Abstract
Coevolutionary processes that drive the patterns of host–parasite associations can be deduced through congruence analysis of their phylogenies. Feather lice and their avian hosts have previously been used as typical model systems for congruence analysis; however, such analyses are strongly biased toward nonpasserine hosts in the temperate zone. Further, in the Afrotropical region especially, cospeciation studies of lice and birds are entirely missing. This work supplements knowledge of host–parasite associations in lice using cospeciation analysis of feather lice (genus Myrsidea and the Brueelia complex) and their avian hosts in the tropical rainforests of Cameroon. Our analysis revealed a limited number of cospeciation events in both parasite groups. The parasite–host associations in both louse groups were predominantly shaped by host switching. Despite a general dissimilarity in phylogeny for the parasites and hosts, we found significant congruence in host–parasite distance matrices, mainly driven by associations between Brueelia lice and passerine species of the Waxbill (Estrildidae) family, and Myrsidea lice and their Bulbul (Pycnonotidae) host species. As such, our study supports the importance of complex biotic interactions in tropical environments.
Keywords: cospeciation, feather lice, host switching, host–parasite associations, passerines, tropical ecology
Associations of lice and their avian hosts in the tropical rainforests of Cameroon are predominantly shaped by host switching. Despite the general incongruence between parasite and host phylogenies, the significant correlation between host and parasite phylogenetic distances suggests the prevalence of host switching to closely related hosts.
1. INTRODUCTION
Resolving the processes that drive the patterns of host–parasite associations is an essential goal of evolutionary parasitology and could contribute to our understanding of parasite distribution and biodiversity. New associations may be established following cospeciation, when host‐specific parasites speciate as a response to speciation of the host. If cospeciation events represent the prevailing source of new host–parasite interactions, the parasite phylogeny should mirror that of the host with respect to both topology and age of the nodes, referred to as Fahrenholz's rule (Eichler, 1948; Farenholz, 1913). On the other hand, parasites may also colonize new hosts via horizontal host switching, which may lead to incongruence in parasite and host phylogenies. While there are a number of potential sources of tree incongruence, for example, sorting events, including parasite extinction, duplication (intrahost speciation), and cohesion (failure to speciate), comparisons of host and parasite phylogenies can be used as a cue for revealing the role of cospeciation and host switching in a given host–parasite system (Page, 2003).
Feather lice represent a convenient, repeatedly used model for cospeciation studies as they are regularly host specific, their entire life cycle takes place on the body of a single host, their survival outside the host is limited, and they are predominantly transmitted vertically between parents and offspring (Price, Hellenthal, Palma, Johnson, & Clayton, 2003). Cospeciation analysis has frequently been applied to feather lice and their avian hosts (de Vienne et al., 2013; Table 1), the results indicating a wide spectrum of potential processes that drive the patterns of host–parasite associations. While incongruences between phylogenies of some feather lice and their hosts suggest that host–parasite associations were mainly established through host switching (e.g., Banks, Palma, & Paterson, 2006; Johnson, Adams, & Clayton, 2002; Weckstein, 2004), phylogenies of other louse groups strongly mirror the phylogenies of their hosts and hence advocate a predominant role for cospeciation (e.g., Page et al., 2004; Paterson, Wallis, Wallis, & Gray, 2000). In addition to differences in the methodological approaches used in cospeciation studies, various parasite species’ life‐history traits may affect the ratio between cospeciation and host switching during the formation of host–parasite associations (Clayton, Bush, & Johnson, 2004). For example, while parasite physiological adaptations to the host apparently support cospeciation (Clayton, Bush, Goates, & Johnson, 2003), phoresis (mechanical transport by louse flies) favors host switching (e.g., Harbison & Clayton, 2011; Johnson et al., 2002). On the other hand, host life‐history traits may affect the frequency and pattern of host switching. According to the “resource tracking hypothesis,” a parasite should switch to a new host on which it can continue to exploit the same resources (Timm, 1983). Exploitation of the new host may be thwarted, however, by a difference between the former and new host that increases with their phylogenetic distance (Engelstädter & Hurst, 2006). The importance of host relatedness has been demonstrated by "natural" experiments, in which lice fail to establish on brood parasites (e.g., cuckoos and indigobirds) despite close contact between the young brood parasites and foster parents in the nest (Balakrishnan & Sorenson, 2007; Brooke & Nakamura, 1998). Difference in body temperature, feather structure, or host immune and behavioral defenses may considerably lower parasite fitness, such that a host switch would result in an evolutionary dead end. Indeed, transfer experiments have shown that lice find it difficult to survive on alien host species (Clayton, Bush, et al., 2003; Tompkins & Clayton, 1999). On the other hand, as lice are parasites with limited dispersal ability, patterns of host shifting will be greatly affected simply by the probability of encountering new hosts (Clayton et al., 2004).
TABLE 1.
Cospeciation analysis of feather lice and their avian hosts
| Parasite | Host | Host speciations accompanied by parasite cospeciation | Significant amount of cospeciation events or phylogenetic congruence | Source |
|---|---|---|---|---|
| Alcedoecus (Ischnocera: Philopteridae) | Halcyoninae (Coraciiiformes) | 4 of 5 (80%) | Catanach et al. (2019) | |
| Alcedofulla (Ischnocera: Philopteridae) | Alcedininae (Coraciiformes) | 5 of 8 (62.5%) | Catanach et al. (2019) | |
| Alcedofulla (Ischnocera: Philopteridae) | Cerylinae (Coraciiformes) | 4 of 6 (66.6%) | † | Catanach et al. (2019) |
| Auricotes, Campanulotes, Coloceras, Physconelloides (Ischnocera: Philopteridae) | Columbiformes | 7 of 19 (36.8%) | † | Johnson and Clayton (2003) |
| Auricotes, Campanulotes, Coloceras, Physconelloides (Ischnocera: Philopteridae) | Columbiformes | 22 of 51 (43.1%) | † | Sweet, Boyd, and Johnson (2016) |
| Austrogoniodes (Ischnocera: Philopteridae) | Sphenisciformes | 4 of 17 (23.5%) | Banks et al. (2006) | |
| Subspecies of Austrophilopterus cancellosus (Ischnocera: Philopteridae) | Ramphastos toucans (Piciformes) | 1 of 10 (10%) | Weckstein (2004) | |
| Paraclisis (Ischnocera: Philopteridae) | Procellariiformes | 9 of 11 (81.8%) | † | Page et al. (2004) |
| Brueelia s.l. (Ischnocera: Philopteridae) | Several orders, mainly Passeriformes | 5 of 24 (20.8%) | Johnson et al. (2002) | |
| Brueelia s.l. (Ischnocera: Philopteridae) | Passeriformes | NA | † | Sweet et al. (2018) |
| Coloceras, Campanulotes, Physconelloides (Ischnocera: Philopteridae) | Columbiformes | 3 of 11 (27.3%) | Sweet et al. (2017) | |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 7 of 19 (36.8%) | † | Johnson and Clayton (2003) |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 3 of 12 (25%) | Clayton and Johnson (2003) | |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 7 of 22 (31.8%) | † | Clayton, Bush, et al. (2003) |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 7 of 27 (25.9%) | † | Johnson, Adams, Page, and Clayton (2003) |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 14 of 51 (27.4%) | † | Sweet et al. (2016) |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 1 of 12 (8.3%) | † | Sweet and Johnson (2016) |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 8 of 11 (72.7%) | † | Sweet et al. (2017) |
| Columbicola (Ischnocera: Philopteridae) | Columbiformes | 1 of 12 (8.3%) | † | Sweet and Johnson (2018) |
| Docophoroides (Ischnocera: Philopteridae) | Procellariiformes | 5 of 8 (62.5%) | Page et al. (2004) | |
| Episbates, Perineus, Harrisoniella (Ischnocera: Philopteridae) | Procellariiformes | 6 of 10 (60%) | Page et al. (2004) | |
| Halipeurus (Ischnocera: Philopteridae) | Procellariiformes | 4 of 4 (100%) | † | Paterson and Banks (2001) |
| Halipeurus (Ischnocera: Philopteridae) | Procellariiformes | 6 of 12 (50%) | Page et al. (2004) | |
| Halipeurus (Ischncoera: Philopteridae) | Procellariiformes | † | Hammer, Brown, Bugoni, Palma, and Hughes (2010) | |
| Paraclisis (Ischnocera: Philopteridae) | Procellariiformes | 9 of 11 (81.8%) | † | Page et al. (2004) |
| Pectinopygus (Ischnocera: Philopteridae) | Pelecaniformes | 10–12 of 17 (59%–71%) | † | Hughes, Kennedy, Johnson, Palma, and Page (2007) |
| Philopteridae (Ischnocera) | Procellariiformes and Sphenisciformes | † | Paterson and Gray (1997) | |
| Philopteridae (Ischnocera) | Procellariiformes and Sphenisciformes | 9 of 10 (90%) | † | Paterson et al. (2000) |
| Philopteridae (Ischnocera) | aquatic birds | 5 of 9 (55.5%) | Johnson, Kennedy, and Mccracken (2006) | |
| Physconelloides (Ischnocera: Philopteroides) | Columbiformes | 8 of 12 (66.7%) | † | Clayton and Johnson (2003) |
| Philopteridae (Ischnocera) | Many bird orders | 6 of 36 (16.7%) | † | de Moya et al. (2019) |
| Physconelloides (Ischnocera: Philopteroides) | Columbiformes | 3 of 10 (30%) | † | Sweet and Johnson (2018) |
| Austromenopon (Amblycera: Meniponidae) | Aquatic birds | 8 of 14 (57%) | † | Marshall (2002) |
| Colpocephalum complex (Phthiraptera: Amblycera) | Several orders of birds | † | Catanach, Valim, Weckstein, and Johnson (2018) | |
| Dennyus (Amblycera: Meniponidae) | Swifts (Apodiformes) | 4 of 6 (67%) | † | Page, Lee, Becher, Griffiths, and Clayton (1998) |
| Dennyus (Amblycera: Meniponidae) | Swifts (Apodiformes) | 13 of 21 (57%) | † | Clayton, Al‐Tamimi, and Johnson (2003) |
| Myrsidea (Amblycera: Meniponidae) | Catharus sp. (Passeriformes) | No congruence | Bueter et al. (2009) | |
| Myrsidea nesomimi (Amblycera: Meniponidae) | Mimus sp. (Passeriformes) | 1 of 6 (16%) | Štefka et al. (2011) |
More cospeciation events or stronger phylogenetic congruence than expected by chance is indicated by a dagger (†). Number of host speciations and accompanied parasite cospeciation are indicated when available as an original publication.
Presently, studies of feather lice and their hosts are strongly biased toward temperate regions. In the tropics, however, strongly dissimilar environments and host life‐history traits may result in different patterns of host–parasite associations. There are several factors that could favor host switching in tropical environment. Higher species diversity in the tropics may increase the probability of encountering new suitable hosts. At the same time, hippoboscid flies, which are known to transfer some louse species, are typically abundant in humid tropical regions (Sweet, Chesser, & Johnson, 2017). Tropical host populations are also typically less dense and abundant than temperate zone ones (e.g., Brown, 2014) and may not represent a reliable or abundant resource. This may favor generalist parasites in the tropics which makes cospeciation less likely (Combes, 2001; Vázquez, Poulin, Krasnov, & Shenbrot, 2005). Lice may also be significantly limited by abiotic factors (Malenke, Newbold, & Clayton, 2011; Moyer, Drown, & Clayton, 2002; Rai & Lakshminarayana, 1980); hence, the high humidity and temperatures of the tropics may increase louse survival off the host, thereby facilitating host switching. Conversely, the stable conditions prevalent in the tropics (i.e., less pronounced seasonality and glacial periods), along with the higher longevity of tropical birds (Snow & Lill, 1974; Wiersma, Muñoz‐Garcia, Walker, & Williams, 2007), could result in tighter parasite–host specialization, which would decrease the success of new host colonization.
The prevailing role of host switching in the tropics for forming feather lice and bird associations is supported by the study of Weckstein (2004), who found frequent host switching between sympatric toucan species in the feather louse subspecies of Austrophilopterus cancellosus. Similarly, Štefka, Hoeck, Keller, and Smith (2011) found that host switching strongly influences host–parasite associations in lineages of Myrsidea nesomimi and their hosts, the Galápagos mockingbirds. However, analogous studies from other tropical regions, or using taxonomically broader tropical feather lice samples, are missing.
In this study, we analyze the coevolutionary processes that drive the patterns of host–parasite associations in two feather louse groups and their hosts in tropical lowland and montane forests in Cameroon (West‐Central Africa). We assess the congruence of parasite and host phylogenies and attempt to find associations that contribute to the cophylogenetic structure.
2. MATERIALS AND METHODS
2.1. Sample collection
Birds were mist‐netted and blood‐sampled at two locations in the Cameroon mountains, a pristine tropical rainforest on the south‐western slopes of Mount Cameroon (4°08′ N 9°07′ E) at elevations of 350, 700 and 2,200 m above sea level (a.s.l.) in November and December 2013 and 2014, and a highly fragmented upper montane forest situated southeast of Big Babanki village in the Bamenda Mountains (6°05′ N 10°19′ E) at elevations of 2,000 and 2,200 m a. s. l. in January and February 2016. Each bird was kept in a new paper bag before parasite collection to prevent cross‐contamination. Lice were collected from the hosts using the “fumigation chamber method” (Clayton & Drown, 2001), followed by manual inspection of the host's head plumage. Lice were stored in ethanol and subsequently classified into genera using morphological criteria (Price et al., 2003).
From the pool of parasites collected, we selected the two most diverse groups of passerine lice within our sample: lice of the genus Myrsidea and the Brueelia complex (including Brueelia s. str., Guimaraesiella, Mirandofures and Sturnidoecus sensu Bush et al. (2016) and Gustafsson and Bush (2017)), each representing one of the two feather lice suborders, that is, Amblycera and Ischnocera, respectively.
Myrsidea lice are host‐specific parasites found predominantly on tropical passerine species (Figure 1), though they were found also on toucans and hummingbirds (Price et al., 2003). Including more than 380 mostly neotropical described species, Myrsidea is one of the most specious phthirapteran genera (Kolencik et al., 2018). They seem to be intolerant to low humidity (Bush et al., 2009), feed on host feathers, and partially utilize host body fluids, including blood (Marshall, 1981).
FIGURE 1.

Cryptospiza reichenovii and its Myrsidea parasite
On the contrary, lice of the Brueelia complex are common in both the tropics and temperate zones, and they are less host‐specific and, in addition to passerines, parasitize other bird groups, including Coraciiformes, Trogoniformes, and Piciformes (Gustafsson & Bush, 2017; Price et al., 2003). So far, over 426 species of this complex have been described (Gustafsson & Bush, 2017). Some Brueelia complex species are also capable of phoresis (horizontal transfer by hitchhiking) on louse flies (Hippoboscidae), which may eventually result in transport between different avian species due to the low specificity of louse flies (Keirans, 1975).
2.2. Molecular methods and species delimitation
Louse DNA was extracted using the Qiagen DNeasy Blood and Tissue Kit (Qiagen), following the manufacturer's protocol. To increase the DNA yield and preserve the parasite's morphological features, each louse was pierced with an entomological pin prior to incubation in proteinase K solution at 56°C for 36 hr. The exoskeleton was then removed and kept as a voucher specimen.
For species delimitation, we used partial sequences of cytochrome c oxidase subunit I (COI) of a single randomly chosen louse individual of each morphologically distinguishable group found on each infected bird. We calculated uncorrected pairwise nucleotide distances in MEGA version 7 (Kumar, Stecher, & Tamura, 2016) and utilized the web version (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) of Automatic Barcode Gap Discovery (ABGD) algorithm (Puillandre, Lambert, Brouillet, & Achaz, 2011) to identify barcoding gaps in the distribution of distances. The barcoding gap separating intra‐ and interspecies distances spanned 0.03–0.17 and 0.02–0.1 in Myrsidea and the Brueelia complex, respectively. Distance matrices, histograms of pairwise nucleotide distances, and COI trees are provided in File S1–S6. According to ABGD results, we classified lice into groups characterized by intragroup COI sequence distances up to 3%. The groups were considered as unique evolutionary units and are hereafter referred to as species. A single individual of each species was used for subsequent cophylogenetic analyses. A description of new species will be given elsewhere (Sychra O., Gajdosova M., Andresova P., Albrecht T. & Munclinger P.,unpublished data).
Partial sequences of COI, wingless (wg), and 18S rDNA were sequenced in lice of both groups. In addition, partial sequences of the elongation factor 1 alpha (EF1α) and hypothetical protein EOG9X3HC5 (hyp) were obtained from Myrsidea and the Brueelia complex, respectively (see Table 2 for primer details). PCR conditions were identical for all loci. Amplification began with 1 min of denaturation at 94°C, followed by 35 cycles of 30 s of denaturation at 92°C, 40 s of annealing at 54°C, and 90 s of elongation at 65°C, the final step comprising 10 min of final extension at 72°C. Owing to amplification problems, we used both original and redesigned forward primers for amplification of 18S rDNA and wingless (Table 2), which resulted in slightly shorter alignments. PCR products were purified using Thermo Fisher CleanSweep™ PCR Purification Reagent (Thermo Fisher Scientific) and Sanger sequenced from both sides using the same primers as for PCR. All sequences are deposited in GenBank under accession numbers MG765475–MG765497, MK031972–MK032011, MK032012–MK032034, and MK315054–MK315114.
TABLE 2.
Primers used for obtaining partial sequences of the elongation factor 1 alpha (EF1α) and hypothetical protein EOG9X3HC5 (hyp) in Myrsidea and Brueelia complex lice
| Locus | Primer name | Primer sequence (5′–3′) | Source |
|---|---|---|---|
| COI | L6625 | CCGGATCCTTYTGRTTYTTYGGNCAYCC | Hafner et al. (1994) |
| COI | H7005 | CCGGATCCACNACRTARTANGTRTCRTG | Hafner et al. (1994) |
| Wingless | Lep‐wg1a | GARTGYAARTGYCAYGGYATGTCTGG | Danforth, Brady, Sipes, and Pearson (2004) |
| Wingless | Lep‐wg2a | ACTICGCARCACCARTGGAATGTRCA | Danforth et al. (2004) |
| Wingless | Wg‐Myr‐F | ATGTCTGGRTCTTGCACGGTGAARAC | This paper |
| 18S rDNA | Ns1 | GTAGTCATATGCTTGTCTC | Barker, Whiting, Johnson, and Murrell (2003) |
| 18S rDNA | Ns2a | CGCGGCTGCTGGCACCAGACTTGC | Barker et al. (2003) |
| 18S rDNA | Ns‐Bru‐F | TGCATGTCTCAGTGCAAGCCGAAT | This paper |
| hyp | BR50‐181L | CTTGARCAATTRCAGAAAAAAGC | Sweet, Allen, and Johnson (2014) |
| hyp | BR50‐621R | GGRTTTTCWGGAGAYCTCATCC | Sweet et al. (2014) |
| EF1α | EF1‐For3 | GGNGACAAYGTTGGYTTCAACG | Danforth and Ji (1998) |
| EF1α | Cho10 | ACRGCVACKGTYTGHCKCATGTC | Danforth and Ji (1998) |
2.3. Genetic diversity and phylogenetic analysis
Sequences of COI, wingless, 18S rDNA, and either EF1α (Myrsidea) or hyp (Brueelia complex) were aligned separately by MAFFT online version 7 (Katoh & Standley, 2013). Secondary structure of 18S rDNA was taken into consideration during alignment construction. A concatenated alignment of 1677 bp (Myrsidea; File S7) and 1616 bp (Brueelia complex; File S8) was obtained from Geneious version 7.1.9 (http://www.geneious.com; Kearse et al., 2012). Optimal genetic models for alignment subsets (each gene and each of the three codon positions of the protein‐coding genes) were assessed using PartitionFinder 1.1.1 (Lanfear, Calcott, Ho, & Guindon, 2012; Table 3). Ricinus sp. collected from Platysteira laticincta and Philopteroides sp. collected from Cinnyris reichenowi were used as out‐groups for Myrsidea and for the Brueelia complex, respectively. Bayesian analysis was conducted using MrBayes version 3.2.6 (Huelsenbeck & Ronquist, 2001; Ronquist & Huelsenbeck, 2003) using the models found by PartitionFinder for particular alignment subsets. Two independent runs were performed, each lasting 2,000,000 generations with two chains, with tree sampling every 100 generations. The first 25% of the sampled trees were discarded as burn‐in. Both runs led to consensus trees with the same topology and almost identical support values (Figure 2). Maximum‐likelihood (ML) phylogenetic approach was applied to louse molecular data using RAxML 8.2.10 (Stamatakis 2014) with GTRGAMMA model and 1,000 bootstrap replicates. Bayesian and maximum‐likelihood analyses resulted in slightly different topologies in both Myrsidea and the Brueelia complex. Hence, we utilized the Bayesian trees, which were better resolved, for cospeciation analyses and ML trees are provided only in Files (S7 and S8). Phylogenies of the avian hosts were obtained as consensus trees generated in Geneious from 2,500 trees taken from the BirdTree database (www.birdtree.org), based on Ericson et al. (2006). The trees were subsequently compared with the recent passerine phylogeny (Oliveros et al., 2019; Selvatti, Gonzaga, & de Moraes Russo, 2015) and taxonomy in the Flux (TIF) checklist, which resulted in a positional correction of Kakamega poliothorax.
TABLE 3.
Models used for alignment subsets
| Alignment | Model | Alignment subset |
|---|---|---|
| Myrsidea | HKY + I+ G | COI 1st position |
| GTR + G | COI 2nd position | |
| K80 + I+G | COI 3rd position | |
| 18S rRNA | ||
| EF1α 3rd position | ||
| HKY + G | Wingless 1st position | |
| EF1α 2nd position | ||
| JC | EF1α 1st position | |
| Wingless 2nd position | ||
| Wingless 3rd position | ||
| Brueelia complex | HKY + I+G | COI 1st position |
| GTR + G | COI 2nd position | |
| hyp 2nd position | ||
| SYM + I | COI 3rd position | |
| Wingless 2nd position | ||
| Wingless 3rd position | ||
| 18S rRNA | ||
| HKY + G | Wingless 1st position | |
| HKY | hyp 1st position | |
| HKY + G | hyp 2nd position |
FIGURE 2.
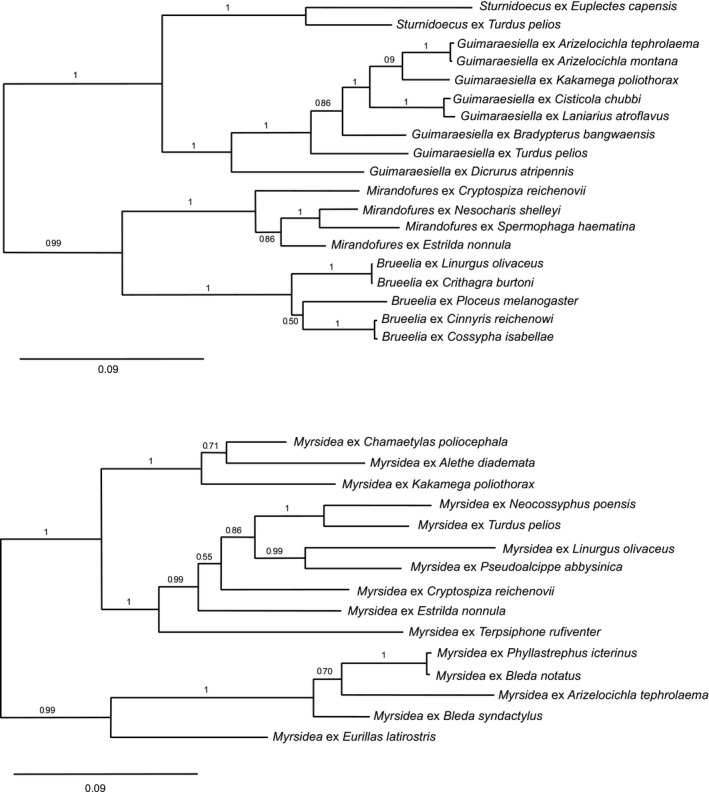
Bayesian phylogenetic trees of Myrsidea (based on COI, wingless, 18S rDNA, and EF1α) and the Brueelia complex (based on COI, wingless, 18S rDNA, and the hypothetical protein‐coding gene). Posterior probabilities are indicated at each node
2.4. Cospeciation analysis
Cophylogenetic history was reconstructed in Jane 4 (Conow, Fielder, Ovadia, & Libeskind‐Hadas, 2010), which accepts multihost parasitism. Jane implements a reconciliation algorithm to find the most optimal scenario of cophylogenetic past. By assigning costs to events which could possibly happen during the host–parasite cophylogenetic history (e. g., cospeciation, sorting events, lineage duplication, host switching, parasite's failure to diverge), Jane finds the least costly scenario that explains the observed situation. Event costs were left as default, that is, cospeciation 0, duplication 1, duplication with host switching 2, loss 1, and failure to diverge 1. The analyses were run for 30 generations with a population size of 1,300. To test whether the reconstructed solution was better than scenarios expected by chance, we compared the cost of the reconstructed scenario with costs of 999 pseudorandom replicates generated using the “random tip mappings” approach. Tanglegrams visualizing host–parasite associations and phylogenies were created in TreeMap3 (Charleston & Robertson, 2002). Codivergence between both groups was further tested using the PACo script (Balbuena, Míguez‐Lozano, & Blasco‐Costa, 2013), using the APE (Paradis, Claude, & Strimmer, 2004) and VEGAN (Dixon, 2003) packages in R version 3.5.1 (R core Team, 2017). PACo is a specific case of Procrustean analysis, which generally assesses the level of congruence between two (or more) ordinations of multivariate data sets. More specifically, PACo is designed to test for congruence between genetic divergence of hosts and parasites. First, we calculated cophenetic distances separately for hosts and parasites based on branch lengths in corresponding phylogenetic trees. Subsequently, principal coordinate analysis (PCoA) with Cailliez correction for negative eigenvalues was applied to extract orthogonal gradients (i.e., PCoA axes) from the two distance matrices. Scores for PCoA axes were used as an input for Procrustean superimposition assessing phylogenetic codivergence between hosts and parasites. Significance of the codivergence was tested by permutations of PCoA‐scaled distances (100,000 random rearrangements with significance level being set a priori as 0.05) as described in Balbuena et al. (2013). We also extracted squared residuals from the PACo fit to assess contributions of individual host–parasite links to the final Procrustean superimposition.
As cophenetic distances were not available for K. poliothorax host species due to correction of its position in the tree, we omitted this species and its parasites from the PACo analysis.
3. RESULTS
In total, 626 birds of 78 passerine species were examined for lice. Thirty‐nine birds were parasitized by Myrsidea lice (prevalence 6.2%) and 52 by lice of the Brueelia complex (prevalence 9.9%; File S12). Parasite loads were relatively low and varied between 1–38 for the Brueelia complex and 1–10 for Myrsidea. The majority of parasite species were found on a single host species; however, 1 of 14 Myrsidea species was found on two bird species, which involved hosts belonging to the same family (Figure 3). More cases of multihost parasites (4 of 15) were found within the Brueelia complex and involved associations with hosts from different families in two cases (Figure 4). One species from the Brueelia complex was even found on hosts of different orders, that is, the Bangwa Warbler (Bradypterus bangwaensis Delacour, 1943) from the Passeriformes and the Yellow‐spotted Barbet (Buccanodon duchaillui Cassin, 1856) from the Piciformes.
FIGURE 3.
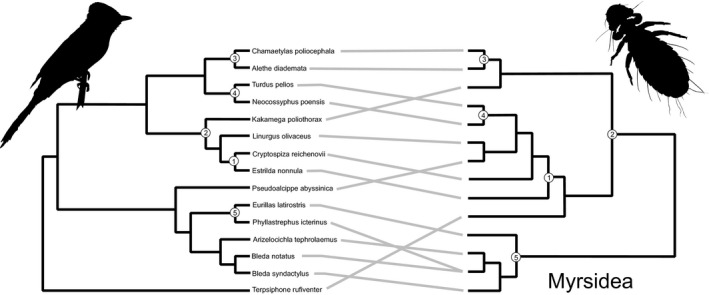
Tanglegram of passerine hosts (left) and Myrsidea parasites (right). The five cospeciation events found in Jane are represented by circles
FIGURE 4.
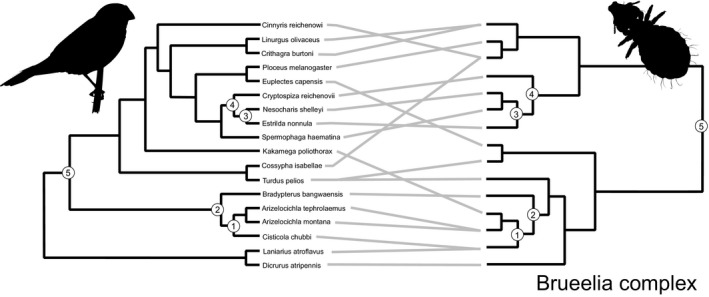
Tanglegram of passerine hosts (left) and Brueelia complex parasites (right). The five cospeciation events found in Jane are represented by circles
Cophylogenetic reconstruction of Myrsidea revealed the most parsimonious scenario to comprise 5 cospeciation events, 0 duplications, 8 host switches, 3 sorting events, and 1 failure to speciate. More than one‐third (36%) of host speciation events were followed by parasite cospeciation (Figure 3); however, almost 9% of random solutions resulted in scenarios with the same or lower overall cost, indicating that the reconstructed solution was not significantly better than solutions created by chance. Codivergence analysis of Myrsidea and its hosts in PACo indicated significant congruence of host and parasite distance matrices (the goodness‐of‐fit value was 14,155.98 with p < .001 based on 100,000 permutations; Figure 5); however, parasites of particular host groups contributed differently to the global codivergence fit (File S11). The association of Bulbuls (Pycnonotidae) and their parasites contributed strongly to the overall congruence pattern.
FIGURE 5.
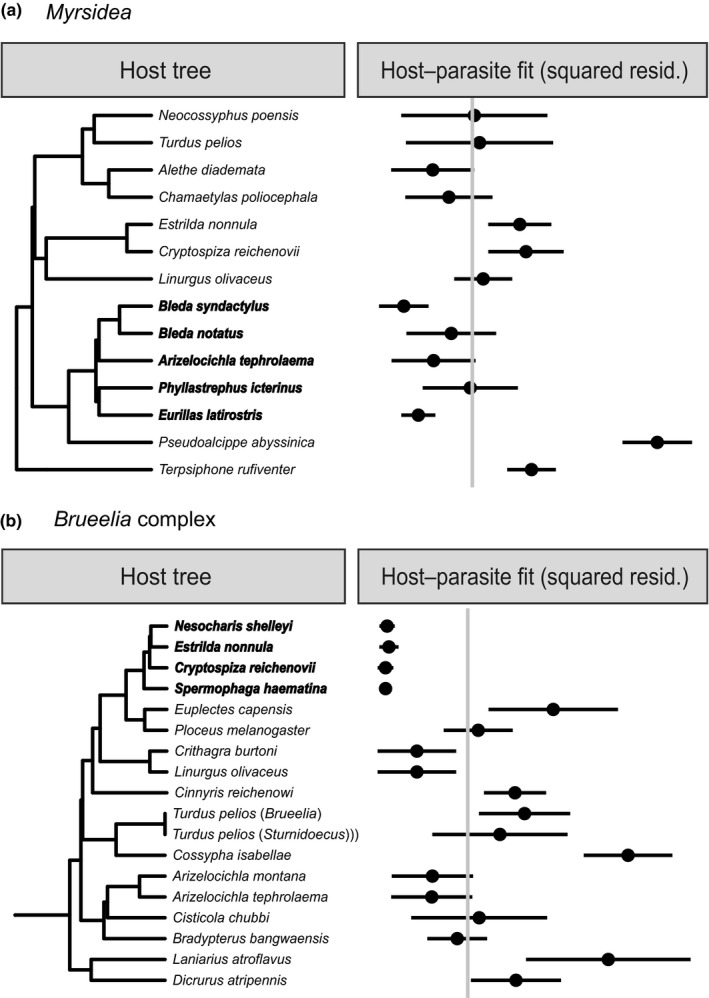
Contribution of individual host–parasite associations to the global codivergence signal based on Procrustes analysis of distance matrices between Myrsidea lice and their hosts (a) and Brueelia complex lice and their hosts (b). Squared residual 95% confidence intervals are shown. The dashed line indicates the median squared residual value. Bulbul (Pycnonotidae) host associations with Myrsidea lice and Waxbill (Estrildidae) host associations with Brueelia complex lice are shown in bold
The most parsimonious scenario found for the Brueelia complex and its hosts comprised 5 cospeciation events, 0 duplications, 9 host switches, 4 sorting events, and 4 failures to speciate (Figure 4). Hence, the frequency of parasite cospeciation (29%) appears to be slightly lower than in Myrsidea, though the overall cost of the scenario was significantly lower than expected by chance (i.e., Jane did not find the same or lower cost in any of 999 randomly permuted samples). There was a significant congruence between host and parasite distance matrices (the goodness‐of‐fit value was 34,205.59 with p < .001 based on 100,000 permutations; Figure 5), with the association between Waxbills (Estrildidae) and their parasites contributing most strongly to the overall congruence pattern (File S9).
4. DISCUSSION
Here, we analyze for the first time the host–parasite associations between lice and their avian hosts in the Afrotropical region. Several species of lice were detected on more than one host species; moreover, it should be noted that our sample was geographically restricted, and hence, the actual number of parasite multihost interactions may have been underestimated. The lower specificity of Brueelia complex lice, which were even found on phylogenetically distant hosts, can be at least partly ascribed to their ability to transfer horizontally between hosts (Keirans, 1975). We also found one Myrsidea species (7%) on two host species. Our study was limited to passerine hosts and lice of two model groups. Moreover, we matched only small fraction of the global diversity of the genus Myrsidea and the Brueelia complex. Deeper analyses of parasite–host interactions, preferably comparing the same groups of lice concurrently in the tropics and temperate regions, are needed to generalize our findings. However, both the multihost interactions and limited number of cospeciation events observed in this study are in good agreement with the general trend of greater parasite richness in the tropics (reviewed in Schemske, Mittelbach, Cornell, Sobel, & Roy, 2009). Under strict cospeciation scenarios, one would expect unique (one‐to‐one) parasite–host associations (Lyal, 1986). However, the number of host switches found in this study was higher than the number of cospeciation events, even though the event costs were set higher for host switching than cospeciation. Thus, our results are in agreement with previous evidence of limited cospeciation between lice and birds in other tropical regions, such as South America (Weckstein, 2004) and the Galapagos (Štefka et al., 2011).
Host switching was prevalent in the most parsimonious scenario for both the Brueelia complex and Myrsidea lice. Frequent host switching of Brueelia species has also been suggested in previous cospeciation analyses (Bueter, Weckstein, Johnson, Bates, & Gordon, 2009; Johnson et al., 2002) and is at least partly explained by horizontal transfer between hosts, enabled by hitchhiking of some Brueelia species on louse flies. However, horizontal transfer can also be mediated by other mechanisms, for example, lice may be transmitted via nest and nest‐site reuse, especially in hole nesters (Timm, 1983; Weckstein, 2004). Indeed, some of the birds in our study (Alethe diademata, Chamaetylas poliocephala and Cossypha isabellae) are known to be hole nesters (del Hoyo, Elliott, & Sargatal, 1997), and there is also evidence of nest and nest‐site reuse in some open nesters, for example, Turdus pelios, Apalis pulchra, and Nesocharis shelleyi (del Hoyo et al., 1997; del Hoyo, Elliott, & Sargatal, 1999, 2003). Additionally, some species (e.g., Cinnyris reichenowi, Cyanomitra olivacea, Estrilda nonnula, and Spermophaga haematina) incorporate feathers from a variety of other species into their nests (del Hoyo, Elliott, & Sargatal, 2003; del Hoyo, Sargatal, & Elliott, 2001). In this context, it should be noted that some Brueelia species have been shown to survive off the host for up to 200 hr (Dumbacher, 1999). Furthermore, the survival of lice during such horizontal transfers may be higher in the tropics due to increased temperature and humidity. Finally, lice may also be transmitted through direct contact between hosts in mixed‐species feeding flocks or at watering places.
The apparent incongruence between parasite and host phylogeny in Myrsidea lice and their hosts appears rather surprising. Myrsidea lice feed partially on blood (Marshall, 1981) and thus come into direct contact with the host's immune system. This may reinforce parasite coadaptation to a particular host and, as a result, lower the possibility of new host colonization. On the other hand, Clayton, Bush, and Johnson (2016) suggested limited cospeciation between lice and passerine hosts due to frequent sympatry with closely related species and the host's small body size. In the latter case, lice cannot maintain sustainable population sizes and thus face the risk of extinction. While cospeciation between passerines and their louse parasites has rarely been studied, the few analyses undertaken thus far mostly show substantial incongruence between their phylogenies (Bueter et al., 2009; Johnson et al., 2002; Štefka et al., 2011; but see Sweet et al., 2018), in accord with our own results. Further, the concept of risk of extinction on small‐bodied hosts fits well with our own findings, which suggest sorting as the prevailing event in the most parsimonious scenarios related to Myrsidea lice.
Despite the general incongruence between parasite and host phylogenies, PACo analysis showed a significant correlation between host and parasite phylogenetic distances, which may be at least partly interpreted through the prevalence of host switching to closely related hosts. The existence of such clade‐limited colonization has already been suggested, for example, in brood parasites of genus Vidua and their passerine hosts (Sorenson, Balakrishnan, & Payne, 2004) or in Monogenoidea (Platyhelminthes) and their Neotropical fish hosts (Braga, Razzolini, & Boeger, 2015). Presumably, limited phylogenetic distances between hosts also reflect sharing of host traits, which allows the parasite to utilize the same resources on a new host. As such, our results appear to be in accord with the “resource tracking hypothesis” (Timm, 1983). Nevertheless, the exact traits that facilitate host shifts remain unknown as related species tend to be similar in morphological, physiological, and behavioral features. On the other hand, congruence appeared to be higher in some host–parasite clades. Similar variation in host–parasite phylogenetic congruence has previously been recorded in Brueelia by Sweet et al. (2018). In our case, the congruence mainly concerned associations between Myrsidea lice and Bulbul (Pycnonotidae) hosts, and Brueelia complex lice and Waxbills (Estrildidae). Species within both these avian families are of similar size and body shape and have similar biology. They are also known to form flocks and sometimes even mixed‐species flocks. While our analysis suggested only one cospeciation event in the Bulbul clade with Myrsidea lice, the majority of host speciations were accompanied by parasite cospeciation in lice from the Brueelia complex and Waxbills. Hence, it would appear that congruence was established through different evolutionary processes in these two parasite–host association groups.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
Magdalena Gajdošová: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Visualization (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Oldřich Sychra: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Writing‐review & editing (equal). Jakub Kreisinger: Formal analysis (equal); Methodology (equal); Resources (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Ondřej Sedláček: Investigation (equal); Resources (equal); Writing‐review & editing (equal). Eric Djomo Nana: Resources (equal); Writing‐review & editing (equal). Tomáš Albrecht: Conceptualization (equal); Funding acquisition (equal); Investigation (equal); Project administration (equal); Resources (equal); Writing‐review & editing (equal). Pavel Munclinger: Conceptualization (equal); Formal analysis (equal); Funding acquisition (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Supervision (equal); Writing‐original draft (equal); Writing‐review & editing (equal).
Supporting information
File S1–S12
ACKNOWLEDGMENTS
We thank the Congo Basin Institute/UCLA for facilitating the research in Cameroon. We also greatly acknowledge the help of our colleagues and local guides in the field and the assistance of Zdeňka Csibreiová in the laboratory. This study was supported by the Czech Science Foundation (GA ČR), project no. 17‐24782S.
Gajdošová M, Sychra O, Kreisinger J, et al. Patterns of host–parasite associations in tropical lice and their passerine hosts in Cameroon. Ecol Evol. 2020;10:6512–6524. 10.1002/ece3.6386
DATA AVAILABILITY STATEMENT
DNA sequence data are deposited in NCBI GenBank under accession numbers MG765475–MG765497, MK031972–MK032011, MK032012–MK032034, and MK315054–MK315114. The alignments, trees, and distance matrices are uploaded as supplements.
REFERENCES
- Balakrishnan, C. N. , & Sorenson, M. D. (2007). Dispersal ecology versus host specialization as determinants of ectoparasite distribution in brood parasitic indigobirds and their estrildid finch hosts. Molecular Ecology, 16(1), 217–229. 10.1111/j.1365-294X.2006.03142.x [DOI] [PubMed] [Google Scholar]
- Balbuena, J. A. , Míguez‐Lozano, R. , & Blasco‐Costa, I. (2013). PACo: A novel procrustes application to cophylogenetic analysis. PLoS ONE, 8(4), e61048 10.1371/journal.pone.0061048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks, J. C. , Palma, R. L. , & Paterson, A. M. (2006). Cophylogenetic relationships between penguins and their chewing lice. Journal of Evolutionary Biology, 19(1), 156–166. 10.1111/j.1420-9101.2005.00983.x [DOI] [PubMed] [Google Scholar]
- Barker, S. C. , Whiting, M. , Johnson, K. P. , & Murrell, A. (2003). Phylogeny of the lice (Insecta, Phthiraptera) inferred from small subunit rRNA. Zoologica Scripta, 32(5), 407–414. 10.1046/j.1463-6409.2003.00120.x [DOI] [Google Scholar]
- Braga, M. P. , Razzolini, E. , & Boeger, W. A. (2015). Drivers of parasite sharing among Neotropical freshwater fishes. Journal of Animal Ecology, 84(2), 487–497. 10.1111/1365-2656.12298 [DOI] [PubMed] [Google Scholar]
- Brooke, M. D. L. , & Nakamura, H. (1998). The acquisition of host‐specific feather lice by common cuckoos (Cuculus canorus). Journal of Zoology, 244(2), 167–173. 10.1111/j.1469-7998.1998.tb00022.x [DOI] [Google Scholar]
- Brown, J. H. (2014). Why are there so many species in the tropics? Journal of Biogeography, 41, 8–22. 10.1111/jbi.12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueter, C. , Weckstein, J. , Johnson, K. P. , Bates, J. M. , & Gordon, C. E. (2009). Comparative phylogenetic histories of two louse genera found on Catharus thrushes and other birds. Journal of Parasitology, 95(2), 295–308. 10.1645/GE-1642.1 [DOI] [PubMed] [Google Scholar]
- Bush, S. E. , Harbison, C. W. , Slager, D. L. , Peterson, A. T. , Price, R. D. , & Clayton, D. H. (2009). Geographic variation in the community structure of lice on western scrub‐jays. Journal of Parasitology, 10–13. 10.1645/GE-1591.1 [DOI] [PubMed] [Google Scholar]
- Bush, S. E. , Weckstein, J. D. , Gustafsson, D. R. , Allen, J. , DiBlasi, E. , Shreve, S. M. , … Johnson, K. P. (2016). Unlocking the black box of feather louse diversity: A molecular phylogeny of the hyper‐diverse genus Brueelia . Molecular Phylogenetics and Evolution, 94, 737–751. 10.1016/j.ympev.2015.09.015 [DOI] [PubMed] [Google Scholar]
- Catanach, T. A. , Johnson, K. P. , Marks, B. D. , Moyle, R. G. , Valim, M. P. , & Weckstein, J. D. (2019). Two lineages of kingfisher feather lice exhibit differing degrees of cospeciation with their hosts. Parasitology, 146(8), 1083–1095. 10.1017/S0031182019000453 [DOI] [PubMed] [Google Scholar]
- Catanach, T. A. , Valim, M. P. , Weckstein, J. D. , & Johnson, K. P. (2018). Cophylogenetic analysis of lice in the Colpocephalum complex (Phthiraptera: Amblycera). Zoologica Scripta, 47(1), 72–83. [Google Scholar]
- Charleston, M. A. , & Robertson, D. L. (2002). Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Systematic Biology, 51(3), 528–535. 10.1080/10635150290069940 [DOI] [PubMed] [Google Scholar]
- Clayton, D. H. , Al‐Tamimi, S. , & Johnson, K. P. (2003). The ecological basis of coevolutionary history In Page R. D. M. (Ed.), Tangled trees: Phylogeny, cospeciation and coevolution (pp. 310–341). Chicago, IL and London, UK: The University of Chicago Press. [Google Scholar]
- Clayton, D. H. , Bush, S. E. , Goates, B. M. , & Johnson, K. P. (2003). Host defense reinforces host–parasite cospeciation. Proceedings of the National Academy of Sciences of the United States of America, 100(26), 15694–15699. 10.1073/pnas.2533751100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, D. H. , Bush, S. E. , & Johnson, K. P. (2004). Ecology of congruence: Past meets present. Systematic Biology, 53(1), 165–173. 10.1080/10635150490265102 [DOI] [PubMed] [Google Scholar]
- Clayton, D. H. , Bush, S. , & Johnson, K. P. (2016). Coevolution of life on hosts: Integrating ecology and history. Chicago, IL: The University of Chicago Press. [Google Scholar]
- Clayton, D. H. , & Drown, D. M. (2001). Critical evaluation of five methods for quantifying chewing lice (Insecta: Phthiraptera). Journal of Parasitology, 87(6), 1291–1301. 10.1645/0022-3395(2001)087[1291:CEOFMF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Clayton, D. H. , & Johnson, K. P. (2003). Linking coevolutionary history to ecological process: Doves and lice. Evolution, 57(10), 2335–2341. 10.1111/j.0014-3820.2003.tb00245.x [DOI] [PubMed] [Google Scholar]
- Combes, C. (2001). Parasitism (pp. 72–73). Chicago, IL: University of Chicago Press. [Google Scholar]
- Conow, C. , Fielder, D. , Ovadia, Y. , & Libeskind‐Hadas, R. (2010). Jane: A new tool for the cophylogeny reconstruction problem. Algorithms for Molecular Biology, 5(1), 16 10.1186/1748-7188-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danforth, B. N. , Brady, S. G. , Sipes, S. D. , & Pearson, A. (2004). Single‐copy nuclear genes recover Cretaceous‐age divergences in bees. Systematic Biology, 53(2). 10.1080/10635150490423737 [DOI] [PubMed] [Google Scholar]
- Danforth, B. N. , & Ji, S. (1998). Elongation factor‐1 alpha occurs as two copies in bees: Implications for phylogenetic analysis of EF‐1 alpha sequences in insects. Molecular Biology and Evolution, 15(3), 225–235. 10.1093/oxfordjournals.molbev.a025920 [DOI] [PubMed] [Google Scholar]
- de Moya, R. S. , Allen, J. M. , Sweet, A. D. , Walden, K. K. , Palma, R. L. , Smith, V. S. , … Johnson, K. P. (2019). Extensive host‐switching of avian feather lice following the Cretaceous‐Paleogene mass extinction event. Communications Biology, 2(1), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vienne, D. M. , Refrégier, G. , López‐Villavicencio, M. , Tellier, A. , Hood, M. E. , & Giraud, T. (2013). Cospeciation vs host‐shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytologist, 198(2), 347–385. 10.1111/nph.12150 [DOI] [PubMed] [Google Scholar]
- del Hoyo, J. , Elliott, A. , & Sargatal, J. (1997). Handbook of the birds of the world (vol. 4). Barcelona, Spain: Lynx edicions. [Google Scholar]
- del Hoyo, J. , Elliott, A. , & Sargatal, J. (1999). Handbook of the birds of the world (vol. 5). Barcelona, Spain: Lynx edicions. [Google Scholar]
- del Hoyo, J. , Elliott, A. , & Sargatal, J. (2003). Handbook of the birds of the world (vol. 7). Barcelona, Spain: Lynx edicions. [Google Scholar]
- del Hoyo, J. , Sargatal, J. , & Elliott, A. (2001). Handbook of the birds of the world (vol. 6). Barcelona, Spain: Lynx edicions. [Google Scholar]
- Dixon, P. (2003). VEGAN, a package of R functions for community ecology. Journal of Vegetation Science, 14(6), 927–930. 10.1111/j.1654-1103.2003.tb02228.x [DOI] [Google Scholar]
- Dumbacher, J. P. (1999). Evolution of toxicity in pitohuis: I. Effects of homobatrachotoxin on chewing lice (order Phthiraptera). The Auk, 116(4), 957–963. [Google Scholar]
- Eichler, W. (1948). Some rules in ectoparasitism. Journal of Natural History, 1(8), 588–598. [Google Scholar]
- Engelstädter, J. , & Hurst, G. D. (2006). The dynamics of parasite incidence across host species. Evolutionary Ecology, 20(6), 603–616. 10.1007/s10682-006-9120-1 [DOI] [Google Scholar]
- Ericson, P. G. , Zuccon, D. , Ohlson, J. I. , Johansson, U. S. , Alvarenga, H. , & Prum, R. O. (2006). Higher‐level phylogeny and morphological evolution of tyrant flycatchers, cotingas, manakins, and their allies (Aves: Tyrannida). Molecular Phylogenetics and Evolution, 40(2), 471–483. 10.1016/j.ympev.2006.03.031 [DOI] [PubMed] [Google Scholar]
- Fahrenholz, H. (1913). Ectoparasiten und Abstammungslehre. Zoologischer Anzeiger, 41, 371–374. [Google Scholar]
- Gustafsson, D. R. , & Bush, S. E. (2017). Morphological revision of the hyperdiverse Brueelia‐complex (Insecta: Phthiraptera: Ischnocera: Philopteridae) with new taxa, checklists and generic key. Zootaxa, 4313(1), 1–443. 10.11646/zootaxa.4313.1.1 [DOI] [Google Scholar]
- Hafner, M. S. , Sudman, P. D. , Villablanca, F. X. , Spradling, T. A. , Demastes, J. W. , & Nadler, S. A. (1994). Disparate rates of molecular evolution in cospeciating hosts and parasites. Science, 265(5175), 1087–1090. [DOI] [PubMed] [Google Scholar]
- Hammer, S. , Brown, R. , Bugoni, L. , Palma, R. L. , & Hughes, J. (2010). On the origin of Halipeurus heraldicus on Round Island petrels: Cophylogenetic relationships between petrels and their chewing lice. Molecular Phylogenetics and Evolution, 55(3), 1111–1120. 10.1016/j.ympev.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Harbison, C. W. , & Clayton, D. H. (2011). Community interactions govern host‐switching with implications for host–parasite coevolutionary history. Proceedings of the National Academy of Sciences of the United States of America, 108(23), 9525–9529. 10.1073/pnas.1102129108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J. P. , & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17(8), 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hughes, J. , Kennedy, M. , Johnson, K. P. , Palma, R. L. , & Page, R. D. (2007). Multiple cophylogenetic analyses reveal frequent cospeciation between pelecaniform birds and Pectinopygus lice. Systematic Biology, 56(2), 232–251. 10.1080/10635150701311370 [DOI] [PubMed] [Google Scholar]
- Johnson, K. P. , Adams, R. J. , & Clayton, D. H. (2002). The phylogeny of the louse genus Brueelia does not reflect host phylogeny. Biological Journal of the Linnean Society, 77(2), 233–247. 10.1046/j.1095-8312.2002.00107.x [DOI] [Google Scholar]
- Johnson, K. P. , Adams, R. J. , Page, R. D. , & Clayton, D. H. (2003). When do parasites fail to speciate in response to host speciation? Systematic Biology, 52(1), 37–47. 10.1080/10635150390132704 [DOI] [PubMed] [Google Scholar]
- Johnson, K. P. , & Clayton, D. H. (2003). Coevolutionary history of ecological replicates: Comparing phylogenies of wing and body lice to Columbiform hosts In Page R. D. M. (Ed.) Tangled trees: Phylogeny, cospeciation and coevolution (pp. 262–286). Chicago, IL and London, UK: The University of Chicago Press. [Google Scholar]
- Johnson, K. P. , Kennedy, M. , & McCracken, K. G. (2006). Reinterpreting the origins of flamingo lice: Cospeciation or host‐switching? Biology Letters, 2(2), 275–278. 10.1098/rsbl.2005.0427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , … Drummond, A. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirans, J. E. (1975). A review of the phoretic relationship between Mallophaga (Phthiraptera: Insecta) and Hippoboscidae (Diptera: Insecta). Journal of Medical Entomology, 12(1), 71–76. 10.1093/jmedent/12.1.71 [DOI] [PubMed] [Google Scholar]
- Kolencik, S. , Sychra, O. , Papousek, I. , Kuabara, K. M. , Valim, M. P. , & Literak, I. (2018). New species and additional data on the chewing louse genus Myrsidea (Phthiraptera: Menoponidae) from wild Neotropical Passeriformes (Aves). Zootaxa, 4418(5), 401–431. 10.11646/zootaxa.4418.5.1 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, R. , Calcott, B. , Ho, S. Y. , & Guindon, S. (2012). PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29(6), 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Lyal, C. H. C. (1986). Coevolutionary relationships of lice and their hosts: A test of Fahrenholz's Rule In Stone A. R., & Hawksworth D. L. (Eds.), Coevolution and systematics (pp. 76–91). Oxford, UK: Claredon Press. [Google Scholar]
- Malenke, J. R. , Newbold, N. , & Clayton, D. H. (2011). Condition‐specific competition governs the geographic distribution and diversity of ectoparasites. American Naturalist, 177(4), 522–534. 10.1086/658176 [DOI] [PubMed] [Google Scholar]
- Marshall, A. G. (1981). The ecology of ectoparasitic insects. London, UK: Academic Press. [Google Scholar]
- Marshall, I. K. (2002). Congruence and cospeciation: Morphological and molecular phylogenetics of the Amblycera (Phthiraptera). Doctoral dissertation. Glasgow, UK: University of Glasgow. [Google Scholar]
- Moyer, B. R. , Drown, D. M. , & Clayton, D. H. (2002). Low humidity reduces ectoparasite pressure: Implications for host life history evolution. Oikos, 97(2), 223–228. 10.1034/j.1600-0706.2002.970208.x [DOI] [Google Scholar]
- Oliveros, C. H. , Field, D. J. , Ksepka, D. T. , Barker, F. K. , Aleixo, A. , Andersen, M. J. , … Faircloth, B. C. (2019). Earth history and the passerine superradiation. Proceedings of the National Academy of Sciences of the United States of America, 116(16), 7916–7925. 10.1073/pnas.1813206116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. D. (2003). Introduction In Page R. D. M. (Ed.), Tangled trees: Phylogeny, cospeciation and coevolution (pp. 1–21). Chicago, IL and London, UK: The University of Chicago Press. [Google Scholar]
- Page, R. D. , Cruickshank, R. H. , Dickens, M. , Furness, R. W. , Kennedy, M. , Palma, R. L. , & Smith, V. S. (2004). Phylogeny of “Philoceanus complex” seabird lice (Phthiraptera: Ischnocera) inferred from mitochondrial DNA sequences. Molecular Phylogenetics and Evolution, 30(3), 633–652. 10.1016/S1055-7903(03)00227-6 [DOI] [PubMed] [Google Scholar]
- Page, R. D. , Lee, P. L. , Becher, S. A. , Griffiths, R. , & Clayton, D. H. (1998). A different tempo of mitochondrial DNA evolution in birds and their parasitic lice. Molecular Phylogenetics and Evolution, 9(2), 276–293. 10.1006/mpev.1997.0458 [DOI] [PubMed] [Google Scholar]
- Paradis, E. , Claude, J. , & Strimmer, K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics, 20(2), 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Paterson, A. M. , & Banks, J. (2001). Analytical approaches to measuring cospeciation of host and parasites: Through a glass, darkly. International Journal for Parasitology, 31(9), 1012–1022. 10.1016/S0020-7519(01)00199-0 [DOI] [PubMed] [Google Scholar]
- Paterson, A. M. , & Gray, R. D. (1997). Host‐parasite co‐speciation, host switching, and missing the boat In Clayton D. H., & Moore J. (Eds.), Host–parasite evolution: General principles and avian models (pp. 236–250). Oxford, UK: Oxford University Press. [Google Scholar]
- Paterson, A. M. , Wallis, G. P. , Wallis, L. J. , & Gray, R. D. (2000). Seabird and louse coevolution: Complex histories revealed by 12S rRNA sequences and reconciliation analyses. Systematic Biology, 49(3), 383–399. 10.1080/10635159950127303 [DOI] [PubMed] [Google Scholar]
- Price, R. D. , Hellenthal, R. A. , Palma, R. L. , Johnson, K. P. , & Clayton, D. H. (2003). Chewing lice: World checklist and biological overview. Illinois Natural History Survey Special Publication 24. x+ 501 pp. [Google Scholar]
- Puillandre, N. , Lambert, A. , Brouillet, S. , & Achaz, G. (2011). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology, 21, 1864–1877. 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- R Core Team (2017). R: A language and environment for statistical computing.Vienna, Austria: R Foundation for Statistical Computing; Retrieved from https://www.R‐project.org/ [Google Scholar]
- Rai, R. K. , & Lakshminarayana, K. V. (1980). A note on the in vitro studies of the chewing‐lice (Phthiraptera). In Workshop on Techniques of Parasitology of the Zoological Survey of India (p. 55). [Google Scholar]
- Ronquist, F. , & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12), 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Schemske, D. W. , Mittelbach, G. G. , Cornell, H. V. , Sobel, J. M. , & Roy, K. (2009). Is there a latitudinal gradient in the importance of biotic interactions? Annual Review of Ecology, Evolution and Systematics, 40, 245–269. 10.1146/annurev.ecolsys.39.110707.173430 [DOI] [Google Scholar]
- Selvatti, A. P. , Gonzaga, L. P. , & de Moraes Russo, C. A. (2015). A Paleogene origin for crown passerines and the diversification of the Oscines in the New World. Molecular Phylogenetics and Evolution, 88, 1–15. 10.1016/j.ympev.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Snow, D. W. , & Lill, A. (1974). Longevity records for some neotropical land birds. The Condor, 76(3), 262–267. 10.2307/1366339 [DOI] [Google Scholar]
- Sorenson, M. D. , Balakrishnan, C. N. , & Payne, R. B. (2004). Clade‐limited colonization in brood parasitic finches (Vidua spp.). Systematic Biology, 53(1), 140–153. 10.1080/10635150490265021 [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štefka, J. , Hoeck, P. E. , Keller, L. F. , & Smith, V. S. (2011). A hitchhikers guide to the Galápagos: Co‐phylogeography of Galápagos mockingbirds and their parasites. BMC Evolutionary Biology, 11(1), 284 10.1186/1471-2148-11-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet, A. D. , Allen, J. M. , & Johnson, K. P. (2014). Novel primers from informative nuclear loci for louse molecular phylogenetics (Insecta: Phthiraptera). Journal of Medical Entomology, 51(6), 1122–1126. 10.1603/ME13218 [DOI] [PubMed] [Google Scholar]
- Sweet, A. D. , Boyd, B. M. , & Johnson, K. P. (2016). Cophylogenetic patterns are uncorrelated between two lineages of parasites on the same hosts. Biological Journal of the Linnean Society, 118(4), 813–828. 10.1111/bij.12771 [DOI] [Google Scholar]
- Sweet, A. D. , Bush, S. E. , Gustafsson, D. R. , Allen, J. M. , DiBlasi, E. , Skeen, H. R. , … Johnson, K. P. (2018). Host and parasite morphology influence congruence between host and parasite phylogenies. International Journal for Parasitology, 48(8), 641–648. 10.1016/j.ijpara.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Sweet, A. D. , Chesser, R. T. , & Johnson, K. P. (2017). Comparative cophylogenetics of Australian phabine pigeons and doves (Aves: Columbidae) and their feather lice (Insecta: Phthiraptera). International Journal for Parasitology, 47(6), 347–356. 10.1016/j.ijpara.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Sweet, A. D. , & Johnson, K. P. (2016). Cophylogenetic analysis of New World ground‐doves (Aves: Columbidae) and their parasitic wing lice (Insecta: Phthiraptera: Columbicola). Molecular Phylogenetics and Evolution, 103, 122–132. 10.1016/j.ympev.2016.07.018 [DOI] [PubMed] [Google Scholar]
- Sweet, A. D. , & Johnson, K. P. (2018). The role of parasite dispersal in shaping a host–parasite system at multiple evolutionary scales. Molecular Ecology, 27(24), 5104–5119. 10.1111/mec.14937 [DOI] [PubMed] [Google Scholar]
- Timm, R. M. (1983). Fahrenholz's rule and resource tracking: A study of host‐parasite coevolution. Chicago, IL: University of Chicago Press. [Google Scholar]
- Tompkins, D. M. , & Clayton, D. H. (1999). Host resources govern the specificity of swiftlet lice: Size matters. Journal of Animal Ecology, 68(3), 489–500. 10.1046/j.1365-2656.1999.00297.x [DOI] [Google Scholar]
- Vázquez, D. P. , Poulin, R. , Krasnov, B. R. , & Shenbrot, G. I. (2005). Species abundance and the distribution of specialization in host–parasite interaction networks. Journal of Animal Ecology, 74(5), 946–955. 10.1111/j.1365-2656.2005.00992.x [DOI] [Google Scholar]
- Weckstein, J. D. (2004). Biogeography explains cophylogenetic patterns in toucan chewing lice. Systematic Biology, 53(1), 154–164. 10.1080/10635150490265085 [DOI] [PubMed] [Google Scholar]
- Wiersma, P. , Muñoz‐Garcia, A. , Walker, A. , & Williams, J. B. (2007). Tropical birds have a slow pace of life. Proceedings of the National Academy of Sciences of the United States of America, 104(22), 9340–9345. 10.1073/pnas.0702212104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1–S12
Data Availability Statement
DNA sequence data are deposited in NCBI GenBank under accession numbers MG765475–MG765497, MK031972–MK032011, MK032012–MK032034, and MK315054–MK315114. The alignments, trees, and distance matrices are uploaded as supplements.


