SUMMARY
Single cell transcriptomics of neocortical neurons have revealed more than one-hundred clusters corresponding to putative cell types. For inhibitory and subcortical projection neurons (SCPNs), there is a strong concordance between clusters and anatomical descriptions of cell types. In contrast, cortico-cortical projection neurons (CCPNs) separate into surprisingly few transcriptomic clusters, despite their diverse anatomical projection types. We used projection-dependent single cell transcriptomic analyses and monosynaptic rabies tracing to compare mouse primary visual cortex CCPNs projecting to different higher visual areas. We find that layer 2/3 CCPNs with different anatomical projections differ systematically in their gene expressions, despite forming only a single genetic cluster. Furthermore, these neurons receive feedback selectively from the same areas to which they project. These findings demonstrate that gene expression analysis in isolation is insufficient to identify neuron types and have important implications for understanding the functional role of cortical feedback circuits.
Keywords: cell types, single-cell RNA sequencing, cortico-cortical projection neurons, feedback circuits, rabies tracing, connectivity, visual cortex
eTOC Blurb
Kim et al. show that neuronal cell groups from within a single transcriptomic cluster can be further separated based on their anatomical projections and connectivity patterns. Connectivity is an important feature for cell type identification. Gene expression alone cannot be used to fully annotate cortical cell types.
INTRODUCTION
The brain is composed of millions of diverse neurons that differ systematically in the genes that they express. For example, mammalian cerebral cortex processes information through two major cell groups: excitatory glutamatergic neurons and inhibitory GABAergic neurons. Since their anatomical description by S. Ramón y Cajal (Ramón y Cajal et al., 1995), heterogeneous neuronal cell types have been extensively characterized and classified through morphology, function, molecular composition, and connectivity (Tremblay et al., 2016; Zeng and Sanes, 2017). The recent explosion of genomic technologies has advanced our ability to characterize cell types based on gene expression at the single cell level, whereby unsupervised clustering methods identify putative cell types based on discretization (Tasic, 2018). These analyses have yielded 111 clusters from the mouse primary visual cortex (Tasic et al., 2018), including 60 inhibitory and 34 excitatory neuron clusters. Excitatory projection neurons are further separated into 11 SCPN (5 pyramidal tract (PT) and 6 corticothalamic (CT)) clusters and 14 CCPN (intratelencephalic (IT)) clusters. For inhibitory neurons and SCPNs these clusters have been shown to have concordance with anatomical descriptions. For example, one of the inhibitory clusters corresponds directly to classically-described “chandelier cells” (Tasic et al., 2018); and SCPN clusters separate anterior lateral motor cortex (ALM) layer 5 (F5) PT neurons that project to the medulla versus the thalamus (Economo et al., 2018). In contrast, CCPN clusters correspond largely to neurons in different layers with no known anatomical correspondence to within-layer clusters.
In mouse primary visual cortex (V1), only two or three transcriptomic clusters have been reported for layer 2/3 (L2/3) excitatory pyramidal neurons (Hrvatin et al., 2018; Tasic et al., 2016; Tasic et al., 2018). In contrast, L2/3 neurons in V1 are known to project to more than 10 different higher cortical areas (Wang and Burkhalter, 2007), with each cell projecting to only a subset of those areas (Han et al., 2018). This would suggest that, based on their different anatomical projections, there are more than two or three cell types (Figure 1A). Furthermore, V1 CCPNs projecting to different cortical areas differ systematically in their visual receptive fields (Glickfeld et al., 2013), and feedback to V1 from different visual areas has preferential functional impacts on V1 cells that match the known functional properties of the different CCPN targets (Huh et al., 2018). Thus, these cell types might also be distinguishable based on their cortical feedback connections. How do we reconcile the larger number of projection-based cell types against the far lower number of genetic clusters? Are the cells that differ in their projections really different cell types? Do these different CCPNs differ in their gene expression or input sources? To address these questions, we characterized the gene expression, connectivity, and laminar distributions of mouse V1 pyramidal neurons that project to different cortical areas.
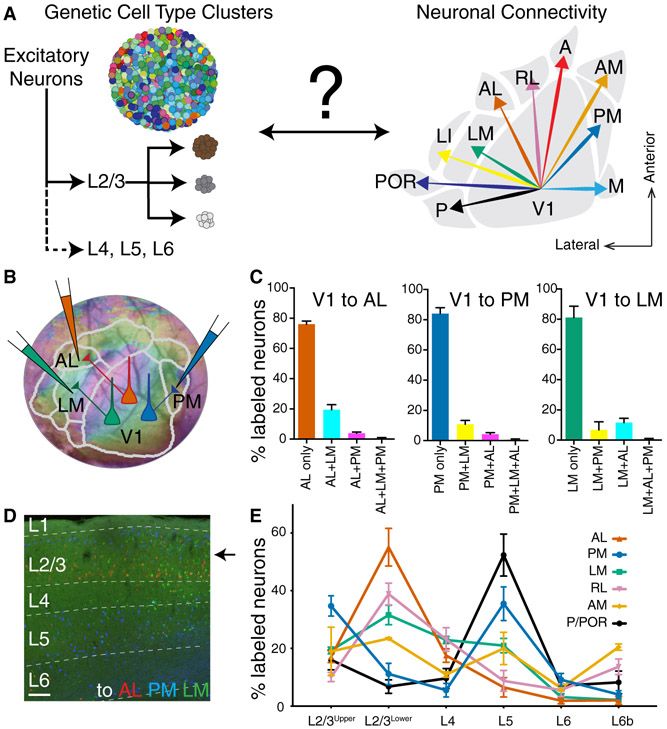
Figure 1. Anatomical Diversity of Cortico-Cortical Projection Neurons in Mouse Primary Visual Cortex.
(A) Schematic of genetic cell types defined by gene expression difference clusters (left) versus anatomical cell types based on long-distance connectivity patterns to higher visual areas (HVAs) in V1 (right). Cartoons describing genetic cell type clusters were adapted and modified from (Poulin et al., 2016). (B) Single neuron retrograde labeling strategy using three different colors of cholera toxin subunit b tracer injections on AL, PM, and LM after area border determination using intrinsic signal imaging. (C) A fraction of V1 neurons projecting to single, double, and triple areas in each case when they have the projections to AL, PM, or LM, respectively. A total of 12791 (V1→AL), 8084 (V1→PM), 2344 (V1→LM) retrogradely labeled neurons were counted per animal (n=3). (D) A coronal section of V1 displaying neurons projecting to AL (red), PM (blue), LM (green) across different cortical layers. (E) Soma locations of V1 neurons are distributed with distinctive laminar patterns depending on their HVA projections (mean ± SEM). Total 55742 retrograde labeled neurons were analyzed in 12 mice. Two-way ANOVA determined that fractions of V1 neurons projecting to HVAs are significantly different between their projection target areas and soma laminar locations (p<0.0001). Tukey’s multiple comparisons tested the significant difference of V1 neuron fraction between projection areas within each layer (Figure S1B and Table S2). Scale bar in panel D, 100μm. A, anterior; AL, anterolateral; AM, anteromedial; LI, interomediolateral; LM, lateromedial; M, medial; P, posterior; PM, posteromedial; POR, postrhinal; RL, rostrolateral.
RESULTS
Anatomical Diversity of Cortico-Cortical Projection Neurons in Mouse Primary Visual Cortex
We first examined the feedforward projection modes of V1 neurons to higher visual areas (HVAs) at the single cell level. We defined mouse visual cortical area borders using intrinsic signal imaging (ISI) (Garrett et al., 2014; Juavinett et al., 2017) and targeted different combinations of neuroanatomical retrograde tracers (cholera toxin subunit B (CTB) conjugated to various fluorophores) to HVAs (Table S1). Connections between V1 and HVAs are retinotopically organized, with systematic topographic relationships along the rostrocaudal and mediolateral axes (Wang and Burkhalter, 2007). Assessment of double and/or triple labeling at retinotopically overlapping locations in V1 allowed assessment of whether cells have collateral projections to more than one of the injected areas. For example, V1 neurons projecting to cortical area AL (anterolateral) (V1→AL), V1 neurons projecting to PM (posteromedial) (V1→PM), and V1 neurons projecting to LM (lateromedial) (V1→LM) were labeled across all the layers by CTB-Alexa Flour 647 (CTB-A647), CTB-Alexa Flour 555 (CTB-A555), and CTB-Alexa Flour 488 (CTB-A488) respectively (colored as red, blue, and green respectively in Figure 1B). The majority of neurons were labeled by a single color CTB (V1→AL 76.01±2.09%, V1→PM 84.08±3.89%, V1→LM 81.14±4.31%), while minor populations of neurons were labeled by two or three colors of CTB (Figures 1C, 1D, and S1A). A previous study using axonal reconstructions from single neurons and RNA barcode-based mapping (MAPseq) showed that more than 88% of V1 CC neurons project to one or two areas among six HVAs (Han et al., 2018). Consistent with these observations, our data show that most V1 neurons targeted retrogradely by three CTB injections project to only one of those three areas. Amongst combinations that resulted in double labeling, AL+LM was most common (19.47±3.29%) while AL+PM was rare (3.84±0.90%), again consistent with projection motifs observed previously (Han et al., 2018). This would suggest that if V1 L2/3 projection neurons do correspond to distinct types, those projecting to AL versus PM would be amongst the best candidates. This possibility is further supported by differences in the visual responses of V1→AL versus V1→PM neurons (Glickfeld et al., 2013).
To assure that the low incidence of double-labeled cells was not due to inefficient tracer uptake, we tested for double labeling after injection of two tracers in the same location. Following injection of both CTB-A488 and CTB-A647 into lateral secondary visual cortex (LM) 98.15% and 99.20% of total labeled cells were co-labeled by both CTB-A488 and CTB-A647 in V1 and lateral posterior (LP) nucleus of thalamus respectively (Figure S1C), demonstrating a high efficiency of uptake.
Cortico-cortical projection neurons exist not only in L2/3 but also in all of the deeper layers down to layer 6 (L6). Therefore, in addition to investigating their projection modes, we also investigated whether V1 neurons projecting to 6 different HVAs (AL, PM, LM, RL, AM, P/POR) differ in their laminar distributions. Interestingly, there exist distinct laminar biases for each population of projection neurons (Figures 1D, 1E, and S1B). For example, V1→AL and V1→RL neurons, which target more anterior HVAs are most abundant in the lower division of L2/3 (L2/3lower) and are rare in layers 5-6. In contrast, V1→PM and V1→P/POR neurons, which target more posterior HVAs are more abundant in the upper division (L2/3upper) than in the lower division of L2/3 and are most abundant in L5. This difference is not likely because V1 neurons project to superficial versus deep layers of HVAs, since both superficial and deep layer CCPNs arborize their axons across all the cortical layers (Harris et al., 2019). In summary, V1 neurons projecting to different higher visual areas differ systematically in their laminar distributions. Despite these biases, it is important to note that there is extensive overlap in the distributions of labeled cells across the full depth of L2/3 and within L5. Of particular relevance, V1→AL and V1→PM projecting neurons, in addition to their functional differences (Glickfeld et al., 2013), differ markedly in the proportions that are located in L5; but within L2/3 they overlap in their distributions (Figure 1D, arrow), despite biases for upper (V1→PM) versus lower L2/3 (V1→AL).
Single Cell Gene Expression Profiles Differ for V1→AL versus V1→PM Neurons
These observations suggested that V1→AL and V1→PM proj ection neurons might be good candidates for assessing whether V1 neurons projecting to different HVAs differ in their gene expression profiles. We therefore performed single nuclei RNA sequencing on these two populations. We first retrogradely labeled VI→AL or VI→PM neurons with nuclear GFP (Sun1-sfGFP) by making ISI-targeted injections of AAVretro-Cre into AL or PM of each R26R-CAG-loxp-stop-loxp-Sun1-sfGFP-Myc mouse (Figure 2A). Three weeks later, V1 was dissected and cut into upper and lower divisions containing L2/3/4 and L4/5/6, respectively (Figure 2A). Dissociated single nuclei from dissected tissues were prepared for fluorescence-activated nuclei sorting (FANS) to collect GFP+ nuclei individually (Figures 2A and S2A-C). cDNA libraries were generated by SMART-Seq v4 Kit and Nextera XT and sequenced by Illumina HiSeq 4000, resulting in mean 1.6 million uniquely mapped reads per nucleus (Table S4). We removed putative glia and classified L2/3, L4, L5+L6 neuronal groups based on the known glia and layer-specific gene marker combinations (Figure S3A). Using L2/3 and L5+L6 neurons, we applied an unsupervised learning method, tSNE, to visualize gene expression relationships across single nuclei (Figure 2B). The resulting tSNE plot illustrates two distinct clusters that correspond to L2/3 and L5+6 neurons. V1→AL and V1→PM did not separate into further clusters within the L2/3 or the L5+6 populations, even in independent clustering analyses of the L2/3 or the L5+6 populations separately (Figure S3C). Interestingly, within the single L2/3 cluster, V1→AL and V1→PM neurons can be seen to be preferentially enriched toward different ends of the cluster as visualized in the tSNE space (Figure 2B).
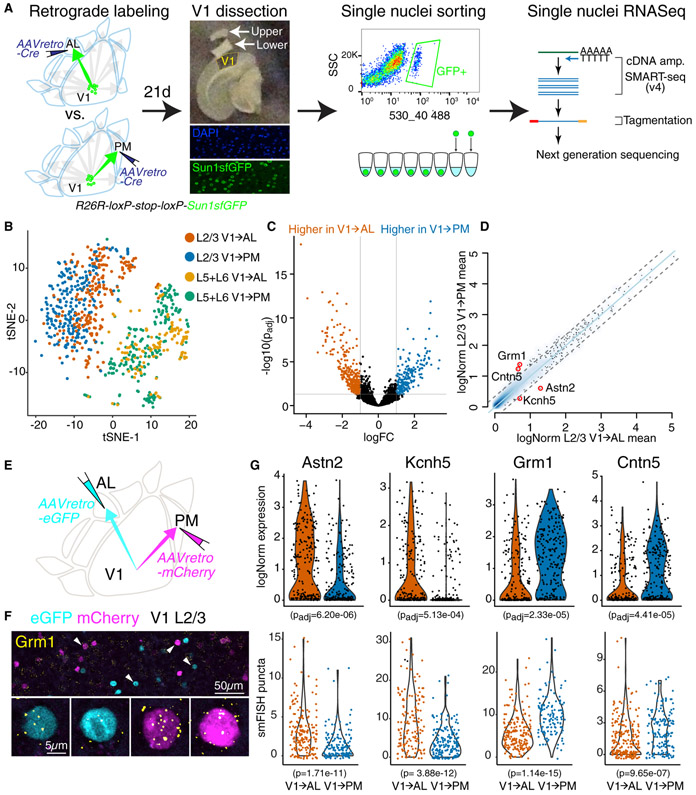
Figure 2. Single Cell Gene Expression Profiles Differ for V1→AL versus V1→PM Neurons.
(A) Schematic of experimental design on projection-based single nuclei RNA sequencing. (B) Unsupervised clustering analysis using tSNE plot based on single nuclei level gene expressions. (C) Volcano plot showing differentially expressed genes between V1→AL and V1→PM neurons. Grey lines indicate significance cutoff: adjusted p-value < 0.05 and logFC (fold change) > 1. (D) Smoothed scatterplot displaying expression differences in V1→AL versus V1→PM neurons. The light blue line showed perfect correlation; grey dashed line indicated 0.5 logFC deviation from the center. Values were averaged Seurat logNorm across all cells from each group. (E) Experimental scheme illustrating projection dependent labeling of V1→AL and VI→PM neurons using AAVretro-eGFP or mCherry. (F) Confocal microscope images of eGFP+ V1→AL and mCherry+ V1→PM neurons with single molecule FISH (smFISH) puncta of Grm1(yellow). Arrowheads indicate the cells shown in lower panels with higher magnification. (G) Violin plots displaying logNorm expression (top) and smFISH puncta count (bottom) of four differentially expressed gene markers per cell on V1→AL and V1→PM types. Total 345, 363, 334, 433 retrogradely labeled neurons for smFISH of Astn2, Kcnh5, Grm1, Cntn5 respectively in three animals (except Kcnh5 for two animals) were counted and analyzed. Adjusted p values for logNorm expressions were determined by multiple testing Benjamini-Hochberg method in Zinbwave-EdgeR, and p values for smFISH puncta were determined by Wilcoxon rank-sum test.
To further validate this result and to identify possible transcriptomic differences between V1→AL and V1→PM neurons, we searched for differentially expressed genes using Zinbwave-EdgeR or Zinbwave-DESeq2, Zero-Inflated Negative Binomial Models for single-cell RNAseq Data (Risso et al., 2018). We found more than 800 genes differentially expressed between L2/3 V1→AL and V1→PM groups (Figure 2C, Tables S5 and S7). Four genes known to have neuronal functions – Astn2, Kcnh5, Grm1, Cntn5 –were selected for further validation with combined retrograde labeling and fluorescence in situ hybridization (FISH) (Figure 2D).
V1→AL and V1→PM neurons were labeled by ISI-targeted AAVretro-nuclear eGFP and AAVretro-nuclear mCherry injections into AL and PM, respectively, in each animal (Figure 2E). Single-molecule FISH revealed differences in expression of all four genes in eGFP+ V1→AL versus mCherry+ V1→PM cells that correspond with the differences observed in the RNAseq data (Figures 2F and 2G). Although there are significant gene expression differences between the populations of V1→AL and V1→PM neurons, there is considerable overlap in the distributions of expression levels observed both with RNAseq and FISH.
V1→AL and V1→PM neurons are more abundant in L2/3lower and L2/3upper, respectively (Figure 1E). To assess whether differences in gene expression might be attributable to these laminar biases, we measured the precise laminar depth of neurons in which smFISH puncta of four genes were quantified. We then identified neurons in L2/3upper that were located 100-200μm from the pia, as there are many interspersed neurons projecting to both AL and PM at this depth. Comparison of these V1→AL and V1→PM neurons revealed that all four genes were still differentially expressed (Figure S4B). Notably, differences between V1→AL and V1→PM transcription profiles do not correspond to previously reported gene expression clusters with unsupervised clustering methods of L2/3 neurons in V1 (Hrvatin et al., 2018; Tasic et al., 2016; Tasic et al., 2018) (Figure S4A). This suggests that an unsupervised clustering method without projection labeling data would not be able to find these differences.
Projection- and Layer-dependent Input Tracing of V1→AL and V1→PM Neurons
We next applied rabies-based projection- and layer-dependent input tracing to investigate whether V1 neurons projecting to HVAs receive distinct feedback inputs across higher cortical areas (Figure 3A). Although the mammalian cortex is comprised of many hierarchically organized areas, cortico-cortical communication is bi-directional. Feedforward connectivity goes from lower to higher cortical areas, while feedback connectivity goes from higher to lower cortical areas (Felleman and Van Essen, 1991). It remains unknown at the cellular level whether feedback to lower visual areas, such as V1, specifically targets neurons according to their feedforward projections. Are feedback connections evenly distributed onto V1 projection neurons or might some more specific connectivity principle exist (Figure 3A, right)? For example, do V1 neurons projecting to a given HVA receive preferential reciprocal feedback from that same area? To assess these possibilities, we first conducted intersectional projection-type and L2/3-specific or L5-specific input tracing using EnvA pseudotyped, G-deleted rabies virus expressing eGFP (EnvA+RVdG-eGFP). We injected AAVretro-FLEXloxp-Flp into AL or PM and AAV8-FLEXfrt-oG/TVAmCherry into V1 of either SepW1-Cre mice, a BAC transgenic line expressing Cre in L2/3 CCPNs, or Tlx3-Cre mice, a BAC transgenic line expressing Cre in L5 CCPNs (Gerfen et al., 2013) after ISI (Figures 3A, 3B, and S5). This results in expression of both the EnvA ligand TVA and optimized rabies glycoprotein (oG (Kim et al., 2016)) exclusively in either L2/3 cells or L5 cells that project to either AL or PM. Fourteen days after AAV injections, we injected EnvA+RVdG-eGFP into V1 to selectively infect the TVA expressing neurons and to allow retrograde monosynaptic spread of the rabies virus. This resulted in labeling of the direct monosynaptic inputs to the defined L2/3 or L5 starter neurons projecting to AL or PM. For postmortem analysis and quantification of eGFP+ rabies-labeled feedback neurons in each of the HVAs, we either cut tangential brain sections that were then mounted, stained and imaged with conventional microscopy, or the intact cortical tissue was optically cleared and then imaged with a lightsheet microscope. eGFP+ input neurons were assigned to the nine different HVAs by aligning surface blood vessels seen in either the top tangential sections or in the light-sheet images to the same blood vessels imaged during the production of ISI-generated area maps (Figures 3B and 3C). Numbers of neurons in each area were then counted.
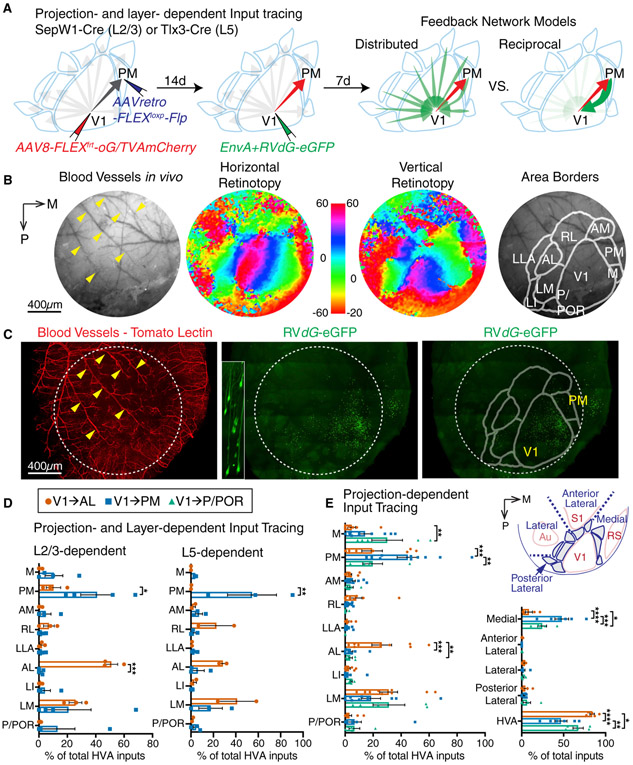
Figure 3. Projection- and Layer-dependent Input Tracing of V1→AL and V1→PM Neurons.
(A) Schematic showing the experimental design on projection- and layer-dependent monosynaptic input tracing by G-deleted rabies (left and middle). Two possible feedback connectivity models depending on projection (right). (B) Intrinsic signal imaging to define higher visual area borders. (C) Reconstruction of rabies-traced and blood vessel stained visual cortex using light sheet microscopy. (D) Summary of long-range feedback inputs onto V1→AL and V1→PM neurons in L2/3 (left) or L5 (right). (E) Summary of long-range feedback inputs onto V1→AL, V1→PM, and V1→P/POR neurons. Values are reported as mean ± SEM. Statistics were calculated from two-way ANOVAs with Sidak’s or Turkey’s post-hoc multiple-comparison tests for panels D and E, respectively. *p<0.05; **p< 0.01; ***p< 0.001; ****p<0.0001. A, anterior; AL, anterolateral; AM, anteromedial; Au, auditory cortex; LI, interomediolateral; LM, lateromedial; M, medial; P, posterior; PM, posteromedial; POR, postrhinal; RL, rostrolateral; RS, retrosplenial cortex; S1, primary somatosensory cortex; V1, primary visual cortex.
We found that feedback projections were strongly biased towards cells projecting to the same area. Both V1→AL and V1→PM L2/3 neurons received almost half of all their HVA inputs from their HVA projection area (50.99±4.47% for V1→AL, 40.97±11.49% for V1→PM), and far fewer inputs from each of the other areas (Figure 3D, left panel). Of particular note, V1→PM L2/3 neurons received virtually no detectable inputs from AL while V1→AL L2/3 neurons received only 10.28±5.08% of their feedback inputs from PM. Given that many V1→AL and V1→PM neurons also have collaterals to LM, it is noteworthy that V1→AL and V1→PM L2/3 neurons also received a considerable fraction of their inputs (25.80±4.53% and 20.76±16.00%, respectively) from LM. Similar results were observed for intersectional projection-type and L5 CCPN-specific input tracing. V1→PM L5 CCPNs received more than half of their HVA inputs from PM (54.47±21.82%), while V1→AL L5 CCPN neurons received majority of their HVA inputs from AL, LM, and RL (28.46±3.29%, 41.16±17.26%, and 22.62±15.95%, respectively). It is of note that V1→AL L5 CCPNs received no detectable inputs from PM while V1→PM L5 CCPNs received only 6.1±5.80% of their inputs from AL (Fig. 3D, right panel).
To determine whether selectivity is a more general property of feedback connections, we assessed inputs to neurons across all V1 layers and also sampled from neurons projecting to a third visual area, P/POR. We injected AAVretro-Cre into AL or PM or P/POR (posterior/postrhinal) and AAV8-FLEX-oG/TVAeGFP into V1 after ISI to prime the starter neurons in a long-distance projection dependent manner. Fourteen days after AAVs injections, we injected EnvA+RdG-mCherry into V1 to label monosynaptic inputs to the projection-defined starter neurons. Collectively, depending on their long-distance HVA projections, V1 cortico-cortical neurons received distinct patterns of brain-wide input not only from HVAs but also from other non-visual cortical areas such as medial regions, including retrosplenial cortex (Figure 3E).
DISCUSSION
In summary, our multi-modal approach reveals that neuronal cell groups from within a single transcriptomic cluster can be further separated based on their anatomical projections and connectivity patterns. These results indicate that connectivity is an important feature for cell type identification and that gene expression alone cannot be used to fully annotate cortical cell types. It should be noted that gene expression differences between V1→AL and V1→PM neurons are not distinct enough for them to separate into discrete clusters. Instead, V1→AL and V1→PM compose a single cluster, displaying continuous variability. Based on retrograde labeling to provide additional knowledge about the anatomy and connectivity of these neurons, we separated them into groups that differ statistically in their gene expression.
We chose to study the two V1 projection neuron populations, VI->AL and V1→PM, because of their functional differences and distinguishable visual receptive fields (Glickfeld et al., 2013). In addition, bifurcation of V1 neurons to AL and PM is the most under-represented projection motif; thus, V1→AL and V1→PM neurons are largely non-overlapping populations in terms of their cortical projections. Future studies should determine gene expression profiles and feedback connectivity of V1 projection neurons targeting other HVAs sharing similar receptive field properties or projection motifs.
Our rabies tracing experiments indicate that cortical feedback connections follow a “like-to-like” rule. This can be inferred from the fact that the functional properties of AL and PM neurons providing feedback to V1 differ and are similar to the functional properties of the corresponding V1→AL and V1→PM neurons (Andermann et al., 2011; Glickfeld et al., 2013; Huh et al., 2018; Marshel et al., 2011). This specificity is likely to underlie the differential functional influences that have been demonstrated for cortical feedback to V1 from AL versus PM (Huh et al., 2018).
We found that specific cortico-cortical projection neurons display different laminar distributions and discrete long-distance feedback depending on their feedforward projections. Future studies should investigate how these specific feedforward and feedback cortico-cortical networks are established during development. L2/3 neurons in V1 share similar birthdates and spatial neurogenic environments during embryogenesis. Given that the neuronal subtypes of L2/3 neurons in V1 have different gene expression profiles in the adult brain, we speculate that genetic programs and activity-dependent mechanisms may interact with each other to establish the observed adult relationships between gene expression and long-distance connections. Future studies will be needed to investigate whether gene expression defines connectivity or whether connectivity, and perhaps the inherent similarity in activity found between neurons of similar circuitry, underlies differences in gene expression. If differential gene expression plays a causal role in directing long-range connectivity from V1 to different HVAs, future experiments might identify candidate genes at the relevant developmental stages and investigate their roles through sets of loss-of-function and gain-of-functional experiments. For the specificity of feedback connections to V1, the available data are more consistent with activity dependent Hebbian mechanisms. Thus, the expected patterns of connectivity would not follow any particular principle except that feedback from areas that are more distinct functionally would provide more distinct feedback. There are several features of our dataset that are consistent with this hypothesis. For example, V1→AL cells receive very little feedback from PM and V1→PM cells receive very little feedback from AL. This is expected from the very different functional properties of AL and PM neurons. But each of these areas receives similar feedback from other areas that are not as distinct functionally. Testing such a hypothesis would require sets of experimental studies manipulating patterns of activity during circuit development.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Edward M. Callaway (callawav@salk.edul
Materials Availability
Reagents generated in this study will be made available on reasonable request. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Edward M. Callaway (callaway@salk.edu)
Data and Code Availability
The single-nuclei RNA sequencing datasets generated in this study are available in the GEO repository under accession number GSE133230. The code necessary to generate the figures in this manuscript will be available from the corresponding author upon reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experimental procedures followed procedures approved by the Salk Institute Animal Care and Use Committee. Knock-in mouse line, R26R-CAG-loxp-stop-loxp-Sun1-sfGFP-Myc (INTACT), and GENSAT BAC transgenic, SepW1-Cre NP39 and Tlx3-Cre PL56 mice, have been previously described (Gerfen et al., 2013; Gong et al., 2007; Mo et al., 2015). Knock-in and transgenic mice were maintained on C57BL/6J backgrounds. C57BL/6J mice have been used as wild-type. All mice were housed with a 12-hour light and 12-hour dark cycle and ad libitum access to food and water. Both male and female mice were used for CTB and rabies tracing experiments, and no obvious differences between sexes were observed. Only male mice were used for RNA sequencing experiments. Specific details of mouse ages are reported in appropriate sections of method details.
METHOD DETAILS
Virus preparation
All AAVs were produced by the Salk Viral Core GT3: AAVretro-EF1a-Cre-WPRE (AAVretro-Cre, 7.20X1012GC/ml), scAAVretro-hSyn1-H2B-eGFP (AAVretro-nuclear eGFP, 1.32X1013 GC/ml), scAAVretro-hSyn1-H2B-mCherry (AAVretro-nuclear mCherry, 3.02X1012 GC/ml), AAVretro-CAG-FLEXloxp-Flp (3.60X1012 GC/ml) , AAV8-CAG-FLEXfrt-TVAmCherry-WPRE (1.85X1014 GC/ml), AAV8-CAG-FLEXfrt-oG-WPRE (5.89X1014 GC/ml), AAV8-CAG-FLEXloxp-oG-WPRE (7.71X1014 GC/ml), AAV8-EF1a-FLEXloxp-eGFP-2A-TVA-WPRE (3.50X1013 GC/ml), AAVretro-nef-lox66/71-tTA (1.77X1012 GC/ml), AAV8-TRE-DIO-OG-WPRE (5.92X1012 GC/ml), AAV8-TRE-DIO-TVAmCherry-WPRE (1.16X1013 GC/ml). pAAV-CAG-FLEXloxp-Flp (Ren et al., 2018) and pAAV-CAG-FLEXfrt-TV AmCherry-WPRE (Schwarz et al., 2015) were gifts from Dr. Liqun Luo. rAAV2-retro capsid (Tervo et al., 2016) was a gift from Dr. Alla Karpova.
EnvA+RVdG-dsRed2 (3.6X107 Infectious Unit (IU)/ml) (Kim et al., 2015) and EnvA+RVdG-eGFP (multiple batches with the titers from 1.14X108 to 9.59X109 TU/ml) were produced.
Intrinsic signal imaging (ISI)
Before tracer or virus injections, cortical visual areas of each mouse were identified by intrinsic signal imaging (Garrett et al., 2014). Mice were anesthetized with isoflurane (2% induction, 1–1.5% surgery) and implanted with a metal frame over the visual cortex for head fixation. Mice were allowed to recover for 1–2 days after head frame implantation. For intrinsic signal imaging experiments, mice were again anesthetized with isoflurane (2% induction) and head fixed in a custom holder. Chlorprothixene (1.25mg/kg) was administered intramuscularly, and isoflurane was reduced to 0.25–0.8% (typically 0.5%) during visual stimulation. Anesthesia levels were titrated to maintain a lightly anesthetized state. Imaging was performed through a thinned skull or implanted window. Retinotopic maps were measured by taking the temporal phase of response to a periodic drifting bar in horizontal and vertical directions. Area borders were identified based on four criteria: 1) Each area must contain the same visual field sign at all locations within the area, 2) Each visual area cannot have a redundant representation of visual space, 3) Adjacent areas of the same visual field sign must have a redundant representation, and 4) An area's location must be consistently identifiable across experiments.
Cholera Toxin Subunit B injection, histology, and analysis
C57BL/6J mice from postnatal day (P) 57 to P95 were used. To label V1 neurons projecting to specific higher visual areas, 0.5% of Cholera Toxin Subunit B (CTB), Alexa Fluor 488, 555, or 647 conjugated (Molecular Probes), were injected to higher visual cortical areas based on functional maps overlaid on the blood vessel pattern, using these blood vessels as landmarks. A craniotomy or a drilled burr hole was made over visual cortex. Injections were done with air pressure either using a picospritzer (General Valve Corp, Fairfield, NJ) or a 1ml syringe with 18G tubing adaptor and tubing. To prevent backflow, the pipette was left in the brain for 5 minutes before and after injection. CTB injections into cortex were 20-50nl for AL, AM, PM, and P/POR, and 40-100nl for LM and RL, with 20-40 PSI for picospritzer or with at least 5 min of constant pressure for syringe injection and a 20-30μm diameter pipette (Conte et al., 2009). After recovery, mice were given water with ibuprofen (30mg/kg) and housed for 4-6 days before tissue harvest.
Brains were harvested after transcardial perfusion using phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Brains were dissected out from skulls and post-fixed with 2% PFA and 15% sucrose in PBS at 4°C overnight, and then immersed in 30% sucrose in PBS at 4°C before sectioning. Using a freezing microtome, 50μm coronal brain sections were cut and stored in PBS with 0.01% sodium azide at 4°C. After DAPI counterstaining, sections were mounted on slides with polyvinyl alcohol mounting medium containing DABCO and allowed to air-dry.
Each section was imaged at 10x using an Olympus BX63 microscope or 10x or 20x using a Zeiss LSM780 confocal microscope with parameters adjusted based on the intensity of expression and background fluorescence. The injection sites for each animal were measured to ensure restriction to small cortical visual areas. For LM and RL injections, animals with injection site diameters greater than 500μm for LM or RL were excluded. For AL, AM, PM, and P/POR, animals with injection site diameters greater than 300μm were excluded.
To quantify single labeled and co-labeled V1 neurons, V1 borders on each section were registered based on the Mouse Brain in Stereotaxic Coordinates atlas (Paxinos et al., 2001), and the subregion of V1 was identified where two or three CTB labeling were overlapped. Each cortical layers were manually registered using the nuclear counterstain (DAPI) channel to visualize layer 4 laminar width, which has higher nuclear density, then layer 2/3, 5, 6, and 6b with distinct nuclear density. Layer 2/3 was sub-divided into halves, layer 2/3upper and layer 2/3lower.Single labeled and co-labeled CTB+ neurons in each layer were manually counted.
Single-nuclei isolation after retrograde labeling
To label individual nuclei of V1 neurons projecting to AL or PM, 30nl of AAVretro-Cre was injected to AL or PM of INTACT male mice (age from P72 to P87) after defining visual area borders using ISI. Animals were given water with ibuprofen (30mg/kg) and housed for three weeks before tissue harvest. After animals were euthanized in a CO2 chamber and decapitated, the brains were dissected out from the skull and submerged in ice-cold PBS. Coronal brain slices (350μm thick) were cut using VF-300 Compresstome™ (Precisionary Instruments, Greenville, NC) and submerged in ice-cold PBS. Subregion of V1 containing GLP+ nuclei in brain slices was micro-dissected out into upper and lower halves under the fluorescent dissection microscope (Olympus SZX16) and transferred to microcentrifuge tubes with ice-cold PBS for single-nuclei isolation. The remaining brain slices after dissection were collected, fixed with ice-cold 4% PFA for 2-4 hours, stained with GFP antibody conjugated to Alexa Fluor 488 (Invitrogen, A-21311), and scanned to validate the correct V1 dissection using Olympus BX63 Microscope.
Single nuclei preparations were performed following the published protocol with modification (Lacar et al., 2016). The dissected samples in 1ml NIM buffer (0.25M sucrose, 25mMKCl, 5mMMgCl2, 10mMTris-HCl (pH7.4), 1mM DL-Dithiothreitol solution (Sigma 646563), 10μl of Protease inhibitor (Sigma P8340), 1.5μ1 of RNasin Plus RNase inhibitor (Promega, N2611), 0.1% TritonX-100, 5μM Hoechst 33342) were homogenized gently using the ice-cold dounce and pestle. After centrifuge at 3000 rpm for 8 min at 4°C, the supernatant was aspirated and gently resuspended in 1ml NIM buffer. After one repeat, the pellet was resuspended in 450μ1 of nuclei storage buffer (0.25M sucrose, 5mMMgC12, lOmM Tris-HCl (pH7.4), ImM DL-Dithiothreitol solution, 9ul of Protease inhibitor), and filtered through 40μM cell strainer on ice. The sample was incubated with 50μ1 of nuclease-free BSA for 1-2 hours at 4°C, then with GFP antibody conjugated with Alexa Fluor 488 (Invitrogen, A-21311) for 1 hour at 4°C.
Fluorescence-activated nuclei sorting of single nuclei was performed using a BD Influx sorter with an 85μm nozzle at 22.5 PSI sheath pressure. Single cell (1 drop single) mode was used for the stringent sorting. GFP+ single nuclei were sorted into 96-well plates preloaded with 2μ1 of lysis buffer (Triton X-100 10%, 0.1μl RNasin Plus RNase inhibitor in H2O). After fast freeze on dry-ice, the plate was stored at −80°C before library preparation.
cDNA amplification and library preparation for RNAseq
Using SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech, 634894), cDNA wasamplifiedwith21 PCR cycle following polyA reverse transcription. The quantity and quality of cDNA was validated using the Agilent 2100 Bioanalyzer. 0.2ng/nl of amplified cDNA was used as input to prepare sequencing libraries using the Nextera XT DNA Library Preparation Kit (Illumina). Multiplexed libraries after normalized concentration were sequenced on Illumina HiSeq 4000 to generate 150bp paired-end reads.
Sequencing data quality control and analysis
After trimming adapter by cutadapt [v2.3] (Martin, 2011), 150 base-pair paired-end reads were aligned to mm10 GENCODE M10 using STAR [v2.5.1b] (Dobin et al., 2013). Sub-sampling was performed to retain half of the total reads from the first 96 samples with twice sequencing depth as the rest samples by Heard DownsampleSam [v1.130] (Picard Toolkit 2019. Broad Institute, GitHub Repository, http://broadinstitute.github.io/picard/; Broad Institute). Uniquely aligned reads mapped across the whole gene body were further quantified by Homer [v4.8] (Heinz et al., 2010). Only one read per position was kept to remove PCR duplicates. 945 cells passed the filter of minium 100K of reads mapped per cell and minimum 1K genes per cell (TablesS4 and S5).
Seurat [v2.3.4] (Butler et al., 2018) was used for data analysis unless specified otherwise. Addditional filtering was performed to get rid of 50 putative glia cells that expressed either one of the molecular markers at specified expression level cutoffs (ApoE: 3.1, Olig2: 1, Sox10: 1, Gfap: 1, S100b: 4.2) based on their distribution of Seurat scaled logNorm values across all cells (Figure S3A). In addition to upper (L2/3/4) versuslower layer (L4/5/6) labeling based on dissection, we defined 134 L4 cells using layer-specific molecular marker combinations, Rorb+(L4/5a), Cux1+ or Cux2+ (L2/3/4), Deptor-(L5) based on their distribution of Seurat scaled logNorm values acrossall remaining cells (Rorb: 0.2, Cux1: −0.2, Cux2: −0.5, Deptor: −0.2) (Figure S3A-B). K-means clustering (k=2) was used to group cells into either L2/3 or L5/6 group. tSNE (t-Distributed Stochastic Neighbor Embedding) was plotted using top Prindpal Component (PC) outputs that captured most of the data variability based on PC elbow plot (top 25 for all cells; top 15for L2/3 or L5/6 cells, respectively). Perplexity of 40 and arandom seed of 7 was used for the final visualization of Figure 2B, whereas perplexity of 20 and a random seed of 3 or 2 were used for FigureS3C left or right plot, respectively. Differential gene expression analyses between V1→AL and V1→PM neuronsin L2/3 or L5/6 groups were performed using Zinbwave [v1.4.1]-EdgeR [v3.20.9] (Tables S5 and S6) and Zinbwave-DESeq2 [v1.20.0] (Tables S7 and S8)(Butler et al2018). The two methods gave comparable results. All data presented in the figures are based on Zinbwave-EdgeR results. Genes expressed in more than 5% of total populations with adjusted p-value < 0.05 and logFC > 0.5 were identified as significantly differentially expressed. All plots were generated using R statistical computing environment (R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria URL https://www.R-proiect.org/)
In vivo validation of gene expression using single molecule fluorescent in situ hybridization (smFISH)
Higher visual area borders of mouse brain were mapped using ISI, and 10nl of scAAVretro-eGFP and 10nl of scAAVretro-mCherry were injected into AL and PM respectively. At 2-3 weeks after virus injections, the animal was sacrificed in a CO2 chamber and brains were dissected out from the skull, and immediately frozen in Tissue-Tek O.C.T Compound on dry ice. Brains were cryo-sectioned coronally with 20μm thickness using Leica CM 1950 and stored at −80°C until use. smFISH was performed using RNAscope Fluorescent Multiplex Assay kit (Advanced Cell Diagnostics). Sections containing eGFP, mCherry, and smFISH in V1 were imaged with 20X or 63X Zeiss Airyscan 880. Imaging was performed with Z-stack with 1μm optical section and tiling. The number of smFISH puncta on eGFP+ or mCherry+ nuclei was manually counted using ImageJ FIJI. Wilcoxon rank-sum test was used for statistical analysis.
Rabies mediated monosynaptic input tracing
For projection-dependent input tracing, the mixture of AAV-FLEX-oG and AAV-FLEX-TVA-2A-eGFP were injected to V1 and AAVretro-Cre to AL, PM, or P/POR of C57BL/6J mice (P45 to P70). For projection- and layer-dependent input tracing, two different viral strategies were used. First, the mixture of AAV-FLEXfrt-oG and AAV-FLEXfrt-TVAmCherry were injected to V1 and AAVretro-FLEXloxp-Flp to AL, PM, or P/POR of SepW1-Cre or Tlx3-Cre mice. Second, the mixture of AAV-TRE-DIO-oG and AAV-TRE-DIO-TVAmCherry were injected to V1 and AAVretro-lox66/71-tTA to AL or PM of SepW1-Cre mice. Detailed information of virus injections on individual mice is found in Table S9. Injection locations in visual cortical areas were based on functional maps overlaid on the blood vessel pattern, using these blood vessels as landmarks. A craniotomy was made over visual cortex. Pressure injections were done with a 1ml syringe with tubing connected to the glass pipette. For landmarks, DiI injections were done outside of HVAs four times. To prevent backflow, the pipette was left in the brain for 5 minutes before and after injection. After recovery, mice were given water with ibuprofen (30mg/kg) and housed for two weeks before rabies injection. 400nl of EnvA+RVdG-dsRed (or eGFP) was injected into V1 after opening the coverslip of the headframe. Animals were housed with ibuprofen for a week before the brain harvest.
Tissue processing and imaging analysis for lightsheet microscopy
Brains were harvested after transcardial perfusion using phosphate-buffered saline (PBS) with heparin (10U/ml) followed by 20ml PBS with 10μg Dylight 647-Lectin (Vectorlabs, DL-1178) per ml PBS. The lectin solution was allowed to incubate for two minutes before resuming perfusion with 4% paraformaldehyde (PFA). Brains were dissected out from skulls and post-fixed with 4% PFA in PBS at 4°C for 16–20 hours, then immersed in PBS at 4°C. Before clearing, the brain was cut subcortically tangential to the injection site in primary visual cortex. Due to microscope hardware limitations, this tissue block was restricted to a 5mm by 5mm by 3mm volume. The tissue block was then cleared using the ScaleSQ method (Hama et al., 2015).
The cleared tissue block was imaged on a Zeiss Z.1 lightsheet microscope using a 5x air objective. Output images were down-sampled by 75% when exporting from the Zen Blue software provided by Zeiss. A custom python script (Puifai Santisakultarm, reorg4terastitcher.py) was then run to organize the image tiles for compatibility with the open source voxel stitching software Terastitcher (Bria and Iannello, 2012). Terastitcher was used to stitch together each Z plane from the lightsheet scan, and the final output resulted in some number of stitched Z planes. The stitched planes were then input into the software Imaris and visualized in three dimensions. The reconstructed lightsheet volume was then free rotated and interpolated in Imaris until the major surface blood vessels were matched in orientation to the ISI blood vessel image. The lightsheet volume was then exported from Imaris as a new series of Z planes which were tangential to the injection site in primary visual cortex. The tangential Z images were then imported into Fiji for analysis and cell counting. The aligned stack of sections was then compared to the blood vessel image from the initial intrinsic signal imaging (ISI) session. Major blood vessels were used as anatomical landmarks to align and warp the ISI blood vessel image to the lightsheet image stack. ISI retinotopy and visual area borders were matched to the aligned ISI blood vessel layer. Cell counting and registration was done in FIJI for each section in the final aligned stack.
Tissue processing and imaging analysis for slide scanner
Brains were harvested after transcardial perfusion using phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Brains were dissected out from skulls and post-fixed with 2% PFA and 15% sucrose in PBS at 4°C for 16-20 hours, then immersed in 30% sucrose in PBS at 4°C. Before sectioning, the brain was cut subcortically tangential to the injection site in primary visual cortex. An initial 250μm section was taken followed by 50μm sections on a freezing microtome and stored in PBS with 0.01% sodium azide at 4°C. To enhance eGFP and dsRed signals, free-floating sections were incubated at 4°C for 16-48 hours with goat anti-GFP (1:1000; Rockland 600-101-215) and rabbit anti-dsRed (1:500; Clontech 632496) primary antibodies in PBS/0.5% normal donkey serum/0.1% Triton-X 100, followed by the appropriate secondary antibodies conjugated with Alexa Fluor 488 or 594 (Molecular Probes). Sections were counterstained with 10μM DAPI in PBS for 30 min to visualize cell nuclei. Immunostained tissue sections were mounted on slides with polyvinyl alcohol mounting medium containing DABCO and allowed to air-dry overnight.
All sections were scanned with a 10x objective on an Olympus BX-63 fluorescent microscope. Scanned images were first processed in Olympus CellSens software using their extended focal imaging algorithm. Subsequent image files were processed and analyzed in NIH ImageJ software (FIJI). Reconstruction of the tissue block was done by aligning and warping serial images using an affine matrix algorithm and radial blood vessels as anatomical landmarks. The aligned stack of sections was then compared to the blood vessel image from the initial intrinsic signal imaging (ISI) session. Major blood vessels were used as anatomical landmarks to align and warp the ISI blood vessel image to the aligned stack. ISI retinotopy and visual area borders were matched to the aligned ISI blood vessel layer. Cell counting and registration was done in FIJI for each section in the final aligned stack. Cells outside the visual areas were assigned into one of four cortical areas, anterior lateral, lateral, posterior lateral, medial (Harris et al., 2019). Two-way ANOVA with Sidak’s multiple comparisons test (for V1→AL and V1→PM of SepW1-Cre or Tlx3-Cre mice) or with Tukey’ s multiple comparisons test (for V1→AL, V1→PM, V1→P/POR) was used for statistical analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
The values of n and what n represents were reported in results and figure legends. Statistical Significance was assessed by two-way ANOVA with Sidak’s or Tukey’s post-hoc multiple-comparison tests using GraphPad Prism 7.0e for Figures 1D, 3D-E, and Table S2, by Wilcoxon rank-sum test using R [v3.5.1] for Figures 2G and S4B (smFISH puncta), and by Zinbwave-EdgeR for Figures 2C, 2G (logNorm expression), and S4A. For Figures 2C and 2G, p-values were adjusted for multiple testing by Benjamini-Hochberg method for Zinbwave-EdgeR pipeline. For all figures, *p<0.05; **p< 0.01; ***p< 0.001; ****p<0.0001.
Supplementary Material
Table S4. Single Nuclei Sequencing Sample Metadata, Related to Figure 2.
Table S5. Differentially Expressed Genes List for V1➔AL and V1➔PM in L2/3 Using Zinbwave- EdgeR, Related to Figure 2.
Table S6. Differentially Expressed Genes List for V1➔AL and V1➔PM in L5 and L6 Using Zinbwave-EdgeR, Related to Figure 2.
Table S7. Differentially Expressed Genes List for V1➔AL and V1➔PM in L2/3 Using Zinbwave- DESeq2, Related to Figure 2.
Table S8. Differentially Expressed Genes List for V1➔AL and V1➔PM in L5 and L6 Using Zinbwave-DESeq2, Related to Figure 2.
Table S9. Virus Injection and Animal Information for Rabies Tracing, Related to Figure 3.
Table S10. Quantification of Rabies Tracing Data, Related to Figure 3.
KEY RESOURCESTABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP, Alexa Flour 488 | Invitrogen | Cat#A-21311; PRID: AB_221477 |
| Goat polyclonal anti-GFP | Rockland | Cat#600-101-215; PRID: AB_218182 |
| Rabbit polyclonal anti-dsRed | Clontech | Cat#632496; PRID: AB_10013483 |
| Bacterial and Virus Strains | ||
| AAVretro-EF1a-Cre-WPRE | Salk Viral Core GT3 | N/A |
| scAAVretro-hSyn1-H2B-eGFP | Salk Viral Core GT3 | N/A |
| scAAVretro-hSyn1-H2B-mCherry | Salk Viral Core GT3 | N/A |
| AAVretro-CAG-FLEXloxp-Flp | Salk Viral Core GT3 | N/A |
| AAV8-CAG-FLEXfrt-TVAmCherry-WPRE | Salk Viral Core GT3 | N/A |
| AAV8-CAG-FLEXfrt-oG-WPRE | Salk Viral Core GT3 | N/A |
| AAV8-CAG-FLEXloxp-oG-WPRE | Salk Viral Core GT3 | N/A |
| AAV8-EF1a-FLEXloxp-eGFP-2A-TVA-WPRE | Salk Viral Core GT3 | N/A |
| AAVretro-nef-lox66/71-tTA | Salk Viral Core GT3 | N/A |
| AAV8-TRE-DIO-oG-WPRE | Salk Viral Core GT3 | N/A |
| AAV8-TRE-DIO-TVAmCherry-WPRE | Salk Viral Core GT3 | N/A |
| EnvA+RVdG-eGFP | Salk Viral Core GT3 | N/A |
| EnvA+RVdG-dsRed2 | Kim et al., 2015 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cholera Toxin Subunit B, Alexa Fluor 488 conjugated | Molecular Probes | Cat#C22841 |
| Cholera Toxin Subunit B, Alexa Fluor 555 conjugated | Molecular Probes | Cat#C22843 |
| Cholera Toxin Subunit B, Alexa Fluor 647 conjugated | Molecular Probes | Cat#C34778 |
| DL-Dithiothreitol | Sigma | Cat#646563; CAS: 3483-12-3 |
| Protease inhibitor | Sigma | Cat#P8340 |
| RNasin Plus RNase inhibitor | Promega | Cat#N2611 |
| Dylight 647-Lectin | Vectorlabs | Cat#DL-1178 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher | Cat#D1306; CAS: 28718-90-3 |
| Hoechst 33342 | Thermo Scientific | Cat#PI62249; CAS: 23491-52-3 |
| Triton X-100 | Thermo Fisher | Cat#AC327371000; CAS: 9002-93-1 |
| Chlorprothixene | Sigma | Cat#C1671; CAS: 6469-93-8 |
| Isoflurane | MWI Veterinary Supply | Cat#502017; CAS: 26675-46-7 |
| Critical Commercial Assays | ||
| SMART-Seq v4 Ultra Low Input RNA Kit | Clontech | Cat#634894 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131-1024 |
| RNAscope Fluorescent Multiplex Assay kit | ACD Biosciences | Cat#320850 |
| Ampure XP beads | Ampure XP beads | Cat#A63880 |
| Deposited Data | ||
| Single-nuclei RNA sequencing datasets | This manuscript | GSE133230 |
| Mouse reference genome mm10 (GENCODE M10) | UCSC Genome Browser | http://genome.ucsc.edu/cgibin/hgGateway?db=mm10 |
| Experimental Models: Organisms/Strains | ||
| Mouse: INTACT: B6.129-Gt(ROSA)26Sortm5(CAGSun1/sfGFP)Nat/MmbeJ | The Jackson Laboratory | JAX: 030952 |
| Mouse: SepW1-Cre NP39 | MMRRC | MMRRC: s037622-UCD |
| Mouse: Tlx3-Cre PL56 | MMRRC | MMRRC: 036547-UCD |
| Mouse: Wildtype: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Oligonucleotides | ||
| RNAscope Mm-Astn2 probe | ACD Biosciences | Cat#523871-C3 |
| RNAscope Mm-Kcnh5 probe | ACD Biosciences | Cat#497691-C2 |
| RNAscope Mm-Grm1 probe | ACD Biosciences | Cat#449781-C2 |
| RNAscope Mm-Cntn5 probe | ACD Biosciences | Cat#567461-C3 |
| Recombinant DNA | ||
| pAAV-CAG-FLEXloxp-Flp | Ren et al., 2018 | N/A |
| pAAV-CAG-FLEXfrt-TVAmCherry-WPRE | Schwarz et al., 2015 | N/A |
| Software and Algorithms | ||
| Cutadapt [v2.3] | Martin, 2011 | https://cutadapt.readthedocs.io/en/v2.3/develop.html |
| STAR [v2.5.1b] | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Picard DownsampleSam [v1.130] | Picard | http://broadinstitute.github.io/picard/ |
| Homer [v4.8] | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Seurat [v2.3.4] | Butler et al., 2018 | https://satijalab.org/seurat/ |
| Zinbwave [v1.4.1]-EdgeR [v3.20.9] | Butler et al., 2018 | https://bioconductor.org/packages/devel/bioc/vignettes/zinbwave/inst/doc/intro.html |
| Zinbwave [v1.4.1]-DESeq2 [v1.20.0] | Butler et al., 2018 | https://github.com/mikelove/zinbwave-deseq2 |
| Terastitcher | Bria and Iannello, 2012 | https://github.com/abria/TeraStitcher |
| reorg4terastitcher.py | Puifai Santisakultarm | N/A |
| FIJI | NIH ImageJ software | https://fiji.sc/ |
| CellSense | Olympus | N/A |
| Zen | Zeiss | N/A |
| Prism 7.0e | GraphPad | N/A |
| R [v3.5.1] | The R Project | https://www.r-project.org/ |
| Customized Matlab code for intrinsic signal imaging | Garrett and Nauhaus et al., 2014 | https://snlc.github.io/ISI/ |
| Imaris | Oxford Instruments | N/A |
| Other | ||
| BD Influx for FANS | BD Biosciences | N/A |
| HiSeq 4000 | Illumina | N/A |
| Picospritzer | General Valve Corp | N/A |
| BX63 microscope | Olympus | N/A |
| LSM780 microscope | Zeiss | N/A |
| VF-300 compresstome™ | Precisionary Instruments | N/A |
| SZX16 dissection microscope | Olympus | N/A |
| Leica CM 1950 cryostat | Leica | N/A |
| Z.1 lightsheet microscope | Zeiss | N/A |
| Airyscan 880 | Zeiss | N/A |
HIGHLIGHTS.
V1 CCPNs display differential gene expression depending on their projections
V1 CCPNs receive feedback inputs from the same areas to which they project
Gene expression analysis in isolation is insufficient to identify neuron types
ACKNOWLEDGEMENTS
We thank all Callaway lab members for discussion, K. Fischer for reading the manuscript, M. Singer and E. Drokhlyansky for RNAseq analysis help during the early phase of the project, B. Jaeger and S. Linker for sharing their single-nuclei preparation protocol, J. Ren and L. Luo for sharing pAAV-CAG-FLEXloxp-Flp and pAAV-CAG-FLEXfrt-TVAmCherry-WPRE plasmids, A. Levine and S. Pfaff for sharing pscAAV plasmid backbone, R. Chinoy for cell counting help. We also thank J. Naughton and J. Marlett at the Salk viral vector core, U. Manor at the Salk advanced biophotonics core, C. Fitzpatrick and C. O’Connor at the Salk flow cytometry core, N. Hah at the Salk next generation sequencing core, M. Shokhirev at the Salk integrative genomics and bioinformatics core and all other staff members. This work was supported by the National Institutes of Health grants EY022577 and MH063912 and by The Brain Research Foundation (E.M.C). The Razavi Newman Integrative Genomics and Bioinformatics Core Facility of the Salk Institute was supported by NIH-NCI CCSG: P30 014195, and the Helmsley Trust. E.J.K. was a Biogen-IDEC Fellow of the Life Science Research Foundation and a recipient of 2012 NARSAD Young Investigator Award from Brain & Behavior Research Foundation.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andermann ML, Kerlin AM, Roumis DK, Glickfeld LL, and Reid RC (2011). Functional specialization of mouse higher visual cortical areas. Neuron 72, 1025–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bria A, and lannello G (2012). TeraStitcher - a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics 13, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte WL, Kamishina H, and Reep RL (2009). The efficacy of the fluorescent conjugates of cholera toxin subunit B for multiple retrograde tract tracing in the central nervous system. Brain Structure and Function 213, 367–373. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut R, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economo MN, Viswanathan S, Tasic B, Bas E, Winnubst J, Menon V, Graybuck LT, Nguyen TN, Smith KA, Yao Z, et al. (2018). Distinct descending motor cortex pathways and their roles in movement. Nature 563, 79–84. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, and Van Essen DC (1991). Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex 1, 1–47. [DOI] [PubMed] [Google Scholar]
- Garrett ME, Nauhaus I, Marshel JH, and Callaway EM (2014). Topography and Areal Organization of Mouse Visual Cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 12587–12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, and Heintz N (2013). GENSAT BAC Cre-Recombinase Driver Lines to Study the Functional Organization of Cerebral Cortical and Basal Ganglia Circuits. Neuron 80, 1368–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickfeld LL, Andermann ML, Bonin V, and Reid RC (2013). Cortico-cortical projections in mouse visual cortex are functionally target specific. Nat Neurosci 16, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, and Gerfen CR (2007). Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci 27, 9817–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa M, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, et al. (2015). ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Han Y, Kebschull JM, Campbell RAA, Cowan D, Imhof F, Zador AM, and Mrsic-Flogel TD (2018). The logic of single-cell projections from visual cortex. Nature 556, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Mihalas S, Hirokawa KE, Whitesell JD, Choi H, Bernard A, Bohn R, Caldqon S, Casal L, Cho A, et al. (2019). Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo R, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh CYL, Peach JP, Bennett C, Vega RM, and Hestrin S (2018). Feature-Specific Organization of Feedback Pathways in Mouse Visual Cortex. Curr Biol 28, 114–120 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juavinett AL, Nauhaus I, Garrett ME, Zhuang J, and Callaway EM (2017). Automated identification of mouse visual areas with intrinsic signal imaging. Nature protocols 12, 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Jacobs MW, Ito-Cole T, and Callaway EM (2016). Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins Cell Rep. [DOI] [PMC free article] [PubMed]
- Kim EJ, Juavinett AL, Kyubwa EM, Jacobs MW, and Callaway EM (2015). Three Types of Cortical Layer 5 Neurons That Differ in Brain-wide Connectivity and Function. Neuron 88, 1253–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Linker SB, Jaeger BN, Krishnaswami S, Barron J, Kelder M, Parylak S, Paquola A, Venepally P, Novotny M, et al. (2016). Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun 7, 11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Garrett ME, Nauhaus I, and Callaway EM (2011). Functional Specialization of Seven Mouse Visual Cortical Areas. Neuron 72, 1040–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3. [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Heard S, Urich MA, Nery JR, Sqnowski TJ, Lister R, et al. (2015). Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ, and Franklin KBJ (2001). The mouse brain in stereotaxic coordinates, 2nd edn (San Diego: Academic Press; ). [Google Scholar]
- Poulin JF, Tasic B, Hjerling-Leffler J, Trimarchi JM, and Awatramani R (2016). Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci 19, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Ramón y Cajal S, Swanson LW, and Swanson N (1995). Histology of the nervous system of man and vertebrates Cerebellum, midbrain, retina, diencephalon, corpus striatum, cerebral cortex - in general and regional, autonomic system Vol. 2 Vol. 2 (Oxford: Oxford University Press; ). [Google Scholar]
- Ren J, Friedmann D, Xiong J, Liu CD, Ferguson BR, Weerakkody T, DeLoach KE, Ran C, Pun A, Sun Y, et al. (2018). Anatomically Defined and Functionally Distinct Dorsal RapheSerotonin Sub-systems. Cell 175, 472–487 e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Perraudeau F, Gribkova S, Dudoit S, and Vert JP (2018). A general and flexible method for signal extraction from single-cell RNA-seq data Nat Commun 9, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, et al. (2015). Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524, 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B (2018). Single cell transcriptomics in neuroscience: cell classification and beyond. Curr Opin Neurobiol 50, 242–249. [DOI] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay R, Lee S, and Rudy B (2016). GABAergic Interneurons in the Neocortex: From Cellular Properties to Circuits. Neuron 91, 260–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, and Burkhalter A (2007). Area map of mouse visual cortex. The Journal of comparative neurology 502, 339–357. [DOI] [PubMed] [Google Scholar]
- Zeng H, and Sanes JR (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci 18, 530–546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. Single Nuclei Sequencing Sample Metadata, Related to Figure 2.
Table S5. Differentially Expressed Genes List for V1➔AL and V1➔PM in L2/3 Using Zinbwave- EdgeR, Related to Figure 2.
Table S6. Differentially Expressed Genes List for V1➔AL and V1➔PM in L5 and L6 Using Zinbwave-EdgeR, Related to Figure 2.
Table S7. Differentially Expressed Genes List for V1➔AL and V1➔PM in L2/3 Using Zinbwave- DESeq2, Related to Figure 2.
Table S8. Differentially Expressed Genes List for V1➔AL and V1➔PM in L5 and L6 Using Zinbwave-DESeq2, Related to Figure 2.
Table S9. Virus Injection and Animal Information for Rabies Tracing, Related to Figure 3.
Table S10. Quantification of Rabies Tracing Data, Related to Figure 3.
Data Availability Statement
The single-nuclei RNA sequencing datasets generated in this study are available in the GEO repository under accession number GSE133230. The code necessary to generate the figures in this manuscript will be available from the corresponding author upon reasonable request.