Abstract
Background
We describe the genomic landscape of circulating tumour DNA (ctDNA) across pathological subtypes of metastatic breast cancer.
Methods
255 clinically annotated patients with ctDNA testing by Guardant360 were stratified into HR+, HER2+, and TNBC cohorts. Frequency and heterogeneity of alterations were reported. Paired ctDNA and tissue sequencing were compared for a subset of patients. The association of ctDNA and metastatic sites of disease on imaging was also assessed.
Findings
89% of patients had at least one ctDNA alteration detected. The most common single nucleotide variants (SNVs) for HR+ patients were PIK3CA, ESR1, and TP53. For HER2+, these were TP53, PIK3CA, and ERBB2 with ERBB2 as the most frequent copy number variant (CNV). For TNBC, the most common SNVs were TP53 and PIK3CA, and the most frequent CNVs were MYC, CCNE1, and PIK3CA. TNBC patients had a significantly higher mutant allele frequency (MAF) of the highest variant compared to HR+ or HER2+ patients (P<0.05). Overall, alterations in PIK3CA, ESR1, and ERBB2 were observed in 39.6%, 16.5%, and 21.6% of patients, respectively. Agreement between blood and tissue was 79–91%. MAF and number of alterations were significantly associated with number of metastatic sites on imaging (P<0.0001).
Interpretation
These data demonstrate the genetic heterogeneity of metastatic breast cancer in blood, the high prevalence of clinically actionable alterations, and the potential to utilise ctDNA as a surrogate for tumour burden on imaging.
Funding
Lynn Sage Cancer Research Foundation, OncoSET Precision Medicine Program, and REDCap support was funded by the National Institutes of Health UL1TR001422.
Keywords: Circulating tumour DNA, cell-free DNA, Next-generation sequencing, Genomics, Metastatic breast cancer
Research in Context.
Evidence before this study
Mutation profiling using tissue next-generation sequencing (NGS) has been well characterised over the last two decades. With the emergence of circulating tumour DNA (ctDNA) as a non-invasive NGS method, there is a critical need to define the mutational landscape in blood and to compare mutational profiles of metastatic breast cancer between blood and tissue. We explored prior work using PubMed with the following terms: “circulating tumour DNA,” “ctDNA,” and “metastatic breast cancer.” Several studies focused on particular resistance alterations, such as PIK3CA or ESR1, which were identified in blood. A few smaller studies compared tissue and blood NGS in metastatic breast cancer. Robust clinical and imaging annotation associated with ctDNA data are lacking in some studies. Furthermore, ctDNA data analysed across breast cancer subtypes (hormone-receptor positive, HER2 positive, and triple negative breast cancer) are limited.
Added value of this study
Here, we present the results of a large (>250 patient), clinically annotated cohort of patients with metastatic breast cancer who underwent ctDNA evaluation. We report the mutational frequencies across subtypes and the frequency of clinically meaningful alterations in PIK3CA, ESR1, and ERBB2. Additionally, there was relatively high agreement between blood and tissue sequencing, and ctDNA content was significantly associated with number of metastatic sites on imaging. These data reinforce the capability of utilising ctDNA to detect tumour heterogeneity across breast cancer subtypes, the high prevalence of clinically actionable alterations, and the association between ctDNA and tumour burden on imaging.
Implications of all the available evidence
ctDNA reflects the tumour landscape in blood to accurately reflect tumour heterogeneity across metastatic breast cancer subtypes. Therefore, ctDNA could serve as a tool to characterise genomic alterations non-invasively, to enrol patients onto targeted precision medicine clinical trials, and to serve as a surrogate for tumour burden on imaging. Further studies are needed to incorporate ctDNA assays into novel clinical trial designs for patient enrolment and stratification and to compare dynamics of serial genomic testing of metastatic breast cancer patients in the blood.
Alt-text: Unlabelled box
1. Introduction
Since the first report of extracellular nucleic acids detected in the peripheral blood in 1948 to the eventual determination that cancer patients had elevated concentrations in DNA that correlated with oncogenic mutations in tissue, liquid biopsies continue to move closer to patient care [1]. These circulating nucleic acids, which were later coined circulating tumour DNA (ctDNA), formed the framework for non-invasive detection of genomic alterations in the peripheral blood. The potential clinical utility of ctDNA assessment has been explored across all stages of disease, including early detection, adjuvant assessment of minimal residual disease, and treatment monitoring and resistance with varying degrees of progress [[2], [3], [4], [5], [6], [7], [8]]. Given the greater tumour volume, number of mutations, and clonal complexity in advanced compared to early stage breast cancer, there is a critical need to define the landscape of ctDNA in metastatic breast cancer.
Based on tissue analysis, there are defined differences in gene expression across the four intrinsic molecular subtypes of early-stage breast cancer: luminal A, luminal B, HER2-enriched, and basal-like [9]. Further, particular genomic profiles confer prognostic information in primary breast cancer [10,11]. Importantly, compared with early stage breast cancer, metastatic breast cancer has greater genomic complexity [12]. This complexity not only varies at baseline tissue sequencing assessment, but specific genomic resistance mutations can emerge in response to the selective pressure of treatment. Examples include the development of ESR1 mutations for patients who develop endocrine resistance, HER2 mutations in HER2 non-amplified disease, and RB1 loss, aberrant FGFR signalling, or CCNE1 overexpression for patients treated with CDK4/6 inhibitors [[12], [13], [14], [15], [16], [17], [18], [19]].
Liquid biopsies are non-invasive tools to explore tumour content in the peripheral blood, including ctDNA, circulating tumour cells (CTCs), exosomes, and/or proteins. Prior studies have explored the concordance between genomic alterations detected in tissue and blood with relatively high agreement, particularly at allele frequencies greater than 1% [20]. There are some technical reasons to account for differences, including spatial and temporal heterogeneity, tumours that do not shed ctDNA, and clonal hematopoiesis of indeterminate potential (CHIP) [20,21]. Prior work has also demonstrated that the amount of tumour released into in the blood increases with stage and that both the detection and genomic heterogeneity of ctDNA in blood correlate with worse clinical outcomes [[22], [23], [24], [25]]. However, the precise degree with which ctDNA content correlates with sites of disease on imaging remains understudied.
Here, we describe the genomic landscape of ctDNA in metastatic breast cancer across pathological subtypes. We further report the prevalence and heterogeneity of clinically actionable alterations, including mutations in PIK3CA, ERBB2, and ESR1. In addition, we evaluate concordance between ctDNA and tissue next-generation sequencing (NGS) for patients with paired sequencing. Finally, we explore the utility of ctDNA assessment as a surrogate for tumour burden on imaging. These analyses aim to define the potential clinical utility of peripheral blood assessment of ctDNA to define the genomic complexity of metastatic breast cancer.
2. Materials and methods
2.1. Patient selection and study design
The Institutional Review Board (IRB) at the Robert H. Lurie Comprehensive Cancer Center of Northwestern University approved the study. Requirement for informed consent was waived per the IRB for this retrospective, de-identified analysis. Studies were performed in concordance with the Health Insurance Portability and Accountability Act and the Declaration of Helsinki. All patients with ctDNA NGS from a single institution (Northwestern University, Chicago, IL) were retrospectively identified from 2015 to 2019. In total, 344 patients and 807 separate ctDNA tests (median 1 per patient) were found. Of these, 255 patients were confirmed Stage IV disease by patient chart review and included in the study. These 255 patients were consecutively enroled in order to limit potential confounding effect. ctDNA reports were reviewed for type and number of alterations, mutant allele frequency (MAF), and timeframe of collection. MAF was defined as the highest allele frequency variant in blood. For patients with more than one blood collection, only the first ctDNA test was included in this study. The electronic medical records were reviewed for all patients and clinical annotation was performed and stored using a secure REDCap database.
2.2. ctDNA sequencing
ctDNA was evaluated using the commercially available Guardant360 assay (Guardant Health, Inc., Redwood City, CA) to evaluate up to 73 cancer-related genes, depending on the date of sequencing [range 54–73 genes] [26,27]. The NGS testing was performed in a CLIA-certified and College of American Pathologists accredited laboratory. ctDNA testing was performed as part of standard clinical care. Two 10 mL of peripheral blood was collected in Streck Cell-Free DNA BCT (Streck, Inc., La Vista, NE) for sample preparation with plasma evaluated for single-nucleotide variants (SNVs), insertions-deletion mutations (indels), fusions, and amplifications of both synonymous and non-synonymous alterations. Previously, the platform was reported to have analytic sensitivity below 0.1% allele fraction, sensitivity of 85.9%, very high specificity (>99.9999%), and clinical sequencing success rate of approximately 99.6% [27].
2.3. Tissue NGS sequencing
Tissue NGS for comprehensive genomic profiling was performed for standard of care and clinical trial enrolment using two commercially available platforms: FoundationOne CDX (Foundation Medicine, Cambridge, MA) or Tempus|xT (Tempus, Chicago, IL). After identifying patients who had ctDNA sequencing, patients with both blood and tissue NGS sequencing were analysed for concordance of detected alterations. Only alterations that were covered in both sequencing platforms were compared.
2.4. ctDNA as a measure of tumour burden
To assess ctDNA as a surrogate for tumour burden on imaging, computed tomography (CT) or positive emission tomography (PET)-CT radiographic reports were reviewed for patients included in the study. The scan in closest proximity to the ctDNA collection date was reviewed for number and sites of metastatic disease. MAF and number of detected ctDNA alterations were associated with metastatic sites of disease detected on imaging. Metastatic sites were categorised into the following: lung, liver, bone, lymph node, soft tissue, and central nervous system. Each category was counted as a single metastatic site (e.g. multiple lymph nodes involved counted as one site of disease).
2.5. Statistical analysis
Clinical and pathological variables were reported using descriptive analyses through frequencies for categorical variables or medians and interquartile range (IQR) for continuous variables. The concordance between alterations in blood and tissue was evaluated using Cohen's kappa coefficient (κ). Associations between ctDNA and sites of disease on imaging and subtypes were tested using the Mann–Whitney U or Kruskal–Wallis as statistically appropriate. High tumour burden defined as 2 or more sites of distant metastasis based on imaging was associated with ctDNA and clinical characteristics using uni- and multivariate logistic regression. Statistical analysis was performed using STATA (StataCorp. (2019) Stata Statistical Software: Release 14.2. College Station, TX: StataCorp LP), JMP (SAS Institute Inc. (2019), version 14. Cary, NC), and R (R Core Team (2019), version 3.6.2. R Foundation for Statistical Computing, Vienna, Austria).
The funding sources had no role in the study design, data collection, data analysis, interpretation, or writing of the manuscript.
3. Results
3.1. Patient characteristics
A total of 255 patients with metastatic breast cancer at the time of ctDNA collection were included in the study (Table 1). The median age of patients in the cohort was 56 years. The cohort consisted of 254 females (99.6%) and 1 male (0.4%). The sample consisted of the following: 124 HR+, HER2- (48.6%), 31 HR-, HER2+ (12.2%), 44 HR+, HER2+ (17.3%), and 56 triple-negative breast cancer (TNBC) patients (22.0%). For the majority of analyses, all HER2+ patients were combined together, regardless of HR status. HR and HER2 status were reassessed through biopsy of a metastatic lesion in 73% of patients. Based on histology, there were 212 patients (83.1%) with invasive ductal carcinoma (IDC), 22 patients (8.6%) with invasive lobular carcinoma (ILC), 7 patients (2.7%) with mixed type, and the remaining 14 patients (5.5%) had unknown histology. Seventy-four patients (30.3%) had inflammatory breast cancer. Median prior lines of treatment in the metastatic setting at the time of ctDNA collection was 3 [IQR 1–5]. With respect to prior treatment, 39% of the patients had received an aromatase inhibitor (AI), 27% had received AI or fulvestrant combined with CDK4/6 inhibitor, and no patients had received prior poly ADP ribose polymerase (PARP) inhibitor therapy.
Table 1.
Patient characteristics. Shown are basic clinical, demographic, and pathologic characteristics for the entire cohort of patients with metastatic breast cancer (N = 255). In addition, sites of disease on imaging in close proximity to collection of circulating tumour DNA are included.
| N | % | |
|---|---|---|
| Age | ||
| <45 | 52 | 20.4 |
| 45–65 | 143 | 56.1 |
| >65 | 60 | 23.5 |
| Subtype | ||
| Luminal-like | 124 | 48.6 |
| HER2 positive | 75 | 29.4 |
| Triple Negative | 56 | 22.0 |
| Liver | ||
| No | 154 | 60.4 |
| Yes | 101 | 39.6 |
| Lung | ||
| No | 168 | 65.9 |
| Yes | 87 | 34.1 |
| Central nervous system | ||
| No | 212 | 83.1 |
| Yes | 43 | 16.9 |
| Bone | ||
| No | 91 | 35.7 |
| Yes | 164 | 64.3 |
| Lymph node | ||
| No | 132 | 51.8 |
| Yes | 123 | 48.2 |
| Soft tissue | ||
| No | 182 | 71.5 |
| Yes | 73 | 28.6 |
| Tumour burden | ||
| Low (1 site) | 74 | 29.0 |
| High (≥ 2 sites) | 181 | 70.0 |
3.2. ctDNA genomic alterations
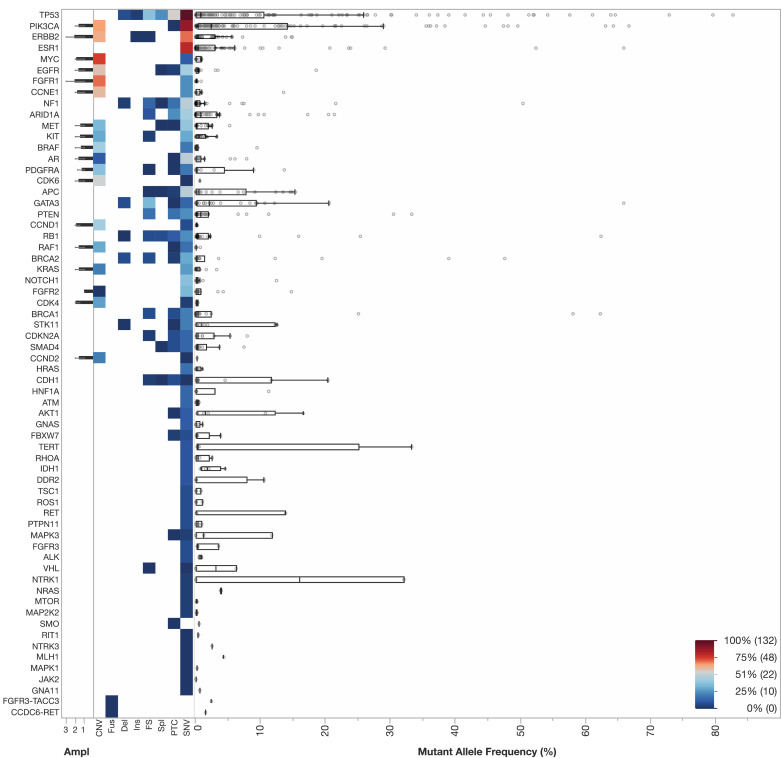
The landscape of genomic alterations in the overall cohort is shown in Fig. 1, including the genes with alterations, types of alterations, and MAF of the highest variant in the blood. 227 patients (89%) had at least one alteration or amplification detected. The median number of alterations was 4 [IQR 1 to 7]. With respect to MAF of the highest variant, the median was 3.8% [IQR 0.6% to 17.5%]. In total, alterations in 60 different genes were observed, as well as two fusions. Across the entire cohort, the most common genes with alterations observed in descending order of frequency were the following: TP53 (189), PIK3CA (144), ERBB2 (77), ESR1 (75), MYC (57), EGFR (55), FGFR1 (49), CCNE1 (42), NF1 (38), and ARID1A (35). CNVs were most common for the following genes: MYC (52), FGFR1 (39), PIK3CA (37), ERBB2 (36), CCNE1 (32), EGFR (31), CDK6 (25), BRAF (21), CCND1 (21).
Fig. 1.
ctDNA landscape in metastatic breast cancer (N: 255). Shown is a heatmap of detected genomic alterations across the entire cohort. The y-axis includes alterations in descending frequency with the most common alterations detected at the following frequencies: TP53 (189), PIK3CA (144), ERBB2 (77), ESR1 (75), MYC (57), EGFR (55), FGFR1 (49), CCNE1 (42), NF1 (38), and ARID1A (35). The following variants were included on the x-axis from left to right: amplifications (ampl), copy number variants (CNV), fusions (fus), deletions (del), insertions (ins), frameshift (fs), splicing variants (Spl), premature termination codons (PTC), and single nucleotide variants (SNV). Boxplots including the interquartile range and outliers for allele frequency are included. Each row represents a gene. A heatmap scale is included in the lower right corner.
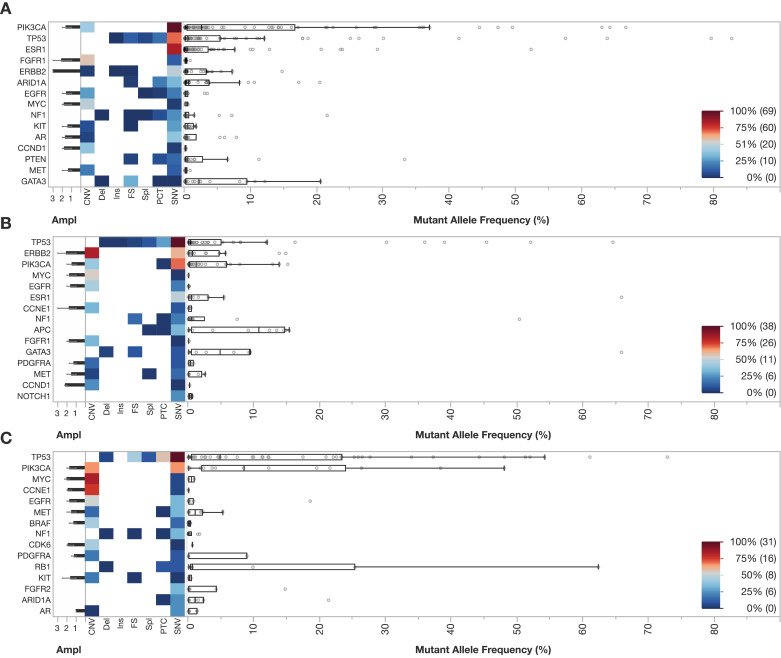
Next, the landscape of alterations was evaluated by subtype obtained from primary pathological reports (HR+, HER2+, or TNBC). The 15 most frequent alterations by subtype were analysed. For HR+ breast cancer, the most common SNVs were PIK3CA (69), ESR1, (64) and TP53 (58) (Fig. 2A). Median allele frequency was highest for mutations in PIK3CA. For HER2+ breast cancer, SNVs were most common in TP53 (38), PIK3CA (23), and ERBB2 (15) with ERBB2 as the most frequently encountered CNV in the cohort (33) (Fig. 2B). For TNBC, the most commonly observed SNVs were TP53 (31) and PIK3CA, (13) and the most frequently observed CNVs were MYC, CCNE1, and PIK3CA (Fig. 2C). Median MAF for HR+, HER2+, and TNBC subtypes were 3.5 (IQR 0.6–16.6), 2.6 (IQR 0.3–8.0), and 9.9 (1.3–27.7), respectively (P<0.05, Kruskal-Wallis equality-of-populations rank test). The landscape of alterations for HR+ vs HR- HER2+ disease is shown in Supplemental Fig. 1. No differences in number of alterations were detected amongst the subtypes: HR+ (median 4, IQR 2–8), HER2+ (median 4.5, IQR 2–7), and TNBC (median 5, IQR 3–7).
Fig. 2.
A–C Landscape of alterations for the 15 most common alterations by pathological subtype (HR+, HER2+, TNBC) (N: 255). Shown is a heatmap of detected alterations with similar design as compared to Fig. 1 for the following cohorts: HR+ (N: 124, Panel A), HER2+ (N: 75, Panel B), and TNBC (N: 56, Panel C). The 5 most common alterations by subtype included PIK3CA (84), TP53 (77), ESR1 (64), FGFR1 (33), and ERBB2 (24) for HR+, TP53 (52), ERBB2 (48), PIK3CA (33), MYC (15), EGFR (15) for HER2+, and TP53 (55), PIK3CA (26), MYC (20), CCNE1 (19), and EGFR (17) for TNBC.
3.3. Evaluation of clinically actionable ctDNA targets
Alterations in PIK3CA, ESR1, and ERBB2 were commonly encountered in the cohort at the following frequencies: 101 (39.6%), 42 (16.5%) and 55 (21.6%), respectively (Fig. 1). For the HR+ HER2- cohort, 37 out of 123 (30.1%) patients had ESR1 mutations. The detection of ESR1 mutations was significantly associated with prior use of AI (P<0.001). With respect to ERBB2, 28 of 55 (50.9%) alterations were amplifications and the remaining 27 of 55 (49.1%) were mutations. For ERBB2 copy number changes, 27/28 (96.4%) of patients with amplifications occurred in HER2+ patients. Notably, one amplification was observed in a HR+ HER2- patient. In contrast, for mutations in ERBB2, 16 were observed in HR+ HER2- (16 of 17 ERBB2 alterations, 94.1%), 8 were encountered in HER2+ patients (8 of 35 ERBB2 alterations, 22.9%), and 3 mutations were found in TNBC patients (3 of 3 ERBB2 alterations, 100%). Importantly, the diversity of alterations detected in the population was broad. In total, 34 unique SNVs were detected in PIK3CA, 31 different SNVs in ERBB2, and 17 different SNVs in ESR1 (Fig. 3). The 3 most common alterations observed in PIK3CA were amplification, H1047R, and E542K. For ERBB2, these included amplification, S310F, and V777L. Finally, the 3 most commonly encountered alterations in ESR1 were D538G, Y537S, and Y537N. No amplifications were detected in ESR1.
Fig. 3.
Gene variant distribution for the main targetable genes in metastatic breast cancer. Variant heatmaps for PIK3CA, ERBB2, and ESR1 are included. Frequency (count) of each variant is shown in the legend in the bottom right corner. Greater diversity of PIK3CA and ERBB2 variants were observed as compared with ESR1. No copy number changes in ESR1 were present in the cohort. The most common single nucleotide variants (SNVs) for PIK3CA, ERBB2, and ESR1 were the following: H1047R, S310F, and D538G, respectively.
Notably, 6 patients had 2 concomitant ESR1 alterations, while 8 patients had 3 or more alterations (Supplemental Table 1). In addition, 30 patients had more than one concomitant PIK3CA alteration, including 15 patients with an amplification combined with an SNV with the remaining patients having multiple SNVs (Supplemental Table 1). Although frequent amplifications were seen in ERBB2, 5 out of 10 patients with concomitant ERBB2 alterations showed a combination of SNVs (Supplemental Table 1).
3.4. Concordance of alterations detected in blood and tissue
Concordance of genomic alterations was compared between blood and tissue for the 10 most commonly observed alterations in the cohort. Genes were only analysed if the particular gene was sequenced in both NGS panels. In total, 105 patients had blood and tissue sequencing in the cohort (54 with Foundation testing, 44 with Tempus testing, 7 with both platforms). Median timeframe between sample collections was 12 weeks (IQR 2.5–64.3) (Table 2). A moderate to high concordance was observed for alterations in TP53 and PIK3CA with a Cohen kappa of 0.5809 and 0.5513, respectively (observed agreement 79.1% and 81.0%, respectively) (Table 2). A higher observed agreement (90.5%) was observed for FGFR1 with a kappa of 0.6313. PIK3CA alterations were detected in tissue, but not ctDNA in 7.6% of cases, while 11.4% of cases were detected in ctDNA only. ERBB2 was discordant in 21% of samples. ESR1 alterations were detected based on tissue biopsy alone in 3.8% of cases, while in 11.4% of cases an alteration was detected in ctDNA, but not tissue.
Table 2.
Concordance of next-generation sequencing (NGS) between blood and tissue for the 10 most common alterations in the cohort. The 10 most common alterations in decreasing frequency were the following: TP53, PIK3CA, ERBB2, ESR1, MYC, EGFR, FGFR1, CCNE1, NF1, and ARID1A. Notably, the 80% agreement for EGFR was confined to wild-type cases, while 95% of the samples classified as mutated through ctDNA were wild-type based on tissue biopsy. Blood and tissue NGS from paired patient samples were compared using Cohen kappa values with >0.7 indicating very high concordance and >0.5 indicating moderate to high concordance.
| Observed agreement | Kappa | 95% C.I. | P | ||
|---|---|---|---|---|---|
| TP53 | 79.1% | 0.5809 | 0.4061 | 0.7147 | < 0.0001 |
| PIK3CA | 81.0% | 0.5513 | 0.360 | 0.6988 | < 0.0001 |
| ERBB2 | 79.1% | 0.3675 | 0.1572 | 0.5596 | 0.0001 |
| ESR1 | 84.8% | 0.4167 | 0.1808 | 0.6167 | < 0.0001 |
| MYC | 80.0% | 0.3860 | 0.1736 | 0.5770 | < 0.0001 |
| EGFR | 80.0% | 0.0541 | −0.1053 | 0.2352 | 0.1421 |
| FGFR1 | 90.5% | 0.6313 | 0.3984 | 0.7929 | < 0.0001 |
| CCNE1 | 83.8% | 0.3014 | 0.0622 | 0.5153 | 0.0001 |
| NF1 | 83.8% | 0.1748 | −0.0131 | 0.4325 | 0.0325 |
| ARID1A | 89.5% | 0.2979 | 0.0611 | 0.5753 | 0.0009 |
3.5. Comparison of ctDNA with imaging evaluation
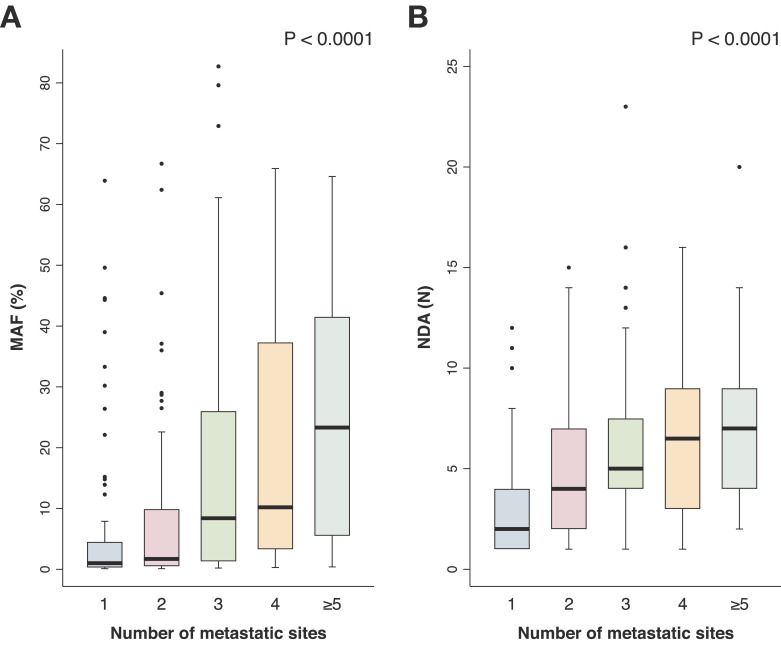
For all patients with available imaging in close proximity to ctDNA evaluation (N = 255), ctDNA MAF of the highest variant and number of detected alterations were associated with the number of metastatic sites observed on imaging (CT or PET-CT). The median timeframe between blood and imaging analysis was 1.6 weeks (IQR 0.71 - 4.57). Sites of metastatic disease were binned into the following groups: 0, 1, 2, 3, 4, ≥5. Higher MAF in ctDNA was significantly associated with a greater number of metastatic sites on imaging in an incremental manner (Mann Whitney U test, P<0.0001) (Fig. 4A). Similarly, a significant finding was observed when analysing the number of detected alterations in ctDNA compared with sites of disease (Mann Whitney U test, P<0.0001) (Fig. 4B), further indicating an association between ctDNA tumour content and heterogeneity as a surrogate marker for tumour burden. In a multivariate model dichotomising number of metastatic sites into low (1 metastatic site) and high (≥2 sites), the number of ctDNA alterations detected was significant (P<0.005, Supplemental Table 2).
Fig. 4.
A,B ctDNA as a reflection of tumour burden on imaging. ctDNA were compared with CT or PET-CT imaging in close proximity with a median of 1.6 weeks (IQR 0.71–4.57) between assessments. There was a significant association between mutant allele frequency (MAF) of the highest variant and number of sites of disease on imaging (Panel A, P<0.0001, Mann Whitney U test) (N: 222). Similarly, a significant association was observed between number of detectable alterations (NDA) and number of metastatic sites (Panel B, P<0.0001, Mann Whitney U test) (N: 227).
4. Discussion
We evaluated the genomic profile and associated clinical data of patients with metastatic breast cancer who underwent ctDNA testing in a large cohort. The goals of the analysis were to identify the landscape of ctDNA across pathological subtypes, to evaluate the prevalence of clinically actionable alterations, to compare blood and tissue NGS for patients with paired sequencing, and to assess similarities between blood and imaging assessment for disease monitoring.
Here, we report several key findings from our analyses. First, ctDNA evaluation was able to capture the landscape of alterations including SNVs, indels, and fusions. Across the cohort, unique alterations were detected in more than 80% of the genes evaluated. Furthermore, the frequency and distribution of genes encountered varied across the HR+, HER2+, and TNBC subtypes with TNBC patients having a statistically higher MAF in our cohort, but not a higher number of alterations. When comparing alterations for patients with paired blood and tissue sequencing, concordance was relatively high with some expected differences between blood and tissue NGS due to biological and technical factors in sampling. The capability of ctDNA evaluation to capture spatial tumour heterogeneity (e.g. reflect tumour from multiple metastatic sites) and the convenience of serial blood sampling reinforce the potential of this technique.
We also reported the frequency of clinically actionable alterations that were encountered in our cohort. For example, in the HR+ cohort, PIK3CA, ESR1, and ERBB2 gene alterations were detected at the following frequencies: 46%, 30%, and 14%, respectively. However, we decided not to report a specific summative statistic, given that the actionability of these alterations is largely dependant on the concurrent drug development pipeline and available drugs at the time of analysis. At present, there is a Food and Drug Administration (FDA) approved targeted agent for PIK3CA (alpelisib), while agents targeting patients with ERBB2 mutations (e.g. neratinib) have demonstrated clinical activity, but no current FDA approval for this indication [17,[28], [29], [30]]. Furthermore, in breast cancer, there is currently only one FDA drug approval linked to a ctDNA companion diagnostic for a PCR-based test to utilise alpelisib with the registry trial including patients with PIK3CA mutations in exons 7, 9, and 20 [28,31]. In contrast, the diversity of alterations observed in our cohort in PIK3CA reinforces the need to explore clinical benefit using drugs across a broader range of SNVs. Data are also emerging that double PIK3CA mutations increase sensitivity to these inhibitors, indicating the clinical importance of assessing for multiple driver mutations in the same gene, concurrently [32,33]. Further work is necessary to identify PIK3CA mutations with decreased sensitivity to drugs targeting the phosphoinositide 3-kinase pathway [34].
The most common genomic alterations encountered in our study were similar to those reported in a large cohort of patients who underwent tissue NGS sequencing with the most commonly observed alterations in TP53 and PIK3CA [12]. The detection of PIK3CA and ESR1 mutations in plasma were also similar to prior ctDNA studies focusing on single resistance mutations [8,14]. Our data also demonstrate the complementary nature of blood and tissue NGS as 11.4% of patients were found to have detectable gene alterations in PIK3CA in ctDNA, but not tissue. With respect to ERBB2, the majority of these mutations (59%) were detected in HR+ patients, indicating the need to assess for resistance mutations in this population. However, our findings also reinforce the current limitations with respect to detecting copy number changes, such as ERBB2, in ctDNA compared to tissue NGS, a finding which is consistent with prior work [20]. Additionally, there are emerging strategies for treating patients who develop endocrine resistance with ESR1 mutations [14,35,36]. Clinical trials for matched molecular targets for many other genomic alterations encountered in our sample are ongoing, including AKT and DNA damage repair pathways, amongst many others.
While our study focused on a baseline ctDNA assessment, the reliable detection of alterations suggests that monitoring alterations in the blood could identify genomic evolution and drug resistance to potentially guide clinical management. Potential clinical utility of ctDNA could include an initial assessment of alterations with known drug targets, such as PIK3CA and ERBB2. In terms of treatment monitoring, a growing body of evidence has identified that changes in specific alterations or tumour burden, as indicated by ctDNA MAF, correlates with treatment response [23,37,38]. This may enable monitoring of specific alterations (e.g. PIK3CA changes in response to alpelisib) or changes in MAF in response to systemic therapy. Here, we add additional insight to this literature by associating findings on imaging with ctDNA assessment. Specifically, MAF (e.g. tumour burden) and genomic heterogeneity (e.g. number of alterations) in ctDNA were both significantly associated with increasing number of metastatic sites on imaging. Therefore, in our cohort, ctDNA evaluation was a reflection of tumour burden on imaging, despite the current inability for this technique to locate specific sites of metastasis. Ongoing work is evaluating how serial ctDNA testing correlates with imaging changes over time and whether particular clinical, pathologic, and genomic profiles in blood may predict sites of metastasis, prior to their occurrence.
There were several limitations of our study. First, this was a retrospective analysis and therefore prone to unknown bias, which we attempted to limit by analysing consecutively enroled patients with ctDNA testing. Second, blood sample collection was obtained per standard clinical care, and there was inherent variability in the ordering practices of providers, although the vast majority of testing was ordered at baseline assessment or at the time of clinical progression. Third, the timeframe of blood and tissue NGS sequencing was not aligned in all cases, which may have introduced greater discordance into these analyses, and paired sequencing data were only available for a subset of patients. In addition, some patients had tissue NGS analysed from the primary tumour site, which may have introduced greater discordance between blood and tissue. Fourth, the findings reported here were based on ctDNA sequencing from a single NGS assay at a single institution, and therefore may not apply to other sequencing platforms, depending on the particular technical specifications of the assay. Furthermore, the inclusion of a relatively large percentage of IBC patients may not be fully generalisable to non-academic referral centers.
In conclusion, our findings demonstrate the landscape of ctDNA in metastatic breast cancer using one of the largest clinically annotated datasets. Collectively, our data reinforce the genetic heterogeneity of metastatic breast cancer that can be captured in blood, concordance between blood and tissue NGS, the high prevalence of clinically actionable alterations, and the potential to utilise ctDNA as a surrogate for tumour burden. Further studies are needed to independently validate these findings, to compare dynamics of serial genomic testing of metastatic breast cancer patients in the blood, and to incorporate ctDNA assays into novel clinical trial designs for patient enrolment and stratification.
Funding sources
Lynn Sage Cancer Research Foundation, OncoSET Precision Medicine Program, and REDCap support was funded in part by a Clinical and Translational Science Award (CTSA) grant from the National Institutes of Health UL1TR001422. The funding sources had no role in the study design, data collection, data analysis, interpretation, or writing of the manuscript.
Declaration of Competing Interest
Andrew A. Davis reports receiving travel support from Menarini Silicon Biosystems, outside the submitted work; Lorenzo Gerratana reports non-financial support from Menarini Silicon Biosystems, personal fees from Lilly, outside the submitted work; Ami N. Shah reports personal fees from Abbvie, personal fees from Taiho, personal fees from Daiichi-Sankyo, outside the submitted work; Amir Behdad reports personal fees from Pfizer, personal fees from Foundation Medicine, personal fees from Bayer, outside the submitted work. Massimo Cristofanilli reports personal fees from Foundation Medicine, personal fees from Lilly, grants and personal fees from Pfizer, personal fees from Cytodyn, personal fees from Sermonix, grants and personal fees from G1Therapeutics, outside the submitted work.
Acknowledgements
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102914.
Appendix. Supplementary materials
References
- 1.Mandel P., Metais P. Les acides nucléiques du plasma sanguin chez l‘homme. C R Seances Soc Biol Fil. 1948;142(3–4):241–243. [PubMed] [Google Scholar]
- 2.Wan J.C.M., Massie C., Garcia-Corbacho J. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 3.Liu M.C., Oxnard G.R., Klein E.A. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:P745–P759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantel K., Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16(7):409–424. doi: 10.1038/s41571-019-0187-3. [DOI] [PubMed] [Google Scholar]
- 5.Heitzer E., Haque I.S., Roberts C.E.S., Speicher M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 6.McDonald B.R., Contente-Cuomo T., Sammut S.J. Personalized circulating tumour DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11(504):eaax7392. doi: 10.1126/scitranslmed.aax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coombes R.C., Page K., Salari R. Personalized detection of circulating tumour DNA antedates breast cancer metastatic recurrence. Clin Cancer Res. 2019;25(14):4255–4263. doi: 10.1158/1078-0432.CCR-18-3663. [DOI] [PubMed] [Google Scholar]
- 8.Buono G., Gerratana L., Bulfoni M. Circulating tumour DNA analysis in breast cancer: is it ready for prime-time? Cancer Treat Rev. 2019;73:73–83. doi: 10.1016/j.ctrv.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Sorlie T., Perou C.M., Tibshirani R. Gene expression patterns of breast carcinomas distinguish tumour subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis C., Shah S.P., Chin S.F. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira B., Chin S.F., Rueda O.M. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertucci F., Ng C.K.Y., Patsouris A. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 13.Toy W., Shen Y., Won H. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fribbens C., O'Leary B., Kilburn L. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016;34(25):2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 15.Chandarlapaty S., Chen D., He W. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016;2(10):1310–1315. doi: 10.1001/jamaoncol.2016.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Razavi P., Chang M.T., Xu G. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438. doi: 10.1016/j.ccell.2018.08.008. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bose R., Kavuri S.M., Searleman A.C. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3(2):224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Formisano L., Lu Y., Servetto A. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun. 2019;10(1):1373. doi: 10.1038/s41467-019-09068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner N.C., Liu Y., Zhu Z. Cyclin E1 expression and palbociclib efficacy in previously treated hormone receptor-positive metastatic breast cancer. J Clin Oncol. 2019;37(14):1169–1178. doi: 10.1200/JCO.18.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chae Y.K., Davis A.A., Jain S. Concordance of genomic alterations by next-generation sequencing in tumour tissue versus circulating tumour DNA in breast cancer. Mol Cancer Ther. 2017;16(7):1412–1420. doi: 10.1158/1535-7163.MCT-17-0061. [DOI] [PubMed] [Google Scholar]
- 21.Razavi P., Li B.T., Brown D.N. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25(12):1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettegowda C., Sausen M., Leary R.J. Detection of circulating tumour DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shatsky R., Parker B.A., Bui N.Q. Next-generation sequencing of tissue and circulating tumour DNA: the UC San Diego moores center for personalized cancer therapy experience with breast malignancies. Mol Cancer Ther. 2019;18(5):1001–1011. doi: 10.1158/1535-7163.MCT-17-1038. [DOI] [PubMed] [Google Scholar]
- 24.Dawson S.J., Tsui D.W., Murtaza M. Analysis of circulating tumour DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 25.Rossi G., Mu Z., Rademaker A.W. Cell-free DNA and circulating tumour cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin Cancer Res. 2018;24(3):560–568. doi: 10.1158/1078-0432.CCR-17-2092. [DOI] [PubMed] [Google Scholar]
- 26.Zill O.A., Banks K.C., Fairclough S.R. The landscape of actionable genomic alterations in cell-free circulating tumour DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24(15):3528–3538. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 27.Odegaard J.I., Vincent J.J., Mortimer S. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24(15):3539–3549. doi: 10.1158/1078-0432.CCR-17-3831. [DOI] [PubMed] [Google Scholar]
- 28.Andre F., Ciruelos E., Rubovszky G. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 29.Ma C.X., Bose R., Gao F. Neratinib efficacy and circulating tumour DNA detection of HER2 mutations in HER2 nonamplified metastatic breast cancer. Clin Cancer Res. 2017;23(19):5687–5695. doi: 10.1158/1078-0432.CCR-17-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyman D.M., Piha-Paul S.A., Won H. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;554(7691):189–194. doi: 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.FDA approves alpelisib for metastatic breast cancer. May 24, 2019. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-alpelisib-metastatic-breast-cancer
- 32.Vasan N., Razavi P., Johnson J.L. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science. 2019;366(6466):714–723. doi: 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito Y., Koya J., Araki M. Landscape and function of multiple mutations within individual oncogenes. Nature. 2020;582:95–99. doi: 10.1038/s41586-020-2175-2. [DOI] [PubMed] [Google Scholar]
- 34.Zardavas D., Phillips W.A., Loi S. PIK3CA mutations in breast cancer: reconciling findings from preclinical and clinical data. Breast Cancer Res. 2014;16(1):201. doi: 10.1186/bcr3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrod A., Fulton J., Nguyen V.T.M. Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer. Oncogene. 2017;36(16):2286–2296. doi: 10.1038/onc.2016.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toy W., Weir H., Razavi P. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7(3):277–287. doi: 10.1158/2159-8290.CD-15-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Leary B., Cutts R.J., Liu Y. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8(11):1390–1403. doi: 10.1158/2159-8290.CD-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang D.H., Ensor J.E., Liu Z.B. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat. 2016;155(1):139–149. doi: 10.1007/s10549-015-3635-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.