Summary
Cytokines were the first modern immunotherapies to produce durable responses in advanced cancer, but their application has been hampered by modest efficacy and limited tolerability1,2. In an effort to identify alternative cytokine pathways for immunotherapy, we found that components of the Interleukin-18 (IL-18) pathway are upregulated on tumor infiltrating lymphocytes (TIL), suggesting that IL-18 therapy could enhance anti-tumor immunity. However, recombinant IL-18 previously failed to demonstrate efficacy in clinical trials3. Here we show that IL-18BP, a high-affinity IL-18 decoy receptor, is frequently upregulated in diverse human and murine tumors and limits the anti-tumor activity of IL-18 in mice. Using directed evolution, we engineered a ‘decoy-resistant’ IL-18 (DR-18), which maintains signaling potential, but is impervious to inhibition by IL-18BP. In contrast to wild-type IL-18, DR-18 exhibits potent anti-tumor efficacy in mouse tumor models by promoting the development of poly-functional effector CD8+ T cells, decreasing the prevalence of exhausted CD8+ T cells expressing TOX, and expanding the pool of stem-like TCF1+ precursor CD8+ T cells. DR-18 also enhances NK cell activity and maturation to effectively treat anti-PD-1 resistant tumors that have lost MHC class I surface expression. These results highlight the potential of the IL-18 pathway for immunotherapeutic intervention and implicate IL-18BP as a major therapeutic barrier.
Cytokines are secreted proteins that provide instructive cues to immune cells and are therefore attractive candidates for use in cancer immunotherapy. However, the clinical application of cytokines has been hampered by their biological pleiotropism, which reduces their therapeutic specificity and can cause toxicities2. A major effort in cytokine research is to engineer “designer” cytokines with tailored biological activities4, enabling precise activation of anti-tumor immune programs. To identify avenues to improve cytokine immunotherapies, we analyzed transcriptional datasets to characterize patterns of cytokine and cytokine receptor expression on CD8+ TILs. We found that IL-18 and the subunits of its receptor (IL-18Rα/Rβ) were enriched in both activated and dysfunctional tumor CD8+ T cells (Extended Data Fig. 1a), suggesting that IL-18 agonism could effectively stimulate anti-tumor responses.
IL-18 is a member of the IL-1 cytokine family and mediates inflammation downstream of the NLRP3 and NLRP1 inflammasomes5. It drives MyD88 signaling through heterodimerization of its receptor subunits IL-18Rα (IL18R1) and IL-18Rβ (IL18RAP). Originally termed Interferon-gamma-inducing-factor (IGIF), IL-18 has been found to stimulate innate lymphocytes and antigen-experienced, but not naive T cells6. Therapeutically, recombinant IL-18 (rIL-18) has been reported to synergize with immune checkpoint inhibitors (ICI)7 and Chimeric Antigen Receptor T (CAR-T) cells in mouse tumor models8. rIL-18 has been administered to patients in clinical trials and found to be safe and well-tolerated9. However, clinical development of rIL-18 has been curtailed by lack of efficacy3. IL-18 is negatively regulated by a decoy receptor called IL-18 binding protein (IL-18BP), a secreted antagonist that binds IL-18 with extremely high affinity (KD < 1nM)10. In patients treated with rIL-18, serum IL-18BP concentrations increased by 10 to 100-fold9,11. Therefore, we hypothesized that IL-18BP produced in the tumor microenvironment (TME) may limit effective rIL-18 immunotherapy as a “secreted immune checkpoint.”
The IL-18 receptor and its decoy IL-18BP are prevalent in the TME
We initially sought to characterize the expression of IL-18 pathway components in mouse tumors. Through immunophenotyping of MC38 and YUMMER1.7 tumors and matched spleens, we found that IL-18Rα expression was widely expressed on NK cells, but dramatically upregulated on tumor CD4+ and CD8+ T cells compared to spleen (Extended Data Fig. 1b–d). Within the T cell compartment, acquisition of IL-18Rα expression was exclusive to antigen-experienced CD44+ T cells (Extended Data Fig. 1e,f). Additionally, examination of IL-18BP expression revealed that both Il18bp transcripts and protein were highly expressed in the TME and further increased by mouse (m) IL-18 treatment in an IFN-γ-dependent fashion (Extended Data Fig. 1g–j).
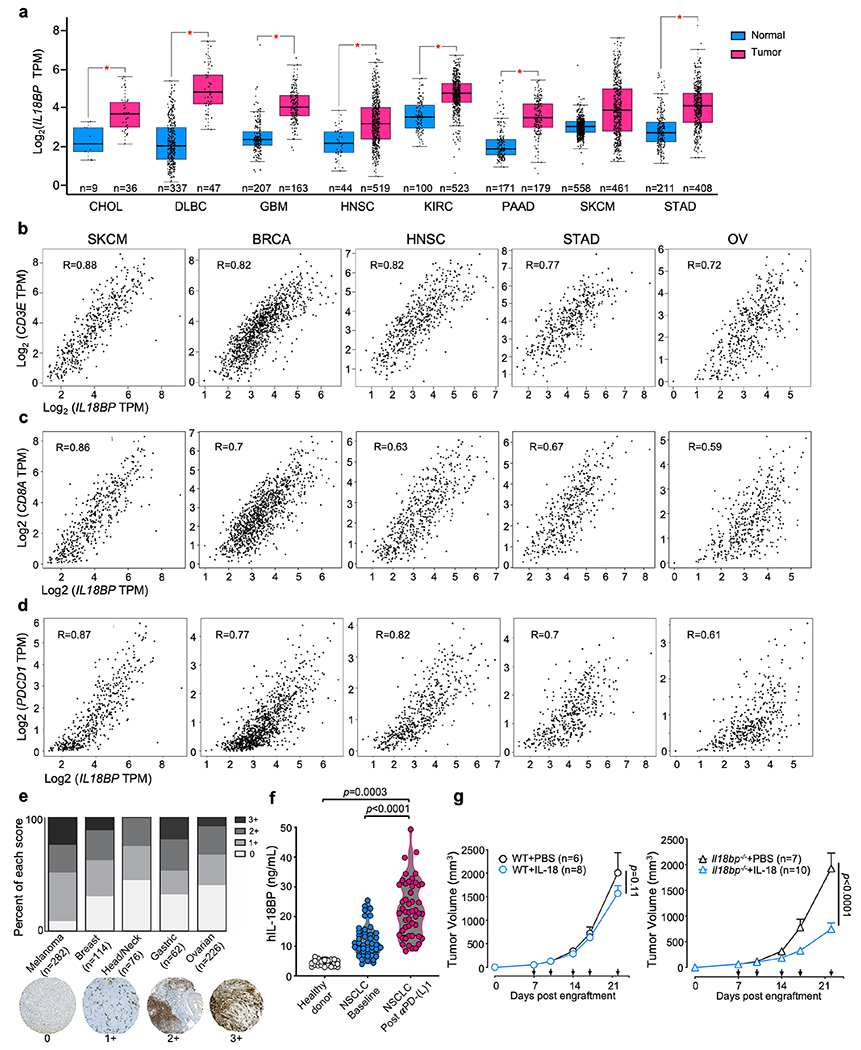
To determine if these results translated to human tumors, we analyzed IL18BP expression in the TCGA database and found increased expression of IL18BP across many tumor types compared to matched normal tissue controls (Extended Data Fig. 2a). Expression of IL18BP strongly correlated with CD3E, CD8A, and PDCD1 (R = 0.59 to 0.88), indicating an association with the presence of activated CD8+ T cells (Extended Data Fig. 2b–d). We confirmed the protein-level expression of IL-18BP in the TME by immunohistochemical staining of tissue microarrays for several tumor types. IL-18BP protein was also elevated in the serum of non-small cell lung cancer patients by ELISA and further increased by anti-PD-L1 treatment (Extended Data Fig. 2e,f).
To assess the functional effect of IL-18BP on IL-18 therapy, we engrafted MC38 tumors into either WT C57BL/6 (WT) or Il18bp−/− mice and administered mIL-18 or vehicle. While mIL-18 exhibited no effect on tumor growth in WT mice, it elicited significant tumor growth inhibition in Il18bp−/− mice (Extended Data Fig. 2g). In aggregate, these data indicate that IL-18BP expression is common in cancer and that it may act as a soluble immune checkpoint.
Engineering a “decoy-resistant” IL-18 (DR-18)
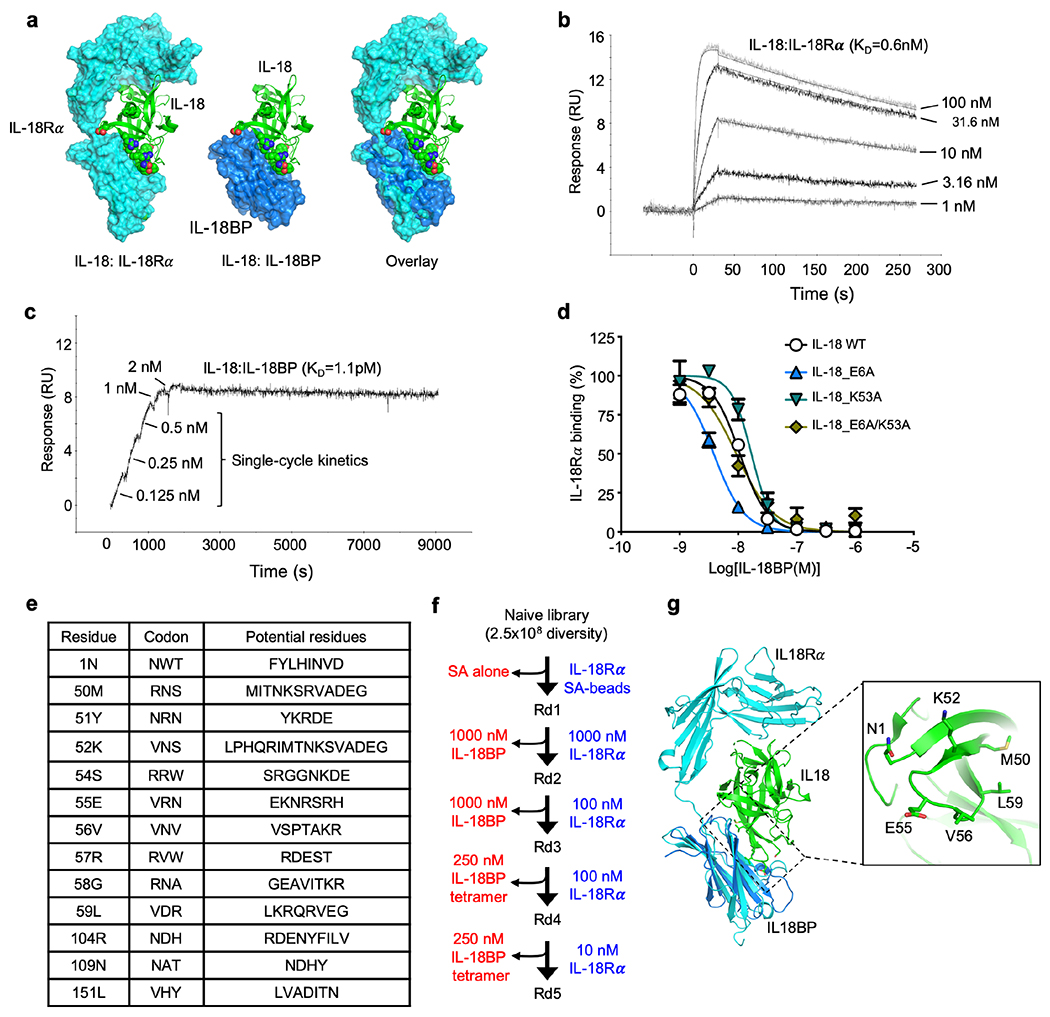
Given the potential limitation of IL-18BP on rIL-18 immunotherapy, we sought to create a “decoy-resistant” IL-18 variant (DR-18) that retains full signaling capacity through the IL-18 receptor, but is impervious to inhibition by IL-18BP (Fig. 1a). This posed an engineering challenge, since IL-18Rα and IL-18BP bind IL-18 at a highly overlapping interface and IL-18BP binds IL-18 with >3 orders of magnitude higher affinity than IL-18Rα (Extended Data Fig. 3a–c). Although point mutations (E6A and K53A) in human (h) IL-18 have been purported to reduce IL-18BP neutralization12, we found that these muteins retained IL-18BP binding without improvements in selectivity towards IL-18Rα (Extended Data Fig. 3d). We therefore used directed evolution with yeast surface display to screen >250 million mIL-18 variants that were randomized at 13 receptor contact positions for those that retained IL-18Rα binding but lacked binding to IL-18BP (Fig. 1a, Extended Data Fig. 3e). After five rounds of selection for IL-18Rα and counter-selection against IL-18BP (Extended Data Fig. 3f), we obtained a population that exclusively bound IL-18Rα (Fig. 1b). Sequencing of the post-round 5 pool revealed 11 unique sequences, from which we created two “consensus sequences”, CS1 and CS2 (Fig. 1c; Extended Data Fig. 3g). We recombinantly expressed these variants and measured their affinities for IL-18Rα and IL-18BP by surface plasmon resonance. All of the selected variants retained IL-18Rα binding, with negligible binding to IL-18BP (KD > 10 μM) (Fig. 1c).
Figure 1. Engineering decoy-resistant IL-18 (DR-18).
(a) Schematic representation of DR-18 (left) and yeast-display directed evolution process (right). (b) Representative histogram assessing IL-18Rα (100 nM, left) and IL-18BP (200 nM, right) staining by flow cytometry of yeast display library after each round of selection. (c) The sequences and dissociation constant (KD) measurements of clones summarized for selected DR-18 variants, with differences for wild-type IL-18 indicated for each mutant at the given amino acid position (top). Green shading highlights converging residues to form consensus sequence (bottom). KD measurements are shown for mIL-18 and DR-18 variants to IL-18Rα and IL-18BP measured by SPR. (--) - not tested, (N.B.D) - no binding detected. (d) Quantification of intracellular IFN-γ staining in splenic NK cells stimulated with IL-18 or CS1 or CS2 in the presence of IL-12. (e) Quantification of intracellular IFN-γ staining in splenic NK cells stimulated with IL-18 or CS1 or CS2 in the presence of IL-12 and varying concentrations of IL-18BP. Data are representative of 3 independent experiments and presented as the mean ± SEM.
To evaluate the functionality of DR-18, we stimulated NK cells ex vivo with either mIL-18 or the DR-18 variants CS1 and CS2 and measured their production of IFN-γ. CS1 had equal potency to IL-18 (EC50 = 74 and 54 pM, respectively), whereas CS2 was ~1.5 logs more potent (EC50 = 2.4 pM; Fig. 1d). We then measured the “decoy-resistance” of the DR-18 variants by fixing the concentration of the variants while titrating increasing amounts of IL-18BP. While mIL-18 was sensitive to IL-18BP (IC50 = 3.6 nM), NK cells stimulated with either CS1 or CS2 maintained robust IFN-γ production irrespective of IL-18BP concentration (Fig. 1e). From these results, we elected to proceed with CS2, hereafter referred to as DR-18, for subsequent studies.
DR-18 elicits potent anti-tumor activity in mouse tumors
We compared the efficacy of DR-18 to mIL-18 in the treatment of syngeneic mouse colorectal and melanoma tumors (Fig 2a, see methods). While treatment with mIL-18 was ineffective, DR-18 treatment produced strong tumor growth inhibition (TGI), enhanced survival, and resulted in complete tumor regression in some mice (Fig. 2b, c; Extended Data Fig. 4a–g). The efficacy of DR-18 was commensurate or superior to anti-PD-1 monotherapy, and the combination of DR-18 and anti-PD-1 produced a synergistic response that resulted in complete tumor regression in most of the treated animals (Fig. 2b,c; Extended Data Fig. 4a,b&e).
Figure 2. DR-18 stimulates anti-tumor T cell responses in immunogenic tumors.
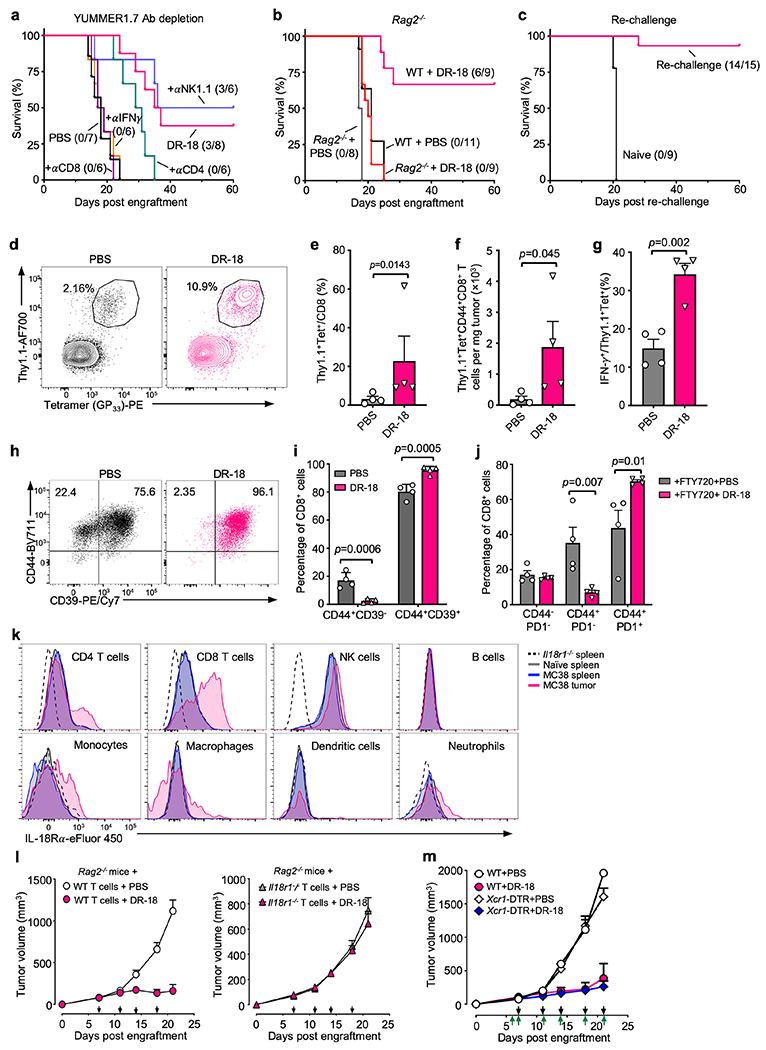
(a) WT mice were subcutaneously (s.c.) engrafted with 0.5x106 MC38 cells. On day 7 (tumor size 50-100 mm3), mice were treated twice weekly with PBS or IL-18 or DR-18 (s.c.), or with anti-PD-1 intraperitoneally (i.p). For depletion/neutralization studies, mice were injected i.p. with either anti-CD4, anti-CD8, anti-NK1.1, or anti-IFN-γ at the indicated time points (red arrows). (b) Mean tumor growth and (c) Kaplan-Meier survival curves of mice bearing MC38 tumors after treatment. Black arrows signify day of treatment. (d) Kaplan-Meier survival curves of mice engrafted with MC38 tumors after indicated depletion/neutralization administration. (e) Kaplan-Meier survival curves of WT or Il18r1−/− mice engrafted with MC38 tumors following treatment. (f) Kaplan-Meier survival curves of Rag2−/− mice reconstituted with bulk CD3+ T cells from either WT of Il18r1−/− donors, subsequently engrafted with MC38 tumors, and treated with PBS or DR-18. (b-f) Data are representative of 3 independent experiments and (b) presented as the mean ± SEM.
To exclude the possibility that DR-18 activity could be attributable to increased IL-18 receptor affinity (as opposed to its independence from IL-18BP), we compared the efficacy of DR-18 (CS2) to the related variant CS1, which has equivalent affinity towards IL-18Rα as mIL-18. CS1 produced similar efficacy to DR-18, whereas even high doses (1 mg/kg) of mIL-18 did not elicit anti-tumor responses (Extended Data Fig. 4h). These results indicate that the anti-tumor efficacy of DR-18 is driven by its independence from IL-18BP.
DR-18 requires adaptive T cell immunity to treat immunogenic tumors
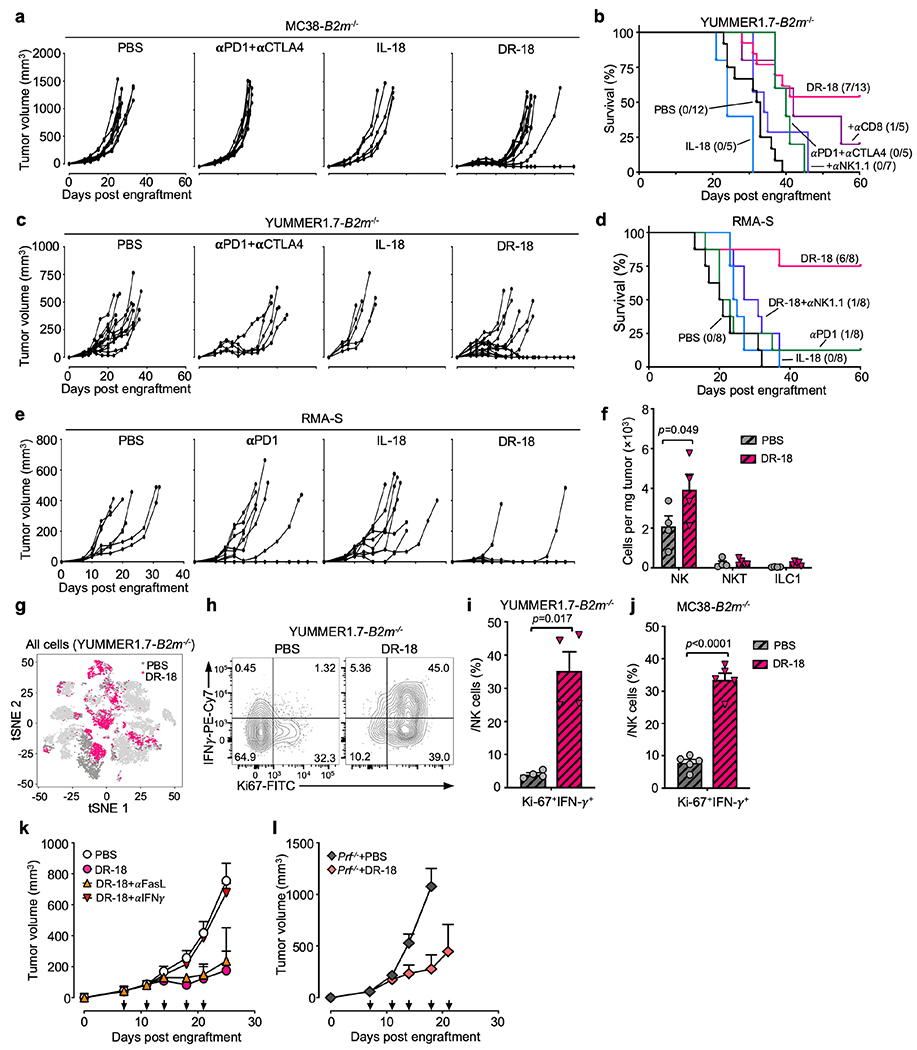
To determine the contribution of particular immune cell populations to DR-18’s efficacy, we performed antibody mediated depletion studies. In MC38 tumors, DR-18’s efficacy was abrogated by depletion of CD8+ T cells and partially inhibited by depletion of NK1.1+ cells (Fig. 2a,d). In YUMMER1.7 tumors, DR-18 efficacy required CD8+ T cells and CD4+ T cells, but not NK1.1+ cells (Extended Data Fig. 5a). Consistent with these results, DR-18 treatment was ineffective towards tumors engrafted in Rag2−/− mice (Extended Data Fig. 5b). In both models, DR-18 activity was dependent on IFN-γ (Fig. 2d, Extended Data Fig. 5a). Similarly, DR-18 treatment was ineffective in Il18r1−/− mice, confirming that DR-18 activity is mediated through the IL-18 receptor (Fig. 2e). To assess the ability of DR-18 to promote memory responses, mice surviving primary MC38 engraftment following DR-18 treatment were re-challenged with MC38; nearly all the mice (14/15) rejected the second tumor inoculation (Extended Data Fig. 5c).
To determine if antigen specific CD8+ T cells are targeted by DR-18 treatment, we adoptively transferred Thy1.1+P14 CD8+ T cells to mice bearing GP33-expressing B16 melanomas and subsequently treated with either DR-18 or vehicle. Following treatment with DR-18, we observed a significant increase in the frequency, number, and functionality (IFN-γ production) of intratumoral P14 CD8+ T cells (Extended Data Fig. 5d–g). Furthermore, analysis of the endogenous CD8+ T cell response (Thy1.1−) revealed that the majority of infiltrating CD8+ TIL were CD44+CD39+ (Extended Data Fig. 5h,i), a phenotype that is associated with tumor antigen-specific cells13. This activity could be localized directly to an effect on intratumoral cells, as DR-18 increased the frequency of activated CD8+ TIL even in the presence of FTY720, an inhibitor of T cell egress from lymphoid tissues (Extended Data Fig. 5j).
Within tumors, IL-18Rα is predominantly expressed on intratumoral T and NK cells, though low levels of expression are present in some myeloid cells (Extended Data Fig. 5k). To determine whether T cells were sufficient to mediate the efficacy of DR-18, we adoptively transferred T cells from either WT or Il8r1−/− donors into Rag2−/− recipients, which we engrafted with MC38 tumors and treated with DR-18 or vehicle. While transfer of WT T cells restored responsivity of Rag2−/− mice to DR-18 treatment, Il18r1−/− T cells conferred no benefit (Fig. 2f, Extended Data Fig. 5l). Furthermore, depletion of XCR1+ cDC1—cells essential for priming anti-tumor CD8+ T cell responses14—did not affect the efficacy of DR-18 during treatment (Extended Data Fig. 5m). In aggregate, these results indicate that DR-18’s direct activity on T cells is sufficient to drive anti-tumor responses.
DR-18 remodels the immune tumor microenvironment
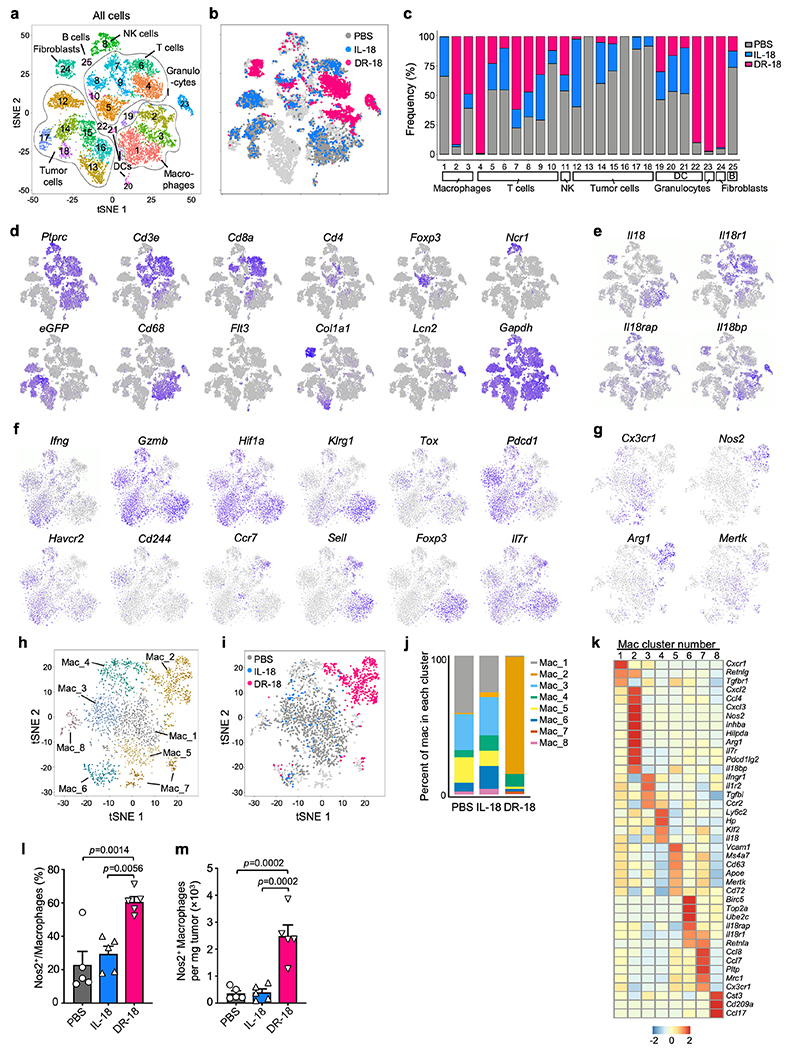
To investigate the consequences of DR-18 treatment on the TME, we performed single cell RNA sequencing (scRNA-seq) of YUMMER1.7 tumors after treatment with DR-18, mIL-18, or vehicle (see methods). Globally, DR-18 induced striking changes in clusters of lymphocytes, macrophages, granulocytes, and fibroblasts (Extended data Fig. 6a–e). Consistent with the antibody depletion studies above, Il18r1 and Il18rap expression was found predominantly on T cells and NK cells. By contrast, most Il18 and Il18bp transcripts were expressed in fibroblasts, granulocytes and macrophages (Extended Data Fig. 6e).
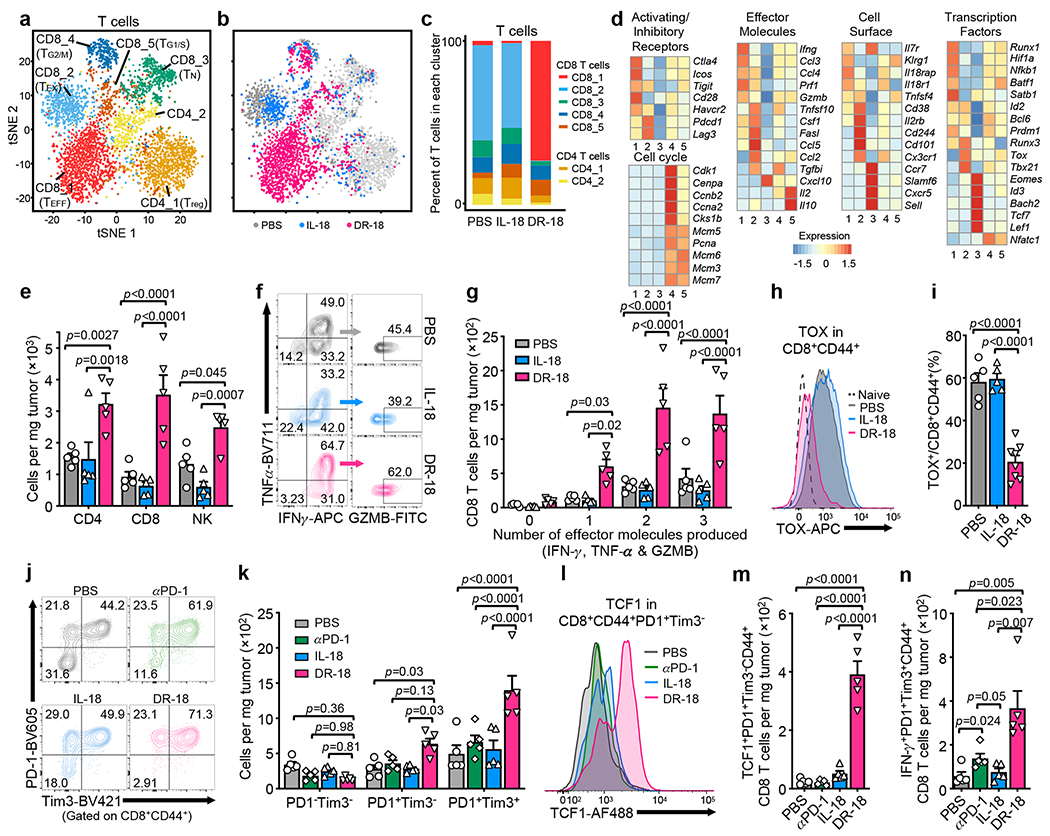
Analysis of Cd3e+ clusters yielded five CD8+ T cell clusters (CD8_1-5) and two CD4+ T cell clusters (CD4_1-2) (Fig. 3a). Cluster CD8_1 was exclusive to and predominant after DR-18 treatment, and was characterized by high levels of effector molecules (Ifng, Prf1, and Gzmb), cell surface proteins (Il8r1 and Klrg1), and co-stimulatory receptors (Fig. 3b–d, Extended Data Fig. 6f), consistent with an effector phenotype (TEFF)15,16. Conversely, most T cells from mIL-18 and vehicle treated tumors were found in cluster CD8_2, which exhibited high expression of the transcriptional regulator of exhaustion, Tox, and Cd101, Cd38, and Cd244 (Fig. 3b–d, Extended Data Fig. 6f), indicative of an exhausted (TEX) phenotype17–19. CD8_3 likely represented naïve-like T cells (TN), expressing lymphoid homing markers and high levels of the Tcf7. CD8_4 and CD8_5 showed expression of cell division genes and transcription factors Tcf7 and Id3, similar to stem-like precursor CD8+ T cells described in chronic viral infection and tumors20–22 (Fig. 3d). Overall, the two CD4 clusters were present at lower frequencies and were differentiated by the expression of Foxp3 in cluster CD4_1 (Fig. 3c; Extended Data Fig. 6f).
Figure 3. DR-18 treatment enhances T cell polyfunctionality and expands stem-like TCF1+ precursor CD8+ T cells.
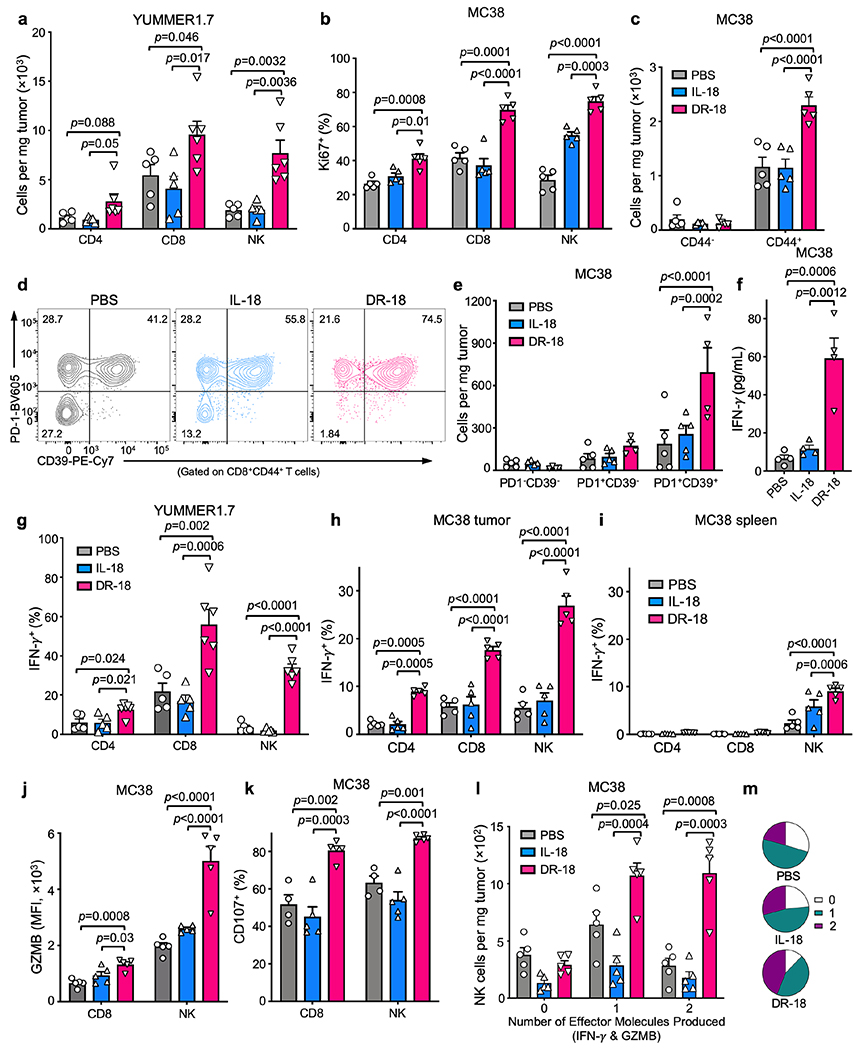
(a) t-SNE projection showing Cd3e positive cells present in the YUMMER1.7 and YUMMER1.7-B2m−/− tumors colored by cluster (see methods). (b) t-SNE projection showing distribution of Cd3e+ cells colored by treatment. (c) Bar plot showing frequency of T cells within each cluster as a function of treatment. (d) Heatmap showing mean expression level of curated gene list in CD8+ T cell clusters from (c). (a-d) Data is from one experiment with n=3 tumors pooled per condition. (e-n) WT mice were subcutaneously engrafted with MC38 tumors and treated twice weekly with either PBS, IL-18, anti-PD-1, or DR-18 day 7 after tumor implantation. Tumors were analyzed on day 15 by flow cytometry. (e) Absolute cell numbers of tumor-infiltrating CD4+ T cells, CD8+ T cells, and NK cells in each treatment group. (f) Representative flow plots and (g) absolute numbers of polyfunctional CD8+ T cells measured by the co-staining of intracellular IFN-γ, TNF-α, and Granzyme B following ex vivo stimulation with PMA/ionomycin. (h) Representative flow plots and (i) percent quantification of intracellular TOX staining in intratumoral CD44+ CD8+ T cells. (j) Representative flow plots and (k) absolute cell numbers of DN (PD-1−Tim3−), SP (PD-1+Tim3−) or DP (PD-1+Tim3+) intratumoral CD44+CD8+ T cells in each treatment group. (l) Representative flow plots and (m) absolute numbers of TCF1+ intratumoral SP CD8+ T cells. (n) Absolute numbers of IFN-γ+ intratumoral DP CD8+ T cells. (e-n) Data are representative of 3 independent experiments with n=5 mice per group and presented as the mean ± SEM. P values were calculated using two-tailed unpaired Student’s t-tests (e,i,m&n) or Two-way ANOVA (g&k).
DR-18 treatment also affected intratumoral myeloid populations (Extended Data Fig. 6g–j). Tumors from vehicle and mIL-18 treated mice primary contained immunosuppressive macrophages expressing Cx3cr1, Apoe, Mrc1 and Mertk23 and monocytes expressing Ccr2 and Ly6c (Extended data Fig. 6g,k). By contrast, DR-18 treated tumors predominantly contained cells from cluster Mac_2, which exhibited high expression of Nos2, Arg1, and Cd274, consistent with a proinflammatory IFN-γ activated signature. We confirmed this phenotype by flow cytometry (Extended Data Fig. 6l,m). Additionally, we observed an increased abundance of granulocytes following DR-18 treatment (Extended Data Fig. 6a,b), similar to effects previously observed with effective ICI treatment24.
DR-18 increases the TIL frequency and effector polyfunctionality
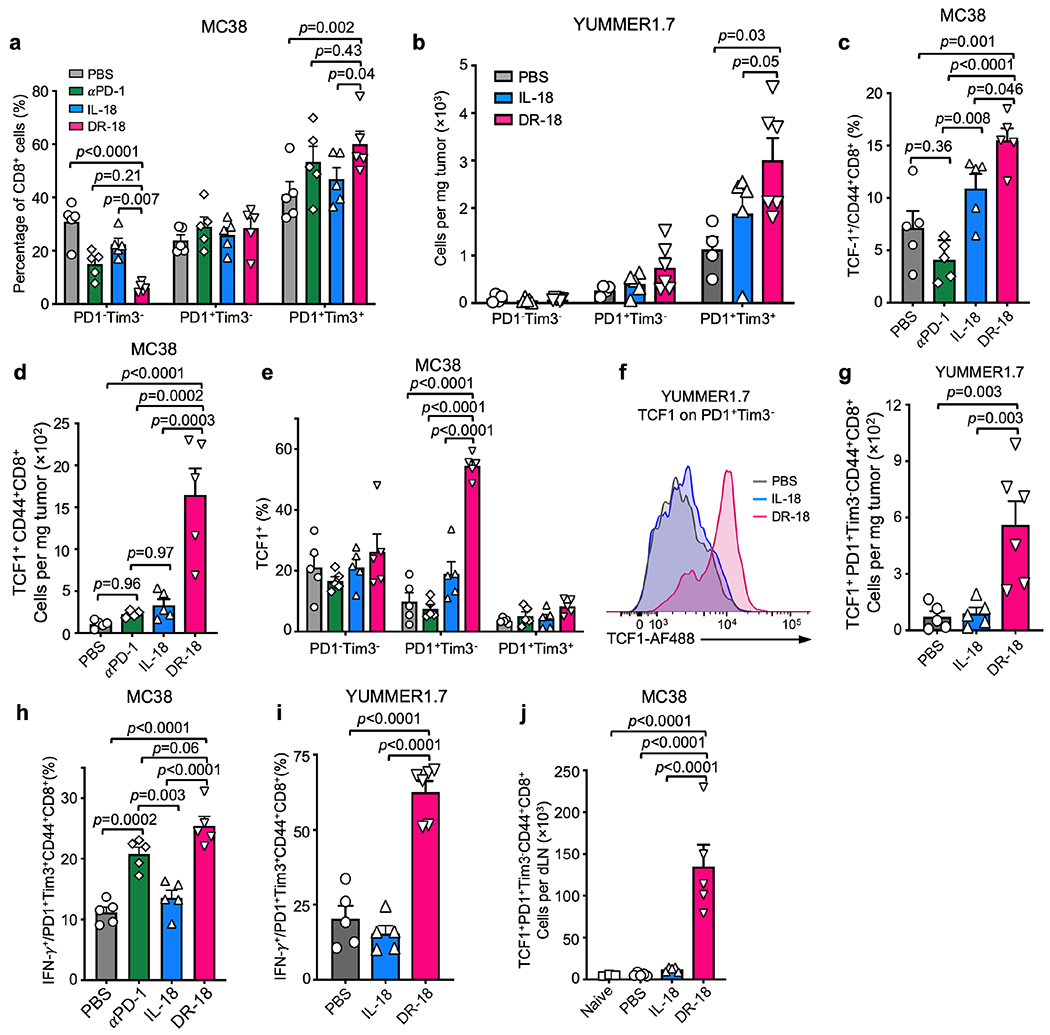
To validate the TIL phenotypes seen by scRNA-seq, we performed immunophenotyping of YUMMER1.7 or MC38 tumors from mice treated with vehicle, mIL-18, or DR-18. We observed an increase in the number and proliferative index (Ki67+) of CD4+ T cells, CD8+ T cells, and NK cells in tumors treated with DR-18 (Fig. 3e, Extended Data Fig. 7a,b), including an increase in CD44+ CD8+ T cells co-expressing CD39 and PD-1 (Extended Data Fig. 7c–e). Intratumoral CD4+ T cells, CD8+ T cells, and NK cells also showed enhanced IFN-γ, Granzyme B, and CD107a staining after DR-18 treatment (Extended Data Fig. 7f–k). Notably, tumors from DR-18 treated mice showed a substantial increase in the number of polyfunctional CD8+ T cells and NK cells (Fig. 3f,g; Extended Data Fig. 7l,m), a feature of an effective anti-tumor response17. Concordant with their increased polyfunctionality, CD8+ T cells from DR-18 treated tumors exhibited lower levels of TOX (Fig. 3h,i).
DR-18 treatment expands intratumoral stem-like TCF1+ CD8+ T cells
Durable immunotherapeutic responses require a stem-like CD8+ T cell precursor population that expresses TCF1 and PD-120,21,25,26. These stem-like cells differentiate into terminal effector cells, which lose TCF1 expression, acquire Tim3, have a greater functional capacity, but are limited in their ability to self-renew. In tumors following vehicle or mIL-18 treatment, the distribution of cells between the PD-1/Tim3 double-negative (DN), PD-1 single-positive (SP), and PD-1/Tim3 double-positive (DP) populations was similar. DR-18 treatment, and to a lesser extent anti-PD-1, shifted the distribution towards the SP and predominantly DP fractions (Fig. 3j,k; Extended Data Fig. 8a,b). Analysis of TCF1 expression revealed that DR-18—but not vehicle, mIL-18, or anti-PD-1— greatly expanded the number of intratumoral CD8+ CD44+ TCF1+ cells, specifically in the SP population (Fig. 3l–m, Extended Data Fig. 8c–g). In the DP population, DR-18 did not affect TCF1 levels but did increase IFN-γ expression, as did anti-PD-1 treatment (Fig. 3n, Extended Data Fig. 8h–i). Interestingly, we observed an expansion of both TCF1+ precursor and PD-1+CD44+ CD8+ T cells in the tumor draining lymph node (tdLN) (Extended Data Fig. 8j,k), suggesting that DR-18 expands a reservoir of stem-like and activated T cells outside the tumor.
DR-18 is efficacious against ICI-refractory tumors that lack surface MHC class I
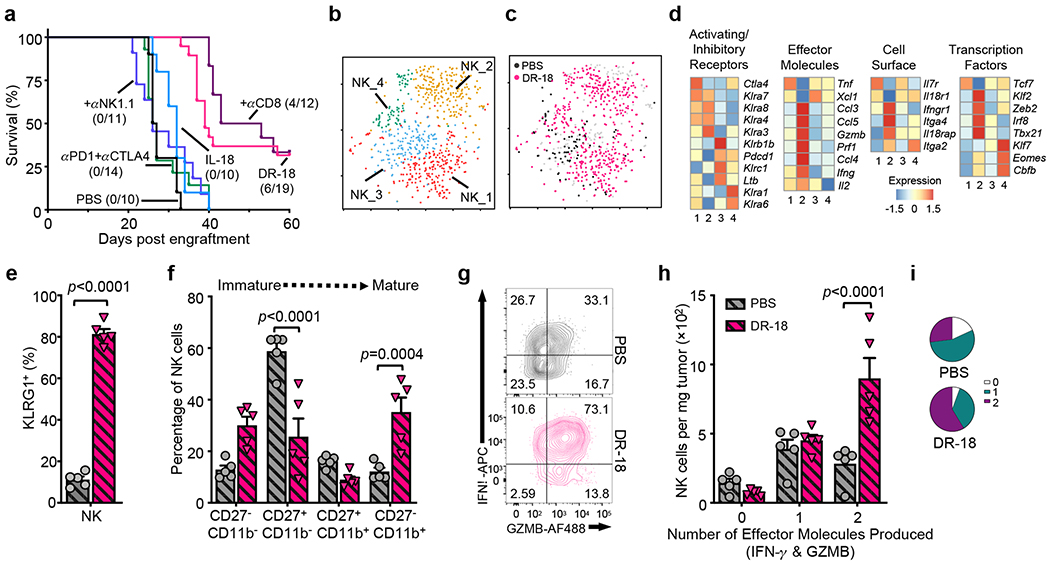
Downregulation or loss of MHC class I is a common feature of human cancers and a resistance mechanism to immunotherapy27. Though MHC class I loss renders tumor cells sensitive to NK mediated cytotoxicity, NK cells readily become exhausted in the TME28. We wondered if DR-18 treatment could restore NK cell function in MHC class I deficient tumors which are refractory to treatment with ICI (Fig. 4a, Extended Data Fig. 9a–e). In MC38-B2m−/−, YUMMER1.7-B2m−/− and RMA-S (tapasin deficient) tumors, DR-18 treatment resulted in tumor regression in 30-75% of mice (Fig. 4a, Extended Data Fig. 9a–e). Antibody depletion studies indicated that NK1.1+ cells were essential for the anti-tumor activity of DR-18 in this setting (Fig. 4a, Extended Data Fig. 9b,d). Analysis of the intratumoral NK1.1+ population after treatment revealed a significant increase in the number of NK cells, with minimal NK/T or ILC1 cells present (Extended Data Fig. 9f).
Figure 4. DR-18 promotes NK cell maturation and polyfunctionality to treat MHC class I deficient tumors.
(a) Kaplan-Meier survival curve of WT mice engrafted s.c. with MC38-B2m−/− tumors and treated twice weekly with PBS, anti-PD-1 plus anti-CTLA-4, IL-18, DR-18, or DR-18 plus anti-CD8 and anti-NK1.1 antibodies. (b,c) t-SNE projection showing Ncr1+ positive cells present in the YUMMER1.7 and YUMMER1.7-B2m−/− tumors colored by (b) unique cluster and (c) by treatment. (d) Heatmaps showing mean expression of curated genes for each of the NK cell clusters shown in (b). (e,f) Quantification of surface (e) KLRG1 and (f) CD27/CD11b staining by percentage on intratumoral NK cells from the indicated treatment groups. (g) Representative flow plots, (h) absolute numbers and (i) frequency of intratumoral polyfunctional NK cells measured by co-staining of intracellular IFN-γ and Granzyme B following ex vivo stimulation with PMA/ionomycin. Data are representative of 3 independent experiments with n=5 mice per group and presented as the mean ± SEM. P values were calculated using two-tailed unpaired Student’s t-tests (e) and Two-way ANOVA (f,h).
scRNA-seq profiling of DR-18 and vehicle treated YUMMER1.7-B2m−/− tumors identified four clusters of Ncr1+ cells (NK_1-4) (Fig. 4b, Extended Data Fig. 9g, see methods). DR-18 treatment uniquely induced NK_2 cluster cells (Fig. 4b,c), which exhibited increased expression of effector molecules (Ifng, Prf1, Gzmb), activating transcription factors (Irf8, Zeb2, Cbfb), and a distinct Ly49 repertoire (Fig. 4d). Flow cytometry analysis showed an increase in NK cell maturation with enhanced proliferative and polyfunctionality following DR-18 treatment (Fig. 4e–i, Extended Data Fig. 9h–j). To probe the mechanism of NK-mediated tumor killing elicited by DR-18, we found that administration of neutralizing IFN-γ antibodies completely ablated DR-18 anti-tumor responses, whereas FasL antagonist antibodies or Perforin deficiency had no effect (Extended Data Fig. 9k,l).
Human IL-18 can be engineered for IL-18BP resistance
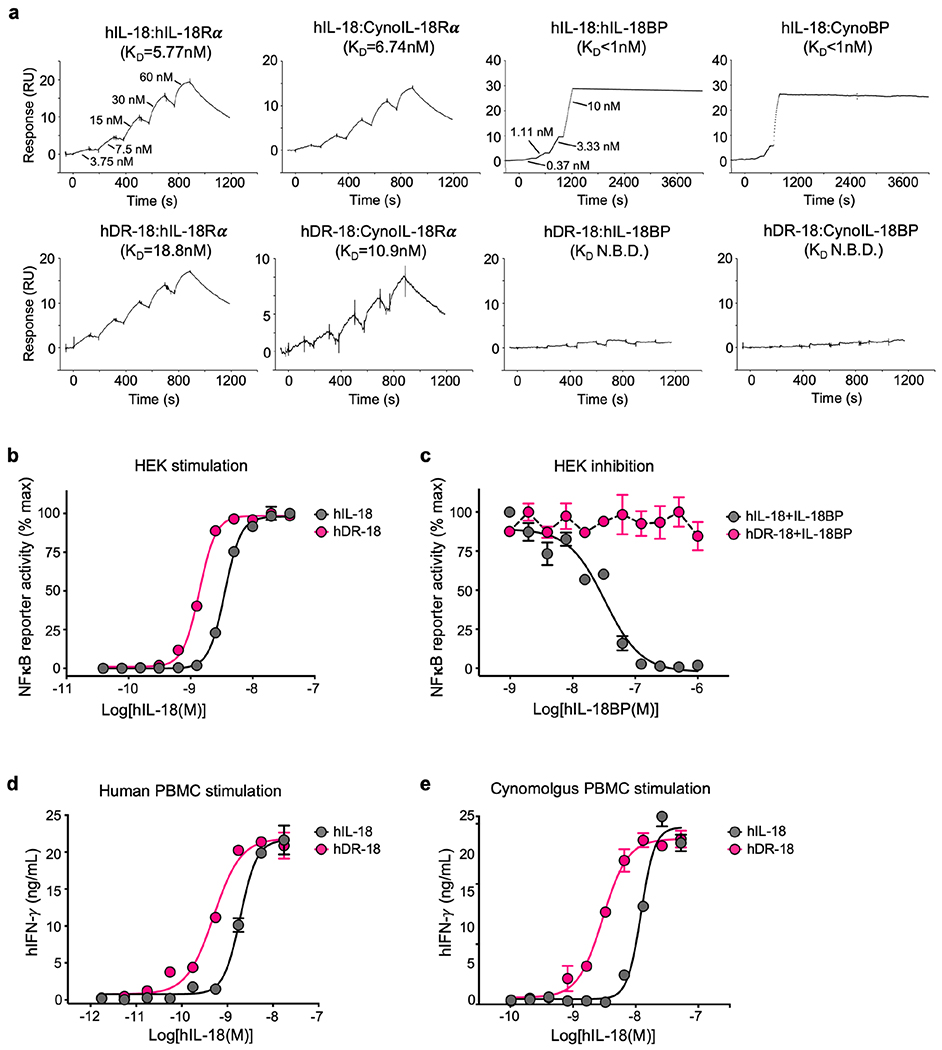
To assess the feasibility of translating the DR-18 approach to human IL-18, we developed a functional human (h) DR-18 through a parallel directed evolution process. hDR-18 demonstrated strong binding by SPR to IL-18Rα, but not IL-18BP, for both human and cynomolgus (Macaca fascicularis) proteins (Extended Data Fig. 10a). Similarly, hDR-18 stimulated NFκB signaling in hIL-18 reporter cells, but was not inhibited by IL-18BP (Extended Data Fig. 10b,c). Finally, we found that hDR-18 elicited IFN-γ production in both human and cynomolgus peripheral blood mononuclear cells (PMBCs) (Extended Data Fig. 10d,e).
Discussion
The powerful activity of DR-18 in mouse tumor models highlights the potential of the IL-18 pathway to enhance anti-tumor immunity. DR-18 expands stem-like TCF1+ precursor CD8+ T cells and biases their differentiation towards polyfunctional TEFF and away from TOX+ TEX. This mechanism appears distinct from the effect of blocking PD-1, which we found augments the function of TEX, but does not significantly affect the numbers of stem-like CD8+ T cells, similar to prior reports21. DR-18 also promotes NK cell responses against ICI-refractory tumors that have lost MHC class I expression, a major resistance mechanism that is not addressed by currently approved immunotherapies.
The efficacy of DR-18 therapy contrasts with prior inferences about IL-18 biology made from mouse genetics and pharmacologic studies with rIL-18. IL-18 is generally not required for tumor immunosurveillance29 and some reports have implicated tumor-promoting roles for IL-1830,31,32,33. However, pleiotropy is a common feature of cytokines, and factors such as dose, schedule, and site-of-action can greatly affect their biological activity. For example, low doses of IL-2 can be used to therapeutically expand immunosuppressive Treg34, yet high-doses stimulate CD8+ T cells for tumor immunotherapy35. Thus, our results do not necessarily contradict these previous findings, but rather highlight aspects of IL-18 pleiotropism that can be optimally tuned for cancer immunotherapy.
Dysregulation of IL-18 is associated with numerous autoinflammatory diseases, which are characterized by increases in systemic concentrations of free IL-18 relative to IL-18BP10. Similarly, a case of biallelic loss of IL18BP was recently associated with fulminant viral hepatitis36. It is therefore conceivable that DR-18 therapy may exhibit more significant toxicities than was seen previously in clinical trials of rIL-18. Nevertheless, our preclinical studies thus far indicate that DR-18 is efficacious and well-tolerated across a wide range of bioactive doses from 0.001 to 1.0 mg/kg.
In summary, our results highlight a crucial role of IL-18BP as a secreted immune checkpoint that fundamentally alters the biological effects of IL-18. The unique mechanism of DR-18 to act on CD8+ TEFF, stem-like TCF1+ CD8+ T cells, and NK cells provides a strong rationale for the clinical development of DR-18 and other IL-18 receptor agonists.
Methods
Patient specimens
Human plasma samples were collected at the University of Navarra in Pamplona, Spain, from two cohorts: cohort #1 included 22 healthy donors and cohort #2 included 52 non-small cell lung cancer (NSCLC) patients who were treated with anti-PD-1 or anti-PD-L1 monoclonal antibodies. Samples from NSCLC patients were collected sequentially at the time of first visit to the clinic (baseline) and after starting anti-PD-1 treatment (approximately 2-3 weeks later). Complete baseline and treatment characteristics of the NSCLC patient are summarized in Supplementary Table 1. The study protocols were approved by the Institutional Review Board at the University of Navarra (approval 11/2010) and all patients provided written informed consent.
Mice
Mice were maintained at Yale University in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC). The following mouse strains were used this study: WT C57BL/6 (Charles River, #C57BL/6NCrl or Jackson Labs, C57BL/6J #000664), Balb/c (Charles River, # BALB/cAnNCrl), Il18bp−/− and Il18r1−/− (Yale, R. Flavell), Rag2−/− (Yale, D. Schatz), XCR1DTR and TCRP14 Transgenic (Yale, A. Iwasaki) and Perforin−/− (Yale, M. Bosenberg). Experiments were conducted using age- and gender-matched mice in accordance with approved institutional protocols.
Cell Lines
The following tumor cell lines were used in this study: YUMMER1.7 (Yale, M. Bosenberg)37, MC38 (Yale, M. Bosenberg), RMA-S cell line (Yale, C. Rothlin)28, CT-26 (ATCC, CRL-2638), B16-F10 (ATCC, CRL-6475) and B16-F10GP33 (Yale, A. Iwasaki). YUMMER1.7 cell line was cultured in DMEM/F12 (Thermo Fisher, #11320-033) including L-glutamine and 2.438g/L sodium bicarbonate and supplemented with 1 x non-essential amino acids (Thermo Fisher #11140050), 10 % fetal bovine serum (Thermo Fisher, #16140-071) and 1x Penicillin and Streptomycin (Thermo Fisher, #15140-122). All other cell lines were cultivated in RPMI-1640 (Thermo Fisher, #21870-076) supplemented with 10 % fetal bovine serum and 1x Penicillin and Streptomycin. All cells were cultured at 37 °C, 5% CO2 and kept at low passage (~3-5 passages) once obtained from vendors or collaborators.
MC38-B2m−/− and YUMMER1.7- B2m−/− cell lines were generated with CRISPR/Cas9 technology. Briefly, B2m guides (forward: CACCGAGTATACTCACGCCACCCAC; reverse: AAACGTGGGTGGCGTGAGTATACTC) targeting exon 2 of the B2m gene were cloned into a Cas9-single guide RNA (sgRNA) expression vector (PX458, Addgene #48138) with a GFP reporter. The plasmid was transiently transfected into each parental line using Lipofectamine 3000 (Fisher Scientific, #L3000-001) according the manufacturer’s instructions. 48 hours after transfection, GFP positive cells were sorted (BD FACS Aria) as single cells into 96-well plates. B2m deletion was confirmed separately in each clone using flow cytometry showing lack of MHC class I surface staining following 12 hours of in vitro incubation with recombinant murine IFN-γ (Peprotech Cat #315-05) at 100 U/mL. Single cell derived clones of each line that exhibited similar morphology, in vitro growth characteristics, and in vivo tumor formation characteristics to the parental lines were selected for experiments.
Stable H2B-GFP expressing YUMMER1.7 and YUMMER1.7-B2m−/− lines were generated and used for single cell RNA sequencing experiments. Briefly, lentivirus was produced using pCMV-VSV-G (Addgene #8454) and psPAX2 (Addgene, #8454) with an H2B-GFP plasmid (Addgene, #25999) in 293T cells. 293T cells were transfected with 10 μg of a 1:2:4 (VSV-G: PAX-2: H2B-GFP) mixture of plasmid DNA in X-treme GENE 9 transfection reagent (Sigma, #6365779001) according to the manufacturer’s instructions in Opti-MEM (Thermo, #31985088). After overnight incubation, the media was replaced with normal growth media (DMEM) and incubated for 48 hours; at which time the media was harvested and filtered through a 0.45 μM filter. Untitered viral supernatant was applied to YUMMER1.7 and YUMMER1.7-B2m−/− lines for 48 hours. Single GFP+ tumor cells were sorted into individual wells of 96 well plates by FACS (BD FACS Aria). Single cell derived clones of YUMMER1.7 and YUMMER1.7-B2m−/− expressing GFP that exhibited similar morphology, in vitro growth characteristics, an in vivo tumor formation characteristic to the parental lines were selected for experiments.
ELISA
IL-18BP and IFN-γ ELISAs were performed using Human IL-18 BPa Quantikine ELISA Kit (R&D system, #DBP180), mouse IL-18BPd DuoSet ELISA kit (R&D system, DY122-05), mouse IFN-γ ELISA MAX™ Deluxe kit (Biolegend, #430804), human IFN-γ Quantikine ELISA Kit (R&D system, #DIF150), and primate IFN-γ DuoSet ELISA (R&D system, #DY961) according to the manufacturers’ instructions.
Immunohistochemistry
Human tumor tissue microarrays (TMAs) were obtained from the Yale Tissue Microarray Facility. IL-18BP immunohistochemical staining of the TMAs was performed by the Yale Dermatopathology laboratory using anti-IL-18BP antibody clone EP1088Y (Abcam) and was previously validated38,39. The melanoma TMA was stained with Azure blue so that melanin (turned green) could be differentiated from the DAB chromagen (brown). All scorable tumor cores were included in this analysis. Melanoma TMA (YTMA-192) contained 282 scorable tumor cores. Breast cancer TMA (YTMA-353) contained 114 scorable tumor cores. Head and neck cancer TMA (YTMA-305) contained 76 scorable tumor cores. Gastric cancer TMA (YTMA-141) contained 62 tumors scorable tumor cores. Ovarian TMA (YTMA-264) contained 226 scorable tumor cores. Where available, cell lines and normal tissue on the TMAs were used as controls. Scoring was performed by a board-certified pathologist (M. Bosenberg) in a blinded fashion. Cores were scored as negative (0) or positive (either 1+, 2+, or 3+).
Mouse IL-18BP immunohistochemical staining was performed on Il18bp−/− spleen, WT spleen (IL-18 treated) and tumor (MC38) using anti-IL-18BP antibody clone EP1088Y (Abcam). Tissue was fixed in 4% PFA overnight on ice. Post-fixation samples were embedded in paraffin and sectioned at 5 μm prior to staining. The number of IL-18BP positive cells per high power field was quantified in representative sections from each condition.
mRNA quantification
Whole blood and tumor samples were harvested in Trizol and total RNA was extracted using the RNeasy kit (Qiagen, #Q74104) according to the manufacturer’s instructions. The total RNA was reverse transcribed using Oligo(dT) primers and Maxima H Minus Reverse Transcriptase (Thermo Fisher, #EP0752). Il18bp expression was assayed by real-time PCR using iQ SYBR® Green Supermix (Bio-rad, #1708880) with primers (forward: GAGGGCCACACAAGTCGC; reverse: GCTGGGCCAGAATGATGTGA) on an ABI 7900HT real-time PCR system. Actb (β-actin) was used as a control (forward: AGGTGACAGCATTGCTTCTG; reverse: GCTGCCTCAACACCTCAAC). All values were normalized to expression levels in naive mouse spleen. Measurements were performed in triplicate for each sample.
Protein expression, purification and biotinylation
The mature form of murine IL-18 (amino acids 37-193) and associated variants were assembled as gene blocks (Integrated DNA Technologies, IDT) and cloned into a pTZ-SH vector for expression of N-terminal SUMO-tagged and C-terminal hexahistidine-tagged proteins in E. coli BL21 (DE3) Rosetta strain (Fisher Scientific, #70954-3). Protein expression was induced with 0.5 mM IPTG at 16°C for 20 hr. The fusion proteins were first purified using a Ni-NTA column (Fisher Scientific, #P188223), followed by removal of the SUMO tag with the SUMO protease Ulp1. Afterward, the protein solution was buffer-exchanged to remove imdizaole and re-applied to a second Ni-NTA column to remove the free SUMO tag. The eluted protein was concentrated and separated from aggregates by gel-filtration (Column SEC70, Bio-rad). Protein from the mono-disperse peak was pooled and loaded on a final Ni-NTA column for endotoxin removal with 0.2% Triton X-114 at 4°C. Finally, the eluted protein was buffer exchanged into sterile, endotoxin free PBS using a PD-10 column (GE Healthcare) and flash-frozen in liquid nitrogen for long-term storage at −80°C.
The mouse IL-18Rα ectodomain (amino acids 19-329) and IL-18 binding protein (IL-18BP, amino acids 31-194) were expressed by transient transfection of Expi293 cells (Thermo Fisher). Both sequences were cloned into the pEZT_D_Lux vector with an N-terminal H7 signal peptide and a C-terminal AviTag and hexahistidine tag. Plasmids were transfected into Expi293 cells by ExpiFectamine 293 Transfection Kit (Thermo Fisher, #A14524) per the manufacturer’s instructions. Cells were harvested 3-5 days after transfection. Proteins were captured from cell supernatant via Ni-NTA chelating resin and further purified by size exclusion chromatography (Column SEC70, Bio-rad) into a final buffer of HEPES buffered saline (HBS; 10 mM HEPES, pH 7.5, 150 mM NaCl).
For protein biotinylation, proteins were expressed with a C-terminal biotin acceptor tag (AviTag)-GLNDIFEAQKIEWHE. After Ni-NTA chromatography, protein biotinylation was carried out at room temperature for 2 hours with soluble BirA ligase enzyme in 0.1 mM Bicine (pH 8.3), 10 mM ATP, 10 mM magnesium acetate, and 0.5 mM biotin (Avidity, #BIO500). Biotinylated proteins were then purified by gel-filtration as described above. Biotinylation efficiency was assessed using an SDS/PAGE streptavidin-shift assay.
Yeast display of IL-18
The nucleotide sequence of the mature form of murine IL-18 (amino acids 37-193) was cloned into an N-terminal displayed vector, pYAL, and displayed on the surface of yeast strain Saccharomyces cerevisiae EBY100. Yeast were maintained and expanded in liquid synthetic dextrose medium with casamino acids (SDCAA) at 30°C and then induced to express IL-18 in liquid synthetic glucose medium with casamino acids (SGCAA) at 20 °C for 24-48 hours. The displayed protein level was verified by staining with a C terminal Myc tag antibody (Cell Signaling Technology, #3739S). Biotinylated IL-18Rα and IL-18BP binding was detected using a fluorescent streptavidin phycoerythrin secondary and quantified by flow cytometry using a Sony SA3800 flow cytometer.
Mouse IL-18 library construction and selection
Thirteen residues in IL-18 which were in contact with both IL-18Rα and IL-18BP were identified by aligning the structure of hIL-18:hIL-18Rα:hIL-18Rβ complex [Protein Data Bank (PDB) ID 3WO4] to the structure of hIL-18:vIL-18BP complex (PDB ID 3F62). A library randomizing these residues was constructed using assembly PCR with the degeneration primers. The PCR products were further amplified with primers containing homology to the pYAL vector and co-electroporated into EBY100 competent yeast together with linearized pYAL vector. The resulting library was later measured to contain 4.0 × 108 transformants.
Transformed yeast were recovered and expanded in SDCAA medium at 30 °C, induced by 1:10 dilution into SGCAA medium and cultured at 20 °C for 24-48 h. The appropriate numbers of induced yeast were used in each round to ensure at least 10-fold coverage of the expected diversity, and not less than 108 cells. All selection steps were carried out at 4 °C using PBE buffer (PBS with 0.5% BSA and 2 mM EDTA). For round 1, the yeast library was counter-selected with anti-Cy5/Alexa Fluor 647 microbeads (Miltenyi, #130-091-395) with a LS MACS column (Miltenyi, #130-042-401) to remove non-specific binders. Positive selection was performed by labeling yeast with 1μM biotinylated IL-18Rα, followed by magnetic selection with Alexa Fluor 647 microbeads and the LS MACS column. For round 2, counter-selection reagent was changed to 1μM biotinylated IL-18BP while the IL-18Rα concentration was kept at 1μM. For rounds 3-5, selection was performed by incubating yeast with Alexa Fluor647-conguated IL-18Rα at concentrations of 100 nM (round 3), 100 nM (round 4), or 10 nM (round 5) in the presence of 250 nM pre-formed and biotin-capped IL-18BP:SA-PE tetramers. IL-18 display levels were determined by staining with Alexa Fluor 488-conjugated anti-Myc (Cell Signaling Technologies, #2279S). Yeast were selected by FACS sorting with a Sony SH800 cell sorter by excluding IL-18BP (PE) binders and gating the top 1% of display-normalized IL-18Rα binders. After each round of selection, recovered yeast were expanded in SDCAA medium at 30 °C overnight and later induced at 20 °C by a 1:10 dilution into SGCAA medium for 24-48 h.
Surface Plasmon Resonance
SPR experiments were conducted using a Biacore T100 and carried out at 25 °C. Interactions were measured using either conventional multiple-cycle programs or a single-cycle kinetics program. Mouse, human, or cynomolgus biotinylated IL-18Rα or IL-18BP were immobilized onto a Biacore biotin capture chip (Series S CAP sensor chip, GE Healthcare) to yield a Rmax of ∼50 RU (IL-18Rα) or ~10 RU (IL-18BP). Measurements were made with half-log dilutions of the IL-18 variants in HBS-P+ buffer (10 mM Hepes pH 7.4, 150 mM NaCl, 0.005% surfactant P20). The surface was regenerated by three 60-s injections of regeneration buffer [3/4 (v/v) 8M guanidine hydrochloride +1/4 (v/v) 1M sodium hydroxide]. Experiments were performed in multiple channels for duplicate measurements (F2-1 and F4-3). All data were analyzed with the Biacore T100 evaluation software version 2.0 with a 1:1 Langmuir binding model.
Isolation of lymphocytes
Spleens were dissociated using a 70 μM cell strainer and red blood cells were lysed using ACK buffer (Thermo Fisher, #A1049201). Lymph node were dissociated using a 70 μM cell strainer. Tumors were cut into small pieces in the presence of 3 mL RPMI-1640 supplemented with 1% FBS. Collagenase IV (Gibco, #17104019, final concentration 1mg/ml) and Dnase I (Roche, #10104159001, final concentration 0.2mg/ml) were added and samples were incubated at 37°C for tissue digestion. After 30 minutes of digestion, 6 mL of RPMI-1640 with 10% FBS was added to neutralize protease activity and tumor tissues were forced through 70 μM cell strainers to prepare single-cell suspensions. Cells were then washed twice and resuspended in 1-3 mL of RPMI-1640 with 1% FBS media for downstream analysis. Cell concentrations were counted by a Beckman Coulter particle counter.
Ex vivo stimulation of splenocytes and PBMCs
For the mouse IL-18/DR-18 stimulation assay, approximately 1 × 106 mouse splenic lymphocytes were stimulated ex vivo with a range of concentrations of mouse IL-18 or DR-18 from 0.00316 to 316 ng/ml. For the mouse IL-18BP inhibition assay, 1 × 106 splenic lymphocytes were stimulated ex vivo with a fixed concentration of IL-18 (1 nM) or DR-18 (0.5 nM) and range of IL-18BP concentrations (0.01 to 100 nM). All stimulations were performed in the presence of mIL-12 (10 ng/ml, Peprotech, #210-12) and protein transport inhibitor (1:200, BD Golgiplug, #554724) for 4 hours in RPMI containing 10% fetal bovine serum at 37°C. Cells cultured with IL-12 alone were used as negative controls, and PMA/Ionomycin was used as a positive control. After 4 hours, IFN-γ expression was measured on NK cells by flow cytometry, gating on CD3−/NK1.1+. Data were normalized by subtracting the background MFI from the unstimulated control and defining the largest mean value as 100%.
For human and cynomolgus PBMC stimulation assays, approximately 0.6 × 106 human (PPA, #15-00012) or 0.3 × 106 cynomolgus (BioIVT, #M3-010-C20M) PBMCs were stimulated ex vivo with gradient of human IL-18 or DR-18 ranging from 0.0316 to 1000 ng/ml. All stimulations were performed in the presence of hIL-12 (10ng/ml, Peprotech, #200-12H) for 12 hours in RPMI containing 10% fetal bovine serum. IFN-γ production after stimulation were measured by corresponding human and cynomolgus ELISA kits per the manufacturers’ instructions.
Human IL-18 reporter assay
The IL-18 HEK-Blue assay (InvivoGen, #hkb-hmil18) was performed according to the manufacturer’s instructions. Briefly, 5× 104 cells were seeded into each well of a 96 well plate and stimulated with 0-100 ng/mL of hIL-18 or DR-18 for 24 hours at 37 °C and 5 % CO2. 40 μL of cell culture supernatant was then taken from each well and mixed with 160 μL QUANTI-Blue media in a 96 well plate, incubated for 3 hours at 37 °C and 5 % CO2, and then read in a microplate reader at 655 nm.
Tumor treatment studies
The number of tumor cells engrafted were as follows: 0.5×106 MC38 cells, 0.5×106 YUMMER1.7 cells, 0.5×106 MC38-B2m−/− cells, 1×106 YUMMER1.7-B2m−/− cells, 1×106 RMA-S cells, 0.25×106 CT26 cells, 0.25×106 B16-F10 cells, and 0.25×106 B16-F10GP33 cells. Tumors were engrafted subcutaneously into the flanks of 8-10-week-old age matched female or male mice. YUMMER1.7 and YUMMER1.7-B2m−/− were only implanted into male mice, as this cell line was derived from a male mouse. Treatment was initiated when mean tumor size was between 50-100 mm3 (usually at day 7 post engraftment); mice with tumors less than 30mm3 or greater than 150 mm3 were excluded from randomization. Remaining mice were randomized into designated groups to ensure an approximately equal average tumor size. Mice were then treated with the designated test articles by intraperitoneal injection twice weekly for a total of 3-5 doses (as indicated in the text). Pilot dose-finding studies with MC38 tumors indicated that DR-18 treatment resulted in tumor growth inhibition (TGI), tumor regression, and clearance at doses as low as 10 μg/kg and at schedules as infrequent as administration once every two weeks, with 0.32 mg/kg given bi-weekly representing the maximally efficacious regimen. Test articles were diluted in sterile PBS and dosed as follows: anti-PD-1 (RMP1-14, Bio X Cell) 8 mg/kg, anti-CTLA-4 (9H10, Bio X Cell) 8 mg/kg, IL-18 0.32 mg/kg and DR-18 0.32 mg/kg administered twice weekly. Control groups were treated with sterile PBS or isotype control antibodies. Tumor growth was tracked twice weekly by caliper measurement. Tumor volume was calculated using volume = 0.5×length×width×width. Mice were euthanized when tumors reached endpoints [volume greater than or equal to 1000 mm3(MC38, CT26, and B16-F10) or 500mm3 (YUMMER1.7 and RMA/S), or volume greater than or equal to 500 mm3 (MC38, CT26, and B16-F10) or 250mm3 (YUMMER1.7 and RMA/S) plus tumor ulceration]. Survival analyses reflect this endpoint.
For immune cell depletion (CD4/CD8/NK) and effector molecule neutralization (IFN-γ/FasL) studies, CD8a (2.43, Bio X cell or TIB210, Bio X cell), CD4 (GK1.5, Bio X cell), NK1.1 (PK136, Bio X Cell), IFN-γ (R4-6A2, Bio X or XMG1.2, Bio X cell), and FasL (MFL3, Bio X cell) antibodies were used. Antibodies were administered by intraperitoneal injection starting on day 6 (one day prior to therapy initiation) and were continued twice weekly for the duration of the experiment. 8 mg/kg per treatment was used for all depleting antibodies. Lymphocyte depletions were confirmed in peripheral blood lymphocytes by flow cytometry with the following antibodies CD8a (53-6.7), CD4 (RM4-5) and NKp46 (29A1.4).
For tumor re-challenge studies, mice exhibiting complete tumor regression as a result of DR-18 treatment were re-inoculated subcutaneously with twice the initial dose of MC38 tumor cells (1×106) 30 days after the initial tumors were cleared. As a control, naive C57BL/6J mice were challenged at the same time. Tumor growth and survival were monitored twice weekly as stated above for up to 60 days.
For ablation of cDC1 studies, WT and XCR1DTR were injected i.p, with 25 ng Diphtheria Toxin (DT) (#150, List Biological Lab) per gram of body weight on day 6 post tumor engraftment. To maintain DT ablation, mice received 100 ng DT per gram of body weight twice weekly after initial DT injection. cDC1 depletion was confirmed by flow cytometry.
For FTY720 experiments, FTY720 (#S5002, Selleck Chemicals) was reconstituted in water (10 mg/mL) and diluted in PBS. WT mice were treated i.p. with 3 mg/kg starting on day 6 (one day prior to therapy initiation) and continued twice weekly together with DR-18 treatment for the duration of the experiment. FTY720 efficacy was confirmed by measuring the reduction of CD3+ T Cells in the blood.
Adoptive transfer experiments
CD3+ Naïve T cells were purified from the spleen and lymph nodes of WT or Il18r1−/− mice using a mouse pan-naïve T cell isolation kit (Stemcell, #19848) by magnetic based bead sorting. Purified T cells were then transferred into Rag2−/− mice (9 million cells per recipient) by retro-orbital intravenous injection. Tumor treatment studies were then conducted as described above.
Immunophenotyping of tumor engrafted mice
Mice with MC38, MC38-B2m−/−, YUMMER1.7, and YUMMER1.7-B2m−/− tumors for immunophenotyping were engrafted and treated the same way as for tumor growth studies. Mice were euthanized 24 hours after the 3rd dose of saline, anti-PD-1, IL-18, or DR-18 (day 15 post engraftment), or at indicated time point, and spleens, draining lymph nodes, and tumors were harvested for analysis. Tumors were dissociated for analysis as previously described. Cell surface staining of single-cell suspensions from spleens and tumors was performed using fluorophore-conjugated antibodies (BD Biosciences, eBioscience, BioLegend, Tonbo, & R&D Systems; see below). All samples were stained with Zombie Yellow (Biolegend, #423104) to identify dead cells and incubated with Fc receptor blocking antibody (Fisher Scientific, #553142). Intracellular staining was performed by fixing and permeabilizing with the eBioscience Foxp3/Transcription Factor Staining Set (Thermo Fisher, #00-5523-00) or Cytofix/Cytoperm kit (BD Biosciences # 554714). For endogenous cytokine production measurement, single cell suspension of splenocytes and tumor cells were incubated with Golgi-plug (1:500, BD Biosciences, # 555029) for 4 hours at 37 °C. For polyfunctional cytokine production, single cell suspensions of splenocytes and tumor cells were stimulated with a PMA/Ionomycin cocktail (1:200, Biolegend, # 423304) and incubated for 4 hours at 37 °C. Cell cultured in media alone (without stimulation) was used as a negative control.
Cell staining was performed using the following fluorophore-conjugated antibodies: CD45 (30-f11), CD8α (53–6.7), CD4 (RM4-5), TCRβ (H57-597), CD3e (17A2), CD19 (ID3), NK1.1 (PK136), IL-18Rα (P3TUNYA), B2m (A16041A), Ki67 (16A8), KLRG1 (2F1), IFN-γ (XMG1.2), GZMB (QA16A02), TNFα (MP6-XT22), CD107a (1D4B), PD-1 (RMP1-30 & 29F.1A12), Tim3 (RMT3-23), CD44 (IM7), CD62L (MEL-14), CD103 (2E7), CD11b (M1/70), CD27 (LG.3A10), Eomes (Dan11mag), Foxp3 (FJK-16s), CD200R1 (OX-110), CD49b/DX5 (DX5), F4/80 (BM8.1), CD64 (X54-5/7.1), CD11c (N418), Ly6C (HK1.4), Ly6G (1A8), I-A/I-E (M5/114.15.2), CD90.1 (OX-7), CD39 (24DMS1), NOS2 (CXNFT), TCF1 (C63D9) and TOX (REA473). Fluorescence spectra were acquired using a LSRII flow cytometer and analyzed by FlowJo (Version 10). For flow cytometry analysis, naive mice splenocytes or fluorescent minus one (FMO) staining was used for gating.
Single cell RNA sequencing sample preparation
Biological replicates from n=3 pooled mice were processed for each experimental condition from YUMMER1.7 and YUMMER1.7-B2m−/− tumors. Biological replicates were then pooled together at the single cell suspension stage with equivalent number of cells from each replicate. The following populations were sorted purified: P1: GFP−CD45+CD3+ (T cells), P2: GFP−CD45+CD3− (non-T immune cells), P3: GFP+/−CD45−CD3− (tumor and stromal cells). P1, P2, and P3 for each sample were then mixed back together at a 2:1:1 ratio, respectively. 5000 cells from each of the mixed sorted samples for each condition were loaded onto the 10x Genomics Chromium System. Library preparation was performed using 10x Genomics reagents according to the manufacturer’s instructions and was performed by the Yale Center for Genome Analysis (YCGA) and passed QC. Libraries were sequenced using an Illumina HiSeq 4000 (one library/lane) at the YCGA.
Single cell RNA sequencing analysis
Samples were processed using the Cellranger software suite commands cellranger mkfastq for processing raw call files into fastq files. Cellranger count was used to align reads to a custom mm10 reference modified to include eGFP (marking tumor cells), to filter reads, and to generate a cell-by-gene matrix for each sample library. Libraries were aggregated using cellranger aggr without normalization to generate a single cell-by-gene matrix. Based on Gapdh expression, the top 14000 cells ranked by nUMI were retained for analysis. The Seurat package for R v.2.3.440 was used to process the matrix and perform downstream analysis. Expression values were log-normalized with a scaling factor of 104, and the 2509 most variable genes were detected and used for further analysis with the FindVariableGenes function. Values were scaled to number of UMIs and percent mitochondrial genes, and principle component analysis (PCA) was performed on the most variable genes. The FindClusters command was used to perform a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm using a resolution of 1.0, and tSNE dimensional reduction was calculated on the first 50 principle components to visualize data. Clusters consisting of cells with low/null expression of Gapdh and Eno1(non-cells), or co-expression of cell type exclusive markers (doublets) such as Cd3e and Cd68 were removed from further analysis by the SubsetData command, and variable genes were re-identified, data were re-scaled and PCA clustering and tSNE were re-run as described. Clusters containing the following cell types were identified using cell type markers: Tumor cells (eGFP), Myeloid cells (Cd68), Natural Killer (NK) cells (Ncr1), T-cells (Cd3e), Neutrophils (Lcn2), and subsets of these groups were identified by markers noted in heatmaps (Extended Data Fig. 6d). Cell type assignments for each cluster were verified by comparing with ImmGen datasets41. T cells, NK cells, and myeloid cells were subsetted and re-analyzed separately as described above. Cluster frequencies by library were normalized to number of cells per library and column plots were generated using ggplot2 v. 3.2.0 (Extended Data Fig. 6c). Gene expression t-SNE plots were plotted using ggplot2 v 3.2.0. For heatmaps, mean scaled expression values of each gene were calculated per cluster and plotted using pheatmap v 1.0.12 with values scaled by row (gene). Cell cycle scoring was performed using the Seurat CellCycleScoring command using mouse gene sets orthologous to previously described human gene sets42.
Analysis of TCGA data
IL18BP expression in individual cancer versus counterpart normal tissues was analyzed using TCGA cancer databases. Median and mean values were calculated. Human IL18BP mRNA differentiated expression, correlation with CD3E, CD8A and PDCD1 data for multiple cancers and matched normal tissues were obtained from TCGA and the GTEx projects and analyzed by webserver GEPIA2. The original microarray data was normalized by cancer browser (http://xena.ucsc.edu/welcome-to-ucsc-xena/) and then analyzed using R. The error bars in figures represent standard error of the mean (SEM).
Statistical analysis
Statistical analyses were conducted with R v.3.4.0 and Prism 8 (GraphPad Software). Pearson’s r was calculated with the R function cor (). Ordinary one-way or two-way ANOVA with Tukey’s multiple comparisons test, or a two-tailed paired or unpaired Student’s t-test (labeled in figure legend) were used to determine statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). The mean and SEM are presented in the figures. The error bars represent the SEM.
Extended Data
Extended Data Figure 1. Regulation of the IL-18 signaling axis in the tumor microenvironment.
(a) Distribution of intratumoral CD8+ T cells transcripts by dysfunction and activation gene scores, adapted from Singer et al43. Blue points mark a curated list of cytokine and cytokine receptors. Red points mark Il18, Il18r1, Il18rap. (b-f) WT C57BL/6 (WT) mice were subcutaneously (s.c.) engrafted with indicated murine syngeneic tumor. Tumor and spleen were harvest and analyzed by flow cytometry at day 7 post engraftment. (b) Representative flow plots and (c) quantification of percentage of IL-18Rα expression on splenic and intratumoral CD4+ T cells (NK1.1−, CD3+, TCRβ+, CD4+), CD8+ T cells (NK1.1−, CD3+, TCRβ+, CD8+), NK cells (CD3−, TCRβ−, NK1.1+, CD200R1−) from WT mice engrafted with MC38. (d) Quantification of percentage of IL-18Rα expression on splenic and intratumoral CD4+ T cells, CD8+ T cells, and NK cells from WT mice engrafted with YUMMER1.7. (e) Representative flow plots and (f) quantification of surface CD44 and IL-18Rα expression on CD8+ T cell by percentage in MC38 tumor bearing mice spleen and tumor. (g) Normalized quantification of Il18bp by qPCR in blood and tumor lysate assessed at day 7 post s.c. engraftment of WT mice with MC38 tumors, treated with either PBS, IL-18 or anti-IFN-γ+IL-18. (h) Quantification of plasma IL-18BP protein level by ELISA at day 7 post s.c. engraftment of WT mice with MC38 tumors, treated with either PBS or IL-18. (i) Representative immunohistochemistical staining for IL-18BP from l18bp−/− mice spleen, WT spleen, MC38 tumor, or MC38 tumor treated with one dose of IL-18, and assessed on day 7. Scale bar, 25 μm. (j) Quantification of IL-18BP+ cells per high power field in representative sections from each group indicated in (i). (b-f,i) Data are representative of 2 independent experiments with n=5 mice per group. (g,h&j) Data are pooled of 2 independent experiments with n=5 mice per group. P values were calculated using Two-way ANOVA (c,d&f) or two-tailed unpaired Student’s t-tests (g,h&j), and data are presented as the mean ± SEM.
Extended Data Figure 2. IL-18BP level is elevated in human cancers and correlated with T cell infilitration.
(a) Expression of IL18BP transcripts in normal (blue) or cancer (red) tissues from the TCGA database. CHOL, cholangiocarcinoma; DLBC, diffuse large B cell lymphoma; GBM, glioblastoma multiforme; HSNC, head and neck squamous carcinoma; KIRC, kidney renal clear cell carcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma (*P<0.01). (b-d) Correlation of IL18BP expression with T cell markers CD3E (b), CD8A (c), and PDCD1 (d) from the TCGA database for SKCM (n=558), BRCA (breast adenocarcinoma, n=1085), HNSC (n=44), STAD (n=221), and OV (ovarian cancer, n=426). (e) Frequency of IL-18BP immunohistochemistry staining levels in human tumor tissue microarrays. Each sample was scored as negative (0) or positive (1+, 2+, or 3+). Representative images are shown for each staining level. (f) Quantification of plasma IL-18BP protein level by ELISA from healthy donors (n=22) and NSCLC patients (n=52) at baseline prior to treatment and at the time of all the following CT-scan after receiving treatment with anti-PD-(L)1 (n=52). (g) Representative mean tumor growth of WT (left) and Il18bp−/− (right) mice s.c. engrafted with MC38 tumors and treated with PBS or IL-18. Data is representative of 3 independent experiments with n=5 mice per group. P values were calculated using One-way ANOVA (a,f) or Two-way ANOVA (g), and data are presented as the mean ± SEM.
Extended Data Figure 3. Related to Figure 1. Generation of Decoy-Resistant IL-18.
(a) Structural alignment of hIL-18 (green): hIL-18Rα/Rβ (cyan) complex (PDB ID 3WO4) with hIL-18: vIL-18BP (blue) complex (PDB ID 3F62). (b-c) Representative surface plasmon resonance (SPR) sensorgrams of murine WT IL-18 (b) binding to IL-18Rα or IL-18BP (c). IL-18Rα measurements were conducted using a conventional multiple cycle program, whereas IL-18BP measurements were conducted using a single-cycle program. (d) Dose-response curves of IL-18BP protein antagonizing IL-18Rα in complex with indicated IL-18 and mutants (E42A, K89A & E42A/K89A). Experiments were performed in duplicates (n=2). (e) Table showing randomized positions of murine IL-18 to create DR_18, with the corresponding degenerate codon and the potential amino acid at each position. (f) Summary of the experimental design for directed evolution and yeast selection process to generate DR-18. Yeast libraries were selected for IL-18Rα binding and counter selected against IL-18BP using MACS (Round 1 & 2) and subsequently FACS (Round 3, 4 & 5). Blue text (right side) indicates positive selection reagent, and red text (left side) shows the counter-selection reagent. (g) Structural representation of DR-18 mutation positions in IL-18Rα and IL-1BP binding overlap region. Side chains from a minimized set of mutations up to 6 consensus residues (1N, 50M, 52K, 55E, 56V and 59L) are displayed as stick models. (b-d) Data is representative of 2 independent experiments, and data are presented as the mean ± SEM.
Extended Data Figure 4. Related to Figure 2. Anti-tumor efficacy of DR-18 in syngeneic tumor models.
WT mice were s.c. engrafted with 0.5x106 MC38 or YUMMER1.7 tumor cells. On day 7 post engraftment, when tumor size reached 50-100 mm3, mice were treated twice weekly with PBS, 0.32 mg/kg IL-18 or DR-18. (a) Representative spider plots depicting tumor growth in WT mice s.c engrafted with MC38 treated with PBS, IL-18, DR-18, anti-PD-1, IL-18+anti-PD-1, or DR-18+anti-PD-1. (b) Representative mean tumor growth and (e) Kaplan-Meier survival curves of WT mice s.c. engrafted with YUMMER1.7 treated with PBS, IL-18, DR-18, anti-PD-1, IL-18+anti-PD-1, or DR-18+anti-PD-1. Arrows signify day of treatment. (c) Mean tumor growth and (f) Kaplan-Meier survival of WT Balb/c mice s.c. engrafted with 0.25x106 CT-26 colon carcinoma cells and treated with PBS, anti-PD-1, IL-18, or DR-18. (d) Mean tumor growth and (g) and Kaplan-Meier survival of WT mice s.c. engrafted with 0.25x106 B16-F10 melanoma cells and treated with PBS, IL-18, or DR-18 starting at day 7. (h) Mean tumor growth of WT mice s.c. engrafted with MC38 and treated with PBS, IL-18, CS1 or CS2 (DR-18), with the indicated doses. (a-d & h) Data are representative of 3 independent experiments with n=5 mice per group. (e-g) Data are summary of 3 (e) or 2 (f-h) independent experiments with n=5 mice per group. Data are presented as the mean ± SEM.
Extended Data Fig. 5. Related to Figure 2. DR-18 acts on antigen-specific T cells in the tumor.
(a) Kaplan-Meier survival curves of mice engrafted with YUMMER1.7 tumors treated with PBS, DR-18, or DR-18 with either anti-CD4, anti-CD8, anti-NK1.1, or anti-IFN-γ. Treatment parameters were the same as for MC38 (Fig. 2a, see methods). (b) Kaplan-Meier survival curves of WT or Rag2−/− mice s.c. engrafted with MC38 tumors and treated with PBS or DR-18. (c) Kaplan-Meier survival curves of mice re-challenged with 1.0x106 MC38 tumor cells after successful DR-18 treatment compared to naïve mice engrafted with 1.0x106 MC38 cells for the first time. (d-i) 2.0x106 Thy1.1+ P14 CD8 T cells were adoptively transferred into WT mice, which were then s.c. engrafted with 0.25x106 B16F10-gp33 tumor cells one day later. Mice were then treated twice weekly with either PBS or DR-18 (0.32 mg/kg) once tumors reached 50-100 mm3 for a total of 3 treatments. (d) Representative flow plots, (e) quantification of frequency, and (f) absolute cell of antigen-specific tumor infiltrating P14 CD8+ T cells, as defined by Thy1.1+ and Gp33-tetramer. (g) Percentage of IFN-γ on intratumoral antigen-specific (Thy1.1+Tetramer+) P14 CD8+ cells after PBS (n=4) or DR-18 (n=4) treatment. (h) Representative flow plots and (i) quantification of CD44 and CD39 expression on endogenous (Thy1.1−Tetramer−) tumor-infiltrating CD8+ T cells. (j). Quantification of CD44 and PD-1 expression of tumor infiltration of CD8+ T cells from WT mice engrafted with MC38 treated with PBS or DR-18 in combination with FTY720. (k) Representative flow cytometry plots of IL-18Rα expression on CD4+ T cells (CD45+, NK1.1−, CD3+, TCRβ+, CD4+), CD8+ T cells (CD45+, NK1.1−, CD3+, TCRβ+, CD8+), NK cells (CD45+, CD3−, TCRβ−, NK1.1+), B cells (CD45+, CD3−, TCRβ−, NK1.1−, CD19+), monocytes (CD45+, CD64+, CD11b+, Ly6C+), macrophages (CD45+, CD64+, MHCII+, F4/80+), dendritic cells (CD45+, CD64−, CD11c+), and neutrophils (CD45+, CD64−, CD11b+, Ly6G+) from MC38 tumors (n=5), matched spleens (n=5), or un-engrafted WT spleens (n=3), or Il18r1−/− spleens (n=3). (l) Representative mean tumor growth of MC38 tumors implanted into Rag2−/− mice that were adoptively transferred with T cells isolated from WT (n=5) or Il18r1−/− (n=8) mice and treated with PBS or DR-18. Black arrows signify day of treatment. (m) Representative mean tumor growth of MC38 tumors implanted into WT (n=4) and XCR1DTR (n=5) mice treated with PBS or DR-18 in combination with DT. Green arrows signify DT treatment regimen. (d-j) Data are representative of 2 independent experiments and presented as the mean ± SEM. P values were calculated using one-tailed (e,f), two-tailed (g) unpaired Student’s t-tests or Two-way ANOVA (i,j).
Extended Data Fig. 6. Related to Figure 3. Single Cell Transcriptomic Landscape of the tumor microenvironment in DR-18 treated tumors.
WT mice were s.c. engrafted with 0.5x106 YUMMER1.7 cells. On day 7 post engraftment, mice were treated twice weekly with PBS, IL-18 or DR-18 (s.c.). On day 15, tumors were harvest and sorted and 5000 cells per condition were used for scRNA-seq analysis. (a) t-SNE projection showing all cells present in YUMMER1.7 and YUMMER1.7-B2m−/− tumors, colored by cluster. Cells from the YUMMER1.7-B2m−/− experiments are shown in light grey and are discussed later. (b) t-SNE projection showing distribution of cells colored by treatment condition (Dark grey: PBS; Blue: IL-18; Magenta: DR-18). (c) Bar plot showing the percentage of cells in each cluster by treatment. (d) t-SNE projection of all cells analyzed showing expression of genes supporting cell-type assignments. (e) t-SNE projections of all cells analyzed showing expression of IL-18 pathway-related genes. (f) t-SNE projections showing expression of selected genes in computationally isolated Cd3e-positive T cells. (g) t-SNE projections of Cd68+ cells, showing expression of Cx3cr1, Nos2, Arg1, and Mertk. (h) t-SNE projection of Cd68+ cells, colored by graph-based cluster. (i) t-SNE projection of myeloid cell distribution as a function of treatment condition. (j) Quantification of the frequency of macrophages within each cluster identified in (h). (k) Heatmap showing differentially expressed genes in Cd68+ cells by clusters identified in (h). (l) Quantification of the frequency and (m) absolute numbers of Nos2+ macrophages from MC38 engrafted tumors following PBS, IL-18 or DR-18 treatment. (l,m) Data are representative of 3 independent experiments with n=5 mice per group and presented as the mean ± SEM. P values were calculated using One-way ANOVA.
Extended Data Figure 7. Related to Figure 3. DR-18 treatment enhances lymphocyte effector function.
WT mice were s.c. engrafted with 0.5x106 MC38 or YUMMER1.7 cells. At day 7 post engraftment mice were treated twice weekly i.p. with either PBS, IL-18, or DR-18. Tumors were harvested and analyzed at day 15 post engraftment. (a) Absolute cell number of tumor-infiltrating CD4+ T cells, CD8+ T cells, NK cells from YUMMER1.7 tumors in each treatment group. (b) Quantification of intracellular Ki-67+ staining by percentage of indicated intratumoral lymphocyte population. (c) Absolute cell number of antigen-experienced (CD44+) CD8+ T cells from MC38 tumors in each treatment group. (d) Representative flow plots of PD-1 and CD39 staining on intratumoral CD44+ CD8+ T cells from MC38 tumors in indicated treatment groups. (e) Absolute cell number of PD-1−CD39−, PD-1+CD39− or PD-1+CD39+ CD44+CD8+ T cells from (d) in each treatment group. (f) Quantification of IFN-γ protein level from tumor lysate of indicated treatment groups by Luminex. (g) Quantification of intracellular IFN-γ staining by percentage of indicated intratumoral lymphocyte populations from YUMMER1.7 tumors. (h,i) Quantification of intracellular IFN-γ staining by percentage in tumor (h) or spleen (i) for each indicated intratumoral lymphocyte population from mice engrafted with MC38. (j) Quantification of intracellular Granzyme B staining by MFI and (k) Quantification of CD107 staining in intratumoral CD8+ T cells and NK cells from MC38 tumors in the indicated treatment groups. (l) Absolute number and (m) frequency of polyfunctional NK cells measured by the co-staining of intracellular IFN-γ and Granzyme B following ex vivo stimulation with PMA/ionomycin. Data are representative of 3 independent experiments with 5 mice per group. P values were calculated using two-tailed unpaired Student’s t-tests (a), One-way ANOVA (b,f,g,j&k) or Two-way ANOVA (c,e,h,i&l) and data are presented as the mean ± SEM.
Extended Data Figure 8. Related to Figure 3. DR-18 promotes intratumoral TCF1+ precursor CD8+ T cells.
(a) Frequency of surface PD-1 and Tim-3 staining on MC38 intratumoral CD44+CD8+ T cells in indicated treatment groups. (b) Absolute cell number of DN (PD-1−Tim3−), SP (PD-1+Tim3−) or DP (PD-1+Tim3+) CD8+ T cells from YUMMER1.7 tumors in each treatment group. (c) Frequency of intracellular TCF1 in intratumoral CD44+CD8+ T cells in MC38 tumors in each treatment group. (d) Absolute cell number of TCF1+CD44+CD8+ T cells from MC38 tumors in each treatment group. (e) Frequency of intracellular TCF1 staining in MC38 intratumoral CD44+CD8+ T cells subsets based on PD-1 and Tim3 surface expression. (f) Representative histograms of intracellular TCF1 staining in intratumoral SP PD1+Tim3−CD44+CD8+ T cells from YUMMER1.7 tumors. (g) Absolute cell number of TCF1+ SP PD1+Tim3− CD44+CD8+ T cells from YUMMER1.7 tumors in each treatment group. (h,i) Frequency of intracellular IFN-γ in DP (PD-1+Tim3+) CD44+CD8+ T cells from MC38 (h) and YUMMER1.7 (i) tumors in each treatment group. (j) Absolute cell number of TCF1+ in SP (PD-1+Tim3−) CD44+CD8+ T cells in draining lymph node from MC38 tumors in each treatment group. Data are representative of 3 independent experiments with 5 mice per group. P values were calculated using One-way (c,d,g-j) or Two-way ANOVA (a,b&e) and data are presented as the mean ± SEM.
Extended Data Figure 9. Related to Fig. 4. DR-18 is effective towards MHC Class I deficient tumors.
(a) Representative spider plots showing tumor growth in WT mice s.c. engrafted with MC38-B2m−/− tumors treated with PBS, anti-PD-1+anti-CTLA-4, or IL-18 or DR-18. (b) Kaplan-Meier survival curves and (c) representative spider plots of WT mice s.c. engrafted with YUMMER1.7-B2m−/− tumors treated twice weekly with PBS, anti-PD-1+anti-CTLA-4, IL-18, DR-18, or DR-18 plus anti-CD8 and anti-NK1.1 antibodies. Depleting antibodies were given the same schedule as MC38-B2m−/− (see methods). (d) Kaplan-Meier survival curves and (e) spider plots showing tumor growth of WT mice s.c. engrafted with 1×106 RMA-S tumor cells and treated with PBS, anti-PD-1, IL-18, DR-18 or DR-18+ anti-NK1.1. Spider plot. (a-e) Data is representative of 2 independent experiments and survival curve data are pooled from 2 independent experiments. (f) Absolute cell number of intratumoral NK cells (NK1.1+CD3−Eomes+DX5+CD200R−), NKT cells (NK1.1+CD3+) and ILC1 cells (NK1.1+CD3−Eomes−DX5−CD200R+) in WT mice s.c engrafted with MC38-B2m−/− tumors in indicated treatment groups. (g) t-SNE projection of different clusters in YUMMER1.7-B2m−/− tumors as a function of treatment. YUMMER1.7 tumors are shown in light grey (discussed in Fig. 3, see methods). (h) Representative flow cytometry plots of IFNγ and Ki67 staining on intratumoral NK cells from YUMMER1.7-B2m−/− tumors treated with PBS or DR-18. (i,j) Frequency of Ki67+IFNγ+ NK cells from YUMMER1.7-B2m−/− (i) and MC38-B2m−/− (j) tumors treated with PBS or DR-18. (k) Representative mean tumor growth of WT mice s.c. engrafted with MC38-B2m−/− tumors and treated with PBS, DR-18, or DR-18 plus anti-FasL or anti-IFN-γ. (l) Representative mean tumor growth of Perforin−/− mice s.c. engrafted with MC38-B2m−/− tumors and treated with PBS (n=3) and DR-18 (n=5) as indicated. (h-l) Data are representative of 2 independent experiments with 5 mice per group. P values were calculated using two-tailed unpaired Student’s t-test, and data are presented as the mean ± SEM.
Extended Data Figure 10. Ex vivo characterization of a human DR-18 variant.
(a) SPR sensorgrams of human DR-18 (hDR-18) binding to human or cynomolgus macaque (Macaca fascicularis) IL-18Rα and IL-18BP. (b) Concentration-response curves of hIL-18 or hDR-18 on HEK Blue IL-18R reporter cells. (c) Titration of hIL-18BP on a fixed concentration of hIL-18 (1 ng/mL) or hDR-18 (0.1 ng/mL) on HEK Blue IL-18R reporter cells. (d) Concentration-response curves of IFN-γ stimulation by hIL-18 or hDR-18 on human peripheral blood mononuclear cells (PBMCs) or (e) cynomolgus macaque PBMCs. IFN-γ concentration is measured by ELISA. Data are representative of at least 2 independent experiments with n=2 for (b-e) and presented as the mean ± SEM.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge all members of the Ring and Bosenberg labs for helpful advice and technical assistance and Ewa Folta-Stogniew for assistance with SPR. We thank Dr. Akiko Iwasaki for mice and reagents. Cartoon in figure 1a was created with BioRender.com. This work was supported by grants from the National Cancer Institute Immuno-Oncology Translation Network (U01CA233096; to A.M.R. and M.W.B.), Gabrielle’s Angel Foundation (to A.M.R.), and the Blavatnik Fund for Innovation at Yale (to A.M.R.). A.M.R. is additionally supported by an NIH Director’s Early Independence Award (DP5OD023088), the Pew-Stewart Scholars Program, and the Robert T. McCluskey Foundation. M.F.S. is supported by a Miguel Servet contract from Instituto de Salud Carlos III, Fondo de Investigacion Sanitaria (Spain). W.D. is supported by a Career Development Award from the Dermatology Foundation. O.W is supported by a NSF Graduate Research Fellowship (1752134) and by a NIH training grant (T32AI055403). The T100 Biacore instrumentation used was supported by NIH Award S10RR026992-0110.
Footnotes
Competing Interests
A.M.R., T.Z., and S.F. are named inventors of a patent application that describes the DR-18 molecule. A.M.R. is the founder of Simcha Therapeutics, the commercial licensee of DR-18, and holds equity in the immuno-oncology companies Forty-Seven Inc., ALX Oncology, and Medicenna Therapeutics. W.D. and M.W.B. serve as consultants for Eli Lilly. W.D. has research funding from Pfizer for unrelated work.
Reporting Summary
Further information on research design will be made available in the Nature Research Reporting Summary linked to this article.
Data Availability
All data generated during this study are available within the paper. The scRNA-seq data were deposited on Gene Expression Omnibus (GSE146609).
References
- 1.Atkins MB, Kunkel L, Sznol M & Rosenberg SA High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J Sci Am 6 Suppl 1, S11–14 (2000). [PubMed] [Google Scholar]
- 2.Waldmann TA Cytokines in Cancer Immunotherapy. Cold Spring Harb Perspect Biol 10, doi: 10.1101/cshperspect.a028472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarhini AA et al. A phase 2, randomized study of SB-485232, rhIL-18, in patients with previously untreated metastatic melanoma. Cancer 115, 859–868, doi: 10.1002/cncr.24100 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Spangler JB, Moraga I, Mendoza JL & Garcia KC Insights into cytokine-receptor interactions from cytokine engineering. Annu Rev Immunol 33, 139–167, doi: 10.1146/annurev-immunol-032713-120211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mantovani A, Dinarello CA, Molgora M & Garlanda C Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity 50, 778–795, doi: 10.1016/j.immuni.2019.03.012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo L, Junttila IS & Paul WE Cytokine-induced cytokine production by conventional and innate lymphoid cells. Trends Immunol 33, 598–606, doi: 10.1016/j.it.2012.07.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Z et al. Augmentation of Immune Checkpoint Cancer Immunotherapy with IL18. Clin Cancer Res 22, 2969–2980, doi: 10.1158/1078-0432.CCR-15-1655 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Hu B et al. Augmentation of Antitumor Immunity by Human and Mouse CAR T Cells Secreting IL-18. Cell Rep 20, 3025–3033, doi: 10.1016/j.celrep.2017.09.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson MJ et al. Clinical and biological effects of recombinant human interleukin-18 administered by intravenous infusion to patients with advanced cancer. Clin Cancer Res 12, 4265–4273, doi: 10.1158/1078-0432.CCR-06-0121 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Dinarello CA, Novick D, Kim S & Kaplanski G Interleukin-18 and IL-18 binding protein. Front Immunol 4, 289, doi: 10.3389/fimmu.2013.00289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson MJ et al. A dose-escalation study of recombinant human interleukin-18 using two different schedules of administration in patients with cancer. Clin Cancer Res 14, 3462–3469, doi: 10.1158/1078-0432.CCR-07-4740 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SH et al. Site-specific mutations in the mature form of human IL-18 with enhanced biological activity and decreased neutralization by IL-18 binding protein. Proc Natl Acad Sci U S A 98, 3304–3309, doi: 10.1073/pnas.051634098 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simoni Y et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579, doi: 10.1038/s41586-018-0130-2 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Salmon H et al. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity 44, 924–938, doi: 10.1016/j.immuni.2016.03.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apetoh L et al. Consensus nomenclature for CD8(+) T cell phenotypes in cancer. Oncoimmunology 4, e998538, doi: 10.1080/2162402X.2014.998538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Best JA et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol 14, 404–412, doi: 10.1038/ni.2536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wherry EJ & Kurachi M Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15, 486–499, doi: 10.1038/nri3862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott AC et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274, doi: 10.1038/s41586-019-1324-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo H et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc Natl Acad Sci U S A 116, 12410–12415, doi: 10.1073/pnas.1905675116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtulus S et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1(−)CD8(+) Tumor-Infiltrating T Cells. Immunity 50, 181–194 e186, doi: 10.1016/j.immuni.2018.11.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller BC et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336, doi: 10.1038/s41590-019-0312-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudson WH et al. Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1(+) Stem-like CD8(+) T Cells during Chronic Infection. Immunity 51, 1043–1058 e1044, doi: 10.1016/j.immuni.2019.11.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray PJ et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20, doi: 10.1016/j.immuni.2014.06.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gubin MM et al. High-Dimensional Analysis Delineates Myeloid and Lymphoid Compartment Remodeling during Successful Immune-Checkpoint Cancer Therapy. Cell 175, 1014–1030 e1019, doi: 10.1016/j.cell.2018.09.030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeannet G et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A 107, 9777–9782, doi: 10.1073/pnas.0914127107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gattinoni L et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 15, 808–813, doi: 10.1038/nm.1982 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sade-Feldman M et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 8, 1136, doi: 10.1038/s41467-017-01062-w (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ardolino M et al. Cytokine therapy reverses NK cell anergy in MHC-deficient tumors. J Clin Invest 124, 4781–4794, doi: 10.1172/JCI74337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Street SE, Trapani JA, MacGregor D & Smyth MJ Suppression of lymphoma and epithelial malignancies effected by interferon gamma. J Exp Med 196, 129–134, doi: 10.1084/jem.20020063 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura K et al. Dysregulated IL-18 Is a Key Driver of Immunosuppression and a Possible Therapeutic Target in the Multiple Myeloma Microenvironment. Cancer Cell 33, 634–648 e635, doi: 10.1016/j.ccell.2018.02.007 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Terme M et al. IL-18 induces PD-1-dependent immunosuppression in cancer. Cancer Res 71, 5393–5399, doi: 10.1158/0008-5472.CAN-11-0993 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Vidal-Vanaclocha F et al. IL-18 regulates IL-1beta-dependent hepatic melanoma metastasis via vascular cell adhesion molecule-1. Proc Natl Acad Sci U S A 97, 734–739, doi: 10.1073/pnas.97.2.734 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park CC et al. Evidence of IL-18 as a novel angiogenic mediator. J Immunol 167, 1644–1653, doi: 10.4049/jimmunol.167.3.1644 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Hirakawa M et al. Low-dose IL-2 selectively activates subsets of CD4(+) Tregs and NK cells. JCI Insight 1, e89278, doi: 10.1172/jci.insight.89278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg SA IL-2: the first effective immunotherapy for human cancer. J Immunol 192, 5451–5458, doi: 10.4049/jimmunol.1490019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belkaya S et al. Inherited IL-18BP deficiency in human fulminant viral hepatitis. J Exp Med, doi: 10.1084/jem.20190669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods references
- 37.Wang J et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res 30, 428–435, doi: 10.1111/pcmr.12591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carbotti G et al. The IL-18 antagonist IL-18-binding protein is produced in the human ovarian cancer microenvironment. Clin Cancer Res 19, 4611–4620, doi: 10.1158/1078-0432.CCR-13-0568 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Lin L et al. Interleukin-37 expression and its potential role in oral leukoplakia and oral squamous cell carcinoma. Sci Rep 6, 26757, doi: 10.1038/srep26757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420, doi: 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heng TS, Painter MW & Immunological Genome Project, C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094, doi: 10.1038/ni1008-1091 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196, doi: 10.1126/science.aad0501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer M et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 166, 1500–1511 e1509, doi: 10.1016/j.cell.2016.08.052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.