Significance
Staphylococcus aureus infections impose an immense burden on the healthcare system. To establish a successful infection in a hostile host environment, S. aureus must coordinate its gene expression to respond to a wide array of challenges. This balancing act is largely orchestrated by the transcriptional regulatory network. Here, we present a model of 29 independently modulated sets of genes that form the basis for a segment of the transcriptional regulatory network in clinical USA300 strains of S. aureus. Using this model, we demonstrate the concerted role of various cellular systems (e.g., metabolism, virulence, and stress response) underlying key physiological responses, including response during blood infection.
Keywords: Staphylococcus aureus, transcriptional regulatory network, virulence, metabolism
Abstract
The ability of Staphylococcus aureus to infect many different tissue sites is enabled, in part, by its transcriptional regulatory network (TRN) that coordinates its gene expression to respond to different environments. We elucidated the organization and activity of this TRN by applying independent component analysis to a compendium of 108 RNA-sequencing expression profiles from two S. aureus clinical strains (TCH1516 and LAC). ICA decomposed the S. aureus transcriptome into 29 independently modulated sets of genes (i-modulons) that revealed: 1) High confidence associations between 21 i-modulons and known regulators; 2) an association between an i-modulon and σS, whose regulatory role was previously undefined; 3) the regulatory organization of 65 virulence factors in the form of three i-modulons associated with AgrR, SaeR, and Vim-3; 4) the roles of three key transcription factors (CodY, Fur, and CcpA) in coordinating the metabolic and regulatory networks; and 5) a low-dimensional representation, involving the function of few transcription factors of changes in gene expression between two laboratory media (RPMI, cation adjust Mueller Hinton broth) and two physiological media (blood and serum). This representation of the TRN covers 842 genes representing 76% of the variance in gene expression that provides a quantitative reconstruction of transcriptional modules in S. aureus, and a platform enabling its full elucidation.
The pathogen Staphylococcus aureus causes a variety of human diseases, ranging from skin and soft tissue infections to infective endocarditis and pneumonia (1). The pathogen can also thrive as part of the commensal microbiome in the anterior nares of healthy patients (2). S. aureus adaptation to many different host environments is enabled, in part, by the underlying transcriptional regulatory network (TRN) that can alter the physiological state of the cell to match the unique challenges presented by each environment (3–5). Such adaptations require coordinated expression of genes in many cellular subsystems, such as metabolism, cell wall biosynthesis, stress response, virulence factors, and so forth. Therefore, a complete understanding of the S. aureus response to different environments necessitates a thorough understanding of its TRN. However, since S. aureus is predicted to have as many as 135 transcriptional regulators (6), with many more potential interactions among them, a bottom-up study of its global TRN becomes intractable.
To address this challenge, we previously introduced an independent component analysis (ICA)-based framework in Escherichia coli that decomposes a compendium of RNA-sequencing (RNA-seq) expression profiles to determine the underlying regulatory structure (7). An extensive analysis of module detection methods demonstrated that ICA outperformed most other methods in consistently recovering known biological modules (8). The framework defines independently modulated sets of genes (called i-modulons) and calculates the activity level of each i-modulon in the input expression profile. ICA analysis of expression profiles in E. coli have been used to describe undefined regulons, link strain-specific mutations with changes in gene expression, and understand rewiring of TRN during adaptive laboratory evolution (ALE) (7, 9). Given the deeper insights it provided into the TRN of E. coli, we sought to expand this approach to the human pathogen S. aureus. To elucidate the TRN features in S. aureus, we compiled 108 high-quality RNA-seq expression profiles for community-associated methicillin-resistant S. aureus (CA-MRSA) strains LAC and TCH1516. Decomposition of these expression profiles revealed 29 independently modulated sets of genes and their activity levels across all 108 expression profiles. Furthermore, we show that using the new framework to reevaluate the RNA-seq data accelerates discovery by: 1) Quantitatively formulating TRN organization, 2) simplifying complex changes across hundreds of genes into a few changes in regulator activities, 3) allowing for analysis of interactions among different regulators, 4) connecting transcriptional regulation to metabolism, and 5) defining previously unknown regulons.
Results
ICA Extracts Biologically Meaningful Components from Transcriptomic Data.
We generated 108 high-quality RNA-seq expression profiles from CA-MRSA USA300 isolates LAC and TCH1516 and two additional ALE-derivatives of TCH1516. To capture a wide range of expression states, we collected RNA-seq data from S. aureus exposed to various media conditions, antibiotics, nutrient sources, and other stressors (Dataset S3). The samples were then filtered for high reproducibility between replicates to minimize noise in the data (SI Appendix, Fig. S1A). The final dataset contained 108 samples representing 43 unique growth conditions, which have an average R2 = 0.98 between replicates. Using an extended ICA algorithm (7), we decomposed the expression compendium into 29 i-modulons. An i-modulon contains a set of genes whose expression levels vary concurrently with each other, but independently of all other genes not in the given i-modulon. Akin to a regulon (10), an i-modulon represents a regulatory organizational unit containing a functionally related and coexpressed set of genes under all conditions considered (Fig. 1A). While regulons are determined based on direct molecular methods (e.g., chromatin immunoprecipitation sequencing [ChIP-seq], RNA immunoprecipitation-ChIP, gene-knockouts, and so forth), i-modulons are defined through an untargeted ICA-based statistical approach applied to RNA-seq data that is a reflection of the activity of the transcriptional regulators (Materials and Methods). However, beyond regulons, i-modulons can also describe other genomic features, such as strain differences and genetic alterations (e.g., gene knockout) that can lead to changes in gene coexpression (7, 9). The outcome of this approach is a biologically relevant, low-dimensional mathematical representation of functional modules in the TRN that reconstruct most of the information content of the input RNA-seq compendium (SI Appendix, Fig. S1B).
Fig. 1.
ICA decomposition of S. aureus USA300 RNA-seq database. (A) An i-modulon is a set of genes that are coexpressed and encode products with shared functions. The PyrR i-modulon, for example (Middle column), is predicted to be under control of pyrR repressor and contains genes that encode enzymes in pyrimidine biosynthesis (purple) and purine salvage (blue) pathway (Right column). The genes in two different pathways are contraregulated (arrows). (B) Activity levels of i-modulons are calculated for all conditions (Upper bar chart), allowing for sample specific (e.g., in three different media) comparison of each i-modulon (boxplot). The activity of all i-modulons are centered around CAMHB base condition, and therefore all i-modulons have mean activity of 0 in this condition. The centerline of the boxplot represents median value, the box limits represent Q1 and Q3, and the whiskers represent the minimum and maximum values. (C) A treemap indicating the names and the size of the i-modulons. The i-modulons are named after the transcription factors whose predicted regulons have highest overlap with the given i-modulon, or based on the shared functionality of genes (e.g., autolysins, translation, B-lactam resistance) in i-modulon if no known regulator was identified. The number in the parenthesis shows the number of genes in a given i-modulon. An i-modulon with low or no correspondence with any of the known features is labeled as Unc-1. BLR, β-lactam resistance; SNFR i-modulon consists of genes with altered expression in the SNFR strain.
Such formulation also quantitatively captures complex behaviors of regulators, such as contraregulation of multiple genes by the same regulator, coregulation of the same gene by multiple regulators, and coordinated expression of multiple organizational units (i-modulons) in various conditions (SI Appendix, Fig. S1 C and D). Therefore, this model enables simultaneous analysis of TRNs at both the gene and genome-scale. ICA also reconstructs the activity of the i-modulons in the samples, which represents the collective expression level of the genes in the i-modulon. Each sample in the dataset can be reconstructed as the summation of the activity of the 29 i-modulons, which makes the transcriptional state in each condition more explainable. Conversely, each i-modulon has a computed activity in every sample, allowing for easy comparisons of i-modulon activities across samples, that in turn reflect the activity of the corresponding transcriptional regulator (Fig. 1B). The reported activity levels are log2 fold-change from the base condition: Growth in cation adjust Mueller Hinton broth (CAMHB). We compared the gene sets in the 29 enriched i-modulons against previously predicted S. aureus regulons in the Reg-Precise database and other regulons described in various publications (Dataset S4). I-modulons with statistically significant overlap (false-discovery rate [FDR] < 1e-05) with a previously predicted regulon were named after the transcription factor associated with the regulon (Materials and Methods). We also manually identified i-modulons that consisted of genes with shared functions (e.g., autolysins, translation) or those that corresponded to other genomic features, such as plasmids, prophages, or strain-specific differences. Taking these data together, we identified 15 metabolic, 6 functional, 3 virulence, 4 stress response-associated, and 1 strain-associated (SNFR) i-modulons (Fig. 1C and Dataset S5). Of the 29 enriched i-modulons, only 1 remains uncharacterized (Dataset S6). In total, the 29 i-modulons consist of 752 unique genes, 90 of which are enriched in more than 1 i-modulons.
ICA Disentangles Complex Change in the Transcriptome.
Differential expression analysis of S. aureus in different environmental conditions can yield hundreds of genes that have significantly altered expression levels, hindering meaningful interpretation. Decomposition of the expression profile into biologically meaningful i-modulons instead allows us to gain a comprehensive understanding of the change in the transcriptome through the activities of few regulators. To demonstrate this capability, we explored the difference in expression profiles of S. aureus grown in two different media, CAMHB, the standard bacteriologic medium for routine antimicrobial susceptibility testing worldwide, and the common physiologically relevant mammalian tissue culture medium RPMI-1640, supplemented with 10% Luria broth (RPMI+10%LB) to support growth kinetics similar to CAMHB. Over 800 genes spanning more than a dozen clusters of orthologous groups (11) categories were differentially expressed between the two media (SI Appendix, Fig. S2A).
Conversely, there were 15 i-modulons with statistically significant differential activation (Fig. 2A). Most differentially activated i-modulons were involved in metabolism (CodY, PurR, guanine-responsive i-modulon [GR], Gal/Man, Rex, MntR, PyrR, LacR, CcpA-1, CcpA-2, Urease). The last four i-modulons were those with functions in virulence (Vim-3, SaeR), Translation, and the Phi-Sa3 phage-specific i-modulon. Concurrent activation of the CodY, PurR, and GR i-modulons in RPMI+10%LB indicates that this media presents a guanine-limited environment, as activity of all three transcription factors decrease in response to falling cellular concentrations of various forms of guanine derivatives (12–15). Consistent with this hypothesis, we also saw decreased activity of the Translation i-modulon in RPMI+10%LB. Down-regulation of translation machinery often occurs during the stringent response, where cellular GTP is depleted as it is rapidly converted to ppGpp (12, 16–18). Similarly, activation of the MntR i-modulon points to manganese starvation in RPMI+10%LB, and the decreased activity of two i-modulons associated with carbon catabolite repressor CcpA (CcpA-1and CcpA-2) reflects a glucose replete environment (19). Analysis of spent media using HPLC confirmed that S. aureus was actively uptaking glucose in RPMI+10%LB while no glucose was detected in CAMHB (SI Appendix, Fig. S2B). The shift in activity of i-modulons between the two media suggests that compared to the bacteriologic medium CAMHB, RPMI+10%LB presents an environment poor in purines (specifically guanine) and manganese but rich in the carbon source glucose.
Fig. 2.
Differential activation of i-modulons in different media conditions. (A) I-modulons from the LAC strain with statistically significant (P < 0.05) differential activation in CAMHB versus RPMI+10%LB. (B) Addition of glucose reduced the activity of CcpA-1 in CAMHB (blue bars). Conversely, replacing glucose with maltose led to higher CcpA-1 activity in RPMI+10%LB. CcpA-2 activity did not change in response to glucose concentration (red bars). (C) The bar plot shows the activity level of GR i-modulon, which contains the genes under the control of guanine riboswitch (xpt and pbuG). Although many different conditions can affect the GR i-modulon activity (blue bar), it sharply decreases when guanine is added to the media. Addition of adenine has no effect. Black dots in B and C represent values from individual samples and error bars represent SD. (D) External validation of Agr and PurR i-modulon activity in the respective agr and purR mutants.
Next, we designed two validation experiments to ensure that the activity level of i-modulons reflect expected outcomes. To this end, we chose three i-modulons to validate—CcpA-1, CcpA-2, and GR—for ease of modifying their activities with supplementation of glucose and purines, respectively. CcpA is the carbon catabolite repressor in S. aureus that controls central carbon metabolism and carbon source utilization (20, 21). Its activity level is indirectly modulated by cellular glucose concentration, although it can also be altered by other glucose-independent signals (22, 23). CcpA transcriptional effects are captured in two i-modulons, CcpA-1 and CcpA-2, which contain 73 and 19 genes, respectively. Both i-modulons had far lower activity in RPMI+10%LB compared to CAMHB. However, the addition of 2 g/L glucose only led to reduced activity of the CcpA-1 i-modulon in CAMHB, closely matching its activity in RPMI+10%LB (Fig. 2B). Similarly, replacement of glucose with maltose in RPMI+10%LB led to increased activity of the CcpA-1 i-modulon. The change in glucose concentration, however, had little effect on the activity level of the CcpA-2 i-modulon, suggesting that the CcpA-1 i-modulon represents direct glucose-responsive CcpA activity, where as the CcpA-2 i-modulon may reflect its glucose-independent activity.
In addition to CcpA activity, we also confirmed the activity of the GR i-modulon. The GR i-modulon contains genes involved in the purine salvage pathway (xpT, pbuX), peptide transport (oppB), and LAC-specific virulence factor ssl11. The two genes in the salvage pathway have been previously demonstrated to be under the control of the guanine riboswitch in S. aureus strain NRS384 (15). The presence of this riboswitch was confirmed using the online RiboSwitch Finder (SI Appendix, Fig. S2 C and D) (24); no riboswitches were detected for the other two genes. The activity of the i-modulon was attenuated by guanine supplementation (25 μg/mL), while the addition of adenine had no effect, demonstrating a guanine-specific activity of the i-modulon (Fig. 2C).
We additionally validated activities of Agr and PurR i-modulons using publically available expression-profiling datasets (GSE18793 and GSE132179) (25, 26). These datasets include expression profiles comparing wild-type USA300 strains to their isogenic agr and purR mutants. As a form of external validation, we did not incorporate these data into the model. Instead we projected the expression data onto the model to convert the gene-expression levels to i-modulon activity levels (Materials and Methods). Compared to their respective wild-types, PurR i-modulon had the largest increase in activity in purR::bursa strain and Agr i-modulon showed the largest drop in activity in the strain with a disrupted agr system, demonstrating that the model can capture activities of these i-modulons in the conditions not included in the model (Fig. 2D).
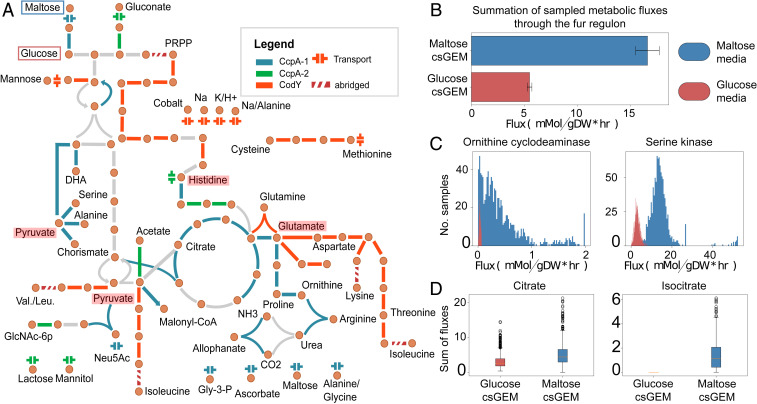
Integration of i-modulons with genome-scale metabolic models (GEMs) reveal systems-level properties of metabolic regulation. GEMs are knowledge-bases reconstructed from all known metabolic genes of an organism, systematically linking metabolites, reactions, and genes (27). Integration of i-modulons with these metabolic models allows us to probe the interaction between the regulatory and metabolic networks. To visualize this cross-talk at the systems level, we overlaid the i-modulons onto central metabolism and amino acid metabolism pathways of the S. aureus metabolic reconstruction iYS854 (Fig. 3A) (28). The CcpA-1 and CodY i-modulons dominate regulation of the genes in these metabolic subsystems of S. aureus. The two CcpA i-modulons controlled many of the genes in carbon metabolism. The genes required for the tricarboxylic acid (TCA) cycle were found primarily in the CcpA-1 i-modulon, with the exception of genes encoding fumarase and malate dehydrogenase. Additionally, the CcpA-1 i-modulon contained genes required for degradation of gluconeogenic amino acids (serine, histidine, and alanine) and secondary metabolites (chorismate and N-acetyl-neuraminic acid). Also included were genes encoding two key gluconeogenic enzymes, phosphoenolpyruvate carboxy kinase and fructose-1,6-bisphosphatase. Genes involved in transport of alternate carbon sources were also present.
Fig. 3.
Regulation of central metabolism and its interaction with other metabolic subsystems. (A) Overlay of i-modulons onto the map of central metabolism and amino acid metabolism of S. aureus. The two main regulators of these metabolic subsystems, CcpA (blue/green) and CodY (orange), control central carbon and nitrogen metabolism, respectively. These two i-modulons intersect at key metabolic nodes: Pyruvate, histidine, and glutamate (highlighted in red). Entry points of sugars used in the next section, glucose and maltose, are highlighted with red and blue boxes, respectively. (B) Activity of reactions associated with the Fur i-modulon in the presence of different carbon sources, maltose and glucose. The bars represent sum of median sampled fluxes through reactions catalyzed by enzymes in the Fur i-modulon. Unexpected increase in Fur i-modulon activity when carbon source was switched from glucose to maltose is recapitulated through metabolic modeling. (C) Reactions associated with the Fur i-modulon with the largest increase in simulated flux in glucose media. (D) Calculated proxy for intracellular metabolite concentrations.
In contrast to catabolic CcpA-regulated genes, the i-modulon associated with CodY regulation was dominated by genes participating in biosynthesis of amino acids lysine, threonine, methionine, cysteine, histidine, and branched chain amino acids (BCAA) isoleucine, leucine, and valine (13). Regulation of interconversion between glutamine and glutamate (gltA), a key component of nitrogen balance and assimilation, was also a part of the CodY i-modulon. While the two i-modulons (CcpA-1, CodY) did not share any genes, they intersected at some key metabolite nodes in central metabolism, including pyruvate, glutamate, histidine, and arginine. Genes in the CcpA-1 i-modulon encode enzymes that generate pyruvate from amino acids and use the pyruvate to generate energy through fermentation, synthesize glucose via gluconeogenesis, or synthesize fatty acids via malonyl-coA. On the other hand, enzymes encoded by genes in the CodY i-modulon redirect pyruvate to instead synthesize BCAA (isoleucine, leucine, and valine). Similarly, glutamate is directed toward the urea cycle by CcpA-1 and toward biosynthesis of the aspartate family amino acids by CodY. While genes required for catabolism of histidine are in the CcpA-1 i-modulon, genes encoding histidine biosynthesis is instead part of the CodY i-modulon. Interestingly, while CcpA regulates l-arginine synthesis from l-proline (via putA and rocD), CodY-regulated genes involved in l-arginine synthesis from l-glutamate (argJ and argB). This dichotomy, combined with the observation that isoleucine affects CodY-dependent repression (13), explains why none of argJBCF were expressed in JE2 ccpA::tetL in CDM, a BCAA-rich medium (29). It is likely that CodY-and CcpA-mediated repression are both active in CDM. By controlling the expression of these key metabolic genes, CcpA-1and CodY i-modulons can readily redirect the fluxes through different metabolic subsystems.
GEMs Compute Flux-Balanced State that Reflect Regulatory Actions of CcpA.
Metabolic network reconstructions can be converted into genome-scale models that allow for the computation of phenotypic states (30). We can compute the optimal flux through the metabolic network using flux-balance analysis (FBA) (28). In particular, we can compute the metabolic state that is consistent with nutrient sources in a given environment to support optimal bacterial growth. In the previous CcpA i-modulon validation experiment, we observed that changing the carbon source from glucose to maltose in RPMI+10%LB also led to an unexpected spike in activity of the iron-responsive Fur i-modulon (SI Appendix, Fig. S3). To investigate whether there was a possible metabolic role explaining the increase in Fur activity, we generated two condition-specific GEMs (csGEMs), starting with iYS854 (28). For both csGEMs, we computed the state of the metabolic network that supports growth in RPMI+10%LB, with either glucose or maltose as the main carbon source (Materials and Methods). We assumed that CcpA-1 repression was active only when glucose was the main glycolytic nutrient source (and the corresponding set of reactions was shut off). Reaction fluxes across the network were then sampled using FBA, assuming that the bacterial objective was biomass production (31). Sampling accounts for different network flux distributions that can achieve the same optimal solutions (i.e., identical biomass production rates).
Under these conditions, the sum of sampled fluxes through reactions associated with the Fur i-modulon was significantly higher in maltose media (Kolmogorov–Smirnov test, P < 0.01, statistics > 0.9), confirming that the spike in Fur activity could be a result of metabolic flux rewiring (Fig. 3B). In particular, fluxes through serine kinase (sbnI, a precursor metabolic step of staphyloferrin B biosynthesis) and ornithine cyclodeam-inase (sbnB) were significantly increased (Fig. 3C). These changes came as a result of flux rewiring away from deactivated metabolic steps. For example, due to arginase (rocF) deactivation, the flux through half of the urea cycle and ornithine cyclodeaminase was lower. Similarly, serine deaminase (sdaB)—located two metabolic steps downstream of serine kinase—was deactivated due to simulated down-regulation of genes in the CcpA-1 i-modulon, and flux through phosphoglycerate dehydrogenase, serine kinase, and phosphoserine phosphatase was decreased. We computed the sum of fluxes producing each metabolite as a proxy for intracellular concentrations and found that the calculated values were significantly larger in maltose media for 68 metabolites, including ammonium, glutamate, and isocitrate. The majority of the TCA cycle was shut off in the glucose-specific GEM (due to simulated repression of citB, icd, odhA, sdhABCD, and sucCD), and therefore the concentration proxy for isocitrate was essentially null while that of citrate was not (Fig. 3D). Previous studies have shown that citB deletion results in increased intracellular concentration of citrate (32). Apart from being an intermediate in the TCA cycle, citrate can be utilized in the model as a precursor to staphyloferrin A and staphyloferrin B biosynthesis (which are included in the Fur i-modulon), or it can be converted back to oxaloacetate and acetate via citrate lyase. All three routes were part of the solution space, with citrate lyase carrying the largest median flux. Taken together, these modeling simulations suggest that utilizing maltose instead of glucose induces metabolic flux rewiring toward reactions associated with the Fur i-modulon.
An i-Modulon Details Possible Scope and Functions of Sigma Factor σS.
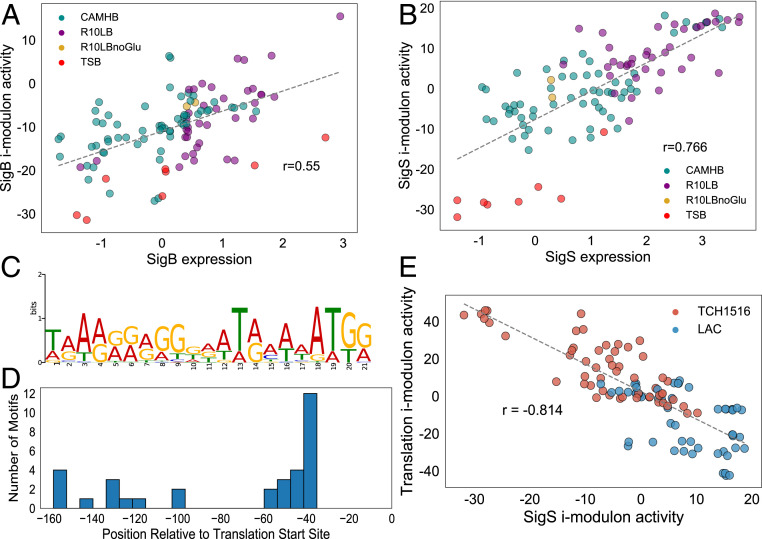
Global stress response in S. aureus is modulated by the alternate sigma factor σB (33, 34). Alhough two other alternate sigma factors, σS and σH, have been recognized in this organism, their exact functions and full regulon are not as well understood (35, 36). We identified two i-modulons that correspond to sigma factors σB and σS. The SigB i-modulon contained genes encoding σB (sigB), anti-σB (rsbW), and anti-σB antagonist (rsbV). The activity of SigB i-modulon was correlated with sigB expression (Pearson R = 0.55, P = 8.2e-11) (Fig. 4A), with the highest activation in stationary phase (OD600 = 1). Furthermore, a conserved 29-bp motif was enriched from 28 unique regulatory regions of SigB i-modulon genes (Methods and Materials and SI Appendix, Fig. S4A). As the regulatory role of σB has been previously explored in detail (34, 37–40), we focused here on the less understood regulatory role of σS. Although σS is important for both intracellular and extracellular stress response, its full regulon has yet to be defined (36, 41). ICA identified a large i-modulon with 137 genes, including sigS itself (which encodes σS). As with the SigB i-modulon, expression of the sigS gene correlated to activity of the ICA-derived SigS i-modulon (Pearson R = 0.77, P = 4.26e-22) (Fig. 4B). Previous studies have shown that CymR represses sigS expression and therefore may lead to its decreased activity (42). We confirmed this relationship as the SigS i-modulon activity was anticorrelated with the CymR i-modulon activity (Pearson R = −0.68, P = 8.23e-10) (SI Appendix, Fig. S4B).
Fig. 4.
Profiling alternate sigma factor S. The expression levels of sigB (A) and sigS (B) genes and the activity levels of their respective i-modulons show strong positive correlation. (C) The regulatory region (150 bp upstream of the first gene in operon) of genes in the SigS i-modulon contained a conserved purine-rich motif. (D) The positions (relative to transcription start site) of the enriched motif within the regulatory sites of genes in the SigS i-modulon. For many genes in the SigS i-modulon, the motif was present 35 bp upstream of the translation start site. (E) Greed vs. fear trade-off is reflected in the activity of the Translation (greed) and SigS (fear) i-modulons. LAC showed increased propensity for fearful bet-hedging strategy while TCH1516 relied on a greedier strategy.
To further characterize σS, we looked for conserved motifs in the regulatory regions of the genes in the i-modulon and found a 21-bp purine-rich motif (E-value = 7.7e-8) in the regulatory region of at least 56 genes in the SigS i-modulon (Fig. 4C). Comparisons against a known prokaryotic motif database revealed that the S. aureus σS motif was most similar to that of the σB (MX000071) motif in Bacillus subtilis (E-value = 1.62e-02) (Materials and Methods). Next, we analyzed the distance between the center of the motif and the transcription initiation site. For most genes, the motif was present at or around 35 bp upstream of the translation start site, although motifs were also found further upstream (Fig. 4D). Of the 137 genes in the i-modulon, only 56 (41%) had an assigned function in the reference genome, further highlighting our limited understanding of σS functionality. However, many of the annotated gene products were key factors in controlling cellular state. These included factors regulating virulence (sarA, sarR, sarX), antimicrobial resistance (cadC, blaI), metabolism (arcR, argR), cell wall biogenesis (vraRST), biofilm formation (icaR), and DNA damage repair (recX). Genes encoding proteins critical for stress response, such as universal stress protein (Usp), toxin MazF, competence proteins ComGFK, and cell division protein were also present.
The SigS i-modulon also plays a critical role in the so-called “fear vs. greed” trade-off in S. aureus. Previously described in E. coli, this trade-off describes the allocation of resources toward optimal growth (greed) versus allocation toward bet-hedging strategies to mitigate the effect of stressors in the environment (fear) (7, 43). This balance is reflected in the transcriptome composition as an inverse correlation between the activities of the stress-responsive SigS i-modulon and the Translation i-modulon (Fig. 4E). Unlike E. coli, however, this relationship was independent of growth rate, as growth rate had weak correlation with Translation i-modulon expression activity (Pearson R = 0.094, P = 0.514). Interestingly, mapping this trade-off highlighted a possible difference in survival strategy between the two USA300 strains. TCH1516 tended toward a greedy strategy with high Translation i-modulon activity, while LAC was more likely to rely on bet-hedging or fear.
ICA Reveals Organization of Virulence Factor Expression.
ICA captured systematic expression changes of several genes encoding virulence factors. Previous studies described over half a dozen transcription factors with direct or indirect roles in regulation of virulence factor expression in S. aureus (44). The number of regulators, and their complex network of interactions, make it extremely difficult to understand how these genes are regulated at a genome scale. In contrast, ICA identified only three i-modulons (named Agr, SaeR, and Vim-3) that were mostly composed of virulence genes (Fig. 5A). The activity level of Agr had extremely low correlation with that of SaeR and Vim-3, suggesting that Agr may have only limited cross-talk with the other two i-modulons in our conditions (SI Appendix, Fig. S5A). However, the activity levels of SaeR and Vim-3 were negatively correlated (Pearson R = −0.57, P = 8.6e-11). As the two i-modulons contain different sets of virulence factors, the negative correlation points to a shift in the virulence state where S. aureus may adopt different strategies to thwart the immune system. Collectively, the three virulence i-modulons revealed coordinated regulation of 65 genes across the genome. These results suggest that the complexity behind virulence regulation can be decomposed into discrete signals and the virulence state of S. aureus can be defined as a linear combination of these signals.
Fig. 5.
Global regulation of virulence factors. (A) The three virulence i-modulons (SaeR, Agr, Vim-3) and the genomic positions of the genes in their respective i-modulons are mapped. The signals encode over 25 virulence factor-associated genes. (B) PurR i-modulon activity is highly correlated with virulence i-modulon SaeR. (C) Challenge with low pH, linezolid, and mupirocin leads to strong activation of agr in exponential growth phase. Interestingly, this activation is stronger than that induced by stationary phase (OD600 = 1.0). Activation of agr was much weaker under all other experimental conditions considered (Upper bar chart). (D) Coactivation of Phi-Sa3 i-modulon with virulence i-modulon Vim-3.
The SaeR i-modulon contained 27 genes, including the genes for the SaeRS two-component system. The activity level of this i-modulon strongly correlated with the expression level of saeRS, further supporting the idea that the genes in this i-modulon are regulated (directly or indirectly) by SaeRS (Pearson R = 0.80, P = 1.38e-25). Furthermore, the virulence genes chp, coa, ssl11, sbi, map, lukA, and scn, previously reported to be under the control of SaeRS (45), were also found in this i-modulon. The activity of SaeR i-modulon was strongly associated with purine metabolism. PurR, the transcription factor that regulates the genes of purine biosynthesis, has been recently implicated in regulation of virulence factors (25, 46). Consistent with this observation, the activity level of the SaeR i-modulon correlated well (Pearson R = 0.77, P = 8.9e-23) with the activity of the PurR i-modulon (Fig. 5B). Thus, SaeR may act as a bridge between virulence and metabolism.
Similarly, the Agr i-modulon contained the agrABCD genes involved in regulation of the quorum-sensing agr regulon (47, 48). As most of our samples were collected during early- to midexponential growth phase, the Agr i-modulon remained inactive in these conditions (SI Appendix, Fig. S5B). Only acidic conditions (pH 5.5) and treatment with translation inhibitors linezolid and mupirocin activated Agr during exponential growth (Fig. 5C). Both pH- and translation inhibition-dependence of agr expression have been previously reported (49–52). Unexpectedly, the Agr i-modulon was activated to a much greater extent by these factors than high cell density (OD 1.0), for which its role in quorum sensing is extensively characterized.
The Vim-3 virulence i-modulon consisted of genes required for siderophore and heme utilization (sbnABC, hrtAB), capsule biosynthesis (cap8a, capBC, cap5F), and osmotic tolerance (kdpA, betAT, gbsA). The Vim-3 i-modulon had maximal activity under a hyperosmotic condition introduced by 4% NaCl and when grown to stationary phase (OD 1.0) in CAMHB (SI Appendix, Fig. S5C). The increased expression of capsule biosynthesis genes have been shown to be responsive to change in osmotic pressure as well as iron starvation, which is consistent with the inclusion of iron scavenging and osmotic tolerance genes in the i-modulon with the capsular biosynthesis genes (53, 54).
We further identified a prophage Phi-Sa3–associated i-modulon as a putative i-modulon required for virulence. The Phi-Sa3 i-modulon consists of genes in the Phi-Sa3 prophage and several genes encoding DNA replication and repair enzymes. Excluded from the i-modulon were the virulence factors that were horizontally acquired along with the phage (scn and chp) (55), which now fell under the control of SaeR. Of the four phages in S. aureus strain Newman, Phi-Sa3 is the only prophage that is unable to generate complete viral particles when challenged with DNA damaging agent mitomycin (56). However, evidence suggests that this prophage is still active in USA300 strains and its genes are expressed during lung infection, where it may play a role in establishing infection (57). Corroborating this hypothesis, we found that the activity of the Phi-Sa3 i-modulon correlated highly with the Vim-3 i-modulon (Pearson R = 0.62, P = 9.9e-13) (Fig. 5D). As the Phi-Sa3 i-modulon does not contain any virulence genes, the phage itself may play an accessory role in establishing virulence.
ICA model Provides a Platform for In Vivo Data Interpretation.
Transcriptomic models based on ICA can also be used to interpret new in vivo and ex vivo expression profiles, leading to greater clarity when compared to analysis with a graph-based TRN model (SI Appendix, Supplementary Note 1). Expression profiling data can be projected onto the i-modulon structure of the TRN, derived from our dataset, to convert the values from gene-expression levels to i-modulon activity levels (Materials and Methods). This projection can supplement gene differential expression analysis by identifying regulators that are driving the large changes in gene expression often seen in vivo.
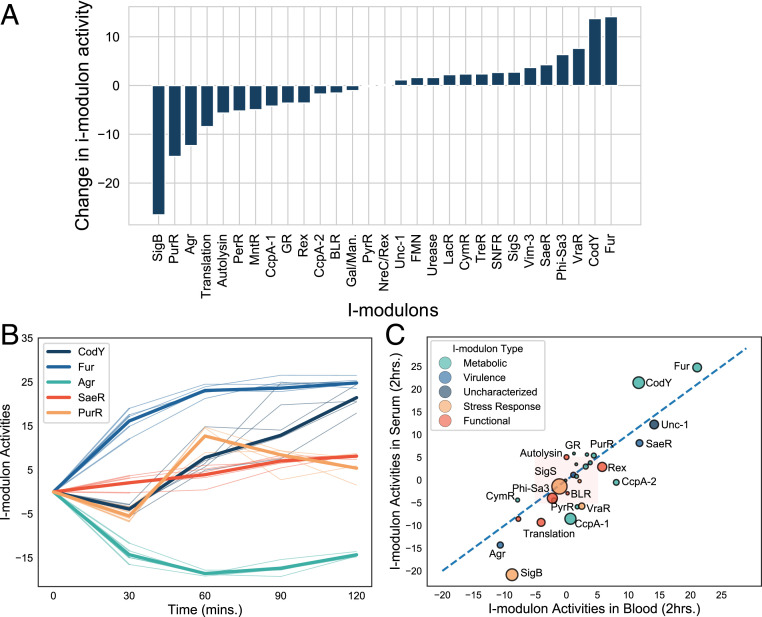
We projected microarray data (GSE61669) taken at 24 h postinfection from a rabbit skin infection model (58). After 24 h, 1,232 differentially expressed genes were reported. Projection of the data on to the model showed that these changes in differential expression are being driven by simultaneous activation of CodY and Fur i-modulons and inactivation of SigB, PurR, Agr, and Translation i-modulons (Fig. 6A).
Fig. 6.
ICA analysis of in vivo and ex vivo data. (A) Change in i-modulon activities at 24 h postinfection in a rabbit skin infection model. (B) Activity levels of select i-modulons in serum over the 2-h time period. The thick line represents the mean activity across all replicates and the thin line represents activity in each individual replicate (n = 4). Activity levels were around the inoculum values, (C) Comparison of i-modulon activity between serum and blood and 2-h time point. The dashed red line is the 45° line; i-modulons below the line have higher activity in blood and those above the line have higher activity in serum. Red shaded area contains i-modulons with less than fivefold change in activity in both conditions.
In time-course data, projecting expression data onto the model can also help us understand the dynamics of different regulators during infection. We projected previously published time-course microarray data collected from S. aureus USA300 LAC grown in tryptic soy broth (TSB), human blood, and serum (59). Bacteria grown to an exponential phase in TSB was used as inoculum for all samples; we used this as our new base condition for the projected data. Therefore, all i-modulon activity levels in this set represent log2 fold-change in activity from this base condition. Once transferred to serum, the activities of Fur and CodY i-modulons in serum increased dramatically, with Fur being activated immediately after exposure to serum while CodY activated slowly over time to reach a similar level as Fur by 2 h (Fig. 6B). The large change in activity coupled with the sizeable number of genes in each i-modulon (80 and 45 genes in CodY and Fur, respectively) indicates that S. aureus reallocates a considerable portion of its transcriptome to reprogram amino acid and iron metabolism in serum. PurR and SaeR activity also increased, although their magnitude of change was dwarfed by the changes in activities of CodY and Fur. On the other hand, Agr activity declined and remained low over the 2-h period. Because agr positively regulates a number of virulence genes, dynamic changes in its activity level could be expected in serum. Consistent with the model prediction, previous studies have demonstrated that agr transcription is dampened in human serum due to sequestration of autoinducing peptide by human apolipoprotein B (60, 61).
We next calculated the differences in i-modulon activities in blood and serum at the final 2-h time point. Fur, CodY, PurR, SaeR, and Agr had similar activity levels in both blood and serum (Fig. 6B). Therefore, the activities of these regulators are likely governed by the noncellular fraction of the blood. I-modulons PyrR, SigB, Translation, VraR, CcpA-1, and CcpA-2 had higher activity levels in blood than in the serum (Fig. 6C). Glucose concentration in blood is lower than in serum, which likely explains the shift in CcpA-1 activity (62). The lower glucose concentration relieves CcpA-mediated repression of its regulon, leading to higher expression. The shift in the PyrR i-modulon also corroborates previous study, which demonstrated that S. aureus strain JE2 (a derivative of LAC) requires more pyrimidine when growing in blood than in serum (63). The signals or cues driving the change in activity of the other i-modulons (SigB, Translation, VraR, and CcpA-2) remains unknown. Overall, the i-modulon analysis revealed that during acute infection, CodY and Fur play key roles in rewiring the S. aureus metabolism in serum and blood when compared to TSB, while SaeR (and not Agr) drives the virulence gene expression. In addition, SigB, Translation, and VraR i-modulons are uniquely activated by the cellular fraction of the blood and may thus be responding to unique stresses they impart. However, these observations are limited as the baseline for comparisons for most of these analyses were in vitro growth in TSB. Although the differentially activated i-modulons may point to important roles that each of the associated regulators play during acute infection, further analysis is still required to understand their relative contribution. The model is also limited in that it is currently blind to the regulators that are not captured in any of the 29 i-modulons. This limitation will be alleviated over time as we incorporate more RNA-seq data that is being generated at an ever-increasing pace.
Discussion
Here, we described an ICA-based method to elucidate the organization of the modules in TRN in S. aureus USA300 strains. Using this method, we identified 29 independently modulated sets of genes (i-modulons) and their activities across the sampled conditions. This framework for exploring the TRN provides three key advantages over traditional methods, especially when working with nonmodel organisms: 1) The method provides an explanatory reconstruction of the TRN; 2) it is an untargeted, and therefore unbiased, approach; and 3) the approach utilizes expression profiling data, an increasingly ubiquitous resource.
First, i-modulons quantitatively capture the complexities of transcriptional regulation and enable a new way to systematically query the transcriptome. By recasting the data in terms of explanatory i-modulons, we gained a deeper understanding of large changes in transcription profiles between CAMHB bacteriologic media and the more physiologically relevant mammalian tissue culture-based media RPMI+10%LB. The analysis reduced the number of features needed to capture most of the information in the transcriptome from hundreds of genes to 15 i-modulons. Additionally, quantified activity levels of i-modulons also enabled integration of regulatory activity with metabolic models and revealed coordination between metabolic and regulatory networks. Such reduction in complexity and the integration of different aspects of S. aureus biology (e.g., virulence, metabolism, stress response, and so forth) will be crucial to understanding the mechanisms that enable successful infection in vivo.
Second, this method presents a platform for untargeted, global analysis of the TRN. Due to its untargeted nature, we also identified two key virulence features of S. aureus. ICA revealed coordinated regulation of genes in capsule biosynthesis, osmotic tolerance, and iron starvation (Vim-3 i-modulon). Both capsule formation and siderophore scavenging are important in nasal colonization (64, 65). Similarly, growth of S. aureus in synthetic nasal medium (SNM3) increases the expression of genes required for osmotolerance. Therefore, theVim-3 i-modulon may represent a concerted regulation of genes required for successful nasal colonization. We also identified the Phi-Sa3 phage i-modulon, whose activity level correlated with that of the Vim-3 i-modulon. The Phi-Sa3 i-modulon did not include the virulence genes (e.g., sak, scn, and others) that were acquired with the phage, suggesting that phage-replication genes were expressed independently of the virulence genes. Given that its activity was correlated with Vim-3, this phage may also play an important role in nasal colonization.
Finally, ICA uses RNA-seq data to extract information about the TRN, making it more accessible to nonmodel organisms, including S. aureus. Reconstructing the TRN with traditional methods is highly resource intensive, as they require targeted antibodies or specialized libraries of plasmids containing all transcription factors of interest (66). While these approaches have given us great insights into TRNs of model organisms like E. coli (10), such comprehensive data are not available for most microbes. Several studies have attempted to circumvent this by comparing the expression profiles of wild-type S. aureus strains with their counterpart with either knocked out or constitutively active transcription factors. However, these approaches often overestimate the regulatory reach of the transcription factor, as such genetic changes can trigger the differential expression of genes not directly under the regulator’s control. By identifying i-modulons consisting of independently regulated sets of genes, the ICA-based method improves on these approaches as it able to segregate specific regulator targets (7). While many expression profiles are required to build such a model, a rapidly growing number of expression profiles are already publicly available on the Gene Expression Omnibus. Indeed, utilizing only RNA-seq data, we predicted the previously unknown regulon of stress-associated sigma factor σS and its possible roles in biofilm formation and general stress response. With the growing number of available expression profiles, such characterizations can be extended to other undefined or poorly defined regulons. Therefore, in the absence of a comprehensive set of targeted antibodies against S. aureus transcription factors, reanalyzing the publicly available database with ICA could be used to further reconstruct its TRN.
We have shown that ICA-based decomposition can be utilized to build a quantitative and explanatory model of S. aureus TRN from RNA-seq data. Application of this model enabled us to query metabolic and regulatory cross-talk, discover new potential regulons, find coordination between metabolism and virulence, and unravel the S. aureus response during growth in blood. Due to this versatility, this model and other models generated through this framework may prove to be a powerful tool in any future studies of S. aureus and other nonmodel organisms.
Materials and Methods
RNA Extraction and Library Preparation.
S. aureus USA300 isolates LAC, TCH1516, and ALE derivatives of TCH1516 (SNFR and SNFM) were used for this study. The growth conditions and RNA preparation methods for data acquired from Choe et al. has been previously described (67). Detailed growth conditions, RNA extraction, and library preparation methods for other samples have also been already described (68). Briefly, an overnight culture of S. aureus was used to inoculate a preculture and were grown to midexponential growth phase (OD600 = 0.4) in respective media (CAMHB, RPMI+10%LB, or TSB). Once in midexponential phase, the preculture was used to inoculate the media containing appropriate supplementation or perturbations. Samples were collected at ODs and time points indicated in the metadata (>Dataset S3). All samples were collected in biological duplicates, originating from different overnight cultures. Samples for control conditions were collected for each set to account for batch effect.
Determining Core Genome with Bidirectional BLAST Hits.
To combine the data from the two strains, core genome-containing conserved genes between the LAC (GenBank: CP035369.1and CP035370.1) and TCH1516 (GenBank: NC_010079.1, NC_012417.1, and NC_010063.1) were first established using bidirectional BLAST hits (69). In this analysis, all protein sequences of CDS from both genomes are BLASTed against each other twice with each genome acting as reference once. Two genes were considered conserved (and therefore part of the core genome) if 1) the two genes have the highest alignment percent to each other than to any other genes in the genome, and 2) the coverage is at least 80%.
RNA-Seq Data Processing.
The RNA-seq pipeline used to analyze and perform QC/QA has been described in detail previously (68). Briefly, the sequences were aligned to respective genomes, LAC or TCH1516, using Bowtie2 (70, 71). The samples from ALE derivatives, SNFM and SNFR, were aligned to TCH1516. The aligned sequences were assigned to ORFs using HTSeq-counts (72). Differential expression analysis was performed using DESeq2 with a P-value threshold of 0.05 and an absolute fold-change threshold of 2 (73). To create the final counts matrix, counts from conserved genes in LAC samples were represented by the corresponding ortholog in TCH1516. The counts for accessory genes were filled with 0s if the genes were not present in the strain (i.e., LAC-specific genes had counts of 0 in TCH1516 samples and vice versa). Finally, to reduce the effect of noise, genes with average counts per sample <10 were removed. The final counts matrix with 2,581 genes was used to calculate transcripts per million (TPM).
Computing Robust Components with ICA.
The procedure for computing robust components with ICA has been described in detail previously (7). Log2(TPM + 1) values were centered to strain-specific reference conditions and used as input of ICA decomposition. These conditions are labeled: “USA300_TCH1516_U01-Set000_CAMHB_Control_1”, “USA300_TCH1516_U01-Set000_CAMHB_Control_2” for TCH1516; and “USA300_LAC_U01-Set001_CAMHB_Control_1”, “USA300_LAC_U01-Set001_CAMHB_Control_2” for LAC. Next, Scikit-learn (v0.19.0) implementation of the FastICA algorithm was used to calculate independent components with 100 iterations, convergence tolerance of 10-7, log(cosh(x)) as contrast function and parallel search algorithm (74, 75). The number of calculated components were set to the number of components that reconstruct 99% of variance as calculated by principal component analysis. The resulting S-matrices containing source components from the 100 iterations were clustered with Scikit-learn implementation of the DBSCAN algorithm with ε of 0.1, and minimum cluster seed size of 50 samples (50% of the number of random restarts). If necessary, the component in each cluster was inverted such that the gene with the maximum absolute weighting the component was positive. Centroids for each cluster were used to define the final weightings for S and corresponding A matrix. The whole process was repeated 100 times to ensure that the final calculated components were robust. Finally, components with activity levels that deviated more than five times between samples in the same conditions were also filtered out.
Determining Independently Modulated Sets of Genes.
ICA enriches components that maximize the nongaussianity of the data distribution. While most genes have weightings near 0 and fall under Gaussian distribution in each component, there exists a set of genes whose weightings in that component deviate from this significantly. To enrich these genes, we used Scikit-learn’s implementation of the D’Agostino K2 test, which measures the skew and kurtosis of the sample distribution (76). We first sorted the genes by the absolute value of their weightings and performed the K2 test after removing the gene with the highest weighting. This was done iteratively, removing one gene at a time, until the K2 statistic falls below a cutoff. We calculated this cutoff based on sensitivity analysis on agreement between enriched i-modulon genes and regulons inferred by RegPrecise (77). For a range of cutoff (between 200 and 600), we ran the iterative D’Agostino K2 test on all components and checked for statistically significant overlap of i-modulons with the regulons predicted by RegPrecise using Fisher’s exact test. For i-modulons with significant overlap, we also calculated precision and recall. The cutoff of 280, which led to the highest harmonic average between precision and recall (F1-score), was chosen as the final cutoff.
Designating Biological Annotations to i-Modulons.
To designate proper annotations to i-modulons, we first compiled a dataset containing previously predicted features, such as regulons, genomic islands, and plasmids. The regulons in the datasets were inferred by either the RegPrecise algorithm and by RNA-seq analysis of transcription factor knockout strains or strains with constitutively active transcription factors (Dataset S4) (67, 78–81). Genomic islands were determined by the online IslandViewer4 tool (82) and phages were identified with PHASTER (83). For studies using different strains of S. aureus orthologs for TCH1516 and LAC were determined using bidirectional BLAST hits. The enriched genes in i-modulons were compared against this dataset for significant overlap using Fisher’s exact test with an FDR of 10 to 5. With this analysis 15 i-modulons were enriched with high confidence (precision ≥ 0.5, recall ≥ 0.2) and 7 were enriched with low confidence. Additionally, i-modulons containing genes with shared functions (e.g., Translation and B-lactam resistance) were annotated manually (Dataset S5).
Differential Activation Analysis.
Distribution of differences in i-modulon activities between biological replicates were first calculated and a log-norm distribution was fit to the differences. In order to test statistical significance, absolute value of difference in activity level of each i-modulon between the two samples was calculated. This difference in activity was compared to the log-normal distribution from above to get a P value. Because differences and P value for all i-modulons were calculated, the P value was further adjusted with Benjamini–Hochberg correction to account for multiple hypothesis testing problem. Only i-modulons with change in activity levels greater than 5 were considered significant.
Motif Enrichment and Comparison.
Genes were first assigned to operons based on operonDB (84, 85). For i-modulon specific motif enrichments, a 150-bp segment upstream of all of the genes in the i-modulons were collected. To avoid enriching ribosome binding sites, the segment started from 15 bp upstream of the translation start site. For genes in minus strand, the reverse complement of the sequence was used instead. If genes were part of an operon, then only the segment in front of the first gene in the operon was used. Motifs and their positions were enriched from these segments using the online Multiple Em for Motif Elicitation (MEME) algorithm (86, 87). The following default parameters we reused: -dna -oc -mod zoops -nmotifs 3 -minw 6 -maxw 50 -objfun classic -revcomp -markov_order 0. Enriched motifs were compared to combined prokaryotic databases CollecTF, Prodoric (release 8.9), and RegTransBase (v4) using TomTom (88–91). The parameters for TomTom were as follows: -oc -min-overlap 5 -mi 1 -dist pearson -evalue -thresh 10.0.
Metabolic Modeling.
We modeled growth in RPMI supplemented with iron, manganese, zinc, and molybdate by setting the lower bound to the corresponding nutrient exchanges in iYS854 to −1 mmol/gDW/h (the negative sign is a modeling convention to allow for the influx of nutrients) (28), and −13 mmol/gDW/h for oxygen exchange (as measured experimentally). Additionally, to account for the utilization of heme by S. aureus terminal oxidases, we removed heme A from the biomass reaction and added as a reactant in the cytochrome oxidase reaction with the stoichiometric coefficient obtained from the biomass reaction (92). Next, we constructed two csGEMs to compare two conditions with: 1) d-glucose as the main glycolytic source and 2) maltose as an alternative carbon source. In the first condition, we set the lower bound to d-glucose exchange to −50 mmol/gDW/h. Assuming that in the presence of d-glucose, ccpA mediates the repression of multiple genes (22, 29), we set the upper and lower bounds of the reactions encoded by genes of the ccpA i-modulon to 0. Specifically, we only turned off the set of 44 reactions obtained by running the “cobra.manipulation.find_gene_knockout_reactions()”command from the cobrapy package (93), feeding it the model and the 52 modeled genes which form part of the CcpA-1 i-modulon. As such, we implemented a method similar to the switch-based approach (94, 95), in which the boolean encoding for the gene-reaction rule is taken into account (i.e., isozymes, and protein complexes). Shutting down all of the reactions yielded a model which could not simulate growth. We thus gap-filled the first csGEM with one reaction (AcCoa carboxylase, involved in straight chain fatty acid biosynthesis). To simulate the second condition in which maltose serves as the main glycolytic source, we set the lower bound of maltose exchange to −50 mmol/gDW/h and blocked d-glucose uptake. No regulatory constraints were added. FBA was implemented with the biomass formation set as the functional network objective, and fluxes were sampled in both csGEMs 1,000 times using the “cobra.sampling.sample” command. To normalize flux values across conditions, we divided all fluxes by the simulated growth rate. We compared the flux distribution of each reaction in the two csGEMs using the Kolmogorov–Smirnov nonparametric test, yielding 93 reactions with significantly differing flux distributions (P < 0.001) having a statistic larger than 0.99. To identify whether there is a metabolic basis for the difference the Fur i-modulon stimulation between conditions, we identified a set of 34 reactions encoded by the 41 modeled genes which are partof the Fur i-modulon (again using the switch-based approach).
Targeted HPLC (HPLC).
For glucose detection, samples were collected every 30 min and filtered as described above. Growth media was syringe-filtered through 0.22-μm disk filters (Millex-GV, Millipore Sigma) to remove cells. The filtered samples were loaded onto a 1260 Infinity series (Agilent Technologies) HPLCy (HPLC) system with an Aminex HPX-87H column (Bio-Rad Laboratories) and a refractive index detector. The system was operated using ChemStation software. The HPLC was run with a single mobile phase composed of HPLC grade water buffered with 5 mM sulfuric acid (H2SO4). The flow rate was held at 0.5 mL/min, the sample injection volume was 10 μL, and the column temperature was maintained at 45 °C. The identities of compounds were determined by comparing retention time to standard curves of glucose. The peak area integration and resulting chromatograms were generated within ChemStation and compared to that of the standard curves in order to determine the concentration of each compound in the samples.
Microarray Data Analysis and Projection.
All microarray data were downloaded from the Gene Expression Omnibus repository (GSE25454, GSE61669, and GSE18793) and processed with the Affy package in R to get gene-expression level (59, 96). The GSE25454 dataset consists of microarray data from samples grown to exponential phase in TSB (TSB 0 h) and transferred to either blood, serum, or TSB. Samples were then collected every 30 min for 2 h. The data were centered on the TSB 0 h time point. The GSE61669 dataset consists of expression profile from 24-h rabbit skin infection. These data were centered on the expression profile from the inoculum. Finally, the GSE18793 expression profile consists of data comparing wild-type LAC and its isogenic agr mutant. These data were centered around the wild-type expression profile. Data projection was used to convert centered gene-expression values to i-modulon activity level as described previously (7).
Data and Code Availability.
All RNA-seq data used to build the model have been deposited to the Sequence Read Archive (SRA). The normalized log TPM, and the calculated S and A matrix of the model can be found in Datasets 7–9, respectively. The data accession numbers can be found in Dataset S3 and refs. 67, 68, and 97–99. Both glucose and maltose models were deposited to BioModels with accession numbers MODEL2005290002 and MODEL2005290001. Their sampled fluxes are also available on Datasets S1 and S2. Custom code of ICA analysis can be found on Github (https://github.com/SBRG/precise-db).
Supplementary Material
Acknowledgments
This research was supported by NIH National Institute of Allergy and Infectious Diseases Grant 1-U01-AI124316.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing data reported in this paper have been deposited in the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra (accession numbers listed in Dataset S3). The glucose and maltose models were deposited to BioModels, https://www.ebi.ac.uk/biomodels/ (accession nos. MODEL2005290002 and MODEL2005290001.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008413117/-/DCSupplemental.
References
- 1.Tong S. Y. C., Davis J. S., Eichenberger E., Holland T. L., Fowler V. G. Jr., Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krismer B., Weidenmaier C., Zipperer A., Peschel A., The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 15, 675–687 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Dastgheyb S. S., Otto M., Staphylococcal adaptation to diverse physiologic niches: An overview of transcriptomic and phenotypic changes in different biological environments. Future Microbiol. 10, 1981–1995 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goerke C., Wolz C., Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int. J. Med. Microbiol. 300, 520–525 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Burian M., Wolz C., Goerke C., Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS One 5, e10040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibarra J. A., Pérez-Rueda E., Carroll R. K., Shaw L. N., Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14, 126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sastry A. V. et al., The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat. Commun. 10, 5536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saelens W., Cannoodt R., Saeys Y., A comprehensive evaluation of module detection methods for gene expression data. Nat. Commun. 9, 1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand A. et al., Adaptive evolution reveals a tradeoff between growth rate and oxidative stress during naphthoquinone-based aerobic respiration. Proc. Natl. Acad. Sci. U.S.A. 116, 25287–25292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos-Zavaleta A. et al., RegulonDB v 10.5: Tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 47, D212–D220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin M. Y., Makarova K. S., Wolf Y. I., Koonin E. V., Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King A. N. et al., Guanine limitation results in CodY-Dependent and -independent alteration of staphylococcus aureus physiology and gene expression. J. Bacteriol. 200, e00136-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pohl K. et al., CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191, 2953–2963 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hove-Jensen B. et al., Phosphoribosyl diphosphate (PRPP): Biosynthesis, enzymology, utilization, and metabolic significance. Microbiol. Mol. Biol. Rev. 81, e00040-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kofoed E. M. et al., De novo guanine biosynthesis but not the Riboswitch-regulated purine salvage pathway is required for staphylococcus aureus infection in vivo. J. Bacteriol. 198, 2001–2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaca A. O., Colomer-Winter C., Lemos J. A., Many means to a common end: The intricacies of (p)ppgpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kriel A. et al., Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol. Cell 48, 231–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivatsan A., Wang J. D., Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Horsburgh M. J. et al., MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44, 1269–1286 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Sadykov M. R. et al., CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology 157, 3458–3468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halsey C. R. et al., Amino acid catabolism in staphylococcus aureus and the function of carbon catabolite repression. MBio 8, e01434-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidl K. et al., Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9, 95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leiba J. et al., A novel mode of regulation of the Staphylococcus aureus catabolite control protein A (CcpA) mediated by Stk1 protein phosphorylation. J. Biol. Chem. 287, 43607–43619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengert P., Dandekar T., Riboswitch finder—A tool for identification of riboswitch RNAs. Nucleic Acids Res. 32, W154–W159 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sause W. E et al., The purine biosynthesis regulator PurR moonlights as a virulence regulator in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 116, 13563–13572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung G. Y. C., Wang R., Khan B. A., Sturdevant D. E., Otto M., Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiele I., Palsson B. Ø., A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seif Y. et al., A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types. PLoS Comput. Biol. 15, e1006644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuxoll A. S. et al., CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog. 8, e1003033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Brien E. J., Monk J. M., Palsson B. O., Using genome-scale models to predict biological capabilities. Cell 161, 971–987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feist A. M., Palsson B. O., The biomass objective function. Curr. Opin. Microbiol. 13, 344–349 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y. et al., Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111, E4981–E4990 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horsburgh M. J. et al., sigmaB modulates virulence determinant expression and stress resistance: Characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184, 5457–5467 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basu A., Shields K. E., Eickhoff C. S., Hoft D. F., Yap M. N. F., Thermal and nutritional regulation of ribosome hibernation in staphylococcus aureus. J. Bacteriol. 200, e00426-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morikawa K. et al., Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 8, e1003003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller H. K. et al., The extracytoplasmic function sigma factor σS protects against both intracellular and extracytoplasmic stresses in Staphylococcus aureus. J. Bacteriol. 194, 4342–4354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenz U. et al., The alternative sigma factor sigma B of Staphylococcus aureus modulates virulence in experimental central venous catheter-related infections. Microbes Infect. 10, 217–223 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Tuchscherr L. et al., Sigma factor SigB is crucial to mediate staphylococcus aureus adaptation during chronic infections. PLoS Pathog. 11, e1004870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senn M. M. et al., Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus. J. Bacteriol. 187, 8006–8019 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamber S., Schwartzman J., Cheung A. L., Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect. Immun. 78, 3637–3646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mäder U. et al., Staphylococcus aureus transcriptome architecture: From laboratory to infection-mimicking conditions. PLoS Genet. 12, e1005962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burda W. N. et al., Investigating the genetic regulation of the ECF sigma factor σS in Staphylococcus aureus. BMC Microbiol. 14, 280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utrilla J. et al., Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution. Cell Syst. 2, 260–271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenul C., Horswill A. R., Regulation of staphylococcus aureus virulence. Microbiol. Spectr. 6, GPP3-0031-2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Q., Yeo W. S., Bae T., The SaeRS two-component system of Staphylococcus aureus. Genes (Basel) 7, 81 (2016). [Google Scholar]
- 46.Goncheva M. I. et al., Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat. Commun. 10, 775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novick R. P. et al., The agr P2 operon: An autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248, 446–458 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Novick R. P., Geisinger E., Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Regassa L. B., Betley M. J., Alkaline pH decreases expression of the accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 174, 5095–5100 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinrick B. et al., Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186, 8407–8423 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regassa L. B., Novick R. P., Betley M. J., Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60, 3381–3388 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joo H. S., Chan J. L., Cheung G. Y. C., Otto M., Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54, 4942–4944 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pöhlmann-Dietze P. et al., Adherence of Staphylococcus aureus to endothelial cells: Influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect. Immun. 68, 4865–4871 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J. C., Takeda S., Livolsi P. J., Paoletti L. C., Effects of in vitro and in vivo growth conditions on expression of type 8 capsular polysaccharide by Staphylococcus aureus. Infect. Immun. 61, 1853–1858 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verkaik N. J. et al., Immune evasion cluster-positive bacteriophages are highly prevalent among human Staphylococcus aureus strains, but they are not essential in the first stages of nasal colonization. Clin. Microbiol. Infect. 17, 343–348 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Bae T., Baba T., Hiramatsu K., Schneewind O., Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 62, 1035–1047 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Jones M. B. et al., Genomic and transcriptomic differences in community acquired methicillin resistant Staphylococcus aureus USA300 and USA400 strains. BMC Genomics 15, 1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malachowa N., Kobayashi S. D., Sturdevant D. E., Scott D. P., DeLeo F. R., Insights into the Staphylococcus aureus-host interface: Global changes in host and pathogen gene expression in a rabbit skin infection model. PLoS One 10, e0117713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malachowa N. et al., Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6, e18617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peterson M. M. et al., Apolipoprotein B Is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe 4, 555–566 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall P. R. et al., Nox2 modification of LDL is essential for optimal apolipoprotein B-mediated control of agr type III Staphylococcus aureus quorum-sensing. PLoS Pathog. 9, e1003166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tonyushkina K., Nichols J. H., Glucose meters: A review of technical challenges to obtaining accurate results. J. Diabetes Sci. Technol. 3, 971–980 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Connolly J. et al., Identification of staphylococcus aureus factors required for pathogenicity and growth in human blood. Infect. Immun. 85, e00337-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Riordan K., Lee J. C., Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17, 218–234 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stubbendieck R. M. et al., Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl. Environ. Microbiol. 85, e02406-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minch K. J. et al., The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 6, 5829 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choe D. et al., Genome-scale analysis of methicillin-resistant Staphylococcus aureus USA300 reveals a tradeoff between pathogenesis and drug resistance. Sci. Rep. 8, 2215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poudel S. et al., Characterization of CA-MRSA TCH1516 exposed to nafcillin in bacteriological and physiological media. Sci. Data 6, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Overbeek R., Fonstein M., D’Souza M., Pusch G. D., Maltsev N., The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U.S.A. 96, 2896–2901 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Highlander S. K. et al., Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7, 99 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pedregosa F. et al., Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 75.Hyvärinen A., Oja E., Independent component analysis: Algorithms and applications. Neural Netw. 13, 411–430 (2000). [DOI] [PubMed] [Google Scholar]
- 76.D’Agostino R. B., Belanger A., A suggestion for using powerful and informative tests of normality. Am. Stat. 44, 316–321 (1990). [Google Scholar]
- 77.Ravcheev D. A. et al., Inference of the transcriptional regulatory network in Staphylococcus aureus by integration of experimental and genomics-based evidence. J. Bacteriol. 193, 3228–3240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyle-Vavra S., Yin S., Jo D. S., Montgomery C. P., Daum R. S., VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 83–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuroda M. et al., Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49, 807–821 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Delauné A. et al., The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 80, 3438–3453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falord M., Mäder U., Hiron A., Débarbouillé M., Msadek T., Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 6, e21323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bertelli C. et al.; Simon Fraser University Research Computing Group , IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arndt D. et al., PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ermolaeva M. D., White O., Salzberg S. L., Prediction of operons in microbial genomes. Nucleic Acids Res. 29, 1216–1221 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pertea M., Ayanbule K., Smedinghoff M., Salzberg S. L., OperonDB: A comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 37, D479–D482 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bailey T. L., Elkan C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994). [PubMed] [Google Scholar]
- 87.Bailey T. L. et al., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kiliç S., White E. R., Sagitova D. M., Cornish J. P., Erill I., CollecTF: A database of experimentally validated transcription factor-binding sites in bacteria. Nucleic Acids Res. 42, D156–D160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eckweiler D., Dudek C. A., Hartlich J., Brötje D., Jahn D., PRODORIC2: The bacterial gene regulation database in 2018. Nucleic Acids Res. 46, D320–D326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kazakov A. E. et al., RegTransBase—A database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 35, D407–D412 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S., Quantifying similarity between motifs. Genome Biol. 8, R24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hammer N. D., Schurig-Briccio L. A., Gerdes S. Y., Gennis R. B., Skaar E. P., CtaM is required for menaquinol oxidase aa3 function in staphylococcus aureus. MBio 7, e00823-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebrahim A., Lerman J. A., Palsson B. O., Hyduke D. R., COBRApy: Constraints-based reconstruction and analysis for python. BMC Syst. Biol. 7, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Becker S. A., Palsson B. O., Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 4, e1000082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hyduke D. R., Lewis N. E., Palsson B. Ø., Analysis of omics data with genome-scale models of metabolism. Mol. Biosyst. 9, 167–174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gautier L., Cope L., Bolstad B. M., Irizarry R. A., affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004). [DOI] [PubMed] [Google Scholar]
- 97.Poudel S., Staphylococcus aureus LAC RNAseq. NCBI SRA. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA470935/. Deposited 10 May 2018.
- 98.Poudel S., S.aureus USA300 LAC Vancomycin. NCBI SRA. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA526539/. Deposited 11 March 2019.
- 99.Poudel S., USA300 strain RNAseq. NCBI SRA. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA634715. Deposited 24 May 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data used to build the model have been deposited to the Sequence Read Archive (SRA). The normalized log TPM, and the calculated S and A matrix of the model can be found in Datasets 7–9, respectively. The data accession numbers can be found in Dataset S3 and refs. 67, 68, and 97–99. Both glucose and maltose models were deposited to BioModels with accession numbers MODEL2005290002 and MODEL2005290001. Their sampled fluxes are also available on Datasets S1 and S2. Custom code of ICA analysis can be found on Github (https://github.com/SBRG/precise-db).