Abstract
Background
High rates of dihydroartemisinin–piperaquine (DHA–PPQ) treatment failures have been documented for uncomplicated Plasmodium falciparum in Cambodia. The genetic markers plasmepsin 2 (pfpm2), exonuclease (pfexo) and chloroquine resistance transporter (pfcrt) genes are associated with PPQ resistance and are used for monitoring the prevalence of drug resistance and guiding malaria drug treatment policy.
Methods
To examine the relative contribution of each marker to PPQ resistance, in vitro culture and the PPQ survival assay were performed on seventeen P. falciparum isolates from northern Cambodia, and the presence of E415G-Exo and pfcrt mutations (T93S, H97Y, F145I, I218F, M343L, C350R, and G353V) as well as pfpm2 copy number polymorphisms were determined. Parasites were then cloned by limiting dilution and the cloned parasites were tested for drug susceptibility. Isobolographic analysis of several drug combinations for standard clones and newly cloned P. falciparum Cambodian isolates was also determined.
Results
The characterization of culture-adapted isolates revealed that the presence of novel pfcrt mutations (T93S, H97Y, F145I, and I218F) with E415G-Exo mutation can confer PPQ-resistance, in the absence of pfpm2 amplification. In vitro testing of PPQ resistant parasites demonstrated a bimodal dose–response, the existence of a swollen digestive vacuole phenotype, and an increased susceptibility to quinine, chloroquine, mefloquine and lumefantrine. To further characterize drug sensitivity, parental parasites were cloned in which a clonal line, 14-B5, was identified as sensitive to artemisinin and piperaquine, but resistant to chloroquine. Assessment of the clone against a panel of drug combinations revealed antagonistic activity for six different drug combinations. However, mefloquine-proguanil and atovaquone–proguanil combinations revealed synergistic antimalarial activity.
Conclusions
Surveillance for PPQ resistance in regions relying on DHA–PPQ as the first-line treatment is dependent on the monitoring of molecular markers of drug resistance. P. falciparum harbouring novel pfcrt mutations with E415G-exo mutations displayed PPQ resistant phenotype. The presence of pfpm2 amplification was not required to render parasites PPQ resistant suggesting that the increase in pfpm2 copy number alone is not the sole modulator of PPQ resistance. Genetic background of circulating field isolates appear to play a role in drug susceptibility and biological responses induced by drug combinations. The use of latest field isolates may be necessary for assessment of relevant drug combinations against P. falciparum strains and when down-selecting novel drug candidates.
Keywords: Drug combination, Exonuclease, Malaria, PfCRT, Piperaquine resistance, Plasmepsin
Background
In 2018, an estimated 228 million cases of malaria resulted in approximately 405,000 deaths globally [1]. Artemisinin-based combination therapy (ACT), composed of potent, short-acting artemisinin derivatives and longer-acting partner drugs, provide the first-line drug treatment for uncomplicated Plasmodium falciparum infection for most malaria endemic areas. However, the spread of drug resistance will require that current anti-malarial drugs be replaced with newer chemotherapeutic agents [2] or more effective drug combinations. Within a few years of dihydroartemisinin-piperaquine (DHA–PPQ) being recommended by the World Health Organization (WHO) as first-line therapy for uncomplicated P. falciparum malaria [3], the emergence of DHA–PPQ resistance was reported in Cambodia [4–7], Vietnam [8] and more recently in Thailand [9], where DHA–PPQ is still the first-line ACT for P. falciparum treatment in most provinces.
Artemisinin (ART) resistance is characterized by delayed parasite clearance times [10, 11] and is associated with mutations in the propeller domain of P. falciparum Kelch-13 gene (pfkelch13) [12–15]. ART resistance can also be confirmed by the Ring-stage Survival Assays (RSA) (in vitro RSA0−3h and ex vivo RSA) [12, 16]. The characterization of PPQ resistance with conventional ex vivo/in vitro dose response assay has been challenging, yielding wide IC50 values [5, 17]. The PPQ survival assay (PSA) has been developed, with a survival rate of more than 10% defining a PPQ resistant phenotype [18]. Genetic markers proposed as modulators of PPQ resistance include P. falciparum multidrug resistance 1 (pfmdr1) (PF3D7_0523000) [19–21], P. falciparum plasmepsin 2 (pfpm2) (PF3D7_1408000) [20–23], P. falciparum exonuclease (pfexo) (PF3D7_1362500) [20, 23, 24], and specific mutations on P. falciparum chloroquine resistance transporter (pfcrt) (PF3D7_0709000) [18, 25–27]. An association between in vitro PPQ-resistant isolates and single-copy pfmdr1 was found [6, 7, 18, 28]; however, not all single copy pfmdr1 isolates demonstrate in vitro PPQ resistance. Ansbro et al. [29] have shown that pfmdr1 amplifications were absent in all Cambodian P. falciparum samples from 2014 to 2015, which could reflect limited use of mefloquine during that period [20]. Two independent genome-wide association studies (GWAS) showed that amplification of the pfpm2 gene was associated with reduced PPQ sensitivity [20, 21]. Targeted gene disruption of either pfpm2 or pfpm3 in P. falciparum 3D7 background accounted for a slight decrease in PPQ IC50 values, and a significant increase in sensitivity to PPQ in a modified PSA [30]. However, the overexpression of pfpm2 and pfpm3 in the 3D7 genetic background did not alter the sensitivity of P. falciparum to PPQ, suggesting that the increase in pfpm2 copy number alone is not the sole modulator of PPQ resistance [31]. Silva et al. [32] have utilized gene editing and chemical inhibition to demonstrate that pfpm2 amplification contributed to PPQ resistance and that the background of the engineered parasites was necessary to gain a bimodal dose–response, in which a second peak of survival for a subset of the parasite is detected at higher PPQ concentrations [22]. It was also suggested that the initial selection of plasmepsin and pfmdr1 copy number variations developed a genetic background important for novel pfcrt mutations to emerge [32].
GWAS confirmed recrudescences from DHA–PPQ treatment failures identified a non-synonymous single nucleotide polymorphism (SNP) E415G substitution on an exonuclease encoding gene [20]. However, the role of E415G-Exo in mediating PPQ resistance, in the absence of other markers of resistance, is unclear. Specific novel mutations of the pfcrt gene have been shown to be associated with PPQ resistance since parasites with a variant of the Dd2 pfcrt allele, either T93S, H97Y, F145I, I218F, M343L, or G353V, have higher median PSA survival rates than those harboring the wildtype Dd2 allele [18, 26, 27], and a C350R substitution in the pfcrt gene resulted in decreased susceptibility to PPQ [33]. Dhingra et al. [27] showed that T93S and I218F-PfCRT mutations have increased in the past 5 years in Southeast Asia although showing an insignificant fitness cost compared to F145I-parasites.
To evaluate the proposed molecular markers of PPQ resistance associated with PPQ in vitro susceptibility, 17 parasite isolates were collected and successfully culture-adapted. Isolates were analysed for copy number variation of pfmdr1, pfpm2 and SNPs of pfkelch13, pfexo, and pfcrt, and correlated with survival assays and drug susceptibility phenotypes. In addition, drug combination assays were utilized to complete isobolographic analysis on circulating P. falciparum parasites and to establish in vitro values and susceptibilities for various drug combinations.
Methods
Culture adaptation and maintenance of Cambodian parasites
The P. falciparum samples were collected from areas of documented multidrug resistance in Cambodia (Clinicaltrials.gov NCT02297477) [34]. Culture adaptation of the parasites was performed by thawing cryopreserved material containing infected red blood cells that had been mixed with glycerol mixture solution. Parasites were maintained in fresh human erythrocytes (O+) in RPMI-1640 medium (Sigma), containing 15% AB+ human serum (heat inactivated and pooled) and supplemented with 25 mM HEPES, 25 mM sodium bicarbonate, and 0.1 mg/mL gentamycin. Human blood products (erythrocytes and serum) were obtained from the Thai Red Cross. Cultures were placed in modular incubator chambers and gassed with 5% CO2, 5% O2, 90% N2 gas and incubated at 37 °C.
Drug resistance genotyping
Parasite genomic DNA were extracted from 200 µL of whole blood using EZ1 DNA blood kit with automated EZ1 Advanced XL purification system or DNeasy® Blood and Tissue (QIAGEN, Valencia, CA, USA) as per the manufacturer’s instructions and stored at − 20 °C. T100TM Thermal Cycler (Bio-Rad Laboratories) was employed to evaluate the propeller domain of the P. falciparum kelch13 (pfkelch13) (amino acid residues 442–727) [12, 35], and P. falciparum exonuclease (pfexo) SNP at a codon corresponding to amino acid position 415 [20], while Master Cycler Nexus Gradient (Eppendorf) was used to detect the pfcrt SNP at codon corresponding to amino acid positions 93, 97, 145, 218, 343, 350, and 353 [26, 33]. Primers used to identify pfkelch13, pfexo, and pfcrt SNPs are shown in Additional file 1: Table S1. P. falciparum reference DNAs from 3D7 and W2 clones (Malaria Research & Reference Reagent Resource, Manassas, VA) were used as positive controls, and all samples were performed in duplicate.
Copy number variation assay
To determine copy numbers of pfmdr1 and pfpm2 gene, real-time quantitative PCR (qPCR) was performed on genomic DNA as previously described [21, 36, 37] with some modifications. For pfmdr1, amplification reactions were performed according to the TaqMan Real-time PCR methods using ABI 7500 Real-time PCR system (Applied Biosystems) with 200 nM of each forward and reverse primer (Additional file 2: Table S2) and 2 ng of DNA template while Rotor-Gene Q (QIAGEN, Valencia, CA) using Type-it® HRM™ kit was employed for pfpm2 [21]. The primers used were as previously described to amplify the following loci: pfmdr1 (PF3D7_0523000) and pfpm2 (PF3D7_1408000), respectively. For the housekeeping gene, β-tubulin (PF3D7_1008700), β-tubulin forward and reverse primers were designed and used as a reference control for all experiments with the same validated PCR conditions as target primers. Plasmodium falciparum 3D7 and Dd2 were used as references for single and multiple copy number of pfmdr1, respectively. All samples including the references clones were performed in duplicate. The average copy number values for each genes were calculated using 2−ΔΔCt method where ΔΔCt is [Ctpfmdr1 orpfpm2 − Ctpf β-tubulin] sample − [Ctpfmdr1 orpfpm2 − Ctpf β-tubulin] 3D7. Parasites with copy number greater than 1.5 copies for pfmdr1 [36] and 1.6 copies for pfpm2 [21] were interpreted to contain multiple copies of each gene.
In vitro 72-h drug susceptibility by Histidine rich protein 2 (HRP2)
Drug susceptibility test using HRP-2 ELISA to measure 50% or 90% inhibitory concentration (IC50 and IC90) was performed following previously published methods [17, 38]. Dried drug-coated plates containing antimalarial drugs dihydroartemisinin (DHA), artesunate (AS), mefloquine hydrochloride (MQ), quinine sulfate hydrate (QN), chloroquine diphosphate (CQ), lumefantrine (LUM), piperaquine tetraphosphate (PPQ), atovaquone (ATQ), doxycycline (DOX), artemisone (ATM), and cycloguanil (CYC) were prepared as described in Chaorattanakawee et al. [17, 39]. In vitro drug susceptibility testing was carried out for control reference clones (W2, D6, C2B) (Malaria Research & Reference Reagent Resource, Manassas, Vermont, USA), as described previously [39]. IC50s and IC90s were estimated by nonlinear regression analysis using GraphPad Prism version 6.0 program.
To determine a bimodal-dose response curve, the PPQ concentration (2.44 to 100,000 ng/mL) and the dilution series were increased from 8 to 24 points, according to the previously published report [22]. Culture-adapted clinical isolates were prepared in the similar manner as in in vitro drug susceptibility testing. The synchronized rings were grown for 72 h in the presence of different concentrations of PPQ (24-point dilution) in 96-well plates at 1.5% haematocrit, 0.5% starting parasitaemia in 0.5% Albumax RPMI 1640. Growth at 72 h was measured by HRP-2 ELISA. Assays were carried out in three biological replicates and the control reference clone W2 was tested along with each culture-adapted clinical isolate.
Ring-staged survival assay (RSA)
In vitro RSA0–3 h was performed on 0–3 h post-invasion rings obtained from selected culture-adapted clinical isolates following published methods [16] with slight modifications. The in vitro RSA0–3 h tests were carried out in sequence numeric order by the study team that was blinded to the results of molecular markers of drug resistance. Briefly, parasite cultures were tightly synchronized using 5% d-sorbitol and 75% Percoll to obtain 0 to 3-h post-invasion rings which were adjusted to 0.5–1% starting parasitaemia with a 2% haematocrit in culture media. (0.5% Albumax RPMI 1640 with 2.5% AB serum), and cultured in a 48-well microplate with 700 nM DHA and 0.1% DMSO in separate wells for growth control. The culture plate was then incubated for 6 h at 37 °C in modular incubator chambers and gassed with 5% CO2, 5% O2 and 90% N2 gas. Cells were then washed once, resuspended in drug-free medium, and cultured further for 66 h. Susceptibility to DHA was assessed microscopically on thin films by estimating the percentage of viable parasites, relative to control (% survival rate). Parasites were counted from 10,000 red blood cells, and two separate individuals served as independent slide reader. In case of difference greater than 20%, slides were examined by a third microscopist blinded to the results. For the controls, the RSA0–3 h was also performed on P. falciparum reference clones W2 (ART-sensitive control), IPC-4884 and IPC-5202 (BEI Resources, NIAID, NIH, USA) as ART-resistant control lines with reported % RSA survival value of 6.2% and 88.2%, respectively [16]. A survival rate > 1% was deemed resistant for RSA.
Piperaquine survival assay (PSA)
PSAs were performed on culture-adapted clinical isolates with 0–3 h ring-stage parasite cultures following a previously published method [18] in sequence numeric order by the study team that was blinded to the results of molecular markers. Briefly, parasite cultures were tightly synchronized using 5% d-sorbitol and 75% Percoll to obtain 0 to 3-h post-invasion. Synchronized ring parasites at 0.5–1% starting parasitaemia and 2% haematocrit were incubated with 200 nM PPQ or 0.5% lactic acid in water at 37 °C for 48 h in a 48-well microplate. The cultures were then washed once, re-suspended in drug-free medium, and cultured further for 24 h. Susceptibility to PPQ was assessed microscopically on thin films by estimating the percentage of viable parasites in the similar manner as RSA. A survival > 10% was deemed resistant to PPQ.
In vitro cloning of Cambodian P. falciparum
Parasite clones were obtained by the combination of limiting dilution cloning and plaque assay by plating a calculated 0.3 parasite per well in flat-bottomed 96-well microplate wells as described [40]. Wells containing single plaques were subsequently expanded into round-bottomed wells. The parasite clones were genotypically and phenotypically characterized. Once established, all clones were maintained in medium without any drug.
Drug combination assay
Fixed ratio combinations of various antimalarial drugs were performed as previously described with some modifications [41, 42]. Stock solutions of the drugs were prepared at 1 mg/mL in 70% ethanol for DHA, MQ, CQ, tafenoquine (TQ), proguanil (PG), and pyronaridine (PND), in 0.5% lactic acid for PPQ, and in dimethyl sulfoxide for ATQ. The solutions for each drug were combined in ratios of 1 + 1, 1 + 3, 3 + 1, 1 + 4, 4 + 1, and 1 + 2; with each drug also tested alone. 50 µL of single and combination drug solutions were then introduced into 96-well plates to give a row with two-fold serial dilutions. 200 µL of parasite culture with a final parasitaemia of 0.5% in a 2% haematocrit were added, and the test plates were incubated for 72 h at 37°C in modular incubator chambers and gassed with 5% CO2, 5% O2 and 90% N2 gas. Parasite growth was measured by HRP2 drug susceptibility testing as described above. The individual 50% fractional inhibitory concentrations (FIC50) were determined as previously described [43]. Isobolograms were constructed by plotting the FIC50 of drug A against the FIC50 of drug B for each of the six drug ratios. A concave curve indicated synergy, a straight line represented additivity and a convex curve indicated antagonism. To obtain numeric values for the interaction, results were expressed as the sum of the FIC50A and FIC50B. The sum FIC50 (ΣFIC50) values indicate the kinds of interaction as follows: synergism when ΣFIC50 ≤ 0.5; toward synergism when ΣFIC50 < 1; additive when ΣFIC50 = 1; toward antagonism when ΣFIC50 > 1; antagonism when ΣFIC50 ≥ 2 to 4. The IC50s of each drug in the test combination were standardized by allocating the value of 1 to each drug that was tested alone and prorated values for each fixed concentration ratio.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6.0 (GraphPad Software, Inc., San Diego, CA, USA). In vitro parasite susceptibility to each test drug was expressed as mean IC50s for all samples. The difference of the IC50 values between groups was assessed by nonparametric Kruskal–Wallis, Mann–Whitney or Dunn’s multiple comparison tests, as appropriate. Statistical significance was defined as a p value of < 0.05.
Results
Molecular genotypes of culture-adapted clinical isolates
Seventeen P. falciparum clinical isolates were successfully culture-adapted and underwent genotypic profiling. The parasite isolates were classified into 4 groups based on the molecular markers. Group 1, parasite isolates 14 and 17, contained K13-, Exo- and PfCRT-wild-type with a single copy of pfpm2. Group 2, parasite isolates 13, harboured C580Y-K13 mutation, Exo- and PfCRT-wild-type with a single copy of pfpm2. Group 3, parasite isolates 4 and 9, had C580Y-K13, E415G-Exo, and PfCRT mutations with a single copy of pfpm2. Group 4, parasite isolates 1, 2, 3, 5, 6, 7, 8, 10, 11, 12, 15, and 16, carried C580Y-K13, E415G-Exo, and PfCRT mutations with multiple copies of pfpm2 (Table 1).
Table 1.
Molecular genotyping of in vitro parasite samples
| Sample ID |
pfmdr1 CN |
K13 C580Y |
pfpm2 CN |
Exo E415G |
PfCRT | Group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T93S | H97Y | F145I | I218F | M343L | C350R | G353V | ||||||
| 1 | 0.72 | Y | 2.85 | G | T | Y | F | I | M | C | G | 4 |
| 2 | 0.96 | Y | 1.80 | G | T | H | I | I | M | C | G | 4 |
| 3 | 0.88 | Y | 2.13 | G | T | H | I | I | M | C | G | 4 |
| 4 | 0.90 | Y | 0.84 | G | T | H | F | F | M | C | G | 3 |
| 5 | 0.91 | Y | 2.39 | G | T | H | F | F | M | C | G | 4 |
| 6 | 0.89 | Y | 1.83 | G | T | H | I | I | M | C | G | 4 |
| 7 | 0.84 | Y | 1.85 | G | T | H | F | F | M | C | G | 4 |
| 8 | 0.89 | Y | 2.73 | G | T | H | I | I | M | C | G | 4 |
| 9 | 0.93 | Y | 1.48 | G | T | H | I | I | M | C | G | 3 |
| 10 | 0.96 | Y | 2.05 | G | T | H | I | I | M | C | G | 4 |
| 11 | 0.78 | Y | 2.22 | G | S | H | F | I | M | C | G | 4 |
| 12 | 0.80 | Y | 2.35 | G | T | H | F | I | M | C | G | 4 |
| 13 | 0.89 | Y | 1.00 | E | T | H | F | I | M | C | G | 2 |
| 14 | 0.84 | C | 1.04 | E | T | H | F | I | M | C | G | 1 |
| 15 | 0.80 | Y | 2.71 | G | T | H | F | F | M | C | G | 4 |
| 16 | 0.82 | Y | 2.48 | G | T | H | F | F | M | C | G | 4 |
| 17 | 0.66 | C | 1.08 | E | T | H | F | I | M | C | G | 1 |
Italic indicates either mutations or multiple copy number. A cut-off copy number of 1.5 and 1.6 are used to define pfmdr1and pfpm2 multiple copy number. Group 1 is for parasites containing WT-K13, WT-Exo, WT-PfCRT, and single copy number (CN) pfpm2. Group 2 is for parasites containing C580Y-K13, WT-Exo, WT-PfCRT, and single CN pfpm2. Group 3 is for parasites harboring C580Y-K13, E415G-Exo, Mut-PfCRT, and single CN pfpm2. Group 4 is for parasites having C580Y-K13, E415G-Exo, Mut-PfCRT, and multiple CN pfpm2
Correlation of ART- and PPQ-resistance markers with RSA and PSA phenotypes
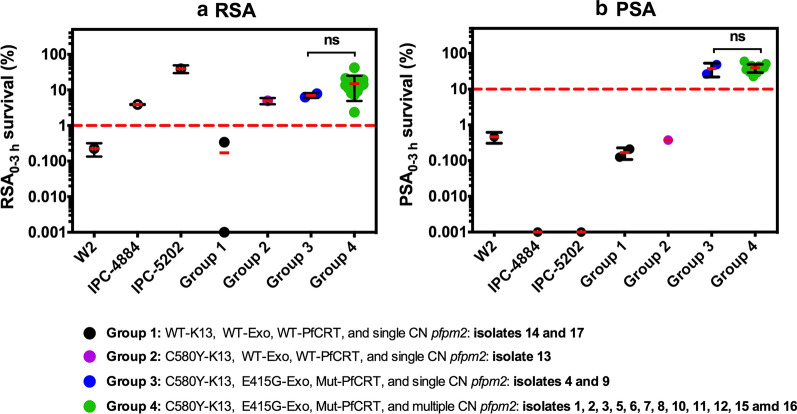
The phenotypic analysis was performed to determine whether the molecular markers were associated with ART- and PPQ-resistance. Parasites without the C580Y-K13 mutation (Group 1) exhibited % RSA survival rate of less than 1 (a cut-off for ART resistance), while parasites with the C580Y-K13 mutation (Group 2–-4) all had % survival rate of greater than 1, clearly demonstrating the correlation between the C580Y-K13 marker and ART resistance (Fig. 1a, Additional file 3: Table S3).
Fig. 1.
Survival assay for ART- and PPQ-resistance. a In vitro RSA0-3h survival rates for standard laboratory-adapted clones (W2 for an ART-sensitive control, IPC-4884 and IPC-5202 for ART-resistance control) and culture-adapted clinical isolates. The dashed line represents the 1% survival rate cut-off that differentiates ART-resistance (≥ 1%, red-dashed line) from ART-sensitive (< 1%) parasites in RSAs. b In vitro PPQ survival assay (PSA0–3 h) survival rates for standard laboratory-adapted clones (W2, IPC-4884 and IPC-5202 for PPQ-sensitive parasites) and culture-adapted clinical isolates. The dashed line represent the 10% survival rate cut-off that distinguishes PPQ-resistance (≥ 10%) from PPQ-sensitive (< 10%, red dashed line) parasites in PSAs. Two biological replicates were performed and survival rates are presented as mean ± S.D. Significance was determined using Mann–Whitney U test. Group 3 and Group 4 are compared. ns is not significant (p ≥ 0.05). Zero values of % survival rate were plotted as 0.001% in logarithmic scale
The relationship between pfpm2 copy number (CN), the E415G-Exo and novel PfCRT mutations and the % PSA survival of the parasites was assessed (Fig. 1b, Additional file 3: Table S3). PPQ-sensitive parasites P. falciparum W2, IPC-4884, and IPC-5202 were used as controls. Parasites in Group 1 and 2, lacking PPQ-resistance markers (pfpm2 multiple CN, the E415G-Exo and novel PfCRT mutations) showed % PSA survival of less than 10, indicative of PPQ-sensitivity. Parasites from Group 3 and 4, which contained E415G-Exo and PfCRT mutations, exhibited % PSA survival higher than 10, indicative of PPQ-resistance. Although parasites from group 3 had a single copy of pfpm2 and parasites from group 4 had multiple copies of pfpm2, there were no observed differences in % PSA survival between these two groups.
In vitro drug susceptibility and cell morphology of culture-adapted clinical isolates
To get a better understanding of cross-resistance, the parasites were tested against a panel of antimalarial drugs (Fig. 2 and Additional file 4: Table S4). The samples were categorized into PPQ-resistant (PPQ-R) (red bars, Groups 3 and 4) and PPQ-sensitive (PPQ-S) (blue bars, Groups 1 and 2) isolates. PPQ-S and PPQ-R parasites had similar IC50 values for AS, DHA, DOX, and ATQ. However, the PPQ-R parasites had higher IC50 for CYC and lower IC50 values for LUM, QN, CQ, and MQ. This suggests reciprocal drug resistant pattern for PPQ and the following drugs, LUM, QN, CQ, and MQ. To assess whether the PPQ resistant field isolates exhibited a second peak of survival around 0.1–10 μM (Bimodal dose–response) as was shown by Bopp et al. [22] in Cambodian parasites, the starting PPQ concentration was increased from 0.5 to 50 μM and the dilution series extended from 12 to 24 points (Fig. 3). PPQ-R parasites exhibited the second peak of survival around 78–20,000 ng/ml (or 0.08–21 µM), indicating a bimodal dose–response curve. Unlike PPQ-R parasites, PPQ-S parasites did not show the second peak, and their dose–response curves were similar to that of the reference clone W2.
Fig. 2.
In vitro P. falciparum susceptibility to multiple antimalarial drugs. Mean ± S.D IC50 values were calculated from 72-h dose–response assays for drugs designated in a–i. White bars represent standard laboratory-adapted clones, while blue and red bars indicate clinical-adapted parasites with PPQ sensitive or PPQ resistance, respectively. Three biological replicates were carried out for each sample. Statistically significant differences relative to isolate 17 are indicated with one (0.05 > p > 0.01) and two (p < 0.01) asterisks
Fig. 3.
PPQ dose–response curves and cell morphology for selected culture-adapted clinical isolates. Increasing the starting concentration and number of data points (24 points) for HRP2 ELISA dose–response curve provided a bimodal distribution of parasite response to PPQ exposure for PPQ-resistant parasites. The PPQ-sensitive W2 parasite (black line) is shown alongside the culture-adapted clinical isolates. The PPQ-sensitive clinical adapted parasites are shown in blue lines, whereas the PPQ-resistant clinical adapted parasites are represented in red lines. Data are shown as mean values from three biological replicates with S.D for isolates. Cell morphology of selected culture-adapted clinical isolates shows that isolate number 9 revealed distended, translucent DV (the red arrow) similar to the engineered parasite with PfCRT variants [26]. The scale bar, 50 µm
Ross et al. [26] has shown previously that pfcrt-edited Dd2 parasites developed a distended and translucent DV phenotype during the development from mid-trophozoites to mid-schizonts. This trait was specific to the pfcrt-edited Dd2 with F145I-, M343L- and G353V-PfCRT mutations and a single copy of pfpm2 but not observed in the PPQ-resistant Cambodian lines PH1008-C (multiple copies of pfpm2 and M343L-PfCRT) or PH1263-C (multiple copies of pfpm2 and H97Y-PfCRT) [26]. To validate if this phenotype could be observed with PPQ resistant parasites collected from the field, schizont morphology was examined. Among the parasites analysed only isolate 9 exhibited the swollen and translucent DV (Fig. 3). This isolate contained F145I-PfCRT and E415G-Exo mutations but a single copy of pfpm2. Parasite isolate 4 had a similar pattern of PPQ-resistance markers (except I218F-PfCRT instead of F145I-PfCRT) but no distended DV was observed. Parasites carrying both an F145I-PfCRT mutation and multiple copies of pfpm2 (such as parasite isolates 3 and 6) did not show an altered DV morphology. This is similar to the observation found in the PPQ-resistant Cambodian lines PH1008-C and PH1263-C [26].
Cloning of Cambodian P. falciparum isolate
Clinical isolates of P. falciparum are a genetically heterogeneous population of parasites. To obtain stable strains of the parasites for long term experiments, a rapid method of cloning was developed using a combination of limiting dilution and plaque assay [40]. Several attempts were carried out to clone 8 Cambodian P. falciparum isolates (isolates 3, 4, 6, 9, 12, 14, 15, and 17) and while all of the selected samples generated a single plaque after 7 days, only clones from isolate 14 could be expanded. After 1 month, four clones from isolate 14 were established including 14-B5, 14-C6, 14-C7, and 14-F5. These clones were genotypically and phenotypically characterized and compared to parent isolate and standard lab clones P. falciparum 3D7 and W2 (Additional file 5: Figure S1). All 4 clones possessed the identical genotypes to the parent isolate, and the RSA and PSA survival assays reflected sensitivity to ART and PPQ (Additional file 5: Figure S1).
Drug susceptibility profiles of the clones and parent isolate are illustrated in Fig. 4 and Additional file 6: Table S5. Compared to 3D7 (CQ-sensitive), all clones and the parent isolate exhibited high IC50s toward CQ similar to that of W2 (CQ-resistance). MQ IC50s for the clones were much higher than for W2 isolate (MQ-sensitive), and comparable to IC50 of D6 (MQ-resistance, IC50-MQ = 130.8 ± 15.96 nM). Collectively, based on the drug susceptibility profile and survival assays all four clones (14-B5, 14-C6, 14-C7, and 14-F5) were classified as ART- and PPQ-sensitive, but CQ-resistant and having reduced MQ sensitivity.
Fig. 4.
Drug susceptibility of P. falciparum Cambodian isolate 14 before and after cloning against DHA, MQ, CQ, and PPQ. The W2 and 3D7 strains of P. falciparum were used as controls. Statistically significant differences relative to 3D7 and isolate 14 are indicated in black and red asterisks, respectively with 0.05 > p > 0.01 for one asterisk and p < 0.01 for two asterisks. Abbreviation, ns is for not significant difference (p ≥ 0.05)
Drug combination testing of P. falciparum 14-B5
Since a new clone from parasite field isolates was successfully obtained, this clone was evaluated for sensitivity against a diverse array of drug combinations utilizing the HRPII-ELISA to establish synergistic, additive, and antagonistic in vitro anti-malarial drug interactions. As validation controls, the fixed ratio combinations of DHA–PPQ, CQ–CQ, and ATQ–PG were first tested against P. falciparum 3D7, W2, D6, C2B, and IPC-5202 strains as well as 14-5B clone. Table 2 and Additional file 7: Table S6 represent summary of drug interaction and the ΣFIC50s of tested fixed drug ratio combination, respectively. DHA–PPQ revealed antagonistic interactions, as reported previously [44]. CQ–CQ, serving as an experimental drug combination control, showed the additive interaction while ATQ–PG revealed a synergistic interaction as previously reported by Co et al. [42].
Table 2.
Summary of drug interaction in asexual stages of different P. falciparum strains
| Drug Combination | P. falciparum strain | |||||
|---|---|---|---|---|---|---|
| 3D7 | W2 | D6 | C2B | IPC-5202 | 14-B5 | |
| DHA–PPQ | Antagonistic | Antagonistic | Antagonistic | Antagonistic | Antagonistic | Antagonistic |
| CQ–CQ | Additive | Additive | Additive | Additive | Additive | Additive |
| ATQ–PG | Synergistic | Synergistic | Synergistic | Synergistic | Synergistic | Synergistic |
| PND–ATQ | Antagonistic | Antagonistic | Antagonistic | Toward Antagonistic | Antagonistic | Antagonistic |
| MQ–ATQ | Toward Antagonistic | Antagonistic | Toward Antagonistic | Toward Synergistic | Toward Antagonistic | Antagonistic |
| TQ–ATQ | Toward Synergistic | Toward Antagonistic | Toward Synergistic | Toward Synergistic | Toward Antagonistic | Antagonistic |
| PND–PG | Toward Antagonistic | Toward Synergistic | Additive | Toward Synergistic | Toward Synergistic | Antagonistic |
| MQ–PG | Toward Synergistic | Toward Synergistic | Toward Synergistic | Toward Synergistic | Toward Synergistic | Toward Synergistic |
| TQ–PG | Synergistic | Toward Antagonistic | Toward Synergistic | Toward Synergistic | Additive | Antagonistic |
ΣFIC50 (50% Fractional Inhibitory Concentrations), synergism when ΣFIC50 ≤ 0.5; toward synergism when ΣFIC50 < 1; additive when ΣFIC50 = 1; toward antagonism when ΣFIC50 > 1; antagonism when ΣFIC50 ≥ 2 to 4. The values show the mean ± S.D of 3 independent assays for each parasite line
DHA dihydroartemisinin, CQ chloroquine, MQ mefloquine, PPQ piperaquine, ATQ atovaquone, PG proguanil, TQ tafenoquine, PND pyronaridine
Potential novel combinations with either ATQ or PG were tested. When PND or MQ were combined with ATQ, antagonism/toward antagonism, except for MQ–ATQ in the C2B strain (Table 2) was observed. In TQ–ATQ combination, responses varied across strains although only one line, 14-B5 clone, revealed antagonistic interaction. When combined with PG, both PND and TQ showed different responses across strains, with 14-B5 showed antagonism between TQ–PG. All of the tested parasites revealed toward synergistic drug interaction against MQ–PG combination.
Discussion
In this study, the in vitro drug sensitivity and molecular profiles of P. falciparum isolates that were collected in Cambodia, a location well known as the epicenter of drug resistance are reported. The samples were collected at the time when PPQ resistance has been widespread. The CQ and ART resistant strains of P. falciparum likely originated in Cambodia, not far from location where these field isolates were collected, thus having unique genetic background. Twenty-six isolates were aimed to in vitro culture but only 17 isolates could be successfully cultured. Most of the analysed samples contained C580Y-K13 mutations and were labelled as ART-R based on RSA results. There may be more than a single pathway to ART resistance with other markers of resistance [45–47]; nonetheless the C580Y-K13 mutation is entrenched in this region consistent with observations for the field isolates included in this analysis. In addition, based on molecular markers and in vitro PSA testing, 14 of 17 P. falciparum samples were piperaquine-resistant. Isolates with all three PPQ resistance markers: amplification of pfpm2, presence of E415G-Exo and PfCRT mutations (T93S, H97Y, F145I, and I218F) showed a decrease in PPQ sensitivity as was expected. However, parasites carrying only the E415G-Exo and PfCRT mutations also resulted in PPQ resistant phenotype, without the requirement for pfpm2 amplification as was previously reported in the literature [20–22]. While increased pfpm2 copy number is accepted as a marker of PPQ resistance [20, 22, 24, 48, 49], molecular surveillance for PPQ resistance should not rely solely on the pfpm2 copy number amplification, as this may result in PPQ resistance being undetected in areas where PPQ resistance may be driven by other mutations, involving E415G-Exo and PfCRT mutations, in the absence of pfpm2 amplification. Since multiple copies of pfpm2 are commonly found in concert with PfCRT mutations, it has been suggested that pfpm2 amplification might help overcome the fitness cost of PfCRT variants by increasing the rate of hemoglobin degradation, and quick sequestration of reactive haem into haemozoin [33] and therefore improving the likelihood of PPQ resistant strains being maintained in the population. All analysed parasites held a single copy of pfmdr1, indicating that this variant should not be used as PPQ resistance marker. The treatment with DHA–PPQ was common when the field isolates were collected and most likely resulted in low MQ pressure, thereby promoting the loss of pfmdr1 amplifications in the parasites [20]. However, the limitation of this analysis is the lack of direct attribution of clinical outcome to the presence of molecular markers and in vitro drug sensitivity data.
Bimodal dose–response curves documented from Cambodian parasites collected in 2011 were also observed in the PPQ-R parasites consistent with reports by Bopp et al. [22]. The results suggested that bimodal dose–response curves can be observed in all PPQ resistant parasites regardless of PPQ resistance markers and the presence of the secondary peak in dose response curve might be another indicator of PPQ-R and was confirmed to be present in the field isolates. It has also been demonstrated that P. falciparum lines with PfCRT mutations in (C101F or L272F) result in the swelling of the parasite’s food vacuole and increased susceptibility to chloroquine and other quinoline antimalarials [50]. The mutations in PfCRT may interfere with the transport of the natural substrates out of the food vacuole, resulting in increased osmotic pressure. This phenotype was also observed in Dd2 parasites expressing the PfCRT mutations F145I, M343L, and G353V, [26]. In this study, the parasite isolate 9 which contained PfCRT-F145I mutation and pfpm2 single copy exhibited a swollen and translucent DV phenotype, while parasite isolates 3 and 6, that had PfCRT mutations and multiple copies of pfpm2, did not show the characteristic of swollen DVs. On the other hand, parasite isolate 4 containing I218F-PfCRT mutation and single copy pfpm2 did not show a swollen DV. This result might imply that not all novel PfCRT mutations exhibit a swollen DV phenotype, depending on the location of the mutated amino acids. Clinical implication and causative mechanisms of this finding still remain to be elucidated.
PPQ-resistant parasites were shown to be more susceptible to QN, CQ, MQ and LUM, similar to the engineered parasites expressing the novel PfCRT mutations [26]. This may have enabled a successful switch of first-line treatment from DHA–PPQ to artesunate–mefloquine by the Cambodian government in 2016. However, in this study, the IC50s against CYC seemed to be higher in PPQ resistant isolates (mean values ranging from 5470 nM to 30,585 nM) than in PPQ sensitive isolates (mean values ranging from 716 to 1435 nM). Overall, parasites demonstrated high level CYC resistance (geometric mean pretreatment ex vivo IC50, 2204 nM) [34]. PPQ-R isolates from Cambodia had greater susceptibility to LUM which may be a promising drug combination that should be explored in future studies. Further study is required to understand the implications of increased IC50 of CYC in the PPQ resistant parasites.
With the current decline of efficacy of partner drugs in the available artemisinin-based combinations, there is pressing need to evaluate novel compounds and new anti-malarial combinations against currently circulating field isolates. By culture-adapting field isolates, four clones (14-B5, 14-C6, 14-C7, and 14-F5) were obtained, and can be used as P. falciparum lines for HRP2-ELISA drug combination susceptibility testing to help quantify the contribution of different drug components on risk of treatment failure. Presented data confirmed findings by others who used the SYBR green I-based fluorescence (MSF) assay [42] and [3H]-hypoxanthine incorporation method [44] to demonstrate a synergistic interaction for ATQ–PG, an additive effect for CQ–CQ, and an antagonistic interaction for DHA–PPQ. It is interesting to observe that the 14-B5 clone tracks very similar to W2 against every tested drug combination except for PND–PG in which W2 is towards synergistic but 14-B5 is antagonistic. Of the drug combination tested, MQ–PG combination provided at least mild synergistic interactions against all the tested parasites including the 14-B5 line, where other drug combinations displayed antagonism. These findings demonstrate the importance of using currently circulating parasite isolates for evaluating the drug combinations as responses may be different based on unique genetic backgrounds. The diversity of parasites and variations in drug interactions that were observed among isolates may provide new insights into the outcomes of clinical studies in Cambodia.
Conclusion
The assessment of molecular markers associated with ART and PPQ resistance provides valuable information on how parasites have responded to DHA–PPQ exposure. This study shows that P. falciparum harbouring either pfpm2 amplification or novel PfCRT mutations with E415G-exo mutation display PPQ resistant phenotype. The increased copy number of pfpm2 may not be required for PPQ resistant phenotype. The cloned Cambodia parasite exhibit varied sensitivities to in vitro drug combinations. Genetic background of circulating field isolates is important for assessment of drug combinations and the use of the latest field isolates may be necessary for assessment of relevant drug combinations against P. falciparum resistant strains.
Supplementary information
Additional file 1: Table S1. The oligonucleotides of each primers P. falciparum for drug-resistant markers testing, including pfkelch13 (covering amino acid positions 539 to 580), pfexo (amino acid position 415), and pfcrt (at amino acid positions 93, 97, 145, 218, 343, 350, and 353) performed by T100™ Thermal Cycler.
Additional file 2: Table S2. The oligonucleotides of each primers for copy number variation of P. falciparum multidrug resistance gene 1 (pfmdr1) and P. falciparum plasmepsin 2 (pfpm2) with β-tubulin, the reference gene, performed by ABI 7500 Real-time PCR system.
Additional file 3: Table S3. Mean ± S.D of % Survival rate of Ring-stage Survival Assay (RSA) and Piperaquine Survival Assay (PSA).
Additional file 4: Table S4. IC50s of asexual stage P. falciparum parasites for different antimalarial drugs (mean ± S.D). N/A is not-applicable.
Additional file 5: Figure S1. Characterization of a new field clone of P. falciparum isolate. (A) Sequencing chromatogram of pfexo gene confirmed the amino acid at position 145 of Exonuclease from P. falciparum Cambodian isolate 14 is glutamic acid (E), which is coded by a codon GAG. (B) Table represents molecular genotyping of P. falciparum Cambodian isolate 14 before and after cloning. The copy number of pfmdr1 and pfpm2 less than 1.5 and 1.6 indicates a single copy of the gene, respectively, while % RSA and % PSA cut off less than 1 and 10 represents the ART- and PPQ-sensitive, respectively. N.D stands for not determined.
Additional file 6: Table S5. Comparison of the in vitro susceptibility of Plasmodium falciparum Cambodian isolate 14 before and after cloning as well as P. falciparum 3D7 and W2 exposed to established antimalarial agents. IC50 values report the mean ± S.D from at least 3 experiments. Statistically significant difference relative to 3D7 is indicated with one asterisk (0.05 > p > 0.01). N/A is non-applicable.
Additional file 7: Table S6. In vitro drug combination assay for 9 combinations in asexual stages of different P. falciparum strains. DHA, dihydroartemisinin; CQ, chloroquine; MQ, mefloquine; PPQ, piperaquine; ATQ, atovaquone; PG, proguanil; TQ, tafenoquine; PND, pyronaridine. ΣFIC50 (50% Fractional Inhibitory Concentrations), synergism when ΣFIC50 ≤ 0.5; toward synergism when ΣFIC50 < 1; additive when ΣFIC50 = 1; toward antagonism when ΣFIC50 > 1; antagonism when ΣFIC50 ≥ 2 to 4. The values show the mean ± S.D of 3 independent assays for each parasite line.
Acknowledgements
The following reagents were obtained through BEI Resources, NIAID, NIH: Plasmodium falicaprum, Strain IPC-4884 (MRA-1238) and IPC-5202 (MRA-1240), contributed by Didier Ménard. The authors would like to thanks the clinical and laboratory staff who made the study possible. The view expressed in this article are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense, U.S. or Cambodian Governments.
Disclaimer
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation/publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
Abbreviations
- ACT
Artemisinin-based combination therapy
- AS
Artesunate
- ATM
Artemisone
- ATQ
Atovaquone
- ART
Artemisinin
- Cas
CRISPR associated protein
- CN
Copy number
- CQ
Chloroquine
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CYC
Cycloguanil
- DHA
Dihydroartemisinin
- DHFR
Dihydrofolate reductase
- DOX
Doxycycline
- DV
Digestive vacuole
- ELISA
Enzyme-linked immunosorbent assay
- Exo
Exonuclease
- FIC
Fractional inhibitory concentration
- GWAS
Genome-wide association studies
- HEPES
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- HRP
Histidine rich protein
- K13
Kelch-13
- LUM
Lumefantrine
- MQ
Mefloquine
- PfCRT
P. falciparum chloroquine resistance transporter
- Pfmdr1
P. falciparum multidrug resistance 1
- PG
Proguanil
- PM
Plasmepsin
- PND
Pyronaridine
- PPQ
Piperaquine
- PQ
Primaquine
- PSA
PPQ survival assay
- QN
Quinine
- qPCR
Quantitative real-time polymerase chain reaction
- RPMI
Rosewell Park Memorial Institute
- RSA
Ring-stage survival assay
- SNP
Single nucleotide polymorphism
- TQ
Tafenoquine
- WHO
World Health Organization
Author’s contributions
NB and BAV conceived, designed and executed the study. PL, PG, WK, PV, MDS, MMF, CL, PLS, NCW, DLS, and MW conceived, designed and supported the study. CT, CP, WF, and KK performed cell culture, cloning, and drug susceptibility assays. PS, CC and KK performed molecular genotyping. NB analyzed the samples and all authors assisted in interpreting the data. NB wrote the first draft. All authors edited, reviewed final manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Naval Advanced Medical Development Program, Washington, DC, USA and Defense Malaria Assistance Program (DMAP). The funding source had no role in the analysis or interpretation of data, preparation of the manuscript or the decision to publish.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
All participants or guardians provided written consent and samples were collected under approval from the Cambodian National Ethics Committee for Health Research and Walter Reed Army Institute of Research Institutional Review board.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12936-020-03339-w.
References
- 1.WHO . World malaria report. Geneva: World Health Organization; 2019. [Google Scholar]
- 2.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO . Guidelines fro the treatment of malaria. 3. Geneva: World Health Organization Press; 2015. [PubMed] [Google Scholar]
- 4.Saunders DL, Vanachayangkul P, Lon C, Program USAMMR, National Center for Parasitology, Entomology, Malaria Control, Royal Cambodian Armed Forces Dihydroartemisinin–piperaquine failure in Cambodia. N Engl J Med. 2014;371:484–485. doi: 10.1056/NEJMc1403007. [DOI] [PubMed] [Google Scholar]
- 5.Leang R, Taylor WR, Bouth DM, Song L, Tarning J, Char MC, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in Western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis. 2015;15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 7.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phuc BQ, Rasmussen C, Duong TT, Dong LT, Loi MA, Menard D, et al. Treatment failure of dihydroartemisinin/piperaquine for Plasmodium falciparum malaria, Vietnam. Emerg Infect Dis. 2017;23:715–717. doi: 10.3201/eid2304.161872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis. 2019;19:952–961. doi: 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Artemisinin Resistance in Cambodia 1 Study C: evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 11.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fairhurst RM. Understanding artemisinin-resistant malaria: what a difference a year makes. Curr Opin Infect Dis. 2015;28:417–425. doi: 10.1097/QCO.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 15.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in vitro and ex vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaorattanakawee S, Saunders DL, Sea D, Chanarat N, Yingyuen K, Sundrakes S, et al. Ex vivo drug susceptibility testing and molecular profiling of clinical Plasmodium falciparum isolates from Cambodia from 2008 to 2013 suggest emerging piperaquine resistance. Antimicrob Agents Chemother. 2015;59:4631–4643. doi: 10.1128/AAC.00366-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duru V, Khim N, Leang R, Kim S, Domergue A, Kloeung N, et al. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med. 2015;13:305. doi: 10.1186/s12916-015-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veiga MI, Ferreira PE, Malmberg M, Jornhagen L, Bjorkman A, Nosten F, et al. pfmdr1 amplification is related to increased Plasmodium falciparum in vitro sensitivity to the bisquinoline piperaquine. Antimicrob Agents Chemother. 2012;56:3615–3619. doi: 10.1128/AAC.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis. 2017;17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bopp S, Magistrado P, Wong W, Schaffner SF, Mukherjee A, Lim P, et al. Plasmepsin II-III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun. 2018;9:1769. doi: 10.1038/s41467-018-04104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parobek CM, Parr JB, Brazeau NF, Lon C, Chaorattanakawee S, Gosi P, et al. Partner-drug resistance and population substructuring of artemisinin-resistant Plasmodium falciparum in Cambodia. Genome Biol Evol. 2017;9:1673–1686. doi: 10.1093/gbe/evx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thanh NV, Thuy-Nhien N, Tuyen NT, Tong NT, Nha-Ca NT, Dong LT, et al. Rapid decline in the susceptibility of Plasmodium falciparum to dihydroartemisinin-piperaquine in the south of Vietnam. Malar J. 2017;16:27. doi: 10.1186/s12936-017-1680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrawal S, Moser KA, Morton L, Cummings MP, Parihar A, Dwivedi A, et al. Association of a novel mutation in the Plasmodium falciparum chloroquine resistance transporter with decreased piperaquine sensitivity. J Infect Dis. 2017;216:468–476. doi: 10.1093/infdis/jix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kumpornsin K, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun. 2018;9:3314. doi: 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhingra SK, Small-Saunders JL, Menard D, Fidock DA. Plasmodium falciparum resistance to piperaquine driven by PfCRT. Lancet Infect Dis. 2019;19:1168–1169. doi: 10.1016/S1473-3099(19)30543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim P, Dek D, Try V, Eastman RT, Chy S, Sreng S, et al. Ex vivo susceptibility of Plasmodium falciparum to antimalarial drugs in western, northern, and eastern Cambodia, 2011-2012: association with molecular markers. Antimicrob Agents Chemother. 2013;57:5277–5283. doi: 10.1128/AAC.00687-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ansbro MR, Jacob CG, Amato R, Kekre M, Amaratunga C, Sreng S, et al. Development of copy number assays for detection and surveillance of piperaquine resistance associated plasmepsin 2/3 copy number variation in Plasmodium falciparum. Malar J. 2020;19:181. doi: 10.1186/s12936-020-03249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee A, Gagnon D, Wirth DF, Richard D. Inactivation of plasmepsins 2 and 3 sensitizes Plasmodium falciparum to the antimalarial drug piperaquine. Antimicrob Agents Chemother. 2018;62:e02309–e02317. doi: 10.1128/AAC.02309-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loesbanluechai D, Kotanan N, de Cozar C, Kochakarn T, Ansbro MR, Chotivanich K, et al. Overexpression of plasmepsin II and plasmepsin III does not directly cause reduction in Plasmodium falciparum sensitivity to artesunate, chloroquine and piperaquine. Int J Parasitol Drugs Drug Resist. 2018;9:16–22. doi: 10.1016/j.ijpddr.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva M, Calcada C, Teixeira M, Veiga MI, Ferreira PE. Multigenic architecture of piperaquine resistance trait in Plasmodium falciparum. Lancet Infect Dis. 2020;20:26–27. doi: 10.1016/S1473-3099(19)30689-9. [DOI] [PubMed] [Google Scholar]
- 33.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA. 2015;112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wojnarski M, Lon C, Vanachayangkul P, Gosi P, Sok S, Rachmat A, et al. Atovaquone-proguanil in combination with artesunate to treat multidrug-resistant P. falciparum malaria in Cambodia: an open-label randomized trial. Open Forum Infect Dis. 2019;6:1–14. doi: 10.1093/ofid/ofz314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talundzic E, Okoth SA, Congpuong K, Plucinski MM, Morton L, Goldman IF, et al. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molecular Module. Copy number estimation of P. falciparum pfmdr1. v1.1. WWARN Procedure. 2011.
- 38.Chaorattanakawee S, Tyner SD, Lon C, Yingyuen K, Ruttvisutinunt W, Sundrakes S, et al. Direct comparison of the histidine-rich protein-2 enzyme-linked immunosorbent assay (HRP-2 ELISA) and malaria SYBR green I fluorescence (MSF) drug sensitivity tests in Plasmodium falciparum reference clones and fresh ex vivo field isolates from Cambodia. Malar J. 2013;12:239. doi: 10.1186/1475-2875-12-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaorattanakawee S, Lon C, Jongsakul K, Gawee J, Sok S, Sundrakes S, et al. Ex vivo piperaquine resistance developed rapidly in Plasmodium falciparum isolates in northern Cambodia compared to Thailand. Malar J. 2016;15:519. doi: 10.1186/s12936-016-1569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas JA, Collins CR, Das S, Hackett F, Graindorge A, Bell D, et al. Development and application of a simple plaque assay for the human malaria parasite Plasmodium falciparum. PLoS ONE. 2016;11:e0157873. doi: 10.1371/journal.pone.0157873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fivelman QL, Adagu IS, Warhurst DC. Modified fixed-ratio isobologram method for studying in vitro interactions between atovaquone and proguanil or dihydroartemisinin against drug-resistant strains of Plasmodium falciparum. Antimicrob Agents Chemother. 2004;48:4097–4102. doi: 10.1128/AAC.48.11.4097-4102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Co EM, Dennull RA, Reinbold DD, Waters NC, Johnson JD. Assessment of malaria in vitro drug combination screening and mixed-strain infections using the malaria Sybr green I-based fluorescence assay. Antimicrob Agents Chemother. 2009;53:2557–2563. doi: 10.1128/AAC.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berenbaum MC. A method for testing for synergy with any number of agents. J Infect Dis. 1978;137:122–130. doi: 10.1093/infdis/137.2.122. [DOI] [PubMed] [Google Scholar]
- 44.Muangnoicharoen S, Johnson DJ, Looareesuwan S, Krudsood S, Ward SA. Role of known molecular markers of resistance in the antimalarial potency of piperaquine and dihydroartemisinin in vitro. Antimicrob Agents Chemother. 2009;53:1362–1366. doi: 10.1128/AAC.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee A, Bopp S, Magistrado P, Wong W, Daniels R, Demas A, et al. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar J. 2017;16:195. doi: 10.1186/s12936-017-1845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demas AR, Sharma AI, Wong W, Early AM, Redmond S, Bopp S, et al. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc Natl Acad Sci USA. 2018;115:12799–12804. doi: 10.1073/pnas.1812317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robert MG, Foguim Tsombeng F, Gendrot M, Mosnier J, Amalvict R, Benoit N, et al. Absence of a high level of duplication of the plasmepsin II gene in Africa. Antimicrob Agents Chemother. 2018;62:e00374-18. doi: 10.1128/AAC.00374-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert MG, Tsombeng FF, Gendrot M, Diawara S, Madamet M, Kounta MB, et al. Baseline ex vivo and molecular responses of Plasmodium faciparum isolates to piperaquine before implementation of dihydroartemisinin-piperaquine in Senegal. Antimicrob Agents Chemother. 2019;63:e02445-18. doi: 10.1128/AAC.02445-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pulcini S, Staines HM, Lee AH, Shafik SH, Bouyer G, Moore CM, et al. Mutations in the Plasmodium falciparum chloroquine resistance transporter, PfCRT, enlarge the parasite’s food vacuole and alter drug sensitivities. Sci Rep. 2015;5:14552. doi: 10.1038/srep14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The oligonucleotides of each primers P. falciparum for drug-resistant markers testing, including pfkelch13 (covering amino acid positions 539 to 580), pfexo (amino acid position 415), and pfcrt (at amino acid positions 93, 97, 145, 218, 343, 350, and 353) performed by T100™ Thermal Cycler.
Additional file 2: Table S2. The oligonucleotides of each primers for copy number variation of P. falciparum multidrug resistance gene 1 (pfmdr1) and P. falciparum plasmepsin 2 (pfpm2) with β-tubulin, the reference gene, performed by ABI 7500 Real-time PCR system.
Additional file 3: Table S3. Mean ± S.D of % Survival rate of Ring-stage Survival Assay (RSA) and Piperaquine Survival Assay (PSA).
Additional file 4: Table S4. IC50s of asexual stage P. falciparum parasites for different antimalarial drugs (mean ± S.D). N/A is not-applicable.
Additional file 5: Figure S1. Characterization of a new field clone of P. falciparum isolate. (A) Sequencing chromatogram of pfexo gene confirmed the amino acid at position 145 of Exonuclease from P. falciparum Cambodian isolate 14 is glutamic acid (E), which is coded by a codon GAG. (B) Table represents molecular genotyping of P. falciparum Cambodian isolate 14 before and after cloning. The copy number of pfmdr1 and pfpm2 less than 1.5 and 1.6 indicates a single copy of the gene, respectively, while % RSA and % PSA cut off less than 1 and 10 represents the ART- and PPQ-sensitive, respectively. N.D stands for not determined.
Additional file 6: Table S5. Comparison of the in vitro susceptibility of Plasmodium falciparum Cambodian isolate 14 before and after cloning as well as P. falciparum 3D7 and W2 exposed to established antimalarial agents. IC50 values report the mean ± S.D from at least 3 experiments. Statistically significant difference relative to 3D7 is indicated with one asterisk (0.05 > p > 0.01). N/A is non-applicable.
Additional file 7: Table S6. In vitro drug combination assay for 9 combinations in asexual stages of different P. falciparum strains. DHA, dihydroartemisinin; CQ, chloroquine; MQ, mefloquine; PPQ, piperaquine; ATQ, atovaquone; PG, proguanil; TQ, tafenoquine; PND, pyronaridine. ΣFIC50 (50% Fractional Inhibitory Concentrations), synergism when ΣFIC50 ≤ 0.5; toward synergism when ΣFIC50 < 1; additive when ΣFIC50 = 1; toward antagonism when ΣFIC50 > 1; antagonism when ΣFIC50 ≥ 2 to 4. The values show the mean ± S.D of 3 independent assays for each parasite line.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.