
Figure 6.
COL3A1 promoted the metastatic ability of TNBC cells. (A) Western blot was used to detect COL3A1 expression in MDA-MB-231 and MDA-MB-468 cells transfected with the si-NC or the si-COL3A1. α-tubulin was used as a loading control. (B) Kaplan–Meier analysis for the DMFS of COL3A1 in TNBC patients using KM-plotter online database. (C,D) The migration and invasion of MDA-MB-231 and MDA-MB-468 with transient COL3A1-KD was detected by transwell assays (left panels). Relative fold change was shown as the proportion of the number of control cells transfected with si-NC (right panels). Original magnification, 100×. (E) The adhesion ability of MDA-MB-231 and MDA-MB-468 after transient COL3A1-KD was evaluated by adhesion assay (left panels). Relative fold change was shown as the proportion of the number of control cells transfected with si-NC (right panels). Original magnification, 100×. *P < 0.05, **P < 0.01. Error bars represent the mean ± SD of three independent experiments.
Reduced COL3A1 m6A Modification by METTL3 Inhibition Leads to Poor Prognosis in TNBC Patients
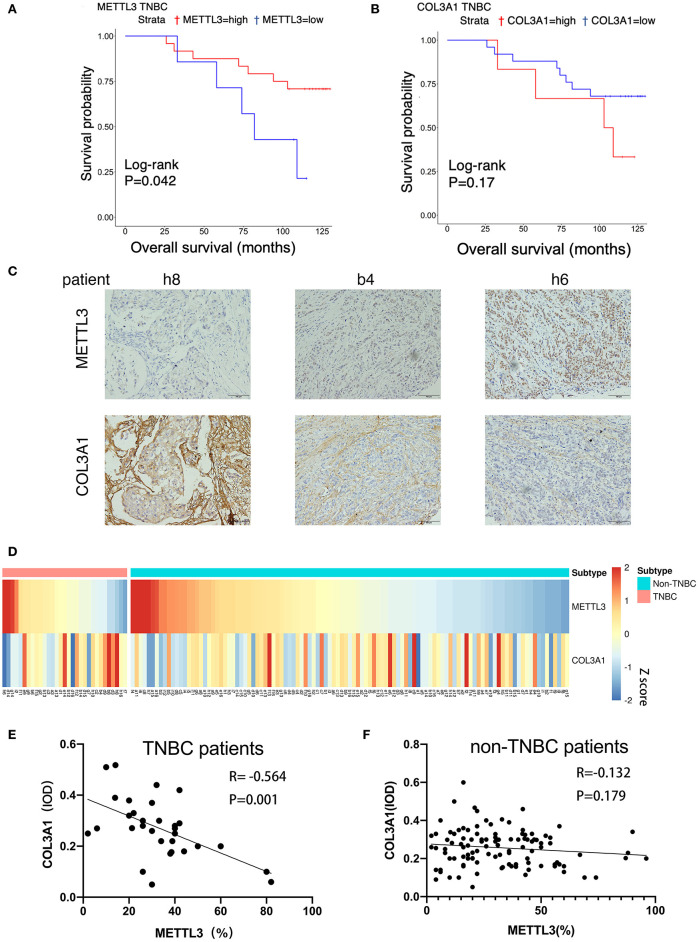
In order to verify the effects of METTL3 and COL3A1 on the prognosis of breast cancer patients in vivo, the expression of METTL3 and COL3A1 was investigated by immunohistochemistry using TMA sections containing 31 TNBC patients and 109 Non-TNBC patients. The results of survival analysis showed that low METTL3 expression was related to short overall survival (OS) (Figure 7A) in TNBC but not in non-TNBC (Figure S2A). The OS of TNBC with COL3A1-high expression was shorter than that with low expression (Figure 7B), which may not be statistically significant because of the small number of TNBC cases. In non-TNBC, COL3A1 has the opposite trend, and patients with high expression have longer OS (Figure S2B). Finally, the relationship between METTL3 and COL3A1 in breast cancer was analyzed with Pearson correlation analysis (Figures 7C,D). The results showed that the expression level of COL3A1 was negatively correlated with METTL3 expression in TNBC patients (R = −0.564, P = 0.001; Figure 7E); however, the expression level of METTL3 and COL3A1 had no significant relationship in the 109 NTNBC patients (R = −0.132, P = 0.170; Figure 7F). This result further confirmed that the metastasis-inhibition function of METTL3 by negatively regulating COL3A1 expression was TNBC specific.
Figure 7.
The correlation between METTL3 and COL3A1 in breast cancer patients. (A,B) Kaplan–Meier analysis for the OS of METTL3 and COL3A1 in TNBC patients of the TMA sections. (C) The expression of METTL3 and COL3A1 detected by IHC in the representative samples of breast cancer. h8, low expression of METTL3 and high expression of COL3A1. b4, middle expression of METTL3 and COL3A1. h6, high expression of METTL3 and low expression of COL3A1. Original magnification, 200×. (D) Heatmap of the expression level of METTL3 and COL3A1 protein in breast cancer patients. (E,F) Correlation of METTL3 and COL3A1 in human TNBC patients (n = 31) and non-TNBC patients (n = 109), respectively, in TMA sections.
Discussion
In this study, by analyzing the prognostic role of m6A modulators (METTL3, METTL14, WTAP, FTO, and ALKBH5) in breast cancer using on-line databases, we found that only METTL3 played an important role in TNBC metastasis. The expression of METTL3 in breast cancer tissues was lower than that in normal tissues, and METTL3 was a protective factor of DMFS in TNBC. Using TNBC cell lines, it was confirmed that METTL3 could inhibit metastasis by increasing the level of m6A modification, and COL3A1 was identified as one of the possible target genes of METTL3. Furthermore, reduced expression of METTL3 was proved to be able to contributes the potential of mobility of triple-negative breast cancer cells by m6A methylation-mediated COL3A1 up-regulation.
The role of m6A modification that is mainly regulated by methyltransferases and demethylases, is complicated and specific in various cancers. A bioinformatics analysis study involving 33 cancers showed that m6A modulators are closely related to both the activation pathway and inhibition pathway of cancer; the distribution of m6A modifications varies widely among different cancers; even for the same type of cancer, the prognostic function of m6A was not consistent within each sub-type (15). Several studies also exhibit the complicated roles of m6A modulators in the development of breast cancer. It was reported that FTO, a key m6A demethylase, was up-regulated and significantly associated with poor prognosis in breast cancer (8); FTO-reduced m6A modification could promote breast cancer cell proliferation and metastasis by inhibiting BNIP3 expression (11). Similarly, METTL14 overexpression or ALKBH5 silence could also inhibit the growth and migration of breast cancer cell line, MDA-MB-231 (16). The opposite result was also reported such that the deficiency of METTL3 could inhibit the proliferation of breast cancer cell line MCF-7, by m6A-level-decreasing-mediated Bcl-2 up-regulation (10). In this study, using online database, we analyzed the prognostic role of five m6A modulators in breast cancer, especially in TNBC, the sub-type with the worst prognosis and the greatest potential for metastasis, and found that METTL3 is the most critical in TNBC, that METTL3 occurred at low expression in TNBC, and was a protective factor of DMSF. The results of the TMA section also confirm the protective effect of METTL3 on the overall survival of TNBC patients. These results are consistent with some previous researches (16), while contradictory to the other researches focused on proliferation (10, 17). This difference might be due to the different subtypes of the breast cancer cell lines used. It should be taken into consideration that those previous study had mainly used non-TNBC cell lines. Meanwhile, the conclusion of previous studies was based on the result of cellular level investigation and lack of clinical specimen validation. Therefore, although the trend in m6A modification levels was consistent, different regulators might eventually cause opposite effects by regulating different target genes; an m6A modulator might also execute different functions in each sub-type due to the heterogeneity of cancer. The function of METTL3 and other m6A modulators in other sub-types of breast cancer warrants further investigation in the future.
Widely distributed in eukaryotes, RNA methylation modification occurs in thousands of genes (18). METTL3 is known to achieve its biological effects by increasing the m6A modification level of target genes, which leads to various effects on target genes, such as faster degradation of target gene mRNA, increase in target gene translation efficiency, or accurate cell localization of target genes (2–5). Among them, the mechanism of accelerating the rate of degradation of target gene mRNA is most widely investigated. Therefore, METTL3 might have many target genes in TNBC, and the inhibitory effect of METTL3 on TNBC metastasis might also be achieved by affecting multiple target genes together. In this study, by expression correlation analysis and methylation search, COL3A1 was identified as the target gene candidate of METTL3. Collagen type III alpha 1 chain (COL3A1), which encodes the pro-alpha 1 chains of type III collagen, previously was reported to be associated with malignant potential of breast cancer (19). To date, no specific mechanism has been reported for COL3A1 regulation. In this study, we proved that knocking down METTL3, while reducing the methylation of m6A, also eventually up-regulated the expression level of COL3A1. Validation of clinical specimens indicated that this relationship appears to be only in TNBC patients. Thus, this study demonstrated that METTL3 and COL3A1 might only play a significant role in the TNBC subtype. Certainly, there must be multiple target genes of METTL3 that play the same role. COL3A1 may also have modification sites different from those provided by online databases. Further MeRIP-sequence is needed to clarify the mechanisms of METTL3 in metastasis inhibition of TNBC in the future and the specific modified sites of COL3A1. Considering that the mRNA level of COL3A1 has changed, we speculate that the change of m6A level may affect the degradation rate of COL3A1 mRNA in TNBC cells. In this case, the position of m6A seems to be more likely to be distributed in the 3′UTR region of mRNA (2). But this speculation still needs further experimental proof.
Collagen, the most abundant component of extracellular matrix (ECM) in the tumor micro-environment, is known to be able to contribute to tumor progression (20). Collagen could promote the metastasis and proliferation of cancer by increasing the accumulation of integrin, which leads to phosphorylation of focal adhesion kinase and activation of extracellular signal-regulated kinase (21). COL3A1, which encodes pro-alpha1 chains of type III collagen, could form homotrimeric fibrils to play its role. Except for normal localization in connective tissues, COL3A1 was also found to be highly expressed in various cancers including bladder cancer, glioblastoma, and gastric cancer (22–24). In breast cancer, it was reported that stromal COL3A1 expression was significantly increased from benign breast tumors to malignant breast tumors (18). Another study has shown that when Pirfenidone, an anti-fibrotic drug, was applied to breast cancer to investigate its possible role on tumor microenvironment normalization, the level of COL3A1 was down-regulated, thereby inhibiting the TGFβ signaling pathway. That causes the reduction of extracellular matrix components, which significantly increases vascular function and perfusion, and increases the anti-tumor efficacy of doxorubicin (25). Therefore, these studies showed that COL3A1 played an important role in the development of breast cancer. In addition, it was reported that COL3A1 up-regulation cause extracellular matrix changes and reduced tumor perfusion, while the hypoxic micro-environment caused by hypoperfusion was considered to be the main reason for forcing cancer cells to metastasize (26). Therefore, the reduction of tumor perfusion caused by up-regulation of COL3A1 was likely to be one of the reasons for the increased ability of TNBC cells to metastasis. In this study, we demonstrated that COL3A1, which was up-regulated by the reduced expression of METTL3, could contribute to TNBC metastasis. The molecular mechanism of COL3A1 in promoting TNBC metastasis warrants further investigation.
In summary, this study not only revealed that, among m6A modulators, only METTL3 played an important role in TNBC metastasis, but also demonstrated that the low expression of METTL3-reduced m6A modification could promote TNBC metastasis by up-regulating its target gene, COL3A1. Our results provided sufficient evidence of the important epigenetic role in the development of TNBC and allowed a more comprehensive understanding of the mechanism of tumor metastasis. METTL3 and its target gene COL3A1 might have the potential to become novel biomarkers for TNBC prognostic prediction and new targets for TNBC therapy.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.cancer.gov/?TCGA-BRCA</b>.
Ethics Statement
This study was approved by the Ethics Committee of Shanghai Outdo Biotechnology Company (YB M-05-02), and all patients have signed informed consent. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YS and CZ analyzed and interpreted the data regarding the m6A modulators and DMFS of breast cancer patients, performed most experiments and were major contributor in writing the manuscript and contributed equally to this work. YJ and DW performed partial qRT-PCR experiment. BB analyzed the correlation analysis between METTL3 and other genes. JF helped evaluated the expression of METTL3 in IHC. ST helped with the making of heatmap. XQ and YL provided guidance on interpreting the results. XC and YT designed the study and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- ALKBH5
AlkB family member 5
- COL3A1
collagen type III alpha 1 chain
- DMFS
the distance-metastasis-free survival
- ECM
cell-extracellular matrix
- FC
fold change
- GO
Gene Ontology
- GTEx
the Genotype Tissue Expression
- IHC
Immunohistochemistry
- KD
knockdown
- METTL3
methyltransferase-like 3
- METTL14
methyltransferase-like 14
- MYH11
myosin heavy chain 11
- NC
negative control
- OE
overexpression
- PVDF
polyvinylidene difluoride
- qRT-PCR
quantitative real-time
- TCGA
the Cancer Genome Atlas
- TNBC
triple-negative breast cancer
- TMA
The tissue microarray
- WTAP
Wilms tumor 1-associated protein.
Footnotes
Funding. This research was supported by National Natural Science Foundation of China (No. 81672605), The Key Research and Development Program of Liaoning Province (2018225060), Science and Technology Plan Project of Liaoning Province (2019-ZD-777), Science and Technology Plan Project of Shenyang city (19-112-4-099), and Science and Technology Plan Project of Liaoning Province (No. 2013225585).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01126/full#supplementary-material
References
- 1.DeSantis CE, Bray F, Ferlay J, Lortet-Tieulent J, Anderson BO, Jemal A. International variation in female breast cancer incidence and mortality rates. Cancer Epidemiol Biomark Prev. (2015) 24:1495–506. 10.1158/1055-9965.EPI-15-0535 [DOI] [PubMed] [Google Scholar]
- 2.Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m6a methylation in cancer. Cancer Res. (2019) 79:1285–92. 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- 3.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. (2018) 28:507–17. 10.1038/s41422-018-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. (2019) 75:379–89. 10.1007/s13105-019-00690-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhao JC. Update: mechanisms underlying N6-Methyladenosine modification of eukaryotic mRNA. Trends Genet. (2016) 32:763–73. 10.1016/j.tig.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Li F, Peng Y, Fang J, Zhou J. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med. (2020) 9:1877–89. 10.1002/cam4.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. (2019) 512:479–85. 10.1016/j.bbrc.2019.03.093 [DOI] [PubMed] [Google Scholar]
- 8.Cai J, Yang F, Zhan H, Situ J, Li W, Mao Y, et al. RNA m6A Methyltransferase METTL3 promotes the growth of prostate cancer by regulating hedgehog pathway. Onco Targets Ther. (2019) 12:9143–52. 10.2147/OTT.S226796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. (2018) 48:838–46. 10.1159/000491915 [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. (2020) 722:144076. 10.1016/j.gene.2019.144076 [DOI] [PubMed] [Google Scholar]
- 11.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. (2019) 18:46. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Naure. (2014) 505:117–20. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m6a reader YTHDC1 regulates mRNA splicing. Mol Cell. (2016) 61:507–19. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 14.Ma J-Z, Yang F, Zhou C-C, Liu F, Yuan Y-H, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology. (2017) 65:529–43. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X, et al. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol Cancer. (2019) 18:137. 10.1186/s12943-019-1066-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. (2019) 19:326. 10.1186/s12885-019-5538-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer letter. (2018) 415:11–9. 10.1016/j.canlet.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 18.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485:201–6. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Resnick MB, Lu S, Hui Y, Brodsky AS, Yang D, et al. Collagen type III α1 as a useful diagnostic immunohistochemical marker for fibroepithelial lesions of the breast. Hum Pathol. (2016) c57:176–81. 10.1016/j.humpath.2016.07.017 [DOI] [PubMed] [Google Scholar]
- 20.Hasebe T. Tumor-stromal interactions in breast tumor progression–significance of histological heterogeneity of tumor-stromal fibroblasts. Expert Opin Ther Targets. (2013) 17:449–60. 10.1517/14728222.2013.757305 [DOI] [PubMed] [Google Scholar]
- 21.Paolillo M, Schinelli S. Extracellular matrix alterations in metastatic processes. Int J Mol Sci. (2019) 20:4947. 10.3390/ijms20194947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi S, Tian B. Identification of biomarkers associated with progression and prognosis in bladder cancer via co-expression analysis. Cancer Biomark. (2019) 24:183–93. 10.3233/CBM-181940 [DOI] [PubMed] [Google Scholar]
- 23.Gao YF, Zhu T, Chen J, Liu L, Ouyang R. Knockdown of collagen α-1(III) inhibits glioma cell proliferation and migration and is regulated by miR128-3p. Oncol Lett. (2018) 16:1917–23. 10.3892/ol.2018.8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie K, Shi L, Wen Y, Pan J, Li P, Zheng Z, et al. Identification of hub genes correlated with the pathogenesis and prognosis of gastric cancer via bioinformatics methods. Minerva Med. (2020) 111:213–25. 10.23736/S0026-4806.19.06166-4 [DOI] [PubMed] [Google Scholar]
- 25.Polydorou C, Mpekris F, Papageorgis P, Voutouri C, Stylianopoulos T. Pirfenidone normalizes the tumor microenvironment to improve chemotherapy. Oncotarget. (2017) 8:24506–17. 10.18632/oncotarget.15534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christner PJ, Ayitey S. Extracellular matrix containing mutated fibrillin-1 (Fbn1) down regulates Col1a1, Col1a2, Col3a1, Col5a1, and Col5a2 mRNA levels in Tsk/+ and Tsk/Tsk embryonic fibroblasts. Amino Acids. (2006) 30:445–51. 10.1007/s00726-005-0265-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.cancer.gov/?TCGA-BRCA</b>.