Significance
Emergence of the pathogen Klebsiella pneumoniae (particularly of carbapenem-resistant strains) is considered a dire threat to public health. Resistance and virulence determinants may favor the emergence of untreatable infections. Understanding the mechanisms involved in the pathogenesis and epidemicity of K. pneumoniae is essential for managing outbreaks and developing treatments. Here we identify a CUP system (Kpi) and infer the epidemiology of Kpi+ K. pneumoniae in Europe. We demonstrate a direct link between Kpi presence and K. pneumoniae persistence in the hospital environment. Adherence of the bacterium to different cell types enables host colonization, favoring nosocomial outbreaks and spread of infection. Kpi appears to play a key role in the host–pathogen interaction and is associated with the worldwide-disseminated ST-15 clone.
Keywords: Klebsiella pneumoniae, GI tract colonization, ST-15 high-risk clone, pathogenesis, chaperone-usher pili system
Abstract
Control of infections caused by carbapenem-resistant Klebsiella pneumoniae continues to be challenging. The success of this pathogen is favored by its ability to acquire antimicrobial resistance and to spread and persist in both the environment and in humans. The emergence of clinically important clones, such as sequence types 11, 15, 101, and 258, has been reported worldwide. However, the mechanisms promoting the dissemination of such high-risk clones are unknown. Unraveling the factors that play a role in the pathobiology and epidemicity of K. pneumoniae is therefore important for managing infections. To address this issue, we studied a carbapenem-resistant ST-15 K. pneumoniae isolate (Kp3380) that displayed a remarkable adherent phenotype with abundant pilus-like structures. Genome sequencing enabled us to identify a chaperone-usher pili system (Kpi) in Kp3380. Analysis of a large K. pneumoniae population from 32 European countries showed that the Kpi system is associated with the ST-15 clone. Phylogenetic analysis of the operon revealed that Kpi belongs to the little-characterized γ2-fimbrial clade. We demonstrate that Kpi contributes positively to the ability of K. pneumoniae to form biofilms and adhere to different host tissues. Moreover, the in vivo intestinal colonizing capacity of the Kpi-defective mutant was significantly reduced, as was its ability to infect Galleria mellonella. The findings provide information about the pathobiology and epidemicity of Kpi+ K. pneumoniae and indicate that the presence of Kpi may explain the success of the ST-15 clone. Disrupting bacterial adherence to the intestinal surface could potentially target gastrointestinal colonization.
The global spread of carbapenem-resistant Enterobacteriaceae (CRE) is a major threat in healthcare settings as these bacteria cause infections associated with high mortality, primarily due to delays in the administration of appropriate empirical therapy and the limited treatment options available (1–3). In this era of antibiotic resistance, Klebsiella pneumoniae is a particularly dangerous multidrug-resistant (MDR) pathogen as it rapidly acquires resistance to all known antibiotics, especially carbapenems (last-line class of antibiotics), and it is thus becoming more and more difficult to treat (4). Moreover, K. pneumoniae is the most common carbapenem-resistant Enterobacteriaceae and one of the most common pathogens causing nosocomial infections. Examination of genomic and epidemiological data from 1,649 K. pneumoniae isolates collected from 244 hospitals in 32 countries across Europe, showed that harboring a carbapenemase is the main cause of carbapenem resistance in diverse phylogenetic backgrounds (5). However, the majority of carbapenemase-positive K. pneumoniae isolates belong to only four clonal lineages: Sequence types (STs) 11, 15, 101, 258/512, and their derivatives (5). Antimicrobial resistance and pathogenic efficacy are likely to be key factors in the success of these worldwide-disseminated high-risk K. pneumoniae clones.
The high incidence of K. pneumoniae infections is promoted by the ability of this pathogen to colonize the gastrointestinal (GI) tract, which is linked to subsequent infection (6, 7). GI colonization thus represents an important reservoir of strains involved in nosocomial infections (8) and which can potentially cause outbreaks. In this context, the successful fight against carbapenem-resistant K. pneumoniae infections should not only focus on antimicrobial resistance but also on the virulence factors that mediate the infectivity of the species. In addition to the advances in the application of carbapenemase inhibitors to treat these infections (9), monoclonal antibodies that act on K. pneumoniae are also being developed and are showing promise as new therapeutic approaches or strategies (10, 11). In this respect, preclinical studies have demonstrated that the antibody KP3, which binds the type 3 fimbrial subunit, MrkA, may contribute to providing protection against MDR K. pneumoniae (10–12). Targeting adherence-related virulence factors is a promising strategy as these factors may determine the capacity of a colonizing strain to cause infection. K. pneumoniae expresses several fimbrial surface structures in order to adhere to abiotic and biotic surfaces. Chaperone-usher pili (CUP) systems are the most abundant adhesive structures in K. pneumoniae and consist of one usher, at least one chaperone, and at least one fimbrial subunit. As CUP operons may have different numbers of chaperones and fimbrial subunits (13), they are classified on the basis of the usher protein into α-, β-, γ-, κ-, π-, and σ-fimbrial clades. Moreover, the γ-fimbrial clade is divided into γ1-, γ2-, γ3-, and γ4-fimbriae on the basis of specific operon features (14).
Up to nine different CUP systems have been identified in a single strain, apparently conferring the capacity to attach to different surfaces (15, 16). Almost all K. pneumoniae isolates possess both classic types of CUP systems: Type 1 (fim) and type 3 (mrk) pili. Type 1 pili play a role in bacterial adherence to human mucosal or epithelial surfaces; although type 3 pili also attach to cell surfaces, they are considered important enhancers of biofilm formation (17). Type 1 pili are key pathogenic factors in K. pneumoniae urinary tract infections (UTIs); however, their involvement in intestinal colonization or lung infection is unclear. It has recently been reported that deletion of the fim operon in an uropathogenic Escherichia coli strain leads to significant defects in colonization (18). However, intestinal colonization and lung infection caused by K. pneumoniae are not affected by either type 1 or type 3 pili (19, 20). In this study, we identified a CUP system in an epidemic carbapenem-resistant K. pneumoniae strain. We demonstrate that this pilus system is involved in biofilm formation and adherence, as well as in the intestinal colonizing capacity of the pathogen. The system may therefore be considered a key pathogenic factor promoting the dissemination of K. pneumoniae. Analysis of a large and epidemiologically diverse K. pneumoniae population revealed that all ST-15 isolates share the same Kpi operon identified in the outbreak-related Kp3380 strain. Thus, the presence of Kpi may explain the success of the ST-15 clone.
Results
Clinical Impact of a Carbapenem-Resistant K. pneumoniae Strain.
At A Coruña University Hospital from 2013 to 2018, patients with positive CRE clinical samples were routinely screened for intestinal colonization of CRE for infection control purposes and according to local guidelines. During the 6-y-long study, 5,340 K. pneumoniae nonduplicate isolates were detected in clinical and colonization samples in the Microbiology Department. Approximately a 13% (682 of 5,340) of these isolates were positive for OXA-48 carbapenemase, thereby dramatically increasing the prevalence of carbapenem-resistant isolates. Genotyping showed that most of the isolates belonged to an ST-15 clone, which was initially isolated from 15 patients in 2013 and spread rapidly, causing a large outbreak despite the implementation of strict control measures. We tested intestinal colonization in 462 of 682 patients infected or colonized with the OXA-48–producing K. pneumoniae strain. A 93.3% of them were positive for this epidemic clone. To understand the success of the ST-15 clone, we conducted an in-depth characterization of the associated virulence functions.
The Epidemic Clone Kp3380 Is Highly Adherent.
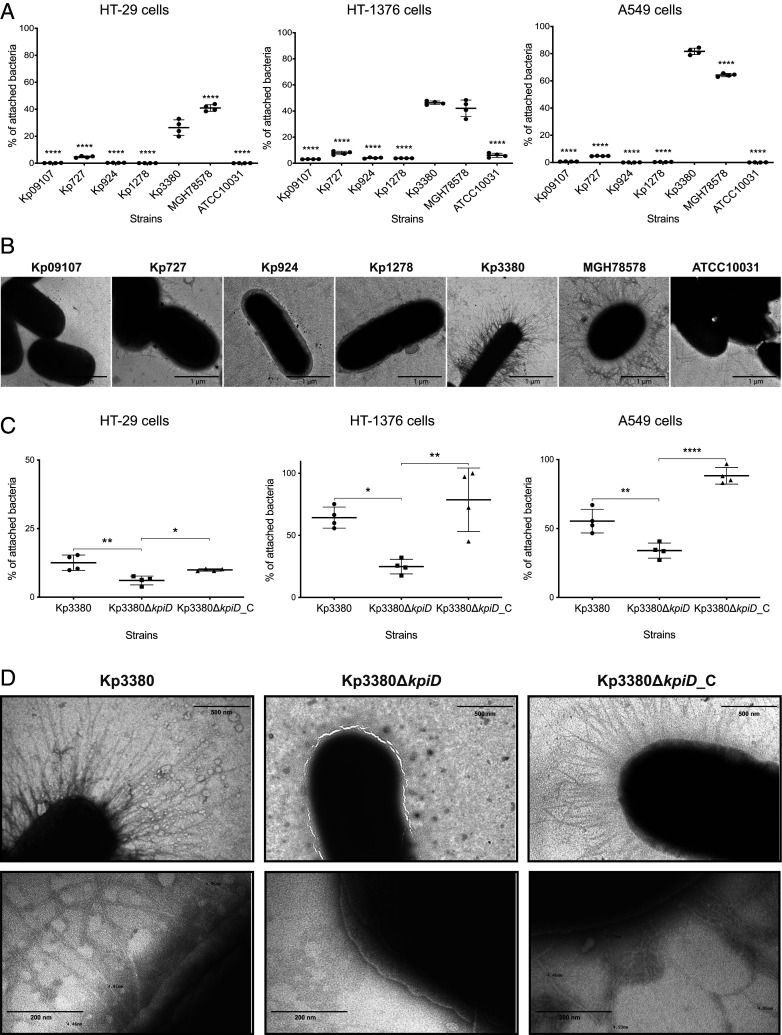
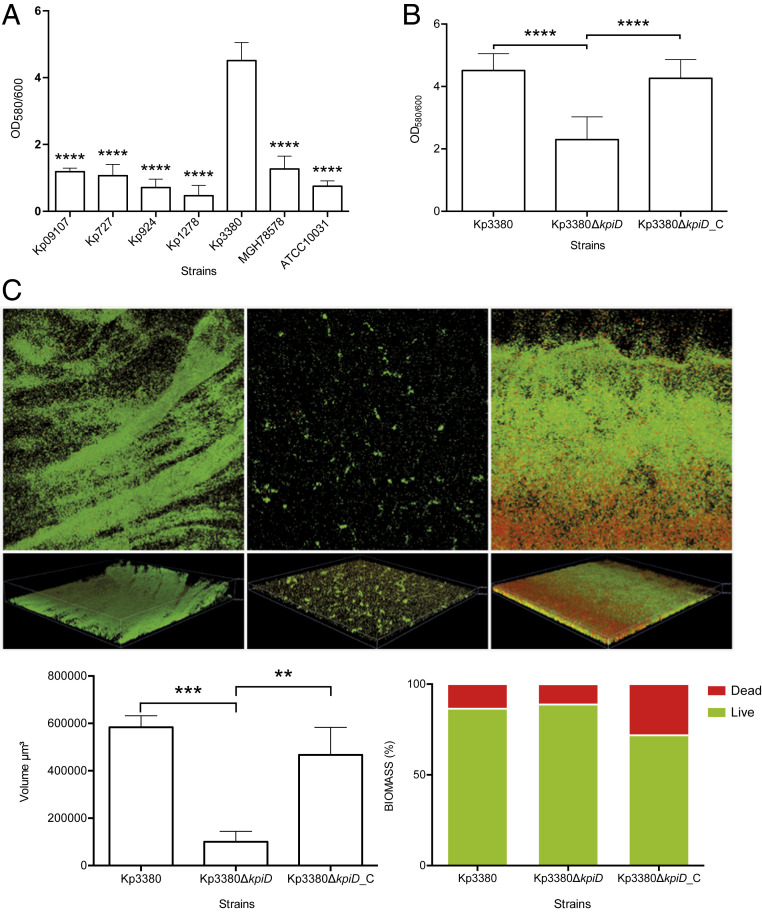
The Kp3380 isolate was used as a representative clone of the above-mentioned outbreak. We used eukaryotic epithelial cells (HT-29 colorectal cells, HT-1376 bladder cells, and A549 alveolar cells) to analyze the adherence phenotype of the clinical strains Kp3380, Kp09107, Kp924, Kp727, Kp1278, along with the reference strains MGH78578 and ATCC10031, which represent different host cells targeted by K. pneumoniae. The data collected (Fig. 1A) revealed that Kp3380 and MGH78578 had a higher capacity for adherence than the other strains tested (P < 0.0001). Similarly, strain Kp3380 demonstrated a hyperbiofilm-producing phenotype, reaching an OD580/600 ratio of ∼4.5 (as assessed by Crystal violet staining), which is significantly different (P < 0.0001) from that of most of the strains assayed (i.e., OD580/600 ratio between 0.4 and 1.3) (Fig. 2A). We wondered whether the differences in adherence shown by these strains could be explained by bacterial surface structures used in cell adhesion processes. To address this question, we cultured strains under the same conditions used to infect the human epithelial cells and examined the cells by transmission electron microscopy (TEM). Microscopic observation of the surface appendages produced by these strains revealed quantitative and qualitative differences between the strains analyzed (Fig. 1B). We observed a relationship between the presence of abundant pilus-like structures on the bacterial surface and a highly adherent phenotype.
Fig. 1.
Adherence phenotype and TEM images of K. pneumoniae. (A and C) Quantification of the adherence of K. pneumoniae strains to HT-29 intestinal colorectal cells, HT-1376 bladder cells, and A549 alveolar cells. Bacterial adherence is reported as the percentage of attached bacteria relative to the number of bacteria used to infect the eukaryotic cells (assumed to be 100%). The data represent four independent biological replicates. ANOVA was applied to the data to indicate significant differences between the groups, and a post hoc Tukey’s test was used to determine the difference between each group and the respective control (Kp3380 in A and Kp3380ΔkpiD in C). Statistical significance is indicated (*P ≤ 0.05, **P ≤ 0.01, and ****P ≤ 0.0001). Values shown are means, and error bars indicate SDs. (B and D) TEM images of K. pneumoniae strains. Micrographs were taken at 30,000× (B), 60,000× (D, Upper), and 250,000× (D, Lower) magnification. Bars indicate the scale.
Fig. 2.
Analysis of biofilm formation of K. pneumoniae isolates. (A and B) Quantification of biofilm formation by Crystal violet staining. Six independent biological replicates were analyzed. ANOVA was applied to the data to indicate significant differences between the groups, and a post hoc Tukey’s test was used to determine the difference between each group and the respective control (Kp3380 in A and Kp3380ΔkpiD in B and C). Statistical significance is indicated (**P ≤ 0.01, ***P ≤ 0.001, and ****P < 0.0001). The values shown are means, and error bars indicate SDs. (C) CLSM images of K. pneumoniae Kp3380, Kp3380ΔkpiD and Kp3380ΔkpiD_C isolates (Upper). Bacteria were stained using the BacLight LIVE/DEAD viability kit. Live cells fluoresce in green and dead cells fluoresce in red. Original magnification 200×. Quantification of fluorescence (live/dead) for each strain is represented Lower Left. Biofilm biomass (live/dead) is represented as a percentage Lower Right.
Characterization of a CUP System in Kp3380.
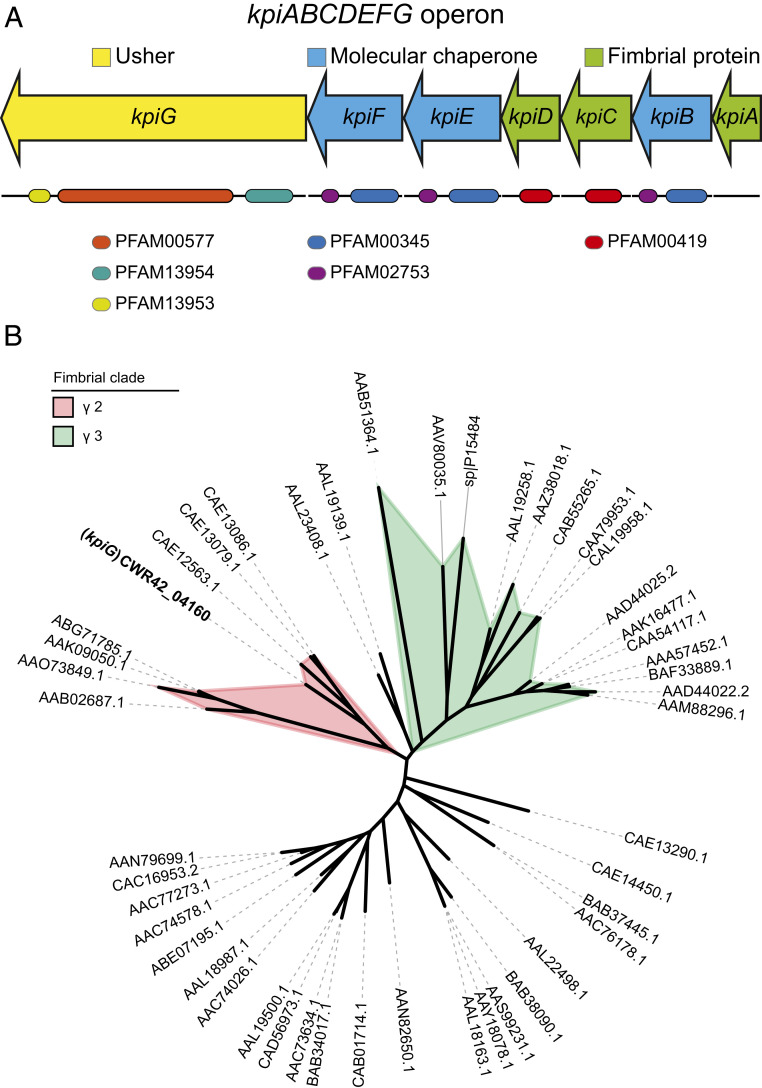
Protein coding genes involved in adherence were analyzed relative to the fully sequenced, annotated genome of clinical isolate Kp3380 (21). Comparative genome analysis enabled us to identify a region of ∼6,500 bp containing seven genes, which are thought to be involved in adherence. In silico analysis of the genomic region revealed that it may code for a polycistronic operon. RT-PCR analysis of total RNA demonstrated that genes CWR42_04160 to CWR42_04190 are cotranscribed as a single operon. Intergenic fragments amplified from cDNA match those observed when genomic DNA was used as a template (SI Appendix, Fig. S1). The operon, named here kpiABCDEFG, consists of one usher gene (kpiG), three molecular chaperone genes (kpiB, kpiE, and kpiF), and three fimbrial genes (kpiA, kpiC, and kpiD) (Fig. 3A). The function of each gene was predicted on the basis of amino acid homologies with other fimbrial proteins. The protein structure of each gene product was predicted using the RaptorX server. Each protein belonging to the Kpi pilus system was used as a query to search for related protein structures included in the Protein Database. The results are summarized in SI Appendix, Table S1. Protein structure modeling revealed that the Kpi pilus system is structurally related to CUP system type 1 (SI Appendix, Table S1). However, the phylogenetic analysis based on the usher (KpiG) amino acid sequence comparisons (SI Appendix, Table S2) showed that Kpi is included in the phylogenetic clade γ2 (Fig. 3B).
Fig. 3.
Characterization of the CUP system Kpi in K. pneumoniae. (A) Genetic organization of the kpiABCDEFG operon. Arrows indicate the location and direction of gene transcription, and colors indicate functions. Functional domains are pointed. (B) ML tree of the usher amino acid sequences described by Nuccio and Baumler (14) (SI Appendix, Table S2) and the usher (KpiG) sequence identified in the Kp3380 genome (GenBank accession code PITM00000000.1). Graphic representation was generated using iTOL v5.1.1.
A more detailed functional analysis of each protein of the Kpi system was conducted using the InterPro and HHpred servers. Three conserved usher domains were identified in KpiG: PF00577 (amino acids 203 to 767), PFAM13954 (amino acids 37 to 182), and PFAM13953 (amino acids 776 to 842) (Fig. 3A). HHpred analysis revealed that all fimbrial proteins contain the conserved domains COG3539 and PFAM00419. However, the domain PFAM00419 was only found in KpiC and KpiD (amino acids 54 to 205 and 38 to 191, respectively) when using InterPro server (Fig. 3A). All three chaperones (KpiB, KpiE, and KpiF) were found to contain two conserved chaperone domains: PFAM00345 (amino acids 24 to 154, 4 to 172, and 3 to 163, respectively) and PFAM02753 (amino acids 176 to 235, 194 to 253, and 186 to 243, respectively) (Fig. 3A). Comparative functional and sequencing analysis of the proteins of the Kpi and the γ2-fimbriae Fas (14) revealed that KpiD might be the major fimbrial subunit as it shares the highest sequence identity (21.76% identity and 96% coverage) with FasA. All three chaperones of the Kpi system (KpiB, KpiE, and KpiF) only share sequence identity with FasB (22.41% identity and 98% coverage, 24.12% identity and 85% coverage, 21.63% identity, and 91% coverage, respectively).
The Kpi Fimbriae System Is Closely Associated with the High-Risk Clone K. pneumoniae ST-15.
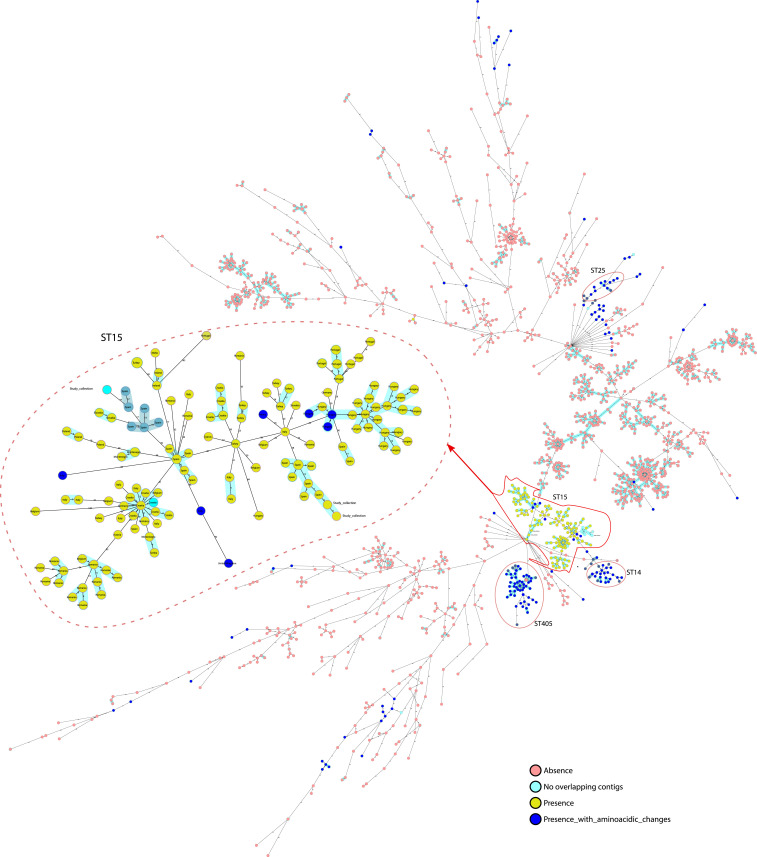
We analyzed the genomes of 1,649 K. pneumoniae isolates collected from 32 European countries as part of the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE), in order to evaluate the distribution of the Kpi system in K. pneumoniae. The Kpi system was detected in 19.1% (315 of 1,649) of isolates belonging to 41 different STs. In addition, 46% (145 of 315 isolates) of Kpi+ K. pneumoniae belonged to ST-15 (Fig. 4), and all of the ST-15 isolates tested positive for the Kpi system. A total of 132 (91.1%) ST-15 isolates shared exactly the same operon with Kp3380 (100% coverage and 100% identity) (Fig. 4). Seven (4.8%) ST-15 isolates had several amino acidic changes, and in six (4.1%) of the ST-15 isolates the whole operon could not be evaluated because of a gap in the genome sequence (between two different contigs) (Fig. 4). The Kpi system was also detected in 16.8% (53 of 315 isolates), 9.5% (30 of 315 isolates), and 4.7% (15 of 315 isolates) of K. pneumoniae isolates belonging to ST-405, ST-14, and ST-25, respectively (Fig. 4). However, the kpiABCDEFG operon from ST-14, ST-25, and ST-405 isolates displayed several amino acid changes relative to the Kp3380 strain (Fig. 4).
Fig. 4.
International spread of Kpi+ K. pneumoniae isolates. The phylogenetic tree includes 1,649 K. pneumoniae isolates collected during the EuSCAPE survey. The colors of the isolates indicate absence/presence of the kpiABCDEFG operon and whether or not the operon has amino acidic changes or the sequence is incomplete. Blue shadow indicates that isolates are genetically related.
Examination of the distribution of ST-15 isolates showed that this clone was involved in outbreaks occurring in Hungary, Croatia, Romania, and Spain (Fig. 4). In addition, the presence of Kpi was evaluated in all of the genomes of K. pneumoniae deposited in GenBank (7,950 genomes), in order to confirm that all ST-15 isolates are positive for the system. Of these genomes, 4.7% (377 of 7,950) of the isolates belonged to ST-15. In addition, 98.1% (370 of 377) of the isolates were Kpi+ and shared the same operon with Kp3380 (100% coverage and 98 to 100% identity). In 1.89% (7 of 377) of the ST-15 isolates, the whole operon could not be evaluated, because of a gap in the genome sequence (between two different contigs). Thus, although the percentage of ST-15 K. pneumoniae isolates in all of the genomes available in GenBank was lower, the results are consistent with those obtained with the EuSCAPE collection.
The Presence of the Kpi Fimbriae System Is Associated with Adherence.
DNA-based multiple comparison of the operon kpiABCDEFG in isolates Kp09107 (ST-101), Kp727 (ST-405), Kp924 (ST-11), Kp1278 (ST-15), Kp3380 (ST-15), and MGH78578 (ST-38) revealed that only Kp3380 and MGH78578 have a complete, apparently functional operon. Analysis of the nucleotide identity of Kpi in MGH78578 revealed several amino acid changes in the operon relative to the Kp3380 strain. Although strain Kp727 has a complete operon, it has multiple stop codons in the kpiD gene. Strain Kp1278 only had the kpiG gene, which codes for the usher protein. Neither of these genes was found in strains Kp924 or Kp09107.
We also analyzed the most common adhesive appendages in K. pneumoniae: That is, the type 1 and type 3 fimbriae (pili), in all of the above-mentioned strains. We found that all strains have intact type 1 (fim) and type 3 (mrk) loci.
We observed a correlation between the presence of the Kpi system and a highly adherent phenotype. Interestingly, we observed a nonadherent phenotype in the Kp727 strain, which harbors a complete operon with multiple stop codons in the kpiD gene. Therefore, deletion of the fimbrial protein KpiD seems to be crucial for the functional inactivation of the Kpi system.
Effect of kpiD Gene Deletion on Attachment to Eukaryotic Cells.
Considering that the fimbrial protein KpiD plays an important role in the production of functional fimbriae in the Kp727 K. pneumoniae strain and it might be the major subunit of the Kpi system, we deleted the kpiD gene to investigate the role of Kpi fimbriae in bacterial functions. A kpiD-defective knockout strain (Kp3380ΔkpiD) was constructed and the parental kpiD gene was introduced into Kp3380ΔkpiD for complementation.
The interactions between Kp3380 (and derivative strains) and eukaryotic cells were also examined. For this purpose, different host cells frequently targeted by K. pneumoniae—including colorectal cells (HT-29), bladder cells (HT-1376), and alveolar cells (A549)—were infected with Kp3380, Kp3380ΔkpiD, and the complemented strain (Kp3380ΔkpiD_C) to determine their adherent phenotype and thus demonstrate the role of the kpiD gene in adherence. This approach revealed that significantly fewer Kp3380ΔkpiD bacteria were recovered from infected cells than from cells infected with Kp3380 bacteria, regardless of the type of host cells tested. Absence of the kpiD gene reduced the number of bacteria attached to HT-29, HT-1376, and A549 eukaryotic cells by, respectively, ∼50% (P ≤ 0.01), 60% (P ≤ 0.05), and 40% (P ≤ 0.01) (Fig. 1C). The parental phenotype was restored in the complemented strain (Kp3380ΔkpiD_C) (Fig. 1C).
Considering that Kp3380 loses its highly adherent phenotype when the kpiD gene is deleted, we wondered whether the production of bacterial surface structures in this mutant strain would be affected. TEM images were obtained from the bacterial surface of the wild-type strain and its derivative strains, cultured under the same culture conditions used to infect the eukaryotic cells. Strain Kp3380 and the Kp3380∆kpiD-complemented strain produced abundant pilus-like structures around the cell surface (Fig. 1D). In contrast, TEM revealed that there were no pilus-like structures around the mutant cells (Fig. 1D).
Deletion of the kpiD Fimbriae Gene Reduces Biofilm Formation.
Two different approaches were used to test the effect of kpiD deletion on biofilm formation. The Crystal violet assay revealed that inactivation of the kpiD gene significantly reduced biofilm production (P ≤ 0.0001) (Fig. 2B). Complementation of the knockout strain with the parental allele (Kp3380ΔkpiD_C) restored the wild-type phenotype (Fig. 2B). Biofilms produced by the strains of interest were also evaluated by confocal laser-scanning microscopy (CLSM) and digital imaging. Fig. 2C shows biomass measurements as well as representative 3D renderings of the biofilms. Quantification using reconstructions of confocal sections revealed minimal differences between live and dead bacteria in comparisons of biofilms formed by the wild-type strain and the knockout strain. However, the Kp3380ΔkpiD strain formed significantly less biofilm, which was also structurally altered (Fig. 2C). Complementation of the mutant strain (Kp3380ΔkpiD_C) restored the parental phenotype. The biofilm formed by the complemented strain showed a structure similar to that of Kp3380.
Kp3380∆kpiD Is Attenuated In Vivo.
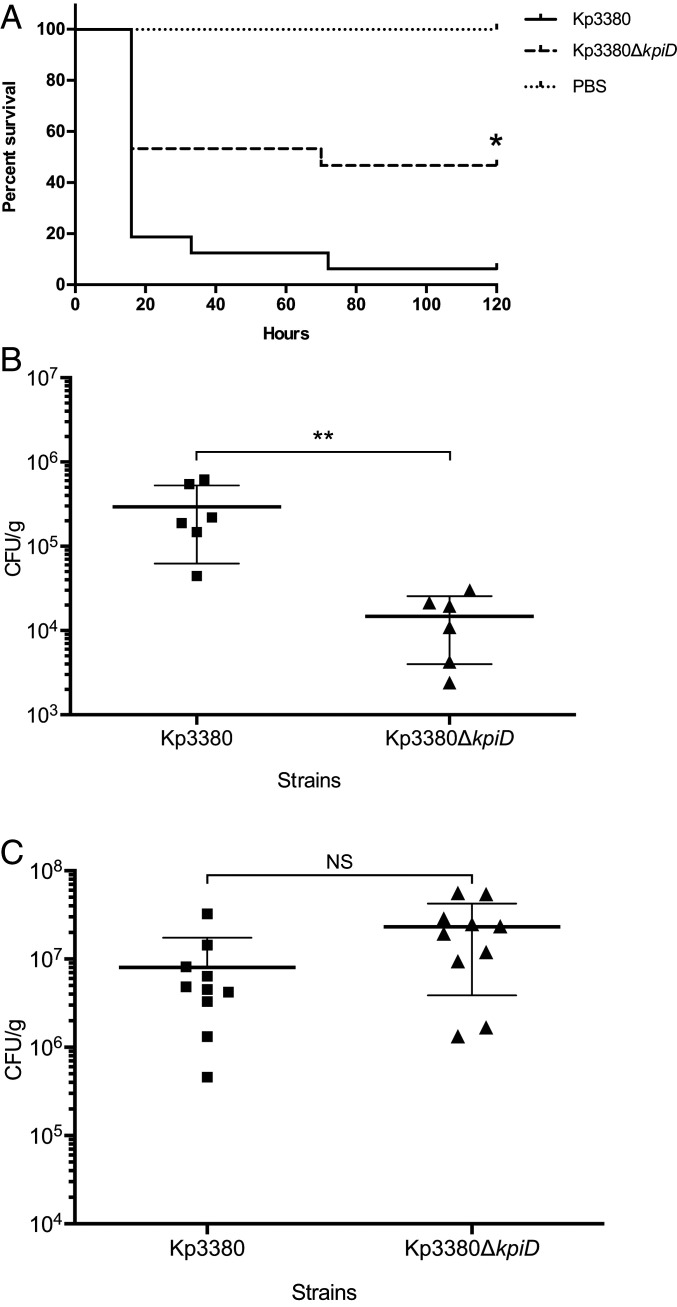
A Galleria mellonella survival assay (16 larvae per group) was used to assess the role of the kpiD gene in the virulence of K. pneumoniae Kp3380. The survival study (Fig. 5A) showed that 84% of the larvae infected with the parental strain died after 5 d of infection, while 50% of the larvae infected with the kp3380∆kpiD mutant survived. The ability of the mutant strain to infect and kill G. mellonella was therefore significantly affected (P = 0.0096), relative to that of the wild-type strain. Larvae injected with sterile PBS were used as negative controls.
Fig. 5.
Role of the kpiD gene in K. pneumoniae virulence. (A) G. mellonella larvae were injected with Kp3380 or Kp3380ΔkpiD bacteria and survival was monitored for 5 d. Larvae injected with PBS were used as negative controls. The log-rank (Mantel–Cox) test was used to compare the survival distributions of two groups: The larvae infected with Kp3380 and the larvae infected with Kp3380ΔkpiD. The statistical significance is indicated (*P = 0.0096). (B) Intestinal colonization of mice with the Kp3380 parental strain or the Kp3380ΔkpiD mutant was achieved by oral gavage with ∼106 bacteria. The bacterial burden in intestinal segments (cecum and colon) was determined 72 h postinoculation. The Mann–Whitney test was used to determine significant differences between the two groups. The statistical significance is indicated (**P = 0.0022). Values are means, and bars indicate the SD. (C) Mice were intratracheally infected with ∼6 × 108 of the Kp3380 parental strain or the Kp3380ΔkpiD mutant. The number of bacteria in lung homogenates was determined 24 h postinfection. The Mann–Whitney test was used to determine significant differences between the groups. Not statistically significance differences were observed (NS). Values are means, and bars indicate the SDs.
The parental phenotype was not successfully restored in the complemented strain, as it did not retain the complementation plasmid in the in vivo model (59% of bacteria did not harbors the plasmid at 15 h postinfection).
Role of Kpi Fimbriae System in Kp3380 Intestinal Colonization and Lung Infection.
Considering the role of the Kpi fimbriae system in in vitro adherence to HT-29 colorectal cell and A549 alveolar cells, we used in vivo mice models to investigate the role the system plays in both intestinal colonization and lung infection. The in vivo colonization ability of strain Kp3380 lacking the kpiD gene was investigated. Mice pretreated with streptomycin (six per group) (Fig. 5B) were individually inoculated (via oral route) with either the parental strain Kp3380 (3.8 × 106 CFU in 100 μL PBS) or the mutant strain Kp3380∆kpiD (4.1 × 106 CFU in 100 μL PBS). The fecal bacterial burden 24 h after colonization was 2.33 × 107 CFU/mL for Kp3380 and 5.5 × 106 CFU/mL for Kp3380∆kpiD. Colonized mice were killed on day 3 after inoculation to enable determination of the intestinal bacterial burden. The levels of bacteria were significantly lower (P = 0.0022) in the cecum and colon of mice colonized with the mutant strain than in the corresponding organs of mice colonized with the wild-type strain (Fig. 5B). Mice orally inoculated with PBS were used as negative controls, and no bacteria were recovered from intestinal segments of these mice. The parental phenotype was not successfully restored in the complemented strain, as it did not retain the complementation plasmid in the in vivo model (45% of bacteria did not harbors the plasmid on day 3 after inoculation).
Similarly, mice were intratracheally infected with the Kp3380 and its kpiD-defective mutant to assess the in vivo virulence of this strain in a lung infection model. At 24 h postinfection the mice infected with Kp3380∆kpiD had a similar lung burden as the mice infected with the parental strain (Fig. 5C).
Discussion
The severity, prevalence, and morbidity of carbapenem-resistant K. pneumoniae infections manifest the urgent need for new strategies to manage and treat these serious infections. A large number of MDR and extensively drug-resistant K. pneumoniae strains belonging to specific clonal groups that have emerged and have become globally disseminated show an enhanced capacity to cause outbreaks worldwide. Clonal group 15, consisting of ST-14 and ST-15, is one of the most significant outbreak-causing clonal groups. Understanding the epidemic traits of these highly disseminated successful clones is essential to enable identification of specific targets for developing preventive measures and treatment (4). Antimicrobial resistance and virulence are the main factors favoring the successful emergence of these high-risk clones.
A number of virulence factors have been demonstrated to promote K. pneumoniae infectivity (17). In this study, we used an outbreak-related ST-15 K. pneumoniae isolate to identify potential pathogenic targets in K. pneumoniae. Phenotypic analysis revealed that this strain has a more adherent phenotype than other strains belonging to different STs from outbreaks in other hospitals in Spain. Electron microscopy data revealed a relationship between the presence of abundant pili on the surface of Kp3380 and its adherent phenotype.
Comparative genome analysis enabled us to identify a CUP system (Kpi) in Kp3380, which was absent or functionally affected in the other strains tested. Thus, we hypothesized that the adherent phenotype observed in Kp3380 may be determined by expression of this system. Proteins encoded by the kpiABCDEFG operon were found to have a fim-like structure, with the exception of KpiA, which is not structurally related to any crystallized protein. However, the Kpi system does not share the same gene organization. Unlike the Fim system, Kpi belongs to the largely unexplored γ2 fimbrial clade. Like the Kpi system, operons belonging to γ2-fimbriae have three chaperone genes (14) and share similar gene organization. Comparative functional and sequencing analysis of Kpi and the best-studied member of the γ2-fimbriae [i.e., Fas in E. coli (14)] revealed that these systems share common characteristics. The fimbrial protein KpiD is possibly the major subunit. KpiA share the functional annotation characteristics with FasG; therefore, we hypothesized that this protein could be the tip adhesin. However, KpiA is shorter than FasG and is not structurally related to any crystallized protein, which pointed out that additional analyses are required to elucidate the function of this protein. Unlike the Fas system, all three chaperones of the Kpi system contain the two chaperone domains: PFAM00345 and PFAM02753. Although Kpi is phylogenetically related to the Fas system, it is phylogenetically more closely related to the uncharacterized member plu0268 from Photorhabdus luminescens (14).
Several CUP systems, including the type 1 and type 3 fimbriae (pili) (15–17), have been reported to be involved in adherence in K. pneumoniae, and we therefore wondered whether the ability of Kp3380 to form biofilms and to attach to eukaryotic cells was due to the presence of the Kpi system.
Although the Kpi pilus system is structurally related to the type 1 pilus system, it plays a role in the interaction of Kp3380 with both abiotic and biotic surfaces. Like their Fim (7) and Kpg (15) counterparts, Kpi is needed for adherence to epithelial surfaces. However, unlike Fim fimbriae, the Kpi system has been demonstrated to be required for the attachment of Kp3380 to different types of epithelial cells, such as HT-29 intestinal cells, A549 alveolar cells, and HT-1376 bladder cells. Similarly, the ability of the Kp3380 strain to form biofilms has been shown to be impaired when the fimbrial protein KpiD was deleted. A number of CUP systems have been reported to play a role in biofilm formation, highlighting the role of type 3 (mrk) pili as a strong biofilm promoter (22–26). Type 1 (fim) pili, along with type 3 (mrk) pili, are involved in biofilm formation on urethral tract catheters (23–27). Up to eight usher-type pili have been identified in the LM21 K. pneumoniae strain, among which Kpa, Kpg, and Mrk also play a role in biofilm formation (15). Similarly, Kpc fimbriae have been shown to increase biofilm formation activity in the K. pneumoniae NTUH-K2044 strain (16).
In view of the fact that Kpi is involved in adherence to colorectal cells, and that the first step in most infections caused by K. pneumoniae is colonization of the patient’s GI tract, we used an in vivo animal model to investigate the role of the fimbriae in intestinal colonization. The Fas system is involved in adherence to piglet intestinal cells in E. coli (14). Other adhesive structures have been reported to confer binding to intestinal cells in K. pneumoniae, suggesting that they play a role in GI tract colonization (28–30). However, the role of type 1 and type 3 fimbriae in intestinal colonization remains controversial. It has been reported that neither type of pilus affects the ability of K. pneumoniae to colonize or infect the lungs (20, 27, 31). In contrast to previous reports, it has been claimed that murine intestinal colonization is impaired in both mrk-defective LM21 and fim-defective LM21 strains (15). It has recently been reported that down-regulation of type 1 fimbriae in K. pneumoniae NTUH-K2044, together with the lack of type VI secretion systems, is important for bacterial intestinal colonization (32).
We have demonstrated here that the colonizing capacity of the KpiD-defective Kp3380 strain was significantly reduced, indicating that the fimbriae are required for GI tract colonization. However, we did not observe attenuation of the virulence of the Kp3380∆kpiD mutant strain in an acute murine pneumonia infection model. The observed differences in these two in vivo models may be due to the different host environment. Bacteria use distinct strategies when interacting with hosts, and the molecular mechanisms used by K. pneumoniae to colonize the GI tract or to cause acute infections, such as pneumonia, may differ widely. Adherence-related virulence factors (such as Kpi) in K. pneumoniae may play a crucial role in colonization while other virulence factors may have a more important role in acute infection models. As colonization with K. pneumoniae is closely associated with subsequent infection, our findings may be important for understanding the underlying mechanisms of disease progression. Kpi+ K. pneumoniae may have an advantage that enables them to persist in humans and in the hospital environment, thus favoring progression of the outbreak, despite intensive intervention protocols. Indeed, we found that this CUP system is closely associated with clonal group 15, which includes widely disseminated high-risk clones of K. pneumoniae causing outbreaks worldwide (4). Therefore, the Kpi system may at least partly explain the superior capacity of these clones to spread and cause outbreaks and may represent a specific target for controlling the emergence of these successful high-risk clones.
Finally, as the ability of the mutant strain to infect and kill G. mellonella was significantly affected, along with its role in adherence and biofilm formation, we suggest that Kpi plays a role in the virulence of K. pneumoniae Kp3380.
It is worth pointing out that, contrary to what we found in the in vitro assays, plasmid-based complementation of the Kpi system did not restore wild-type function in the in vivo models, despite roughly half of the bacteria harboring the complementation plasmid.
A possible explanation for this might be that the plasmid-carrying bacteria are outcompeted by plasmid-free bacteria, due to the fitness cost associated with plasmid expression (33, 34), particularly under the growth-limiting conditions found in the host environment. This fact, along with the observed loss of complementation plasmid, might be decisive in the plasmid-bearing bacteria being unable to develop the infection properly, leading to the complementation failure.
In summary, we identified a CUP system associated with the worldwide-disseminated high-risk clone K. pneumoniae ST-15. Our findings provide evidence that this system positively contributes to K. pneumoniae adherence, biofilm formation, and virulence in specific models. The information acquired regarding the pathobiology and epidemiology of Kpi+ K. pneumoniae indicates that the Kpi system could be targeted in order to interrupt host pathogen-interactions that promote dissemination of the K. pneumoniae ST-15 clone.
Materials and Methods
Screening of Carbapenemase-Producing K. pneumoniae.
Infection prevention and control interventions in our hospital include screening of CRE colonized patients when a positive clinical sample for CRE is detected (35). First, intestinal colonization (rectal swabs) is analyzed for patients infected with CRE (clinical sample positive for CRE). Then, screening of patients admitted to the same room, as well as those patients admitted to the same department, are also included in the screening, according to the infection control protocols of our hospital. Patients from whom CRE have been isolated from a clinical sample are placed in strict contact isolation until two consecutive rectal swabs are negative for CRE. These patients are then discharged from contact isolation measures. None of the authors has had any contact with patients. The samples were handled under strict confidentiality criteria without any possibility of relating a bacterial isolate to any patient.
Multilocus Sequence Typing Analysis.
Multilocus sequence typing (MLST) was performed using seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpob, and tonB) as previously described (36). Sequences of amplified genes were used to obtain the MLST profile using the K. pneumoniae MLST database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Bacterial Strains, Plasmids, and Culture Conditions.
The bacterial strains and plasmids used in this study are listed in SI Appendix, Table S3. Strain Kp3380 was selected from a collection of 682 K. pneumoniae outbreak-related isolates obtained between February 2013 and October 2018. Isolates were selected on the basis of reduced susceptibility to ertapenem. All isolates were positive for OXA-48 carbapenemase, as determined by the Xpert Carba-R test run on a Genexpert platform (Cepheid). Strains were grown in Luria Bertani (LB) broth (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) or on LB agar (LB broth supplemented with 20 g/L of agar). Hygromycin and apramycin were used to select transformant strains at concentrations of 200 µg/mL and 150 µg/mL, respectively. Gentamicin and kanamycin were used at a concentration of 50 µg/mL. The strains were routinely grown at 37 °C with shaking and stored at −80 °C in LB broth with 10% glycerol.
RNA Extraction and RT-PCR.
To demonstrate the polycistronic nature of the operon, the cDNA was obtained from RNA samples with the iScrip cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s recommendations. RNA was obtained using the High Pure RNA Isolation Kit (Roche). RNA samples were treated with DNase I and purified with RNeasy MiniElute Cleanup Kit (Quiagen). The cDNA was amplified with GoTaq.
For DNA polymerase (Promega), pairs of primers were designed to be complementary to the 3′ end of every gene and the 5′ end of the next one. Genomic DNA was used as a template for the positive control and total RNA was used as a template for the negative control of the amplification. The primers and probes used are listed in SI Appendix, Table S4.
Bioinformatics Analysis.
Rapid Annotation with Subsystem Technology (RAST) (https://rast.nmpdr.org/) was used for comparative analysis of K. pneumoniae genomes. Functional annotation of proteins of the CUP system KpiABCDEFG was assessed using InterPro and HHpred (37, 38). Protein structure modeling of the CUP system KpiABCDEFG in K. pneumoniae was predicted using RaptorX (RaptorX: Protein Structure and Function Prediction Powered by Deep Learning, raptorx.uchicago.edu/).
Phylogenetic Analyses.
Phylogenetic analysis based on the usher amino acid sequence was used to classify the kpiABCDEFG fimbrial operon. A set of reference proteins (SI Appendix, Table S2) was used to establish the γ1, γ2, and γ3 groups (14).
A hidden Markov model (HMM) was built using the seed alignment of the PF00577 family from Pfam (usher family), with hmmbuild in the HMMER v3.0 package (39). The usher sequence set (KpiG) and the proteins listed in SI Appendix, Table S2 were aligned against the HMM by using hmmalign in the HMMER v3.0 package, with the outformat phylip option (39). The resulting alignment was used in the phylogenetic analysis. Maximum-likelihood (ML) phylogeny was constructed using RAxML v8.0 (40) and the LG model of amino acid substitution rates with empirical amino acid frequencies and the γ-model of rate heterogeneity (–m PROTGAMMALGF option). A rapid Bootstrap analysis and search for the best scoring ML tree was performed (–f a option) with 1,000 repetitions (the random number seed for the parsimony inferences was –p 12345 and for rapid bootstrapping, –x 12345). The best tree graphic representation was generated using iTOL v5.1.1 (41).
BLASTn (42) was used to evaluate the presence/absence of the CUP system in K. pneumonia strains. The sequence of the CUP system was used as a query against all of the K. pneumoniae sequences available in the National Center for Biotechnology Information database (in October 2019) and against 1,649 K. pneumoniae strains isolated in 32 European countries. The presence and absence of the kpiABCDEFG sequence were identified using megablast and filtered considering a coverage of between 34% and 100% and an identity of between 99.51% and 100%.
A core-genome MLST (cg-MLST) was performed to describe the distribution of the CUP system in a large population of K. pneumoniae. The cg-MLST relies on species-specific schemes with a fixed number of chromosomal target genes. Thus, 2,538 targets provided by SeqSphere+3.5.0 (Ridom) were used to compare K. pneumoniae isolates from this study with K. pneumoniae strains described in the EuSCAPE project (5), and data on the presence/absence of the CUP system were included to examine its distribution.
Construction of the Knockout Strain.
The kpiD gene was deleted from Kp3380 by a previously described method (43). Briefly, the pIJ773 plasmid was used as a template to amplify the apramycin resistance cassette flanked by FRT sites with primers that included 5′ extensions with homology for the kpiD gene (SI Appendix, Table S4). For chromosomal gene replacement, the PCR product was electroporated into Kp3380 previously transformed with the plasmid pACBSR-Hyg, which expresses the λ-Red system and a hygromycin selection marker. Bacteria were incubated overnight on LB Agar supplemented with apramycin, and correct insertion of the apramycin cassette was confirmed by PCR. The Kp3380 derivative strain was grown on LB agar with apramycin (150 μg/mL), at 37 °C for 3 d, and then incubated on LB agar with apramycin (150 μg/mL) and LB agar with hygromycin (200 μg/mL), overnight at 37 °C, to test for loss of the helper plasmid. For removal of resistance markers, cells were transformed with pFLP-Hyg and grown overnight at 30 °C. The mutants were then incubated at 43 °C in order to remove the apramycin resistance cassette. Apramycin-susceptible mutants were selected to confirm the kpiD gene deletion by PCR and sequencing. The level of expression of each gene in the kpiABCDEFG operon was determined. Results confirmed that only the kpiD gene expression was null in the Kp3380∆kpiD strain. The primers used in this procedure are listed in SI Appendix, Table S4.
Complementation of the Mutant Derivative Kp3380∆kpiD Strain.
The fimbrial gene kpiD was PCR-amplified with specific oligonucleotides (SI Appendix, Table S4) from the Kp3380 strain. The amplified DNA was purified, digested with SmaI and SacI (Thermo Fisher Scientific), and then ligated (T4 DNA ligase, Thermo Fisher Scientific) into a similarly digested expression pUCP24 vector under control of the tetracycline gene promoter. The tetracycline gene promoter was first amplified from the pWH1266 plasmid, digested with BamHI and SmaI endonucleases (Thermo Fisher Scientific) and then cloned into the pUCP24 vector. The construct was checked by PCR analysis with specific primers (SI Appendix, Table S4). The derivative pUCP24/kpiD plasmid was incorporated into the Kp3380∆kpiD mutant strain by electroporation for complementation studies. Transformant strains were selected on agar plates with gentamicin (50 µg/mL).
Bacterial Adhesion to Human Epithelial Cells.
Adhesion abilities of the K. pneumoniae strains were determined following the previously described procedure (44, 45), with some modifications. Three cell lines were used in this experiment, including A549 human alveolar epithelial cells, HT-29 human colorectal epithelial cells, and HT-1376 human urinary bladder epithelial cells. The A549 cells were grown in the presence of 5% CO2 at 37 °C, on DMEM (Sigma-Aldrich) supplemented with 10% heat-inactivated FBS (Sigma-Aldrich) and 1% penicillin-streptomycin (P/S, Gibco). McCoy’s 5A Medium 1× (Gibco) supplemented with 10% FBS, 1% P/S, and 1% Glutamax (Gibco) and Minimum Essential Medium (MEM, Sigma-Aldrich) supplemented with 10% FBS, 1% P/S, 1% Glutamax, and 1% MEM nonessential amino acid Solution (Sigma-Aldrich) were used to grow HT-29 and HT-1376, respectively. Cells were washed, trypsinized, and transferred to 24-well plates (Corning Costar TC-Treated Multiple Well Plates) to produce a monolayer of ∼105 cells per well. After incubation for 24 h at 37 °C, confluent monolayers were washed twice with saline solution and once with modified HBSS (mHBSS, HBSS without glucose). For preparation of inoculum, 100 mL of fresh LB Broth in a 250-mL glass Erlenmeyer flask was inoculated with 1 mL of the overnight culture and then grown at 37 °C with shaking at 180 rpm until the culture reaches OD600 of 0.1. Bacteria cells were pelleted by centrifugation (4,000 × g, 15 min), washed twice with PBS, and resuspended in mHBSS (1:10 dilution). The cells were then infected with 105 bacteria per well and incubated for 3 h in mHBSS at 37 °C. The infected monolayers were washed three times with saline solution and lysed in 500 µL of 0.5% sodium deoxycholate (Sigma-Aldrich). Dilutions of the lysates were plated onto LB agar and incubated at 37 °C for 24 h. In order to determine the percentage of bacteria that attached to A549, HT-29 or HT-1376 cells, colony-forming units (CFU) were counted 24 h later. The percentage of attached bacteria was compared with the total number of infecting bacteria. Four independent biological replicates were analyzed. ANOVA was used to indicate significant differences between the groups, and a post hoc Tukey’s test was used to determine the difference between each group and the respective control. Kp3380 was used as control group when analyzing data from panel A. Kp3380ΔkpiD was used as a control group when analyzing data from panel C.
Quantitative Biofilm Assay.
Biofilm formation was quantified following a previously described procedure (46), with some modifications. K. pneumoniae strains were grown on LB broth overnight at 37 °C with shaking. Overnight cultures were diluted 1:100 and inoculated in 48-well flat bottom polystyrene microtiter plates (Corning Costar TC-Treated Multiple Well Plates). Samples were incubated at 37 °C for 24 h under static conditions, and biofilm formation was visualized by staining with 0.1% Crystal violet. Bacterial growth was measured at OD600 to estimate total cell biomass. Biofilm formation was quantified (OD580) after being solubilized with 30% acetic acid. The amount of biofilm formed was determined from the OD580/OD600 ratio, to compensate for variations due to differences in bacterial growth. Six independent biological replicates were analyzed. ANOVA was used to indicate significant differences between the groups, and a post hoc Tukey’s test was used to determine the difference between each group and the respective control. Kp3380 was used as the control group when analyzing data from panel A. Kp3380ΔkpiD was used as the control group when analyzing data from panel B.
TEM.
Bacterial cells were grown under the same conditions used to infect epithelial cells. Freshly prepared carbon-coated, nitrocellulose substrate TEM grids were placed substrate side down, directly on top of the drop containing the bacterial suspension and allowed to sit covered for 5 min. Grids were removed and negatively stained with 5 mL of 1.5% (wt/vol) ammonium molybdate for 5 min. The grids were then blotted with filter paper and allowed to dry. Images were captured at 120 kv with a JEOL JEM-2010. Images of three grids (100 mesh) were obtained for each strain. Representative images of bacterial cells were taken after evaluation of the whole grid. Experiments were performed in duplicate for each strain.
CLSM.
Biofilm architecture related to the selected strains was studied in four-well microslides (Ibidi) as previously described (47). Briefly, the slides were placed in an inclined position (∼45°) in an incubator to form a liquid-air interface and after incubation for 24 h at 37 °C, unfixed planktonic cells were removed by rinsing with saline (0.85% NaCl). The bacterial viability within biofilms was determined using the BacLight LIVE/DEAD bacterial viability kit (Molecular Probes). A series of optical sections was obtained with a Nikon A1R confocal microscope; the excitation wavelengths were 488 nm (green) and 561 nm (red), and 500- to 550-nm and 570- to 620-nm emission filters, respectively, were used. Images were captured at random at the liquid-air interface with a 20× Plan Apo (numerical aperture [NA], 0.75) objective. Reconstructions of confocal sections and quantitative measurements were performed using NIS-Elements software, v3.2. Four independent biological replicates were analyzed. ANOVA was used to indicate significant differences between the groups, and a post hoc Tukey’s test was used to determine the difference between each group and the respective control (Kp3380ΔkpiD).
G. mellonella Virulence Assay.
The virulence of Kp3380 strain and its derivative strains (Kp3380ΔkpiD and Kp3380ΔkpiD_C) was evaluated using a G. mellonella survival assay conducted according to the previously described protocol (33). Briefly, G. mellonella larvae were injected with 10 μL of a suspension containing ∼2 × 102 CFU/larva in groups of 16 larvae. For preparation of inoculum, 100 mL of fresh LB Broth in a 250-mL glass Erlenmeyer flask was inoculated with 1 mL of the overnight culture and then grown at 37 °C with shaking at 180 rpm until the culture reaches OD600 of 0.7. Bacteria cells were pelleted by centrifugation (4,000 × g, 15 min), washed twice, and resuspended in PBS (1:1,000 dilution). One group was infected with the wild-type strain Kp3380 and the two other groups were infected with the Kp3380ΔkpiD and Kp3380ΔkpiD_C strains. Larvae injected with an equivalent volume of sterile PBS were used as controls. The infected and control larvae were incubated at 37 °C in darkness, and death was monitored at 8-h intervals during 5 d. The resulting survival curves were plotted using the Kaplan–Meier method. The presence of the complementation plasmid in the complemented strain Kp3380ΔkpiD_C was tested at 15 h postinfection by PCR. The log-rank (Mantel–Cox) test was used to compare the survival distributions of two groups: The larvae infected with Kp3380 and the larvae infected with Kp3380ΔkpiD.
In Vivo Murine Models.
The role of the Kpi pilus system in intestinal colonization and lung infection was assessed using two experimental mouse models.
The intestinal colonization capacity of the K. pneumoniae strains was assessed using a previously described murine colonization model (18), with some modifications. Six-week-old female BALB/c mice were randomly distributed in cages with one mouse per cage. The mice were previously treated with a single dose of streptomycin (1 g/kg in 100 μL water by oral gavage), to clear the GI tract of colonizing bacteria, and then they were individually colonized with Kp3380, Kp3380∆kpiD, or Kp3380∆kpiD_C (∼4 × 106 CFU in 100 μL PBS). For preparation of inoculum 100 mL of fresh LB Broth in a 250-mL glass Erlenmeyer flask was inoculated with 1 mL of the overnight culture and then grown at 37 °C with shaking at 180 rpm until the culture reached OD600 of 0.7. Bacteria cells were pelleted by centrifugation (4,000 × g, 15 min), washed twice, and resuspended in PBS. After 24 h, 0.3 g of fecal sample was collected and the bacterial burden was determined in order to confirm intestinal colonization with the strains of interest. Three days after inoculation, the mice were killed and intestinal segments (cecum and colon) were processed under aseptic conditions. Intestinal segments were weighed before homogenization and plated on LB supplemented with kanamycin at 50 mg/mL to determine the bacterial burden. The presence of the complementation plasmid in the complemented strain Kp3380ΔkpiD_C was tested on day 3 after inoculation by PCR.
The pneumonia model procedure was previously described (33). Briefly, mice anesthetized with an oral suspension of sevoflurane (Zoetis, #NADA 141–103) were intratracheally inoculated via the oral cavity with an endoscope. A 40-μL bacterial suspension (2 × 107 CFU per mouse) prepared (as described above) in sterile saline solution and 10% porcine mucin (wt/vol; Sigma) mixed at a 1:1 ratio was used to establish pneumonia. Mice that died in the first 4 h postinfection were not analyzed further. The mice were killed 20 h after infection and processed under aseptic conditions. The lung tissue was plated on LB agar to determine the bacterial burden. The Mann–Whitney test was applied to the data to determine any significant between-group differences.
All mice were maintained in the specific pathogen-free facility at the Technology Training Centre of the A Coruña University Hospital Complex (Spain).
Ethics.
Animal experiments were carried out with the approval of and in accordance with regulatory guidelines and standards established by the Animal Ethics Committee of the A Coruña University Hospital Complex (project code P82). This committee follows the recommendations of the National Committee for the Replacement, Refinement, and Reduction of Animal Research.
Data Availability.
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials. TEM and CLSM images are openly available in Zenodo at https://zenodo.org/record/3806241#.Xuy4v2hKhk4.
Supplementary Material
Acknowledgments
The authors thank Dr Fidel Madrazo for help with transmission electron microscopy and confocal laser-scanning microscopy assays. This research was supported by Projects p-01216A and IJCI-2016-29524 (to A.P.), funded by the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC) and the Minestry of Economy and Competetiveness (MINECO), respectively. It was also supported by Projects PI11/01034 (to M.P.), PI14/00059 and PI17/1482 (to M.P. and A.B.), and PI18/00501 (to G.B.), included in the National Plan for Scientific Research, Development and Technological Innovation 2013-2016 and funded by the Instituto de Salud Carlos III (ISCIII) and Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/006) cofinanced by European Development Regional Fund “A way to achieve Europe” and operative program Intelligent Growth 2014-2020. Grant BFU2016-77835-R of the MINECO (to A.R.) also supported this research. E.G. was financially supported by the SEIMC project. J.C.V.-U. was financially supported by the PFIS (Contratos Predoctorales de Formación en Investigación en Salud) program (F18/00315); J.A.V. was financially supported by IN607A 2016/22; M.M.-G. was financially supported by a Clara Roy grant (SEIMC); A.B. was financially supported by the Miguel Servet program (ISCIII, Spain); B.K.R.-J. was financially supported by Marie S. Curie Action SaPhaDe project (MSCA-IF-GF-836754); and A.P. was financially supported by the Juan de la Cierva program (MINECO, IJCI-2016-29524).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The transmission electron microscopy and confocal laser-scanning microscopy images are openly available in Zenodo at https://zenodo.org/record/3806241#.Xuy4v2hKhk4.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921393117/-/DCSupplemental.
References
- 1.European Centre for Disease Prevention and Control (ECDC) , Rapid Risk Assessment: Carbapenem-Resistant Enterobacteriaceae-8 April 2016, (ECDC, Stockholm, 2016). [Google Scholar]
- 2.WHO , WHO publishes list of bacteria for which new antibiotics are urgently needed (WHO Media Center, 2017). https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. Accessed 31 October 2019.
- 3.Frieden T., Antibiotic Resistance Threats in the United States, 2013, (Centers for Disease Control and Prevention, 2013), Vol. 114. [Google Scholar]
- 4.Navon-Venezia S., Kondratyeva K., Carattoli A., Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275 (2017). [DOI] [PubMed] [Google Scholar]
- 5.David S. et al.; EuSCAPE Working Group; ESGEM Study Group , Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919–1929 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin R. M. et al., Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. MSphere 1, e00261-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin R. M., Bachman M. A., Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8, 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorrie C. L. et al., Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calfee D. P., Recent advances in the understanding and management of Klebsiella pneumoniae. F1000 Res. 6, 1760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q. et al., Target-agnostic identification of functional monoclonal antibodies against Klebsiella pneumoniae multimeric mrka fimbrial subunit. J. Infect. Dis. 213, 1800–1808 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Wang Q. et al., Anti-MrkA monoclonal antibodies reveal distinct structural and antigenic features of MrkA. PLoS One 12, e0170529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickey S. W., Cheung G. Y. C., Otto M., Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 16, 457–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busch A., Waksman G., Chaperone-usher pathways: Diversity and pilus assembly mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1112–1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nuccio S.-P., Bäumler A. J., Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khater F. et al., In silico analysis of usher encoding genes in Klebsiella pneumoniae and characterization of their role in adhesion and colonization. PLoS One 10, e0116215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C. C., Huang Y. J., Fung C. P., Peng H. L., Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 156, 1983–1992 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Paczosa M. K., Mecsas J., Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spaulding C. N. et al., Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 546, 528–532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Struve C., Bojer M., Krogfelt K. A., Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect. Immun. 77, 5016–5024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Struve C., Bojer M., Krogfelt K. A., Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 76, 4055–4065 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gato E. et al., Draft genome sequences of two epidemic OXA-48-producing Klebsiella pneumoniae clinical strains isolated during a large outbreak in Spain. Genome Announc. 6, e00026-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy C. N., Clegg S., Klebsiella pneumoniae and type 3 fimbriae: Nosocomial infection, regulation and biofilm formation. Future Microbiol. 7, 991–1002 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Stahlhut S. G., Struve C., Krogfelt K. A., Reisner A., Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol. Med. Microbiol. 65, 350–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilksch J. J. et al., MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7, e1002204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson J. G., Murphy C. N., Sippy J., Johnson T. J., Clegg S., Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 193, 3453–3460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroll C., Barken K. B., Krogfelt K. A., Struve C., Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10, 179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosen D. A. et al., Molecular variations in Klebsiella pneumoniae and Escherichia coli FimH affect function and pathogenesis in the urinary tract. Infect. Immun. 76, 3346–3356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Martino P., Livrelli V., Sirot D., Joly B., Darfeuille-Michaud A., A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 64, 2266–2273 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favre-Bonte S., Darfeuille-Michaud A., Forestier C., Aggregative adherence of Klebsiella pneumoniae to human intestine-407 cells. Infect. Immun. 63, 1318–1328 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darfeuille-Michaud A. et al., R-plasmid-encoded adhesive factor in Klebsiella pneumoniae strains responsible for human nosocomial infections. Infect. Immun. 60, 44–55 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B., Zhao Y., Liu C., Chen Z., Zhou D., Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 9, 1071–1081 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Hsieh P. F., Lu Y. R., Lin T. L., Lai L. Y., Wang J. T., Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219, 637–647 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Álvarez-Fraga L. et al., Pneumonia infection in mice reveals the involvement of the feoA gene in the pathogenesis of Acinetobacter baumannii. Virulence 9, 496–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porse A. et al., Genome dynamics of Escherichia coli during antibiotic treatment: Transfer, loss, and persistence of genetic elements in situ of the infant gut. Front. Cell. Infect. Microbiol. 7, 126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oteo J., Bou G., Chaves F., Oliver A., Microbiological methods for surveillance of carrier status of multiresistant bacteria. Enferm. Infecc. Microbiol. Clin. 35, 667–675 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Diancourt L., Passet V., Verhoef J., Grimont P. A. D., Brisse S., Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmermann L. et al., A completely reimplemented MPI Bioinformatics Toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Jones P. et al., InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mistry J., Finn R. D., Eddy S. R., Bateman A., Punta M., Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Huang T. W. et al., Capsule deletion via a λ-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res. Notes 7, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rumbo-Feal S. et al., Contribution of the A. baumannii A1S_0114 gene to the interaction with eukaryotic cells and virulence. Front. Cell. Infect. Microbiol. 7, 108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Álvarez-Fraga L. et al., Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 7, 443–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomaras A. P., Dorsey C. W., Edelmann R. E., Actis L. A., Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Remuzgo-Martínez S. et al., Biofilm formation and quorum-sensing-molecule production by clinical isolates of Serratia liquefaciens. Appl. Environ. Microbiol. 81, 3306–3315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials. TEM and CLSM images are openly available in Zenodo at https://zenodo.org/record/3806241#.Xuy4v2hKhk4.