Summary
Rapid spread of coronavirus disease 2019 (COVID-19) is ravaging the globe. Since its first report in December 2019, COVID-19 cases have exploded to over 14 million as of July 2020, claiming more than 600,000 lives. Implementing fast and widespread diagnostic tests is paramount to contain COVID-19, given the current lack of an effective therapeutic or vaccine. This review focuses on a broad description of currently available diagnostic tests to detect either the virus (SARS-CoV-2) or virus-induced immune responses. We specifically explain the working mechanisms of these tests and compare their analytical performance. These analyses will assist in selecting most effective tests for a given application, for example, epidemiology or global pandemic research, population screening, hospital-based testing, home-based and point-of-care testing, and therapeutic trials. Finally, we lay out the shortcomings of certain tests and future needs.
Subject Areas: Virology, Diagnostic Equipment, Infection Control in Health Technology, Analytical Chemistry
Graphical Abstract
Virology; Diagnostic Equipment; Infection Control in Health Technology; Analytical Chemistry
Introduction
The importance of diagnostic tests for population management is nowhere clearer than with the current coronavirus disease 2019 (COVID-19) pandemic. South Korea, for example, rapidly validated and deployed test kits by private companies (Lee and Lee, 2020), examining over 200,000 individuals within the first 7 weeks of COVID-19 (Shim et al., 2020). Ensuing isolation and contact tracing enabled the country to slow down disease spread without imposing severe social and economic lockdown. A similar success was observed in Germany, which performed nearly 120,000 tests a day. The United States in contrast lagged behind, performing about 5,000 tests in the first 7 weeks in the entire country, and likely missed the critical window to contain the outbreak (Bialek et al., 2020). Centralized testing by the US Centers for Disease Control and Prevention (US-CDC) and other health care organizations have given way to automated testing using commercial platforms. As of July 2020, over 110 commercial molecular tests have received the Emergency Use Authorization (EUA) from US Food and Drug Administration (FDA), while over 300 antibody-based tests, which do not require FDA clearance, flooded the market (Food and Drug Administration, 2020; Foundation for Innovative New Diagnostics, 2020). The diagnostic performance of available tests varies widely, which could lead to confusing results (Maxmen, 2020) and sometimes ill-advised policies.
This technical review aims to evaluate key diagnostic technologies for COVID-19 disease. We focus on (1) explaining the underlying working principles of major tests, (2) comparing their analytical parameters and potential limitations, and (3) surveying initial performance with clinical samples. We envision that this comparative report will help clinicians, researchers, and health care agencies to evaluate the standards of existing tests as well as further improve them for the future.
COVID-19 Etiology
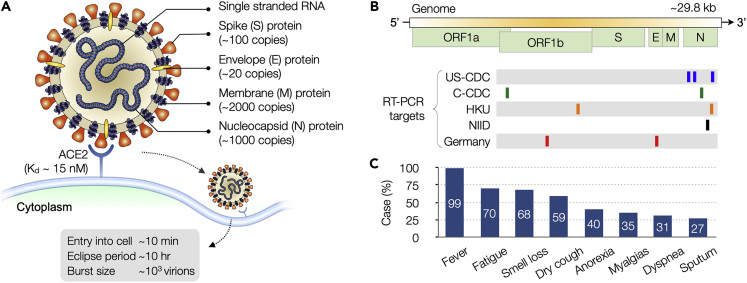
COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; Figure 1A). It is a single-strand RNA virus belonging to the beta family of coronaviruses, which also include SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus OC43 (HCoV-OC43), and human coronavirus HKU1 (HCoV-HKU1). The viral envelope of SARS-CoV-2 consists of a lipid bilayer. Petal-shaped spikes are composed of heavily glycosylated type I glycoproteins (S protein) and anchored on the envelope along with membrane (M) and envelop (E) proteins. Inside the envelope resides a ribonucleoprotein (RNP) core, which comprises the RNA genome and a single species of nucleocapsid (N) protein. SARS-CoV-2 genome (29.8 kb) codes for 10 genes to produce 26 proteins (Figure 1B) (Chen et al., 2020). The genes are arranged in the order 5′-replicase-S-E-M-N-3′, with genes for accessory proteins interspersed among structural ones (S, E, M, N). About two-thirds of the entire RNA is occupied by the polymerase gene, which has two overlapping open reading frames (ORFs), 1a and 1b. The entry of the virus to host cells is mediated by S protein upon recognition of the peptidase domain of angiotensin-converting enzyme 2 (ACE2) by S1 subunit (Wrapp et al., 2020), followed by viral and cellular membrane fusion through S2 subunit (Yan et al., 2020b; Lai et al., 2020). It has been reported that SARS-CoV-2 S protein binds to ACE2 with higher affinity than that of SARS-CoV (Wrapp et al., 2020).
Figure 1.
SARS-CoV-2 Virus and COVID-19
(A) The virus is enveloped and spherical (~120 nm in diameter), with petal-shaped surface spikes (~20 nm long). Key structural proteins, spike (S), envelope (E), and membrane (M), are anchored on the viral envelop. The nucleocapsid (N) protein, together with the genomic RNA, forms a helical nucleocapsid inside the envelop. The virus enters host cells through the binding of S protein to angiotensin-converting enzyme 2 (ACE2) on the cell surface.
(B) SARS-CoV-2 virus contains a positive-sense, single-stranded RNA genome. The organization of genome is 5′-leader-UTR (untranslated region)-replicase-S-E-M-N-3′-UTR-poly (A) tail. The open reading frame (ORF) 1a and 1b encode the replicase. Location of target genes for select COVID-19 RT-PCR tests are shown. Many tests target N gene, because sequence conservation of this gene within the coronavirus genus is low.
(C) Patients with COVID-19 display symptoms similar to those of common cold and influenza, and in some cases are asymptomatic. Diagnostics must be confirmed through highly specific molecular tests. US-CDC, United States Center for Disease Control and Prevention; C-CDC, Chinese Center for Disease Control and Prevention; HKU, Hong Kong University; NIID, National Institute of Infectious Diseases (Japan). (A) Adapted with permission from Ref (Graham et al., 2013). Copyright 2013 Nature Publishing Group. (B) Adapted with permission from Ref (Jung et al., 2020).
SARS-CoV-2 can infect humans and a small number of animals (Zhou et al., 2020). Human-to-human transmission is through droplets or direct contact (Andersen et al., 2020). Molecular tests are critical for COVID-19 diagnosis, as its symptoms (e.g., fever, fatigue, dry cough, breathing difficulties) overlap with those of common cold and influenza (Figure 1C). Like other viral tests, different molecular targets are available. Viral nucleic acids or proteins can be used to detect the presence of virus in patients to diagnose acute infection. Detecting antibodies that are generated by hosts against the virus can tell the history of a past infection and immunity gained to the disease. Selecting specific targets would determine the proper assay formats and technologies (Table 1) that are detailed in the following sections. Up-to-date information on FDA-cleared commercial tests is available at https://csb.mgh.harvard.edu/covid.
Table 1.
Comparison of Diagnostic Tests of COVID-19
| Intended Use | Methods | Description | Operating Temperature | Analytical Sensitivity | Specificity | Assay time | Ref. |
|---|---|---|---|---|---|---|---|
|
RT-qPCR |
|
Thermal cycling | 0.14 copy/μL | 96%–100% | 2–4 h | (Corman et al., 2020) |
| ddPCR |
|
Thermal cycling | 0.02 copy/μL | 94.9% | 1 h | (Suo et al., 2020) | |
| RT-LAMP |
|
60°C–65°C | 4.8 copy/μL | 99% | 15–60 min | (Zhang et al., 2020b) | |
| RT-RPA |
|
37°C–42°C | 0.2 copy/μL | NA | 30 min | (Xia and Chen, 2020) | |
| RT-NEAR |
|
55°C–59°C | 0.13 copy/μL | 100% | <15 min | (Abbott, 2020) | |
| DETECTR |
|
62°C for RT-LAMP 37°C for LbCas12a |
10 copy/μL | PPV: 95% NPV: 100% |
30 min | (Broughton et al., 2020) | |
| STOP |
|
60°C for RT-LAMP 60°C for AapCas12b |
2 copy/μL | 100% | 70 min | (Joung et al., 2020) | |
|
ELISA |
|
Ambient | 100 pg/mL | 99.3% | 3–5 h | (Freeman et al., 2020) |
|
RDT |
|
Ambient | Qualitative | 84.2%–100% | <15 min | (Liu et al., 2020c) |
| ELISA |
|
Ambient | 100 pg/mL | 67%–98.6% | 3–5 h | (SinoBiological, 2020) | |
| Neutralization test |
|
Ambient | 0.22 copy/μL | 100% | 2–3 days | (Tan et al., 2020) |
RT, reverse transcription; PCR, polymerase chain reaction; qPCR, quantitative PCR; ddPCR, digital droplet PCR; LAMP, loop-mediated isothermal amplification; RPA, recombinase polymerase amplification; NEAR, nicking endonuclease amplification reaction; DETECTR, DNA endonuclease-targeted CRISPR transreporter; STOP, SHERLOCK Testing in One Pot; NA, not available for clinical samples; PPV, positive predictive value; NPV, negative predictive value; ELISA, enzyme=linked immunoabsorbent assay; RDT, rapid diagnostic test.
Nucleic Acid Amplification Tests
Nucleic acid amplification tests (NAATs) for COVID-19 diagnostics are designed to detect unique viral RNA sequences in N, E, S, or RNA-dependent RNA polymerase (RdRp) genes. The viral genome of original SARS-CoV-2 was sequenced (Chen et al., 2020) and released in January 2020 (Wuhan-Hu-1, GenBank: MN908947.3), enabling fast development of COVID-19 NAATs. Since then different strains have been sequenced many times providing (1) a clearer picture of mutations and conserved sites and (2) global evolution of different strains. Current NAAT primers and reagents are developed based on this information. NAATs offer a high accuracy; after taking samples and transporting them to laboratories, results are typically obtained within a couple of hours with a limit of detection (LOD) down to 0.02 copy/μL (Suo et al., 2020). As such, NAATs are recommended for acute disease detection even when the patients have mild or non-specific symptoms (e.g., fever, cough). A number of different NAATs are available for COVID-19 diagnosis.
Sample Collection and Transport
Sample collection and storage is an important pre-analytical factor affecting the overall assay performance. The US-CDC guidelines list upper and lower respiratory tract specimens, such as nasopharyngeal (NP) or oropharyngeal (OP) swabs, sputum, lower respiratory tract aspirates, bronchoalveolar lavage, and nasopharyngeal wash/aspirate or nasal aspirate (CDC, 2020a). Alternative sources include saliva (To et al., 2020), anal swabs (Zhang et al., 2020a), urine and stool (Xie et al., 2020a), tears, and conjunctival secretions (Xia et al., 2020). For initial diagnoses, the US-CDC recommends collecting an upper respiratory specimen, prioritizing the NP swab, although OP swabs remain an acceptable specimen type (CDC, 2020b).
Swabs are the most widely used tools for sample collection and considered as FDA Class I-exempt medical devices. As for materials, synthetic fiber (e.g., nylon, polyester filaments) swabs with plastic shafts should be used. Calcium alginate swabs or swabs with wooden shafts should be avoided because they may contain substances that inactivate some viruses and can inhibit PCR testing (CDC, 2020b). For high viral yields, sample collection with a flocked swab is preferred (Daley et al., 2006). The rapid spread of COVID-19, however, has resulted in shortages of NP swabs because of unprecedented high demands. Responding to this bottleneck, an open-development consortium started developing 3D-printed NP swabs that can be mass produced (Callahan et al., 2020). The team tested different designs and materials and validated promising candidates in clinical trials. These efforts led to FDA-registered test swabs with superior or equivalent efficacy to flocked swabs (Callahan et al., 2020).
For specimen transport and storage, the swab material should be placed in a sterile tube filled with viral transport medium (VTM) and kept refrigerated (2–8°C) for up to 72 h after collection. If a delay in testing or shipping is expected, specimens should be stored at −70°C or below. The US-CDC- and World Health Organization (WHO)-recommended VTM is based on Hanks-balanced salt solution (HBSS) and contains heat-inactivated fetal bovine serum and antibiotics (gentamycin and amphotericin B). VTM shortage has also been experienced during the COVID-19 pandemic, impairing local and regional capacity for diagnosis (Radbel et al., 2020). Radbel et al. tested phosphate-buffered saline (PBS) as a potential alternative to VTM (Radbel et al., 2020). Using clinical endotracheal secretion samples (n = 16), the authors evaluated the stability of the PCR signal from three viral targets (N, ORF1ab, and S genes) when samples were stored in these media at room temperature for up to 18 h. The test results were similar between PBS and VTM-based storages, which may establish PBS as a cost-effective media for short-term preservation of specimens. The study, however, used tracheal secretions from mechanically ventilated patients. Futher valdiations with NS swabs are needed.
COVID-19 RT-PCR Test
Reverse transcription polymerase chain reaction (RT-PCR) was the first method developed for COVID-19 detection and is the current gold standard (Corman et al., 2020). WHO adopted its version of RT-PCR test and implemented it in different countries (Sohrabi et al., 2020). In the United States, the CDC developed its own standards (CDC, 2020a).
RT-PCR assay starts with extracting RNA from clinical specimens. Several commercial kits are recommended by the US-CDC for this process (Table 2). These kits are based on solid-phase extraction using silica substrates; negatively charged nucleic acids selectively bind to positively charged silica surface in the presence of chaotropic ions. Following wash steps, adsorbed nucleic acids are eluted with low-salt solution. To remove DNA contamination, the eluate is treated with DNase, followed by heat treatment (15 min, 70°C) to inactivate the DNase. Using magnetic beads coated with silica can facilitate sample handling, eliminating the need for centrifugation. Several extraction platforms indeed employ magnetic actuation for automation of high-throughput sample processing (Ali et al., 2017). Next, the extracted viral RNA is mixed with reagents containing target gene primers, probes, and RT-PCR master mix and amplified. Depending on the probe design, PCR products can be detected during the amplification process (quantitative PCR, qPCR) or after its completion.
Table 2.
Commercial RNA Extraction Kits
| Manufacturer and Kit Name | Catalog No. | |
|---|---|---|
| QIAGEN | QIAmp DSP Viral RNA Mini Kit | 50 extractions (61,904) |
| QIAamp Viral RNA Mini Kit | 50 extractions (52,904); 250 extractions (52,906) | |
| EZ1 Advanced XL DSP Virus Kit | 48 extractions (62,724) Buffer AVL (19073) EZ1 Advanced XL DSP Virus Card (9018703) |
|
| EZ1 Advanced XL Virus Mini Kit v2.0 | 48 extractions (955134) Buffer AVL (19073) EZ1 Advanced XL Virus Card v2.0 (9018708) |
|
| QIAcube QIAamp DSP Viral RNA Mini Kit | 50 extractions (61,904) | |
| QIAcube QIAamp Viral RNA Mini Kit | 50 extractions (52,904); 250 extractions (52,906) | |
| Roche | MagNA Pure LC Total Nucleic Acid Kit |
192 extractions (03,038 505 001) |
| MagNA Pure Compact Nucleic Acid Isolation Kit I |
32 extractions (03,730 964 001) | |
| MagNA Pure 96 DNA and Viral NA Small Volume Kit |
576 extractions (06,543 588 001) External Lysis Buffer (06,374 913 001) |
|
| bioMérieux | NucliSENS easyMAG Instrument EMAG Instrument Automated magnetic extraction system Reagents sold separately. Both instruments use the same reagents and disposables, with the exception of tips. |
easyMAG Magnetic Silica (280133) easyMAG Lysis Buffer (280134) easyMAG Lysis Buffer, 2 mL (200292) easyMAG Wash Buffers 1, 2, 3 (280130, 280131, 280132) easyMAG Disposables (280135) Biohit Pipette Tips (easyMAG only) (280146) EMAG1000μL Tips (418922) easyMAG Disposables (280135) Biohit Pipette Tips (easyMAG only) (280146) EMAG1000μL Tips (418922) |
Analytical accuracy of COVID-19 RT-PCR relies primarily on the primer design. Due to high genomic similarity among different coronavirus species, identifying unique gene sequences is important to eliminate cross-reactivity. Viral targets are selected from E, N, S, and Orf1ab regions of SARS-CoV-2 genome (Figure 1B), and human RNAse P (RP) is used for internal positive control (Jung et al., 2020). Table 3 shows selected primer-probe sets announced by WHO. According to the initial comparison of these probes, US-CDC 2019-nCoV_N2, 2019-nCoV_N3, and Japanese NIID_2019-nCOV_N primer sets were highly sensitive for the N gene, and the Chinese-CDC ORF1ab panel was sensitive for the ORF1ab gene (Jung et al., 2020). The N3 assay manufactured by the US-CDC, however, encountered false-positive issues and was removed from the US-CDC diagnostic panel in March 2020 (CDC, 2020a). A study by the US-CDC showed that removing the N3 assay had negligible effects on sensitivity to detect SARS-CoV-2 (Lu et al., 2020c). Targeting only N1 and N2 also simplified the overall test, increasing throughput and reducing cost (Lu et al., 2020c). As more commercial and laboratory-developed RT-PCR tests become available, it is increasingly critical to evaluate their performance using a common standard. The Foundation for Innovative New Diagnostics (FIND), in partnership with WHO, is now conducting independent evaluations of SARS-CoV-2 molecular tests (Foundation for Innovative New Diagnostics, 2020).
Table 3.
Primers and Probes for RT-PCR COVID-19 Diagnostics
| Target | Country | Name | Type | Sequence (5’ → 3′) |
|---|---|---|---|---|
| N | US-CDC | 2019-nCoV_N1-F | F | GAC CCC AAA ATC AGC GAA AT |
| 2019-nCoV_N1-R | R | TCT GGT TAC TGC CAG TTG AAT CTG | ||
| 2019-nCoV_N1-P | P | ACC CCG CAT TAC GTT TGG TGG ACC | ||
| 2019-nCoV_N2-F | F | TTA CAA ACA TTG GCC GCA AA | ||
| 2019-nCoV_N2-R | R | GCG CGA CAT TCC GAA GAA | ||
| 2019-nCoV_N2-P | P | ACA ATT TGC CCC CAG CGC TTC AG | ||
| 2019-nCoV_N3-F | F | GGG AGC CTT GAA TAC ACC AAA A | ||
| 2019-nCoV_N3-R | R | TGT AGC ACG ATT GCA GCA TTG | ||
| 2019-nCoV_N3-P | P | AYC ACA TTG GCA CCC GCA ATC CTG | ||
| Charité Germany |
N_Sarbeco_F | F | CAC ATT GGC ACC CGC AAT C | |
| N_Sarbeco_R | R | GAG GAA CGA GAA GAG GCT TG | ||
| N_Sarbeco_P | P | ACT TCC TCA AGG AAC AAC ATT GCC A | ||
| Chinese-CDC | CCDC-N-F | F | GGG GAA CTT CTC CTG CTA GAA T | |
| CCDC-N-R | R | CAG ACA TTT TGC TCT CAA GCT G | ||
| CCDC-N-P | P | TTG CTG CTG CTT GAC AGA TT | ||
| Hong Kong University |
HKU-N-F | F | TAA TCA GAC AAG GAA CTG ATT A | |
| HKU-N-R | R | CGA AGG TGT GAC TTC CAT G | ||
| HKU-N-P | P | GCA AAT TGT GCA ATT TGC GG | ||
| NIID Japan | NIID_2019-nCOV_N_F2 | F | AAA TTT TGG GGA CCA GGA AC | |
| NIID_2019-nCOV_N_R2 | R | TGG CAG CTG TGT AGG TCA AC | ||
| NIID_2019-nCOV_N_P2 | P | ATG TCG CGC ATT GGC ATG GA | ||
| E | Charité Germany |
E_Sarbeco_F1 | F | ACA GGT ACG TTA ATA GTT AAT AGC GT |
| E_Sarbeco_R2 | R | ATA TTG CAG CAG TAC GCA CAC A | ||
| E_Sarbeco_P1 | P | ACA CTA GCC ATC CTT ACT GCG CTT CG | ||
| RdRp | Charité Germany |
RdRp_SARSr-F | F | GTG ARA TGG TCA TGT GTG GCG G |
| RdRp_SARSr-R | R | CAR ATG TTA AAS ACA CTA TTA GCA TA | ||
| RdRp_SARSr-P2 | P | CAG GTG GAA CCT CAT CAG GAG ATG C | ||
| ORF1 | Chinese-CDC | CCDC-ORF1-F | F | CCC TGT GGG TTT TAC ACT TAA |
| CCDC-ORF1-R | R | ACG ATT GTG CAT CAG CTG A | ||
| CCDC-ORF1-P | P | CCG TCT GCG GTA TGT GGA AAG GTT ATG G | ||
| Hong Kong University |
HKU-ORF1-F | F | TGG GGY TTT ACR GGT AAC CT | |
| HKU-ORF1-R | R | AAC RCG CTT AAC AAA GCA CTC | ||
| HKU-ORF1-P | P | TAG TTG TGA TGC WAT CAT GAC TAG | ||
| RNAse P (control) | US-CDC | RP-F | F | AGA TTT GGA CCT GCG AGC G |
| RP-R | R | GAG CGG CTG TCT CCA CAA GT | ||
| RP-P | P | TTC TGA CCT GAA GGC TCT GCG CG |
RT-PCR offers both high accuracy and throughput. The LOD was reportedly down to 4–8 copies of the virus upon amplification of Orf1ab, E, and N genes at 95% confidence intervals (Xia et al., 2020; Han et al., 2020; Zou et al., 2020), and multiple assays can be carried out in a parallel format (e.g., 384-well plate). Specificity of a test is enhanced by targeting multiple loci. Indeed, the US-CDC diagnostic recommends the use of two targets (N1, N2) in N gene and RP as a control; WHO recommends E gene assay for first-line screening and further confirming positive cases with RdRp gene assay. The metric for COVID-19 diagnosis is the cycle threshold (Ct). A Ct value less than 40 is clinically reported as PCR positive. Viral RNA loads become detectable as early as day 1 of symptom onset and peak within a week. The positivity declines by week 3 and subsequently becomes undetectable (Sethuraman et al., 2020). RT-PCR tests are usually performed in centralized laboratories due to the requirement of dedicated equipment, trained personnel, and stringent contamination control. Establishing efficient logistics for sample transfer and securing reagents are critical to minimize delays in assay turnaround. Proper sample preprocessing (e.g., sample collection, RNA extraction) is also key to reduce false-negatives (Ai et al., 2020; Xie et al., 2020b).
Digital PCR Based SARS-CoV-2 Detection
Digital PCR enables the absolute quantification of target nucleic acids. The method partitions samples into large numbers of small (∼nanoliters) reaction volumes, ensuring that each partition contains a few or no target sequence per Poisson's statistics (Baker, 2012). Following PCR, amplification-positive partitions are counted for quantification. Among various partitioning methods (e.g., microwell plates, capillaries, oil emulsion, miniaturized chambers), droplet digital PCR (ddPCR) is the most widely used method with commercial systems available (Hindson et al., 2013). ddPCR has higher sensitivity (∼10−2 copy/μL) than conventional PCR, which makes it possible to detect very low viral loads. For example, when pharyngeal swab samples from patients with COVID-19 who were convalescing were compared, ddPCR detected viral RNA (Chinese CDC sequences) in 9 of 14 (64.2%) RT-PCR-negative samples (Dong et al., 2020). In another ddPCR application, researchers tracked treatment progress by analyzing clinical samples collected at different dates. ddPCR reported decrease in viral load as treatment proceeds, whereas RT-PCR showed sporadic appearance of positive results. Viral loads of specimens collected from different locations of the same patient were compared as well: the load was the highest in pharyngeal samples, lower in stool samples, and the lowest in serum (Lu et al., 2020a).
COVID19-NAATs Based on Isothermal Amplification
Applying isothermal amplification enabled the development of point-of-care (POC) COVID-19-NAATs. This amplification technique uses specialized DNA polymerases with the capacity of strand displacement; the polymerases can push their way in and unzip a double-strand DNA as they synthesize a complementary strand. Importantly, the reaction takes place at a fixed temperature, removing thermal cycling steps and thereby simplifying device design. Various isothermal amplification methods have been adapted to detect SARS-CoV-2 RNA targets (Zhang et al., 2020b; Yu et al., 2020; Lu et al., 2020b; Zhu et al., 2020). Analytical sensitivities of those isothermal amplification methods were shown to be comparable to that of RT-PCR, but with shorter assay time (<1 h).
Isothermal NAATs have unique applications in POC COVID-19 diagnostics, providing fast results without need for specialized equipment (Foo et al., 2020; Yan et al., 2020a). Practical considerations. however, still position RT-PCR as the principal method: (1) RT-PCR has been a gold standard over decades and has a well-developed supply chain for reagents and equipment; (2) RT-PCR is simpler in the primer design and requires fewer additives, which brings down the cost per test; (3) in clinical laboratories where large batches of samples are processed, RT-PCR easily makes up for the speed advantage of isothermal NAATs; and (4) RT-PCR is license free with most patents expired, whereas major isothermal NAATs are proprietary products.
Loop-Mediated Isothermal Amplification
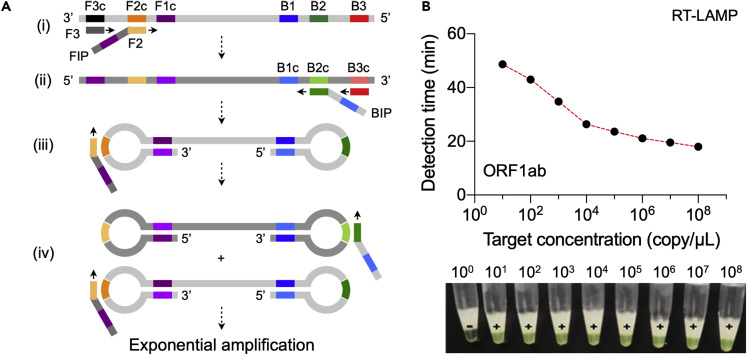
Loop-mediated isothermal amplification (LAMP) uses 4 or 6 primers, targeting 6–8 regions in the genome, and Bsm DNA polymerase (Notomi et al., 2015). As the reaction starts, pairs of primers generate a dumbbell-shaped DNA structure, which subsequently functions as the LAMP initiator (Figure 2A). The method can generate ∼109 DNA copy within an hour, and the reaction takes place at constant temperature between 60°C and 65°C (Ménová et al., 2013). The enzyme is resistant to inhibitors in complex samples, making it possible to use native samples (blood, urine, or saliva) with minimal processing. LAMP reaction produces magnesium pyrophosphate as a by-product, which can be exploited for visual readout of the assay using metal-sensitive indicators or pH-sensitive dyes. FDA-approved LAMP tests are already available for Salmonella and Cytomegalovirus detection (Yang et al., 2018; Schnepf et al., 2013).
Figure 2.
LAMP-Based COVID-19 Test
(A) LAMP mechanism. (i) The reaction mix contains dNTPs, DNA polymerase with high displacement activity, and pairs of primers (F1-F1c; F2-F2c; F3-F3c; B1-B1c; B2-B2c; B3-B3c). Forward inner primer (FIP) binds to the F2c region in the target and is extended. F3 primer also hybridizes to the F3c region and is extended, displacing the FIP-linked complementary strand. (ii) The displaced single-stranded DNA serves as a template for extension reactions by backward inner primer (BIP) and B3 primer. (iii) The BIP-linked DNA self-anneals and forms a dumbbell-like structure that initiates subsequent rounds of amplification. FIP binds and opens up the loop at the 3′-end converting the dumbbell shape into a stem-loop structure through its extension toward 5′ end. (iv) The extended strand from (iii) forms a new loop and serves as a template for BIP binding and extension. Both these products become the seed for the exponential amplification.
(B) RT-LAMP-based detection of ORF1ab gene. The amplification process monitored via turbidimeter readings at 650 nm or visual observation of calcein-mediated color change from orange to green. The target concentration is inversely proportional to detection time and the color change.
Reproduced with permission from Ref (Yan et al., 2020a). Copyright 2020 European Society of Clinical Microbiology and Infectious Diseases.
Designing primer sets is a key challenge when developing COVID19-LAMP assays, as multiple pairs of primers are required for a given target sequence and the melting temperature of these primers should match with the optimum working temperature of the DNA polymerase (Notomi et al., 2000). Fortunately, online software (Primer Explorer V5) is available to facilitate the process (Eiken Chemical Co. Ltd, 2020).
Most studies reported primer sets targeting regions of SARS-CoV-2 ORF1a and N genes (Yu et al., 2020; Lu et al., 2020b; Park et al., 2020; Yan et al., 2020a). Using these sets, the typical assay run-time was ∼1 h, and the LOD in the order of 10 copy/μL (Figure 2B). Zhu et al. reported analytical sensitivity of 100% for 33 SARS-CoV-2-positive oropharynx swab samples and 96 SARS-CoV-2-negative samples (Zhu et al., 2020). Importantly, the entire reaction could be performed in one pot (RT-LAMP) by using a master mix containing reverse transcriptase (e.g., NEB WarmStart Colorimetric LAMP 2X Master Mix). The total assay time, however, could be > 1 h (90–150 min) when manual sample handling steps are included (e.g., RNA extraction). Another drawback is the difficulty in multiplexing. With each target requiring 4–6 primers, increasing target numbers could easily complicate the primer design and the chance of primer-primer interactions.
Nicking Endonuclease Amplification Reaction
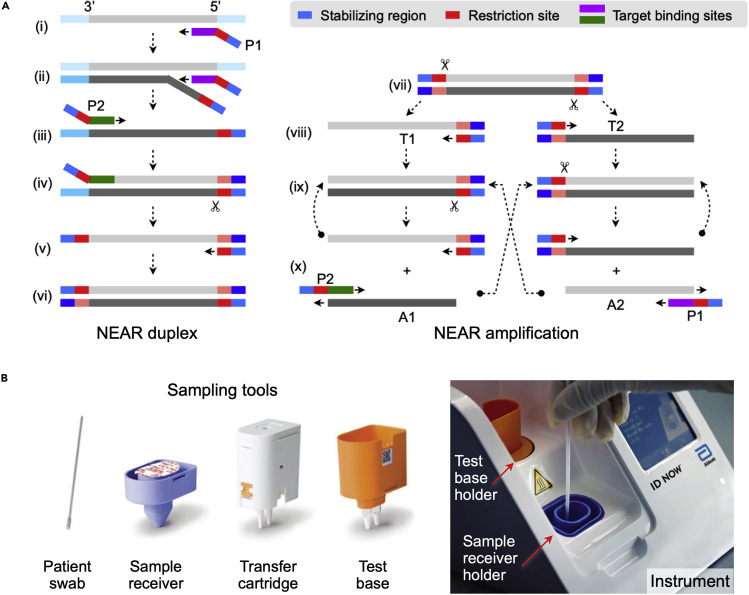
Nicking endonuclease amplification reaction (NEAR) uses both strand-displacement DNA polymerase (e.g., Bst polymerase) and nicking endonuclease enzymes to exponentially amplify short oligonucleotides (Wang et al., 2018b). Figure 3A shows the two-step working mechanism. First, nicking primers (P1, P2), each containing a restriction or nicking site, a stabilizing region, and a binding sequence, are mixed with a sample. Primer binding, displacement extension, and nicking action produce double-stranded DNA with restriction sites at both ends (NEAR amplification duplex; Figure 3A, left). Next, nicking enzymes cleave the restriction sites of the duplex, making two free-ended templates (T1, T2; Figure 3A, right) that are not stable due to elevated temperature (55°C) and are ready to dissociate (Ménová et al., 2013). Each template undergoes repeated polymerization and single-strand cleavage, which results in the amplification of products (A1, A2). These products also hybridize with primers (A1-P2; A2-P1) and contribute to successive amplification in a bidirectional manner until the depletion of reaction mixture components. In this way, thousands of copies could be produced from one restriction side, which makes NEAR a unique technique with the highest amplification efficiency. A major drawback of NEAR is the formation of non-specific products, which limits the sensitivity and raises the threshold of detection methods. The issue can be mitigated by finding the optimal reaction condition (e.g., nicking enzyme concentration, reaction temperature, Mg2+ concentration) (Wang et al., 2018b).
Figure 3.
NEAR-Based COVID-19 Test
(A) Left, NEAR duplex formation. (i) The reverse primer (P1) binds to the target region and is extended. (ii) A second P1 binds to the same target and is extended, displacing the first extended strand. (iii) Primer P2 binds to the released strand and is extended, creating a double-stranded nicking enzyme recognition site. (iv) Nicking enzyme (indicated by scissors) binds and nicks the downstream of the recognition sequence. (v) Polymerase synthesizes complementary sequence off the cleaved site. (vi) The final product is a double-stranded DNA with restriction sites at both ends. Right, exponential amplification. (vii) Nicking enzyme binds and nicks the NEAR duplex at both restriction sides, making two templates (T1, T2). (viii) Free ends of templates are extended. (ix) Repeated nicking and polymerization steps start. (x) Cleaved complexes are regenerated, whereas amplified products (A1, A2) anneal to primers (P2, P1), resulting in bidirectional extension and creating duplexes.
(B) ID NOW COVID-19 system by Abbott. Disposable tools (left) minimize hands-on processes. Patient swab (nasal, nasopharyngeal, or throat) is eluted in the sample receiver containing elution/lysis buffer. After 10 s mixing, the mixture is manually transferred to the test base holder (via transfer cartridge) that contains lyophilized NEAR agents. Heating, agitation, and detection by fluorescence are performed automatically by the instrument. The assay detects SARS-CoV-2 RdRp gene. Adapted with permission from Ref (Nie et al., 2014).
Copyright 2014, American Society for Microbiology.
Abbott Laboratories adopted the NEAR technique and rolled out a compact, integrated diagnostic system, ID NOW (Figure 3B). The system comes with a convenient cartridge for sample processing. Total hands-on time is 2 min, and the total assay time <15 min. The company already has ID NOW tests for Group A Streptococcus and influenza on the market (Wang et al., 2018a; Nie et al., 2014), which helped the rapid introduction of ID NOW COVID-19. The test was designed to detect a sequence in RdRp regions of SARS-CoV-2 genome, and the reported LOD was 0.125 copy/μL. The assay received FDA-EUA for COVID-19 diagnostics.
Recombinase Polymerase Amplification
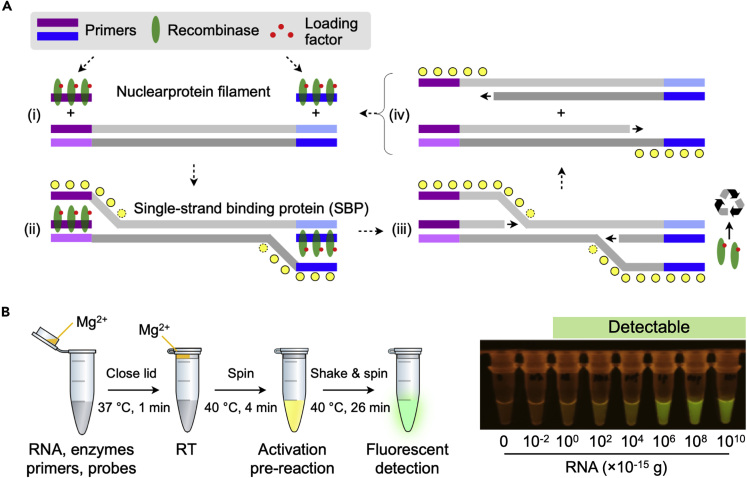
Recombinase polymerase amplification (RPA) borrows its concept from homologous DNA recombination to amplify double-stranded DNA (Lobato and O'Sullivan, 2018; Li et al., 2018). In this process (Figure 4A), primers first bind to recombinase to form nucleoprotein filaments. These complexes search for homologous sequences in the target DNA and invade the cognate sites. Subsequently, the recombinase disassembles the nucleoprotein-bonded strand and the DNA polymerase executes the strand-displacing extension. During this process, the displaced strand is stabilized by single-stranded binding proteins, and the released recombinases become available to form new nucleoprotein filaments that will be used for further cycles. At the end of this process, double-stranded DNA target is exponentially duplicated.
Figure 4.
RPA-Based COVID-19 Test
(A) RPA mechanism. RPA reaction mix contains recombinase, primers, loading factors, and single-stranded binding proteins. (i) The recombinase binds to primers in the presence of loading factors, forming nucleoprotein filaments. (ii) This complex binds to complementary sequences in the target DNA, forming D loop structure, and initiates strand exchange. Single-stranded binding proteins stabilize the displaced DNA stands. (iii) Recombinase disassembles from the nucleoprotein filament to be re-used for subsequent amplification cycles. (iv) DNA polymerase extend primers, separating parallel strands to form duplexes. Repeated cycle of this process enables exponential amplification.
(B) RT-RPA assay developed for COVID-19 diagnostics. Extracted RNA sample is mixed with RT-RPA reaction mixture. RPA activator (Mg2+) is loaded inside the lid of the vial. RT is performed at 37°C (1 min), and then the vial is spun to introduce Mg2+ into the reaction mixture. The reaction vial is heated to 40°C (4 min) for initial RPA activation. After shake and spin, the reaction is let to proceed for additional 26 min at 40°C. The reaction product is then detected via green fluorescence excited by blue light.
Reproduced with permission from Ref (Xia and Chen, 2020).
RPA has been widely used for POC infection diagnostics. It requires only a pair of primers like NEAR but can be carried out at lower temperature (37–42°C) and therefore is more suitable for one-spot assay design (Ménová et al., 2013). All RPA reagents are commercially available through TwistDx (a subsidiary of Abbott), even in a lyophilized pellet format. The company also supplies probe kits for different detection methods (e.g., gel electrophoresis, real-time fluorescent detection, lateral flow strip). Compared with LAMP, RPA is much faster (20 min) but might produce non-specific amplification due to simpler primer design.
For COVID-19 detection, Xia et al. designed RPA primers targeting regions of N gene (Xia and Chen, 2020). Typical RPA reagents were mixed with transcriptase and RNase inhibitor to enable one-spot RNA reverse transcription (Figure 4B). Amplified targets were then detected using commercial fluorescent or lateral flow probe kits. The reaction time was about 30 min, and the LOD was 0.2 copy/μL. The results, however, were limited to using synthetic RNA rather than analyzing extracted viral RNA samples.
CRISPR-Based Detection
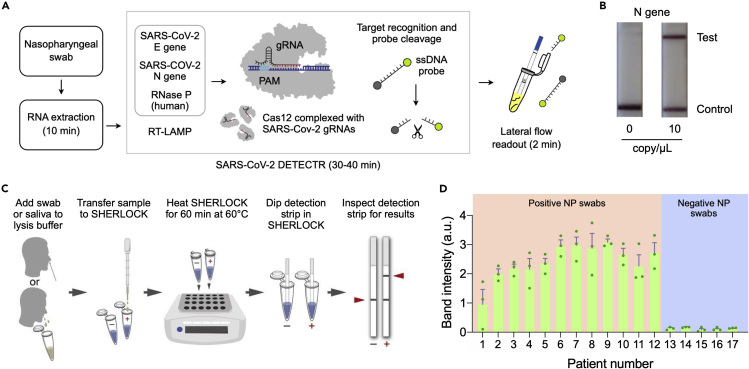
Clustered regularly interspaced short palindromic repeats (CRISPR) systems offer new ways to amplify analytical signal with the precision down to single-nucleotide variants (Kellner et al., 2019; Gootenberg et al., 2017; Aman et al., 2020). Most advanced form of these assays use Cas12a (CRISPR-associated protein 12a) or Cas13a (CRISPR-associated protein 13a) enzymes, exploiting collateral cleavage of single-stranded DNA (Cas12a) or RNA (Cas13a) by these nucleases. In one method, termed SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) (Gootenberg et al., 2017), RNA targets are first amplified via RT-RPA and the amplified DNAs are transcribed to target RNA. CRISPR RNA (crRNA)-Cas13a complex then binds and cleaves target RNA. Non-target RNA probes conjugated with a fluorescent dye (F) and quencher (Q) pair are also cleaved by the complex to provide a fluorescent signal. Similarly, the DETECTR (DNA endonuclease-targeted CRISPR trans reporter) method uses a crRNA-Cas12a complex to recognize amplified DNA targets (Chen et al., 2018). Binding of the crRNA-Cas12a complex to target DNA induces indiscriminate cleaving of non-target FQ-DNA reporters.
Broughton et al. applied the Cas12a method for COVID-19 detection (Figure 5A). The assay was designed to detect regions in E and N genes of SARS-CoV-2, and human RNase P gene as a control. Target genes were amplified via RT-LAMP and recognized by crRNA-LbCas12a complex, which cut DNA reporter probes (Figure 5B). Using synthetic in-vitro transcribed (IVT) SARS-CoV-2 RNA gene targets, the authors reported the LOD of 10 copy/μL. The assay was complete in 45 min, and the analytical signal was read out with lateral flow strips (Broughton et al., 2020). Metsky et al. designed a Cas13-based COVID-19 test (Metsky et al., 2020). The study used machine learning algorithms to generate multiplex panels (67 assays) to identify SARS-related coronavirus species. The assay amplified target RNA via RT-RPA, which was then transcribed to RNA for recognition by crRNA-LwaCas13 conjugates.
Figure 5.
CRISPR-Based COVID-19 Test
(A) Schematic of DETECTR coupled with lateral flow readout. RNA targets extracted from nasopharyngeal swabs are amplified by RT-LAMP. Cas12a complexes, pre-incubated with guide RNAs (gRNAs), recognize target DNA and cleave single-stranded DNA (ssDNA) probes for signal generation.
(B) The intact ddDNA reporters are captured on the control line, whereas the cleaved reporter is captured on the test line. Lateral flow results for the DETECTR are shown for N gene at 0 and 10 copy/μL.
(C) Schematic of SHERLOCK Testing in One Pot (STOP) test. A nasopharyngeal swab or saliva is transferred to the lysis buffer. Lysate is then added to SHERLOCK master mix, and the mixture is heated for 60 min at 60°C. Test results are read out using lateral flow strips (2 min).
(D) Twelve positive and five negative nasopharyngeal (NP) swab samples were analyzed by STOP. The assay made correct diagnosis of these samples. The data were displayed as mean ± standard deviation from three independent experiments.
(A and B) Adapted with permission from Ref (Broughton et al., 2020). Copyright 2020 Nature Publishing Group. (C and D) Adapted with permission from Ref (Joung et al., 2020).
Several drawbacks, however, limit practical use of these assays. The reported methods still require nucleic acid amplification to achieve high sensitivity; CRISPR techniques offer a signal transduction mechanism after such amplification. The assays also involve extra hands-on processes. crRNA-Cas complexes need to be mixed separately and incubated (30 min, 37°C) before each test, and amplified nucleic acids should be mixed with these complexes. In comparison, most isothermal NAATs for COVID-19 already offer one-pot amplification and detection. Overcoming these issues, Joung et al. introduced a one-step approach, SHERLOCK Testing in One Pot (STOP), which integrated LAMP amplification with CRISPR-mediated detection (Figure 5C) (Joung et al., 2020). The authors found that Cas12b from Alicyclobacillus acidiphilus (AapCas12b) retained sufficient activity in the same temperature range of LAMP. They further identified the optimal combination of primers and guide sequence and screened 94 additives to improve the thermal stability of the one-pot reaction. After the assay, the signal was detected with lateral flow reporter devices. The reported LOD was about 2 copy/μL (N gene) using SARS-CoV-2 genome standards spiked into pooled healthy saliva or nasopharyngeal swabs. The assay was validated with clinical nasopharyngeal swab samples (Figure 5D); STOP correctly diagnosed 12 COVID-19 positive and 5 negative patients of 3 replicates. The assay time was about 70 min using lateral flow readout.
COVID-19 Immunoassays
Immunoassays detect the presence of virus-specific antigens or antibodies against virus (Figure 6A). NAATs are ideally suited to diagnose viral infection during its initial phase; immunoassays, particularly antibody tests, can allow for the detection of ongoing or past infection, promoting the better understanding of the transmission dynamics. Immunoassays can also augment NAATs to reduce false-negative results (Racine and Winslow, 2009; Louie et al., 2004); antigens and antibodies are more stable than RNA, and therefore less susceptible to degradation during transport and storage.
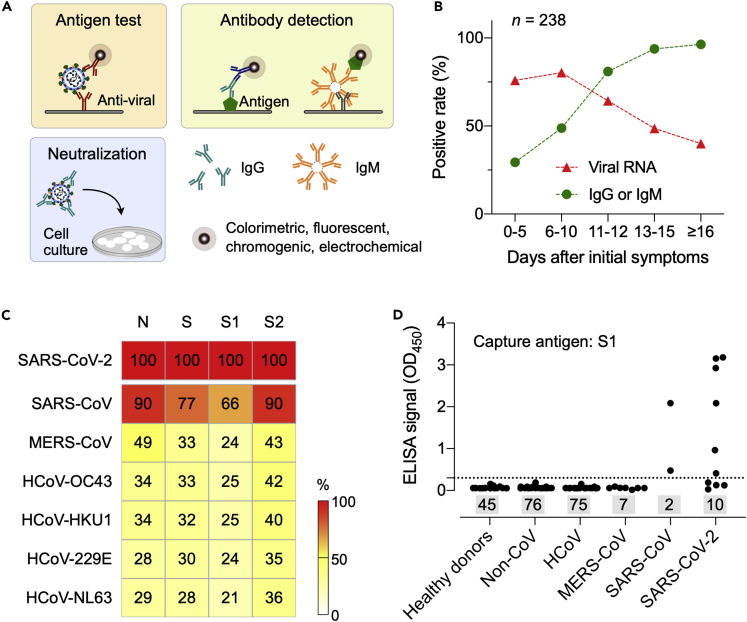
Figure 6.
Immunoassay Design for COVID-19 Detection
(A) Principles of different types of immunoassays. Antigen tests directly capture viral proteins or the whole virus, whereas in antibody tests, viral antibodies (e.g., IgG, IgM) generated from host immune response are captured by synthetic viral antigens or anti-human antibodies. Both tests use a reporter probe for signal generation. Virus neutralization tests check whether a specimen contains effective antibodies that can prevent viral infection on cells.
(B) Positive rates of viral RNA and antibodies (IgG or IgM) were detected in 238 patients with COVID-19 who were at different disease stages. Note that the antibody positive rates were low in the first 5 days after initial onset of symptoms and then rapidly increased as the disease progressed. Adapted with permission from Ref (Liu et al., 2020a).
(C) Similarity of coronavirus S and N proteins. Protein domains from different coronaviruses were compared with those of SARS-CoV-2 (top row, 100% concordance). S1 and S2 are subunits of S. Note that S1 has the least degree of similarity.
(D) Evaluation of S1 ELISA. SARS-CoV-2 S1 protein was used as a capture agent. Serum samples from healthy donors and patients either with non-CoV respiratory, HCoV, MERS-CoV, SARS-CoV, or SARS-CoV-2 infections were analyzed. S1 ELISA showed no cross-reactivity with non-SARS serum samples. The dotted horizontal line indicates ELISA cutoff values, and the sample numbers are inside shaded rectangles. OD, optical density.
(C and D) Adapted from Ref (Okba et al., 2020).
Serologic Tests
These are typically blood-based tests to detect host-derived antibodies against virus. Previous SARS epidemics showed that viral-specific immunoglobulin M (IgM) appears within a week of infection, followed by the production of IgG for long-term (∼2 years) immunity (Wu et al., 2007). Immunological data for COVID-19 have yet to emerge, but a recent study on 214 patients (Hubei, China) indicated a similar early pattern: IgM positivity was higher than that of IgG during initial days of disease onset, and then dropped in about 1 month (Liu et al., 2020b). Another study on 238 patients (Hubei, China) compared the positive rates of RT-PCR and serologic tests (Figure 6B) (Liu et al., 2020a). Antibody positive rates (IgG, IgM, or both) were 29.4% (5/17) in the first 5 days of symptom onset, and then increased to 81% (17/21) after day 10. Conversely, RT-PCR test had initial positive rate of 75.9% (41/54), which dropped to 64.3% after day 11. These results point to the potential utility of serologic tests, not for diagnosing acute COVID-19, but rather as a wide screening tool. For example, by testing antibodies among the general public through random sampling (i.e., serosurvey), public health agencies can estimate the true size of infection (prevalence) and its fatality rate. Serologic tests could also be an assessment tool to decide whether individuals can return to their social contacts.
Developing serologic tests critically relies on producing suitable viral antigens or recombinant proteins to capture host antibodies. Based on previous data on SARS-CoV, it is likely that S and N proteins would be the main immunogens among the four structural proteins (i.e., S, E, M, N proteins) (Okba et al., 2020; Meyer et al., 2014), but SARS-CoV-2 antigenic candidates should be evaluated for their specificity against most common human coronaviruses (HCoV-OC43, HCoV-HKU1, HCoV-229E, HCoV-NL63) and zoonotic ones (SARS-CoV, MERS-CoV). Okba et al. analyzed the similarity of S and N proteins among these coronaviruses and found that S1 subunit in the SARS-CoV-2 S protein has the least overlap with other coronaviruses (Figure 6C) (Okba et al., 2020). The authors further assessed N and S1 ELISA using serum samples from healthy donors as well as from patients infected with non-CoV, HCoV, MERS-CoV, SARS-CoV, or SARS-CoV-2 (Figure 6D). S1 ELISA showed high specificity against healthy, non-CoV, HCoV, and MERS-CoV cohorts, whereas N ELISA was more sensitive in detecting antibodies from patients with mild COVID-19. Differentiating SARS-CoV-2 and SARS-CoV samples was not possible due to cross-reactivity. However, it was noted that the human population with SARS-CoV antibodies is expected to be small: SARS-CoV has not circulated since 2003 and a previous study reported the waning of SARS-CoV antibodies to an undetectable level (21 out of 23 samples) in 6 years after infection (Tang et al., 2011). Considering these results, S1 and N proteins are likely the most suitable antigens for COVID-19 serologic tests.
Rapid Diagnostic Tests
Rapid diagnostic tests (RDTs) are based on host antibody detection on a nitrocellulose membrane. Easy to operate and portable, these tests are suited for POC analyses of fingerprick blood, saliva, or nasal swab fluids. Samples are dropped on a loading pad and transferred via capillary motion. During this flow, antibodies in the sample bind to nanoparticles, and the whole complex is captured downstream at designated spots on the membrane by anti-human antibodies (Figure 7A). The final results are usually displayed as colored lines for naked eye detection: a control line confirming test reliability and test line(s) indicating the presence of target antibodies. Most assays use gold nanoparticles for signal generation, whereas carbon or colored latex nanoparticles are alternative labeling candidates.
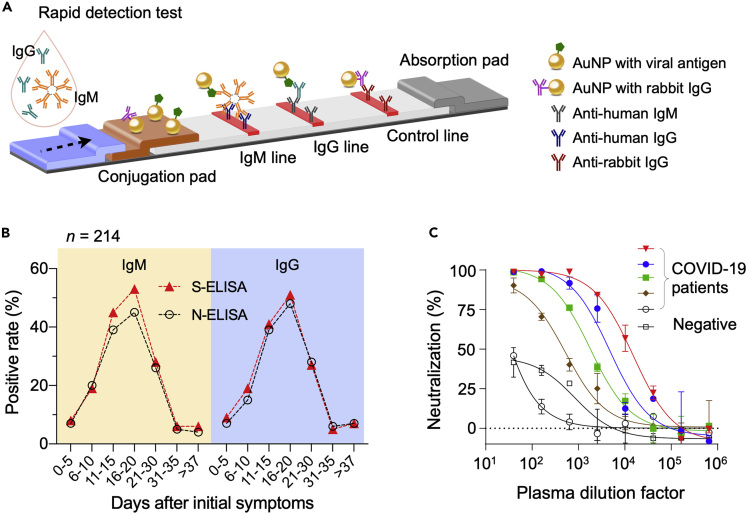
Figure 7.
Examples of COVID-19 Immunoassays
(A) Schematic of a rapid detection test (RDT) device. The sample, dropped on a loading pad, flows through the device via capillary effect and wet colloidal gold nanoparticles (AuNPs) loaded in the conjugation pad. AuNPs tagged with viral antigen bind to IgM and IgG antibodies, and the complexes are captured in the downstream by pre-spotted anti-human IgM and IgG antibodies. AuNP conjugated with non-human IgG antibodies are captured by appropriate antibodies to generate a control signal.
(B) Two types of ELISA were compared. One used recombinant SARS-CoV-2 N protein as a capture antigen (N-ELISA), and the other, recombinant SARS-CoV-2 S protein (S-ELISA). Serum samples from 214 patients with COVID-19 were tested. Overall, S-ELISA showed higher detection rate than N-ELISA.
(C) Plasma samples from patients (n = 5) who recovered from COVID-19 were used for virus neutralization tests. In a concentration-dependent manner, all five plasma inhibited the infection of 293T/ACE2 cells by SARS-CoV-2 pseudo virus. Plasma from a healthy donor was used as a negative control. The median percentage of neutralization is shown from duplicate measurements. Data points represent the median percentage of neutralization. Error bars indicate standard deviation from duplicate measurements.
(B) Adapted with permission from Ref (Liu et al., 2020b). Copyright 2020 American Society for Microbiology. (C) Reproduced with permission from Ref (Wu et al., 2020).
Initial studies reported high analytical sensitivity (86%–89%) and specificity (84.2%–98.6%) of RDTs (Liu et al., 2020c; Li et al., 2020). Test accuracies, however, vary significantly among different commercial vendors. As more companies are racing to develop serologic RDTs (>100 companies as of May, 2020), the need for rigorous vetting is increasing. FDA and FIND are currently conducting independent evaluation on selected products (Foundation for Innovative New Diagnostics, 2020).
Enzyme-Linked Immunosorbent Assay
ELISA is a laboratory-based test with high sensitivity and throughput. It typically uses a multi-well plate coated with viral proteins. Blood, plasma, or serum samples from patients are introduced to these wells for antibody capture and then washed. Subsequently, secondary antibodies labeled with enzymes are added, which catalyzes signal generation. The assay format can be adapted for different detection modalities, including colorimetric, fluorescent, and electrochemical methods (Figure 6A). The analytical sensitivity is down to picomolar (pM) ranges, and the typical assay time is 2–5 h (Weissleder et al., 2020; Younes et al., 2020).
Like COVID-19 RDTs, identifying effective viral antigens is an important factor in ELISA development. One study compared three ELISA sets, each using N protein, S1 subunit (S protein), or receptor binding domain (RBD; S protein) as a viral antigen (Okba et al., 2020). The RBD and N ELISA tests were shown to be more sensitive than S1 ELISA in detecting antibodies from mildly infected COVID-19 patients (Okba et al., 2020), but the cohort (n = 3 for COVID-19 infection) was too limited to draw conclusions. In another study, sera from 214 patients with COVID-19 were subjected to N and S ELISAs to detect IgG and IgM (Liu et al., 2020a). In this study, S-based ELISA showed slightly higher sensitivity than N-based ELISA (Figure 7B). These seemlingly conflicting findings are likely resolved in near future, as more blood samples from larger population become available.
Virus Neutralization Test
Virus neutralization test (VNT) is a gold standard to assess whether an individual has active antibodies against a target virus. In this assay, serial dilutions of test serum (or plasma) are prepared, typically in a 96-well plate, and incubated with a set amount of infectious virus. The mixture is then inoculated on to susceptible cells (e.g., VeroE6) and cultured for 2–3 days. The test results are typically read out via microscopy for evidence of viral cytopathic effect; neutralizing antibodies would block virus replication to let cells grow. Plaque reduction neutralization test (PRNT) as a type of VNT is used for counting plaque-forming units on the agar- or carboxymethyl cellulose-coated cell layer, whereas focus reduction neutralization test (FRNT) relies on immunocolourimetric-based analysis for calculating neutralizing antibody titers. Suthar et al. compared the efficiency of PRNT and FRNT assays for RBD-specific IgG responses that patients with COVID-19 developed 6 days after the PCR diagnosis and found a strong correlation between those tests (Suthar et al., 2020). In another study, Wang et al. used PRNT to evaluate the human monoclonal antibody, 47D11, that binds to S-RBD and can neutralize both SARS-CoV-2 and SARS-CoV (Wang et al., 2020). Although highly specific, VNT is time-intensive and requires specialty laboratories (e.g., biosafety level 3 facilities for COVID-19). As such, these tests are primarily used for vaccine and therapeutic developments.
Several groups have developed pseudovirus-based neutralization assays (PBNAs) using pseudovirus (PSV) as a safer (biosafety level 2) surrogate to SARS-CoV-2 virus (Nie et al., 2020; Wu et al., 2020). Wu et al. generated PSV by incorporating SARS-CoV-2 S protein into the envelop of vesicular stomatitis virus pseudotypes. These PSVs were used for VNTs with plasma samples from patients with COVID-19 who recovered (Wu et al., 2020). Convalescent plasma from patients with COVID-19 inhibited SARS-CoV-2 infection (Figure 7C) and did not cross-react with SARS-CoV pseudovirus. The study also showed that titers of neutralizing antibodies reached their peak at 10 to 15 days after disease onset and remained stable thereafter. Interestingly, about 30% recovered patients (n = 175) showed low levels of neutralizing antibodies; this observation may have implications when applying and interpreting serologic tests to detect past COVID-19 infection.
Antigen Detection Assay
This assay detects the presence of viral proteins (antigens) through a conventional immunocapture format (Figure 6A). Viral antigens can be detected when the virus is actively replicating, which makes this assay type highly specific. The assay, however, has a suboptimal sensitivity, generally requiring sufficient antigen concentrations in samples. Data from influenza antigen tests (Bruning et al., 2017) showed a sensitivity of 61% and a specificity of 98%. Potential use of antigen assays thus could be as a triage test to rapidly identify patients who are likely to have COVID-19, reducing or eliminating the need for lengthy molecular confirmatory tests. Monoclonal antibodies against the N protein of SARS-CoV-2 have been generated, and several rapid test kits are under development (Cheng et al., 2020).
Future Developments
Aggressive testing and isolation measures have started blunting the first wave of COVID-19 in some countries. From these experiences, lessons are emerging for new diagnostics and surveillance policies that will better prepare us for the potential next waves (Fineberg, 2020). For the diagnostic aspect, we identify the following needs to be addressed.
Reducing Sampling Errors
Most current NAATs have analytical sensitivities and specificities around 95% or higher under ideal circumstances and when performed by skilled operators. Yet, in clinical practice, the sensitivity drops precipitously to 60%–70% (Ai et al., 2020; Lassaunière et al., 2020), necessitating re-testing that causes loss of valuable time in symptomatic patients (Weissleder et al., 2020). The likely reason for this discrepancy is swabbing efficiency of nasopharyngeal, oral, sputum, and bronchial samples. Some countries have instituted dual testing of nasopharyngeal and sputum/throat samples to increase the accuracy. Systematic research is needed to evaluate the efficacy of the swab material and RNA yields.
Developing Fast, Cost-Effective Antigen Tests
There is a need to develop rapid, antigen-based COVID-19 tests for a number of reasons, one of them being to reduce the use of lengthy RT-PCR. The development of NAATs was a reasonable emergency decision, considering NAATs' high analytical sensitivity and the short lead time in assay development. However, NAATs are generally process intensive, susceptible to contamination, and expensive. Antigen-based tests, on the other hand, could be a niche tool for cost-effective POC diagnosis at primary care settings. Such systems have already been developed for influenza (CDC, 2020c). For example, the rapid influenza diagnostic tests (RIDTs), which detect the presence of influenza A and B viral nucleoprotein antigens, can identify patients with flu with high specificity. RIDT-positive patients can receive necessary care after this quick (<20 min) test, and only negative samples need to be routed for laboratory molecular analyses. With effective COVID-19 antigen tests, a similar triaging strategy can be implemented to ease the demand for molecular tests.
Establishing Effective Serologic Tests
As we transition to the flattening phase of COVID-19, the need for serologic tests will increase. Individuals, who have recovered from the disease or been asymptomatic, can use these tests to make informed decision on social activities; population-wide serologic screening will allow governments to learn the true extent of infections. A major issue with current serological tests is their high variability (Lassaunière et al., 2020), and often low sensitivity and specificity. Comparison of different test kits, virtually all based on lateral flow assay format, has shown that some perform much better (Whitman et al., 2020), which is presumably due to affinity reagents used. Identifying and synthesizing most immunogenic, high-affinity viral antigens is a critical step to improve diagnostic accuracy. Equally important is to conduct interference challenges to check how drugs, medications, and coagulation status affect serological testing outcomes.
Serial Tracking with Digital Health
Infection with SARS-CoV-2 is highly dynamic with viral titers and antibody levels changing over time in asymptomatic and symptomatic patients. Serial testing is necessary to identify patients before irreversible complications occur as well as to confirm full recovery. Accumulated data will further inform the duration of SARS-CoV-2 immunity and help us setting cutoffs for antibody positivity. Integrating POC tests with digital networks will facilitate implementing such tasks. Patients at home, for example, can log in their test results and symptoms and receive telemedicine feedback, which would be a cost-effective, safer caring model for stable or recovering patients. Digital services will also allow public health agencies to gather data from large population to track disease transmission in real time.
Setting up Global Standards
New COVID-19 tests are approved based on their analytical validity, with sensitivity and specificity measured on manufacturers' own artificial samples. However, significant performance deviations have been reported from independent testings (Foundation for Innovative New Diagnostics, 2020). This situation demands developing global reference standards (e.g., pseudovirus, viral nucleic acids, viral antigen, antibodies) to enable objective inter-test comparison. Also necessary is to establish guidelines (e.g., sensor specifications, cost, accuracy) per different test purpose, for example, diagnosing acute infections in hospitals or long-term care facilities, at-home monitoring, and population survey. These efforts will provide benefits to both clinical and research communities, motivating technical innovations.
New Biosensors
We should accelerate the development of new diagnostic methods. In particular, novel transducer technologies, such as nanoplasmonics, ion-gate transistors, and optical resonators, have exquisite sensitivities and potentially enable direct viral detection. Several exciting systems have been reported: a graphene-based transistor with LOD 2.4 × 102 viruses/mL (Seo et al., 2020) and a plasmonic photothermal sensor that detected the RdRp target down to 0.22 pM (Qiu et al., 2020). Pursuing these approaches would be critical to transcending current NAATs and realizing rapid, on-site diagnostics.
Resource Availability
Lead Contact
Hakho Lee, Center for Systems Biology, Massachusetts General Hospital Research Institute, Boston, MA 02114, USA (hlee@mgh.harvard.edu).
Materials Availability
Not applicable.
Data and Code Availability
Not applicable.
Acknowledgments
T.K. is grateful to the Swiss National Science Foundation for the Postdoc Mobility fellowship (No. P400PM_180788/1). H.L. is supported in part by U.S. NIH Grants R01CA229777, R21DA049577, U01CA233360, US DOD-W81XWH1910199, W81XWH1910194, and MGH Scholar Fund.
Author Contributions
Conceptualization, T.K., R.W., and H.L.; Visualization, T.K. and H.L.; Writing, T.K., R.W., and H.L.
References
- Abbott ID Now Covid-19. 2020. https://www.alere.com/en/home/product-details/id-now-covid-19.html
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N., Rampazzo R.C.P., Costa A.D.T., Krieger M.A. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. Biomed. Res. Int. 2017;2017:9306564. doi: 10.1155/2017/9306564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman R., Mahas A., Mahfouz M. Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth. Biol. 2020;9:1226–1233. doi: 10.1021/acssynbio.9b00507. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Digital PCR hits its stride. Nat. Methods. 2012;9:541–544. [Google Scholar]
- Bialek S., Bowen V., Chow N., Curns A., Gierke R., Hall A., Hughes M., Pilishvili T., Ritchey M., Roguski K. Geographic differences in COVID-19 cases, deaths, and incidence — United States, February 12–April 7, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:465–471. doi: 10.15585/mmwr.mm6915e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning A.H.L., Leeflang M.M.G., Vos J.M.B.W., Spijker R., de Jong M.D., Wolthers K.C., Pajkrt D. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin. Infect. Dis. 2017;65:1026–1032. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan C.J., Lee R., Zulauf K.E., Tamburello L., Smith K.P., Previtera J., Cheng A., Green A., Azim A.A., Yano A. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00876-20. Published online May 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. 2020. https://www.fda.gov/media/134922/download
- Center for Disease Control and Prevention Interim Guidelines for Clinical Specimens for COVID-19. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html
- Center for Disease Control and Prevention Rapid Influenza Diagnostic Tests. 2020. https://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: a narrative review. Ann. Intern. Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley P., Castriciano S., Chernesky M., Smieja M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J. Clin. Microbiol. 2006;44:2265–2267. doi: 10.1128/JCM.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. medRxiv. 2020 doi: 10.1101/2020.03.14.20036129. Posted online March 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg H.V. Ten weeks to crush the curve. N. Engl. J. Med. 2020;382:e37. doi: 10.1056/NEJMe2007263. [DOI] [PubMed] [Google Scholar]
- Foo P.C., Najian A.B.N., Muhamad N.A., Ahamad M., Mohamed M., Yean C.Y., Lim B.H. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: a comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotech. 2020;20:1–15. doi: 10.1186/s12896-020-00629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration Emergency Use Authorization. 2020. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization#covidinvitrodev
- Foundation for Innovative New Diagnostics SARS-CoV-2 diagnostics: performance data. 2020. https://www.finddx.org/covid-19/dx-data/
- Freeman B., Lester S., Mills L., Rasheed M.A.U., Moye S., Abiona O., Hutchinson G.B., Morales-Betoulle M., Krapinunaya I., Gibbons A. Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and serosurveillance. bioRxiv. 2020 doi: 10.1101/2020.04.24.057323. Posted online April 25, 2020. [DOI] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Luo Q., Mo F., Long L., Zheng W. SARS-CoV-2 RNA more readily detected in induced sputum than in throat swabs of convalescent COVID-19 patients. Lancet Infect. Dis. 2020;20:655–656. doi: 10.1016/S1473-3099(20)30174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson C.M., Chevillet J.R., Briggs H.A., Gallichotte E.N., Ruf I.K., Hindson B.J., Vessella R.L., Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M.W., Kim N.G., Yu X., Li J., Walker B.D. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020 doi: 10.1101/2020.05.04.20091231. Posted online May 8, 2020. [DOI] [Google Scholar]
- Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim S., Park E.C., Park D., Lee J.-H., Byeon C.W. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.02.25.964775. Posted online February 27, 2020. [DOI] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C.Y., Fomsgaard A., Krogfelt K.A., Jørgensen C.S. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.09.20056325. Posted online April 10, 2020. [DOI] [Google Scholar]
- Lee D., Lee J. Testing on the move: South Korea’s rapid response to the COVID-19 pandemic. Transport. Res. Interdiscip. Perspect. 2020;5:100111. doi: 10.1016/j.trip.2020.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Macdonald J., von Stetten F. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. 2018;144:31–67. doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. Published online February 27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu W., Zheng Y., Jiang X., Kou G., Ding J., Wang Q., Huang Q., Ding Y., Ni W. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect. 2020;22:206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S. Evaluation of nucleocapsid and spike protein-based enzyme-linked Immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. Published online March 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu Y., Diao B., Ren F., Wang Y., Ding J., Huang Q. Diagnostic indexes of a rapid IgG/IgM combined antibody test for SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.03.26.20044883. Posted online March 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato I.M., O'Sullivan C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt Chem. 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie J.K., Hacker J.K., Mark J., Gavali S.S., Yagi S., Espinosa A., Schnurr D.P., Cossen C.K., Isaacson E.R., Glaser C.A. SARS and common viral infections. Emerg. Infect. Dis. 2004;10:1143–1146. doi: 10.3201/eid1006.030863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiken Chemical Co., Ltd PrimerExplorer V5.0. 2020. https://primerexplorer.jp/e/
- Lu R., Wang J., Li M., Wang Y., Dong J., Cai W. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis, treatment monitoring and criteria for discharge. medRxiv. 2020 doi: 10.1101/2020.03.24.20042689. Posted online March 30, 2020. [DOI] [Google Scholar]
- Lu R., Wu X., Wan Z., Li Y., Zuo L., Qin J., Jin X., Zhang C. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 2020:1–4. doi: 10.1007/s12250-020-00218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxmen A. The researchers taking a gamble with antibody tests for coronavirus. Nature. 2020 doi: 10.1038/d41586-020-01163-5. Published online April 21, 2020. [DOI] [PubMed] [Google Scholar]
- Ménová P., Raindlová V., Hocek M. Scope and limitations of the nicking enzyme amplification reaction for the synthesis of base-modified oligonucleotides and primers for PCR. Bioconjug. Chem. 2013;24:1081–1093. doi: 10.1021/bc400149q. [DOI] [PubMed] [Google Scholar]
- Metsky H.C., Freije C.A., Kosoko-Thoroddsen T.-S.F., Sabeti P.C., Myhrvold C. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. bioRxiv. 2020 doi: 10.1101/2020.02.26.967026. Posted online March 2, 2020. [DOI] [Google Scholar]
- Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S., Roth R.B., Stiles J., Mikhlina A., Lu X., Tang Y.W., Babady N.E. Evaluation of Alere i Influenza A&B for rapid detection of influenza viruses A and B. J. Clin. Microbiol. 2014;52:3339–3344. doi: 10.1128/JCM.01132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Mori Y., Tomita N., Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J. Microbiol. 2015;53:1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., Maeng J.S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Racine R., Winslow G.M. IgM in microbial infections: taken for granted. Immunol. Lett. 2009;125:79–85. doi: 10.1016/j.imlet.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbel J., Jagpal S., Roy J., Brooks A., Tischfield J., Sheldon M., Bixby C., Witt D., Gennaro M.L., Horton D.B. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is comparable in clinical samples preserved in saline or viral transport medium. J. Mol. Diagn. 2020;22:871–875. doi: 10.1016/j.jmoldx.2020.04.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf N., Scieux C., Resche-Riggon M., Feghoul L., Xhaard A., Gallien S., Molina J.M., Socié G., Viglietti D., Simon F. Fully automated quantification of cytomegalovirus (CMV) in whole blood with the new sensitive Abbott Real Time CMV assay in the era of the CMV international standard. J. Clin. Microbiol. 2013;51:2096–2102. doi: 10.1128/JCM.00067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Shim E., Tariq A., Choi W., Lee Y., Chowell G. Transmission potential and severity of COVID-19 in South Korea. Int. J. Infect. Dis. 2020;93:339–344. doi: 10.1016/j.ijid.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SinoBiological Antigen Detection Assay. 2020. https://www.sinobiological.com/research/virus/sars-cov-2-antigen-detection-assay
- Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., Vanderheiden A., Nyhoff L., Davis C.W., Adekunle O. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020;1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Chen M.I., Hu Z., Young B.E., Tan Y., Yi Y., Lye D.C., Anderson D.E., Wang L. A SARS-CoV-2 surrogate virus neutralization test (sVNT) based on antibody-mediated blockage of ACE2-spike (RBD) protein-protein interaction. Res. Square. 2020 doi: 10.21203/rs.3.rs-24574/v1. Posted online April 23, 2020. [DOI] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa149. Published online February 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Deng J., Tang Y.W. Profile of the Alere i Influenza A & B assay: a pioneering molecular point-of-care test. Expert Rev. Mol. Diagn. 2018;18:403–409. doi: 10.1080/14737159.2018.1466703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Qian C., Wu H., Qian W., Wang R., Wu J. Technical aspects of nicking enzyme assisted amplification. Analyst. 2018;143:1444–1453. doi: 10.1039/c7an02037f. [DOI] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12:eabc1931. doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., Rathore U., Goldgof G.M., Whitty C., Woo J.M. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv. 2020 doi: 10.1101/2020.04.25.20074856. Posted online May 17, 2020. [DOI] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.P., Wang N.C., Chang Y.H., Tian X.Y., Na D.Y., Zhang L.Y., Zheng L., Lan T., Wang L.F., Liang G.D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. Posted online April 20, 2020. [DOI] [Google Scholar]
- Xia S., Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discov. 2020;6:37. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Tong J., Liu M., Shen Y., Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020;92:589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Jiang L., Huang G., Pu H., Gong B., Lin H., Ma S., Chen X., Long B., Si G. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Domesle K.J., Ge B. Loop-mediated isothermal amplification for Salmonella detection in Food and feed: current applications and future directions. Foodborne Pathog. Dis. 2018;15:309–331. doi: 10.1089/fpd.2018.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D.W., AL-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12:582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Li X., Liu X., Ye S., Han H., Dong X., Li X., Li J. Rapid colorimetric detection of COVID-19 coronavirus using a reverse tran-scriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform: iLACO. medRxiv. 2020 doi: 10.1101/2020.02.20.20025874. Posted online February 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 doi: 10.1101/2020.02.26.20028373. Posted online February 29, 2020. [DOI] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S. Reverse transcription loop-mediated isothermal amplification combined with nanoparticles-based biosensor for diagnosis of COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.17.20037796. Posted online March 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.