Abstract
Background
MYH14 gene mutations have been suggested to be associated with nonsyndromic/syndromic sensorineural hearing loss. It has been reported that mutations in MYH14 can result in autosomal dominant nonsyndromic deafness-4A (DFNA4).
Methods
In this study, we examined a four-generation Han Chinese family with nonsyndromic hearing loss. Targeted next-generation sequencing of deafness genes was employed to identify the pathogenic variant. Sanger sequencing and PCR-RFLP analysis were performed in affected members of this family and 200 normal controls to further confirm the mutation.
Results
Four members of this family were diagnosed as nonsyndromic bilateral sensorineural hearing loss with postlingual onset and progressive impairment. A novel missense variant, c.5417C > A (p.A1806D), in MYH14 in the tail domain of NMH II C was successfully identified as the pathogenic cause in three affected individuals. The family member II-5 was suggested to have noise-induced deafness.
Conclusion
In this study, a novel missense mutation, c.5417C > A (p.A1806D), in MYH14 that led to postlingual nonsyndromic autosomal dominant SNHL were identified. The findings broadened the phenotype spectrum of MYH14 and highlighted the combined application of gene capture and Sanger sequencing is an efficient approach to screen pathogenic variants associated with genetic diseases.
Keywords: MYH14, Mutation, Hearing loss, Targeted next-generation sequencing, Family
Background
The MYH14 gene, also known as nonmuscle heavy chain II C (NMHCII-C), encodes one of the myosin members. In motility processes, such as cytoskeleton rearrangement, organelle translocation and ion channel gating, MYH14 is indicated to play a major role in the directed movement of cell components along actin filaments by ATP hydrolysis to generate force [1–3]. MYH14 (NMHCII-C) has been characterized recently, but there is limited information on its biological functions. The MYH14 gene is located on the chromosome 19q13.33, contains 41 exons and encodes a protein with 1995 amino acids, which contains a myosin head region, two IQ domains, an N-terminal myosin domain, and a C-terminal myosin tail [4]. A polar structure is formed by the dimerization of heavy chains. The N-terminal domain has two globular heads that are ATP-binding and actin-binding regions and are essential for motility, while the helical C-terminus is a singular rod-like tail that can polymerize molecules into bipolar filaments in nonmuscle and muscle cells [5]. The MYH14 gene was reported to be related to peripheral neuropathy, myopathy, hoarseness and hearing loss (PNMHH) [6], and the MYH14 protein is expressed widely within cochlear tissues, such as the organ of Corti, spiral prominence epithelium, stria vascularis, and cochlear duct. Low expression was detectable in the Reissner’s membrane and spiral ligament, but no expression was detected in vestibular epithelia in mice [7]. MYH14 plays a part in neurogenesis and maintenance of apical cell junctions in epithelial cells within the cochlea [8, 9]. The expression of MYH14 in mice and humans is generally higher in adults than in adolescents [4, 10, 11].
Hereditary sensorineural hearing loss (SNHL) can be classified into nonsyndromic and syndromic hearing loss. 75% of hereditary SNHL cases are nonsyndromic. Eighty percent of nonsyndromic SNHL is in an autosomal recessive (AR) inheritance pattern and tends to result in severe SNHL with prelingual onset, whereas autosomal dominant (AD) inherited deafness usually leads to mild and flat downsloping hearing loss with postlingual onset [12, 13]. Pathogenic variants of the MYH14 gene can cause either syndromic or nonsyndromic SNHL and can be identified as the pathogenic factor DFNA4A [7, 14–16]. To date, missense and nonsense variants of the MYH14 gene have been reported in families with mild to severe degrees of AD SNHL, which usually manifests with milder hearing loss and later onset than AR inherited hearing loss. A recent study showed the MYH14 gene may be an essential gene related to nonsyndromic AD SNHL [17]. However, the physiological link between MYH14 mutations and sensorineural hearing loss is still unclear.
In the current study, we investigated pathogenic variants in a four-generation Han Chinese family suffering from nonsyndromic sensorineural hearing loss with postlingual onset. We screened 127 genes known to be related to deafness by gene capture and next-generation sequencing and identified the novel variant c.5417C > A (p.A1806D) in MYH14 on the tail domain of NMH II C. By reporting novel pathogenic variants, the phenotypic spectrum of the MYH14 gene could be broadened in the field of hereditary hearing loss.
Methods
Subjects
Fifteen members from a four-generation Han Chinese family were enrolled in this study, and four of them were diagnosed as sensorineural hearing loss with postlingual onset and progressive impairment. All participants agreed to undergo clinical examinations as well as audiometric and vestibular function evaluations, including otoscopy, tympanometry, pure-tone audiometry (PTA), speech recognition score (above the hearing threshold of 30 dB HL) evaluation, auditory brainstem response (ABR) evaluation, distortion product otoacoustic emission (DPOAE) recording, a vestibular bithermal caloric test, and ocular/cervical vestibular evoked myogenic potential (o/c VEMP) evaluation. High-resolution computed tomography (HRCT) to image the temporal bone and brain MRI were used to define cochleovestibular malformation. The hearing loss onset of members IV-1 and IV-2 at the ages of 3 and 1 was evaluated by behavior observation audiometry. The degree of SNHL was classified into five grades, namely, normal hearing (PTA ≤ 25 dB HL) and mild (26 ~ 40 dB HL), moderate (41 ~ 60 dB HL), severe (61 ~ 80 dB HL), and profound hearing loss (PTA ≥81 dB HL), based on the average PTA threshold applied at 250, 500, 1000, 2000, 4000, and 8000 Hz. The study protocol was permitted by the Ethics Committee of Shandong Provincial ENT Hospital (XYK20140101), and adhered to the Declaration of Helsinki principles. Informed written consent was obtained from each subject or, in the case of minors, from their parents.
Targeted gene capture and next-generation sequencing
To investigate the pathogenic mutations in this family, a genomic DNA (gDNA) purification kit (Axygen, San Francisco, CA) was applied to extract gDNA from peripheral blood by the manufacturer’s instructions. Targeted deafness gene capture and next-generation sequencing were performed in the probands to screen 127 genes [18] (Table S1 in Supplementary Material) related to nonsyndromic and syndromic hearing loss by BGI (Beijing Genomics Institute, Shenzhen, China) through a standardized next-generation capture sequencing platform [19]. All exons and the surrounding ±10 bp in the flanking intronic regions of the 127 deafness genes were sequenced. An E210 DNA-shearing devise (Covaris S2, Massachusetts, USA) was employed to fragment the whole gDNA. The library fragment sizes were mainly distributed within 250 ~ 300 bp. Adapter ligation, end repair and adenylation were performed for library preparation in accordance with the standard Illumina protocols. An Illumina HiSeq2000 analyzer was used to capture targeted DNA fragments [20], and image analysis, base calling, and error estimation were carried out to generate primary data by the Illumina Pipeline (version 1.3.4). Using BWA MultiVision software, reads were aligned to the NCBI37/hg19 assembly. SOAPsnp software was used as a reference for recorded SNPs, indels and splice variants. The other variant types, including CNVs and complicated genomic rearrangements, were not included. Mutations were detected by the GATK Indel Genotyper. Databases including NCBI dbSNP, the 1000 Genomes database, the HapMap database and BGI’s own databases were used as references [21]. The pathogenic variations were scanned using the 1000 Genomes database (Phase I) (http://www.1000genomes.org) and the HapMap database (combined Phases II and III). The American Medical Genetics and Genomics Guide were referred to interpret and filtered the data based on a simple AD inheritance pattern by keeping only the heterozygous variants. Remaining variants were then filtered using a low likelihood of adverse functionality on the basis of the mutation type, as assessed with pathogenicity prediction tools, including Mutation Taster, SIFT and PolyPhen 2. The American College of Medical Genetics and Genomics (ACMG) guidelines were used to define the DNA variants.
Mutation verification
PCR amplification and Sanger sequencing were carried out to verify candidate variants. As previously described [22], PCR was executed in a 50 μl reaction mixture. The forward and reverse primers were 5′- GATGGTCTCGTGGACTTAT-3′ and 5′-TGTGGAGGTCACCTTTCT-3′ for the MYH14 c.5417C > A mutation and 5′- TTGGTGTTTGCTCAGGAAGA-3′ and 5′-GGCCTACAGGGGTTTCAAAT-3′ for the GJB2 c.109G > A mutation. PCR products were purified and sequenced by the ABI 3730XL Genetic Analyzer. Analysis of sequencing data was performed by DNASTAR sequence analysis software. PCR-RFLP was employed to confirm the c.5417C > A (p.A1806D) mutation in the MYH14 gene. Primers flanking the candidate mutation were designed to amplify a 512-bp PCR product, which was then digested into 308-bp and 204-bp fragments by the restriction enzyme Eco130I (Thermo Scientific, USA). The digested products were analyzed through electrophoresis by using a 2% agarose gel. MYH14 mRNA (RefSeq NM_001077186.2) was used to align the sequences as a reference by Lasergene SeqMan software. Multiple-sequence alignment was conducted to perform phylogenetic analysis by ClustalW2 software. The Multiple sequences aligned included NP_001070654.1 (Homo sapiens), NP_001094160.1 (Rattus norvegicus), NP_001258467.1 (Mus musculus), XP_014980128.1 (Macaca mulatta), XP_003316592.1 (Pan troglodytes), XP_023988172.1 (Physeter catodon), and XP_010813541.1 (Bos taurus).
Results
Clinical findings
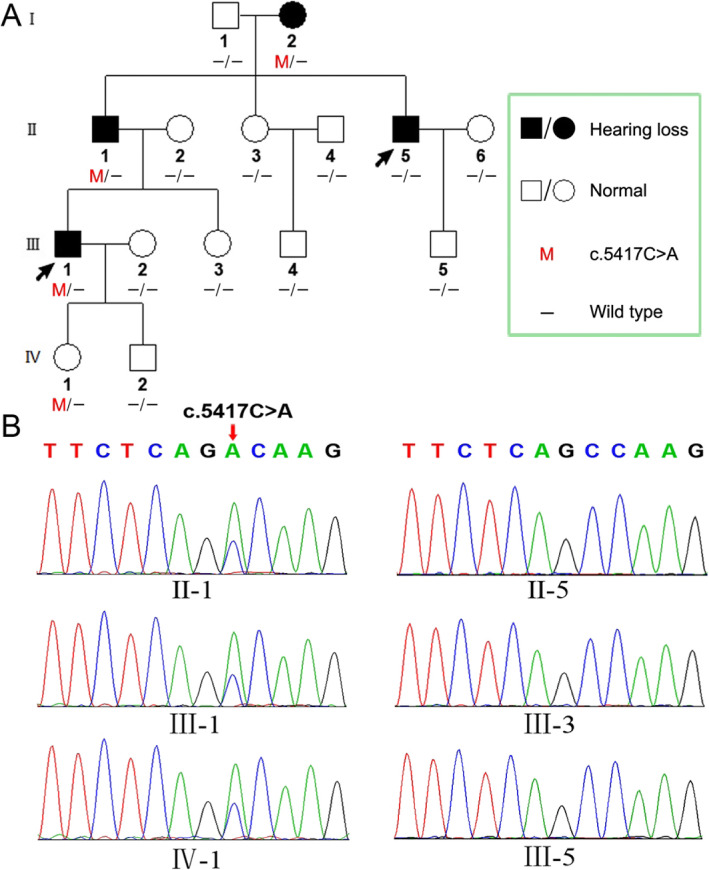
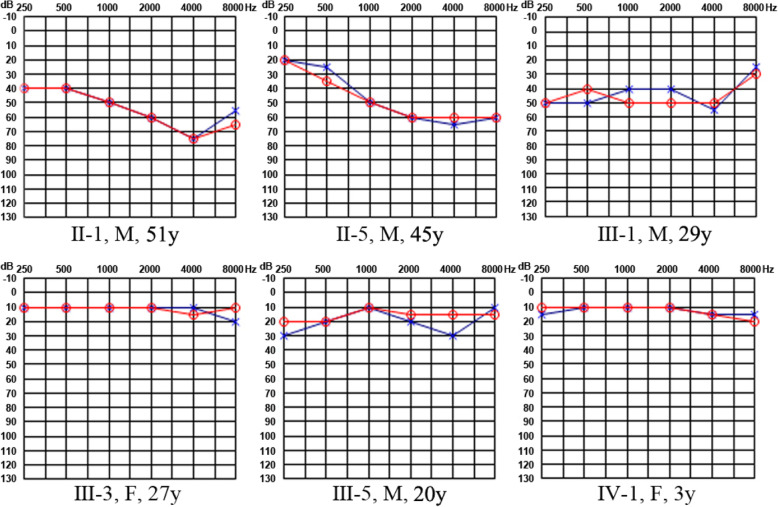
A four-generation Chinese family with 15 members from Shandong Province, China, was enrolled in the study. The family members of II-5 and III-1 were probands, and the pedigree is shown in Fig. 1 according to the statements of the participants. Four members of this Han Chinese family suffered from similar symptoms of tinnitus and hearing loss and presented bilateral and symmetric hearing impairment with postlingual onset and progressive impairment. The age of onset widely ranged from teens to thirties. The PTA results for the members with symptoms showed that moderate sensorineural hearing loss affected almost all frequencies (Fig. 2).
Fig. 1.
a The genealogical tree of the Chinese pedigree. The probands are indicated by arrows. b the DNA sequencing profile shows the c.5417C > A mutation in the MYH14 in II-1, III-1 and IV-1
Fig. 2.
Bilateral pure tone audiograms from individuals in this family. Individual IV-1 was examined by behavior observation audiometry (circles in the audiograms represent the right air conduction thresholds, and crosses, the left ear)
One of the probands (patient III-1) in this family was a 30-year-old male suffering from bilateral nonsyndromic hearing impairment that started in his teens and developed progressively. The PTA performed in our hospital at the age of 29 years showed moderate and flat downsloping SNHL in both ears. The maximum speech recognition scores (SRSs) were 36% in the left and 32% in the right ear. The vestibular function tests showed coordination, and III-1 never complained of vertigo or dizziness. Another proband (II-5) had bilateral nonsyndromic hearing loss with onset in his thirties. However, he has worked in a glassware factory since his twenties and was exposed to noise more than 8 h every day over 10 years.
Constant binaural tinnitus was a common symptom among these patients. The tinnitus volume was at 5 ~ 10 dB HL, which was higher than the auditory threshold of 3 ~ 6 kHz. Family member II-1 had bilateral nonsyndromic hearing loss with onset in his thirties, and member I-2 was affected by progressive hearing impairment since her twenties. The two affected members failed to pass the DPOAE test bilaterally at most or all frequencies, and their ABR results were consistent with the PTA results, which revealed moderate sensorineural hearing loss (Fig. 2). Vestibular tests showed no obvious dysfunction in II-1 and II-5. The clinical examinations are listed in Table 1. The unaffected participants’ examinations and evaluations were normal, and no clinical syndromes were identified.
Table 1.
Clinical features of individuals with sensorineural hearing loss in this family
| Examinations | II-1 | II-5 | III-1 |
|---|---|---|---|
| PTA threshold (dB HL) | 56.25 / 58.75 (left/right) | 50.00 / 51.25 (left/right) | 56.25 / 58.75 (left/right) |
| SRS (%) | 64 / 64 (left/right) | 92 / 88 (left/right) | 36 / 32 (left/right) |
| Tympanometry | “A” type | “A” type | “A” type |
| ABR threshold (dB nHL) | 70 (both) | 70/60 (left/right) | 60 (both) |
| DPOAE | Absent (both) | Absent (both) | Absent (both) |
| Cochlear microphone | No elicited | No elicited | No elicited |
| Vestibular bithermal calorid test | Normal | Normal | Labyrinth reactivity lower (8.4°/s) |
| cVEMP | Normal | Normal | No wave |
| oVEMP | Low-amplitude in both ears | Low-amplitude in left and normal in right | N1 and P1 waves recorded only in right ear |
| Optic nerve electroretinogram | Normal | Normal | Normal |
| Temporal bone HRCT | Normal | Normal | Normal |
| Brain MRI | Normal | Normal | Normal |
Targeted gene capture and next-generation sequencing
To identify the pathogenic variants in this Chinese family, targeted gene capture sequencing was performed in the two probands (II-5 and III-1). All variants were filtered through NCBI dbSNP, the HapMap database, the 1000 Genomes database and the in-house databases. The c.5417C > A mutation in MYH14 and c.109G > A mutation in GJB2 were detected. Next, Sanger sequencing was used to screen these two candidate variants in 15 family members (Supplementary Table 2). Based on the inheritance pattern and characteristics of deafness, combined with the results of Sanger sequencing, one heterozygous missense mutation c.5417C > A (p.A1806D) was identified, which cosegregated with hearing impairment in this family and was inherited in an autosomal dominant pattern. The c.5417C > A (p.A1806D) variant, located in the tail domain of the MYH14 gene, causes a change from alanine to aspartic acid at codon 1806. The deleterious and pathogenic aspects of the mutation c.5417C > A (p.A1806D) are listed in Table 2.
Table 2.
Characteristics of the MYH14 variant, analysis of predicted protein structure and disease-causing effects
| Variation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Exon | Nucleotidea | Amino acida | Type | Status | SIFT | PolyPhen 2 | Mutation Taster | 1000G | DVD | Clinvar | LOVD3 |
| MYH14 | 39 | c.5417C > A | p.A1806D | missense | Heter | Damaging | Damaging | Disease causing | – | – | – | – |
c variation at cDNA level, Clinvar Clinvar Database, DVD Deafness Variation Database, 1000G 1000 Genomes, Heter heterozygote, LOVD3 Leiden Open Variation Database, p variation at protein level, MYH14 myosin heavy chain 14 (NM_001077186), PolyPhen 2 Polymorphism Phenotyping v2, SIFT sorts intolerant from tolerant
aAll nucleotide and amino acid are abbreviated according to the International Union of Pure and Applied Chemistry
A novel missense mutation identified in MYH14
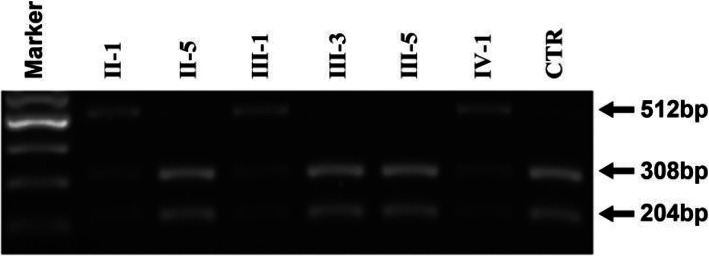
Sanger sequencing was employed to verify this candidate mutation in all the family members. We found that three affected adults (I-2, II-1, III-1) and the three-year-old daughter (IV-1) of patient III-1 harbored this heterozygous variant c.5417C > A (p.A1806D). None of the other subjects carried this variant (as shown in Fig. 1). Considering the age of onset of hearing loss in this family, the three-year-old daughter of patient III-1 could have normal hearing despite carrying the pathogenic variant. Additionally, consistent with our previous diagnosis, the hearing impairment of patient II-5 was caused by noise exposure, not by genetic mutation. To test the frequency of the c.5417C > A (p.A1806D) variant in the general population, Sanger sequencing was performed in 200 people with normal hearing, and no mutation was found at this site. Subsequently, we further analyzed the presence of the c.5417C > A (p.A1806D) mutation by RFLP. As shown in Fig. 3, the PCR products from normal members were digested into two fragments (308 bp and 204 bp), while the PCR products from the affected members (II-1, III-1) and the daughter (IV-1) of III-1 were digested into three fragments (308 bp, 204 bp and 512 bp) because of the presence of the c.5417C > A (p.A1806D) mutation. Figure 4 lists MYH14 alignments from different species, including Homo sapiens, Rattus norvegicus, Mus musculus, Macaca mulatta, Pan troglodytes, Physeter catodon and Bos taurus. The high conservation of alanine at position 1806 in MYH14 demonstrates that it might play a key role in the biological function, and a variant at this amino acid position could be pathogenic.
Fig. 3.
PCR-RFLP results confirms the identification of c.5417C > A mutation in the MYH14 gene. 512-bp PCR products around c.5417C region were digested with Eco130I and analyzed by electrophoresis through a 2% agarose gel stained with ethidium bromide
Fig. 4.
The protein alignment shows conservation across seven species. The mutation c.5417C > A occurred at evolutionarily conserved amino acids
Discussion
MYH14 is involved in many motility processes, such as organelle translocation, cytoskeleton rearrangement and ion channel gating. The protein is more abundant in adult tissues, with an approximately six-fold increase between E12.5 and E16.5 in the mouse cochlear duct [11, 23]. It has an active role in cellular remodeling in the epithelium of cochlear sensory cells. Inhibition of the MYH14 gene can lead to defects in extension of the cochlear duct, which may be the physiological mechanism of hearing impairment in subjects who carry MYH14 gene mutations [24]. Mutations in the MYH14 gene can result in peripheral neuropathy, hoarseness, myopathy, and hearing loss [6].
In our study, we examined a four-generation Han Chinese family presenting with autosomal dominant nonsyndromic deafness and identified a novel missense mutation, c.5417C > A (p.A1806D), in the MYH14 gene by using targeted gene capture and Sanger sequencing. Patient III-1 and his father (II-1) and grandmother (I-2) suffered from bilaterally symmetric SNHL with postlingual onset at ages ranging from teens to thirties, and their hearing loss gradually accelerated with aging. All three affected members were identified as carrying this novel variant c.5417C > A (p.A1806D) in the MYH14 gene, but this variant was absent in 200 normal controls and all the other unaffected family members except for patient III-1’s three-year-old daughter (IV-1). Considering the late onset of SNHL in her family, we will closely follow-up with the three-year-old girl. Interestingly, the SRS of III-1 was much worse than that of II-1. This finding might be related to the age of onset of hearing loss, which exhibited a slow progression. We should follow-up with these individuals from the family.
Patient II-5 worked in a glassware factory and was exposed to noise for more than 8 h every day over 10 years since his twenties. His PTA hearing test showed an increase in medium-high frequencies but normal levels in low frequencies bilaterally, and the SRS values were 92% for the left side and 88% for the right side, which also suggested noise-induced hearing loss. The syndromic features associated with the brain, heart and kidney in this family were excluded.
Although there were no clinical symptoms of vestibular dysfunction in any of the members of this Chinese family, the functional evaluation in proband III-1 showed vestibular impairment. Previous studies reported that there was no detectable expression of MYH14 in vestibular epithelia in mice [7]. Therefore, despite the presence of a missense mutation in MYH14, patient III-1 did not show any clinical vestibular symptoms, which could be due to functional compensation of the vestibular system during the long course of disease.
Table 3 shows all the previously reported pathogenic or likely pathogenic variants in the MYH14 gene that cause mild to severe degrees of nonsyndromic and progressive SNHL with postlingual onset, as well as the novel variant identified in the current study. Interestingly, among these missense mutations, only one mutation has been reported to lead to syndromic hearing loss, but the others cause nonsyndromic hearing loss [26]. In this study, we demonstrated a new missense mutation in MYH14 that could have led to moderate nonsyndromic SNHL with postlingual onset in a Han Chinese family. There are some East Asian alleles reported in gnomAD, indicating that this mutation may be an allele in Asians that causes hearing loss. The evolutionary conservation of the alanine residue at codon 1806 (A1806D) in MYH14 indicates to us that the variant could be pathogenic and cosegregated with the disease in this family. PCR-RFLP was performed to further confirm the results in all the family members and 200 individuals with normal hearing. However, the mechanism by which the c.5417C > A (p.A1806D) variant is associated with hearing loss is not clear. Further functional researches are required to analyze the pathophysiologic mechanism underlying the auditory defects caused by the variant in MYH14 by constructing a transgenic mouse model.
Table 3.
MYH14 gene mutations related with sensorineural hearing loss
| Exon | Mutationa | Amino acid change | Type of variant | Reference | DVD | ClinVar | LOVD3 |
|---|---|---|---|---|---|---|---|
| E-1 | c.20 C > A | S7X | Nonsense | Donaudy(2004) [7] | + | + | + |
| E-2 | c.73 C > T | G25X | Nonsense | Kim(2016) [17] | – | – | – |
| E-2 | c.359 C > T | S120L | Missense | Yang(2010) [16] | + | + | + |
| E-2 | c.466C > T | S120L | Missense | Yang(2005) [3] | – | – | – |
| E-3 | c.541 G > A | A181T | Missense | Qing (2014) [25] | + | – | + |
| E-3 | c.572 A > G | A191G | Missense | Kim(2016) [17] | – | – | – |
| E-9 | c.1126G > T | G376C | Missense | Donaudy(2004) [7] | – | – | – |
| E-12 | c.1314del | P438L | Missense | – | – | – | + |
| E-16 | c.2176 C > A | R726S | Missense | Donaudy(2004) [7] | – | – | + |
| E-19 | c.2299C > A | A767S | Missense | – | + | + | – |
| E-20 | c.2569A > C | L857G | Missense | – | – | – | + |
| E-22 | c.2594C > T | T865 M | Missense | Qing (2014) [25] | – | – | + |
| E-22 | c.2692A > C | L898G | Missense | – | + | – | – |
| E-23 | c.2717C > T | T906M | Missense | – | + | – | – |
| E-23 | c.2798G > T | A933L | Missense | – | – | – | + |
| E-23 | c.2822G > T | R941L | Missense | Choi(2011) [6, 26] | – | – | – |
| E-24 | c.2921G > T | A974L | Missense | – | + | + | – |
| E-24 | c.2926C > T | L976F | Missense | Donaudy(2004) [7] | – | – | + |
| E-24 | c.2971G > A | G991L | Missense | – | – | + | – |
| E-25 | c.3049C > T | L1017P | Missense | – | + | + | – |
| E-33 | c.4780G > A | G1594L | Missense | – | – | – | + |
| E-34 | c.4885C > T | A1629C | Missense | – | – | – | + |
| E-35 | c.4903G > A | G1635L | Missense | – | + | – | – |
| E-36 | c.5008C > T | A1670C | Missense | Vona (2014) [27] | + | – | – |
| E-39 | c.5417C > A | A1806D | Missense | this study | - | - | - |
| E-41 | c.5893G > T | G1965X | Nonsense | - | - | - | + |
| E-43 | c.6016G > T | G2006T | Missense | – | + | – | – |
c variation at cDNA level, Clinvar Clinvar Database, DVD Deafness Variation Database, LOVD3 Leiden Open Variation Databas, MYH14 myosin heavy chain 14 (NM_001077186)
aAll nucleotide and amino acid are abbreviated according to the International Union of Pure and Applied Chemistry
Conclusion
This study identified the novel and potentially pathogenic heterozygous missense variant c.5417C > A (p.A1806D) in the MYH14 gene; this variant is responsible for postlingual nonsyndromic SNHL in a four-generation Han Chinese family. The findings also highlighted the combined application of gene capture and Sanger sequencing is an efficient approach to screen pathogenic variants associated with genetic diseases such as autosomal dominant nonsyndromic deafness.
Supplementary information
Additional file 1: Table 1. Summary of the 127 targeted deafness genes.
Additional file 2: Table 2. Candidate variants were verified in the family using Sanger sequencing.
Acknowledgements
We are grateful to the family members for their contributions to this research.
Authors’ contributions
All authors contributed significantly to this research. MW and YZ collected the information. YZ and FZ carried out the experiments. XB analyzed the data. MW, ZF, XB, and HW conceived and designed the study. MW, YZ, XB, and HW drafted and refined the manuscript. All authors read and approved the final manuscript.
Funding
In this study, clinical examination, laboratory testing and data analysis were supported by grants from the National Natural Science Foundation of China (81670942, 81470704, 81670932). The manuscript preparation was mainly supported by the Key Technology Research and Development Program of Shandong (2019GSF108248).
Availability of data and materials
The raw datasets generated and analysed during the current study are under restricted access from the Key Technology Research and Development Program of Shandong. However, they are available from the corresponding author on reasonable request.
The datasets generated and/or analysed during the current study are available in NCBI dbSNP (https://www.ncbi.nlm.nih.gov/snp/?term=), 1000 Genomes (http://www.1000genomes.org), HapMap database (https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html), BGI’s own databases (BGI Inc., Shenzhen, China), Clinvar Database (https://www.ncbi.nlm.nih.gov/), Deafness Variation Database (http://deafnessvariationdatabase.org/gene/MYH14), and Leiden Open Variation Database (https://databases.lovd.nl/shared/genes/MYH14). The accession numbers corresponding to some of the datasets used in this study and obtained from NCBI (National Center for Biotechnology Information) include: NM_001077186.2, NP_001070654.1, NP_001094160.1, NP_001258467.1, XP_014980128.1, XP_003316592.1, XP_023988172.1, XP_010813541.1 and the NCBI37/hg19 assembly (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/).
Ethics approval and consent to participate
The study protocol was permitted by the Ethics Committee of Shandong Provincial ENT Hospital, Shandong University (XYK20140101), and adhered to the Declaration of Helsinki principles. Informed written consent was obtained from each subject or, in the case of minors, from their parents.
Consent for publication
Written consent was obtained from all the participants or their guardians for the publication of this study.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mingming Wang and Yicui Zhou contributed equally to this work.
Contributor Information
Xiaohui Bai, Email: baixiaohui@sdu.edu.cn.
Haibo Wang, Email: whboto11@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01086-y.
References
- 1.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288(46):33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X, Billington N, Shu S, Yu SH, Piszczek G, Sellers JR, Korn ED. Effect of ATP and regulatory light-chain phosphorylation on the polymerization of mammaliannonmuscle myosin II. Proc Natl Acad Sci U S A. 2017;114(32):E6516–E6525. doi: 10.1073/pnas.1702375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang T, Pfister M, Blin N, Zenner HP, Pusch CM, Smith RJH. Genetic heterogeneity of deafness phenotypes linked to DFNA4. Am J Med Genet. 2005;139(1):9–12. doi: 10.1002/ajmg.a.30989. [DOI] [PubMed] [Google Scholar]
- 4.Leal A, Endele S, Stengel C, Kathrin H, Loetterle J, Barrantes R, et al. A novel myosin heavy chain gene in human chromosome 19q13.3. Gene. 2003;312:165–171. doi: 10.1016/s0378-1119(03)00613-9. [DOI] [PubMed] [Google Scholar]
- 5.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496(1):3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 6.Choi BO, Kang SH, Hyun YS, Kanwal S, Park SW, Koo H, et al. A complex phenotype of peripheral neuropathy, myopathy, hoarseness, and hearing loss is linked to an autosomal dominant mutation in MYH14. Hum Mutat. 2011;32(6):669–677. doi: 10.1002/humu.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Stazio MD, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am J Hum Genet. 2004;74(4):770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebrahim S, Fujita T, Millis BA, Kozin E, Ma X, Kawamoto S, et al. NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry. Curr Biol. 2013;23(8):731–736. doi: 10.1016/j.cub.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wylie SR, Chantler PD. Myosin IIC: a third molecular motor driving neuronal dynamics. Mol Biol Cell. 2008;19(9):3956–3968. doi: 10.1091/mbc.E07-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buxton DB, Golomb E, Adelstein RS. Induction of nonmuscle myosin heavy chain II-C by butyrate in RAW 264.7 mouse macrophages. J Biol Chem. 2003;278(17):15449–15555. doi: 10.1074/jbc.M210145200. [DOI] [PubMed] [Google Scholar]
- 11.Golomb E, Ma X, Jana SS, Preston YA, Kawamoto S, Shoham NG, et al. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J Biol Chem. 2004;279(4):2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- 12.Hilgert N, Smith RJ, Camp GV. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2–3):189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shearer AE, Hildebrand MS, Smith RJH. GeneReviews®. 1999. Hereditary Hearing Loss and Deafness Overview. [Google Scholar]
- 14.Bearer EL, Chen AF, Chen AH, Li Z, Mark HF, Smith RJ, Jackson CL. 2E4/Kaptin (KPTN) - a candidate gene for the hearing loss locus, DFNA4. Ann Hum Genet. 2000;64(Pt 3):189–196. doi: 10.1046/j.1469-1809.2000.6430189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mhatre AN, Li J, Chen AF, Yost CS, Smith RJ, Kindler CH, Lalwani AK. Genomic structure, cochlear expression, and mutation screening of KCNK6, a candidate gene for DFNA4. J Neurosci Res. 2004;75(1):25–31. doi: 10.1002/jnr.10839. [DOI] [PubMed] [Google Scholar]
- 16.Yang R, Li H, Zhan CX, Mao HY, Zhan TL, Zhu ZF, et al. C.359T>C mutation of the MYH14 gene in two autosomal dominant non-syndromic hearing impairment families with common ancestor. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27(3):259–262. doi: 10.3760/cma.j.issn.1003-9406.2010.0.005. [DOI] [PubMed] [Google Scholar]
- 17.Kim SJ, Lee S, Park HJ, Kang TH, Sagong B, Baek JI, et al. Genetic association of MYH genes with hereditary hearing loss in Korea. Gene. 2016;591(1):177–182. doi: 10.1016/j.gene.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Xiao Y, Zhang F, Han Y, Li J, Xu L, et al. Novel compound heterozygous mutations in MYO7A gene associated with autosomal recessive sensorineural hearing loss in a Chinese family. Int J Pediatr Otorhinolaryngol. 2016;83:179–185. doi: 10.1016/j.ijporl.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Wang M, Xiao Y, Zhang F, Zhou Y, Li J, et al. A novel nonsense mutation of POU4F3 gene causes autosomal dominant hearing loss. Neural Plast. 2016;2016:1512831. doi: 10.1155/2016/1512831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang T, Wei X, Chai Y, Li L, Wu H. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85. doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Y, Luo J, Zhang F, Li J, Han Y, Zhang D, et al. A novel mutation in PAX3 associated with Waardenburg syndrome type I in a Chinese family. Acta Otolaryngol. 2016;136(5):439–445. doi: 10.3109/00016489.2015.1132846. [DOI] [PubMed] [Google Scholar]
- 22.Bai X, Lv H, Zhang F, Liu J, Fan Z, Xu L, et al. Identification of a novel missense mutation in the WFS1 gene as a cause of autosomal dominant nonsyndromic sensorineural hearing loss in all-frequencies. Am J Med Genet B Neuropsychiatr Genet. 2014;164 A(12):3052–3060. doi: 10.1002/ajmg.a.36760. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto N, Okano T, Ma X, Adelstein R, Kelley M. Myosin II regulates extension, growth and patterning in the mammalian cochlear duct. Development. 2009;136(12):1977–1986. doi: 10.1242/dev.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D, et al. Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res. 1991;69(2):530–539. doi: 10.1161/01.res.69.2.530. [DOI] [PubMed] [Google Scholar]
- 25.Qing J, Yan D, Zhou Y, Liu Q, Wu W, Xiao Z, et al. Whole-exome sequencing to decipher the genetic heterogeneity of hearing loss in a Chinese family with deaf by deaf mating. PLoS One. 2014;9(10):e109178. doi: 10.1371/journal.pone.0109178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyadurai S, Arnold WD, Kissel JT, Ruhno C, Mcgovern VL, Snyder PJ, et al. Variable phenotypic expression and onset in MYH14 distal hereditary motor neuropathy phenotype in a large, multigenerational north American family. Muscle Nerve. 2017;56(2):341–345. doi: 10.1002/mus.25491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vona B, Müller T, Nanda I, Neuner C, Hofrichter MA, Schröder J, et al. Targeted next-generation sequencing of deafness genes in hearing-impaired individuals uncovers informative mutations. Genet Med. 2014;16(12):945–953. doi: 10.1038/gim.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table 1. Summary of the 127 targeted deafness genes.
Additional file 2: Table 2. Candidate variants were verified in the family using Sanger sequencing.
Data Availability Statement
The raw datasets generated and analysed during the current study are under restricted access from the Key Technology Research and Development Program of Shandong. However, they are available from the corresponding author on reasonable request.
The datasets generated and/or analysed during the current study are available in NCBI dbSNP (https://www.ncbi.nlm.nih.gov/snp/?term=), 1000 Genomes (http://www.1000genomes.org), HapMap database (https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html), BGI’s own databases (BGI Inc., Shenzhen, China), Clinvar Database (https://www.ncbi.nlm.nih.gov/), Deafness Variation Database (http://deafnessvariationdatabase.org/gene/MYH14), and Leiden Open Variation Database (https://databases.lovd.nl/shared/genes/MYH14). The accession numbers corresponding to some of the datasets used in this study and obtained from NCBI (National Center for Biotechnology Information) include: NM_001077186.2, NP_001070654.1, NP_001094160.1, NP_001258467.1, XP_014980128.1, XP_003316592.1, XP_023988172.1, XP_010813541.1 and the NCBI37/hg19 assembly (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/).