Abstract
Characterization of foodborne pathogens including Salmonella species allows for the determination of their relationship and/or relatedness with others. This study characterized Salmonella enterica (S. enterica) isolated from five meat types (mutton, beef, chevon, guinea fowl, and local chicken) obtained from Tamale metropolis, Ghana. The S. enterica were characterized phenotypically (n = 44) based on their antibiotic resistance pattern with the disc diffusion method and genetically (n = 16) using whole‐genome sequencing (WGS) as well as with bioinformatic analysis for the prediction of their clonal and phylogenomic relationship. Of the 225 meat samples examined, 107 (47.56%) were positive for S. enterica. Mutton was the most contaminated meat type and the least was local chicken. The 44 S. enterica isolates exhibited five different antibiotic patterns with multiple antibiotic resistance (MAR) index ranging from 0.13 to 0.63. Resistant to only erythromycin was most common and was exhibited by 34 isolates (77.27%). Four isolates were resistant to four different antibiotics (TeAmpSxtECro) with a percentage of 9.09%, while two isolates (4.55%) were resistant to none of the antibiotics. The sequenced S. enterica isolates consisted of 7 serovars and 8 clonal lineages with the S. enterica subsp. enterica serovar Hato (ST5308) being the predominate strain. Phylogenomic analysis showed that the isolates clustered according to their serovars and sequence types (clonal lineages). However, further metadata insights coupled with the phylogenomics revealed a complex intraspread of multiple S. enterica subsp. enterica serovars in diverse meat sources in areas in Tamale which is very worrying for infection management. In summary, our study provides useful insights into S. enterica in meat reservoirs obtained from Tamale metropolis, Ghana.
Keywords: antibiotics, characterization, meat, phylogenomic analysis, Salmonella species
Meat samples were contaminated by Salmonella and aerobic bacteria. Salmonella species exhibited some resistance to antibiotics. The Salmonella isolates were genetically diverse.
![]()
1. INTRODUCTION
Meats are major component of human diets and serve as an excellent source of protein. Other nutrients including fats (omega‐3‐ polyunsaturated fatty acids), minerals (iron, magnesium, potassium, selenium sodium, and zinc), and vitamins (vitamin A, vitamin E, B6, B12, niacin, thiamine, and riboflavin) can also be found in meats (Ahmad, Imran, & Hussain, 2018; America Meat Science Association, 2016). They are consumed worldwide by all races except vegetarians and people who have deliberately refuse to eat meats due to welfare concerns and/or love for animals. The consumption of meats has been associated with the risk of foodborne infections and illnesses.
Salmonellae are gram‐negative facultative anaerobe bacteria that have been associated with foodborne infections (Wallace & Hammack, 2013). For instance, a recent foodborne outbreak suspected to be caused by Salmonella Dublin was linked to the consumption of ground beef (Centers for Disease Control & Prevention, 2019). Other Salmonella infections have been linked to pork, 30% hospitalizations with 0 death (Centers for Disease Control & Prevention, 2015), mechanically separated chicken, 22% hospitalizations with 0 death (Centers for Disease Control & Prevention, 2014) and chicken, 33% hospitalizations with 0 death (Centers for Disease Control & Prevention, 2013). A total of 91,662 human confirmed cases of Salmonella infections were reported in 2017 by the European Union, with 43.1% hospitalizations and 0.25% case fatality (European Food Safety Authority, 2018). Saba et al. (2013) indicated that data on Salmonella infections in most developing and underdeveloped countries is scared.
Similarly, reported incidences of Salmonella infections in Ghana are limited if not unavailable, but the organism has been found in various meats including, beef, chevon, mutton, and pork (Adzitey, 2015; Adzitey, Nsoah, & Teye, 2015; Adzitey, Teye, & Anachinaba, 2015; Danikuu, 2004). The treatment of Salmonella infections relies on the use of antibiotics. Meanwhile, resistance of Salmonella to antibiotics is a treat to public health and a concern worldwide. Salmonella isolated from various meat samples have been demonstrated to be resistant to one or more antibiotics such as amoxycillin/clavulanic acid, ampicillin, chloramphenicol, sulfamethoxazole/trimethoprim, tetracycline, vancomycin, and others (Adzitey, 2015; Arslan & Eyi, 2010; Ejo, Garedew, & Alebachew, 2016).
Characterization of foodborne pathogens has some importance including the determination of their history, relationship, and closeness. This intend helps to predict their characteristics, properties, or behavior from others. Characterization of foodborne pathogens at the phenotypic and genotypic levels have been achieved using serotyping, antibiotic profiling, whole‐genome sequencing, multilocus sequencing typing, pulsed‐field gel electrophoresis, repetitive extragenic palindromic, among others (Adhikari et al., 2010; Adzitey, Deli, & Ali, 2015; Adzitey, Saba, & Deli, 2014; Jaja, Bhembe, Green, Oguttu, & Muchenje, 2019).
Report on the characterization and comparison of Salmonella from various meat sources (mutton, beef, chevon, guinea fowl, and local chicken) in the Tamale metropolis is scare. Therefore, this study was carried out to characterize Salmonella enterica isolated from various meat types using antibiotic resistance, and phylogenomic analyses.
2. MATERIALS AND METHODS
2.1. Study area
The study was conducted in the Tamale metropolis, Ghana. The metropolis lies in between latitude 9°16 and 9°34 North and longitudes 0°36 and 0°57 West (Ghana Statistical Service, 2014). The Tamale metropolis shares boundaries with Sanarigu District to the west and north, Mion District to the east, East Gonja to the south, and Central Gonja to south‐west. It is the capital town of the Northern Region of Ghana and the third most populace town in Ghana (Ghana Statistical Service, 2014).
2.2. Samples examined
Two hundred and twenty‐five (225) samples made of beef, chevon, mutton, local chicken, and guinea fowl were examined. Forty‐five (45) samples each of the various meat types were randomly sampled from both traditional and close modern markets between the hours of 10:00–14:00 GMT. An area of 10 cm2 was swabbed using sterile cotton swabs. The swab samples were transported in an ice chest containing ice block and were analyzed immediate on reaching the laboratory.
2.3. Analysis of meat samples for Salmonella enterica
A slightly modified method of the Food and Drug Administration‐Bacteriological Analytical Manual was used (Adzitey, Nsoah, et al., 2015; Wallace & Hammack, 2013). Briefly, swab samples were pre‐enriched in 10 ml Buffered Peptone Water (BPW) and incubated at 37°C for 24 hr. Then, 0.1 ml aliquots were transferred into 10 ml Rappaport and Vassiliadis (RV) and Selenite Cystine (SC) broths. Samples in RV broths were incubated at 42°C for 24 hr while samples in SC broths were incubated at 37°C for 24 hr (enrichment). After which 0.1 ml of the aliquots were streaked on Xylose Lysine Deoxycholate and Brilliant Green agars and incubated at 37°C for 24–48 hr. Presumptive Salmonella colonies were picked, purified, Gram stained and subjected to the following biochemical tests; growth characteristics on triple sugar iron, lysine iron and Simon citrate agars, and urease production. Salmonella isolates were confirmed by Latex Agglutination Kit for Salmonella (Oxoid Limited). All media used were also purchased from Oxoid Limited.
2.4. Analysis of meat samples for microbial load
Microbial load determination was done as describe by Maturin and Peeler (2001) and Adzitey, Ekli, and Abu (2019). Swabs were dipped into 10 ml of 1% BPW and thoroughly agitated. Serial dilutions (10−1–10−5) were made in 9 ml BPW using 1 ml, and 100 µl dilution spread plated onto Plate Count Agar (PCA; Oxoid Limited) plates. The plates were incubated at 37°C for 24 hr and colonies counted using a colony counter.
2.5. Phenotypic antibiotic susceptibility testing
The disk diffusion method of Bauer, Kirby, Sherris, and Turk (1966) was used for antibiotic susceptibility testing of 44 pure S. enterica isolates against the following antibiotics; ampicillin (10 μg), ceftriaxone (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), erythromycin (15 µg), gentamicin (10 μg), suplfamethoxazole/trimethoprim (22 μg), and tetracycline (30 μg). The purified Salmonella species were inoculated in Trypticase Soy Broth (TSB; Oxoid Limited) and incubated at 37°C for 18 hr. The turbidity was adjusted to 0.5 McFarland standard using sterile TSB and spread plated on Müller Hinton Agar (MHA; Oxoid). Four antibiotic disks were placed on the surface of the MHA at a distance to avoid overlapping of inhibition zones. The plates were incubated at 37°C for 24 hr. After incubation, the inhibition zones were measured, and the results interpreted using the Clinical Laboratory Standard Institute (2017). Multiple antibiotic resistance (MAR) index was calculated and interpreted according to Krumperman (1983) using the formula: a/b, where “a” represents the number of antibiotics to which a particular isolate was resistant and “b” the total number of antibiotics tested.
2.6. Genomic sequence, assembly annotation and bioinformatic analysis of Salmonella enterica
Sixteen S. enterica were randomly selected, sequenced, assembled, and annotated as described by Tay et al. (2019).
2.7. WGS‐based molecular typing and phylogenomic analyses of Salmonella enterica
Multilocus sequence typing (MLST) was performed in silico using the WGS data online platform MLST v2.0 (https://cge.cbs.dtu.dk/services/MLST/) from the assembled genomes which also predicted the allelic profiles of the seven housekeeping genes of S. enterica. The reference Salmonella online platform, SeqSero v1.0 (www.denglab.info/SeqSero) was used to infer the serotypes of the isolates.
A phylogenetic tree was also constructed for all the genomes to determine the relatedness of the S. enterica strains using CSI Phylogeny‐v1.4 (https://cge.cbs.dtu.dk/services/CSIPhylogeny/), an online service which identifies SNPs from WGS data, filters and validates the SNP positions, and then infers phylogeny based on concatenated SNP profiles. A bootstrapped with 100 replicates indicator was applied to identify recombined regions and provide the phylogenetic accuracy in groups with little homoplasy. The Figtree was used to edit and visualize the phylogenetic tree. The phylogeny was visualized alongside annotations for isolate demographics (source and area of collection) and WGS in silico molecular typing (serovar and sequence type) metadata using Phandango (Hadfield et al., 2017).
2.8. Accession numbers
The raw read sequences and the assembled whole‐genome contigs have been deposited in GenBank under the project number PRJNA484344.
2.9. Statistical analysis
Data obtained for S. enterica was analyzed using binary logistic generalized linear model of Statistical Package for Service Solutions Program version 20.0; and test for statistical difference was done using wald chi‐square. Means were separated at 5% significancet level. Microbial load was analyzed using GenStat Release 12 Edition; and Analysis of Variance was used to test the significant difference at p < .05.
3. RESULTS
3.1. Distribution of Salmonella enterica and microbial load in the various meat types
The distribution of S. enterica and microbial load in the meats is shown in Table 1. S. enterica were found in beef 19 (42.22%), chevon 22 (48.89%), guinea fowl 20 (44.44%), local chicken 13 (28.89%), and mutton 33 (73.33%). The contamination of mutton by S. enterica was significantly higher (p < .05) than the rest of the meat types. The presence of S. enterica in local chicken was significantly lower (p < .05) than chevon but not guinea fowl and beef. Contamination of beef, chevon, and guinea fowl by S. enterica did not differ (p > .05) from each other.
TABLE 1.
Distribution of Salmonella enterica and microbial load in various meat types sold at the Tamale metropolis, Ghana
| Sample | No. of samples examined | No. (%) positive | Microbial load (log cfu/cm2) |
|---|---|---|---|
| Beef | 45 | 19 (42.22) | 3.36 |
| Chevon | 45 | 22 (48.89) | 4.03 |
| Guinea fowl | 45 | 20 (44.44) | 3.33 |
| Local chicken | 45 | 13 (28.89) | 4.34 |
| Mutton (Lamb) | 45 | 33 (73.33) | 4.9 |
| Overall | 225 | 107 (47.56) | 3.99 |
Abbreviations: No., number of samples positive for Salmonella enterica.
The microbial load was 3.36, 4.03, 3.33, 4.34, and 4.90 log cfu/cm2 for beef, chevon, Microbial load did not differ significantly (p = .212) among the various meat samples.
3.2. Phenotypic antibiotic susceptibility testing of Salmonella enterica isolates
The phenotypic antibiotic characterization of the 44 S. enterica isolates is shown in Tables 2 and 3. The S. enterica isolates were highly resistant to erythromycin (93.18%) but susceptible to ampicillin (79.55%), ciprofloxacin (97.73%), chloramphenicol (93.18%), gentamicin (79.55%), suplfamethoxazole/trimethoprim (90.91%), and tetracycline (84.09%).
TABLE 2.
Phenotypic antibiotic susceptibility of Salmonella enterica in various meat types sold at the Tamale metropolis, Ghana
| Antimicrobial | % Resistance | % Intermediate resistance | % Susceptibility |
|---|---|---|---|
| Ampicillin (AMP) 10 µg | 11.36 | 9.09 | 79.55 |
| Ciprofloxacin (CIP) 5 µg | 0.00 | 2.27 | 97.73 |
| Ceftriaxone (CRO) 30 µg | 13.64 | 20.45 | 65.91 |
| Chloramphenicol (C) 30 µg | 0.00 | 6.82 | 93.18 |
| Erythromycin (E) 15 µg | 93.18 | 0.00 | 6.82 |
| Gentamicin (CN) 10 µg | 4.55 | 15.91 | 79.55 |
| Sulphamethoxazole/trimethoprim (SXT) 22 µg | 9.09 | 0.00 | 90.91 |
| Tetracycline (TE) 30 µg | 9.09 | 6.82 | 84.09 |
| Overall % | 17.61 | 7.67 | 74.72 |
TABLE 3.
Antibiotic resistance profile and multiple antibiotic resistance index of individual Salmonella enterica isolated from various meat types sold at the Tamale metropolis, Ghana
| No. | Salmonella code | Source | Antibiotic resistant profile a | Number of antibiotics | MAR index |
|---|---|---|---|---|---|
| 1 | NB10 | Beef | AmpECro | 3 | 0.38 |
| 2 | AB11 | Beef | Cro | 1 | 0.13 |
| 3 | AC2 | Chevon | E | 1 | 0.13 |
| 4 | AC3 | Chevon | E | 1 | 0.13 |
| 5 | AC5 | Chevon | E | 1 | 0.13 |
| 6 | AM2 | Mutton | E | 1 | 0.13 |
| 7 | AM3 | Mutton | E | 1 | 0.13 |
| 8 | NC2 | Chevon | E | 1 | 0.13 |
| 9 | NC6 | Chevon | E | 1 | 0.13 |
| 10 | CB1 | Beef | E | 1 | 0.13 |
| 11 | CB5 | Beef | E | 1 | 0.13 |
| 12 | CB14 | Beef | E | 1 | 0.13 |
| 13 | CC3 | Chevon | E | 1 | 0.13 |
| 14 | CC5 | Chevon | E | 1 | 0.13 |
| 15 | CC8 | Chevon | E | 1 | 0.13 |
| 16 | CM1 | Mutton | E | 1 | 0.13 |
| 17 | CM7 | Mutton | E | 1 | 0.13 |
| 18 | CM11 | Mutton | E | 1 | 0.13 |
| 19 | NC13 | Chevon | E | 1 | 0.13 |
| 20 | NM1 | Mutton | E | 1 | 0.13 |
| 21 | CG1 | Guinea fowl | E | 1 | 0.13 |
| 22 | CG4 | Guinea fowl | E | 1 | 0.13 |
| 23 | CG15 | Guinea fowl | E | 1 | 0.13 |
| 24 | NLC9 | Local chicken | E | 1 | 0.13 |
| 25 | NLC13 | Local chicken | E | 1 | 0.13 |
| 26 | SG14 | Guinea fowl | E | 1 | 0.13 |
| 27 | TG5 | Guinea fowl | E | 1 | 0.13 |
| 28 | TG14 | Guinea fowl | E | 1 | 0.13 |
| 29 | TG15 | Guinea fowl | E | 1 | 0.13 |
| 30 | NLC8 | Local chicken | E | 1 | 0.13 |
| 31 | SLC10a | Local chicken | E | 1 | 0.13 |
| 32 | SLC10b | Local chicken | E | 1 | 0.13 |
| 33 | SLC10c | Local chicken | E | 1 | 0.13 |
| 34 | TLC7a | Local chicken | E | 1 | 0.13 |
| 35 | TLC7b | Local chicken | E | 1 | 0.13 |
| 36 | TLC7c | Local chicken | E | 1 | 0.13 |
| 37 | AB7 | Beef | ECn | 2 | 0.25 |
| 38 | AC3 | Chevon | ECn | 2 | 0.25 |
| 39 | AM10 | Mutton | None | 0 | 0.00 |
| 40 | NB8 | Beef | None | 0 | 0.00 |
| 41 | NB2 | Beef | TeAmpSxtECro | 5 | 0.63 |
| 42 | NM7 | Mutton | TeAmpSxtECro | 5 | 0.63 |
| 43 | NM14 | Mutton | TeAmpSxtECro | 5 | 0.63 |
| 44 | SG15 | Guinea fowl | TeAmpSxtECro | 5 | 0.63 |
Ampicillin (Amp) 10 μg; ciprofloxacin (Cip) 5 µg; ceftriaxone (Cro) 30 µg; gentamicin (Cn) 10 µg; erythromycin (E) 15 µg; sulphamethoxazole/trimethoprim (Sxt) 22 µg; tetracycline (Te) 30 µg.
The multiple antibiotic (MAR) index ranged from 0.13 (resistant to one antibiotic) to 0.63 (resistant to five antibiotics). The 44 S. enterica isolates were resistance to zero (4.55%), one (79.55%), two (4.55%), three (2.27%), and five (9.09%) antibiotics. They exhibited five different resistance patterns thus, AmpECro (1 isolate), Cro (1 isolate), E (34 isolates), ECn (2 isolates), and TeAmpSxtECro (4 isolates). Multidrug resistant that is resistant to 3 or more different classes of antibiotics was observed in five (11.36%) of the isolates.
3.3. Phylogenomic analysis and metadata insights
The phylogenetic relationship and epidemiological distribution of the S. enterica isolate from meat samples in Tamale are depicted in Figure 1. The isolates generally clustered according to their serovars and sequence types (clonal lineages). Of note, there were 7 clades (grouped A–G) and 1 subclade (A1, ST3899) which collaborated with the 8 clonal lineages. This affirms the ability of WGS‐based typing methods to accurately differentiate between isolates using appropriately curated databases.
FIGURE 1.
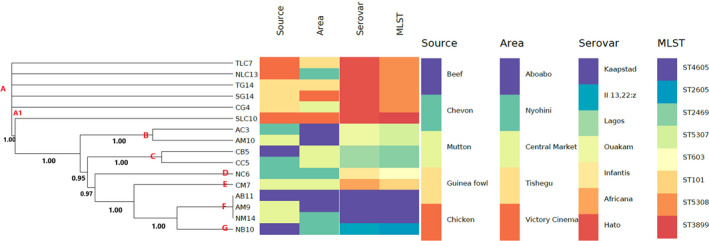
The whole‐genome MLST phylogenetic branch and metadata (WGS in‐silico molecular typing [serovar and sequence type] and Demographics [source and area]) coupled using Phandango in isolated Salmonellae enterica strains (n = 16) isolated from meats in the Tamale metropolis of Ghana
4. DISCUSSION
Results from this study indicated that, meat samples obtained from the Tamale metropolis were contaminated with S. enterica. Therefore, eating undercooked meats can serve as sources of Salmonella infections. Contamination of meats by S. enterica was highest in mutton and least in local chicken. This is not surprising since it was observed during sample collection that the environment where local chickens were processed was neater than where mutton and other ruminants were processed. Cattle, sheep, and goats are normally processed into beef, chevon, and mutton, respectively, in the Tamale abattoir which lacks all the equipment required for a modern abattoir. Furthermore, butchers slaughtered animals on the floor and do not adhere to strict hygienic slaughter and personal hygiene. Guinea fowls and local chickens are normally processed by individuals in an open market or by the roadside. Most often, guinea fowls and local chickens are sold directly after processing at the processed point, while beef, chevon, and mutton are transported to from the abattoir to the market for sale. They are transported from the abattoir to the market by taxis, motor kings®, motorbikes, bicycles, and sometimes meat vans that are not well maintained. The meats are transported openly or covered with plastic rubber. The selling points are either at the roadside, in traditional open or close modern markets. All of these expose meats to microorganisms blow by dust, wind, or smoke from cars. The contamination of the various meat types by S. enterica can also be attributed to the tables on which meats are placed for sale and the knives used for cutting meats (Adzitey, Nsoah, et al., 2015). The S. enterica could have cross‐contaminated the meats from the gastrointestinal tracts. Adzitey, Sulleyman, and Kum (2020) also found that meat sellers in the Tamale metropolis do not sterilize their knives and majority (48%) of them sold meat on open tables, which expose meats to flies, dusts, smoke from vehicles, and other contaminants. The contamination of beef samples by Salmonella species was higher (75%) in the Techiman municipality (Adzitey, Nsoah, et al., 2015) and lower (31%) in the Tamale metropolis (Adzitey, 2015) of Ghana as compared to this study. Arslan and Eyi (2010) found Salmonella species in 29.3% of poultry meat and 16% of beef samples in Bolu, Turkey. The prevalence results for poultry meat were similar to this study but a higher prevalence of Salmonella was detected in beef samples in this study. In Gondar, Ethiopia, Salmonella species were detected in 12% of raw meat which was lower than that of this study (Ejo et al., 2016).
The intrinsic characteristics of meats such as nutrient composition, pH, water activity, and temperature promote the growth of microorganisms. Meat is a good medium for the growth of microorganisms including S. enterica because it rich in protein, lipids, and other nutrients which microorganisms use for their growth (Prescott, Harley, & Klein, 2002). Salmonella spp. have also been reported to grow at a temperature range of 5°C–47°C (optimum of 35°C–37°C), pH range of 4–9 (optimum of 6.5–7.5) and a water between of 0.99 and 0.94 (Dodd, Aldsworth, Stein, Cliver, & Riemann, 2017). Meat has a neutral pH and a water activity of between 0.98 and 0.99 (Meter Food, 2017). The pH and water activity of meat favors the growth of S. enterica. Also the meats sampled were warm and sold under ambient temperature, thus providing favorable environment for the growth of S. enterica and other microorganisms. The microbial load (<106) observed in this study is considered satisfactory (Center for Food Safety, 2014). Numerically, it was highest in mutton and lowest in guinea fowl. The scalding of guinea fowl could have contributed to the lower microbial load observed as compared to mutton. Microbial load for local chicken was also expected to be lower than that of beef and chevon but this was not observed. The presence of microbial load in the meat samples examined means that lapses occurred during the processing of the meat samples as the muscle of a healthy living animal is essentially sterile. A higher microbial load ranging from 3.99–6.19 log cfu/cm2 in fresh guinea fowls (Adzitey, Teye, et al., 2015) and 4.75–6.31 log cfu/cm2 for fresh beef (Anachinaba, Adzitey, & Teye, 2015) was reported in the Bolgatanga municipality, Ghana. Soepranianondo, Wareham, Budiarto, and Diyantoro (2019) reported a lower microbial load of 1.62 log cfu/g in beef samples collected from slaughterhouses in East Java, Indonesia as compared to this study. However, a higher microbial load (5.40–8.35 log cfu/g) in comparison with this study was reported by Jahan, Mahbub‐E‐Elahi, and Siddique (2015) in beef obtained from markets of Sylhet Sadar in Banladesh.
The resistance of S. enterica to some of the antibiotics used in this study can be attributed to the use of these antibiotics for the treatment of cattle, goats, guinea fowls, local chickens, and sheep during their production. Reports from other researchers have shown that farm animals are sources of antibiotic resistant Salmonella strains (Founou, Amoako, Founou, & Essack, 2018; Jaja et al., 2019; Nair, Venkitanarayanan, & Johny, 2018; Wang et al., 2019) that can be transferred to humans. Adzitey, Nsoah, et al. (2015) also found that Salmonella species of beef origin were highly resistant to Erythromycin (75.56%), but susceptible to ciprofloxacin (100%), gentamicin (86.67%), ceftriaxone (73.33%), and sulfamethoxazole/trimethoprim (68.89%). Arslan and Eyi (2010) reported that Salmonella species of poultry and beef origin were susceptible to ampicillin (63.6 vs. 0.0), ciprofloxacin (81.8 vs. 100), chloramphenicol (59.1 vs. 91.7), gentamicin (72.7 vs. 91.7), sulfamethoxazole/trimethoprim (63.6 vs. 91.7), and tetracycline (31.8 vs. 58.3), respectively.
Similarly, to the current study Adzitey, Nsoah, et al. (2015) reported MAR index range of 0.11–0.67 for Salmonella species isolated from beef. They also found that the Salmonella species exhibited multiple antibiotic resistance and 23 different resistance patterns. Jaja et al. (2019) reported MAR index of 0.67–0.93 and 0.60–0.93 for Salmonella species isolated from meats collected from formal meats sector and informal slaughter points, respectively. Other researchers including Arslan and Eyi (2010) and Ejo et al. (2016) have also reported multidrug resistance Salmonella strains of meat origin. Arslan and Eyi (2010) indicated that 62% of Salmonella strains exhibited multiple resistance to three or more antimicrobial agents. Ejo et al. (2016) showed that 20% Salmonella isolates were resistant to one antimicrobial, while 80% were resistant to two or more antimicrobials. Jaja et al. (2019) found a high prevalence of multidrug‐resistant S. enterica isolates in meats and indicated that there is a high risk associated with the consumption of contaminated meat. Resistant Salmonella species can contaminate carcasses, processing equipment, and other foods which pose a risk for public and animal health.
The tree analysis coupled with metadata revealed useful insights with regard to the diversity of serovars clones in meat sources and area of collection (Figure 1). For instance; meat sources; beef, chevon, and mutton contained different serovars of S. enterica isolates which were clonally distinct (Figure 1). This finding corroborated with other studies reported worldwide specifically in Europe (Müller, Jansen, Grabowski, & Kehrenberg, 2018), Africa (Thomas et al., 2020), and Asia (Yang et al., 2019) which isolated different serovars S. enterica in food samples. However, all the guinea fowl and chicken samples contained the same serovar; S. enterica subsp. enterica serovar Hato strain which predominately belonged to the ST5308 clone (Figure 1). Interestingly, Fagbamila et al. (2017) also reported the presence of Hato serovar in chicken layer farms in Nigeria.
With respect to the area of collection; there was clonal spread of the S. enterica subsp. enterica serovars in some areas in Tamale irrespective of the meat source. For example; the Kaapstad serovar (ST4605) was found in both beef and mutton samples in Aboabo while the Ouakam serovar (ST5307) were also isolated in chevon and mutton in the same area. More so, beef and chevon from the central market area contained the S. enterica subsp. enterica serovar Lagos (ST2469) strain. A similar trend was also observed in guinea fowl and chicken from both Victory Cinema and Tishegu area. This complex intraspread of multiple S. enterica subsp. enterica serovars in diverse meat sources in areas in Tamale is very worrying for infection management. A combination of WGS data, demographics, and graphical visualization should be applied to offer vital insights and increase confidence during molecular epidemiological investigations (Amoako et al., 2019).
5. CONCLUSION
Overall, 107 (47.56%) Salmonella species and 3.99 log cfu/cm2 microbial load were detected in the meat samples. Mutton (lamb) was the most contaminated source. Phenotypic characterization revealed a high resistance to erythromycin but susceptibility (≥90) to ciprofloxacin, chloramphenicol, and sulfamethoxazole/trimethoprim. Phylogenomic analysis showed that the isolates clustered according to their serovars and sequence types (clonal lineages). However, further metadata insights coupled with the phylogenomics revealed a complex intraspread of multiple S. enterica subsp. enterica serovars in diverse meat sources in areas in Tamale which is very worrying for infection management. In summary, our study provides useful insights into S. enterica in meat reservoirs obtained from Tamale metropolis, Ghana which warrants an urgent action to curb this possible threat.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ETHICAL APPROVAL
The study did not involve any human or animal testing.
INFORMED CONSENT
Verbal consent was obtained from all meat sellers.
ACKNOWLEDGMENTS
The authors are grateful to the University for Development Studies for making available a microbiology laboratory for this work. The sequencing was supported by Commonwealth Science Conference (CSC) Follow‐on Travel Grants (CSC\R1\170022) and Nanyang Technological University Research Initiative. We also acknowledged Dr. Moon Tay Yue Feng and Prof. Jorgen Schlundt of Nanyang Technological University Food Technology Centre (NAFTEC) for their assistance with the sequencing.
Adzitey F, Teye GA, Amoako DG. Prevalence, phylogenomic insights, and phenotypic characterization of Salmonella enterica isolated from meats in the Tamale metropolis of Ghana. Food Sci Nutr. 2020;8:3647–3655. 10.1002/fsn3.1647
REFERENCES
- Adhikari, B. , Besser, T. E. , Gay, J. M. , Fox, L. K. , Hancock, D. D. , & Davis, M. A. (2010). Multilocus variable‐number tandem‐repeat analysis and plasmid profiling to study the occurrence of blaCMY‐2 within a pulsed‐field gel electrophoresis‐defined clade of Salmonella enterica serovar Typhimurium. Applied and Environmental Microbiology, 76, 69–74. 10.1128/AEM.00210-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzitey, F. (2015). Prevalence of Escherichia coli and Salmonella spp. in beef samples sold at Tamale metropolis, Ghana. International Journal of Meat Science, 5, 8–13. 10.3923/ijmeat.2015.8.13 [DOI] [Google Scholar]
- Adzitey, F. , Deli, R. A. , & Ali, G. R. R. (2015). Characterization of Salmonella Typhimurium, Salmonella Enteritidis and Salmonella Albany isolated from chickens and ducks using random amplified polymorphic DNA (RAPD)‐PCR. Journal of Microbiology, Immunology and Biotechnology, 2, 15–21. [Google Scholar]
- Adzitey, F. , Ekli, R. , & Abu, A. (2019). Prevalence and antibiotic susceptibility of Staphylococcus aureus isolated from raw and grilled beef in Nyankpala community in the Northern Region of Ghana. Cogent Food and Agriculture, 5, 1671115 10.1080/23311932.2019.1671115 [DOI] [Google Scholar]
- Adzitey, F. , Nsoah, J. K. , & Teye, G. (2015). Prevalence and antibiotic susceptibility of Salmonella species isolated from beef and its related samples in Techiman Municipality of Ghana. Turkish Journal of Agriculture ‐ Food Science and Technology, 3, 644–650. 10.24925/turjaf.v3i8.644-650.399 [DOI] [Google Scholar]
- Adzitey, F. , Saba, C. S. S. , & Deli, R. A. (2014). Characterization of Salmonella Typhimurium, Salmonella Enteritidis and Salmonella Albany isolated from chickens and ducks using repetitive extragenic palindromic (Rep)‐PCR. Ghana Journal of Science, Technology and Development, 1, 1–10. [Google Scholar]
- Adzitey, F. , Sulleyman, K. W. , & Kum, P. K. (2020). Knowledge and practices of meat safety by meat sellers in the Tamale metropolis of Ghana. Food Protection Trends, 40, 40–47. [Google Scholar]
- Adzitey, F. , Teye, G. A. , & Anachinaba, I. A. (2015). Microbial quality of fresh and smoked guinea fowl meat sold in the Bolgatanga Municipality, Ghana. Asian Journal of Poultry Science, 9, 165–171. 10.3923/ajpsaj.2015.165.171 [DOI] [Google Scholar]
- Ahmad, R. S. , Imran, A. , & Hussain, M. B. (2018). Nutritional composition of meat. Retrieved from https://www.intechopen.com/books/meat‐science‐and‐nutrition/nutritional‐composition‐of‐meat, Accessed on 01/11/2019. [Google Scholar]
- America Meat Science Association . (2016). Nutrients in meat. Retrieved from https://meatscience.org/TheMeatWeEat/topics/meat‐in‐the‐diet/nutrients‐in‐meat, Accessed on 01/06/2017. [Google Scholar]
- Amoako, D. G. , Somboro, A. M. , Abia, A. L. K. , Allam, M. , Ismail, A. , Bester, L. , & Essack, S. Y. (2019). Genomic analysis of methicillin‐resistant Staphylococcus aureus isolated from poultry and occupational farm workers in Umgungundlovu District, South Africa. Science of the Total Environment, 670, 704–716. 10.1016/j.scitotenv.2019.03.110 [DOI] [PubMed] [Google Scholar]
- Anachinaba, I. A. , Adzitey, F. , & Teye, G. A. (2015). Assessment of the microbial quality of locally produced meat (beef and pork) in Bolgatanga Municipal of Ghana. Internet Journal of Food Safety, 17, 1–5. [Google Scholar]
- Arslan, S. , & Eyi, A. (2010). Occurrence and antimicrobial resistance profiles of Salmonella species in retail meat products. Journal of Food Protection, 73, 1613–1617. 10.4315/0362-028X-73.9.1613 [DOI] [PubMed] [Google Scholar]
- Bauer, A. W. , Kirby, W. M. M. , Sherris, J. C. , & Turk, M. (1966). Antibiotic susceptibility testing by a standardized single disc method. American Journal of Clinical Pathology, 45, 493–496. [PubMed] [Google Scholar]
- Center for Food Safety . (2014). Microbiological guidelines for food (for ready‐to‐eat food in general and specific food items. Retrieved from https://www.cfs.gov.hk/english/food_leg/files/food_leg_Microbiological_Guidelines_for_Food_e.pdf, Accessed 20/07/2019.
- Centers for Disease Control and Prevention . (2013). Multistate outbreak of Salmonella Heidelberg infections linked to chicken. Retrieved from https://www.cdc.gov/salmonella/heidelberg‐02‐13/index.html, Accessed on 11/11/2019.
- Centers for Disease Control and Prevention . (2014). Outbreak of Salmonella Heidelberg infections linked to Tyson Brand mechanically separated chicken at a correctional facility. Retrieved from https://www.cdc.gov/salmonella/heidelberg‐01‐14/index.html, Accessed on 11/11/2019.
- Centers for Disease Control and Prevention . (2015). Multistate outbreak of multidrug‐resistant Salmonella I 4,[5],12:i:‐ and Salmonella Infantis infections linked to pork. Retrieved from https://www.cdc.gov/salmonella/pork‐08‐15/index.html, Accessed on 11/11/2019.
- Centers for Disease Control and Prevention . (2019). Outbreak of Salmonella infections linked to ground beef. Retrieved from https://www.cdc.gov/salmonella/dublin‐11‐19/index.html, Accessed on 11/11/2019.
- Clinical and Laboratory Standards Institute . (2017). Performance standards for antimicrobial susceptibility testing: 27th edition informational supplement M100–S27. Wayne, PA: CLSI. [Google Scholar]
- Danikuu, M. F. (2004). Isolation and serotyping of Salmonella from slaughtered food animals in the Kumasi metropolis. Retrieved from http://ir.knust.edu.gh/handle/123456789/1936, Accessed on 11/11/2019.
- Dodd, C. E. R. , Aldsworth, T. , Stein, R. A. , Cliver, D. O. , & Riemann, H. P. (2017). Foodborne diseases, third edition. Cambridge, MA: Academic Press. Imprint: ISBN 978‐0‐12‐385007‐2. [Google Scholar]
- Ejo, M. , Garedew, L. , Alebachew, Z. , & Worku, W. (2016). Prevalence and antimicrobial resistance of Salmonella isolated from animal‐origin food items in Gondar, Ethiopia. Biomed Research International, 2016, 4290506, 10.1155/2016/4290506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority . (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2017. European Food Safety Authority Journal, 16, 5500 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbamila, I. O. , Barco, L. , Mancin, M. , Kwaga, J. , Ngulukun, S. S. , Zavagnin, P. , … Muhammad, M. (2017). Salmonella serovars and their distribution in Nigerian commercial chicken layer farms. PLoS ONE, 12, e0173097 10.1371/journal.pone.0173097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Founou, L. L. , Amoako, D. G. , Founou, R. C. , & Essack, S. Y. (2018). Antibiotic resistance in food animals in Africa: A systematic review and meta‐analysis. Microbial Drug Resistance, 25, 648–665. 10.1089/mdr.2017.0383 [DOI] [PubMed] [Google Scholar]
- Ghana Statistical Service . (2014). 2010 Population and Housing Census, District Analytical Report, Tamale Metropolis. Retrieved from http://www2.statsghana.gov.gh/docfiles/2010_District_Report/Northern/Tamale%20Metropolitan.pdf, Accessed on 26/05/2019.
- Hadfield, J. , Croucher, N. J. , Grater, R. J. , Abudahab, K. , Aanensen, D. M. , & Harris, S. R. (2017). Phandango: An interactive viewer for bacterial population genomics. Bioinformatics, 34, 292–293. 10.1093/bioinformatics/btx610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan, F. , Mahbub‐E‐Elahi, A. T. M. , & Siddique, A. B. (2015). Bacteriological quality assessment of raw beef sold in Sylhet Sadar. The Agriculturists, 13, 09–16. 10.3329/agric.v13i2.26654 [DOI] [Google Scholar]
- Jaja, I. F. , Bhembe, N. L. , Green, E. , Oguttu, J. , & Muchenje, V. (2019). Molecular characterisation of antibiotic‐resistant Salmonella enterica isolates recovered from meat in South Africa. Acta Tropica, 190, 129–136. 10.1016/j.actatropica.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high‐risk sources of fecal contamination of food. Applied and Environmental Microbiology, 46, 165–170. 10.1093/ajcp/45.4_ts.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maturin, L. , & Peeler, J. T. (2001). Bacteriological analytical manual, chapter 3: Aerobic plate count. Retrieved from https://www.fda.gov/food/laboratory‐methods‐food/bam‐aerobic‐plate‐count, Accessed on 25/08/2017. [Google Scholar]
- Meter Food . (2017). How water activity and pH work together to control microbial growth. Retrieved from https://www.metergroup.com/food/articles/how‐water‐activity‐and‐ph‐work‐together‐to‐control‐microbial‐growth/, Accessed on 11/04/2020.
- Müller, A. , Jansen, W. , Grabowski, N. T. , & Kehrenberg, C. (2018). Characterization of Salmonella enterica serovars recovered from meat products legally and illegally imported into the EU reveals the presence of multiresistant and AmpC‐producing isolates. Gut Pathogens, 10, 40 10.1186/s13099-018-0268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, D. V. T. , Venkitanarayanan, K. , & Johny, A. K. (2018). Antibiotic‐resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods, 7, 167 10.3390/foods7100167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, L. M. , Harley, J. P. , & Klein, D. A. (2002). Food and industrial microbiology (pp. 125–964). New York, USA: Mc Graw‐ Hill Companies, Inc. [Google Scholar]
- Saba, C. K. , Escudero, J. A. , Herrera‐Leon, S. , Porrero, M. C. , Suarez, M. , Dominguez, L. , … Gonzalez‐Zorn, B. (2013). First identification of Salmonella Urbana and Salmonella Ouakam in humans in Africa. The Journal of Infection in Developing Countries, 7, 691–695. 10.3855/jidc.3548 [DOI] [PubMed] [Google Scholar]
- Soepranianondo, K. , Wardhana, D. K. , Budiarto, & Diyantoro. (2019). Analysis of bacterial contamination and antibiotic residue of beef meat from city slaughterhouses in East Java Province, Indonesia. Veterinary World, 12, 243–248. 10.14202/vetworld.2019.243-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, M. Y. F. , Adzitey, F. , Sultan, S. A. , Tati, J. M. , Seow, K. L. G. , & Schlundt, J. (2019). Whole‐genome sequencing of nontyphoidal Salmonella enterica isolates obtained from various meat types in Ghana. Microbial Resource Announcement, 8, e00033‐19 10.1128/MRA.00033-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, K. M. , de Glanville, W. A. , Barker, G. C. , Benschop, J. , Buza, J. J. , Cleaveland, S. , … Crump, J. A. (2020). Prevalence of Campylobacter and Salmonella in African food animals and meat: A systematic review and meta‐analysis. International Journal of Food Microbiology, 315, 108382 10.1016/j.ijfoodmicro.2019.108382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, H. A. , & Hammack, T. S. (2013). Salmonella in bacteriological analytical manual. Retrieved from http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/UCM070149, Accessed on 01/06/2017. [Google Scholar]
- Wang, X. , Biswas, S. , Paudyal, N. , Pan, H. , Li, X. , Fang, W. , & Yue, M. (2019). Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Frontiers in Microbiology, 10, 985 10.3389/fmicb.2019.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Wu, Q. , Zhang, J. , Huang, J. , Chen, L. , Wu, S. , … Wei, X. (2019). Prevalence, bacterial load, and antimicrobial resistance of Salmonella serovars isolated from retail meat and meat products in China. Frontiers in Microbiology, 10, 2121 10.3389/fmicb.2019.02121 [DOI] [PMC free article] [PubMed] [Google Scholar]


