Significance
Honey bee colony defense is an emergent trait composed of individual aggressive responses. Here, we investigated the relationship between individual genotype, colony allele frequency, and aggression in individual bees. Our findings show that the colony-level defense response strongly correlates with colony-level allele frequency in a way that can be used to identify causative genomic regions. Importantly, we were able to validate a key associated region as also being under selection. As very similar allele frequency correlations are observed in both soldier and forager bees, we conclude that group genetics is more important than individual genetics in this case, giving further insight into the relationships between “nature,” “nurture,” and behavioral evolution.
Keywords: behavioral genetics, GWAS, aggression
Abstract
For social animals, the genotypes of group members affect the social environment, and thus individual behavior, often indirectly. We used genome-wide association studies (GWAS) to determine the influence of individual vs. group genotypes on aggression in honey bees. Aggression in honey bees arises from the coordinated actions of colony members, primarily nonreproductive “soldier” bees, and thus, experiences evolutionary selection at the colony level. Here, we show that individual behavior is influenced by colony environment, which in turn, is shaped by allele frequency within colonies. Using a population with a range of aggression, we sequenced individual whole genomes and looked for genotype–behavior associations within colonies in a common environment. There were no significant correlations between individual aggression and specific alleles. By contrast, we found strong correlations between colony aggression and the frequencies of specific alleles within colonies, despite a small number of colonies. Associations at the colony level were highly significant and were very similar among both soldiers and foragers, but they covaried with one another. One strongly significant association peak, containing an ortholog of the Drosophila sensory gene dpr4 on linkage group (chromosome) 7, showed strong signals of both selection and admixture during the evolution of gentleness in a honey bee population. We thus found links between colony genetics and group behavior and also, molecular evidence for group-level selection, acting at the colony level. We conclude that group genetics dominates individual genetics in determining the fatal decision of honey bees to sting.
Western honey bees (Apis mellifera L.) are attractive to a variety of predators because their hives contain large quantities of nutrients, notably tens of kilograms of high-carbohydrate honey and tens of thousands of lipid- and protein-rich larvae and adults. To protect these resources, honey bees have evolved a sophisticated system of social defense that involves communication and division of labor (1). Some individuals act as guards, patrolling the hive entrance and releasing alarm pheromones when they encounter an intruder, while others act as soldiers that respond first to an alarm by flying out of the hive to sting the intruder, dying in the process. Aggression varies in intensity among honey bee ecotypes, linked to ecological differences and human selection for gentler bees (2).
In honey bees, the genetic basis of aggression has been most extensively examined in relation to the Africanization of domestic populations in the Western Hemisphere resulting from the introduction and subsequent hybridization with a highly aggressive African honey bee (AHB) subspecies Apis mellifera scutellata (3–5). As an emergent group response, colony defense is a complex phenotype modulated by external environmental effects but also, indirect genetic effects derived from the interactions between individuals within a colony (6–8). Direct genetic associations have implicated large genomic regions (3, 9), and phenotype assessment has shown that colony defense has a strong heritable component (5, 10) with a degree of parental bias in expression (11, 12).
For many behavioral traits in animals and humans, individual genotypic variants typically account for only a small percentage of trait variation, and heritability is limited (13). These factors complicate the mechanistic analysis of behavior using genetic and genomic tools and have provoked a variety of evolutionary (14) and systems biology (15) hypotheses. Social behaviors are especially sensitive to the environment, yet the social environment is at least partly determined by the genotypes of other individuals in a social group (16). Such feedback effects (17) can complicate analyses, specifically for genome-wide association studies (GWAS) (18, 19). Indirect effects within groups also have profound implications for evolutionary selection, potentially leading to social phenotypes that are displayed and selected at the group level (6, 8, 20). We thus hypothesized that the influence of the genes of other colony members would be an important determinant of aggression in individual bees.
To test this hypothesis, we designed a modified GWAS that uses colony, rather than individual, measurements of genotype and phenotype and compared its performance with conventional methods. We used gentle African honey bees (gAHBs) from Puerto Rico, which show reduced aggression in defense of their colonies relative to other AHBs in Western countries. Although much less aggressive than the ancestral A. m. scutellata, gAHBs retain both genetic (21) and phenotypic diversity, despite a restricted allele set relative to other AHB populations due to a genetic bottleneck and recent (ca. 1994 to 2006) soft selective sweep (21).
Results
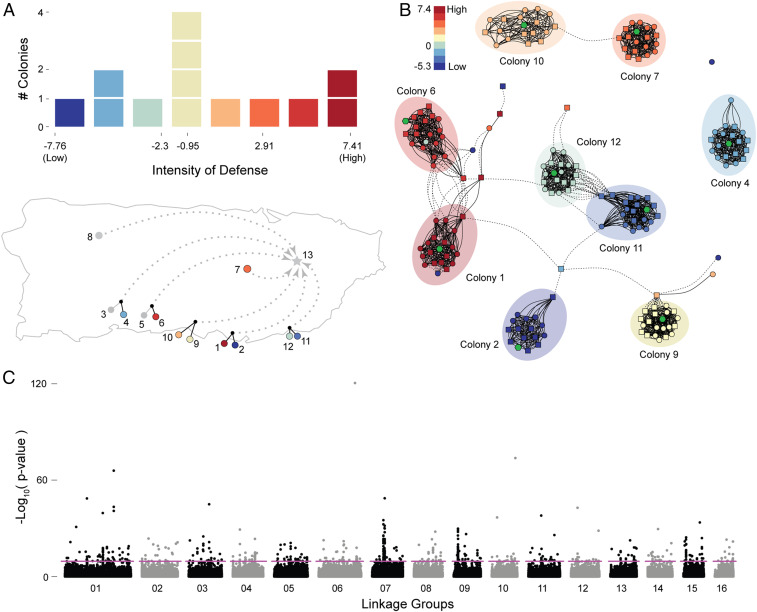
We quantified colony response (n = 9) through simulated attack and behavioral profiling of honey bee colonies, adapting established methodology (3, 9). Measures of overall aggressive response mounted by each colony in defense varied by colony (Fig. 1A and Dataset S1: colony), as is known. After the simulated attack, we collected soldiers who responded by stinging and nonaggressive bees who continued to forage (foragers) during the attack. We sequenced the genomes of 9 to 10 soldiers and 9 to 10 foragers from each colony. All colonies were in a common environment, each headed by a naturally mated queen unrelated to the others. Variant calling from the 177 genomes resulted in ∼3 million single-nucleotide polymorphisms (SNPs).
Fig. 1.
Genome-wide associations of aggression at the group level. (A) Distribution of colony aggression phenotypes derived using dimensional reduction of response to experimental threat and postthreat behavioral screening assay for all colonies used in the study; the map illustrates the original geographic source for all of the colonies, with colors highlighting intensity of response or exclusion (if gray). (B) Network diagram of kinship structure derived from a sparse matrix of coefficient of relationships (r) retaining only the degrees of relationship above first cousin (0.125). Circle vertices identify foragers, square vertices identify soldiers, and color of vertices matches colony response from A. Green hexagons identify queen samples, and edges highlight kinship associations within (solid) and between (dotted) colonies. Grouping was achieved with the Fruchterman–Reingold algorithm. (C) Association of colony-level MAF across the genome to colony aggression phenotype; the dashed line shows significance threshold (Bonferroni corrected α = 7.67E−10).
Association Analysis of Individual and Group Genetics and Phenotypes.
A conventional case-control GWAS was performed to detect associations between individual genotype and phenotype (i.e., soldier/forager status) (22). Our estimate of narrow-sense heritability, derived from the genotype matrix (23) for individual-level aggression (h2gSNP = 0.31, 95% CI [0.03, 0.56]), is comparable with previous population genetic studies of honey bees (3, 5, 24). However, we did not identify any significant associations with specific loci (SI Appendix, Fig. S1).
We next examined correlation between this SNP set and colony-level phenotype derived from two measures of colony aggression (SI Appendix, Fig. S2) (1, 3). The estimate of heritability for this colony aggression phenotype was approximately double the estimate for individual-level aggression (h2gSNP = 0.63, 95% CI [0.50, 0.75]). We then calculated the per-colony minor allele frequency (MAF) at each polymorphic locus, again using the genome sequences of the same individuals. Importantly, we determined that for the most significant associations, the correlations were very similar for soldiers and foragers from the same colony (SI Appendix, Fig. S3). Soldiers and foragers from each colony were thus combined, giving n = 19 to 20 diploid individuals per colony. We used this measure of genotype in a modified group-level GWAS to test for allelic associations with colony aggression phenotype. Essentially, this analysis consisted of multivariate regression of colony MAF for each haplotype block against colony aggression score, with covariates for population structure. This is an adaptation of methods developed for pooled sequencing studies (25–27).
Using group-level association, we detected 1,172 SNPs (Dataset S1: marker) whose colony allele frequency was significantly associated with colony aggression phenotype (Bonferroni threshold α = 3.35E−10) (Fig. 1C). The strength of the genotype to phenotype association was surprising, but simulations (SI Appendix, Fig. S4 and Dataset S2) confirmed that strongly collinear relationships can produce such highly significant results even with small sample sizes. A strong signal of association was present between the group allele frequency and group phenotype, showing a large number of association peaks throughout the genome.
Given the strength of the observed association signals, from just a small number of colonies, it is likely that a subset of associated loci was responsible. The remainder of the significant peaks would, therefore, be present due to chance correlation with one or more large-effect alleles, which is likely given the small number of colonies. To test this hypothesis, we added the most statistically significant SNP, an isolated SNP on Linkage Group (LG) 06 (Fig. 1C), in the analysis as a covariate to the group GWAS. A Manhattan plot of all significant SNPs from Fig. 1C with and without the highlighted SNP as a covariate (SI Appendix, Fig. S5) reveals that most of these loci are strongly correlated with this SNP. This result suggests that this SNP, or any of the other highly significant loci, can explain most of the association between aggression and group allele frequency.
To investigate the biological significance of the statistically significant, aggression-associated SNPs and to try to determine which were more likely to be causative, we identified 253 genes within 509 group aggression-associated haplotype blocks. Annotations via homology with Drosophila melanogaster were possible for 178 of the 253 genes (Dataset S1: gene). The genes on this list were significantly enriched (Bonferroni corrected α = 1.00E−7) for gene ontology terms associated with immunoglobulin (Ig) domains. Genes in the Ig domain cluster were found on several different chromosomes but occurred consistently within the most significant peaks of association (Fig. 1C). Ig domain genes are involved in an axon guidance pathway associated with brain development and implicated in human aggression via association studies (28). Among these 178 genes were two members of the defective proboscis extension response (dpr) family of Ig domain genes, primarily defined as encoding sensory receptors and also associated with behavior in Drosophila (29). One strong peak on LG 07, containing 77 significant SNPs in 28 distinct haplotype blocks (Fig. 1C), is centered on the locus LOC724823 on LG07, an ortholog of dpr4. Variations in the genes in this linkage block on LG 07 are thus plausible candidates for biological association with variation in colony aggression, although no nonsynonymous SNPs are present within the population in the coding region of dpr4 itself. However, the location of a large-effect aggression locus in this region of LG 07 is consistent with a previous statistical genetic analysis of a different honey bee colony population (5), which identified an important aggression Quantitative Trait Loci (QTL), known as Sting1, on LG 07.
The strongest correlate we found of the relative aggression of a colony was colony allele frequency, with similar correlations between whole-colony allele frequency and the allele frequencies of either soldiers or foragers alone (SI Appendix, Fig. S3). This was surprising because colony patriline structure (derived from queen polyandry) can significantly influence the probability of any given bee becoming a soldier or forager (1, 2). Kinship analysis results support the presence of strong colony and patriline genetic structure in our study population (Fig. 1B and SI Appendix, Fig. S6), which was corrected for in all our GWAS analyses. Consistent with a prior study (9), we observed influence of genetic relatedness on behavioral role in some, but not all, colonies (SI Appendix, Fig. S6B). As this raised the possibility of geographic variation in behavior, we confirmed that colony location prior to establishing the common garden design did not substantially alter the result of the colony GWAS (SI Appendix, Fig. S7). The strong correlation observed between aggression and group genotype (but not individual genotype) provides strong evidence of indirect genetic effects, where the intensity of aggression is determined by the group allele frequencies more than by individual genetics.
Selection and Admixture at the Loci Associated with Group Aggression.
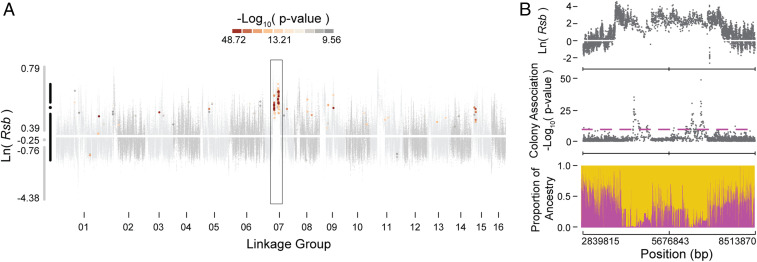
To further determine which of the colony-level association peaks play a deterministic role in aggression, we investigated signatures of selection. If one or more of the aggression-associated loci identified above have a large influence on the level of colony aggression, we would expect them to be under selection during the recent evolution of gentleness in the gAHB population (21). This is the case; 142 of 509 aggression-associated haplotype blocks also showed significant (α < 0.05) signatures of selection (Fig. 2A). Aggression-associated loci are thus significantly enriched for loci under selection (P = 2.37E−29). All 142 associated haplotypes showed a significant bias (α = 0.001) toward positive selection in gAHB relative to aggressive AHB (Fig. 2A). The haplotype blocks under selection include all or part of 60 aggression-associated genes (Fig. 1 and Dataset S1: gene). The peaks on LG 07, where the dpr4 gene is located, while not the most statistically significant in the Manhattan plot, stand out with the strongest signatures of selection, with a number of strongly selected SNPs within an ∼2 megabase region of the chromosome. This peak contains the aforementioned dpr4 ortholog but also shows significant selection and association in several other nearby genes (Dataset S1). The signatures of selection were determined from an independent sample (21), thus also providing confirmatory evidence for a role of the loci in determining aggression.
Fig. 2.
Signatures of selection and admixture in loci associated with colony aggression. (A) Gray dots show selection signatures [the natural log ratio of decay in LD, ln(Rsb)] across the genome for variants between gAHBs and AHBs (data from ref. 21). Those that show both statistically significant selection and statistically significant association between colony-level genotype and colony aggression phenotype in the present study are colored according to significance of the group genotype–phenotype association. The association and selection studies used different samples of individuals. Adjacent to the y axis, simplified box plots for all of the haplotype blocks (gray) and those with overlap (black) identify the extremes, interquartile range, and median of the ln(Rsb) distribution. The dark rectangle highlights a span of notable, significant overlap in LG 07. (B) Region within the rectangle of A, expanded to show selection, association, and admixture. (Top) The ln(Rsb) signal from Fig. 2A. (Middle) The colony genome-wide association signal from Fig. 1C. (Bottom) The proportional contribution of EHB (yellow) and AHB (magenta) reference populations from ref. 14. Proportional contribution was derived via RFMix ancestry determination analysis and ranges from zero to one (y axis). The x axes for all three plots correspond to base pair coordinates of the span within LG 07.
The gAHB population resulted from a soft selective sweep in Puerto Rico that gave rise to a gentler AHB (21). We also conducted an admixture analysis to determine the source background for the alleles of the markers in question. The multiple association peaks detected on LG 07 (Fig. 2A) that also exhibited a significant signature of selection appear to be of European honey bee (EHB) ancestry (Fig. 2B). These results suggest that the evolution of gentle behavior in gAHB involved selection for loci retained from EHB on LG 07.
Discussion
Our results indicate that variation in aggression in honey bees, both at the individual and group levels, has a significant heritable component. As seen in other association studies of aggression-related behavior (30), our individual genotype–phenotype association analysis (SI Appendix, Fig. S1) shows limited power to detect association. This is likely the result of our relatively small, outbred population of 177 individuals lacking sufficient power to detect the small influence of many genes on the phenotype.
By contrast, extremely strong associations were found when we investigated association between group-level phenotype and colony allele frequencies, despite a sample size of only nine colonies in our final analysis. In addition, the heritability of the group-level phenotype (0.63) is unusually strong for a behavioral trait (13, 19) and is more heritable when compared with individual phenotypes, even though the group-level heritability measures the degree to which variation in individual genomes accounts for variation in group phenotype. This raises the possibility that the additional power and heritability observed when measuring the group aggression phenotype may be partly a result of this phenotype being a more accurate measure of individual mean aggression than individual stinging. Almost all of the signal from group allele frequency covaries across all of the 1,172 significant SNPs, so although the group phenotype is clearly genetically determined to a substantial degree, it is not possible to ascribe it to a single locus or variant. However, allele frequencies of the peak variants on LG 07 show strong and consistent overlap in both association and selection analyses (Fig. 2B) and strong enrichment for EHB alleles. This LG 07 region is located nearby, and may be coincident with, the locus previously described as the QTL Sting1 (5).These results suggest that the key determinants of variation in aggression in this population are located on LG 07, especially genes related to brain development and sensory responsiveness in the dpr family of Ig domain genes.
We identified two possible explanations for the tight correlation between the loci that are associated with aggression in the group-level analysis. First, several loci act together to form an oligogenic trait and are strongly correlated by inheritance, despite being unlinked and on different chromosomes. This would require an unlikely mechanism (e.g., selection specifically for males carrying this combination of alleles [worker bees do not reproduce directly]). Second, all of these loci are correlated by random chance, and just one or a subset of them is controlling aggression at the colony level in this population. Given the small number of colonies studied here, we judge this second explanation to be more likely. Moreover, as stated above, the peak on LG 07 is the most likely candidate for a large-effect locus affecting aggression at the group level, based on its strong statistical significance, large number of significant flanking SNPs in linkage disequilibrium (LD), and strong selection signal. We therefore propose that the LG 07 locus is a single large-effect locus controlling a large proportion of the variation in aggression within this population.
Surprisingly, the genetic mechanisms controlling individual and group aggression appear to be distinct. The individual analysis shows peaks that do not meet the Bonferroni significance threshold but would likely become significant with a larger population. However, those peaks are mostly different from those for the group phenotype. For example, the LG 07 peak with the strong association (Fig. 1) and signature of selection (Fig. 2) does not even appear as an association peak below the level of significance in the individual GWAS (SI Appendix, Fig. S1). These results underscore the fact that as a highly social animal, the honey bee has evolved a response to colony threats that involves the integration of multiple behavioral systems to detect and attack intruders, implemented by multiple groups of individuals. Not only is the influence of genetics on individual decisions to sting less strong, but the aggression phenotype is only visible in nonreproductive individuals; thus, individual selection pressure is unlikely to be as strong on loci determining aggression at the individual level.
In addition to likely playing an important role in influencing the underlying neurobiology of aggression, the locus underlying the signal on LG 07 also appears to be playing an important role in the evolution of gentleness in the Puerto Rico population. The very strong selection pressure observed at loci strongly associated with group-level aggression indicates that allele frequency at this locus has been shaped by group (colony-level) selection. Most likely, one or more alleles on LG 07 from the preexisting EHB population on the island were selected for during the evolution of gentle behavior (21). Through recombination and forced outcrossing characteristic of honey bees, multiple adaptive haplotypes would have been favored in a soft selective sweep as the gentle EHB alleles at this locus came to dominate the population. It also is intriguing to note that the selection signature on this chromosome lifts above the baseline for a substantial region of around 2 Mb (Fig. 2A), which suggests the possibility that a large structural polymorphism affecting several genes is responsible for the observed evolutionary change in group behavior. Ongoing genome sequencing projects for gAHB and other honey bee lines will likely resolve this possibility.
Our approach has revealed associations that indicate indirect effects of group (colony) genotype on behavior. Such effects have been predicted in social insects (6) and observed in other systems, such as the influence of peer genotypes on the propensity of humans to smoke (31), although they are rarely as strong as we observed in this study. We showed that such group influences can be identified, and shown to be highly heritable, by group-level association studies. We also demonstrate that further evidence for a role of these genomic correlates of group phenotypes in determining behavior can be obtained from signatures of selection and admixture data. Selection for the aggression trait in bees must occur at the group level, in this case a kinship-based group, as the phenotype is not displayed in reproductive individuals. A soft selective sweep for one or more loci conferring gentle behavior, inherited from a gentle ancestor population (EHB), therefore likely occurred via group-level selection.
Our findings also add to the long-running “nature vs. nurture” debate, as the “nurture” (colony environment) of the bees appears to be the strongest factor in determining aggression. However, we characterized the different colony environments using the genetic composition of the colony and showed that the aggression levels of the colonies are strongly correlated to the frequency of specific alleles in the colony, regardless of the behavior of the individuals concerned (SI Appendix, Fig. S1). Thus, nurture in turn was determined by “nature” in this case, as previously described in the indirect genetic effects literature (16, 17). In the case of the gAHB system, we were able to show that colony-level aggression is strongly heritable and strongly linked to specific genes, one of which is under strong selection for a gentle (EHB) region of the genome. Despite the complexities of using honey bee as a genetic model, therefore, it can be a compelling system for studying genetic determination of group behaviors.
Materials and Methods
Phenotyping and Collection.
Thirteen colonies from eight locations were sampled across Puerto Rico (Fig. 1A). As colony aggression is known to be influenced by colony size (1, 3, 24), we ensured that colony size was similar for all colonies sampled. They each occupied a single 10-frame Langstroth hive box, with 8 to 10 honeycomb frames covered with adult bees, yielding estimates of 16,000 to 20,000 bees per colony following past standards (3). All colonies were transported to the Gurabo Agricultural Field Station, University of Puerto Rico (except for the one already there). Colonies were >1 m apart from each other. Following transportation, colonies were allowed a period of at least 2 wk to acclimate to the new location. A final inspection of the colony was made 10 d before the first phenotyping assay was conducted to confirm population size and presence of queen and brood.
To measure individual phenotype, we adapted assays previously described (3, 9). We placed a string of 5 × 5-cm black suede patches 21 cm from the entrance. After in place, we disturbed the colony by rhythmically striking the top cover with a piece of cinder block 40 times. The gentleness of many of our colonies necessitated such an intensity of disturbance to elicit a sufficient number of aggressive responders. During response, bees left the hive to sting the black suede patches; although slightly distinct from the flag assay (3), this approach allowed for the speedy and accurate collection of bees in the act of stinging. For 120 s after the start of the disturbance, we aspirated and flash froze bees in the act of stinging (soldiers). At the end of the 120 s, we rapidly collected the suede patches, placing them in a sealable plastic bag and removing them from the area entirely. We then restricted the hive entrance and used talcum powder to dust bees that continued to emerge for an additional 30 s. The hive entrance was then opened, and the colony left undisturbed for 30 min, after which the entrance was sealed; we collected those talcum powder-dusted bees returning to the colony that also had clearly visible pollen loads (foragers). As these were bees present inside their hives during the disturbance, we assume they perceived the disturbance but did not respond with defensive behavior.
Only two colonies were sampled each day, with the two colonies sampled being at least 3 m apart to minimize the chance that disturbance to one would affect the other. Our scheduling also assured that colonies adjacent to those sampled on a given day were undisturbed for > 48 h when possible. Upon completion of collection, samples were transported in liquid nitrogen to the laboratory of T.G. at the University of Puerto Rico, Rio Piedras, and transferred to a −80 °C freezer.
We also took two phenotypic measurements of all colonies sampled above. First, by counting the number of stings in each set of black suede patches, we obtained a measure of the intensity of colony response. This was done after collecting soldiers during individual phenotype assay. Second, 2 wk later, we examined and scored colonies with an established behavioral ranking system (3, 32, 33). The assay provides a subjective rank score on a scale of one to four for four behaviors measured at the colony level: 1) running on the comb, 2) hanging from the comb, 3) flying around the hive, and 4) propensity to sting (3, 33). Behaviors 1 and 2 are proxies for general activity within the colony, while behavior 3 is a stronger expression of arousal. Behavior 4 is different from the measure of colony stinging behavior obtained during the first test because it measures a response to a lower level of disturbance and focuses on likelihood rather than intensity. The final score is a sum of the independent behavioral scores and arrives at a rank score with a range of 4 to 16. To simplify ensuing analyses, the rank scale was shifted to a range of 1 to 12. In summary, the 12 colonies were ranked in their overall aggression according to the above criteria.
We examined the relationship between our two measures of colony phenotype and combined them to form a unified colony phenotype (Fig. 1A and SI Appendix, Fig. S2). The rank of the number of stings on the flags was correlated with the cumulative rank score from the second behavioral assay (Kendall’s tau, n = 13, z = 0.38, P value = 0.044) (SI Appendix, Fig. S2A). Using these two rank scores, we derived our combined colony phenotype vector by applying a multidimensional scaling (MDS) analysis (SI Appendix, Fig. S2B). The approach arrived at two dimension vectors, with the first dimension highly correlated with intensity (rank value) across the two measures (dimension 1 vs. mean of rank scores, Kendall’s tau, n = 13, z = 4.76, P value < 0.001; dimension 2 vs. mean of rank scores, Kendall’s tau, n = 13, z = −0.24, P value = 0.807) (SI Appendix, Fig. S2C) and dimension 2 correlated with consistency in rank value between the assays (dimension 1 vs. difference of rank scores, Kendall’s tau, n = 13, z = 0.18, P value = 0.855; dimension 2 vs. difference of rank scores, Kendall’s tau, n = 13, z = −4.71, P value < 0.001) (SI Appendix, Fig. S2D).
After sampling and phenotyping was completed, we collected and flash froze queens from 11 of 13 colonies (two were queenless and excluded from further analysis). A total of 12 queens were collected, with 2 sampled from one colony that was in the midst of colony fission preparations (colony 6).
All worker and queen samples were individually stored in microcentrifuge tubes in dry ice in preparation for shipping. Samples were transported to the Carl R. Woese Institute for Genomic Biology by A.A. inside a dry shipper (CXR500 Cryogenic Shipper; Taylor-Wharton America). DNA (see below) was shipped to BGI in Shenzhen, China.
Sample Selection.
In addition to the exclusion of colonies 5 and 13 (queenless), we also excluded colony 8 as it showed signs of having undergone colony fission during the interval between the first (individual) and second (colony) phenotyping sessions. Colony 6 was not excluded as both queens were collected, and the mother queen of the workers was ascertained through ovary dissections (34). For the remaining 10 colonies, we retained >100 individual foragers and soldiers. A total of 20 per group were selected for sequencing along with their corresponding queens. In addition to the above-mentioned behavioral criteria, all selected foragers carried pollen loads and bore traces of talcum powder, and all soldiers showed clearly anatomical evidence of having stung (i.e., absence of sting apparatus). The initial sample size for genome sequencing was 210:200 workers (10 foragers and 10 soldiers from each of 10 colonies) and 10 queens.
Sequencing and Variant Calling.
Libraries of 250-bp insert size were constructed and sequenced for each sample using the BGI-Seq 500 platform, resulting in ∼5 Gbp data for each sample. The protocol repaired the DNA fragment ends by T4 DNA Polymerase and T4 Polynucleotide Kinase, ligated adapters via T4 DNA ligase, and filled in adapters with Bst Warmstart Polymerase. Then, libraries were purified with the QiaQuick purification kit (QIAGEN) in silica columns. Purified DNA libraries were amplified with AmpliTaq Gold polymerase (Applied Biosystems) and quantified by Qubit fluorometer. Sequencing was performed with the BGI-Seq 500 platform with paired end 100 bp reads.
Sequencing files were aligned to the most recent honey bee assembly, Amel HAv3.1 (https://www.ncbi.nlm.nih.gov/assembly/GCF_003254395.2), and variant calling was conducted using the Sentieon DNaseq workflow (https://support.sentieon.com/manual/DNAseq_usage/dnaseq/). Reads were aligned using the Burrows–Wheeler algorithm (Sentieon bwa). Resulting alignment files were sorted and deduplicated followed by indel realignment and variant calling (via Sentieon Haplotyper) resulting in a **.VCF file for each of our 210 samples. Joint genotyping was conducted on this set of files (Sentieon genotyper) to arrive at our final multisample **.VCF file.
Initial filters removed indels, multiallelic variants, and those variants in unplaced scaffolds or the mitochondrial genome. Remaining genomic biallelic SNPs were filtered by joint quality measures and dataset representation. Representation was assessed on a per-sample and per-marker basis utilizing the combined proportion of missing calls and low coverage calls across the sample by SNP matrix. These criteria identified three samples with excessive missingness (samples 6.4.6, 10.4.6, and 12.2.6), with most of the SNPs showing <10 reads that confirmed the genotype. These also were excluded from further analysis, leaving 197 worker and 10 queen genome sequences. A similar consideration was applied to identify poorly represented SNP markers. Low coverage for a specific SNP was defined as an instance where the reported genotype was confirmed by less than five reads. Using this method, we retained only those markers both present and adequately confirmed by coverage in at least 80% (168/210) of our sample set.
Genome-Wide Associations.
We used two association strategies. For association with individual phenotype, we applied a standard GWAS analysis with a binary phenotype (soldier vs. forager). For association with the colony-level phenotype, we correlated the per-colony MAF with Dimension 1 (D1) from our MDS analysis (SI Appendix, Fig. S2), described below.
Prior to association analyses, we first established independence of markers through pruning using LD. This provided a set of SNPs that are generally independent. For the LD-pruning step, we utilized the snpgdsLDpruning() function in the R package SNPRelate using the D` metric with an LD threshold of 0.30 and MAF of 0.20 (25). This reduced set of markers was used to estimate kinship components and construct the genetic relatedness matrix (GRM).
Using the GENESIS package in ref. 22, we did not find significant population structure in our dataset beyond the clear kinship groups represented by the colonies (Fig. 1B). The analysis did identify that colony 3 did not show a concordant pattern of relationship between members; even the colony 3 queen did not seem to be related to the workers collected from that colony, as estimated by the coefficient of relationship (r). Unsure whether the workers were the offspring of the queen (and thus, compromising genotyping), we excluded colony 3 from the rest of the analyses for a final sample size of 177 worker and 9 queen genomes.
To account for variation due to kinship, we first derived the GRM using the pcrelate() function. The procedure juxtaposes a principal component analysis (PCA) of samples with the matrix resulting from the Kinship-based Inference for GWAS (KING)-robust kinship coefficient estimator to arrive at a GRM that accounts for both population substructure as well as patterns of admixture (35, 36). This resulted in our final matrix, used to derive covariates in the GWAS by the corresponding GENESIS functions.
For the individual-level phenotype association, we implemented the GENESIS quasilikelihood approximation. The approach tested our model across each of the ∼3 million SNPs in our dataset. This resulted in a per-SNP score and P value describing significance of the association for the genotype (SI Appendix, Fig. S1).
For the colony-level phenotype association, we used allele frequency as a measure of group genotype within the colony. Our approach was to use the per-individual genotype to identify the minor allele at each polymorphic locus across the entire sample set. We then grouped samples by colony and for each marker, calculated the frequency of that minor allele within each colony. This resulted in a vector of nine allele frequencies, one per colony, for each individual SNP in our dataset.
To account for confounding errors due to genetic similarities between colonies, we implemented an approach that mirrors established methodology applied in pooled sequencing studies (25–27). Briefly, we treated our colony MAF as a pooled sample and filtered our dataset to the same LD-pruned SNPs used in kinship estimation of individual samples. This we used to derive a matrix of covariance of allele frequencies between the nine colonies. The resulting matrix was further reduced using PCA to extract the first Principal Component (PC) vector accounting for the largest amount of variation in genetic similarities between colonies.
To formally test for correlations, we used a per-SNP linear regression using the function
where y is our vector of colony phenotype, X is a vector of per-colony allele frequency for the specific SNP tested, and G is the first principal component of the per-colony covariance matrix.
In our model, the G variable was derived from M representing a matrix of MAF ordered colony × SNP. This matrix was mean centered, its cross-product derived (MM′), and then, divided by the sum of expected SNP variances. The resulting product matrix contained the colony × colony covariance in MAF. The G variable is the first eigenvector from the single-value decomposition of this matrix.
For each marker in our dataset, we fit our full model and a null model containing only the relationships between phenotype and structure covariates (y = 1 + G) and then used a likelihood ratio test to derive significance. In this way, the colony-level comparison parallels estimates of significance applied in the individual-level analysis.
After candidate correlations were established, we also examined the distributions of genotypes between behavioral groups (soldiers and foragers) within each colony (SI Appendix, Fig. S6). To test for possible differences between behavioral groups within each colony, we derived MAF for each behavioral group for the focal SNPs in our top five peaks of association and examined the goodness of fit using the function derived from the colony-wide model fit (SI Appendix, Fig. S3). For each colony, we conducted a PCA (SI Appendix, Fig. S6A) and applied an iterative k-means clustering method on the first two resulting PCs to arrive at the optimal number of clusters (k) in each colony. For every colony, optimal k was defined via the elbow method using the within-group total sum of squares. Resulting genetic clusters likely correspond to patrilineal assemblages within each colony (honey bee queens are highly polyandrous). We then tested whether behavioral groups were differentially distributed across these genetic clusters for each colony using a Fisher’s exact test (SI Appendix, Fig. S6B).
Concordance with Regions under Selection.
We tested for concordance of alleles identified in the group-level GWAS identified in the present study and genomic regions showing signatures of selection in a previously published paper (21) as follows. We aligned the data in ref. 21 to the newest assembly of the honey bee genome (37) and conducted variant calling. After obtaining variant calls, we applied the same stringent filters described above and then utilized the data to 1) derive haplotype blocks and 2) examine signals of selection within gAHB. Haplotype blocks (1) were estimated using the approach described in ref. 21, where all three populations in that study (EHB, AHB, and gAHB) were provided as input to the GERBIL (38) algorithm to arrive at the most conservative LD-linked spans of variants.
For the selection 2 assessment, we used the dataset to calculate the ratio of the area of decay in LD surrounding each marker (Rsb) (39) between the gAHB and AHB populations. A high ratio corresponds to positive selection or population bottleneck, while a low ratio corresponds to negative or balancing selection (39). For the present analysis, we constrained the comparison only between gAHB and AHB, as we were interested in novel selection (positive or negative) arising in gAHB that was not inherited from the AHB population.
This combined approach provided us with 1) spans of LD-linked markers and 2) a per-marker measure of selection. To assess overlap with our signal of association, we first localized the signals of selection 2 in ref. 21 to their corresponding haplotype blocks (1). This subset of haplotype blocks was then directly overlapped with those markers showing significant correlations between colony genotype and colony defensive response.
P Value Simulations.
In our group-level GWAS (between colony MAF and colony aggression), we obtained extremely small P values (roughly on the order of 10−10 to 10−50) for a number of loci. These low P values are surprising in light of our small sample size of n = 9 colonies; it would seem unlikely that n = 9 observations contain enough information to give such highly significant results.
We conducted simulations (Dataset S2) that illustrate that P values on the order of 10 to 50 are indeed possible in this type of analysis (SI Appendix, Fig. S4). The key reason is that the amount of information in a sample is determined not only by the number of samples but also, by the amount of variability in the phenotype that cannot be explained by the genotype. In our data, genotype contains more information than usual (as it is a continuous measurement of colony allele frequency), and this residual variance appears to be very low, so even a sample of size n = 9 contains a great deal of information.
Admixture Analysis.
From prior research (21), we know the gAHB population represents an admixed composite with contributions from several ancestral sources. To assess contribution of these sources across the genomes of our samples, we used the algorithm RFMix (40). Effectively, this approach sections the genome into noncontiguous windows and examines likelihood contributions from reference populations to target populations. Our dataset was the target population, and we used the AHB and EHB samples in ref. 21 as reference populations. This method resulted in a per-SNP proportion of contribution from each of the reference populations within our samples. Using the haplotype blocks, we grouped these data, resulting in a per-haplotype block summary of correlation, selection, and ancestry (Fig. 2B).
Data Availability.
Genomic datasets and pertinent metadata are freely available via appropriate repositories including the National Center for Biotechnology Information Short Read Archive (BioProject ID no. PRJNA557446). Code files for individual and colony GWAS can be found in GitHub (https://github.com/AAvalos82/Avalos-PanEtAl2020_GWAS) and accompanying data in Dryad (https://doi.org/10.5061/dryad.q573n5tg8).
Supplementary Material
Acknowledgments
We thank G. Diaz and F. Noel for assistance during sample collection and trait measurements; C.J. Fields, G. Rendon, and the rest of the High Performance Computing for Biology team for technical assistance; and B. Harpur for providing archival QTL marker data. This work was supported by National Science Foundation (NSF) Grants 15-501 1547830 (to A.A.), HRD 1736019 (to A.R.L. and T.G.), Puerto Rico Science, Technology and Research Trust 2020-00139 (to A.R.L. and T.G.), and NSF Division of Environmental Biology 1826729 (to A.R.L. and T.G.) and National Institutes of Health Grant R01GM117467 (to G.E.R. and N. Goldenfeld). This project was also supported, in part, by Strategic Priority Research Program of the Chinese Academy of Science Grant XDB13000000; Lundbeckfonden Grant R190-2014-2827 (to G.Z.); and funding from the Carl R. Woese Institute for Genomic Biology at the University of Illinois (to M.E.H.).
Footnotes
The authors declare no competing interest.
Data deposition: Genomic datasets and pertinent metadata are freely available via appropriate repositories including the National Center for Biotechnology Information Short Read Archive (BioProject ID no. PRJNA557446). Code files for individual and colony GWAS can be found in GitHub (https://github.com/AAvalos82/Avalos-PanEtAl2020_GWAS) and accompanying data in Dryad (https://doi.org/10.5061/dryad.q573n5tg8).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922927117/-/DCSupplemental.
References
- 1.Breed M. D., Guzmán-Novoa E., Hunt G. J., Defensive behavior of honey bees: Organization, genetics, and comparisons with other bees. Annu. Rev. Entomol. 49, 271–298 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Seeley T. D., The Lives of Bees: The Untold Story of the Honey Bee in the Wild, (Princeton University Press, 2019). [Google Scholar]
- 3.Hunt G. J., Guzmán-Novoa E., Fondrk M. K., Page R. E. Jr., Quantitative trait loci for honey bee stinging behavior and body size. Genetics 148, 1203–1213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzmán-Novoa E., Hunt G. J., Uribe J. L., Smith C., Arechavaleta-Velasco M. E., Confirmation of QTL effects and evidence of genetic dominance of honeybee defensive behavior: Results of colony and individual behavioral assays. Behav. Genet. 32, 95–102 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Hunt G. J. et al., Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften 94, 247–267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linksvayer T. A., Direct, maternal, and sibsocial genetic effects on individual and colony traits in an ant. Evolution 60, 2552–2561 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Linksvayer T. A., Fondrk M. K., Page R. E. Jr., Honeybee social regulatory networks are shaped by colony-level selection. Am. Nat. 173, E99–E107 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Linksvayer T. A., The Molecular and Evolutionary Genetic Implications of Being Truly Social for the Social Insects, (Elsevier Ltd., ed. 1, 2015). [Google Scholar]
- 9.Breed M. D., Robinson G., Page R. E., Division of labor during honey bee colony defense. Behav. Ecol. Sociobiol. 27, 395–401 (1990). [Google Scholar]
- 10.Collins A., Rinderer T., Heritabilities and correlations for several characters in the honey bee. J. Hered. 75, 135–140 (1984). [Google Scholar]
- 11.Guzman-Novoa E. et al., Paternal effects on the defensive behavior of honeybees. J. Hered. 96, 376–380 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Gibson J. D., Arechavaleta-Velasco M. E., Tsuruda J. M., Hunt G. J., Biased allele expression and aggression in hybrid honeybees may be influenced by inappropriate nuclear-cytoplasmic signaling. Front. Genet. 6, 343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okbay A. et al.; LifeLines Cohort Study , Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClellan J., King M. C., Genetic heterogeneity in human disease. Cell 141, 210–217 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Boyle E. A., Li Y. I., Pritchard J. K., An expanded view of complex traits: From polygenic to omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dick D. M., Gene-environment interaction in psychological traits and disorders. Annu. Rev. Clin. Psychol. 7, 383–409 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bijma P., The quantitative genetics of indirect genetic effects: A selective review of modelling issues. Heredity 112, 61–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam V. et al., Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20, 467–484 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Brinker T., Bijma P., Vereijken A., Ellen E. D., The genetic architecture of socially-affected traits: A GWAS for direct and indirect genetic effects on survival time in laying hens showing cannibalism. Genet. Sel. Evol. 50, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodnight C. J., Multilevel selection theory and evidence: A critique of gardner, 2015. J. Evol. Biol. 28, 1734–1746 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Avalos A. et al., A soft selective sweep during rapid evolution of gentle behaviour in an Africanized honeybee. Nat. Commun. 8, 1550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gogarten S. M. et al., Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics 35, 5346–5348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., Zeng J., Goddard M. E., Wray N. R., Visscher P. M., Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet. 49, 1304–1310 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Rinderer T., Bee Genetics and Breeding, (The University of Michigan, 1986). [Google Scholar]
- 25.Manichaikul A. et al., Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashraf B. H., Jensen J., Asp T., Janss L. L., Association studies using family pools of outcrossing crops based on allele-frequency estimates from DNA sequencing. Theor. Appl. Genet. 127, 1331–1341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cericola F. et al., Optimized use of low-depth genotyping-by-sequencing for genomic prediction among multi-parental family pools and single plants in perennial ryegrass (Lolium perenne L.). Front. Plant Sci. 9, 369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernàndez-Castillo N., Cormand B., Aggressive behavior in humans: Genes and pathways identified through association studies. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 171, 676–696 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Goldman T. D., Arbeitman M. N., Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 3, e216 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shorter J. et al., Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. U.S.A. 112, E3555–E3563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sotoudeh R., Harris K. M., Conley D., Effects of the peer metagenomic environment on smoking behavior. Proc. Natl. Acad. Sci. U.S.A. 116, 16302–16307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giray T. et al., Genetic variation in worker temporal polyethism and colony defensiveness in the honey bee, Apis mellifera. Behav. Ecol. 11, 44–55 (2000). [Google Scholar]
- 33.Avalos A., Rodríguez-Cruz Y., Giray T., Individual responsiveness to shock and colony-level aggression in honey bees: Evidence for a genetic component. Behav. Ecol. Sociobiol. 68, 761–771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rueppell O. et al., Genetic architecture of ovary size and asymmetry in European honeybee workers. Heredity 106, 894–903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng X. et al., A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conomos M. P., Miller M. B., Thornton T. A., Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 39, 276–293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallberg A. et al., A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome-length scaffolds. BMC Genomics 20, 275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimmel G., Shamir R., GERBIL: Genotype resolution and block identification using likelihood. Proc. Natl. Acad. Sci. U.S.A. 102, 158–162 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang K., Thornton K. R., Stoneking M., A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5, e171 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maples B. K., Gravel S., Kenny E. E., Bustamante C. D., RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic datasets and pertinent metadata are freely available via appropriate repositories including the National Center for Biotechnology Information Short Read Archive (BioProject ID no. PRJNA557446). Code files for individual and colony GWAS can be found in GitHub (https://github.com/AAvalos82/Avalos-PanEtAl2020_GWAS) and accompanying data in Dryad (https://doi.org/10.5061/dryad.q573n5tg8).