Abstract
Because of the ongoing pandemic around the world, the mechanisms underlying the SARS-CoV-2-induced COVID-19 are subject to intense investigation. Based on available data for the SARS-CoV-1 virus, we suggest how CoV-2 localization of RNA transcripts in mitochondria hijacks the host cell’s mitochondrial function to viral advantage. Besides viral RNA transcripts, RNA also localizes to mitochondria. SARS-CoV-2 may manipulate mitochondrial function indirectly, first by ACE2 regulation of mitochondrial function, and once it enters the host cell, open-reading frames (ORFs) such as ORF-9b can directly manipulate mitochondrial function to evade host cell immunity and facilitate virus replication and COVID-19 disease. Manipulations of host mitochondria by viral ORFs can release mitochondrial DNA (mtDNA) in the cytoplasm and activate mtDNA-induced inflammasome and suppress innate and adaptive immunity. We argue that a decline in ACE2 function in aged individuals, coupled with the age-associated decline in mitochondrial functions resulting in chronic metabolic disorders like diabetes or cancer, may make the host more vulnerable to infection and health complications to mortality. These observations suggest that distinct localization of viral RNA and proteins in mitochondria must play essential roles in SARS-CoV-2 pathogenesis. Understanding the mechanisms underlying virus communication with host mitochondria may provide critical insights into COVID-19 pathologies. An investigation into the SARS-CoV-2 hijacking of mitochondria should lead to novel approaches to prevent and treat COVID-19.
Keywords: aging, coronavirus, COVID-19, mitochondria, mitochondrial DNA, SARS-CoV
INTRODUCTION
In the Chinese city of Wuhan in December 2019, a new severe acute respiratory syndrome virus (SARS-CoV-2) emerged, which has led to a worldwide pandemic (35, 63). SARS-CoV-2 is a positive single-stranded RNA β-coronavirus. It contains a 29,903-nucleotide-long genome (10). SARS-CoV-2 causes the human disease known as coronavirus disease 2019 (COVID-19). SARS-CoV-2-positive cases are reported in more than 5 million people, resulting in more than 3,247,000 COVID-19-related deaths in 200 countries (source: World Health Organization https://COVID19.who.int/).
The genome organization of SARS-CoV-2 is similar to CoV-1 and other coronaviruses (15). SARS-CoV-2 contains open-reading frames (ORFs) common to all β-coronaviruses. These include the ORF1ab responsible for most of the enzymatic proteins, the surface spike glycoproteins (S), the envelope proteins (E), the membrane proteins (M), and the nucleocapsid proteins (N). Other proteins include nonstructural proteins expressed from ORF3a, ORF6a, ORFF7a, and ORF8a. SARS-CoV-2 also includes ORF10a as part of its genome (29, 31, 59).
Both SARS-CoV-1 and CoV-2 enter into the cell by viral spike (S) proteins binding to cellular receptors and on S protein priming by host cell proteases. Recently, it was demonstrated that like SARS-CoV-1, SARS-CoV-2 transmembrane serine protease 2 (TMPRSS2) primes S protein and uses angiotensin-converting enzyme carboxypeptidase 2 (ACE2) host receptor for entry into the cell (29). Genetic variation analyses in the ACE2 gene showed substantial variation among populations worldwide (9, 55). Thus, ACE2 might impact CoV-2 entry differently in different people. Interestingly, ACE2 variation may also affect mitochondrial function, a mechanism that CoV-2 may use to preferentially infect populations compromised in mitochondrial function due to either variation in ACE2 or chronic diseases such as diabetes in which mitochondrial dysfunction plays a significant role (18, 43, 51).
To identify variations among the ORFs encoded by SARS-CoV-1 and SARS-CoV-2, we analyzed genome diversity between the two coronaviruses from different regions of the world. We report a lack of significant mutational differences between the CoV-1 and CoV-2 and highlight how CoV-2 targeting of host mitochondria by direct transport of viral RNA as well as the RNA transcripts hijacks and manipulates mitochondrial function to facilitate host immune suppression, viral replication, and COVID-19.
SARS-CoV-2 MANIPULATION OF MITOCHONDRIA TO GAIN ENTRY INTO THE CELL
SARS-CoV-1 entry into the cell depends on angiotensin-converting enzyme carboxypeptidase 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2). It has been demonstrated that SARS-CoV-2 also uses the host ACE2 for entry and the TMPRSS2 for S protein priming (29). SARS-CoV-2 uses ACE2 as the entry receptor (68). ACE2 utilization serves as a critical determinant of CoV-2 transmissibility (14, 68, 69). The ACE2 gene is present in the X-chromosome of humans (14).
The ACE2, which cleaves angiotensin II (Ang II) into angiotensin 1–7 (Ang 1–7), regulates mitochondrial functions (51). ACE2-knockout mice exhibit impaired mitochondrial respiration and reduced production of ATP. ACE2 overexpression also restores impaired mitochondrial function. It also regulates mitochondria-localized NADPH oxidase 4, which is known to produce reactive oxygen species (ROS) in the mitochondria (25, 32).
We checked the spatial variations present in the ACE2 gene to see whether some populations carry substitutions that categorize them as more susceptible or resistant to the virus. Consistent with the previous studies (3, 9), we didn’t find any rare variant in the coding region; however, an exonic variant rs2285666 (G8790A) showed a significant difference (P = 2 × 10−16) for European and East Asian populations. The pathogenicity of these variants was studied extensively for type 2 diabetes, coronary artery disease, and hypertension (12, 65, 67). It is interesting to note that the G/G genotype could reduce the expression of ACE2 protein up to 50% (42, 65).
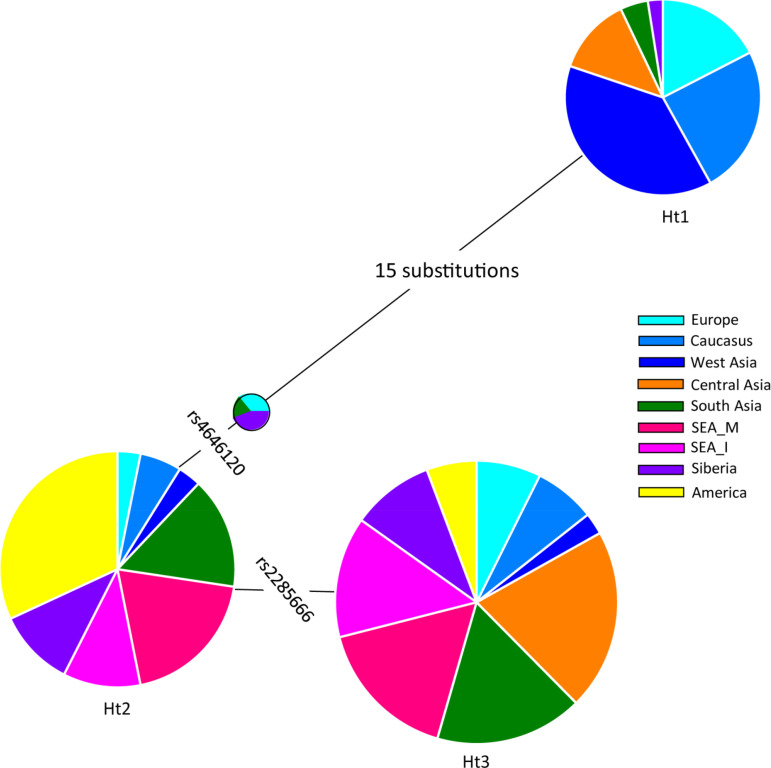
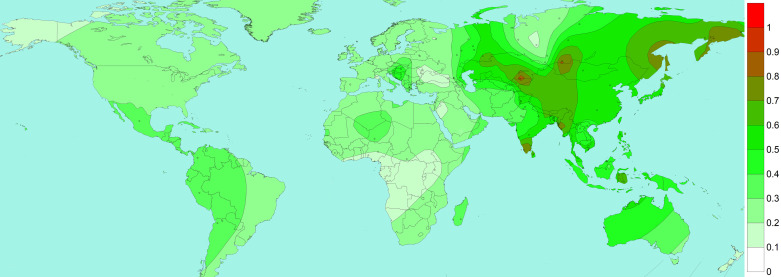
Our haplotype-based analysis of this gene suggested contrasting clustering of South Asian haplotypes with the East and Southeast Asian haplotypes (55). The excessive sharing of this haplotype was in the background of allele rs2285666 (Fig. 1). We obtained three major haplotypes in this gene, where haplotype 1 was predominantly present among West Eurasian populations. This haplotype was distinct from haplotype 2 and haplotype 3 by a long branch of 15 substitutions (Fig. 1). Another allele rs4646120 reduces the haplotype diversity substantially and yields haplotype, which is derived mainly from East Eurasian, South Asian, and American populations. Subsequently, the third haplotype is separated by variant rs2285666 (Fig. 1). Haplotype 3 is majorly composed of East Eurasian, South Asian, and Central Asian populations. The phylogeographic distribution of rs2258666 revealed relatively higher frequency in East Eurasia than in Europe, Africa, and North America (Fig. 2). Among South Asians, its incidence is higher among Kerala and Tamilnadu states of Southern Indian, Himalayan, Northeast Indian, and Bangladeshi tribal populations. A few Central Asian and Siberian tribes also had a higher frequency of this allele. Thus, it is likely that a high rate of rs2285666 allele among populations may modulate the susceptibility for SARS-CoV-2.
Fig. 1.
The most parsimonious structure of major haplotypes present in the angiotensin-converting enzyme carboxypeptidase 2 gene (ACE2). Frequencies of each regional population present in a haplotype are shown in color. Phylogenetic relationships between the observed haplotypes were reconstructed with the NETWORK 5.0 program.
Fig. 2.
Spatial distribution of angiotensin-converting enzyme carboxypeptidase 2 (ACE2) rs2285666 in world populations. Isofrequency maps were generated by using Surfer8 of Golden Software (Golden Software, Inc., Golden, CO), following the Kriging procedure. The isofrequency map illustrates the geographic spread of the respective allele. Data points are shown for each population.
Besides ACE2, TMPRSS2 transmembrane serine protease also allows the entry of coronaviruses into host cells by proteolytic cleaving and activating viral envelope glycoproteins (29). Among the coronaviruses, HCoV-229E, MERS-CoV, SARS-CoV, and SARS-CoV-2 are demonstrated to be dependent on TMPRSS2. Indirect evidence suggests that TMPRSS2 may regulate mitochondrial function via ERRa (estrogen-related receptor-α). ERRα is a ligand-independent nuclear receptor that, together with its coactivator peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), transcriptionally regulates energy homeostasis and mitochondrial functions (66). Alternatively, spliced transcript variants of TMPRSS2 isoforms have been described. It is conceivable that an isoform(s) may localize into the mitochondria and play a direct role in regulating mitochondrial function.
HOST MITOCHONDRIA HIJACKING BY SARS-CoV-2 RNA
Recent evidence indicates that mitochondria play a central role in the host response upon viral infection and immunity (5, 34). Wu et al. (64) compared a broad set of SARS-CoV-2 genomes to the human transcriptome and carried out systematic computational analyses to identify the subcellular signals. Using machine learning models, they determined that the SARS-CoV-2 RNA genome and all subgenomic RNAs were enriched in the host mitochondria and nucleolus. Wu et al. (63) reported that the 5′- and 3′-untranslated regions of CoV-2 contained the distinct mitochondrial localization signals. This analysis provides an interesting hypothesis that needs to be validated experimentally for RNA mode of the hijacking of mitochondria by SARS-CoV-2 (63).
Mitochondrial stress induces the formation of mitochondria-derived vesicles that are involved in the cross-talk between mitochondria and endoplasmic reticulum (ER) (57). Coronavirus replication consists of the formation of double-membrane vesicles (DMVs) derived from ER. These DMVs serve as a site for viral replication and help conceal the virus from host cellular defenses (6, 27, 33). Like viral manipulation of ER, it is plausible that CoV-2 manipulation of mitochondria results in the induction of (double-membrane) mitochondria-derived vesicles (MDVs). It is likely that CoV-2 RNA localization in mitochondria induces mitochondrial dysfunction and increases mitochondria-derived double-membrane vehicles in which the virus can hide and replicate. In this context, it noteworthy that point mutations in the murine coronavirus are known to decrease the number of ER-derived DMV but simultaneously increase the localization of viral protein into the mitochondria (17).
The mitochondrial stress-induced MDV is intimately involved in cross-talk with ER (1). Interestingly, HIV RNA also localizes to host mitochondria and induces mitochondrial dysfunction (54). The exact mechanism of how either mitochondria-localized corona or HIV viral RNA causes mitochondrial dysfunction is unknown. Multiple mechanisms are likely involved. To date, several questions remain unanswered. These include the following. 1) How is the viral RNA imported inside the host mitochondria? 2) Viral nonstructural protein interacts directly with Tomm70, a mitochondrial import receptor (24); however, it is unclear whether this pathway is involved in RNA import into the mitochondria. 3) We do not yet know whether noncoding RNAs encoded by coronavirus inhibit mitochondrial transcription, protein translation, or tRNA processing. 4) Also, we do not know whether there are cassettes of the coronavirus RNA genome that can be read using the mitochondrial amino acid translation code. Finally, we do not yet know whether the formation of ER-derived DMV and mitochondria-derived MDV resemble one another mechanistically and whether mitochondrial MDV indeed serves as a reservoir for CoV-2 replication.
HOST MITOCHONDRIA HIJACKING BY SARS-CoV-2 PROTEINS
Mitochondria function as a platform for innate immune signaling (2, 5, 37). Notably, the host responses against viral infections depend on mitochondrial functions. Indeed, mtDNA itself acts as a danger-associated molecular pattern (DAMP). The mitochondrial outer membrane functions as a platform for signaling molecules (40, 41, 48, 61). With age, mtDNA content and mitochondrial function decline. Mutations in mtDNA are also reported to accumulate with age, resulting in mitochondrial dysfunction, inflammation, and alterations in immune response (41, 53).
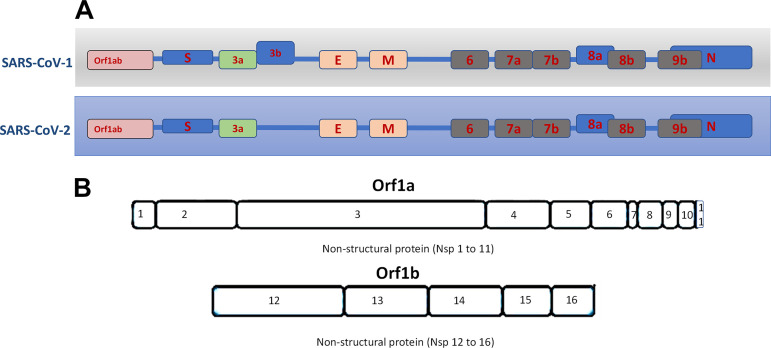
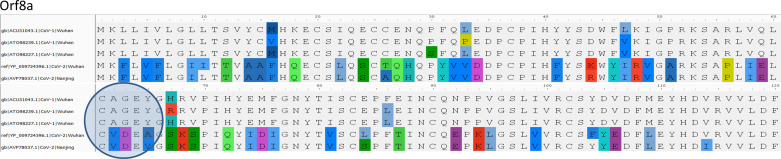
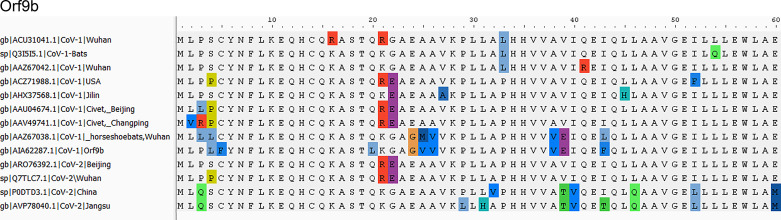
SARS-CoV-1 virus upon infection affects mitochondrial functions, influences its intracellular survival, or evades host immunity. Interestingly, SARS-CoV-1 ORF-9b localizes into host mitochondria, which suppresses innate immunity by manipulating mitochondrial function and mitochondrial antiviral signaling protein (MAVS)/TNF receptor-associated factor (TRAF)3/TRAF6 signaling pathway to host innate immunity (50). Furthermore, SARS-COV-1 ORF3b (23), ORF7a (58), and ORF8a also localize to mitochondria and promote viral replication (13). We compared mitochondria localized CoV-1 ORF with ORFs encoded by the CoV-2 genome. With the exception of ORF3b, SARS-CoV-2 encodes amino acid sequences very similar to SARS-CoV-1 ORFs (ORF7a, -8a, and -9b) proven to be localized to host mitochondria. Notably, CoV-2 lacks ORF3b (Fig. 3).
Fig. 3.
A: overview of open-reading frames (ORFs) present in SARS-CoV-1 and SARS-Cov-2. B: the Orf3b gene is absent in CoV-2. E, envelope proteins; M, membrane proteins.
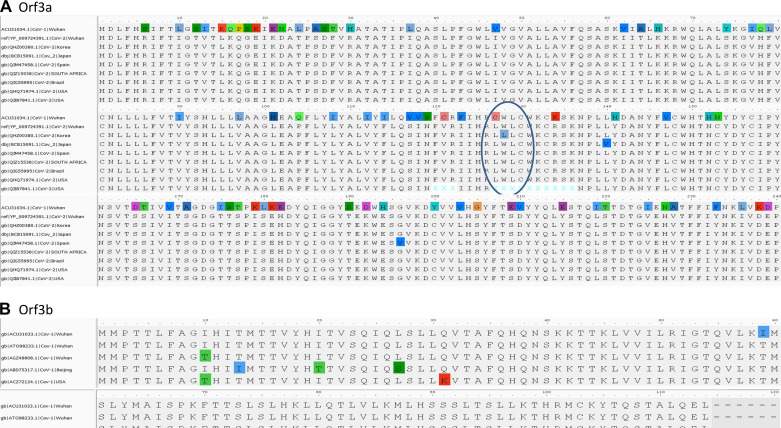
ORF3a is a 275-amino acid protein with one amino acid less in CoV-1. The sequences show 71.64% identity with each other, with 86% similarities. We observed that 174-GTTSPIS-180 aa motif shows an epitope tag, as it formed exposed regions thatallow the protein to open up. The relative surface accessibility for these exposed regions is also unique compared with the CoV-2 (Fig. 4). ORF3b is absent in CoV-2, whereas the 114-amino acid protein shows sequence similarity with the +2 reading frame, which remains untranslated. The translated nucleotide query against the translated nucleotide sequence of CoV-2 showed a significant similarity between them (99%) but across different reading frames (Fig. 4). ORF7a reveals an 87.7% identity between SARS-CoV-2and CoV-1 with one gap (Fig. 5). ORF8a is yet another protein with 68% similarity to SARS-CoV-2 and CoV-1 (Fig. 6). This ORF contains two signature sequences, viz. EDPCP and INCQ. Structurally, these form the β-pleated sheets, and their function is not yet known. ORf9b is absent in CoV-2, even as there are many coiled structures untranslated from various reading frames (Fig. 7), just as in the case of ORF3b. The exposed regions from 32 to 39 amino acids have a high propensity to form coiled structures and could be used to show an inhibitory effect from interferons.
Fig. 4.
Characteristic motif in open-reading frame 3a (Orf3a). A: characteristic LXXC motif in Orf3a from multiple sequence alignment of reported strains. The LWLC is associated with viral expression, whereas CWLC is the motif described in CoV-1. B: conserved CoV-1 sequences of Orf3b. The header of alignment shows the amino acid positions highlighting nonconsensus sequences.
Fig. 5.
Characteristic motif in open-reading frame 7a (Orf7a). A: ADNK motif in Orf7a. B: Orf7b with characteristic LEXQDXX motif with good epitope sites. The header of alignment shows the amino acid positions highlighting nonconsensus sequences.
Fig. 6.
Characteristic motif in open-reading frame 8a (Orf8a). Orf8a shows characteristic CXXE motif as a good epitope site. The header of alignment shows the amino acid positions highlighting nonconsensus sequences.
Fig. 7.
Characteristic motif in open-reading frame 9b (Orf9b). Orf9b shows characteristic DAXX motif as a good epitope site. The header of alignment shows the amino acid positions highlighting nonconsensus sequences.
SARS-CoV-2 NONCODING RNA REGULATION OF HOST MITOCHONDRIAL FUNCTION
Some of the CoV-1 and CoV-2 regions show similarities to that of annotated lncRNAs (NONHSAT239888) and several nonannotated genomic regions (NONHSAT257828, NONHSAT257829, and NONHSAT257830). We hypothesize that this representation and association of lncRNAs in response to virus infection may facilitate mitochondrial hijacking of CoV. A differential expression study determining the function of these sequences is needed to confirm the biological relevance of lncRNA (45).
In a recent publication, Pasquier and Robichon (45) noted that ORF3a includes a 20-base sequence that targets USP30, a mitochondrial ubiquitin-specific peptidase 30, a subunit of ubiquitin ligase complex (FBXO21). The 20 nucleotides present in ORF3a of SARS-CoV-2 target the sequence AAAGATAGAGAAAAGGGGCT found in USP30 transcripts. USP30 is a mitochondrial deubiquitinase involved in mitochondria homeostasis and controls mitophagy. SARS-CoV-2 might affect mitochondria function by altering ubiquitination and contribute to the suppression of immunity in COVID-19 patients.
SARS-CoV-2 PROTEIN INTERACTION WITH HOST MITOCHONDRIAL PROTEINS
A recent study by Gordon et al. (24) mapping the SARS-CoV-2 interactions with host proteins revealed a significant number of mitochondrial proteins interacting with the viral proteins. These include Nsp8 (Fig. 3B) interaction with mitochondrial MRPS2, MRPS5, MRPS25, and MRPS27 ribosomal proteins, ORF9c interaction with mitochondrial NDUFAF1 and NDUFB9, and Nsp7 interaction with mitochondrial NDUFAF2. Notably, both NDUFAF1 and -2 are critical players involved in the assembly of complex I. NDUFB9 is an essential subunit of complex I comprised of more than 40 subunits. Furthermore, viral M protein was described to interact with ATP1B1, ATP6V1A, ACADM, AASS, PMPCB, PITRM1, COQ8B, and PMPCA (Table 1). These proteins are part of the critical metabolic pathways carrying mitochondrial metabolism (24). Indeed, the authors note that approximately 55 genes were mitochondrial, and approximately 105 genes modulated cellular function in response to bioenergetics function. Previously, studies have also reported interaction of Cov-1 Nsp2 interaction with prohibiting protein PHB and Nsp10 interaction with mtDNA-encoded COX II (complex IV) and NADH4L (complex I) (20, 36). The study also identified NDUFA10 as one of the master regulators of CoV-2 pathology (26). Unfortunately, the biological significance of such interaction is lacking. Gordon et al. (24) found CoV-2 interactions with Tomm70. This mitochondrial import receptor plays a critical role in transporting proteins into mitochondria and, more importantly, in modulating anti-viral cellular defense pathways (60). These protein-protein interactions (Table 1) may give coronavirus a local advantage in manipulating mitochondria to suppress anti-viral response and host immunity.
Table 1.
Predicted SARS-CoV-2 RNA and protein interaction with host mitochondrial proteins
| Interacting Mitochondrial Proteins | Ref. No. | |
|---|---|---|
| Viral RNA | ||
| Mitochondria localized | Unknown | 64 |
| Orf1ab | ||
| S | ||
| Orf3a | ||
| E | ||
| M | ||
| Orf6 | ||
| Orf7a | ||
| Orf7b | ||
| Orf8 | ||
| N | ||
| Orf10 | ||
| Viral protein | ||
| Nonstructural proteins | ||
| Nsp2 | PHB | 20 |
| Nsp7 | NDUFAF2 | 24 |
| Nsp8 | MRPS2, MRPS5, MRPS25, MRPS27 | 24 |
| Nsp10 | NADH4L, COX II | 36 |
| Structural protein | ||
| M | ATP1B1, ATP6V1A, ACADM, AAAS, PMPCB, PITRM1, PMPCA, COQ8B | 24 |
| Accessory proteins | ||
| Orf7a | Bcl-XL | 58 |
| Orf8a | Mitochondria localized, interaction unknown | 13 |
| Orf9b | MAVS | 50 |
| Orf9c | NDUFAF1, NDUFB9 | 24 |
ACADM, acyl-coenzyme A dehydrogenase; ATP1B1, sodium/potassium-transporting ATPase subunit-β1; ATP6V1a, V-type proton ATPase catalytic subunit A; COQ8B, coenzyme Q8B; COX II, cytochrome c oxidase subunit II; E, envelope protein; M, membrane protein; MAVS, mitochondrial antiviral signaling protein; MRPS, mitochondrial ribosomal protein S; N, nucleocapsid protein; NDUFAF, NADH:ubiquinone oxidoreductase complex assembly factor; Orf, open-reading frame; PHB, prohibitin; PITRM1, pitrilysin metalloproteinase 1; S, surface spike glycoprotein.
FUNCTIONAL DOMAINS OF VIRAL PROTEINS
To explore the host targeting by the viral proteins, we inspected the subcellular localization of ORFs and observed that ORF7a and ORF8 contain NH2-terminal signal peptide anchors. Although from the predictions they are shown to harbor signal peptides, the attachment of proteins to the extracellular membrane is prenylated. It appears that the combinatorial occurrence of domains and motif is associated with the proteins. For example, ORF3a shows motif exchange and similarity with LWLC associated with viral expression (44). Likewise, ORF7a is related to a motif ADNK, which might have evolved to resemble host protein motifs associated with the respiratory mechanisms in the host cells. These motifs indicate that they are functionally mimicking the binding sites of the proteins in carrying out the interactions. Although the sequence similarity and motifs are strain/sample specific, the superfamily domains these proteins are associated with do not show any particular links to human homologs.
MITOCHONDRIA AND SEX-SPECIFIC MORTALITY
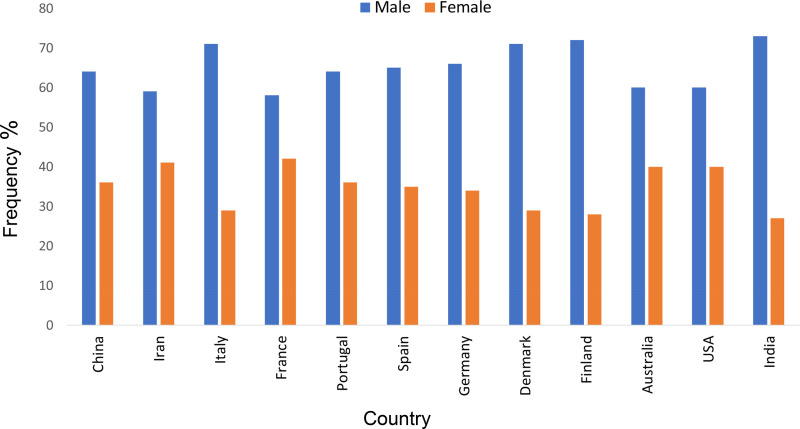
Compared with women, SARS-CoV-2 infection results in a high rate of mortality in older men (Fig. 8). The underlying cause of the high frequency is unknown. Both androgen and estrogen are known to regulate mitochondrial function. TMPRSS2 is induced by androgen and regulated by androgen receptor (16). It is conceivable that a high prevalence of mortality in men is due to androgenic induction of TMPRSS2. Furthermore, androgen receptors as well as estrogen receptors localize the mitochondrial compartment and regulate mitochondrial function. It is plausible that the hormonal regulation of mitochondrial targets and functions differs between men and women, which may contribute to a difference in susceptibility. Other hallmarks of aging, such as changes in epigenetic patterns, exhibiting different expressions in older men than age-matched women (4, 30), may underlie factors involved in sex-specific mortality.
Fig. 8.
COVID-19-related frequency of occurrence among males and females in significant countries. Data from various countries were obtained from their official sources. These include China (http://www.chinacdc.cn/en/), India (https://www.COVID19india.org/), Iran (behdasht.gov.ir), Australia (www.health.gov.au/COVID-19), the United States (coronavirus.gov), and European countries https://www.ecdc.europa.eu/en/COVID-19/sources-updated).
HOST GENETIC DIVERSITY AND SEVERITY OF COVID-19
Differences in mitochondrial genetic makeup contribute to functional differences in the mitochondrial function in tissues, organs, and organisms as a whole. Thus mitochondrial variants have different metabolic capacities, resulting in differences in anti-viral and inflammatory responses (34). Interestingly, the association between mtDNA haplogroups and ICU outcomes has been reported (56). Mitochondrial function declines with age and consistently reduces the level of mtDNA. Studies at the level of mtDNA in plasma of COVID-19-affected people and their ages may help in understanding whether the mtDNA-dependent systemic inflammation is deranged in aged people or whether individuals (or populations), albeit at young ages, may be endowed with mtDNA variants that are abnormally proinflammatory/and prone to being released (56).
AGING HALLMARKS AND MORTALITY
Aging is characterized by a progressive decline in cellular function, resulting in increased susceptibility to age-related morbidity and mortality. Mitochondrial dysfunction is one of the hallmarks of aging that contributes significantly to the physiology of aging as well as the pathophysiology of age-related disorders. In most of the COVID-19 patients, systemic hyper inflammation leads to severe and often lethal outcomes. Aging is also characterized by systemic inflammation described as inflammaging and immunosenescence, an impairment of acquired immunity (7, 22). It is conceivable that the decline in these critical players of aging contributes to the high rate due to CoV-2 infection.
Mitochondrial dysfunction induces senescence (62). Senescent cells are accumulated during aging and acquire the senescence-associated secretory phenotype (SASP), which contributes to inflammaging (19, 22). Senescence impairs macrophages, which protect against infection (22). Macrophages are critical factors in protecting the lung after infection by SARS-CoV-2 (39). It is likely that older individual senescent macrophages are less active in protecting inflammation induced by COVID-19 (28, 39, 47) and cause lethality. These observations support the notion that aging-associated inflammaging and immune senescence compromise robust responses as in young individuals to protect against the SAR-CoV-2 high rate of mortality. Notably, the clinical manifestations in the most severe patients show a cytokine storm in which interleukin-6 (IL-6) values turn out to be significantly high. The extent of inflammaging differs between aged men and woman. High levels of IL-6 are found in older men compared with age-matched women (56). High levels of IL-6 may result in negative outcomes of COVID-19 in older men affected by comorbidity.
Another mechanism involves mtDNA directly to trigger host immunity. In this mechanism, upon viral infection, mtDNA leaks out of the mitochondria, into the cytoplasm, and into the extracellular space and induces innate immunity and inflammation. This is an evolutionarily conserved signaling mechanism induced upon many types of viral infection. Interestingly, high levels of circulating mtDNA are reported in older individuals (46). Older individuals suffering from COVID-19 may release mtDNA, resulting in the onset of a detrimental inflammatory response.
Additional factors that may contribute to age-related mortality include differences in ACE2 expression between young and older individuals. Several studies suggest that ACE2 is downregulated in aging (11, 49), which may contribute to the increased risk of vascular injury and cardiovascular disease affecting older men (49). ACE2 is also downregulated in patients with type 2 diabetic men with severe COVID-19 outcomes (9). In contrast, ACE2 is overexpressed in women compared with men (14).
HOST IMMUNE SUPPRESSION BY MITOCHONDRIAL HIJACKING
Mitochondria serve as a signaling platform for mitochondrial anti-viral signal (MAVS). Mitochondria are involved in inflammation and both innate and adaptive immunity (41). Many single-strand positive RNA viruses similar to SARS-CoV-2 induce an inflammatory response that involves the mitochondrial biogenesis, mitochondrial fusion, fission, and mtDNA release to outside the cell. This phenomenon is described as neutrophil extracellular trap (NET)osis mechanisms (based on NET formation) (8). The release of mtDNA is an ancestral cell stress mechanism that triggers local and systemic trigger of inflammation. Upon cell stress or damage, mtDNA leaks out into the cytoplasm and then outside, where it triggers inflammation and anti-viral response, which involves cytoplasmic AIM2 and/or endosomal/extracellular TLR9. Levels of mtDNA increase with the severity of damage and correlate with the onset of multiorgan failure in patients affected by multiple systemic conditions observed with acute respiratory distress (52).
MITOCHONDRIAL INTERVENTIONS
Presently, a large number of drugs are being repurposed against COVID-19 disease. These include, e.g., remdesivir, lopinavir/ritonavir, and anti-inflammatory agents such as chloroquine and tocilizumab (21, 38). It is likely that drugs that modulate mitochondrial function and inhibit inflammation may help treat patients with COVID-19.
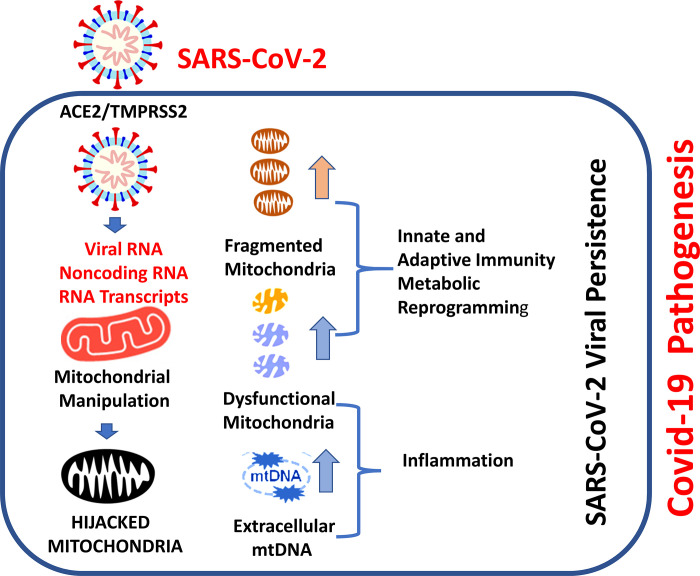
Figure 9 provides the schematic describing the conceptual framework related to SARS-CoV-2 hijacking of mitochondria resulting in COVID-19 disease. A schematic shows how the SARS-CoV-2 enters into the host cell utilizing ACE2, a polymorphic protein that regulates mitochondrial function. Upon entry into the cells, viral RNA and RNA transcript translocate to mitochondria to hijack and manipulate host mitochondria to suppress host immunity. We do not yet know how CoV-2 RNA localized to mitochondria to impact immunity. It is likely that it involves multiple mitochondrial mechanisms at different levels. Conceivably, postinfection noncoding RNA may regulate USP30, a host protein that controls mitochondrial dynamics (fission and fusion). SARS-2-CoV-2 also can manipulate the release of mtDNA, leading to inflammation. Thus, the hijacking of mitochondria may be an essential mechanism in the induction of COVID-19 disease. These observations suggest that drugs that selectively restore mitochondrial function and promote mitochondrial biogenesis may serve as anti-inflammatory agents to prevent or treat COVID-19.
Fig. 9.
Mechanisms involved in SARS-CoV-2 hijacking of host mitochondria. Schematic showing the SARS-CoV-2 entry into the host cell utilizing angiotensin-converting enzyme carboxypeptidase 2 (ACE2), a polymorphic protein that regulates mitochondrial function. Upon entry into the cells, viral RNA and proteins localize to mitochondria. Postinfection noncoding RNA may also regulate host proteins (such as USP30) involved in mitochondrial dynamics. SARS-2-CoV-2 appears to hijack host mitochondria to suppress host immunity by regulating mitochondrial dynamics, mitochondrial function, and mtDNA release. Hijacking mitochondria may be one of the essential mechanisms leading to COVID-19.
GRANTS
K.K.S. is supported by the National Cancer Institute Grant R01CA204430.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.K.S., G.C., and P.S. prepared figures; K.K.S. drafted manuscript; K.K.S., G.C., J.Y.C., and P.S. edited and revised manuscript; K.K.S., G.C., J.Y.C., and P.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Ayam Gupta for help in fine-tuning the figures.
REFERENCES
- 1.Abuaita BH, Schultz TL, O’Riordan MX. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24: 625–636.e5, 2018. doi: 10.1016/j.chom.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnoult D, Soares F, Tattoli I, Girardin SE. Mitochondria in innate immunity. EMBO Rep 12: 901–910, 2011. doi: 10.1038/embor.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asselta R, Paraboschi EM, Mantovani A, Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany NY) 12. In press. doi: 10.18632/aging.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett EL, Richardson DS. Sex differences in telomeres and lifespan. Aging Cell 10: 913–921, 2011. doi: 10.1111/j.1474-9726.2011.00741.x. [DOI] [PubMed] [Google Scholar]
- 5.Biacchesi S, LeBerre M, Lamoureux A, Louise Y, Lauret E, Boudinot P, Brémont M. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol 83: 7815–7827, 2009. doi: 10.1128/JVI.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard E, Roingeard P. Virus-induced double-membrane vesicles. Cell Microbiol 17: 45–50, 2015. doi: 10.1111/cmi.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonafè M, Sabbatinelli J, Olivieri F. Exploiting the telomere machinery to put the brakes on inflamm-aging. Ageing Res Rev 59: 101027, 2020. doi: 10.1016/j.arr.2020.101027. [DOI] [PubMed] [Google Scholar]
- 8.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science 303: 1532–1535, 2004. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, Wen F, Huang X, Ning G, Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov 6: 11, 2020. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 9: 221–236, 2020. [Erratum in Emerg Microbes Infect 9: 540, 2020]. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang SY, Chen YW, Chenier I, Tran SM, Zhang SL. Angiotensin II type II receptor deficiency accelerates the development of nephropathy in type I diabetes via oxidative stress and ACE2. Exp Diabetes Res 2011: 1–12, 2011. doi: 10.1155/2011/521076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaoxin J, Daili S, Yanxin H, Ruwei G, Chenlong W, Yaobin T. The influence of angiotensin-converting enzyme 2 gene polymorphisms on type 2 diabetes mellitus and coronary heart disease. Eur Rev Med Pharmacol Sci 17: 2654–2659, 2013. [PubMed] [Google Scholar]
- 13.Chen CY, Ping YH, Lee HC, Chen KH, Lee YM, Chan YJ, Lien TC, Jap TS, Lin CH, Kao LS, Chen YM. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J Infect Dis 196: 405–415, 2007. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Jiang Q, Xia X, Liu K, Yu Z, Tao W, Gong W, Han JDJ. Individual variation of the SARS-CoV2 receptor ACE2 gene expression and regulation. Preprints 2020030191 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol 92: 418–423, 2020. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Song X, Li Q, Xie L, Guo T, Su T, Tang C, Chang X, Liang B, Huang D. Androgen Receptor-Activated Enhancers Simultaneously Regulate Oncogene TMPRSS2 and lncRNA PRCAT38 in Prostate Cancer. Cells 8: 864, 2019. doi: 10.3390/cells8080864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clementz MA, Kanjanahaluethai A, O’Brien TE, Baker SC. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375: 118–129, 2008. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper JM, Mann VM, Schapira AH. Analyses of mitochondrial respiratory chain function and mitochondrial DNA deletion in human skeletal muscle: effect of ageing. J Neurol Sci 113: 91–98, 1992. doi: 10.1016/0022-510X(92)90270-U. [DOI] [PubMed] [Google Scholar]
- 19.Coppé J-P, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez P-Y, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: 2853–2868, 2008. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornillez-Ty CT, Liao L, Yates JR III, Kuhn P, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol 83: 10314–10318, 2009. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson N, Laydon D, Nedjati Gilani G, Imai N, Ainslie K, Baguelin M, Bhatia S, Boonyasiri A, Cucunuba Perez Z, Cuomo-Dannenburg G. Report 9: Impact of non-pharmaceutical interventions (NPIs) to reduce COVID19 mortality and healthcare demand (Online). London: Imperial College COVID-19 Response Team; https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-9-impact-of-npis-on-covid-19/ [16 March 2020]. [Google Scholar]
- 22.Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann NY Acad Sci 908: 244–254, 2000. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 23.Freundt EC, Yu L, Park E, Lenardo MJ, Xu X-N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol 83: 6631–6640, 2009. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, O’Meara MJ, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Naing ZZC, Zhou Y, Peng S, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Shen W, Shi Y, Zhang Z, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Ramachandran R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Lin Y, Wankowicz SA, Bohn M, Trenker R, Young JM, Cavero D, Hiatt J, Roth T, Rathore U, Subramanian A, Noack J, Hubert M, Roesch F, Vallet T, Meyer B, White KM, Miorin L, Agard D, Emerman M, Ruggero D, García-Sastre A, Jura N, von Zastrow M, Taunton J, Schwartz O, Vignuzzi M, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor S, Fraser JS, Gross J, Sali A, Kortemme T, Beltrao P, Shokat K, Shoichet BK, Krogan NJ. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv 2020.03.22.002386, 2020. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham KA, Kulawiec M, Owens KM, Li X, Desouki MM, Chandra D, Singh KK. NADPH oxidase 4 is an oncoprotein localized to mitochondria. Cancer Biol Ther 10: 223–231, 2010. doi: 10.4161/cbt.10.3.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guzzi PH, Mercatelli D, Ceraolo C, Giorgi FM. Master regulator analysis of the SARS-CoV-2/human interactome. J Clin Med 9: 982, 2020. doi: 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagemeijer MC, Rottier PJ, de Haan CA. Biogenesis and dynamics of the coronavirus replicative structures. Viruses 4: 3245–3269, 2012. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall BM, Balan V, Gleiberman AS, Strom E, Krasnov P, Virtuoso LP, Rydkina E, Vujcic S, Balan K, Gitlin II, Leonova KI, Consiglio CR, Gollnick SO, Chernova OB, Gudkov AV. p16(Ink4a) and senescence-associated β-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging (Albany NY) 9: 1867–1884, 2017. doi: 10.18632/aging.101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8, 2020. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 14: R115, 2013. [Erratum in Genome Biol 14: R115, 2013]. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell 181: 914–921.e10, 2020. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SM, Kim YG, Jeong KH, Lee SH, Lee TW, Ihm CG, Moon JY. Angiotensin II-induced mitochondrial Nox4 is a major endogenous source of oxidative stress in kidney tubular cells. PLoS One 7: e39739, 2012. doi: 10.1371/journal.pone.0039739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6: e226, 2008. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koshiba T. Mitochondrial-mediated antiviral immunity. Biochim Biophys Acta 1833: 225–232, 2013. doi: 10.1016/j.bbamcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents 55: 105924, 2020. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Wang L, Dong C, Che Y, Jiang L, Liu L, Zhao H, Liao Y, Sheng Y, Dong S, Ma S. The interaction of the SARS coronavirus non-structural protein 10 with the cellular oxido-reductase system causes an extensive cytopathic effect. J Clin Virol 34: 133–139, 2005. doi: 10.1016/j.jcv.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong SK, Huang IC, Xu K, Vasilieva N, Murakami A, He Y, Marasco WA, Guan Y, Choe H, Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J 24: 1634–1643, 2005. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu A. China turns Roche arthritis drug Actemra against COVID-19 in new treatment guidelines (Online). https://www.fiercepharma.com/pharma-asia/china-turns-roche-arthritis-drug-actemra-against-covid-19-new-treatment-guidelines [4 March 2020].
- 39.Liu Y, Qu HQ, Tian L, Hakonarson H. Expression pattern of the SARS-CoV-2 receptor ACE2 and TMPRSS2 in the respiratory tract. Preprints 2020050040: 2020. doi: 10.20944/preprints202005.0040.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehta MM, Weinberg SE, Chandel NS. Mitochondrial control of immunity: beyond ATP. Nat Rev Immunol 17: 608–620, 2017. doi: 10.1038/nri.2017.66. [DOI] [PubMed] [Google Scholar]
- 41.Mills EL, Kelly B, O’Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol 18: 488–498, 2017. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 42.Minato T, Nirasawa S, Sato T, Yamaguchi T, Hoshizaki M, Inagaki T, Nakahara K, Yoshihashi T, Ozawa R, Yokota S, Natsui M, Koyota S, Yoshiya T, Yoshizawa-Kumagaye K, Motoyama S, Gotoh T, Nakaoka Y, Penninger JM, Watanabe H, Imai Y, Takahashi S, Kuba K. B38-CAP is a bacteria-derived ACE2-like enzyme that suppresses hypertension and cardiac dysfunction. Nat Commun 11: 1058, 2020. doi: 10.1038/s41467-020-14867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohlke KL, Jackson AU, Scott LJ, Peck EC, Suh YD, Chines PS, Watanabe RM, Buchanan TA, Conneely KN, Erdos MR, Narisu N, Enloe S, Valle TT, Tuomilehto J, Bergman RN, Boehnke M, Collins FS. Mitochondrial polymorphisms and susceptibility to type 2 diabetes-related traits in Finns. Hum Genet 118: 245–254, 2005. doi: 10.1007/s00439-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 44.Pasqualini R, Koivunen E, Ruoslahti E. A peptide isolated from phage display libraries is a structural and functional mimic of an RGD-binding site on integrins. J Cell Biol 130: 1189–1196, 1995. doi: 10.1083/jcb.130.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasquier C, Robichon A. SARS-CoV-2 might manipulate against its host the immunity RNAi/Dicer/Ago system. Does mitochondria collapse upon COVID-10 infection? bioRxiv:1–15, 2020. doi: 10.1101/2020.04.08.031856. [DOI] [Google Scholar]
- 46.Pinti M, Cevenini E, Nasi M, De Biasi S, Salvioli S, Monti D, Benatti S, Gibellini L, Cotichini R, Stazi MA, Trenti T, Franceschi C, Cossarizza A. Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for “inflamm-aging”. Eur J Immunol 44: 1552–1562, 2014. doi: 10.1002/eji.201343921. [DOI] [PubMed] [Google Scholar]
- 47.Rana DR, Dulal S. Therapeutic application of chloroquine in clinical trials for COVID-19. medRxiv: 1–15, 2020. doi: 10.1101/2020.03.22.20040964. [DOI] [Google Scholar]
- 48.Rongvaux A. Innate immunity and tolerance toward mitochondria. Mitochondrion 41: 14–20, 2018. doi: 10.1016/j.mito.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Sato T, Suzuki T, Watanabe H, Kadowaki A, Fukamizu A, Liu PP, Kimura A, Ito H, Penninger JM, Imai Y, Kuba K. Apelin is a positive regulator of ACE2 in failing hearts. J Clin Invest 123: 5203–5211, 2013. doi: 10.1172/JCI69608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, Kehrl JH. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 193: 3080–3089, 2014. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi TT, Yang FY, Liu C, Cao X, Lu J, Zhang XL, Yuan MX, Chen C, Yang JK. Angiotensin-converting enzyme 2 regulates mitochondrial function in pancreatic β-cells. Biochem Biophys Res Commun 495: 860–866, 2018. doi: 10.1016/j.bbrc.2017.11.055. [DOI] [PubMed] [Google Scholar]
- 52.Simmons JD, Lee YL, Pastukh VM, Capley G, Muscat CA, Muscat DC, Marshall ML, Brevard SB, Gillespie MN. Potential contribution of mitochondrial DNA damage associated molecular patterns in transfusion products to the development of acute respiratory distress syndrome after multiple transfusions. J Trauma Acute Care Surg 82: 1023–1029, 2017. doi: 10.1097/TA.0000000000001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh B, Schoeb TR, Bajpai P, Slominski A, Singh KK. Reversing wrinkled skin and hair loss in mice by restoring mitochondrial function. Cell Death Dis 9: 735, 2018. doi: 10.1038/s41419-018-0765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Somasundaran M, Zapp ML, Beattie LK, Pang L, Byron KS, Bassell GJ, Sullivan JL, Singer RH. Localization of HIV RNA in mitochondria of infected cells: potential role in cytopathogenicity. J Cell Biol 126: 1353–1360, 1994. doi: 10.1083/jcb.126.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srivastava Anshika, Pandey Rudra Kumar, Kumar Pramod, Rasalkar Avinash Arvind, Tamang Rakesh, Shrivastava Pankaj, Chaubey Gyaneshwer. Most frequent South Asian haplotypes of ACE2 share identity by descent with East Eurasian populations (Online) https://www.researchsquare.com/article/rs-27310/v1. [DOI] [PMC free article] [PubMed]
- 56.Storci G, Bonifazi F, Garagnani P, Olivieri F, Bonafe M. How studies on inflamm-aging may help to understand and combat COVID-19 pandemic. Preprints 2020040158: 2020. doi: 10.20944/preprints202004.0158.v1 [DOI] [Google Scholar]
- 57.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J 33: 2142–2156, 2014. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan YX, Tan TH, Lee MJR, Tham PY, Gunalan V, Druce J, Birch C, Catton M, Fu NY, Yu VC, Tan YJ. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J Virol 81: 6346–6355, 2007. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. In press. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C, Liu X, Wei B. Mitochondrion: an emerging platform critical for host antiviral signaling. Expert Opin Ther Targets 15: 647–665, 2011. doi: 10.1517/14728222.2011.561321. [DOI] [PubMed] [Google Scholar]
- 61.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity 42: 406–417, 2015. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiley CD, Velarde MC, Lecot P, Liu S, Sarnoski EA, Freund A, Shirakawa K, Lim HW, Davis SS, Ramanathan A, Gerencser AA, Verdin E, Campisi J. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab 23: 303–314, 2016. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature 579: 265–269, 2020. [Erratum in Nature 580: E7, 2020]. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu K, Zou J, Chang HY. RNA-GPS predicts SARS-CoV-2 RNA localization to host mitochondria and intracellula nucleolus. bioRxiv 1–17, 2020. doi: 10.1101/2020.04.28.065201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Y, Li J, Wang C, Zhang L, Qiao H. The ACE 2 G8790A polymorphism: involvement in type 2 diabetes mellitus combined with cerebral stroke. J Clin Lab Anal 31: e22033, 2017. doi: 10.1002/jcla.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Z, Wang Y, Xiao ZG, Zou C, Zhang X, Wang Z, Wu D, Yu S, Chan FL. Nuclear receptor ERRα and transcription factor ERG form a reciprocal loop in the regulation of TMPRSS2:ERG fusion gene in prostate cancer. Oncogene 37: 6259–6274, 2018. doi: 10.1038/s41388-018-0409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang M, Zhao J, Xing L, Shi L. The association between angiotensin-converting enzyme 2 polymorphisms and essential hypertension risk: a meta-analysis involving 14,122 patients. J Renin Angiotensin Aldosterone Syst 16: 1240–1244, 2015. doi: 10.1177/1470320314549221. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 46: 586–590, 2020. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, thereceptor of SARS-CoV-2. bioRxiv 1–15, 2020. doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]