Abstract
Transcription factor Yin Yang 1 (YY1) is upregulated in multiple tumors and plays essential roles in tumor proliferation and metastasis. However, the function of YY1 in breast cancer stemness remains unclear. Herein, we found that YY1 expression was negatively correlated with the overall survival and relapse-free survival of breast cancer patients and positively correlated with the expression of stemness markers in breast cancer. Overexpression of YY1 increased the expression of stemness markers, elevated CD44+CD24− cell sub-population, and enhanced the capacity of cell spheroid formation and tumor-initiation. In contrast, YY1 knockdown exhibited the opposite effects. Mechanistically, YY1 decreased microRNA-873-5p (miR-873-5p) level by recruiting histone deacetylase 4 (HDAC4) and HDAC9 to miR-873-5p promoter and thus increasing the deacetylation level of miR-873-5p promoter. Sequentially, YY1 activated the downstream PI3K/AKT and ERK1/2 pathways, which have been confirmed to be suppressed by miR-873-5p in our recent work. Moreover, the suppressed effect of YY1/miR-873-5p axis on the stemness of breast cancer cells was partially dependent on PI3K/AKT and ERK1/2 pathways. Finally, it was found that the YY1/miR-873-5p axis is involved in the chemoresistance of breast cancer cells. Our study defines a novel YY1/miR-873-5p axis responsible for the stemness of breast cancer cells.
Keywords: YY1, miR-873-5p, HDAC, stemness, breast cancer, PI3K/AKT, ERK1/2, deacetylation
Graphical Abstract
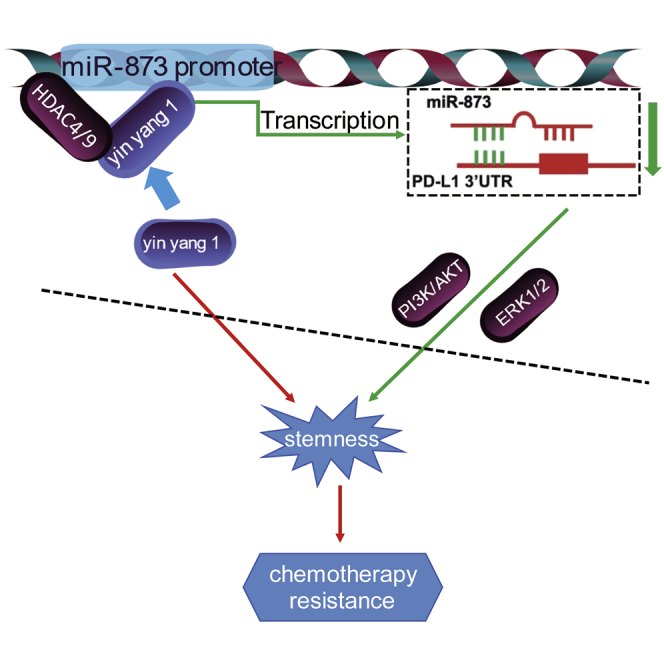
Tumor recurrence and chemotherapy resistance often result from cancer stem cells. Guo et al. define a novel YY1/miR-873-5p axis responsible for the stemness of breast cancer cells. Moreover, the researchers found that the YY1/miR-873-5p axis is involved in the chemoresistance of breast cancer cells. The authors’ research indicates that YY1/miR-873-5p regulatory axis may be a potential molecular target for breast cancer therapy.
Introduction
Breast cancer is the most common cancer for women worldwide with a continuous increase morbidity and mortality.1 Currently, lumpectomy, targeted therapy, and chemotherapy remain the main treatments for breast cancer patients. Among them, the chemotherapy is the only option for triple-negative breast cancer.2 However, tumor recurrence and chemotherapy resistance often result from cancer stem cells (CSCs), which is an urgent problem need to be solved.3 CSCs are a small group of tumor cells with stem cell characteristics and maintain self-renewal and multi-directional differentiation potential through asymmetric division, leading to tumor cell proliferation and promoting tumor heterogeneity.4 At present, the mechanisms underlying CSCs attributes are not well defined.
Yin Yang 1 (YY1), a zinc finger protein, is first identified as a member of the YY family.5,6 YY1 is overexpressed in multiple cancers including breast, pancreatic cancers, and melanoma.7, 8, 9 As a transcription factor, YY1 can either activate or repress gene expression by directly binding to their promoters10 and regulate about 10% mammalian gene set.11 For example, YY1 regulates MYCT1 transcription12 and stabilizes hypoxia-inducible factor HIF-1α in a p53-independent manner.13 Clinical research shows that YY1 expression is positively associated with the expression of multiple CSC markers, including SOX2 and OCT4.14 Previous studies have shown that YY1 overexpression confers cancer cells with chemoresistance in non-Hodgkin’s lymphoma.15,16 However, the function of YY1 in regulating the stemness and chemoresistance of breast cancer cells remains unclear.
MicroRNAs (miRNAs), a kind of non-coding RNAs, inhibit gene expression by directly binding to the 3′ untranslated region (UTR) of the target genes.17 Recent studies have shown that miRNAs, like miR-119a and miR-128, regulate cancer stemness and drug resistance in breast cancer.18,19 miR-873-5p has been shown to inhibit the growth of colon, glioblastoma and lung tumors.20, 21, 22 Cui et al.23 have shown that miR-873-5p attenuates tamoxifen resistance by targeting CDK3 and modulates ERα transcriptional activity in breast cancer cells. We recently indicate that miR-873-5p suppresses the stemness of breast cancer cells through targeting PD-L1, an immune checkpoint, and thus suppressing the PI3K/AKT and ERK1/2 signaling pathways.24 However, the mechanisms through which miR-873-5p is regulated in breast cancer are still not fully understood. In the present study, JASPAR database (http://jaspar.binf.ku.dk/cgi-bin/jaspar_db.pl) and LASAGNA-Search 2.0 (http://biogrid-lasagna.engr.uconn.edu/lasagna_search/) were used to predict the potential transcriptional factors binding to miR-873-5p promoter. We found that there were two putative binding sites of YY1 on miR-873-5p promoter with the highest scores. Thus, we hypothesized that YY1 could directly bind to miR-873-5p promoter and regulate miR-873-5p expression, which is involved in YY1-mediated regulation on the stemness of breast cancer cells.
Here, we showed that YY1 knockdown attenuated the stemness and chemoresistance of breast cancer cells by decreasing miR-873-5p level through recruiting histone deacetylase 4 (HDAC4) and HDAC9 to the promoter of miR-873-5p. Additionally, PI3K/AKT and ERK1/2 signaling were required for the YY1/miR-873-5p axis-mediated regulation of breast cancer cell stemness. Therefore, the YY1/miR-873-5p regulatory axis might serve as a potential therapeutic target in suppressing breast cancer progression.
Results
YY1 Is Significantly Increased and Engaged in Breast Cancer Stemness
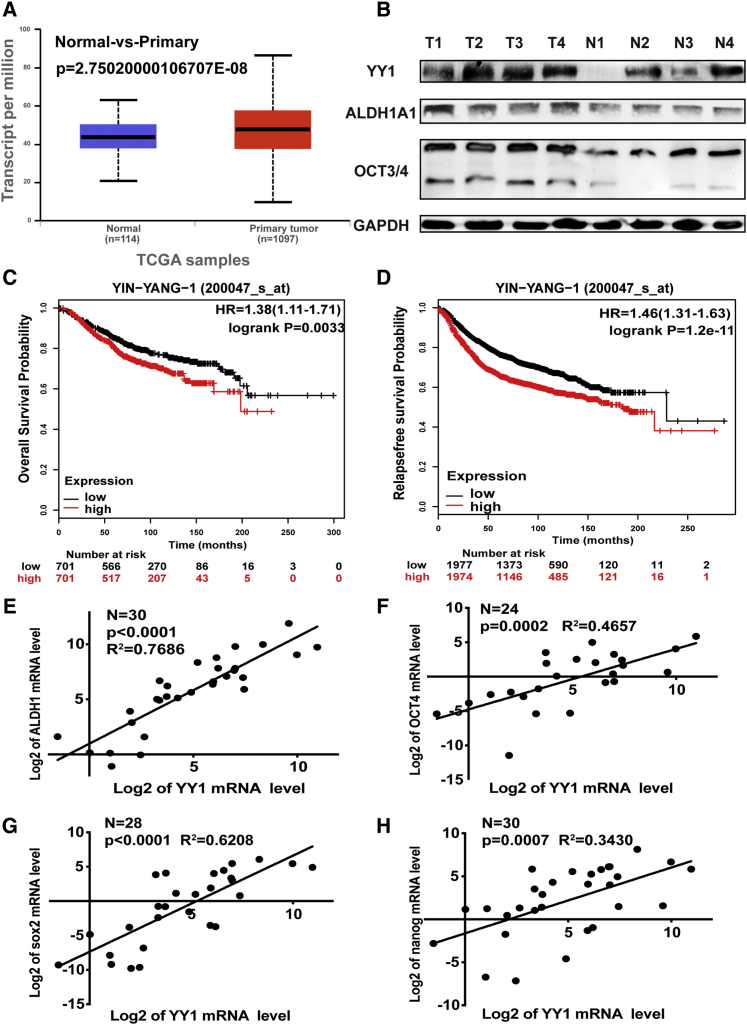
YY1 expression in breast primary tumor and normal adjacent tissues was initially examined using online clinical posited data25 (http://ualcan.path.uab.edu/index.html). As shown in Figure 1A, YY1 expression was significantly increased in breast tumor tissues, and western blot assay displayed a consistent result in fresh breast cancer and normal adjacent tissues (Figure 1B). Meanwhile, KM-plotter analysis26 (http://kmplot.com and https://hgserver1.amc.nl/cgi-bin/r2/main.cgi) from different datasets indicated that YY1 expression was negatively correlated with the overall survival and relapse-free survival of breast cancer patients (Figures 1C and 1D; Figures S1A and S1B). YY1 is also highly expressed in the four basic subtypes of breast cancer, and the high expression of YY1 predicts a poor survival rate (Figure S2). Notably, the expression of YY1 and stemness markers (OCT3/4, ALDH1A1, SOX2, and NANOG) exhibited a positive correlation in clinical samples (Figures 1B and 1E–1H). Additionally, we further evaluated the expression correlation between YY1 and stemness markers using R2: Genomics Analysis and Visualization Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). The expression of stemness markers (ALDH1A1, SOX2, and NANOG) is positively correlated with YY1 expression in breast cancer as well (Figure S1C). These results indicate that YY1 might regulate the stemness of breast cancer cells.
Figure 1.
YY1 Expression Was Positively Correlated with the Expression of Stemness Markers in Breast Cancer Tissues
(A) YY1 expression in breast cancer tissues and normal adjacent tissues via online clinical posited data. (B) Expression of YY1 and CSCs markers (ALDH1A1, OCT3/4) in breast tumor tissues and normal tissues was examined via western blot. Samples were derived from the same experiment and that blots were processed in parallel. (C and D) Correlation between YY1 expression and overall survival (C) and relapse-free survival (D) of breast cancer patients based on online KM-Plotter analysis. (E–H) Correlation between YY1 expression and the expression of ALDH1A1, OCT4, SOX2, and nanog via quantitative real-time PCR in breast cancer tissues. The data are presented as the mean ± SD, n ≥ 3.
YY1 Promotes the Stemness of Breast Cancer Cells
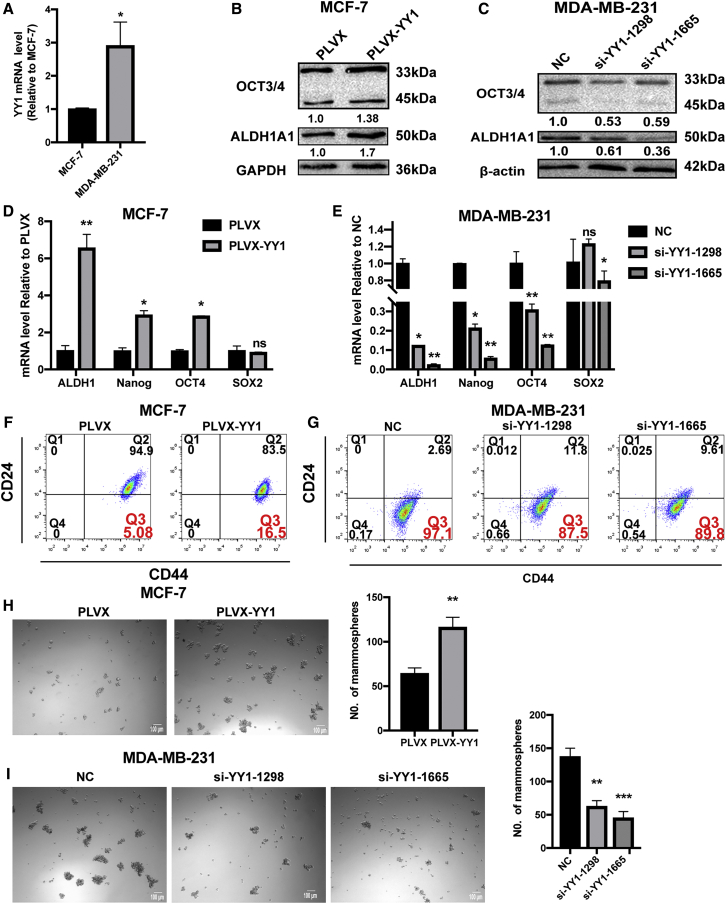
To determine whether YY1 could confer the stemness of breast cancer cells, we examined YY1 expression in MCF-7 and MDA-MB-231 cells, which exhibited a lower and higher stemness, respectively.27, 28, 29 As shown in Figure 2A, YY1 displayed a lower level in MCF-7 cells relative to MDA-MB-231 cells. Then YY1 was overexpressed in MCF-7 cells and knocked down in MDA-MB-231 cells, respectively. The overexpression and knockdown efficiency were confirmed (Figure S3). As expected, the expression of stemness markers was increased by YY1 overexpression, and YY1 knockdown exhibited opposite effects (Figures 2B–2E). Further flow cytometry analysis showed that CD44+/CD24− sub-population with stemness was increased in MCF-7 cells with YY1 overexpression and reduced in MDA-MB-231 cells with YY1 knockdown (Figures 2F and 2G). Additionally, YY1 overexpression improved the capacity of cell spheroid formation, characterized as the increase of spheroid size and number, and YY1 knockdown exerted opposite effects (Figures 2H and 2I). Therefore, these results suggest that YY1 could promote the stemness of breast cancer cells.
Figure 2.
YY1 Promoted the Stemness of Breast Cancer Cells
(A) YY1 mRNA level was detected in MCF-7 and MDA-MB-231 cells. (B and C) The protein expression of stemness markers (ALDH1A1 and OCT3/4) were detected by western blot in MCF-7 cells with or without YY1 overexpression (B) and MDA-MB-231 cells with or without YY1 knockdown (C). Samples were derived from the same experiment and blots were processed in parallel. (D and E) The mRNA levels of stemness markers (ALDH1A1, Nanog, OCT4, and SOX2) were detected via quantitative real-time PCR in MCF-7 cells with or without YY1 overexpression and MDA-MB-231 cells with or without YY1 knockdown. (F–I) CD44+/CD24− population or spheroid formation (50X) was measured in cells depicted in MCF-7 cells with or without YY1 overexpression and MDA-MB-231 cells with or without YY1 knockdown. The data are presented as the mean ± SD, n ≥ 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus PLVX or NC.
YY1 Regulates miR-873-5p Expression through Binding to its Promoter
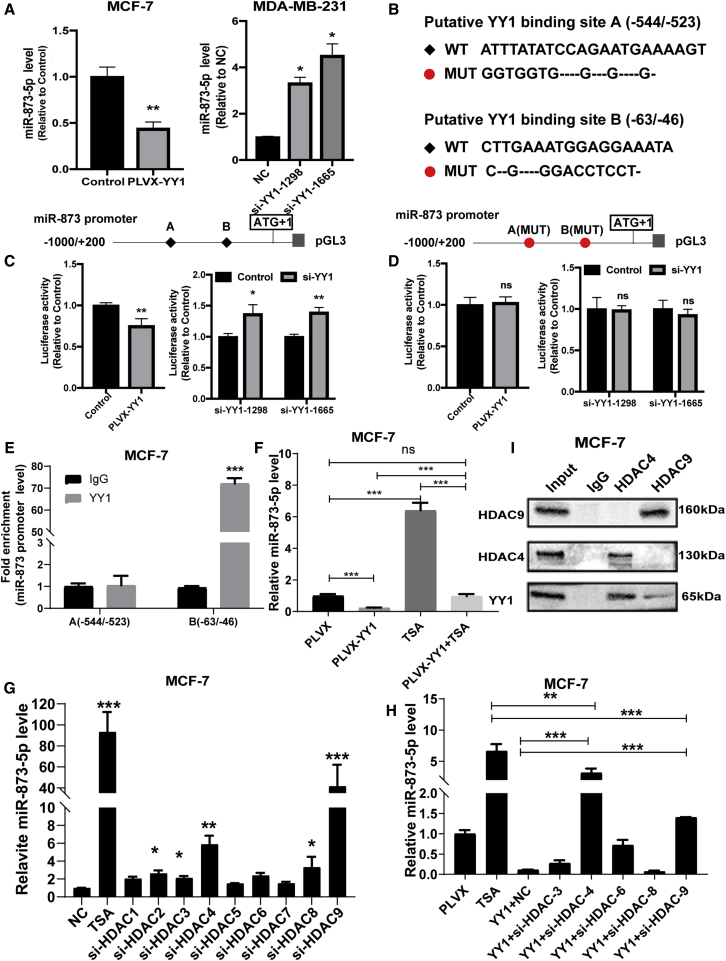
Then we searched the downstream effectors of YY1, and since miRNAs play important roles in regulating cancer progression, here we focused on miRNAs. According to YY1-binding consensus and the prediction by JASPAR database (http://jaspar.binf.ku.dk/cgi-bin/jaspar_db.pl) and LASAGNA-Search 2.0 (http://biogrid-lasagna.engr.uconn.edu/lasagna_search/), there were two putative binding sites of YY1 with the highest scores on miR-873-5p promoter, which has been previously confirmed to suppress breast cancer progression by our group and others.23,24,30 As shown in Figure 3A, YY1 overexpression downregulated miR-873-5p level, while knockdown of YY1 upregulated it. The two potential binding sites of YY1 on miR-873-5p promoter were designated YY1-binding sites A and B located at −544/−523 and −61/−50, respectively. The wild-type (WT) or mutant (MUT) binding sites were shown in Figure 3B. Additionally, luciferase reporter analysis demonstrated that the luciferase activities of WT binding sites A and B were downregulated and upregulated in HEK293T cells with YY1 overexpression and YY1 knockdown, respectively (Figure 3C; Figures S4A and S4B). In contrast, the activity of MUT A and B was unaffected (Figure 3D; Figures S4C and S4D). Chromatin immunoprecipitation (ChIP) analysis obtained the consistent result showing that the region containing the binding site B was remarkably enriched in DNA pulled down by anti-YY1 (Figure 3E). These results indicate that YY1 could directly bind to miR-873-5p promoter.
Figure 3.
YY1 Regulated miR-873-5p Expression through Binding to its Promoter
(A) miR-873-5p level was detected in MCF-7 cells with YY1 overexpression and MDA-MB-231 cells with YY1 knockdown, respectively. (B) Potential WT and MUT binding sites for YY1 on miR-873-5p promoter; “–” means deleted bases. (C) Relative luciferase activity of luciferase reporter plasmids with WT sites was measured in HEK293T cells with YY1 overexpression or knockdown. (D) Relative luciferase activity of luciferase reporter plasmids with MUT sites was determined in HEK293T cells with YY1 overexpression or knockdown. (E) The abundance of miR-873-5p promoter was evaluated by ChIP assay with anti-YY1 and rabbit IgG as control. (F) miR-873-5p level was detected in MCF-7 cells with different treatment as indicated. (G) miR-873-5p level was detected in MCF-7 cells with TSA or siRNAs against of HDAC 1-9 by quantitative real-time PCR, respectively. (H) Cell with transfection of YY1 with or without siRNAs against of HDAC3, HDAC4, HDAC6, HDAC8, or HDAC9. (I) Co-immunoprecipitation (coIP) was performed in MCF-7 cells with anti-HDAC4, anti-HDAC9, and rabbit IgG as control. Samples derive from the same experiment and that blots were processed in parallel. The data are presented as the mean ± SD, n ≥ 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus PLVX or NC.
YY1 Decreases miR-873-5p Expression through Increasing the Deacetylation Level of miR-873-5p Promoter
DNA methylation and histone deacetylation are two critical mechanisms regulating the transcription of genes;31 however, there are no potential methylation sites in miR-873-5p promoter via analyzing CpG islands using CpGFinder (http://www.softberry.com/berry.phtml?topic=cpgfinder&group=programs&subgroup=promoter; Figure S4E). Thus, we assumed that histone deacetylation is engaged in YY1-mediated regulation on miR-873-5p transcription. Additionally, TSA, an inhibitor of HDACs (HDACi), reversed the decrease of miR-873-5p level led by YY1 overexpression (Figure 3F), but had no effect on the endogenous expression of YY1 (Figure S4F). Since HDAC includes HDAC1-9, we further explore which HDACs are engaged in YY1-mediated regulation on miR-873-5p expression, and small interfering RNAs (siRNAs) against HDACs were transfected into breast cancer cells, respectively. It was shown that HDAC3, HDAC4, HDAC6, HDAC8, and HDAC9 siRNA could upregulate miR-873-5p level in both MCF-7 and MDA-MB-231 cells (Figure 3G; Figure S4G). Subsequently, the siRNAs against these five HDACs were co-transfected into MCF-7 cells with YY1 overexpression, as shown in Figure 3H; only HDAC4 and HDAC9 siRNA reversed YY1-induced downregulation of miR-873-5p level in MCF-7 cells. We assumed that YY1 could recruit HDAC4 and HDAC9 to miR-873-5p promoter, coimmunoprecipitation (coIP) analysis revealed that YY1 indeed bound to HDAC4 and HDAC9 (Figure S4H). Conversely, HDAC4 and HDAC9 could interact with YY1, while there was no interaction between HDAC4 and HDAC9 in breast cancer cells (Figure 3I; Figure S4I). In addition, ChIP analysis showed that the region of YY1 binding site B on miR-873-5p was obviously enriched in the DNA pulled down by anti-HDAC4 and anti-HDAC9 (Figure S4J). Thus, these results demonstrate that YY1 could recruit HDAC4 and HDAC9 to miR-873-5p promoter, increase its deacetylation level, and thus decrease miR-873-5p level in breast cancer cells.
MiR-873-5p Is Responsible for YY1-Mediated Regulation on the Stemness of Breast Cancer Cells
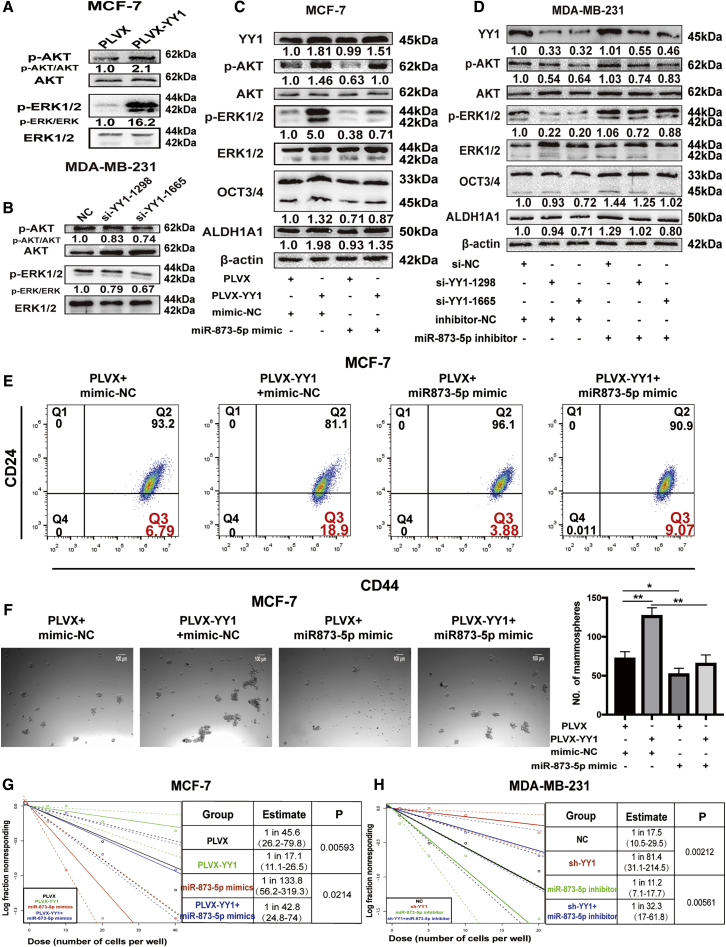
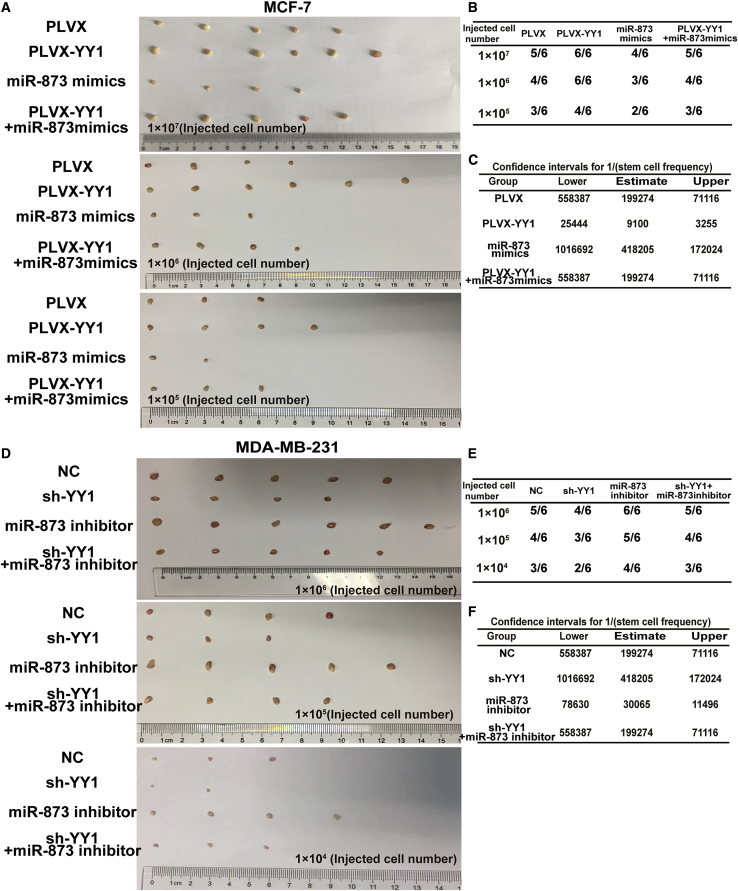
The previous study showed that the expression of stemness markers could be improved in breast CSCs by the activation of PI3K/AKT and ERK1/2 pathways,32 and AKT could also be activated by YY1.33 And we previously indicated that miR-873-5p downregulates the stemness of breast cancer cells by inactivating the PI3K/AKT and ERK1/2 pathways.24 We further explored whether YY1 promoted the stemness of breast cancer cells through miR-873-5p-mediated inhibition on these two signaling pathways. First, we verified that YY1 overexpression indeed increased p-AKT and p-ERK1/2 levels, whereas YY1 knockdown displayed opposite effects (Figures 4A and 4B). MCF-7 cells with YY1 overexpression or MDA-MB-231 cells with YY1 knockdown were transfected with miR-873-5p mimic or miR-873-5p inhibitor, respectively, we found that miR-873-5p overexpression rescued the promoting effects of YY1 overexpression on the stemness and PI3K/AKT and ERK1/2 pathways, characterized as the decrease of stemness marker expression, PI3K/AKT and ERK1/2 signaling activation (Figures 4C and 4D), CD44+/CD24− sub-population (Figure 4E), and cell spheroid formation (Figure 4F). Meanwhile, miR-873-5p inhibitor in MDA-MB-231 cells with YY1 knockdown displayed the opposite effects (Figures S5A and S5B). Furthermore, we assessed the sphere formation capacity of those cells by determining the frequency of sphere-forming cells (SFCF) in those cells with a limited dilution assay, and found that YY1 overexpression showed a high SFCF than untreated group in MCF-7 cells, which was attenuated by miR-873-5p overexpression (Figures 4G and 4H). In front of our results showed that the siRNA of HDAC4 and HDAC9 upregulated the level of miR-873-5p by weakening the inhibition of YY1. As shown in Figures S6A and S6B, TSA or HDAC4 and HDAC9 siRNA rescued the promoting effects of YY1 overexpression on the stemness and PI3K/AKT and ERK1/2 pathways. Further flow cytometry analysis also showed the consistent result that the effect of YY1 overexpression could be partially abrogated by HDAC4/9 knockdown (Figure S6C), and knockdown of HHAC4/9 reduced the stemness breast cancer cells, which was partially reversed by miR-873-5p knockdown (Figure S6D). To sum up, our results demonstrate that YY1 promotes the stemness of breast cancer cells through miR-873-5p in vitro. Then, we constructed MCF-7 cells with YY1 (PLVX-YY1) stable overexpression with or without miR-873-5p overexpression, and MDA-MB-231 cells with YY1 knockdown (shYY1) plus miR-873-5p knockdown or not. It was found that YY1-overexpressed cells showed an increased tumor size and tumor-formation rate (Figures 5A and 5B), while miR-873-5p overexpression decreased the tumor-initiating ability of MCF-7 cells and attenuated the YY1 overexpression-mediated promoting effect on tumor-initiating ability (Figure 5C). Consistently, the knockdown of YY1 remarkably reduced the tumor-initiating potential of MDA-MB-231 cells, which was reversed by miR-873-5p knockdown (Figures 5D–5F). Therefore, these results suggest that YY1 could promote the stemness of breast cancer cells through miR-873-5p.
Figure 4.
MiR-873-5p Was Responsible for YY1-Mediated Regulation on the Stemness of Breast Cancer Cells
(A and B) The protein expression of p-AKT and p-ERK1/2 was detected in MCF-7 cells with or without YY1 overexpression and MDA-MB-231 cells with or without YY1 knockdown. (C and D) Expression of stemness markers (ALDH1A1 and OCT3/4), p-AKT, and p-ERK1/2 was examined by western blot in MCF-7 cells and MDA-MB-231 cells with different treatment, samples derive from the same experiment and that blots were processed in parallel. (E) CD44+/CD24− population was determined via flow cytometry analysis in MCF-7 cells. (F) Spheroid formation (50X) was measured in MCF-7 cells. (G and H) MCF-7 cells with or without YY1 overexpression and MDA-MB-231 cells with or without YY1 knockdown were plated in ultra-Low attachment 96-well plates by a limited dilution assay and cultured for 10–12 days to evaluate the SFCf. Tumor formation rate and stem cell frequency were measured by SFCf. The data are presented as the mean ± SD, n ≥ 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus PLVX or NC.
Figure 5.
miR-873-5p Was Responsible for YY1-Mediated Regulation on the Tumor Formation Ability In Vivo
(A and B) Images of tumors size and number when nude mice were injected with different numbers of MCF-7 cells (PLVX, PLVX-YY1, miR-873-5p mimics, PLVX-YY1 plus miR-873-5p mimics), respectively (A) and the tumor-formation rate was examined (B). (C) The stem cell frequency in theses groups depicted in (A) was calculated using the ELDA analysis. (D and E) Images of tumors size and number when nude mice were injected with different numbers of MDA-MB-231 cells (shNC, shYY1, miR-873-5p inhibitor, shYY1 plus miR-873-5p inhibitor) (D), and the tumor-formation rate was measured (E). (F) The stem cell frequency in theses groups depicted in (D) was calculated using the ELDA analysis.The data are presented as the mean ± SD, n ≥ 3, ∗p < 0.05, ∗∗p < 0.01, versus control group.
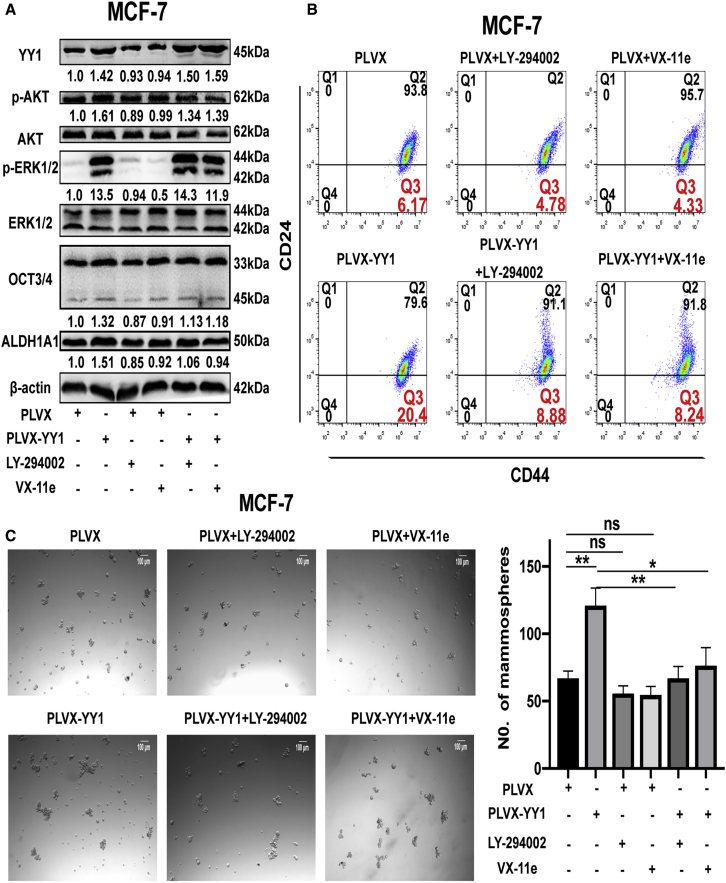
YY1 Promotes the Stemness of Breast Cancer Cells Dependent on PI3K/AKT and ERK1/2 Pathways
Additionally, MCF-7 cells with or without YY1 overexpression were treated with PI3K inhibitor (LY-294002) or ERK1/2 inhibitor (VX-11e), as shown in Figures 6A–6C, the treatment with LY-294002 or VX-11e obviously decreased p-AKT and p-ERK1/2 levels, and attenuated the YY1-induced promoting effects on the expression of stemness markers, CD44+/CD24− sub-population and cell spheroid formation capacity. Thus, our results demonstrate that YY1 promotes the stemness of breast cancer cells at least through miR-873-5p-mediated inhibition of PI3K/AKT and ERK1/2 pathways.
Figure 6.
YY1 Regulated PI3K/AKT and ERK1/2 Pathways Responsible for the Stemness of Breast Cancer Cells
(A) Expression of stemness markers (ALDH1A1 and OCT3/4), p-AKT, and p-ERK1/2 was detected by western blot in MCF-7 cells with different treatments as indicated. Samples were derived from the same experiment and blots were processed in parallel. (B and C) CD44+/CD24− population or spheroid formation (50X) was measured in cells as depicted in (A). The data are presented as the mean ± SD, n ≥ 3, ∗p < 0.05, ∗∗p < 0.01, versus PLVX or PLVX-YY1.
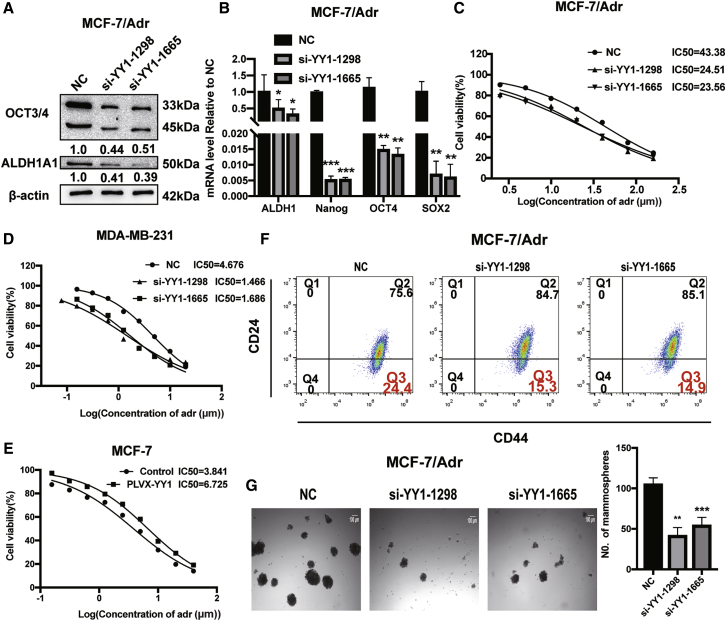
YY1 Knockdown Attenuates Adriamycin Resistance of Breast Cancer Cells through Reducing Cell Stemness
Since CSCs could result in chemoresistance, we further examined whether YY1 regulated adriamycin resistance in adriamycin-resistant MCF-7 cells (MCF-7/Adr). As shown in Figure S7A, the IC50 value of adriamycin in MCF-7/Adr cells was about 10-fold than that in MCF-7 cells. Notably, we found that YY1 expression was higher in MCF-7/Adr cells than MCF-7 cells (Figure S7B). Thus, YY1 was knocked down in MCF-7/Adr cells. The knockdown efficiency was confirmed by western blot and quantitative real-time PCR (Figures S7C and S7D). As shown in Figures 7A and 7B, YY1 knockdown decreased the expression of CSC markers in MCF-7/Adr cells. As expected, YY1 knockdown attenuated adriamycin resistance, evident by the reduced IC50 value (Figure 7C), and YY1 knockdown promoted adriamycin sensitivity in MDA-MB-231 cells (Figure 7D), while YY1 overexpression reduced adriamycin sensitivity of MCF-7 cells (Figure 7E). We also detected CD44+/CD24− sub-population and cell spheroid formation in MCF-7/Adr cells with YY1 knockdown. As illustrated in Figures 7F and 7G, the CD44+/CD24− sub-population and capacity of cell spheroid formation were attenuated by YY1 knockdown. Importantly, the expression of P-gp (a multi-drug resistance protein) was positively regulated by YY1 in breast cancer cells (Figures S7E–S7G). These results suggest that YY1 knockdown attenuates adriamycin resistance of breast cancer cells through reducing cell stemness.
Figure 7.
Knockdown of YY1 Attenuated Breast Cancer Cells Resistance to Adriamycin Treatment through Reducing Cells Stemness
(A and B) Stemness marker expression was examined by western blot and quantitative real-time PCR in MCF-7/Adr cells with or without YY1 knockdown; samples derive from the same experiment and blots were processed in parallel. (C–E) IC50 values of adriamycin in MCF-7/Adr cells with or without YY1 knockdown. (F and G) CD44+/CD24− population or spheroid formation (50X) measured in cells depicted in (A). The data are presented as the mean ± SD, n ≥ 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus MCF-7 or NC.
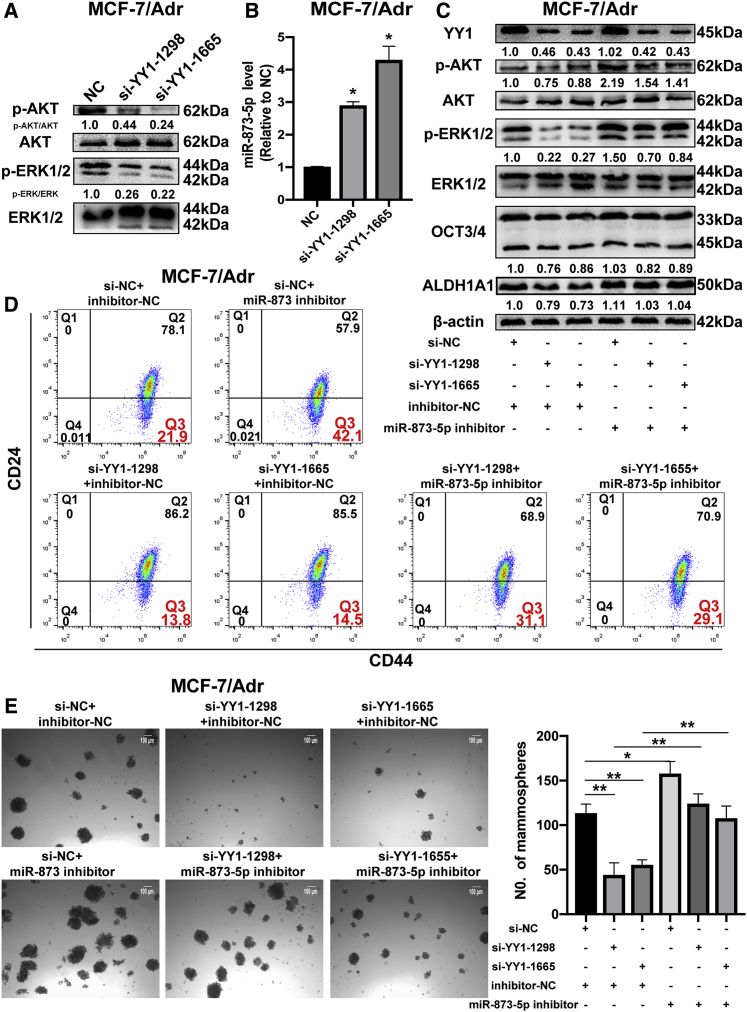
YY1/miR-873-5p Axis Negatively Regulated Chemotherapeutic Sensitivity
We have demonstrated that YY1 regulation on the stemness of breast cancer cells and PI3K/Akt and ERK1/2 pathways were through miR-873-5p. We finally explored whether YY1-mediated chemotherapeutic resistance dependent on miR-873-5p. As expected, we substantiated that YY1 knockdown certainly declined p-AKT, p-ERK1/2, and miR-873-5p levels in MCF-7/Adr cells (Figures 8A and 8B). Meanwhile, MCF-7/Adr cells with YY1 knockdown were co-transfected with miR-873-5p inhibitor and we found that miR-873-5p inhibitor rescued the inhibition of YY1 knockdown on stemness, PI3K/AKT and ERK1/2 pathways, and adriamycin sensitivity, characterized as the increase of expression of stemness markers and re-activation of PI3K/AKT and ERK1/2 signaling (Figure 8C), rescue of CD44+/CD24− sub-population (Figure 8D), the capacity of cell spheroid the formation (Figure 8E), and cell viability (Figure S7H). In addition, miR-873-5p knockdown partially abrogated the promoting effects of YY1 knockdown on the adriamycin sensitivity in MDA-MB-231 cells (Figure S7I), and miR-873-5p overexpression rescued the inhibition of YY1 overexpression on the adriamycin sensitivity in MCF-7 cells (Figure S7J). A consistent result was obtained upon determining the sensitivity of another therapeutic drug, taxol (Figures S8A and S8B). miR-873-5p knockdown or YY1 overexpress reduced the adriamycin and taxol sensitivity, which was also rescued by HDAC4/9 knockdown (Figures S8C–S8F). Meanwhile, miR-873-5p inhibitor attenuated the effects of HDAC4/9 knockdown on the stemness of MCF-7/Adr cells (Figure S8F). Notably, YY1 and miR-873 level displayed an opposite expression model in MCF-7, MDA-MB-231, and MCF-7/Adr cells (Figure S9). Therefore, our results suggest that YY1 could confer chemotherapeutic resistance through miR-873-5p.
Figure 8.
miR-873-5p Was Responsible for YY1-Mediated Regulation on the Stemness of MCF-7/Adr Cells
(A) Protein levels of p-AKT and p-ERK1/2 were determined in MCF-7/Adr cells with YY1 knockdown. (B) miR-873-5p level was detected via quantitative real-time PCR in cells depicted in (A). (C–E) Protein levels of stemness markers (ALDH1A1 and OCT3/4), p-AKT, p-ERK1/2 (C), CD44+/CD24− population (D), and (E) spheroid formation (50X) were examined in MCF-7/Adr cells with different treatment. Samples were derived from the same experiment and blots were processed in parallel. The data are presented as the mean ± SD, n ≥ 3, ∗p < 0.05, ∗∗p < 0.01, versus NC.
Discussion
The present study indicated that YY1 expression was significantly increased in breast cancer tissues, negatively correlated with overall survival and relapse-free survival of breast cancer patients. We have predicted that YY1 is overexpressed in the basic subtypes of breast cancer and that high expression of YY1 has a poor overall survival by the online clinical posited data25 and KM-plotter analysis.26 YY1 overexpression led to stemness-like properties, and YY1 knockdown attenuated the stemness of breast cancer cells. Mechanistically, we proved that YY1 could directly bind to miR-873-5p promoter, downregulate miR-873-5p level through enhancing its deacetylation level, and subsequently activate PI3K/AKT and ERK1/2 pathways, through which YY1 exerted its effects. To the best of our knowledge, this is the first study showing YY1 roles in regulating the stemness of breast cancer cells.
For the past few years, transcription factors have shed new lights on our understanding of the regulation of gene expression and the pathogenesis of various diseases. As a ubiquitously distributed transcription factor, YY1 acts negatively or positively depending on its target genes.11 Previous literatures demonstrate a high YY1 expression in cancer tissues including breast carcinoma34 and melanoma cancer.8 Although YY1 positively regulates BRCA1 and inhibits mammary cancer formation;35 increasing evidence confirms that YY1 regulates various pro-tumorigenic pathways by activating human epidermal growth factor receptor 2 (HER2; ERBB2, neu)7,34 and EGFR.36 Through these pathways, YY1 promotes tumor angiogenesis, metastasis, and cell proliferation. As CSCs lead to the proliferation of tumor cells and the promotion of tumor heterogeneity,4 in the present study, we further explored YY1 roles in breast cancer cell stemness and demonstrated a novel role of YY1 in breast tumorigenesis. We found that YY1 expression was significantly increased and positively related to the expression of stemness markers in breast cancer tissues. However, it is noteworthy that the correlation between YY1 and Sox2 expression was inconsistent in three breast cancer cells (MCF-7/Adr, MCF-7, and MDA-MB-231). For these unexpected results, we assumed that there were diverse regulatory mechanisms in different cells, for instance, YY1 overexpression inhibited Sox2 in pancreatic ductal adenocarcinoma cells.37 Notably, a previous study indicates that stemness marker expression could be improved in CSCs via PI3K/AKT and ERK1/2 pathways activation32 and AKT activation could also be regulated by YY1.33 These are consistent with our results that YY1 contributes to the stemness of breast cancer cells through PI3K/AKT and ERK1/2 pathways.
The YY1-miRNAs interaction has been previously reported, for example miR-381 suppresses YY1 expression in endometrioid endometrial carcinoma cells,38 and miR-34a inhibits invasion and migration by targeting YY1 in esophageal squamous cell carcinoma.39 Recent studies find that YY1 directly binds to miR-10a and consequently downregulates its transcriptional activity.40 Here, we proved that YY1 could directly bind to miR-873-5p promoter and then decreased miR-873-5p level. However, the concrete mechanisms underlying YY1-mediated downregulation on miR-873-5p promoter activity deserve further study.
DNA methylation and histone deacetylation, which are important forms of epigenetic modification, are the main mechanisms underlying gene transcription inhibition.31,41 Promoter methylation of tumor suppressor genes (TSGs) has been found to promote tumorigenesis, such as BRCA1, and PLCD1 and PCDH17 are downregulated in cancer.42,43 Promoter methylation of BNIP3 causes downregulation of BNIP3 level and contributes to acquired sorafenib resistance in human liver cancer cells.44 HDAC can suppress the transcription level of target genes by recruiting oncogenic fusion proteins to specific gene promoters, and finally promote the occurrence and development of cancer.45 For example, PCDH17, a common tumor suppressor gene, transcriptional downregulation is mainly mediated by abnormal histone acetylation in acute leukemia.46 Additionally, ZEB1 represses ER-α expression by forming a ZEB1/DNMT3B/HDAC1 complex on its promoter, which improves ER-α promoter methylation and histone deacetylation.47 In consistent, YY1 can silence antigen-presenting cells (APCs) through recruitment of EZH2 and trimethylation of histone 3 lysine 27 on its promoter.48 Therefore, in the present study, we assumed that YY1 might improve miR-873-5p promoter histone deacetylation or methylation, thus suppress miR-873-5p transcriptional activity. ChIP analysis validated that YY1 just binds to site B of miR-873-5p promoter, which is partially consistent with results obtained by luciferase report analysis. This could be due to the fact that the gene transcription process is not isolated—it takes place in the context of the genomic environment—and transcriptional interference (TI) is broadly defined as the direct negative effect of the transcription of one gene on the second gene located in cis1.49 For example, if two genes are oriented in tandem on the same DNA strand, it is possible for the elongating RNA Polymerase II (Pol II) from the upstream gene to intrude into the promoter region of the downstream genes.50 In the promoter of miR-873-5p, the A site, far from the transcription start site, may be covered by the transcription of the previous gene, and the B site is exposed so it can be bound by transcription factors actually. However, the specific mechanism for the difference in results needs to be further explored. Then we screened and verified that transcription factor YY1 can bind to HDAC4/9 through coIP analysis, this suggests that YY1-mediated downregulation of miR-873-5p transcription may be mediated by HDAC4 and HDAC9. However, HDAC4 and HDAC9 did not exhibit an interaction in breast cancer cells, this indicates that YY1 might interact with HDAC4 and HDAC9, separately but not simultaneously. These results suggest that targeting HDACs might facilitate breast cancer treatment through attenuating the stemness. For example, combined use of HDAC inhibitor with doxorubicin could target both breast CSCs and non-stem breast cancer cells. Histone deacetylase has always been a hot target in the design of drug. Although some of the HDAC inhibitors have been used in the clinical treatment of tumor, the clinical effect of solid tumor treatment is not optimistic. Our study also revealed that HDAC4 and HDAC9 plays an important role in the regulation of breast cancer cell stemness by the YY1/miR-873-5p axis. The regulation of YY1 on miR-873-5p promoter is mediated by HDAC4/9, and TSA or HDAC siRNA can significantly increase the chemotherapeutic sensitivity of breast cancer cells. Therefore, our study also provides a new idea for HDACi in solid tumor treatment. Meanwhile, non-specific HDACi has different degrees of adverse reactions in tumor-clinical trials, and whether single HDACi could overcome these shortcomings in tumor treatment needs further exploration. To sum up, the epigenetic modification involved in deacetylation provides new treatment strategies for tumor-individualized therapy.
Additionally, we showed that YY1 knockdown significantly enhanced chemotherapeutic sensitivity in MCF-7/Adr cells, and this effect was partially dependent on miR-873-5p (Figure S7H). Simultaneously, our results also show that YY1 could confer taxol resistance by the YY1/miR-873-5p axis (Figures S8A and S8B). However, YY1 regulated chemotherapeutic sensitivity in MCF-7 and MDA-MB-231 cells without a significant difference. For these unexpected results, we assumed that there were diverse regulatory mechanisms in different cells. These results suggest that the YY1/miR-873-5p axis and chemotherapeutic resistance might form a positive feedback in breast cancer cells. Despite efforts to develop chemotherapy have been made for killing CSCs in the past decades,51,52 the significant setbacks are presumably due to limited effectiveness in late-stage clinical trials. Aside from toxicity and side effects, the reasons might also be the lack of predictive biomarkers for patients.
In conclusion, our results provide evidence that YY1 regulates the stemness of breast cancer cells by targeting miR-873-5p. This novel YY1/miR-873-5p regulatory axis provides putative targets to develop new strategies for targeting and compromising the stemness of breast cancer, and we hypothesize that a gene expression signature of YY1 and miR-873-5p could predict chemotherapeutic sensitivity in breast cancer patients.
Materials and Methods
Clinical Samples, Cell Culture, and Reagents
Thirty paraffin-embedded breast cancer tissue samples, four fresh breast cancer, and normal adjacent tissues were collected from The Affiliated Cancer Hospital of Zhengzhou University from October 2016 to October 2018. Written informed consent from all patients and approval of the hospital ethics review committees were obtained. Human embryonic kidney HEK293T cells and breast cancer cell lines MCF-7 and MDA-MB-231 were obtained from the ATCC (Manassas, VA, USA) and preserved in our laboratory. Adriamycin-resistant MCF-7 cells (MCF-7/Adr) were purchased from KeyGen BioTECH (Nanjing, China). MCF-7 and HEK293T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (GIBCO). MDA-MB-231 cells were maintained in L15 medium (GIBCO), and MCF-7/Adr cells were cultured in 1640 medium (GIBCO). All the media were supplemented with 10% fetal bovine serum (GIBCO), 80 U/mL penicillin, and 0.08 mg/mL streptomycin. Cells were incubated at 5% CO2 at 37°C. Trichostatin A (TSA), doxorubicin hydrochloride (Adr), and Paclitaxel (taxol) were purchased from Medchemexpress (Monmouth Junction, NJ, USA).
siRNAs, miRNA, Plasmids, Chemical Reagents, and Transfection
siRNAs and negative control (NC) were synthesized by Gene Pharma (Shanghai, China). miRNA mimics and inhibitor, mimics NC and inhibitor NC were synthesized by Biomics Biotechnology (Nantong, China). Sequences of siRNAs were denoted in Table S1. The lentivirus LV3-siYY1(shYY1), LV3-has-miR-873-5p mimics (miR-873-5p mimics), and LV3-has-miR-873-5p inhibitor (miR-873-5p inhibitor) were synthesized by Gene Pharma (Shanghai, China), and used to construct cell line with YY1 stable knockdown (shYY1), miR-873-5p stable knockdown, or overexpression (miR-873-5p inhibitor or mimics). The coding sequences of YY1 were cloned into PLVX-IRES-ZsGreen 1 vector and designated PLVX-YY1. Sequences of primers used for PLVX-YY1 were described previously.53 The sequences of miR-873-5p promoter with YY1 binding sites or mutated binding sites were cloned into the pGL3 vector, respectively. JetPRIME (Polyplus Transfection, France) was used for transfection. 24 h after transfection, cells were treated with PI3K inhibitor (LY-294002) at a concentration of 10 nM or ERK1/2 inhibitor (VX-11e) at a concentration of 1 nM for 24 h. LY-294002 and VX-11e were purchased from APExBIO. After another 48 h, gene expression was measured by quantitative real-time PCR and western blot.
Western Blot
Detailed procedure was described previously.54 Protein in fresh tissues was extracted using Minute Total Protein Extraction Kit (Invent). Antibodies against the following proteins were used: YY1 (1:1,000, Wanleibio), OCT3/4 (1:1,000, Wanleibio), ALDH1A1 (1:1,000, Proteintech), p-AKT-s473 (1:1,000, Proteintech), AKT (1:1,000, Wanleibio), anti-p-ERK1/2 (Thr202/Tyr204; 1:1,000, Cell Signaling Technology), ERK1/2 (1:1,000, Wanleibio), GAPDH (glyceraldehyde-3-phosphate dehydrogenase 1:2,000, Beyotime), and β-actin (1:5,000, Beyotime). Protein expression levels were quantified by density analysis using Quantity One Software and normalized to β-actin or GAPDH.
Quantitative Real-Time PCR
Total RNA from cells was extracted with TRIeasy Total RNA Extraction Reagent TRIeasy (YEASEN, Shanghai, China) following the manufacturer’s recommendation. Total RNA from paraffin section was extracted using RNAprep Pure FFPE Kit (TIANGEN, Beijing, China). cDNA was synthesized by using the M-MLV (Vazyme Biotech, China) following standard protocols. Quantitative real-time PCR was performed by using the Applied Biosystems StepOnePlus system. mRNA levels were normalized to GAPDH. EzOmics SYBR qPCR mix and miR-873-5p qPCR kit were purchased from Biomics. miR-873-5p level was normalized to U6 snRNA. The primer sequences were mentioned in Table S2. The relative gene expression levels were calculated using the 2−ΔΔct method.
Luciferase Reporter Assay
The −544/−523 region of miR-873-5p promoter containing the putative YY1 binding site was designated A site (5′-ATTTATATCCAGAATGAAAAGT-3′), and the −63/−46 region containing the putative YY1 binding site was designated B site (5′-CTTGAAATGGAGGAAATA-3′). The WT or MUT of A and B sites were cloned respectively into the firefly pGL3 luciferase reporter vector. HEK293T cells were co-transfected with recombinant firefly luciferase reporter plasmids (WT or MUT), YY1 or si-YY1 and β-gal reporter control plasmid using JetPRIME. Luciferase activity was measured according to the manufacturer’s protocol and normalized to β-gal activity. All the primer sequences for this experiment were listed in Table S3.
Flow Cytometry Analysis
MCF-7, MDA-MB-231, or MCF-7/Adr cells were seeded into a 6-well plate. When cells confluency reached 60%–80%, cells were transfected with plasmids, miRNAs, or siRNAs. After 48 h, cells were derived from plates with Accutase (Invitrogen). At least 1 × 106 cells were pelleted by centrifugation at 400 × g for 5 min at 4°C. After washing with PBS, the cells were re-suspended with anti-CD44-APC (BD Biosciences) and anti-CD24-PE (BD Biosciences) and finally analyzed on a flow cytometry (BECKMAN). Flow cytometry values have been normalized by subtracting the appropriate isotype control value.
Cell Spheroid Formation Assay
Mammosphere formation assay was performed using MammoCult Human Medium Kit (STEMCELL Technologies, Canada). Totally 3,000 cells were mixed with Complete MammoCult Medium and seeded in 24-well ultra-low attachment plates (Corning) for 7 days. Spheroids were counted and photographed. All images were obtained with a Leica DMI microscope (DE). Cells were plated in ultra-low attachment 96-well plates with a limited dilution assay (1, 5, 10, 20 cells/well) and cultured for 10–12 days to evaluate the SFCf. The number of wells containing spheres was counted, and the SFCf was calculated using the ELDA software (http://bioinf.wehi.edu.au/software/elda/index.html).
MTT Assay
Cells were seeded in 96-well plates at the density of 5,000/well, and treated with different concentrations of adriamycin for 48 h. During the last 3 h, MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide, Amresco’s) was added into the medium at a final concentration of 0.5 mg/mL. Then the medium was removed, and the formazan crystals were dissolved in 150 μL dimethyl-sulfoxide in room temperature for 10 min. Finally, the absorbance was measured using a spectrophotometer (BIO-RAD) at a test wavelength of 490 nm.
ChIP Assay
A ChIP assay was performed using the EpiQuik Chromatin Immunoprecipitation Kit (Catalog # P-2002, Epigentek, USA) following the manufacturer’s protocols and modified according to our previous work.55 Primers flanking the YY1 binding sites on the promoters of miR-873-5p site A (−544/523) and miR-873-5p site B (−63/−46) were used for quantitative real-time PCR. The following antibodies were used: HDAC4 (1:100, Proteintech, China), YY1 (1:100, Cell Signaling Technology, USA), and HDAC9 (1:100, Abcam, USA).
site A F: 5′-GGATCTTCCAGAGATTGTATAAACACTTCCATTCTTTGTTTCC-3′,
site A R: 5′-CTGCCGTTCGACGATTTTGCTTCAGTTTTTTTTTTAATTTTAA-3′;
site B F: 5′-GGATCTTCCAGAGATTGTCTGGGATGCCCACAAAA-3′,
site B R: 5′-CTGCCGTTCGACGACGATTTTCAATAGGAGACTCACAAGTTCCT-3′.
CoIP Assay
MDA-MB-231 cell lysates were prepared by incubating the cells in NP-40 lysis buffer containing protease inhibitor cocktails (1:10,000). Lysates were centrifuged at 12,000 rpm for 10 min at 4°C and incubated with control or specific antibodies for 0.5 h. Add 30 μL protein A/G agarose (Pierce, USA) of each tube at 4°C with constant rotation for 8–12 h. After incubation was performed, the beads were washed 5–6 times by using cold buffer. The precipitated proteins were eluted from the beads by re-suspending the beads in 2× SDS-PAGE loading buffer and boiling for 5 min at 99°C. The boiled immune complexes were subjected to western blotting. The following antibodies were used: HDAC4 (1:100, Proteintech, China), YY1 (1:100, Cell Signaling Technology, USA), HDAC9 (1:100, Santa Cruz Biotechnology, USA), and immunoglobulin G (IgG; 1:100, Cell Signaling Technology, USA).
In Vivo Tumor-Forming Assay
All animal experiments were performed with the approval of Ethics Committee for Animal Experimentation of China Pharmaceutical University. MCF-7 and MDA-MB-231 cells with different treatments were subcutaneously injected at the density of 1 × 107, 1 × 106, 1 × 105 and 1 × 106, 1 × 105, and 1 × 104 cells/tumor, respectively. Mice were euthanized after 8–10 days and tumors were stripped. The ratio of breast CSC was calculated using an ELDA:56 Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda/).
Statistical Analysis
GraphPad Prism 8.0.0 (131) software (GraphPad Software, La Jolla, CA, USA) was used for statistical analysis. The data are presented as the mean ± SD, n ≥ 3. The statistical evaluation for data analysis was determined using an unpaired Student’s test. p <0.05 was considered to be statistically significant.
Author Contributions
Q.G., L.Z., and T.X. designed the research. Q.G., T.W., and L.Z. analyzed the data and wrote the paper. Q.G., T.W., Y.Y., L.Z., Q.Z., and W.Z. performed the research. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 81702957, China); the Medical Science and Technology Research Project of Henan Province (no. LHGJ20190675); the Science and Technology Research Project of Henan Province (no. 202102310158); the Basic Scientific Research Business Expense Project of China Pharmaceutical University (no. 2632020ZD10); the Special Postdoctoral Funding Scheme (no. 2019T120485) and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. All procedures performed in this study were in accordance with the ethical standards of the institutional research ethics committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.06.018.
Contributor Information
Tao Xi, Email: xitao18@hotmail.com.
Lufeng Zheng, Email: zhlf@cpu.edu.cn.
Supplemental Information
References
- 1.Kroenke C.H., Michael Y.L., Poole E.M., Kwan M.L., Nechuta S., Leas E., Caan B.J., Pierce J., Shu X.O., Zheng Y., Chen W.Y. Postdiagnosis social networks and breast cancer mortality in the After Breast Cancer Pooling Project. Cancer. 2017;123:1228–1237. doi: 10.1002/cncr.30440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gradishar W.J., Anderson B.O., Balassanian R., Blair S.L., Burstein H.J., Cyr A., Elias A.D., Farrar W.B., Forero A., Giordano S.H. Invasive Breast Cancer Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2016;14:324–354. doi: 10.6004/jnccn.2016.0037. [DOI] [PubMed] [Google Scholar]
- 3.Wang L., Leite de Oliveira R., Huijberts S., Bosdriesz E., Pencheva N., Brunen D., Bosma A., Song J.-Y., Zevenhoven J., Los-de Vries G.T. An Acquired Vulnerability of Drug-Resistant Melanoma with Therapeutic Potential. Cell. 2018;173:1413–1425. doi: 10.1016/j.cell.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y., Seto E., Chang L.S., Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 6.Seto E., Shi Y., Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 7.Allouche A., Nolens G., Tancredi A., Delacroix L., Mardaga J., Fridman V., Winkler R., Boniver J., Delvenne P., Begon D.Y. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao G., Li Q., Wang A., Jiao J. YY1 regulates melanoma tumorigenesis through a miR-9 ~ RYBP axis. J. Exp. Clin. Cancer Res. 2015;34:66. doi: 10.1186/s13046-015-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J.J., Zhu Y., Yang C., Liu X., Peng Y.P., Jiang K.R., Miao Y., Xu Z.K. Yin Yang-1 increases apoptosis through Bax activation in pancreatic cancer cells. Oncotarget. 2016;7:28498–28509. doi: 10.18632/oncotarget.8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon S., Akopyan G., Garban H., Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 12.Qu S.Y., Sun Y.Y., Li Y.H., Xu Z.M., Fu W.N. YY1 directly suppresses MYCT1 leading to laryngeal tumorigenesis and progress. Cancer Med. 2017;6:1389–1398. doi: 10.1002/cam4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park A., Lee J., Mun S., Kim D.J., Cha B.H., Moon K.T., Yoo T.K., Kang H.G. Identification of Transcription Factor YY1 as a Regulator of a Prostate Cancer-Specific Pathway Using Proteomic Analysis. J. Cancer. 2017;8:2303–2311. doi: 10.7150/jca.19036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufhold S., Garbán H., Bonavida B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J. Exp. Clin. Cancer Res. 2016;35:84. doi: 10.1186/s13046-016-0359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vega M.I., Huerta-Yepez S., Martinez-Paniagua M., Martinez-Miguel B., Hernandez-Pando R., Gonzalez-Bonilla C.R., Gonzales-Bonilla C.R., Chinn P., Hanna N., Hariharan K., Jazirehi A.R. Rituximab-mediated cell signaling and chemo/immuno-sensitization of drug-resistant B-NHL is independent of its Fc functions. Clin. Cancer Res. 2009;15:6582–6594. doi: 10.1158/1078-0432.CCR-09-1234. [DOI] [PubMed] [Google Scholar]
- 16.Vega M.I., Huerta-Yepez S., Jazirehi A.R., Garban H., Bonavida B. Rituximab (chimeric anti-CD20) sensitizes B-NHL cell lines to Fas-induced apoptosis. Oncogene. 2005;24:8114–8127. doi: 10.1038/sj.onc.1208954. [DOI] [PubMed] [Google Scholar]
- 17.Felekkis K., Touvana E., Stefanou Ch., Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–240. [PMC free article] [PubMed] [Google Scholar]
- 18.Celià-Terrassa T., Liu D.D., Choudhury A., Hang X., Wei Y., Zamalloa J., Alfaro-Aco R., Chakrabarti R., Jiang Y.Z., Koh B.I. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat. Cell Biol. 2017;19:711–723. doi: 10.1038/ncb3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y., Yu F., Jiao Y., Feng J., Tang W., Yao H., Gong C., Chen J., Su F., Zhang Y. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin. Cancer Res. 2011;17:7105–7115. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- 20.Gong H., Fang L., Li Y., Du J., Zhou B., Wang X., Zhou H., Gao L., Wang K., Zhang J. miR-873 inhibits colorectal cancer cell proliferation by targeting TRAF5 and TAB1. Oncol. Rep. 2018;39:1090–1098. doi: 10.3892/or.2018.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R.J., Li J.W., Bao B.H., Wu H.C., Du Z.H., Su J.L., Zhang M.H., Liang H.Q. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J. Biol. Chem. 2015;290:8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Sun Z., Zhang Y., Fu T., Liu C., Liu Y., Lin Y. The microRNA-635 suppresses tumorigenesis in non-small cell lung cancer. Biomed. Pharmacother. 2016;84:1274–1281. doi: 10.1016/j.biopha.2016.10.040. [DOI] [PubMed] [Google Scholar]
- 23.Cui J., Yang Y., Li H., Leng Y., Qian K., Huang Q., Zhang C., Lu Z., Chen J., Sun T. MiR-873 regulates ERα transcriptional activity and tamoxifen resistance via targeting CDK3 in breast cancer cells. Oncogene. 2015;34:4018. doi: 10.1038/onc.2015.201. [DOI] [PubMed] [Google Scholar]
- 24.Gao L., Guo Q., Li X., Yang X., Ni H., Wang T., Zhao Q., Liu H., Xing Y., Xi T., Zheng L. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine. 2019;41:395–407. doi: 10.1016/j.ebiom.2019.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V.S.K., Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy Á., Lánczky A., Menyhárt O., Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prat A., Parker J.S., Karginova O., Fan C., Livasy C., Herschkowitz J.I., He X., Perou C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Porath I., Thomson M.W., Carey V.J., Ge R., Bell G.W., Regev A., Weinberg R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L., Xiang C., Li X., Guo Q., Gao L., Ni H., Xia Y., Xi T. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling. J. Hematol. Oncol. 2018;11:72. doi: 10.1186/s13045-018-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng L., Meng X., Li X., Zhang Y., Li C., Xiang C., Xing Y., Xia Y., Xi T. miR-125a-3p inhibits ERα transactivation and overrides tamoxifen resistance by targeting CDK3 in estrogen receptor-positive breast cancer. FASEB J. 2018;32:588–600. doi: 10.1096/fj.201700461RR. [DOI] [PubMed] [Google Scholar]
- 31.Wittenberger T., Sleigh S., Reisel D., Zikan M., Wahl B., Alunni-Fabbroni M., Jones A., Evans I., Koch J., Paprotka T. DNA methylation markers for early detection of women’s cancer: promise and challenges. Epigenomics. 2014;6:311–327. doi: 10.2217/epi.14.20. [DOI] [PubMed] [Google Scholar]
- 32.Almozyan S., Colak D., Mansour F., Alaiya A., Al-Harazi O., Qattan A., Al-Mohanna F., Al-Alwan M., Ghebeh H. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int. J. Cancer. 2017;141:1402–1412. doi: 10.1002/ijc.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q., Wan M., Shi J., Horita D.A., Miller L.D., Kute T.E., Kridel S.J., Kulik G., Sui G. Yin Yang 1 promotes mTORC2-mediated AKT phosphorylation. J. Mol. Cell Biol. 2016;8:232–243. doi: 10.1093/jmcb/mjw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begon D.Y., Delacroix L., Vernimmen D., Jackers P., Winkler R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J. Biol. Chem. 2005;280:24428–24434. doi: 10.1074/jbc.M503790200. [DOI] [PubMed] [Google Scholar]
- 35.Lee M.H., Lahusen T., Wang R.H., Xiao C., Xu X., Hwang Y.S., He W.W., Shi Y., Deng C.X. Yin Yang 1 positively regulates BRCA1 and inhibits mammary cancer formation. Oncogene. 2012;31:116–127. doi: 10.1038/onc.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng H.Y., Chen Y.A., Jen J., Shen P.C., Chen L.M., Lin T.D., Wang Y.C., Hsu H.L. Oncogenic MCT-1 activation promotes YY1-EGFR-MnSOD signaling and tumor progression. Oncogenesis. 2017;6:e313. doi: 10.1038/oncsis.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J.J., Zhu Y., Zhang X.F., Liu D.F., Wang Y., Yang C., Shi G.D., Peng Y.P., Zhang K., Tian L. Yin Yang-1 suppresses pancreatic ductal adenocarcinoma cell proliferation and tumor growth by regulating SOX2OT-SOX2 axis. Cancer Lett. 2017;408:144–154. doi: 10.1016/j.canlet.2017.08.032. [DOI] [PubMed] [Google Scholar]
- 38.Xia B., Li H., Yang S., Liu T., Lou G. MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumour Biol. 2016;37:9157–9167. doi: 10.1007/s13277-016-4805-8. [DOI] [PubMed] [Google Scholar]
- 39.Nie J., Ge X., Geng Y., Cao H., Zhu W., Jiao Y., Wu J., Zhou J., Cao J. miR-34a inhibits the migration and invasion of esophageal squamous cell carcinoma by targeting Yin Yang-1. Oncol. Rep. 2015;34:311–317. doi: 10.3892/or.2015.3962. [DOI] [PubMed] [Google Scholar]
- 40.Mu N., Gu J., Huang T., Zhang C., Shu Z., Li M., Hao Q., Li W., Zhang W., Zhao J. A novel NF-κB/YY1/microRNA-10a regulatory circuit in fibroblast-like synoviocytes regulates inflammation in rheumatoid arthritis. Sci. Rep. 2016;6:20059. doi: 10.1038/srep20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang T.H., Perry M.R., Laux D.E. Methylation profiling of CpG islands in human breast cancer cells. Hum. Mol. Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 42.Xiang T.X., Yuan Y., Li L.L., Wang Z.H., Dan L.Y., Chen Y., Ren G.S., Tao Q. Aberrant promoter CpG methylation and its translational applications in breast cancer. Chin. J. Cancer. 2013;32:12–20. doi: 10.5732/cjc.011.10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tapia T., Smalley S.V., Kohen P., Muñoz A., Solis L.M., Corvalan A., Faundez P., Devoto L., Camus M., Alvarez M., Carvallo P. Promoter hypermethylation of BRCA1 correlates with absence of expression in hereditary breast cancer tumors. Epigenetics. 2008;3:157–163. doi: 10.4161/epi.3.3.6387. [DOI] [PubMed] [Google Scholar]
- 44.Méndez-Blanco C., Fondevila F., Fernández-Palanca P., García-Palomo A., Pelt J.V., Verslype C., González-Gallego J., Mauriz J.L. Stabilization of Hypoxia-Inducible Factors and BNIP3 Promoter Methylation Contribute to Acquired Sorafenib Resistance in Human Hepatocarcinoma Cells. Cancers (Basel) 2019;11:1984. doi: 10.3390/cancers11121984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Licht J.D. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene. 2001;20:5660–5679. doi: 10.1038/sj.onc.1204593. [DOI] [PubMed] [Google Scholar]
- 46.Thanh Nha Uyen L., Amano Y., Al-Kzayer L.F.Y., Kubota N., Kobayashi J., Nakazawa Y. PCDH17 functions as a common tumor suppressor gene in acute leukemia and its transcriptional downregulation is mediated primarily by aberrant histone acetylation, not DNA methylation. Int. J. Hematol. 2019;111:451–462. doi: 10.1007/s12185-019-02799-4. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J., Zhou C., Jiang H., Liang L., Shi W., Zhang Q., Sun P., Xiang R., Wang Y., Yang S. ZEB1 induces ER-α promoter hypermethylation and confers antiestrogen resistance in breast cancer. Cell Death Dis. 2017;8:e2732. doi: 10.1038/cddis.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Zhou L., Lu L., Wang L., Li X., Jiang P., Chan L.K., Zhang T., Yu J., Kwong J. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013;32:3432–3442. doi: 10.1038/onc.2012.360. [DOI] [PubMed] [Google Scholar]
- 49.Shearwin K.E., Callen B.P., Egan J.B. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X., Martin P.G.P., Michaels S.D. BORDER proteins protect expression of neighboring genes by promoting 3′ Pol II pausing in plants. Nat. Commun. 2019;10:4359. doi: 10.1038/s41467-019-12328-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee K.M., Giltnane J.M., Balko J.M., Schwarz L.J., Guerrero-Zotano A.L., Hutchinson K.E., Nixon M.J., Estrada M.V., Sanchez V., Sanders M.E. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Met. 2017;26:633–647. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonuccelli G., Peiris-Pages M., Ozsvari B., Martinez-Outschoorn U.E., Sotgia F., Lisanti M.P. Targeting cancer stem cell propagation with palbociclib, a CDK4/6 inhibitor: Telomerase drives tumor cell heterogeneity. Oncotarget. 2017;8:9868–9884. doi: 10.18632/oncotarget.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S., Kasim V., Kano M.R., Tanaka S., Ohba S., Miura Y., Miyata K., Liu X., Matsuhashi A., Chung U.I. Transcription factor YY1 contributes to tumor growth by stabilizing hypoxia factor HIF-1α in a p53-independent manner. Cancer Res. 2013;73:1787–1799. doi: 10.1158/0008-5472.CAN-12-0366. [DOI] [PubMed] [Google Scholar]
- 54.Zheng L., Li X., Gu Y., Lv X., Xi T. The 3'UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res. Treat. 2015;150:105–118. doi: 10.1007/s10549-015-3298-2. [DOI] [PubMed] [Google Scholar]
- 55.Zheng L., Guo Q., Xiang C., Liu S., Jiang Y., Gao L., Ni H., Wang T., Zhao Q., Liu H. Transcriptional factor six2 promotes the competitive endogenous RNA network between CYP4Z1 and pseudogene CYP4Z2P responsible for maintaining the stemness of breast cancer cells. J. Hematol. Oncol. 2019;12:23. doi: 10.1186/s13045-019-0697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.