Summary
In a short period, many research publications that report sets of experimentally validated drugs as potential COVID-19 therapies have emerged. To organize this accumulating knowledge, we developed the COVID-19 Drug and Gene Set Library (https://amp.pharm.mssm.edu/covid19/), a collection of drug and gene sets related to COVID-19 research from multiple sources. The platform enables users to view, download, analyze, visualize, and contribute drug and gene sets related to COVID-19 research. To evaluate the content of the library, we compared the results from six in vitro drug screens for COVID-19 repurposing candidates. Surprisingly, we observe low overlap across screens while highlighting overlapping candidates that should receive more attention as potential therapeutics for COVID-19. Overall, the COVID-19 Drug and Gene Set Library can be used to identify community consensus, make researchers and clinicians aware of new potential therapies, enable machine-learning applications, and facilitate the research community to work together toward a cure.
Data Science Maturity: DSML 3: Development/Pre-production: Data science output has been rolled out/validated across multiple domains/problems
Graphical Abstract
Highlights
-
•
Collections of drug and gene sets relevant to COVID-19 research
-
•
Detailed comparison of results from six in vitro SARS-CoV-2 drug screens
-
•
Analysis of hits that up- or downregulate the ACE2 expression module
-
•
Machine-learning framework to further prioritize hits and other similar drugs
The Bigger Picture
The COVID-19 pandemic requires rapid response by the research community to develop vaccines and therapeutics. While the development of vaccines may take years, drug repurposing can offer pandemic mitigation much quicker. In vitro drug screening is the first step toward identifying and prioritizing potential safe therapeutics for COVID-19. However, these screens are done by different laboratories across the world using different methods. As a result, these screens produce different lists of hits. Here, we attempted to consolidate the results from these drug screens to find out whether consensus emerges. In addition, we utilized machine-learning methods to further predict and prioritize the validity of the hits from these drug screens. Such analysis identified molecular mechanisms that may explain how some of these drugs interfere with viral replication inside human cells. As more SARS-CoV-2 drug screens are published, a clearer picture of the most promising drug candidates is expected to emerge.
Kuleshov et al. developed a web-based platform that collects and presents drug and gene sets related to COVID-19 research. Analysis of the results from six in vitro drug screens by comparing the overlap among these screens shows that there is some unexpected overlap among them. The authors also use the hits from these screens to develop a machine-learning classifier that further prioritizes the hits and identifies a pharmacological theme that is shared among several hits.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus that causes the coronavirus disease (COVID-19). Globally, there are more than 21.5 million confirmed COVID-19 cases and ∼766,000 reported deaths (as of August 15, 2020). Many biomedical researchers have shifted their efforts to investigate different aspects of the coronavirus COVID-19 pandemic. One area of activity is computationally prioritizing and experimentally testing approved and experimental drugs for repurposing as candidate therapies for COVID-19. Drug-repurposing studies present a promising avenue for quickly offering a treatment because many of these drugs have known safety profiles. So far, drug-repurposing studies can be categorized into two groups, in vitro screens1, 2, 3, 4, 5, 6 and computational predictions. Computational predictions are mostly based on structural biology methods,7, 8, 9, 10 but some are based on network analysis and transcriptomics.11, 12, 13 Few studies have validated top computational predictions in cell-based assays.7,11,12 The lists of drugs mentioned in these studies can be analyzed for consensus while identified drugs can be grouped by their type.
At the same time, many researchers attempt to understand the molecular mechanisms of the SARS-CoV-2 virus life cycle. Much attention has been given to studies that use profiling with mass spectrometry proteomics and phosphoproteomics. These methods identify host proteins that interact with each of the SARS-CoV-2 proteins12 or differentially phosphorylated proteins before and after SARS-CoV-2 infection.14 Another important study produced RNA-sequencing gene expression signatures from various relevant human cell lines, ferret lungs, and human lung biopsies before and after SARS-CoV-2 infection.15 These are just a few examples of the many studies that produce gene sets that can be organized and compared. In the past, we have developed a crowdsourcing project whereby we asked the community to identify gene expression signatures from drug, gene, and disease perturbations.16 The collection of over 6,000 signatures that were collected with the help of more than 70 contributors from around the world enabled us to produce a useful database called CREEDS (https://amp.pharm.mssm.edu/CREEDS/). Similarly, for this project, we developed a crowdsourcing project to integrate drug and gene sets related to COVID-19 research collected with the assistance of the research community. The resource is delivered as a web-based platform that has already been accessed by >1,700 unique users.
Results
Analysis and Visualization of Consensus Drug and Gene Sets
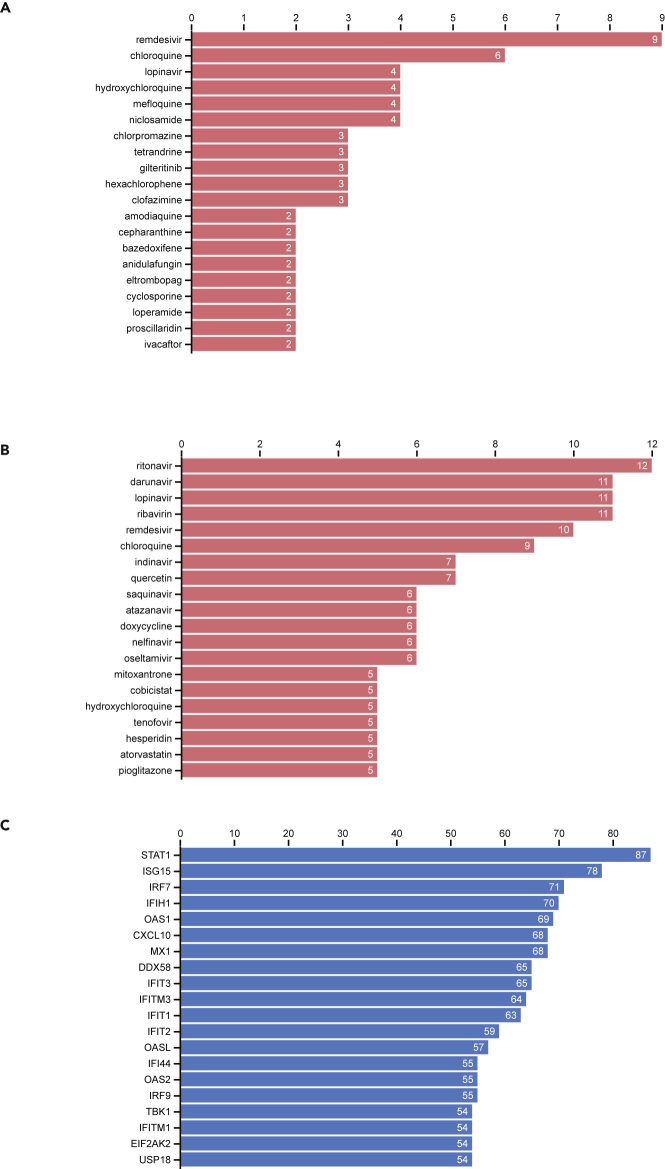
So far, we have collected 173 drug sets composed of 1,620 unique drugs, and 444 gene sets consisting of 18,676 unique human genes. These are presented to users via the COVID-19 Drug and Gene Set Library website in several sortable and searchable tables (Figure 1). The drug sets are subdivided into two categories: experimental (n = 26) and computational (n = 81). The top 20 most frequent drugs and genes across all sets are displayed in Figures 2A–2C. The experimental drugs, with most supportive evidence, are remdesivir, chloroquine, hydroxychloroquine, and mefloquine (Figure 2A). Although hydroxychloroquine, chloroquine, and remdesivir have received a lot of attention by the media and are tested in many clinical trials, mefloquine received far less attention. Mefloquine, just like hydroxychloroquine and chloroquine, is an anti-malaria drug.17 However, it has a different chemical structure and is known to act via different mechanisms. The top 20 most commonly computational predicted drugs include several known antivirals such as ritonavir, darunavir, lopinavir, and ribavirin (Figure 2B). This might be due to their pre-selection as candidates for computational docking. The top 20 most frequently submitted genes are all members of the innate immune response (Figure 2C). These genes include the typical interferon and cytokine response genes observed to be involved in the response of human cells to most pathogens.
Figure 1.
Screenshot from the Landing Page of the COVID-19 Drug and Gene Set Library
Figure 2.
Counts of Library Drugs and Genes
(A) Counts of most common drugs from the collection of experimental studies that reported lists of drugs that inhibit SARS-CoV-2.
(B) Counts of most common drugs from the collection of computational studies that reported lists of drugs that may inhibit COVID-19.
(C) Counts of most common genes from the collection of all gene sets in the library.
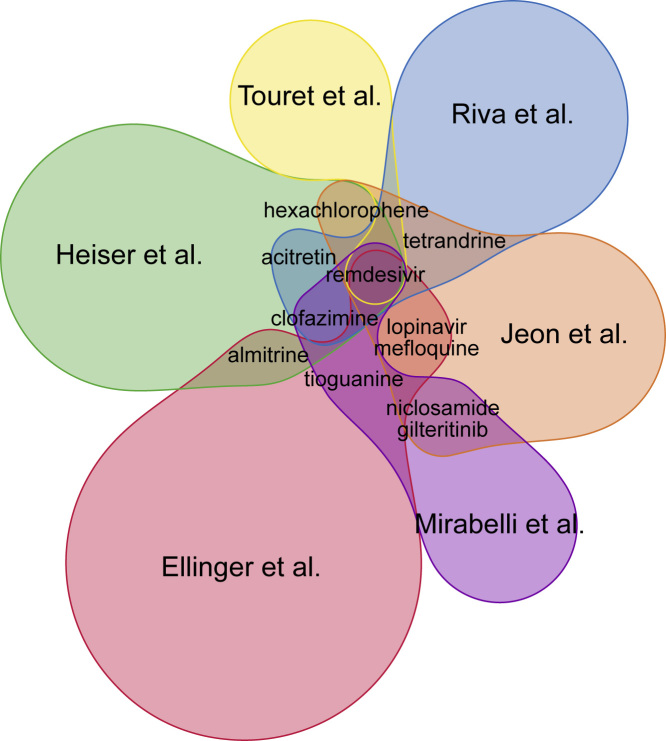
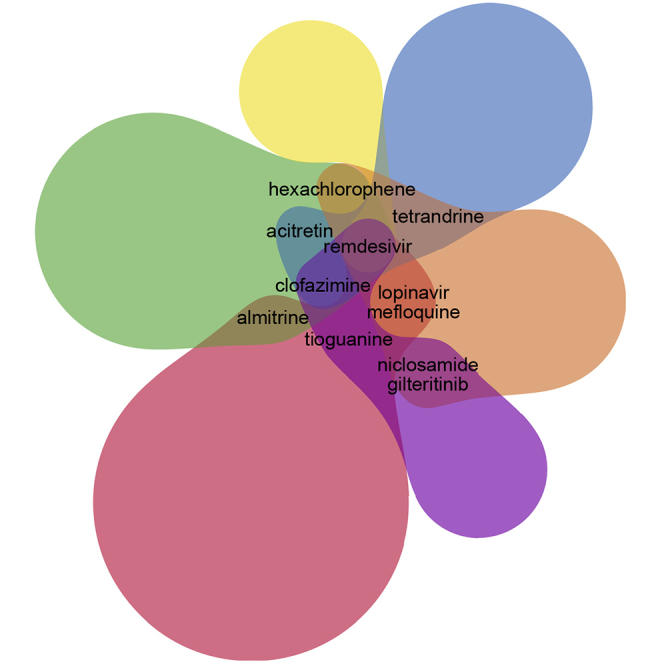
While most of the drug sets in the library are from studies that utilized computational methods, several key studies are from large-scale drug screens that include mostly Food and Drug Administration-approved drugs.1, 2, 3, 4, 5, 6 Using a Venn diagram, we compared the results from these six in vitro SARS-CoV-2 drug screen studies (Figure 3). Overall, there is little overlap across these screens, with only 11 drugs shared across two or more studies (Table 1). Namely, the drugs that appear as hits in more than one screen, in addition to remdesivir and chloroquine, are mefloquine, clofazimine, acitretin, gilteritinib, hexachlorophene, niclosamide, tetrandrine, tioguanine, and almitrine. Clofazimine is the only drug that appeared as a hit in three out of the six screens. Clofazimine is a drug used to treat leprosy, and its mechanisms of action suggest that it interferes with DNA synthesis.18 Acitretin is an anti-inflammatory second-generation retinoid that is used to treat severe psoriasis; it is a metabolite of etretinate.19 Almitrine is a drug that stimulates respiratory respiration by activating receptors of carotid bodies.20 It is used for the treatment of chronic obstructive pulmonary disease,21 and as such, it is relevant to COVID-19 symptoms. It should be noted that remdesivir appears as a hit in all six screens, but it was pre-selected as a positive control in half of the studies.
Figure 3.
Overlap across Six In Vitro Drug-Repurposing Screens for SARS-CoV-2 Inhibitors
Table 1.
Summary of the Six In Vitro COVID-19 Drug Screens Analyzed
| Authors | Journal | Hits | Method | Cells |
|---|---|---|---|---|
| Jeon et al.2 | Antimicrob. Agents Chemother. | 24 | inhibition assay | Vero cells |
| Touret et al.3 | bioRxiv | 12 | inhibition assay | Vero cells |
| Ellinger et al.4 | Research Square | 66 | inhibition assay | Caco-2 |
| Heiser et al.5 | bioRxiv | 36 | image-based assay | HRCE cells |
| Riva et al.6 | bioRxiv | 18 | inhibition assay | Vero cells |
| Mirabelli et al.1 | bioRxiv | 15 | image-based assay | Huh-1 cells |
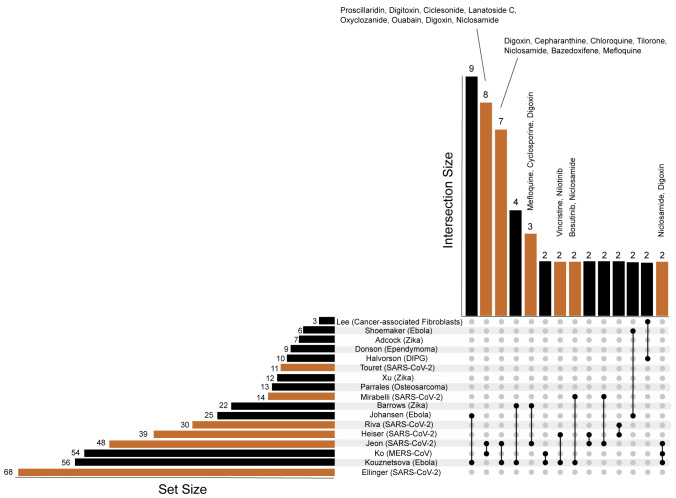
The small overlap among the screens can be due to various reasons including different assay types, cellular contexts, inclusion criteria, original library content, and different laboratory protocols. We carefully reviewed and compared the results from these screens including compounds screened, assays, drug concentrations used in screens, incubation, multiplicity of infection, and hit criteria. These aspects are summarized in Table S1, and the final drug sets from each study are provided in Table S2. This analysis enabled us to compare the IC50 values reported for those drugs that appeared in multiple screens (Tables 2 and S3). Overall, we observe relative consistency of reported IC50 values across screens. We also checked whether the hits from the six COVID-19 screens also appeared as hits in other previously published similar screens for other viruses and other diseases (Figure 4; Tables S4 and S5). We observe that the hits from the Jeon et al. study2 overlap with several other screens that reported potential antivirals for Zika,22 Ebola,23 and MERS.24 This might confirm the potentially good quality of the Jeon et al. screen. Next, we examined whether any of the drugs considered as hits across the six COVID-19 screens contain pan assay interference compounds (PAINS) chemotypes.25 To achieve this we compared the COVID-19 screen hits with a list of PAINS filters downloaded from ChEMBL.26 To check for possible PAINS among the hits, we checked whether any of the hits contain any one of the PAINS substructure chemotypes (Table S6). Six hits, namely eltrombopag, ketoconazole, phenazopyridine, posaconazole, SDZ-62-434, and Z-Leu-Val-Gly-diazomethylketone, out of 195 total hits contain such substructures, although this level of overlap is not statistically significant (Fisher's exact test, p = 0.57).
Table 2.
Compounds that Appear as Hits in Multiple Studies
| Drug | Touret et al.3 IC50 (μM) | Heiser et al.5 IC50 (μM) | Riva et al.6 IC50 (μM) | Ellinger et al.4 IC50 (μM) | Jeon et al.2 IC50 (μM) | Mirabelli et al.1 IC50 (μM) | Overlap |
|---|---|---|---|---|---|---|---|
| Remdesivir | 1.65 | x | 0.62 | 0.76 | 11.41 | 0.10 | 6 |
| Clofazimine | x | x | 0.08 | 3 | |||
| Acitretin | x | x | 2 | ||||
| Almitrine | x | 1.42 | 2 | ||||
| Gilteritinib | 6.76 | 0.22 | 2 | ||||
| Hexachlorophene | x | 0.90 | 2 | ||||
| Lopinavir | 19.11 | 9.12 | 2 | ||||
| Mefloquine | 14.15 | 4.33 | 2 | ||||
| Niclosamide | 0.28 | 0.14 | 2 | ||||
| Tetrandrine | 1.1 | 3 | 2 | ||||
| Tioguanine | 1.71 | 0.022 | 2 |
If available, the IC50 value calculated in each study is shown. Otherwise, the hit is marked by an “x.” Note that different studies use different assays and cell lines to measure dose response.
Figure 4.
UpSet Plot to Visualize the Hits from the Six COVID-19 Screens (Orange) and 11 Similar Non-COVID-19 Screens (Black)
ACE2 Up- or Downregulation Effects of Drug Hits?
To further explore the molecular effects of the positive hits from the six in vitro drug screens and to demonstrate the utility of the collected library, we developed a case study that asks whether the hits from the six screens up- or downregulate genes that are highly co-expressed with the ACE2 gene. ACE2 is the suspected cell surface receptor for SARS-CoV-2,27 and cells that do not express this gene have been shown to be less prone to SARS-CoV-2 infection. Since it is still undetermined whether it is desired to up- or downregulate the ACE2 expression module, we queried drugs from the published in vitro drug screen hits against the library of network-based cellular signatures (LINCS) L1000 data.28 We identified 61 drug hits from the six screens that have been profiled by L1000 assay. There are two drugs that significantly upregulate the ACE2 module (50 genes most correlated with ACE2 based on RNA-sequencing data from the Gene Expression Omnibus [GEO]) and one drug that significantly downregulates these genes after p-value correction (false discovery rate <0.1) (upregulated: homoharringtonine, 5.32 × 10−9; alvocidib, 1.58 × 10−5; downregulated: tazarotene, 5.77 × 10−2). Overall, 33 drugs on average upregulate the ACE2 module and 28 downregulate the module (Figure 5), suggesting that upregulating the ACE2 module might be more protective than harmful, which is counterintuitive. However, the relatively balanced division of drugs that induce or suppress this module makes this assertion inconclusive.
Figure 5.
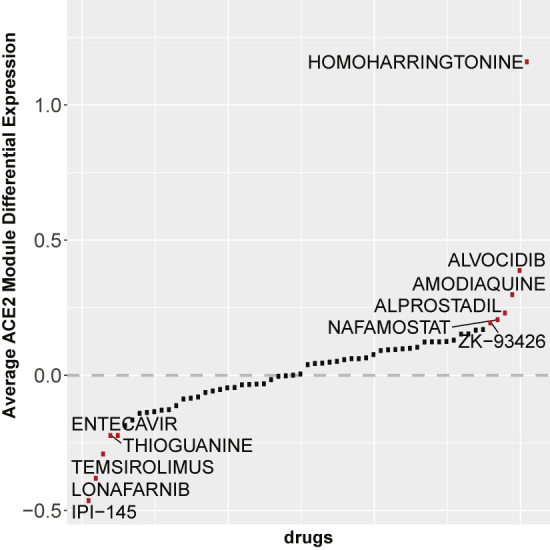
L1000 Profiled Drugs' Effects on the ACE2 Module
Average change in overall expression of the ACE2 co-expression module for 61 drug hits from the six published in vitro screens that also have L1000 profiling gene expression data.
Machine Learning to Rank Hits and Prioritize Other Candidates
The positive hits from the six COVID-19 drug screens can be used to train machine-learning models that can be used to prioritize the hits and suggest additional compounds that strongly share features with these hits. Using gene expression (GE) and chemical structure (CS) features of the hits and additional drugs and small molecules profiled via the L1000 assay, we implemented an Extra Trees (ET) classifier as a model that can be used to predict whether a drug is likely to inhibit SARS-CoV-2 in vitro. The ET classifier was able to predict hits from the six SARS-CoV-2 drug screens with an average area under the receiver-operating characteristic curve (AUROC) of 0.76 across cross-validation splits, suggesting that GE and CS features are overall predictive of the types of compounds that could inhibit SARS-CoV-2 infection (Figures 6A and 6B; Table S7). The lower value for the area under the precision recall curve can be explained by the class imbalance, which causes many non-hits to be ranked above known hits (Tables 3 and 4). Similar training and predictions were done using only GE features as input. In this case, the ET classifier achieved an average cross-validation AUROC of 0.66, which was lower than when CS features were also included but still statistically significant (Figures 6C and 6D; Table S8). It should be noted that the top-ranked predicted drugs are all from the same class of ATPase inhibitor cardiac drugs that have a similar structure and a similar GE signature effect in the L1000 assay. These drugs are over-represented in the Jeon et al. screen,2 so these initial results should be viewed with caution. The classifier also highly ranked lanatoside C, a drug identified as an active compound against MERS-CoV infection.24 This confirms that the machine-learning method could prioritize compounds that were missed by the six drug screens. In sum, this simple machine learning classification model is intended to demonstrate the potential for utilizing the drug sets collected for the library for machine-learning applications.
Figure 6.
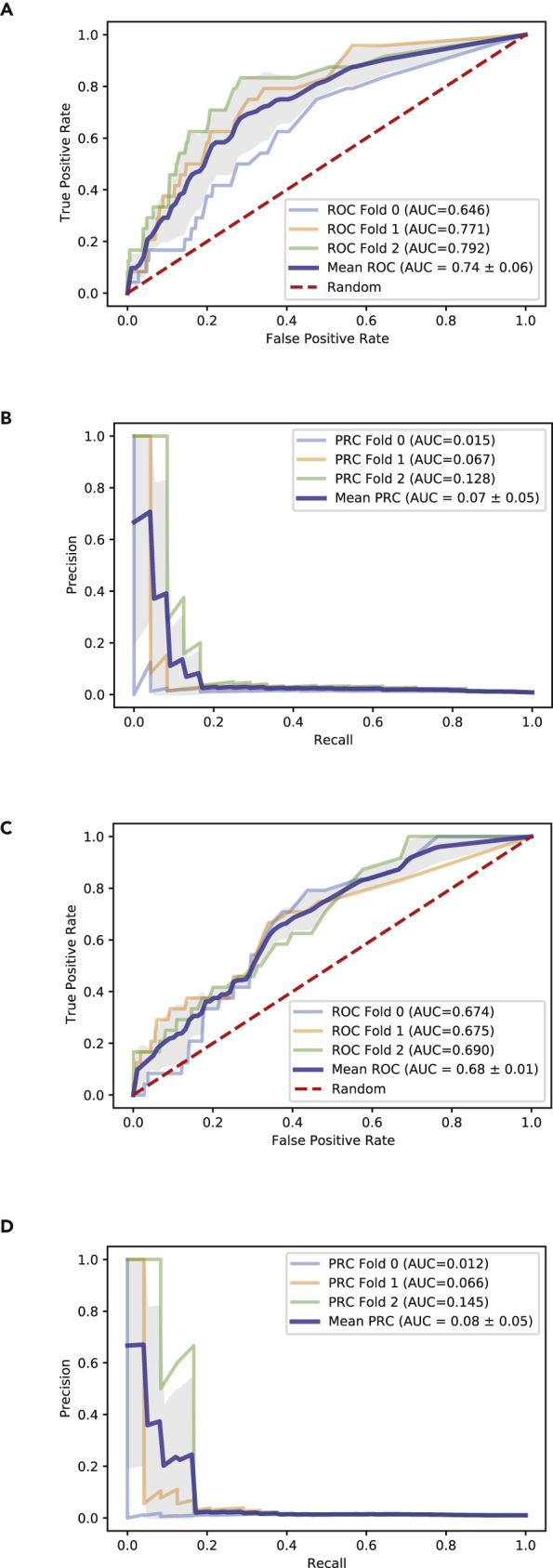
Evaluation of ET Classifiers Ability to Predict SARS-CoV-2 Inhibitors
(A) ROC curve for L1000 + MACCS-based predictions across cross-validation splits.
(B) PR curve for L1000 + MACCS-based predictions across cross-validation splits.
(C) ROC curve for L1000-only predictions across cross-validation splits.
(D) PR curve for L1000-only predictions across cross-validation splits.
Table 3.
Ranked Predictions for Screen Hits Based on L1000 + MACCS Input with p > 0.01
| Broad Pert. ID | Drug | Hit | Prediction Probability |
|---|---|---|---|
| BRD-K23478508 | digoxin | 1 | 0.8677456 |
| BRD-A34806832 | proscillaridin | 1 | 0.61186494 |
| BRD-A68930007 | ouabain | 1 | 0.48673511 |
| BRD-K13514097 | everolimus | 1 | 0.12437698 |
| BRD-K76674262 | omacetaxine mepesuccinate | 1 | 0.03459994 |
| BRD-K88538023 | oxiconazole | 1 | 0.02330089 |
| BRD-A29731977 | 17-hydroxyprogesterone-caproate | 1 | 0.02278362 |
| BRD-K59873006 | digitoxin | 1 | 0.02124448 |
| BRD-K06926592 | tretinoin | 1 | 0.02050656 |
| BRD-A80908310 | cloperastine | 1 | 0.01705306 |
| BRD-K96390176 | calcipotriol | 1 | 0.01589157 |
| BRD-K33882852 | ZK-93423 | 1 | 0.01579197 |
| BRD-K90699611 | acitretin | 1 | 0.01383878 |
| BRD-A10070317 | propranolol | 1 | 0.01347796 |
| BRD-A99117172 | hydroxychloroquine | 1 | 0.01282602 |
| BRD-A50287119 | sirolimus | 1 | 0.01201528 |
| BRD-K15409150 | penfluridol | 1 | 0.01139704 |
| BRD-A62025033 | temsirolimus | 1 | 0.011242 |
| BRD-K74501079 | azithromycin | 1 | 0.01123628 |
| BRD-K87909389 | alvocidib | 1 | 0.01096243 |
| BRD-K68392338 | ZK-93426 | 1 | 0.01075777 |
| BRD-K99964838 | bosutinib | 1 | 0.01062753 |
| BRD-A62184259 | cycloheximide | 1 | 0.01058221 |
| BRD-K12184470 | flunarizine | 1 | 0.01058221 |
| BRD-K17561142 | amiodarone | 1 | 0.01029646 |
| BRD-A64290322 | cyclosporin A | 1 | 0.0101906 |
| BRD-K68246049 | TTNPB | 1 | 0.01013295 |
| BRD-A91699651 | chloroquine | 1 | 0.01005631 |
Table 4.
Ranked Predictions for Top Additional Compounds Based on L1000 + MACCS Input
| Broad Pert. ID | Drug | Hit | Prediction Probability |
|---|---|---|---|
| BRD-A80502530 | cinobufagin | 0 | 0.70859567 |
| BRD-A76528577 | vincristine | 0 | 0.3044745 |
| BRD-K51290057 | SA-792709 | 0 | 0.25357778 |
| BRD-A68202111 | BRD-A68202111 | 0 | 0.1923075 |
| BRD-U19872303 | spiramycin | 0 | 0.186088 |
| BRD-A22783572 | vinblastine sulfate | 0 | 0.18156875 |
| BRD-K04010869 | prostaglandin A1 | 0 | 0.15031656 |
| BRD-K08486545 | cymarin | 0 | 0.14312159 |
| BRD-K01188359 | vinblastine | 0 | 0.12795675 |
| BRD-A57089740 | peruvoside | 0 | 0.12597708 |
| BRD-K67783091 | haloperidol | 0 | 0.10281666 |
| BRD-A44827100 | erythromycin | 0 | 0.10106031 |
| BRD-K36248164 | etretinate | 0 | 0.10086468 |
| BRD-A29322418 | canrenoic acid | 0 | 0.09826209 |
| BRD-K46523383 | pramocaine | 0 | 0.08840484 |
| BRD-A52650764 | ingenol | 0 | 0.08561776 |
| BRD-K80348542 | cephaeline | 0 | 0.08324268 |
| BRD-A29854054 | lorglumide | 0 | 0.0632068 |
| BRD-K03981224 | ethisterone | 0 | 0.06260333 |
| BRD-A90131694 | alclometasone | 0 | 0.06221619 |
| BRD-U66370498 | androstanol | 0 | 0.06103837 |
| BRD-K21667562 | AM 404 | 0 | 0.05919457 |
| BRD-A89434049 | sarmentogenin | 0 | 0.05852038 |
| BRD-A94810754 | ionomycin | 0 | 0.05814178 |
| BRD-A37501891 | BRD-A37501891 | 0 | 0.05203431 |
Discussion
Here we describe a platform created to collect drug and gene sets related to COVID-19 research using various methods of data accrual. Many top-ranked frequent genes that are associated with COVID-19 are part of the interferon pathway. This is consistent with our knowledge that type I (IFN-α, IFN-β) and type III (IFN-λ) interferon systems are the primary defense against viral infections. However, it was suggested that one of the evasion mechanisms by SARS-CoV-2 is to dampen the interferon response.15 It has been hypothesized that hyperinflammation in COVID-19 could drive disease severity and would be amenable to treatment with drugs that reduce inflammation.29,30 However, this remains controversial because the high level of antiviral response could be reflective of increased viral burden rather than an inappropriate host response.31 The most striking result from the meta-analysis applied to the content of the library is the limited overlap across drug screen studies. It is expected that experimental validation of drugs to inhibit SARS-CoV-2 in vitro will be more consistent. The inconsistency across these studies could be due to a need to produce results quickly because of the urgency for discovering potential treatments. Regardless, there is some interesting overlap that cannot be explained by artifacts such as PAINS chemotypes. Hence, there is an expectation that as more similar screens are published, the top most consistent leads will advance to animal models and human trials for further testing. To prioritize compounds that may treat COVID-19, some researchers have used the strategy of finding drugs that modulate genes related to ACE2 GE.32 We found few hits that also highly significantly up- or downregulate the genes most correlated with ACE2. However, it is inconclusive whether up- or downregulation of this module is beneficial. Finally, we have demonstrated how the positive hits across the screens can be pooled to develop machine-learning models that can further prioritize candidates based on direct experimental accumulating evidence about potential SARS-CoV-2 inhibitors.
It should be clear that the consensus analysis results should be viewed with caution. The most common drugs are not necessarily the most efficacious or promising treatments. At the same time, the most common genes may not be the most relevant to furthering COVID-19 research. It should be noted that not all drug sets and gene sets have equal weight in quality and relevancy. A list of computationally predicted drugs is not as useful toward identifying a therapy for COVID-19 when compared with a list of experimentally validated drugs. A list of upregulated genes after SARS-CoV-2 infection of cells may provide more useful information about the virus life cycle when compared with a list of genes returned from a PubMed search using the term SARS. Hence, the users of the data collected for the library should be aware of such limitations. With these limitations in mind, we hope that researchers will be able to better develop or refine their hypotheses by considering the information in the library.
In a period of rapid development of methods and data related to COVID-19 research, it is critical to provide the means to organize the accumulated information in a way that it can be summarized and reused. The COVID-19 Drug and Gene Set Library provides such utility. The library of drug and gene sets can be used to identify community consensus and make researchers and clinicians aware of the developments in new potential therapies as they become available, as well as allow the research community to work together toward a cure for COVID-19. However, it is important to note that while there are now many drugs that show promise in blocking SARS-CoV-2 in vitro, in vivo studies are needed before any of these drugs can be considered real original therapeutics.
Experimental Procedures
Resource Availability
Lead Contact
Further information and requests for digital resources should be directed to and will be fulfilled by the Lead Contact, Avi Ma'ayan (avi.maayan@mssm.edu).
Materials Availability
This study generated The COVID-19 Drug and Gene Set Library website available at: https://amp.pharm.mssm.edu/covid19/.
Data and Code Availability
All data collected for this project is made available via the website https://amp.pharm.mssm.edu/covid19. The data from the site can be accessed via API. The code behind the site is available on GitHub at https://github.com/maayanlab/covid19_crowd_library. The consensus analysis of the drugs that up- or downregulate the ACE2 module is available from https://github.com/maayanlab/covid19l1000. All code and data are provided openly under the Apache License version 2.0. The supporting tables are provided openly at Mendeley Data at https://data.mendeley.com/datasets/mjbygmkdt3/1 https://doi.org/10.17632/mjbygmkdt3.1.
Collecting Drug Sets from Publications that Describe SAR-CoV-2 Drug Screens
Since the emergence of the COVID-19 pandemic, thousands of new publications related to COVID-19 research have emerged in just a few months. We continually surveyed these publications to identify research articles that describe drug screens and manually extracted drug sets from these studies to populate the COVID-19 Drug and Gene Set Library database. We also submitted to the platform published drug sets from historical sources such as those from studies that listed drugs showing antiviral activity for other related viruses. To assist us with developing and maintaining the collection, we have received help from the research community by allowing researchers to upload drug and gene sets to the database. These submissions are manually evaluated before making them publicly available.
Collecting SARS Signatures from GEO with GEO2Enrichr, BioJupies, and GEN3VA
Gene expression signatures resulting from infection of different coronaviruses for different cell types and tissues, with expression data originating from the GEO database, were processed using the GEO2Enrichr33 and BioJupies,34 and stored on the GEN3VA platform.35 The entries were submitted to the COVID-19 crowdsourcing platform, with an upregulated and a downregulated gene set associated with each signature.
Collecting COVID-19-Related Gene Sets with Geneshot
Geneshot36 is a platform that can be used to convert PubMed searches into gene sets. Using Geneshot, gene sets associated with the search terms: SARS, SARS-CoV, MERS-CoV, ACE2, and TMPRSS2 were created using both the AutoRIF and GeneRIF37 options. Additionally, top COVID-19 drug-repurposing candidates reported in recent literature were included as search terms. Predictions of additional genes potentially associated with the genes directly co-mentioned with these terms were also added to the database. These predictions were based on five strategies: co-occurrence via AutoRIF, GeneRIF,37 Enrichr,38 or Tagger,39 and co-expression using data from ARCHS4.40
Developing the COVID-19 Gene and Drug Set Library Website
The COVID-19 Drug and Gene Set Library website has two sortable and searchable tables that list the drug and gene sets. Sorting can be based on the date of submission, alphabetical ordering, or list size. The tables are searchable via metadata terms such as title, authors, and descriptions, as well as via data search for specific drug or gene terms. Users can download each drug or gene set as well as the entire library. In addition, each gene set is provided with the option to perform gene set enrichment analysis with Enrichr,38 while genes are linked to Harmonizome41 for further interrogation. Similarly, drug sets can be analyzed with DrugEnrichr, a drug set enrichment analysis tool. The individual drugs that map to known compounds are linkable to their corresponding DrugBank landing pages.42 The website enables users to submit drug and gene sets related to COVID-19 research by completing a simple form. The form includes a dataset title, a URL source, and a description that explains how the set is relevant to COVID-19 research. The submitter is also provided with mechanisms to add additional metadata terms that can describe the cell type, tissue, organism, and other critical information about the submitted set. Users can specify the category of the additional metadata, allowing for a broad set of expanded annotations for each submitted set. Users can also submit their contact information; this information is kept private, but users can opt-in to make it public. Once a user submits a contribution to the site, their dataset is directed to a review queue in which we manually examine the validity and relevance of the contribution. The reviewing process enables an administrator to approve or reject the submitted set. If approved, the set is added to the database. To make it easy for contributors to submit multiple sets, users can access the site via API. The code behind the site is open source and available at https://github.com/maayanlab/covid19_crowd_library.
Expression Analysis of In Vitro Screen Hits
Drug sets extracted from the six in vitro screens1, 2, 3, 4, 5, 6 were matched to drugs profiled by the L1000 assay available from GEO: GSE92742.28 Average signatures for each drug were computed by taking the Z score mean for each gene. To quantify the average change in expression of genes co-expressed with ACE2, we obtained the top 50 genes that mostly co-express with ACE2 from the ARCHS4 resource.40 We then calculated the mean Z scores of the top 50 correlated genes to ACE2 and compared those values against a distribution calculated from sampling 50 random genes, repeatedly 10,000 times. The p values were calculated against the sampled distribution and corrected for multiple hypothesis testing by applying the Bonferroni correction method. The code behind this analysis is open source and available at https://github.com/maayanlab/covid19l1000.
Identifying Drug Sets from Previously Published Drug Screens for Other Diseases
To identify publications that describe similar in vitro drug screens from other contexts, we followed these steps. (1) We first queried PubMed for studies that contain the term [“drug screen” AND “in vitro”]. (2) The text from these studies was processed such that papers containing a table with drug names were saved for further manual inspection. (3) We then manually selected studies that performed drug screens comparable with the published screens for SARS-CoV-2. The study selection criteria required the identification of in vitro studies that included quantitative measures of many drugs' efficacy against a disease cell-based model.
Machine-Learning Approach to Prioritize Compounds Based on In Vitro Screens
A list of 195 drug hits from the six in vitro screens1, 2, 3, 4, 5, 6 (Table S1) was used as positives for applying a machine-learning method to prioritize these compounds and additional compounds. GE L1000 signatures for 19,777 drugs measuring the response of 978 landmark genes and their associated 166 MACCS molecular fingerprints were obtained from the SEP-L1000 project.43 The binary MACCS key association matrix was TF-IDF normalized to account for the frequency of different chemical structures. The dataset included 19,777 different drugs, of which 96 matched the 195 hits from the drug screens. After removing compounds from the library that appeared to be similar structurally, 8,787 compounds remained, of which 72 were hits. ET classifiers44 were trained to identify drug screen hits from the GE and CS features and evaluated using 3-fold cross-validation. Class weights were set inversely proportional to the class frequencies to address class imbalance. Otherwise, all ET parameters were the default Scikit-learn values.45 Feature selection was performed by recursive feature elimination to use 128 when both GE and CS data were used as features, or 64 features when only GE data were used. Additionally, prediction probabilities were calibrated across cross-validation splits.
Acknowledgments
We would like to thank Akira Mitsui, Russ Altman, Anne Carpenter, Pedro Bellester, and Tudor Oprea for contributing information about missing publications and contributing drug and gene sets to the library. This project is partially funded by NIH U54HL127624 and U24CA224260 awarded to A.M. and NIH grants U24AA025479 and F32AA028148 supporting the work of L.B.F.
Author Contributions
M.V.K., D.J.B.C., and J.E.E. developed the website; D.J.S., A.L., J.H., and A.M. performed the analyses; E.K., A.Bartal, A. Bailey, K.M.J., M.C., A.Z., and L.B.F. contributed drug and gene sets; A.M. initiated and managed the project; all authors contributed to writing the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: July 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.patter.2020.100090.
Supplemental Information
References
- 1.Mirabelli C., Wotring J.W., Zhang C.J., McCarty S.M., Fursmidt R., Frum T., Kadambi N.S., Amin A.T., O'Meara T.R., Pretto-Kernahan C.D. Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19. bioRxiv. 2020 doi: 10.1101/2020.05.27.117184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Touret F., Gilles M., Barral K., Nougairède A., van Helden J., Decroly E., de Lamberellie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. bioRxiv. 2020 doi: 10.1101/2020.04.03.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellinger B., Bojkova D., Zaliani A., Cinatl J., Claussen C., Westhaus S., Reinshagen J., Kuzikov M., Wolf M., Geisslinger G. Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection. Research Square. 2020 doi: 10.21203/rs.3.rs-23951/v1. [DOI] [Google Scholar]
- 5.Heiser K., McLean P.F., Davis C.T., Fogelson B., Gordon H.B., Jacobson P., Hurst B.L., Miller B.J., Alfa R.W., Earnshaw B.A. Identification of potential treatments for COVID-19 through artificial intelligence-enabled phenomic analysis of human cells infected with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.21.054387. [DOI] [Google Scholar]
- 6.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V. A large-scale drug repositioning survey for SARS-CoV-2 antivirals. bioRxiv. 2020 doi: 10.1101/2020.04.16.044016. [DOI] [Google Scholar]
- 7.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 9.Rensi S., Altman R.B., Liu T., Lo Y.C., McInnes G., Derry A., Keys A. Homology modeling of TMPRSS2 yields candidate drugs that may inhibit entry of SARS-CoV-2 into human cells. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12009582. [DOI] [Google Scholar]
- 10.Hijikata A., Shionyu-Mitsuyama C., Nakae S., Shionyu M., Ota M., Kanaya S., Shirai T. Knowledge-based structural models of SARS-CoV-2 proteins and their complexes with potential drugs. FEBS Lett. 2020;594:1960–1973. doi: 10.1002/1873-3468.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing J., Shankar R., Drelich A., Paithankar S., Chekalin E., Dexheimer T., Rajasekaran S., Tseng C.T.K., Chen B. Reversal of infected host gene expression identifies repurposed drug candidates for COVID-19. bioRxiv. 2020 doi: 10.1101/2020.04.07.030734. [DOI] [Google Scholar]
- 12.Ge Y., Tian T., Huang S., Wan F., Li J., Li S., Yang H., Hong L., Wu N., Yuan E. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. bioRxiv. 2020 doi: 10.1101/2020.03.11.986836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alakwaa F.M. Repurposing didanosine as a potential treatment for COVID-19 using single-cell RNA sequencing data. mSystems. 2020;5:e00297-20. doi: 10.1128/mSystems.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouhaddou, M., Memon, D., Meyer, B., White, K.M., Rezelj, V.V., Marrero, M.C., Polacco, B.J., Melnyk, J.E., Ulferts, S., Kaake, R.M., et al. The global phosphorylation landscape of SARS-CoV-2 infection Cell 182, 10.1016/j.cell.2020.06.034 [DOI] [PMC free article] [PubMed]
- 15.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Monteiro C.D., Jagodnik K.M., Fernandez N.F., Gundersen G.W., Rouillard A.D., Jenkins S.L., Feldmann A.S., Hu K.S., McDermott M.G. Extraction and analysis of signatures from the gene expression omnibus by the crowd. Nat. Commun. 2016;7:12846. doi: 10.1038/ncomms12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer K.J., Holliday S.M., Brogden R.N. Mefloquine. A review of its antimalarial activity, pharmacokinetic properties and therapeutic efficacy. Drugs. 1993;45:430–475. doi: 10.2165/00003495-199345030-00009. [DOI] [PubMed] [Google Scholar]
- 18.Yawalkar S., Vischer W. Lamprene (clofazimine) in leprosy. Lepr. Rev. 1979;50:135–144. doi: 10.5935/0305-7518.19790020. [DOI] [PubMed] [Google Scholar]
- 19.Guenther L.C., Kunynetz R., Lynde C.W., Sibbald R.G., Toole J., Vender R., Zip C. Acitretin use in dermatology. J. Cutan. Med. Surg. 2017;21:2s–12s. doi: 10.1177/1203475417733414. [DOI] [PubMed] [Google Scholar]
- 20.López-López J.R., Perez-Garcia M.T., Canet E., Gonzalez C. Effects of almitrine bismesylate on the ionic currents of chemoreceptor cells from the carotid body. Mol. Pharmacol. 1998;53:330–339. doi: 10.1124/mol.53.2.330. [DOI] [PubMed] [Google Scholar]
- 21.Mélot C., Naeije R., Rothschild T., Mertens P., Mols P., Hallemans R. Improvement in ventilation-perfusion matching by almitrine in COPD. Chest. 1983;83:528–533. doi: 10.1378/chest.83.3.528. [DOI] [PubMed] [Google Scholar]
- 22.Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R., Galarza-Muñoz G., McGrath E.L., Urrabaz-Garza R., Gao J. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouznetsova J., Sun W., Martínez-Romero C., Tawa G., Shinn P., Chen C.Z., Schimmer A., Sanderson P., McKew J.C., Zheng W. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg. Microbes Infect. 2014;3:e84. doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko M., Chang S.Y., Byun S.Y., Choi I., d'Alexandry A.L.P.H., Shum D., Min J.Y., Windisch M.P. Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19. bioRxiv. 2020 doi: 10.1101/2020.02.25.965582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 26.Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrián-Uhalte E. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faust S., Horby P., Lim W.S., Emberson J., Mafham M., Bell J.L., Linsell L., Staplin N.D., Brightling C.E., Ustianowski A. Effect of dexamethasone in hospitalized patients with COVID-19: preliminary report. medRxiv. 2020 doi: 10.1101/2020.06.22.20137273. [DOI] [Google Scholar]
- 31.Ritchie A.I., Singanayagam A. Immunosuppression for hyperinflammation in COVID-19: a double-edged sword? Lancet. 2020;395:1111. doi: 10.1016/S0140-6736(20)30691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cava C., Bertoli G., Castiglioni I. In silico discovery of candidate drugs against Covid-19. Viruses. 2020;12:404. doi: 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gundersen G.W., Jones M.R., Rouillard A.D., Kou Y., Monteiro C.D., Feldmann A.S., Hu K.S., Ma'ayan A. GEO2Enrichr: browser extension and server app to extract gene sets from GEO and analyze them for biological functions. Bioinformatics. 2015;31:3060–3062. doi: 10.1093/bioinformatics/btv297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torre D., Lachmann A., Ma'ayan A. BioJupies: automated generation of interactive notebooks for RNA-seq data analysis in the cloud. Cell Syst. 2018;7:556–561. doi: 10.1016/j.cels.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gundersen G.W., Jagodnik K.M., Woodland H., Fernandez N.F., Sani K., Dohlman A.B., Ung P.M.U., Monteiro C.D., Schlessinger A., Ma'ayan A. GEN3VA: aggregation and analysis of gene expression signatures from related studies. BMC Bioinformatics. 2016;17:461. doi: 10.1186/s12859-016-1321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lachmann A., Schilder B.M., Wojciechowicz M.L., Torre D., Kuleshov M.V., Keenan A.B., Ma'ayan A. Geneshot: search engine for ranking genes from arbitrary text queries. Nucleic Acids Res. 2019;47:W571–W577. doi: 10.1093/nar/gkz393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osborne, J.D., Lin, S., Kibbe, W.A., Zhu, L., Danila, M.I., and Chisholm, R.L. (2007) GeneRIF Is a More Comprehensive, Current and Computationally Tractable Source of Gene-Disease Relationships than OMIM. Bioinformatics Core, Northwestern University Technical Report.
- 38.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen L.J. Tagger: BeCalm API for rapid named entity recognition. bioRxiv. 2017 doi: 10.1101/115022. [DOI] [Google Scholar]
- 40.Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma'ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018;9:1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., Ma'ayan A. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016 doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Clark N.R., Ma’ayan A. Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics. 2016;32:2338–2345. doi: 10.1093/bioinformatics/btw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geurts P., Ernst D., Wehenkel L. Extremely randomized trees. Mach. Learn. 2006;63:3–42. [Google Scholar]
- 45.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. J. Machine Learn. Res. 2011;12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data collected for this project is made available via the website https://amp.pharm.mssm.edu/covid19. The data from the site can be accessed via API. The code behind the site is available on GitHub at https://github.com/maayanlab/covid19_crowd_library. The consensus analysis of the drugs that up- or downregulate the ACE2 module is available from https://github.com/maayanlab/covid19l1000. All code and data are provided openly under the Apache License version 2.0. The supporting tables are provided openly at Mendeley Data at https://data.mendeley.com/datasets/mjbygmkdt3/1 https://doi.org/10.17632/mjbygmkdt3.1.