Abstract
Background
Our study aimed to identify a host cytokine biosignature that could distinguish childhood tuberculosis (TB) from other respiratory diseases (OD).
Methods
Cytokine responses in prospectively recruited children with symptoms suggestive of TB were measured in whole blood assay supernatants, harvested after overnight incubation, using a Luminex platform. We used logistic regression models with Least Absolute Shrinkage and Selection Operator (LASSO) penalty to identify the optimal biosignature associated with confirmed TB disease in the training set. We subsequently assessed its performance in the test set.
Findings
Of the 431 children included in the study, 44 had bacteriologically confirmed TB, 60 had clinically diagnosed TB while 327 had OD. All children were HIV-negative. Application of LASSO regression models to the training set (n = 260) resulted in the combination of IL-1ra, IL-7 and IP-10 from unstimulated samples as the optimally discriminant cytokine biosignature associated with bacteriologically confirmed TB. In the test set (n = 171), this biosignature distinguished children diagnosed with TB disease, irrespective of microbiological confirmation, from OD with area under the receiver operator characteristic curve (AUC) of 0•74 (95% CI: 0•67, 0•81), and demonstrated sensitivity and specificity of 72•2% (95% CI: 60•4, 82•1%) and 75•0% (95% CI: 64•9, 83•4%) respectively, with its performance independent of their age group and their age- and sex-adjusted nutritional status.
Interpretation
This novel biosignature of childhood TB derived from unstimulated supernatants is promising. Independent validation with further optimisation will improve its performance and translational potential.
Funding
Steinberg Fellowship (McGill University); Grand Challenges Canada; MRC Program Grant.
Keywords: Children, Tuberculosis, Cytokine, Biosignature, Diagnosis
1. Introduction
With an estimated 1.1 million annual cases and 205,000 deaths, childhood tuberculosis (TB) remains a serious threat to global child health [1]. More than 70% of all childhood TB cases occur in the World Health Organization (WHO) Africa and southeast Asia regions where childhood cases remain underreported due to the well-known difficulties with bacteriological confirmation [2,3]. Childhood TB is paucibacillary and obtaining good quality sputum specimen is a challenge, particularly in the very young [4]. Symptoms of TB in children can mimic other respiratory diseases and the diagnosis relies on clinical, epidemiological and radiological features if there is no microbiological confirmation. Given that microbiological confirmation is achieved in less than 40% of all children commencing TB therapy even in the best of settings [5,6], the development of a non-sputum-based point-of-care (POC) test remains an identified critical need as acknowledged by the WHO [1,7]. Such a test should enable fast and accurate distinction between TB disease and other respiratory infections and be applicable for use at lower levels of the health care system in resource-limited settings [7,8].
Even in the era of the GeneXpert and Xpert Ultra [9,10], challenges remain for TB diagnosis in children, which highlights the fact that better diagnostics for children might have to be based on host immune responses rather than pathogen detection. However, none of the currently available immune-diagnostic tests, including the tuberculin skin test (TST) and interferon (IFN)-γ release assays (IGRA), can distinguish between latent Mycobacterium tuberculosis (M.tb) infection (LTBI) and TB disease, or more importantly distinguish between TB disease and other respiratory infections 11, 12, 13. It has therefore been suggested that a combination of factors, such as antigen-stimulated cytokine biosignatures or gene expression profiles, might offer increased sensitivity and specificity over assays based on a single marker such as IFN-γ [14,15].
To identify a host biosignature based on a panel of secreted cytokines that could distinguish children with TB disease from those with other respiratory infections, we prospectively enroled a cohort of children with symptoms compatible with TB in The Gambia.
2. Materials and methods
2.1. Setting and recruitment procedures
The Childhood TB group of the Medical Research Council Unit The Gambia at the London School of Hygiene and Tropical Medicine (MRCG at LSHTM) prospectively recruited symptomatic children with suspected intrathoracic TB disease from February 2012 to June 2017 as part of a comprehensive childhood TB research programme. Ethical approval for the study was obtained from the Gambia Government/MRC joint ethics committee. All children had documented household exposure to an adult with smear-positive TB. The aims, study setting, screening/recruitment and samples processing procedures for the cohort have been previously described [16]. In brief, following written informed consent obtained from the parent/guardian of each respective child contact, all children underwent systematic community-based screening for symptoms suggestive of TB disease as part of active household contact tracing activities. All children with symptoms suggestive of TB had further clinical evaluation at a dedicated childhood TB clinic. This included symptom review, physical examination, HIV testing, chest radiograph, pathogen detection tests (smear microscopy, Xpert MTB/Rif assay [Cepheid, Sunnyvale, CA, USA] and MGIT™ liquid mycobacterial culture [BD, Sparks, MD, USA]) on relevant clinical samples such as sputum (spontaneous or induced) or gastric aspirates, and a maximum of 5 ml of venous blood sample drawn for host response studies. A TST was also performed as part of the baseline evaluation of the children and a positive result was defined as transverse skin induration ≥10 mm, regardless of bacille Calmette Guerin (BCG) vaccination status, measured 48–72 h after intradermal injection of 0.1 ml of two tuberculin units of purified protein derivative (RT23; Statens Serum Institute, Copenhagen, Denmark) in the volar aspect of the left forearm.
Using data from the baseline clinical evaluation and laboratory investigations of the children and given that all children were identified via household contact tracing, TB disease was defined according to the reporting framework proposed by the WHO comprising bacteriologically confirmed TB and clinically diagnosed TB cases [17], as described in Table 1. All children diagnosed with TB disease were referred for the standard six months TB treatment according to the Gambian paediatric TB treatment guidelines [18], and followed up at the childhood TB clinic at 2-months and 6-months after treatment initiation.
Table 1.
Diagnostic classification according to the revised WHO case definitions.
| Bacteriologically confirmed TB | - Detection of AFB by microscopy of secretions or;- Identification of M. tuberculosis by culture or;- Identification of M. tuberculosis by Xpert. |
|---|---|
| Clinically diagnosed TB* | - does not fulfil criteria for bacteriological confirmation but; - Suggestive appearances on chest X-ray and; - Favourable response to specific antituberculous therapy. +/- Positive tuberculin skin test +/-Suggestive histological appearances on biopsy material. |
Clinically diagnosed TB cases had symptoms and signs suggestive of TB, did not fulfil the criteria for bacteriological confirmation of disease, had suggestive appearance on chest X-ray and failed to respond to empirical broad-spectrum antibiotics. Favourable response to antituberculous therapy was an integral part of the clinical TB diagnosis.
In the absence of bacteriological confirmation or radiological signs of TB disease and resolution of symptoms spontaneously or after treatment with conventional antibiotics, children were diagnosed as having other respiratory diseases but not TB (“other diseases” [OD]). All child contacts were followed up at home by trained field workers at 3-monthly intervals for a year, with repeated symptoms screening and clinic visits if the child became unwell. After exclusion of TB disease, all child contacts aged <5 years received isoniazid preventive therapy (IPT) for six months, home-delivered by field workers [19]. None of the children with OD in this study developed TB disease during follow-up.
2.2. Immunological assays
At baseline clinical evaluation, a whole blood cytokine secretion assay (WBA) was employed in the TB Immunology laboratory at the MRCG at LSHTM using ESAT-6/CFP-10 (EC)-fusion protein (10ug/ml final concentration; kindly provided by Tom Ottenhoff, Leiden University Medical Centre, Leiden, Netherlands), positive (phytohaemagglutinin-L [PHA-L]; Sigma-Aldrich, Gillingham, UK; 10ug/ml final concentration) and negative (RPMI 1640 medium; BioWhittaker, Verviers, Belgium) controls, as previously described [16]. Supernatants were harvested after overnight incubation at 37°C with 5% carbon dioxide and stored at −20°C until cytokine responses were measured by a Luminex platform.
2.2.1. Interferon gamma release assay (IGRA)
To determine evidence of M.tb sensitization, IFN-γ was measured in the supernatants by enzyme-linked immunosorbent assay (ELISA), as previously described [16].
2.2.2. Multiplex cytokine assay (MCA)
Unstimulated (negative control) and EC-stimulated WBA supernatants were analysed for a panel of cytokines, using the commercially available Bio-Rad Human cytokine Th-1/Th-2 27-plex kit (Bio-Rad, USA) according to the manufacturer's instruction. The cytokines assessed included IL-1b, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17, Eotaxin, basic-FGF, G-CSF, GM-CSF, IFN-γ, IL-10, MCP-1 (MCAF), MIP-1α, MIP-1β, PDGF-bb, RANTES, TNF-α and VEGF. All samples were randomly distributed between assay plates to avoid any batch effects. The plates were read on a Bio-Plex 200 analyser with the analyses conducted using Bioplex manager software (version 4.0; Bio-Rad, USA) and a low photomultiplier tube (PMT) setting, as described previously [20].
The laboratory scientists who performed the immunological assays were blinded to all clinical data including results of microbiological investigations and disease status of study subjects, while the childhood TB clinical team were also blinded to the immunological data.
2.3. Statistical analysis
We did not perform sample size calculations. We used a convenience sample of children who were consecutively enroled during the study period. Demographic and clinical characteristics were compared between the three diagnostic outcome groups i.e. bacteriologically confirmed TB, clinically diagnosed TB and other diseases (OD), using Kruskal-Wallis rank test for continuous variables and Chi-square test for categorical variables. We directly analysed the fluorescence intensity (FI) values of the analytes in our multiplex cytokine assay. All the cytokine responses were log2 transformed prior to analysis. The unstimulated and EC-stimulated fluorescence values of each cytokine or chemokine were analysed as separate dependent variables in log2 scale. Differences in analytes expression fluorescence values between the diagnostic groups were evaluated using random-intercept linear regression models, including diagnostic group, cytokines, and the interaction terms between diagnostic group and cytokines as covariates to account for the dependence of the cytokine responses within the study subjects, followed by pairwise Wald tests with Sidak multiple comparisons adjustment [21].
For the identification of a cytokine biosignature, study subjects were randomly selected into training and test sets based on diagnosis. The training set included children with bacteriologically confirmed TB and OD only, and consisted of 70% of children randomly selected within each of those two groups. The test set consisted of the remaining 30% of the children with bacteriologically confirmed TB and those with OD, and all those with clinically diagnosed TB. We used logistic regression with Least Absolute Shrinkage and Selection Operator (LASSO) penalty to determine the optimally discriminant cytokine prediction model associated with bacteriologically confirmed TB disease in the training set [22]. The optimal LASSO penalty (λ), which predicted bacteriologically confirmed TB disease or OD, was determined by a 10-fold cross validation in the training set. We then investigated the predictive accuracy of the selected biosignature by logistic discriminant analysis [23], whereby the training set was used to build our predictive model whose performance was subsequently assessed in the test set. Further details of the statistical analysis, including the use of the LASSO penalty to select an optimal number of biosignatures and use of logistic discriminant analysis to build predictive models, are provided in the supplementary material. Data analyses were done using Stata 16 (StataCorp, College Station, TX, United States).
3. Results
Out of the 7104 actively traced child contacts originally screened for symptoms of TB in the households, 1865 children had symptoms compatible with TB. Complete clinical and laboratory results with defined diagnostic outcome and suitable blood samples were available from 1416 children. Of the 1416 children, all 104 diagnosed consecutively with TB disease (comprising 44 bacteriologically confirmed TB and 60 clinically diagnosed TB cases) and a randomly selected sample of 327 children with OD were included in the study. Therefore, cytokine measurements were conducted in samples from a total of 431 children. Fig. 1 shows the flowchart for recruitment of study subjects according to the Standards for Reporting of Diagnostic Accuracy (STARD) studies [24].
Fig. 1.
STARD Flowchart. Flow diagram of recruitment and diagnostic classification of study subjects from February 2012 to June 2017 according to the Standards for Reporting of Diagnostic Accuracy (STARD) studies. TB=Tuberculosis; WBA= whole blood cytokine secretion assay; IGRA=interferon-gamma release assay; OD=other respiratory diseases but not TB.
Table 2 describes the baseline characteristics of the 431 study participants. Although children with OD were older than the children diagnosed with TB disease, there was no evidence of difference in the distribution of gender and of M.tb sensitization between the groups. All children were HIV-negative.
Table 2.
Baseline characteristics of study subjects stratified by diagnosis.
| Total number (n= 431) | Bacteriologically confirmed TB (n= 44) | Clinically diagnosed TB (n= 60) | OD (n= 327) | P | |
|---|---|---|---|---|---|
| Age in years, median(range) | 429* | 5•2 (0•3–14) | 2•9 (0•3–12) | 6 (0•2–14) | <0•001$ |
| < 2 years old, n (%) | 429* | 12 (27.3) | 20 (33.3) | 50 (15.4) | 0.002# |
| < 5 years old, n (%) | 429* | 26 (59.1) | 47 (78.3) | 155 (48.0) | <0.001# |
| Sex, n (%) | 431 | 0•88# | |||
| Male | 231 | 22 (50•0) | 32 (53•3) | 177 (54•1) | |
| Female | 200 | 22 (50•0) | 28 (46•7) | 150 (45•9) | |
| Weight in Kg, median (range) | 431 | 13•9 (5•5–50•2) | 11•4 (3•9–34•7) | 17•4 (5•2–54•1) | <0•001$ |
| Height in cm, median (range) | 431 | 98•1 (63–166) | 90•6 (58–148) | 114 (60•6–174) | <0•001$ |
| IGRA, n (%) | 431 | 0•11# | |||
| Positive | 125 | 37 (84•1) | 40 (66•7) | 229 (70•0) | |
| Negative | 306 | 7 (15•9) | 20 (33•3) | 98 (30•0) |
Kruskal-Wallis rank test;.
Chi-square test;.
Age was missing for 2 children with OD.
3.1. Patterns of cytokine and chemokine profiles between study phenotypes
Tables 3 and 4 show the results from comparison of the log2 transformed unstimulated and EC-stimulated fluorescence values of the 27 cytokines and chemokines, respectively, between children in the three diagnostic groups. The diagnostic plots of the level-1 and level-2 residuals can be found in the supplementary materials (Supplementary Figs 1 and 2).
Table 3.
Comparison of unstimulated analyte expression fluorescence values between diagnostic groups.
| Cytokine | Clinically diagnosed TB (n = 60) vs. Bacteriologically confirmed TB (n = 44) |
OD (n = 327) vs. Bacteriologically confirmed TB (n = 44) |
||
|---|---|---|---|---|
| Estimated difference$ (95% CI) | P# | Estimated difference$ (95% CI) | P# | |
| IL-1b | 0•02 (−0•89, 0•93) | 0•99 | −0•92 (−1•66, −0•19) | 0•002 |
| IL-1ra | 0•05 (−0•86, 0•95) | 0•99 | −1•26 (−2•00, −0•52) | <0•001 |
| IL-2 | 0•07 (−0•84, 0•98) | 0•99 | −0•41 (−1•15, 0•33) | 0•98 |
| IL-4 | 0•21 (−0•70, 1•12) | 0•99 | −0•31 (−1•05, 0•43) | 0•99 |
| IL-5 | 0•12 (−0•79, 1•03) | 0•99 | −0•18 (−0•92, 0•55) | 0•99 |
| IL-6 | −0•05 (−0•86, 0•96) | 0•99 | −1•40 (−2•14, −0•67) | <0•001 |
| IL-7 | 0•16 (−1•07, 0•75) | 0•99 | −0•42 (−1•15, 0•32) | 0•97 |
| IL-8 | 0•20 (−1•11, 0•71) | 0•99 | −1•21 (−1•95, −0•48) | <0•001 |
| IL-9 | −0•18 (−1•09, 0•73) | 0•99 | −0•01 (−0•75, 0•72) | 0•99 |
| IL-10 | 0•33 (−1•24, 0•58) | 0•99 | −0•09 (−0•82, 0•65) | 0•99 |
| IL-12P70 | 0•11 (−0•80, 1•02) | 0•99 | −0•25 (−0•99, 0•49) | 0•99 |
| IL-13 | 0•08 (−0•83, 0•99) | 0•99 | −0•17 (−0•91, 0•56) | 0•99 |
| IL-15 | 0•11 (−0•80, 1•02) | 0•99 | −0•51 (−1•24, 0•23) | 0•71 |
| IL-17 | 0•28 (−0•63, 1•19) | 0•99 | −0•15 (−0•89, 0•58) | 0•99 |
| EOTAXIN | 0•09 (−0•82, 1•00) | 0•99 | −0•36 (−1•10, 0•38) | 0•99 |
| FGFBasic | 0•07 (−0•84, 0•98) | 0•99 | −0•29 (−1•03, 0•44) | 0•99 |
| GCSF | 0•29 (−0•62, 1•20) | 0•99 | −0•34 (−1•07, 0•40) | 0•99 |
| GMCS-F | 0•06 (−0•85, 0•97) | 0•99 | −0•01 (−0•74, 0•73) | 0•99 |
| IFN-γ | −0•02 (−0•93, 0•89) | 0•99 | −0•58 (−1•31, 0•16) | 0•41 |
| IP-10 | 0•07 (−0•84, 0•99) | 0•99 | −0•97 (−1•70, −0•23) | <0•001 |
| MCP-1 | 0•10 (−0•81, 1•01) | 0•99 | −0•80 (−1•53, −0•06) | 0•02 |
| MIP-1a | 0•04 (−0•87, 0•95) | 0•99 | −1•24 (−1•98, −0•50) | <0•001 |
| MIP-1b | −0•11 (−1•02, 0•80) | 0•99 | −0•43 (−1•16, 0•31) | 0•96 |
| PDGFBB | 0•31 (−0•60, 1•22) | 0•99 | −0•20 (−0•94, 0•53) | 0•99 |
| RANTES | −0•02 (−0•93, 0•89) | 0•99 | 0•002 (−0•74, 0•74) | 0•99 |
| TNFa | 0•18 (−0•73, 1•09) | 0•99 | −0•40 (−1•14, 0•33) | 0•98 |
| VEGF | 0•14 (−0•77, 1•05) | 0•99 | −0•29 (−1•03, 0•45) | 0•99 |
Estimated difference of log-2 transformed responses;.
All 95% confidence intervals (CI) and p-values are adjusted using Sidak correction for the Type I error because of multiple testing; OD = other respiratory diseases but not TB.
Table 4.
Comparison of EC-stimulated analyte expression fluorescence values between diagnostic groups.
| Cytokine | Clinically diagnosed TB (n = 60) vs. Bacteriologically confirmed TB (n = 44) |
OD (n==327) vs. Bacteriologically confirmed TB (n = 44) |
||
|---|---|---|---|---|
| Estimated difference$ (95% CI) | P# | Estimated difference$ (95% CI) | P# | |
| IL-1b | −0•40 (−1•33, 0•52) | 0•99 | −0•29 (−1•04, 0•46) | 0•99 |
| IL-1ra | −0•17 (−1•09, 0•76) | 0•99 | −0•69 (−1•44, 0•06) | 0•12 |
| IL-2 | −0•17 (−1•10, 0•75) | 0•99 | −1•53 (−2•29, −0•78) | <0•001 |
| IL-4 | −0•10 (−1•03, 0•82) | 0•99 | −0•36 (−1•11, 0•39) | 0•99 |
| IL-5 | 0•19 (−0•73, 1•12) | 0•99 | −0•38 (−1•13, 0•37) | 0•99 |
| IL-6 | −0•25 (−1•18, 0•68) | 0•99 | −0•20 (−0•95, 0•55) | 0•99 |
| IL-7 | −0•04 (−0•96, 0•89) | 0•99 | −0•28 (−1•03, 0•47) | 0•99 |
| IL-8 | −0•16 (−1•09, 0•77) | 0•99 | −0•15 (−0•91, 0•60) | 0•99 |
| IL-9 | 0•02 (−0•91, 0•95) | 0•99 | −0•08 (−0•83, 0•67) | 0•99 |
| IL-10 | −0•35 (−1•28, 0•57) | 0•99 | 0•05 (−0•70, 0•80) | 0•99 |
| IL-12P70 | 0•06 (−0•86, 0•99) | 0•99 | −0•43 (−1•18, 0•32) | 0•96 |
| IL-13 | 0•05 (−0•88, 0•98) | 0•99 | −1•57 (−2•33, −0•82) | <0•001 |
| IL-15 | −0•10 (−1•03, 0•82) | 0•99 | −0•45 (−1•20, 0•30) | 0•93 |
| IL-17 | 0•02 (−0•90, 0•95) | 0•99 | −0•33 (−1•08, 0•42) | 0•99 |
| EOTAXIN | 0•03 (−0•92, 0•93) | 0•99 | −0•30 (−1•05, 0•45) | 0•99 |
| FGFBasic | −0•07 (−1•00, 0•85) | 0•99 | −0•43 (−1•18, 0•32) | 0•97 |
| GCSF | −0•27 (−1.20, 0•66) | 0•99 | −0•27 (−1.02, 0•48) | 0•99 |
| GMCS-F | 0•16 (−0•77, 1•08) | 0•99 | −0•74 (−1•49, 0•01) | 0•06 |
| IFN-γ | −0•22 (−1•15, 0•71) | 0•99 | −0•93 (−1•68, −0•18) | 0•002 |
| IP-10 | −0•10 (−1•02, 0•83) | 0•99 | −0•36 (−1•11, 0•39) | 0•99 |
| MCP-1 | −0•23 (−1•15, 0•70) | 0•99 | −0•05 (−0•80, 0•70) | 0•99 |
| MIP-1a | −0•23 (−1•16, 0•70) | 0•99 | −0•10 (−0•85, 0•65) | 0•99 |
| MIP-1b | −0•07 (−1•00, 0•86) | 0•99 | −0•02 (−0•77, 0•73) | 0•99 |
| PDGFBB | 0•25 (−0•68, 1•18) | 0•99 | −0•33 (−1•10, 0•41) | 0•99 |
| RANTES | −0•01 (−0•93, 0•92) | 0•99 | 0•02 (−0•73, 0•77) | 0•99 |
| TNFa | −0•37 (−1•29, 0•56) | 0•99 | −0•45 (−1•20, 0•30) | 0•92 |
| VEGF | −0•32 (−1•25, 0•61) | 0•99 | −0•27 (−1•02, 0•48) | 0•99 |
Estimated difference of log-2 transformed responses;.
All 95% confidence intervals (CI) and p-values are adjusted using Sidak correction for the Type I error because of multiple testing; OD = other respiratory diseases but not TB.
We found no evidence of difference in the unstimulated and EC-stimulated expression fluorescence values of all the 27 cytokines and chemokines between children with bacteriologically confirmed TB and those with clinically diagnosed TB disease. However, there was strong evidence that the unstimulated fluorescence values of IL-1ra, IL-6, IL-8, IP-10, MCP-1 and MIP-1a, and the EC-stimulated fluorescence values of IL-2, IL-13, and IFN-γ were higher in children with bacteriologically confirmed TB compared to children with OD.
3.2. Identification of a host-derived cytokine biosignature for the diagnosis of childhood TB
The result of assignment of study subjects into training and test sets are shown in Supplementary Table 1. In the test set, we pragmatically combined the bacteriologically confirmed and clinically diagnosed TB cases into one group, as we did not find any evidence of difference in the expression fluorescence values of all analytes between the two groups; the binary outcome variable was therefore defined as ‘TB disease’ (comprising bacteriologically confirmed TB and clinically diagnosed TB) compared to OD.
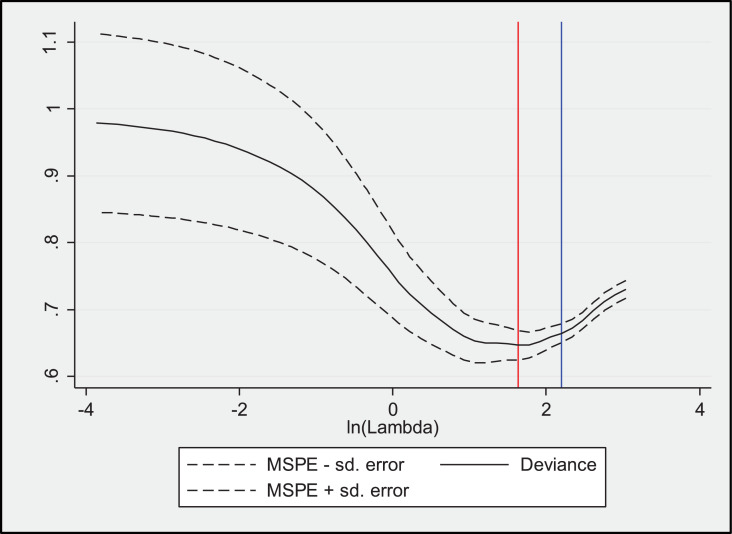
Using the log2 transformed cytokine expression fluorescent values of all 260 children in the training set for logistic regression analyses combined with LASSO penalty, we derived a three-marker model consisting of IL-1ra, IL-7 and IP-10 from unstimulated samples as the minimum but optimally discriminant cytokine biosignature associated with bacteriologically confirmed TB (Table 5; Fig. 2).
Table 5.
Results from logistic LASSO model.
| Selected biosignatures in Log2 scale | LASSO coefficients |
|---|---|
| IL-1ra | 0•134 |
| IL-7 | 0•371 |
| IP-10 | 0•262 |
| Intercept: | −8•94 |
| Optimal lambda with MSPE within one standard error of the minimum loss*: | 8•994 |
| Minimum MSPE or deviance: | 0•664 |
MSPE= Mean-squared prediction error. Minimum loss is the minimum MSPE.
Fig. 2.
Mean-squared prediction error from logistic LASSO model. The red vertical line represents the lambda that minimizes MSPE whereas the blue vertical line represents the largest lambda for which MSPE is within one standard deviation of the minimum MSPE.
As we had earlier shown that there was strong evidence of differences in age and basic anthropometric measurements of children in the three diagnostic groups, we assessed the predictive performance of the 3-marker biosignature by logistic discriminant analyses adjusting the prediction models for age and age- and sex-adjusted nutritional status with the inclusion of age group as a binary variable (< 5 years and ≥ 5 years) and Body Mass Index (BMI)-for-age z score, calculated using the WHO growth standards [25], and interaction terms between the selected biosignatures and BMI-for-age z-scores, as covariates. Six out of the 431 children enroled had very implausible BMI-for-age z-scores in which the absolute values of their z-scores were ≤ −5 or ≥ 5, and two children had missing BMI-for-age z-scores. Thus, a total of 423 children had plausible BMI z-scores and were included in the discriminant analyses. Table 6 shows the predictive performance of the 3-marker cytokine biosignature for TB disease in four different logistic discriminant analysis models built in the training set, and subsequently assessed in the independent test set.
Table 6.
Estimated sensitivity, specificity and AUC from different logistic discriminant models in the training and test datasets.
| Covariates | Training set (n = 260)* |
Test set (n = 171)* |
||||
|---|---|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | AUCa (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | AUCa (95% CI) | |
| Log2 IL1-ra, Log2 IL7, Log2 IP10, age group, BMI-for-age z-score (model M1–1) | 70•0 (50•6, 85•3) | 72•1 (65•8, 77•8) | 0•71 (0•62, 0•80) | 76•4 (64•9, 85•6) | 72•8 (62•6, 81•6) | 0•75 (0•68, 0•81) |
| Log2 IL1-ra, Log2 IL7, Log2 IP10, age group, BMI-for-age z-score, interaction Log2 IL7 and BMI-for-age (model M1–2) | 70•0 (50•6, 85•3) | 72•5 (66•2, 78•2) | 0•71 (0•62, 0•80) | 75•0 (63•4, 84•5) | 73•9 (63•7, 82•5) | 0•74 (0•68, 0•81) |
| Log2 IL1-ra, Log2 IL7, Log2 IP10, age group, BMI-for-age z-score, interaction Log2 IL7and BMI-for-age (model M1–3) | 70•0 (50•6, 85•3) | 72•5 (66•2, 78•2) | 0•71 (0•62, 0•80) | 76•4 (64•9, 85•6) | 72•8 (62•6, 81•6) | 0•75 (0•68, 0•81) |
| Log2 IL1-ra, Log2 IL7, Log2 IP10, age group, BMI-for-age z-score, interaction Log2 IP10 and BMI-for-age (model M1–4) | 73•3 (54•1, 87•7) | 73•8 (67•6, 79•4) | 0•74 (0•65, 0•82) | 72•2 (60•4, 82•1) | 75•0 (64•9, 83•4) | 0•74 (0•67, 0•81) |
423 children (training set: 259; test set: 164) ultimately included in the discriminant analyses: six children (training set: 1; test set: 5) had very implausible BMI-for-age z-scores i.e. z-scores ≤ −5 or ≥ 5, and two children in test set had missing BMI-for-age z-scores. aArea under the receiver operator characteristic curve. Age group is a binary variable i.e. < 5 years old and ≥ 5 years old.
Model M1–4, which includes the 3-marker biosignature, the binary age group, and BMI-for-age z-score with the interaction term between BMI-for-age z-score and log2 IP10, had the most stable diagnostic performance in the training and test sets as assessed by its sensitivity, specificity and AUC. In this model, the 3-marker biosignature distinguished children diagnosed with TB disease, irrespective of microbiological confirmation, from OD in the test set with an AUC of 0•74 (95% CI: 0•67, 0•81) and demonstrated sensitivity and specificity of 72•2% (95% CI: 60•4, 82•1%) and 75•0% (95% CI: 64•9, 83•4%), respectively, with its performance independent of the age group of the children and their age- and sex-adjusted nutritional status. Further details, including the estimated coefficients of components of Model M1–4, and comparison of the log2 mean (standard deviation) for each of the three-markers in the biosignature by age groups, are provided in the Supplementary Results.
The results of an additional six logistic discriminant models (M0–1 to M0–6) based on the same set of 423 children, including only the 3-marker biosignature, age group (< 5 years and ≥ 5 years) and the interaction terms between the cytokine markers, are shown in Supplementary Table 2.
4. Discussion
Using LASSO regression models, we have identified a novel three-marker biosignature of childhood TB consisting of IL-1ra, IL-7 and IP-10. Somewhat surprisingly, this biosignature was identified in samples that had not been stimulated with M.tb antigens, which might therefore make it more suitable for further development as a field test. Using a composite of all children diagnosed with TB disease irrespective of microbiological confirmation in the test set, the 3-marker biosignature distinguished children with TB disease from those diagnosed with other respiratory diseases with an AUC of 0•74 and demonstrated a sensitivity and specificity of 72% and 75% respectively, with its performance independent of their age group and their nutritional status standardized by age and sex.
Two aspects of our study methodology are distinct from most other published studies. First, we used the revised WHO case definitions for our diagnostic classification [17], and not the clinical case definitions proposed by an NIAID/NIH expert panel for classification of intrathoracic TB disease in children [26]. This is because, as discussed in depth by the authors of the expert panel report, their proposed case definitions are not appropriate for studies such as ours that incorporate active investigation of possible TB in children from household case-finding studies since this could influence the likelihood of finding TB disease. The authors of the expert panel report further explained that: (i) active case finding is likely to identify cases at an earlier stage of disease and a much shorter duration of symptoms, compared with children investigated for tuberculosis at the referral level; and (ii) given that the entry point for contact studies is, by definition, a positive history of exposure, it compromises one of the definitions used for clinical classification in the original proposed definitions [26]. Therefore, we employed the use of the WHO case definitions that capture disease within the broad criteria that includes clinical, microbiological and radiological parameters [17], as this is more compatible with the clinical and epidemiological setting of active case finding. Secondly, we focused on the direct statistical analyses of the fluorescence intensities (FI) of our multiplex cytokine assays. This differs from the traditional concentration-based analyses that is mostly reported in the literature but have now been shown to have strong limitations in detecting low- or high-abundance out-of-range analytes [27,28]. Such out-of-range values in concentration-based analysis are frequently imputed by maximum likelihood estimations, extrapolation or substitution thereby increasing the risk of obtaining inaccurate estimations and false conclusions 29, 30, 31, 32, 33, 34. Several studies have shown that fluorescence values do not have out-of-range problems, and that fluorescence-based analysis has higher statistical power than concentration-based analysis, is a better choice for statistical differential analyses and reproducibility, and that background correction is not required [28,33,35,36]. The use of fluorescence intensity values for our discriminant analysis is relatively novel. While this might have allowed us to identify discriminant markers that would otherwise have been missed or not identified in concentration-based analysis, it also calls for further research on how to accurately translate the relative fluorescence intensity analysis into easily measurable thresholds for clinical decisions.
A number of adult studies have described different combinations of cytokines such as TNF-α, IL-12(p40), IL-6, IL-10, IL-18, IL-17, sCD40L, FGF and VEGF as implicated in the immune response against M.tb and/or in distinguishing TB disease from LTBI or OD in TB endemic countries [20,37,38]. Given the report that distinct cytokine expression profiles of CD4+ T-cells are associated with bacterial loads in adults [39], a very different cytokine expression profile could be expected in childhood TB cases, which are paucibacillary and most likely represent progression of primary infection rather than reactivation disease. In line with our observations, gene expression profiling results have also differed between adults [40,41], and children [42].
The three-marker biosignature described in our cohort contains cytokines known to be important in TB immunity. IL-1ra is a naturally occurring competitive inhibitor of the pro-inflammatory effects of IL-1α and IL-1β that is strongly induced by M.tb and encoded by polymorphic genes [43]. Polymorphisms of the IL-1ra gene on chromosome 2 have been shown to be associated with genetic susceptibility to TB amongst Gambians and a higher IL-1ra/IL-1β ratio in response to M.tb indicates the inflammatory profile of an individual [43,44]. IL-7 is a pleiotropic growth factor that binds to the IL-7 receptor (IL-7R) and promotes the generation, expansion and survival of T-cells, and decreases the production of TGF-β [45,46]. Its production was shown to increase in the lung tissues of non-human primates that also showed increased survival following M.tb challenge [47]. The expression profiles of a biomarker signature in adult TB patients in The Gambia showed that the gene expression of IL-7R has statistically significant discriminatory power to classify treated TB patients from untreated TB cases as early as 2 months after treatment [48]. IP-10 is a chemokine that stimulates the activation and migration of natural killer (NK) cells and T-cells to the site of M.tb infection. It is produced by monocytes, macrophages and bronchial epithelium cells in TB patients in response to lipoarabinomannan in the cells wall of virulent strains of M.tb and the expressions can be upregulated by IFN-γ and inhibited by IL-4 [49,50]. A systematic review of 55 articles identified IP-10 as amongst the most promising diagnostic biomarkers for TB disease based on its presence in both mycobacterial antigen-stimulated and unstimulated samples [51].
Several studies that explored the use of procalcitonin and C-reactive protein to distinguish between TB disease and community acquired pneumonia found that these inflammatory markers lack specificity 52, 53, 54. However, results from other studies, including systematic reviews with individual patient data meta-analyses, suggest that procalcitonin and respiratory polymerase chain reaction panel might have some value in guiding duration of antibiotic therapy in patients with acute respiratory tract infections other than TB 55, 56, 57. Few studies have examined cytokine profiles for diagnostic purposes in childhood cohorts, and these were mainly aimed at the distinction between TB disease and LTBI. Tebruegge and colleagues recently reported that an M.tb antigen-specific biosignature, comprising the combination of TNF-α, IL-1ra and IL-10, showed the best discriminatory ability between TB disease and LTBI in paediatric cases [58]. Similar to our finding, Chegou et al. reported that unstimulated levels of IL-1ra and IP-10 and antigen-specific levels of VEGF in Quantiferon (QFT) supernatants may be useful for diagnosing TB disease, and differentiating between TB disease and M.tb infection in children investigated in a high HIV/TB prevalence setting [59]. Dhanasekaran et al. reported that a combination of IL-2 and IL-8 from antigen-stimulated QFT supernatants discriminated TB disease from LTBI with an AUC of 0.70 in children aged less than three years in India [60]. In contrast to these reports from limited paediatric studies which focused on the distinction between LTBI and TB disease, we aimed to derive markers that can distinguish between TB disease and other respiratory diseases, given the clinical need to initiate the appropriate therapies for symptomatic children, and our biosignature was identified exclusively in unstimulated samples with its performance independent of the age group and nutritional status of the children standardized by age and sex.
We found no evidence of difference in the unstimulated and M.tb-specific antigen-stimulated fluorescence values of all the analytes between children with bacteriologically confirmed TB and clinically diagnosed TB cases, supporting our decision to combine these groups in assessing the predictive performance of the biosignature models in the independent test set. Furthermore, we reported with strong statistical evidence that the unstimulated fluorescence values of IL-1ra, IL-6, IL-8, IP-10, MCP-1 and MIP-1a, and the EC-stimulated fluorescence values of IL-2, IL-13, and IFN-γ were higher in children with bacteriologically confirmed TB compared to children with OD.
Whether a new test should serve as a confirmatory diagnostic or triage test depends on its performance characteristics, target population, setting and ease of use amongst other factors that will influence its position within a clinical algorithm; guidelines and target product profiles (TPP) for such tests have been published [7]. These WHO-endorsed TPP criteria recommended minimal targets of 66% sensitivity and 98% specificity for a new diagnostic test for TB in children, and 90% sensitivity and 70% specificity for a triage test. The biosignature in our study correctly detected more than 70% of all childhood TB disease irrespective of microbiological confirmation. This three-marker signature was also identified in unstimulated supernatants and thus could potentially be measured in finger prick blood samples without the need for much laboratory support. The biosignature does not meet minimal specificity TPP target for a diagnostic test and the minimal sensitivity TPP target for a triage test. However, the fact that the specificity of the biosignature meets the minimal specificity TPP target for a triage test together with its potential ease of use suggest that it could serve as a screening or triage test to identify symptomatic children with ‘higher risk’ of TB disease as a first step. Such children could then have follow-up clinical evaluation and confirmatory diagnostic investigations using tests with higher specificity such as the Xpert Ultra assay. This assertion is supported by the consensus of experts who defined the WHO-endorsed TPP criteria for high priority diagnostic needs in TB, which posited that a triage test that is easier to do and can be conducted at lower levels of health care will conceivably identify more children with a higher likelihood of TB disease and could be cost-effective in an implementation strategy even with a fairly good sensitivity relative to a confirmatory test [7].
Our study has some clear limitations. We identified the biosignature in a prospective cohort of HIV-uninfected children with known household exposure who could probably have mild or early-stage TB disease and the signature could perform differently in severely ill children, including HIV-infected children. This setting might therefore not be representative of the children with suspected intrathoracic TB in the broader community. While splitting our dataset into training and test sets was an appropriate statistical method for internal validation of the model, this could have reduced its precision and increased the prediction error of the biosignature. The use of 10-fold cross-validation within the training set will have minimised the likelihood of the model to over-fit the data.
In conclusion, we have identified a three-marker biosignature that distinguished between TB disease and other respiratory diseases in prospectively recruited, symptomatic TB-exposed children with suspected intrathoracic TB. The fact that all the markers in this biosignature were derived from unstimulated supernatants suggests that they could potentially also be measured directly in finger prick blood samples. Validation of this biosignature in independent cohorts derived from more diverse epidemiological contexts, including settings with high HIV/TB burden and in hospitalised children, is now needed. This will provide opportunities to further refine and optimize the performance characteristics of the biosignature to increase its sensitivity, which will increase its benefit as a triage test. Following further validation, this cytokine-based biosignature has the potential to be translated into a field-friendly POC or near-POC test that could enhance the rapid and accurate diagnosis of TB in children.
Research in context
Evidence before this study
Diagnosis of tuberculosis (TB) in children is difficult because childhood TB is paucibacillary and obtaining good quality sputum specimen is a challenge, particularly in the very young. The development of a non-sputum-based point-of-care (POC) test remains an identified critical need as acknowledged by the World Health Organization (WHO). Research into TB biomarkers has gained prominence due to the lack of suitable tests based on detection of Mycobacterium tuberculosis (M.tb), and their potential for translation into a non-sputum-based POC test. We searched PubMed for studies of biosignatures for diagnosis of active tuberculosis in exclusively paediatric study subjects, defined as age < 15 years, published between January 1, 2000 and June 1, 2019. The PubMed search term used was as follows: ((((tuberculosis[ti] OR TB[ti]) AND (child*[Text Word] OR pediat*[Text Word])) ((("biological markers"[mesh] OR biological marker*[Text Word] OR biomarker*[Text Word] OR biosignature*[tw]) NOT (tumour*[Text Word] OR tumour*[tw] OR "tumour markers, biological"[mesh])) OR (miRNA[tw] OR microRNA[tw] OR proteom*[tw] OR transcriptom*[tw] OR immunoassay*[tw] OR immunoassay[mesh] OR LAM[tw] OR lipoarabinomannan*[tw] OR ("immunologic tests"[mesh] AND diagnos*[tw]) OR ((mycolic acid[tw] OR glycolipid*[tw]) AND (diagnos*[tw] OR detect*[tw])) OR (cytokine*[tw] AND diagnos*[tw]))) NOT (animals[mesh] NOT humans[mesh]))). We found that published studies of childhood TB biomarkers were mostly early-stage studies that are heterogenous in study design, types of biomarker and clinical samples. Only few studies have examined cytokine profiles for diagnostic purposes in childhood cohorts, and these were mainly aimed at the distinction between TB disease and latent TB infection (LTBI). In contrast, our study aimed to derive markers that can distinguish between TB disease and other respiratory diseases, given the clinical need to initiate the appropriate therapies for symptomatic children.
Added value of this study
We identified a novel three-marker biosignature that distinguished between TB disease, irrespective of microbiological confirmation, and other respiratory diseases in prospectively recruited TB-exposed children with clinical suspicion of intrathoracic TB, and its performance is independent of the age group of the children and their nutritional status standardized by age and sex. Somewhat surprisingly, this biosignature was identified in samples that had not been stimulated in-vitro with M.tb antigens.
Implications of the available evidence
The fact that all the markers in this biosignature were derived from unstimulated supernatants suggests that they could potentially also be measured directly in finger prick blood samples without the need for antigenic stimulation or much laboratory support. This cytokine-based biosignature is very promising; following independent validation with further optimisation of its performance, it has the potential to be translated into a field-friendly POC or near-POC test that could enhance the rapid diagnosis of TB in children.
Funding sources
This work was supported with funds from a Steinberg Fellowship in Global Health from McGill University awarded to TT, a Grand Challenges Canada (GCC) grant awarded to MP and TT, and principally funded by an MRC program grant awarded to BK (Ref: MR/K011944/1). The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of interests
This study was conducted at the MRC Unit The Gambia at LSHTM. Raw data and laboratory analysis results were extracted at the MRC Unit The Gambia. Data analysis was carried out at the LSHTM. The authors have declared that no competing interests exist. The study was principally funded by an MRC Program grant awarded to Professor Beate Kampmann (Ref: MR/K011944/1). Dr. Togun, Dr. Hoggart and Professor Kampmann have a patent for the Childhood TB diagnostic biomarker signature (MRC Technology ref: A813/3133; TB Biomarkers Script IP Ref: P10280GB).
Author contributions
TT and BK conceptualised and designed the study. UE and AKS coordinated recruitment and clinical evaluation of study subjects. TT, MPG and FM analysed the samples and collected the data. BS coordinated the data management. TT, CJH and SCA did the analysis and generated the estimates. TT drafted the first version of the manuscript with input from MP and BK. All authors contributed to the revision and correction on multiple iterations of the manuscript.
Acknowledgements
The authors wish to acknowledge the MRCG at LSHTM Childhood TB Group field and clinic teams (field workers and supervisors, field and nurse coordinators, etc.) who facilitated the recruitment, screening and follow-up of the study subjects. We also acknowledge the McGill Global Health Programme (McGill GHP) and the entire programme staff. We are particularly grateful to the Gambia National Leprosy and TB Control Programme (NLTBCP), and all the children and their families who participated in this study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102909.
Contributor Information
Toyin Togun, Email: Toyin.Togun@lshtm.ac.uk.
Beate Kampmann, Email: Beate.Kampmann@lshtm.ac.uk.
Appendix. Supplementary materials
References
- 1.World Health Organization (WHO) WHO; Geneva. Switzerland: 2018. Global tuberculosis report 2019.https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf?ua=1 Available from. [Cited 27 Jan 2020] [Google Scholar]
- 2.Dodd P.J., Gardiner E., Coghlan R., Seddon J.A. Burden of childhood tuberculosis in 22 high-burden countries: a mathematical modelling study. Lancet Glob Health. 2014;2(8):e453–e459. doi: 10.1016/S2214-109X(14)70245-1. [DOI] [PubMed] [Google Scholar]
- 3.Nelson L.J., Wells C.D. Global epidemiology of childhood tuberculosis. Int J Tuberc Lung Dis. 2004;8(5):636–647. [PubMed] [Google Scholar]
- 4.Kampmann B., Hemingway C., Stephens A. Acquired predisposition to mycobacterial disease due to autoantibodies to IFN-gamma. J Clin Invest. 2005;115(9):2480–2488. doi: 10.1172/JCI19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marais B.J., Pai M. Recent advances in the diagnosis of childhood tuberculosis. Arch Dis Child. 2007;92(5):446–452. doi: 10.1136/adc.2006.104976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zar H.J., Hanslo D., Apolles P., Swingler G., Hussey G. Induced sputum versus gastric lavage for microbiological confirmation of pulmonary tuberculosis in infants and young children: a prospective study. Lancet. 2005;365(9454):130–134. doi: 10.1016/S0140-6736(05)17702-2. [DOI] [PubMed] [Google Scholar]
- 7.Denkinger C.M., Kik S.V., Cirillo D.M. Defining the Needs for Next Generation Assays for Tuberculosis. J Infect Dis. 2015;211(suppl 2):S29–S38. doi: 10.1093/infdis/jiu821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicol M.P., Gnanashanmugam D., Browning R. A Blueprint to Address Research Gaps in the Development of Biomarkers for Pediatric Tuberculosis. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2015;61(Suppl 3):S164–S172. doi: 10.1093/cid/civ613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacha J.M., Ngo K., Clowes P. Why being an expert – despite xpert –remains crucial for children in high TB burden settings. BMC Infect Dis. 2017;17(1):123. doi: 10.1186/s12879-017-2236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabi I., Rachow A., Mapamba D. Xpert MTB/RIF Ultra assay for the diagnosis of pulmonary tuberculosis in children: a multicentre comparative accuracy study. J Infect. 2018;77(4):321–327. doi: 10.1016/j.jinf.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Velez C.M., Marais B.J. Tuberculosis in children. New Engl J Med. 2012;367(4):348–361. doi: 10.1056/NEJMra1008049. [DOI] [PubMed] [Google Scholar]
- 12.Rangaka M.X., Wilkinson K.A., Glynn J.R. Predictive value of interferon-γ release assays for incident active tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):45–55. doi: 10.1016/S1473-3099(11)70210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollock L., Basu Roy R., Kampmann B. How to use: interferon gamma release assays for tuberculosis. Arch Dis Child Edu Pract Ed. 2013;98(3):99–105. doi: 10.1136/archdischild-2013-303641. [DOI] [PubMed] [Google Scholar]
- 14.Walzl G., Haks M.C., Joosten S.A., Kleynhans L., Ronacher K., Ottenhoff T.H. Clinical Immunology and Multiplex Biomarkers of Human Tuberculosis. Cold Spring Harb Perspect Med. 2014;5(4) doi: 10.1101/cshperspect.a018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobsen M., Mattow J., Repsilber D., Kaufmann S.H. Novel strategies to identify biomarkers in tuberculosis. Biol. Chem. 2008;389(5):487–495. doi: 10.1515/bc.2008.053. [DOI] [PubMed] [Google Scholar]
- 16.Togun T.O., Egere U., Gomez M.P. No added value of interferon-gamma release to a prediction model for childhood tuberculosis. Eur Respir J: Off J Eur Soc Clin Respir Physiol. 2016;47(1):223–232. doi: 10.1183/13993003.00890-2015. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization (WHO) WHO; Geneva, Switzerland: 2013. Definitions and reporting framework for tuberculosis - 2013 revision.www.who.int/iris/bitstream/10665/79199/1/9789241505345_eng.pdf Available from. [Cited 20 September 2019] [Google Scholar]
- 18.Gambian National Leprosy and Tuberculosis Control Programme (NLTP) Department of State for Health, The Gambia; Banjul, The Gambia: 2012. National guidelines for the management of tuberculosis. [Google Scholar]
- 19.Egere U., Sillah A., Togun T. Isoniazid preventive treatment among child contacts of adults with smear-positive tuberculosis in The Gambia. Public Health Action. 2016;6(4):226–231. doi: 10.5588/pha.16.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutherland J.S., de Jong B.C., Jeffries D.J., Adetifa I.M., Ota M.O. Production of TNF-alpha, IL-12(p40) and IL-17 can discriminate between active TB disease and latent infection in a West African cohort. PLoS ONE. 2010;5(8):e12365. doi: 10.1371/journal.pone.0012365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidak Z. Rectangular Confidence Regions for the Means of Multivariate Normal Distributions. J Am Stat Assoc. 1967;62(318):626–633. [Google Scholar]
- 22.Friedman J., Hastie T., Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 23.Albert A., Lesaffre E. MULTIPLE GROUP LOGISTIC DISCRIMINATION. In: Choi SC, editor. Statistical methods of discrimination and classification: pergamon. 1986. pp. 209–224. [Google Scholar]
- 24.Bossuyt P.M., Reitsma J.B., Bruns D.E. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD Initiative. Ann. Intern. Med. 2003;138(1):40–44. doi: 10.7326/0003-4819-138-1-200301070-00010. [DOI] [PubMed] [Google Scholar]
- 25.WHO . World Health Organization; Geneva: 2007. Child growth standards: length/height-for-age, weight-for-age, weight-for-length, weight-for-height and body mass index-for-age: methods and development. 2007. [Google Scholar]
- 26.Graham S.M., Cuevas L.E., Jean-Philippe P. Clinical Case Definitions for Classification of Intrathoracic Tuberculosis in Children: an Update. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2015;61(Suppl 3):S179–S187. doi: 10.1093/cid/civ581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Won J.H., Goldberger O., Shen-Orr S.S., Davis M.M., Olshen R.A. Significance analysis of xMap cytokine bead arrays. Proc. Natl. Acad. Sci. U.S.A. 2012;109(8):2848–2853. doi: 10.1073/pnas.1112599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breen E.J., Tan W., Khan A. The Statistical Value of Raw Fluorescence Signal in Luminex xMAP Based Multiplex Immunoassays. Sci Rep. 2016;6:26996. doi: 10.1038/srep26996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong H.L., Pfeiffer R.M., Fears T.R., Vermeulen R., Ji S., Rabkin C.S. Reproducibility and correlations of multiplex cytokine levels in asymptomatic persons. Cancer Epidemiol Biomark Prev. 2008;17(12):3450–3456. doi: 10.1158/1055-9965.EPI-08-0311. [DOI] [PubMed] [Google Scholar]
- 30.Lubin J.H., Colt J.S., Camann D. Epidemiologic evaluation of measurement data in the presence of detection limits. Environ Health Perspect. 2004;112(17):1691–1696. doi: 10.1289/ehp.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breen E.C., Reynolds S.M., Cox C. Multisite comparison of high-sensitivity multiplex cytokine assays. Clin Vaccine Immunol: CVI. 2011;18(8):1229–1242. doi: 10.1128/CVI.05032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breen E.J., Polaskova V., Khan A. Bead-based multiplex immuno-assays for cytokines, chemokines, growth factors and other analytes: median fluorescence intensities versus their derived absolute concentration values for statistical analysis. Cytokine. 2015;71(2):188–198. doi: 10.1016/j.cyto.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 33.Whitcomb B.W., Schisterman E.F. Assays with lower detection limits: implications for epidemiological investigations. Paediatr Perinat Epidemiol. 2008;22(6):597–602. doi: 10.1111/j.1365-3016.2008.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helsel D.R. Fabricating data: how substituting values for nondetects can ruin results, and what can be done about it. Chemosphere. 2006;65(11):2434–2439. doi: 10.1016/j.chemosphere.2006.04.051. [DOI] [PubMed] [Google Scholar]
- 35.Ballenberger N., Lluis A., von Mutius E., Illi S., Schaub B. Novel statistical approaches for non-normal censored immunological data: analysis of cytokine and gene expression data. PLoS ONE. 2012;7(10):e46423. doi: 10.1371/journal.pone.0046423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.May R.C., Chu H., Ibrahim J.G., Hudgens M.G., Lees A.C., Margolis D.M. Change-point models to estimate the limit of detection. Stat Med. 2013;32(28):4995–5007. doi: 10.1002/sim.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Algood H.M., Chan J., Flynn J.L. Chemokines and tuberculosis. Cytokine Growth Factor Rev. 2003;14(6):467–477. doi: 10.1016/s1359-6101(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 38.Ota M.O., Mendy J.F., Donkor S. Rapid diagnosis of tuberculosis using ex vivo host biomarkers in sputum. Eur Respir J: Off J Eur Soc Clin Respir Physiol. 2014;44(1):254–257. doi: 10.1183/09031936.00209913. [DOI] [PubMed] [Google Scholar]
- 39.Caccamo N., Guggino G., Joosten S.A. Multifunctional CD4(+) T cells correlate with active Mycobacterium tuberculosis infection. Eur. J. Immunol. 2010;40(8):2211–2220. doi: 10.1002/eji.201040455. [DOI] [PubMed] [Google Scholar]
- 40.Berry M.P., Graham C.M., McNab F.W. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaforou M., Wright V.J., Oni T. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 2013;10(10) doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson S.T., Kaforou M., Brent A.J. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370(18):1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkinson R.J., Patel P., Llewelyn M. Influence of polymorphism in the genes for the interleukin (IL)-1 receptor antagonist and IL-1beta on tuberculosis. J. Exp. Med. 1999;189(12):1863–1874. doi: 10.1084/jem.189.12.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellamy R., Ruwende C., Corrah T., McAdam K.P., Whittle H.C., Hill A.V. Assessment of the interleukin 1 gene cluster and other candidate gene polymorphisms in host susceptibility to tuberculosis. Tuber Lung Dis: Off J Int Union Against Tuberc Lung Dis. 1998;79(2):83–89. doi: 10.1054/tuld.1998.0009. [DOI] [PubMed] [Google Scholar]
- 45.Sprent J., Surh C.D. Interleukin 7, maestro of the immune system. Semin Immunol. 2012;24(3):149–150. doi: 10.1016/j.smim.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 46.Terrazzini N., Mantegani P., Kern F., Fortis C., Mondino A., Caserta S. Interleukin-7 Unveils Pathogen-Specific T Cells by Enhancing Antigen-Recall Responses. J. Infect. Dis. 2018;217(12):1997–2007. doi: 10.1093/infdis/jiy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rane L., Rahman S., Magalhaes I. Increased (6 exon) interleukin-7 production after M. tuberculosis infection and soluble interleukin-7 receptor expression in lung tissue. Genes Immun. 2011;12(7):513–522. doi: 10.1038/gene.2011.29. [DOI] [PubMed] [Google Scholar]
- 48.Joosten S.A., Goeman J.J., Sutherland J.S. Identification of biomarkers for tuberculosis disease using a novel dual-color RT-MLPA assay. Genes Immun. 2012;13(1):71–82. doi: 10.1038/gene.2011.64. [DOI] [PubMed] [Google Scholar]
- 49.Riedel D.D., Kaufmann S.H. Chemokine secretion by human polymorphonuclear granulocytes after stimulation with Mycobacterium tuberculosis and lipoarabinomannan. Infect. Immun. 1997;65(11):4620–4623. doi: 10.1128/iai.65.11.4620-4623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauty A., Dziejman M., Taha R.A. The T cell-specific CXC chemokines IP-10, Mig, and I-TAC are expressed by activated human bronchial epithelial cells. J Immunol. 1999;162(6):3549–3558. [PubMed] [Google Scholar]
- 51.John S.H., Kenneth J., Gandhe A.S. Host biomarkers of clinical relevance in tuberculosis: review of gene and protein expression studies. Biomark: Biochem Indic Expo, Response, Suscept Chem. 2012;17(1):1–8. doi: 10.3109/1354750X.2011.628048. [DOI] [PubMed] [Google Scholar]
- 52.Kang Y.A., Kwon S.-.Y., Yoon H.I., Lee J.H., Lee C.-.T. Role of C-reactive protein and procalcitonin in differentiation of tuberculosis from bacterial community acquired pneumonia. Korean J Intern Med. 2009;24(4):337–342. doi: 10.3904/kjim.2009.24.4.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon C., Chaisson L.H., Patel S.M. Diagnostic accuracy of C-reactive protein for active pulmonary tuberculosis: a meta-analysis. Int J Tuberc Lung Dis: Off J Int Union Against Tuberc Lung Dis. 2017;21(9):1013–1019. doi: 10.5588/ijtld.17.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendelson F., Griesel R., Tiffin N. C-reactive protein and procalcitonin to discriminate between tuberculosis, Pneumocystis jirovecii pneumonia, and bacterial pneumonia in HIV-infected inpatients meeting WHO criteria for seriously ill: a prospective cohort study. BMC Infect. Dis. 2018;18(1):399. doi: 10.1186/s12879-018-3303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuetz P., Wirz Y., Sager R. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. Cochrane Database Syst Rev. 2017;10(10) doi: 10.1002/14651858.CD007498.pub3. CD007498-CD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang T., Wang Y., Yang Q., Dong Y. Procalcitonin-guided antibiotic therapy in critically ill adults: a meta-analysis. BMC Infect. Dis. 2017;17(1) doi: 10.1186/s12879-017-2622-3. 514- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moradi T., Bennett N., Shemanski S., Kennedy K., Schlachter A., Boyd S. Use of procalcitonin and a respiratory polymerase chain reaction panel to reduce antibiotic use via an EMR alert. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2019 doi: 10.1093/cid/ciz1042. ciz1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tebruegge M., Dutta B., Donath S. Mycobacteria-Specific Cytokine Responses Detect Tuberculosis Infection and Distinguish Latent from Active Tuberculosis. Am J Respir Crit Care Med. 2015;192(4):485–499. doi: 10.1164/rccm.201501-0059OC. [DOI] [PubMed] [Google Scholar]
- 59.Chegou N.N., Detjen A.K., Thiart L. Utility of host markers detected in Quantiferon supernatants for the diagnosis of tuberculosis in children in a high-burden setting. PLoS ONE. 2013;8(5):e64226. doi: 10.1371/journal.pone.0064226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhanasekaran S., Jenum S., Stavrum R. Identification of biomarkers for Mycobacterium tuberculosis infection and disease in BCG-vaccinated young children in Southern India. Genes Immun. 2013;14(6):356–364. doi: 10.1038/gene.2013.26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.