Significance
Endosymbiotic bacteria are found in diverse fungi, but little is known about how they communicate with their hosts. Some plant pathogenic bacteria use type III-translocated TAL effectors to control host transcription, and TAL-like proteins are encoded in genomes of the fungal endosymbiotic bacterium Mycetohabitans rhizoxinica. In this paper, we present evidence that these proteins are, like TAL effectors, type III-secreted, nuclear-localizing effectors that perturb host transcription and show that one enhances tolerance of the fungal host to cell membrane stress. Our characterization of an effector in a bacterial–fungal symbiosis opens a new door to molecular understanding of these interkingdom partnerships. Our findings also provide insight into the functional diversity and evolution of the TAL effector protein family.
Keywords: symbiosis, Rhizopus microsporus, TAL effector, type III secretion, Btl proteins
Abstract
Symbioses of bacteria with fungi have only recently been described and are poorly understood. In the symbiosis of Mycetohabitans (formerly Burkholderia) rhizoxinica with the fungus Rhizopus microsporus, bacterial type III (T3) secretion is known to be essential. Proteins resembling T3-secreted transcription activator-like (TAL) effectors of plant pathogenic bacteria are encoded in the three sequenced Mycetohabitans spp. genomes. TAL effectors nuclear-localize in plants, where they bind and activate genes important in disease. The Burkholderia TAL-like (Btl) proteins bind DNA but lack the N- and C-terminal regions, in which TAL effectors harbor their T3 and nuclear localization signals, and activation domain. We characterized a Btl protein, Btl19-13, and found that, despite the structural differences, it can be T3-secreted and can nuclear-localize. A btl19-13 gene knockout did not prevent the bacterium from infecting the fungus, but the fungus became less tolerant to cell membrane stress. Btl19-13 did not alter transcription in a plant-based reporter assay, but 15 R. microsporus genes were differentially expressed in comparisons both of the fungus infected with the wild-type bacterium vs. the mutant and with the mutant vs. a complemented strain. Southern blotting revealed btl genes in 14 diverse Mycetohabitans isolates. However, banding patterns and available sequences suggest variation, and the btl19-13 phenotype could not be rescued by a btl gene from a different strain. Our findings support the conclusion that Btl proteins are effectors that act on host DNA and play important but varied or possibly host genotype-specific roles in the M. rhizoxinica–R. microsporus symbiosis.
Bacteria are critical partners in symbiotic interactions with a variety of animals and plants, ranging from bobtail squid to legumes. Many of these associations are well studied and the reciprocal benefits and molecular interactions underlying them well understood. Recently, diverse bacterial–fungal symbioses have been described, with fungal hosts ranging from tree endophytes to human pathogens (1). One such partnership is Mycetohabitans rhizoxinica [family Burkholderiaceae; formerly Burkholderia rhizoxinica (2)] and the mucoromycete Rhizopus microsporus. A rice pathogen and opportunistic human pathogen, R. microsporus can also be found as a free-living soil saprophyte and is used in soy fermentation. M. rhizoxinica produces an antimitotic toxin, rhizoxin, which is critical for the fungus to infect rice seedlings (3).
Though challenging, M. rhizoxinica can be cultured and genetically manipulated independently of its fungal host, making it a useful model. R. microsporus isolates that harbor M. rhizoxinica or other Mycetohabitans spp. require their bacterial partner for asexual and sexual sporulation, while nonhost isolates sporulate without bacteria (4–6). As is true for many Gram-negative bacterial pathogens and mutualists of plants and animals, M. rhizoxinica requires a type III secretion system (T3SS) to invade its host (7, 8). T3SSs inject effector proteins directly into host cells to promote infection (9). Over the past few decades, the T3 effector repertoires of many plant- and animal-associated bacteria have been identified and their modes of action characterized (10).
Despite the demonstrated importance of the T3SS in the Rhizopus–Mycetohabitans partnership, no T3 effector proteins from M. rhizoxinica or any other endohyphal bacterium have been characterized (11). However, homologs of transcription activator-like (TAL) effectors from plant pathogenic Xanthomonas and Ralstonia spp. were identified in the first sequenced M. rhizoxinica genome (12–14). TAL effectors are T3-secreted, sequence-specific, DNA-binding proteins that act as transcription factors once inside plant cells, up-regulating target genes (15). Often these targets are so-called susceptibility genes that contribute to bacterial proliferation, symptom development, or both; examples include sugar transporter, transcription factor, and other genes (15, 16). Outside of plant pathogens, TAL effector-like gene fragments have been found in the marine metagenome, although the organism or organisms of origin remain unknown (17). The limited discovered distribution of this effector family makes its evolutionary origins and functional diversity a mystery.
TAL effectors have four structural components (Fig. 1A): an N-terminal T3S signal (18), N- or C-terminal nuclear localization signals (19–21), a central, repetitive DNA recognition domain (22, 23), and a C-terminal activation domain (19). The DNA recognition domain consists of highly conserved repeats each with two variable amino acids, the repeat variable diresidue (RVD). The repeats each interact with single nucleotides, in a contiguous fashion, with specificity dictated by the RVDs. This one-to-one recognition “code” makes it possible to predict the DNA sequence(s) a TAL effector will bind given its RVD sequence (24, 25). Also, TAL effectors can be synthesized to target specific DNA sequences by putting together repeats with the appropriate RVDs, making these proteins useful for biotechnology (15, 26, 27).
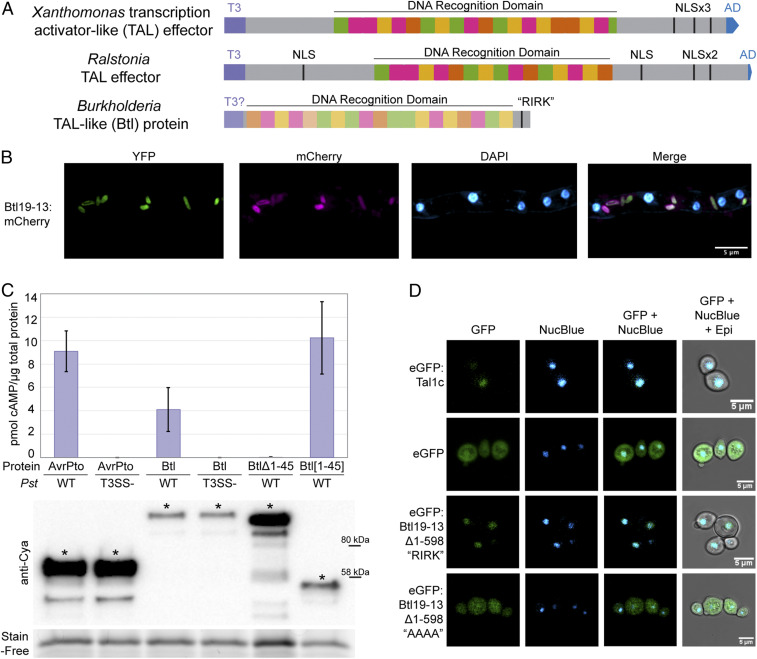
Fig. 1.
Structure, expression, T3 secretion, and nuclear localization of Btl19-13. (A) Domain structure of a Xanthomonas TAL effector, a Ralstonia TAL effector, and a Btl protein. T3, type III secretion signal; T3?, putative type III secretion signal; NLS, nuclear localization signal; “RIRK,” aa sequence of a putative NLS in Btl19-13; AD, activation domain. (B) Confocal microscopy of Mycetohabitans sp. B13 cells constitutively expressing EYFP and expressing Btl19-13:mCherry under the native btl19-13 promoter, within a R. microsporus hypha. DAPI staining of the nuclei is included for reference. (C) Quantification of cAMP by ELISA in Nicotiana benthamiana leaf punches 6 h after infiltration with Pseudomonas syringae pv. tomato DC3000 (Pst) expressing the following: AvrPto:Cya (61 kDa), Btl19-13:Cya (118.6 kDa), Btl19-13:Cya missing the first 45 aa (Btl19-13Δ1-45, 110 kDa), and the first 45 aa of Btl19-13 fused to CyaA (Btl19-13[1-45], 51 kDa). T3SS designates an hrpQ-U mutant incapable of type III secretion (28). Each bar shows the results from three biological replicates with two technical replicates each. Error bars denote SD. The experiment was repeated twice with similar results. Below, a Western blot of leaf homogenates probed with anti-Cya, shown with a Stain-Free loading control (Bio-Rad). Asterisks (*) indicate bands corresponding to the expected size. (D) Confocal microscopy of Saccharomyces cerevisiae cells expressing the indicated proteins and stained with NucBlue to locate nuclei. Protein expression was induced with galactose 24 h prior to imaging. “AAAA” indicates an alanine substitution of the “RIRK” motif in Btl19-13 (residues 692–695).
The initial discovery of TAL effector homologs in Mycetohabitans (then Burkholderia) came just as TAL effectors were garnering interest as engineerable tools for DNA targeting applications such as genome editing. Although their repeats are less highly conserved, the Burkholderia proteins were found to bind DNA targets specifically, wrapping the DNA in a superhelical structure, like TAL effectors (12, 14, 23, 29). In the biochemical studies, different names for the proteins were introduced, including BurrH, and the collective term Bat (Burkholderia TAL-like). Bat follows bacterial gene nomenclature convention but was in use prior to refer to bacteriopsin activator genes. Therefore, we refer to the proteins as Btl (for Burkholderia TAL-like; pronounced “beetle”) and encoding genes as btl. Although the studies to date established Btl proteins as sequence-specific DNA-binding proteins with potential for synthetic biology, none has yet addressed the function(s) of these proteins within the bacterial–fungal symbiosis. The Btl protein structure is truncated at the N- and C-termini compared with canonical TAL effectors and lacks a T3S signal, nuclear localization signals, and an activation domain (Fig. 1A). Thus, even whether Btl proteins act as translocated transcription factors is itself uncertain.
The identification of proteins homologous to TAL effectors in Mycetohabitans spp. presents an opportunity to probe the molecular basis of a fungal–bacterial symbiosis and to better understand the functional diversity of this intriguing family of proteins. In this study, we carried out several experiments to test the hypothesis that Btl proteins function like Xanthomonas TAL effectors, transiting the T3SS and altering host transcription. We also probed whether Btl proteins play any role in establishing the symbiosis or confer a benefit to the fungus. Finally, we explored the prevalence of btl genes across geographically diverse Mycetohabitans spp. and tested whether distinct btl genes found in different isolates might be functionally interchangeable.
Results
The btl Gene in Mycetohabitans sp. B13 Is Expressed Endohyphally.
We identified btl genes in each of the three publicly available genome sequence assemblies of Mycetohabitans spp. that are complete or largely so: Mycetohabitans rhizoxinica HKI 0454/B1 endosymbiont of R. microsporus ATCC 62417 [NCBI accession: PRJEA51915; (13)], Mycetohabitans sp. B13 endosymbiont of R. microsporus ATCC 52813 (NCBI accession: PRJNA303198), and Mycetohabitans sp. B14 endosymbiont of R. microsporus ATCC 52814 (NCBI accession: PRJNA303197). Strain B13 has one intact btl gene with 19 repeats and a small btl gene fragment, whereas strains B1 and B14 each have three btl genes, with varying numbers of repeats and RVD sequences (SI Appendix, Table S1). TAL effector genes likewise occur in different numbers in different strains, at different locations in the genome, and differ primarily in their numbers of repeats and the sequences of the RVDs. For these reasons, orthology is not always possible to ascertain. Indeed, for TAL effectors, duplication and differentiation through point mutations and chimerism via recombination are common. And even for clearly orthologous genes, slight changes in repeat number or RVD sequence can change function by altering the DNA-binding specificity. Thus, rather than attempt an orthology-based nomenclature, we chose to give each btl gene (and Btl protein) across strains a unique yet informative designator. We used the number of repeats in the recognition domain followed by the strain number; for example, the single intact btl gene of strain B13, which has 19 repeats, was named btl19-13. For functional characterization, we chose to focus on this gene and avoid the possible redundancy in strains B1 and B14.
First, to determine whether btl19-13 is expressed, we cloned it on a plasmid under the control of its native promoter and with a C-terminal translational fusion to mCherry. After transforming B13 with this plasmid, we introduced the transformant into the corresponding, cured R. microsporus isolate and assayed for mCherry fluorescence by confocal microscopy. The plasmid we used also harbors a constitutively expressed EYFP (enhanced yellow fluorescent protein) gene to enable independent visualization of the bacterial cells. Btl19-13:mCherry and EYFP signals were evident and coincident within the fungal hyphae (Fig. 1B), indicating that the bacterium expresses btl19-13 during the symbiosis.
Btl19-13 Transits the T3SS of Pseudomonas Syringae.
Our next question was whether Btl19-13 could, like TAL effectors, transit a T3SS and thereby potentially function within the fungal host. In the fluorescence microscopy described above, Btl19-13:mCherry was detectable only in the bacterium, but the fluorophore may have prevented secretion. Computational analysis of the Btl19-13 protein sequence using EffectiveT3 2.0.1 (30) detected with low confidence the presence of a T3S signal in the N terminus. Upstream of btl19-13 we discovered a sequence that aligns with the plant-inducible promoter element found in promoters of genes coregulated with the T3SS in Xanthomonas and Ralstonia spp. (31, 32). The sequence upstream of btl19-13, as well as btl genes in B1 and B14, matches the consensus targeted by the regulator HrpB, TTCG-N16-TTCG (SI Appendix, Table S2) (8).
To test whether Btl19-13 is a T3S substrate, we used an established assay for T3S of plant pathogen effectors in which leaves of the model plant Nicotiana benthamiana are infiltrated with a suspension of Pseudomonas syringae pv. tomato DC3000 (Pst) expressing the candidate effector fused to adenylate cyclase (Cya; the catalytic domain of the Bordetella pertussis toxin CyaA) (33). Conversion of adenosine triphosphate to cyclic adenosine monophosphate (cAMP) by Cya requires calmodulin and can be detected by enzyme-linked immunosorbent assay (ELISA), acting as a reporter of localization into the host cell following T3S (34). We used the Pst effector AvrPto as a positive control and included a Pst T3SS-deficient strain to assess T3SS-dependence of activity (28, 33). Following infiltration, Btl19-13:Cya resulted in increased cAMP when expressed in Pst but not Pst T3SS-, indicating that Btl19-13 is a T3S substrate (Fig. 1C). Furthermore, a 45 amino acid (aa) N-terminal truncation of Btl19-13 abolished cAMP accumulation, while just the N-terminal 45 amino acids fused to Cya (Btl19-13[1:45]:Cya), resulted in increased cAMP, indicating that a sequence within those first 45 amino acids of Btl19-13 is necessary and sufficient for T3S (Fig. 1C). Together, the results presented here support the hypothesis that Btl proteins are T3 effectors.
Btl19-13 Has a Functional Nuclear Localization Signal.
While Btl proteins bind DNA, suggesting they localize to the nucleus, their short C-terminus does not contain any predicted NLSs. However, we identified an NLS-like sequence, “RIRK,” in the C-terminal region of Btl19-13 (residues 692–695) that is also present in the other Btl proteins (SI Appendix, Table S1). Because there is not yet a method to genetically manipulate R. microsporus, in order to assess Btl19-13 subcellular localization, we transformed the yeast Saccharomyces cerevisiae with inducible expression plasmids encoding enhanced green fluorescent protein (eGFP) fusion proteins. When viewed with confocal microscopy, yeast cells induced to produce free eGFP showed diffuse localization of the protein. Conversely, as expected, eGFP-tagged TAL effector Tal1c cloned from Xanthomonas oryzae pv. oryzicola localized to the nucleus (Fig. 1D). However, we were unable to see fluorescence in induced cells transformed with an expression construct for eGFP:Btl19-13 (SI Appendix, Fig. S1), suggesting that the full-length Btl19-13 protein is toxic to yeast cells. We generated a construct encoding an N-terminally truncated derivative, Btl19-13Δ1-598, containing only a little over two repeats of the DNA recognition domain and the entire C-terminal region, with the “RIRK” motif. The fusion protein eGFP:Btl19-13Δ1-598 localized to the nuclei of the yeast cells, demonstrating that Btl19-13 contains at least one functional NLS (Fig. 1D). Localization to the nucleus was abolished when the “RIRK” motif was mutated to alanines (AAAA). The full-length Btl19-13 protein with this same substitution, fused to eGFP, had no apparent cytotoxicity and resulted in diffuse signal in the yeast cells (SI Appendix, Fig. S1). The presence of a functional NLS in Btl19-13, along with the observation that wild-type Btl19-13 appears to be toxic to yeast while a nonnuclear localizing derivative is not, supports the hypothesis that Btl proteins function in the host nucleus.
Btl19-13 Does Not Affect Activity of a Reporter Gene in an In Planta Transient Expression Assay.
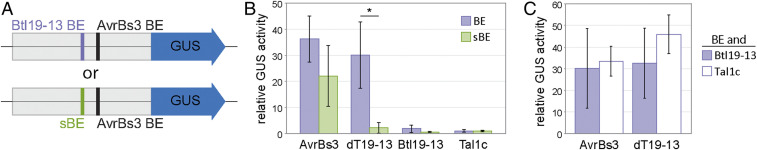
To address whether Btl proteins might directly alter transcription of host genes, as TAL effectors do, we used an Agrobacterium-mediated transient expression-based GUS reporter assay in N. benthamiana that is commonly used to assess TAL effector-dependent expression changes (e.g., refs. 24, 35). We amended a minimal promoter from the pepper Bs3 gene (35, 36) with either a binding element (BE) for Btl19-13 or a scrambled binding element (sBE) (Fig. 2A). We designed the BE based on the Btl19-13 RVD sequence and confirmed by electrophoretic mobility shift assay that Btl19-13 binds it specifically (SI Appendix, Fig. S2). Then, to first test whether Btl19-13 can activate gene expression, each of these promoter constructs was transformed into A. tumefaciens and coinfiltrated with another strain delivering an expression construct for Btl19-13 or one of two control proteins. The control proteins were AvrBs3, the native TAL effector of Xanthomonas euvesicatoria that activates the minimal Bs3 promoter via a separate BE, and Tal1c, which has no predicted BE in the promoter of the reporter constructs (Fig. 2 A and B). For reference, we also tested a designer TAL effector, dT19-13, constructed to bind the Btl19-13 BE (Fig. 2B and SI Appendix, Fig. S3). Unlike dT19-13, which specifically activated the reporter harboring the Btl19-13 BE, Btl19-13 did not alter the activity of either reporter (Fig. 2B). Next, toward testing whether Btl19-13 might be a repressor of transcription, we asked whether Btl19-13 could reduce reporter activity by competing with an activator that binds the same target sequence or one nearby. Using the construct harboring the Btl19-13 BE (and the AvrBs3 BE just downstream), we cointroduced an activator, dT19-13 or AvrBs3, with Btl19-13 or with Tal1c, as a control. Btl19-13 did not significantly reduce GUS activity relative to Tal1c in either combination (Fig. 2C).
Fig. 2.
Effect of Btl19-13 on reporter gene activity in a transient expression assay in N. benthamiana leaves. (A) Schematic of the two reporter constructs used. GUS is driven by a pepper Bs3 minimal promoter harboring a binding element for the Xanthomonas TAL effector AvrBs3 and either a binding element for Btl19-13 (BE) or a scrambled binding element (sBE). (B and C) Fluorometric assays of GUS activity in planta. N. benthamiana leaves were coinfiltrated with Agrobacterium tumefaciens strains delivering one of the two reporter constructs, as indicated, and one (B) or more (C) expression constructs for the following effector proteins: AvrBs3; dT19-13, a designer TAL effector with similar RVD sequence to Btl19-13; Btl19-13; Tal1c, a Xanthomonas TAL effector which has no predicted binding element in the Bs3 promoter. GUS activity was assayed 48 hpi and is shown relative to the corresponding reporter construct with only Tal1c. Each value is an average of three replicates, and the experiment was repeated twice with similar results. An asterisk (*) over two values indicates a significant difference (paired Student’s t test, P < 0.05). Error bars denote SD.
Btl19-13 Contributes to R. Microsporus Tolerance to Cell Membrane Stress.
Having established in heterologous contexts that Btl19-13 is likely an effector that acts in the R. microsporus nucleus, but having not detected any transcription factor activity in the reporter assay, we decided to knock out the btl19-13 gene in strain B13 to directly assess its impact on the symbiosis. We generated a btl19-13 knockout derivative, B13Δbtl19-13 (SI Appendix, Fig. S4A). B13Δbtl19-13 was no different from wild type in its ability to infect the fungus. Furthermore, R. microsporus infected with B13Δbtl19-13 showed no difference in asexual sporulation, could still mate to produce sexual spores, and grew normally on nutrient-rich and -poor media (SI Appendix, Fig. S4 B–E).
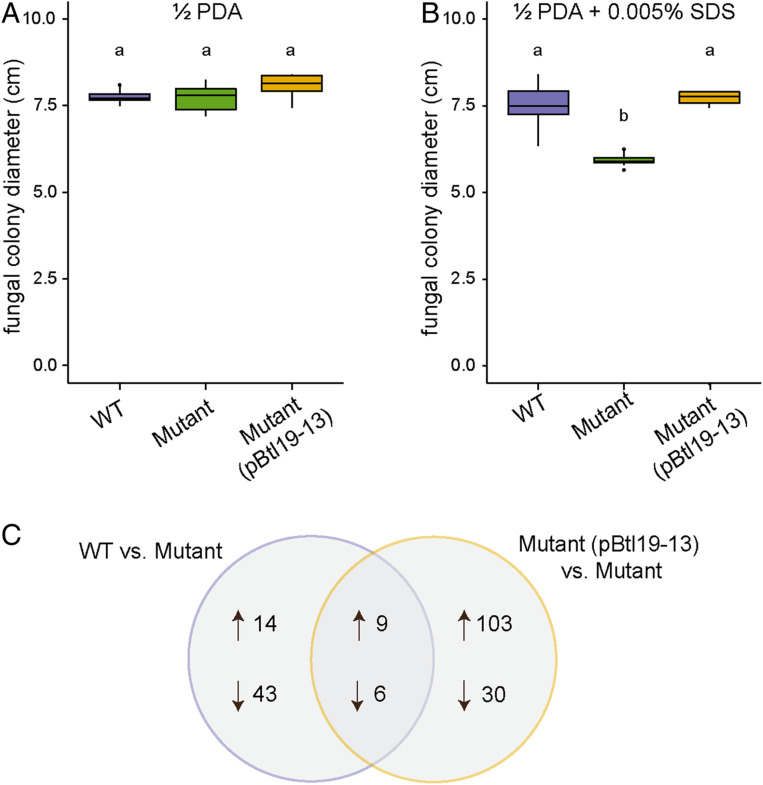
Since the btl19-13 knockout had no gross effect on establishment and maintenance of the host–symbiont interaction, we next explored the hypothesis that Btl19-13 contributes to the symbiosis by increasing tolerance of the fungus to environmental stresses. We measured growth on half-strength potato dextrose agar (1/2 PDA) amended as follows to test specific stresses: with hydrogen peroxide (H2O2) for oxidative stress, with sodium chloride (NaCl) for osmotic stress, and with sodium dodecyl sulfate (SDS) for cell membrane stress. R. microsporus infected with B13Δbtl19-13 grew as well as the fungus infected with wild-type B13 on 500 mM NaCl and on 3 mM H2O2 (SI Appendix, Fig. S4 F and G), but more slowly on media amended with 0.005% SDS (Fig. 3 A and B). The reduced growth in the presence of the detergent was restored to wild-type levels when B13Δbtl19-13 was transformed with a plasmid (pBtl19-13) carrying the cloned btl19-13 gene with its native promoter (620 bp), prior to reinfection of the fungus.
Fig. 3.
Effect of Btl19-13 on R. microsporus cell membrane stress tolerance and global gene expression. (A and B) Colony diameter of R. microsporus infected with wild-type Mycetohabitans sp. B13 (WT), B13∆btl19-13 (mutant), or the mutant strain carrying btl19-13 on a plasmid (pBtl19-13). Fungi were grown at 28 °C on half-strength potato dextrose agar without (A) or with (B) 0.005% SDS for 3 and 6 d, respectively. Data shown represent 10 biological replicates for each bacterial genotype and were analyzed by ANOVA with a post hoc Tukey’s test to determine significance as indicated by lowercase letters (P < 0.001). The experiment was repeated twice and yielded similar results. (C) Venn diagram of the genes that were differentially expressed between R. microsporus infected with wild-type B13 or the complement, B13∆btl19-13(pBtl19-13), and the mutant B13∆btl19-13. Differential expression was determined by RNA sequencing. Data were analyzed with DESeq2 (37) using an adjusted P value < 0.05 as the threshold for significance. Three biological replicates were sequenced, each containing tissue from three plates.
Btl19-13 Alters the R. Microsporus Transcriptome.
To probe the mechanism underlying the contribution of Btl19-13 to cell membrane stress tolerance of R. microsporus, we conducted RNAseq on mycelia infected with B13, B13Δbtl19-13, or the complemented B13Δbtl19-13 strain and carried out pairwise comparisons (38). Fifteen genes were similarly differentially expressed in the comparison between B13 and B13Δbtl19-13 and in the comparison between the complemented strain and B13Δbtl19-13 (Fig. 3C and SI Appendix, Table S3). Fourteen of the fifteen have predicted Btl19-13 BEs within their promoters (SI Appendix, Table S3). However, annotations and pfam analysis did not provide obvious clues to the mechanism by which Btl19-13 contributes to SDS tolerance; all but two of the DE genes encode hypothetical proteins or domains of unknown function (SI Appendix, Table S3).
btl Genes Are Present across Mycetohabitans sp. but can Differ Functionally.
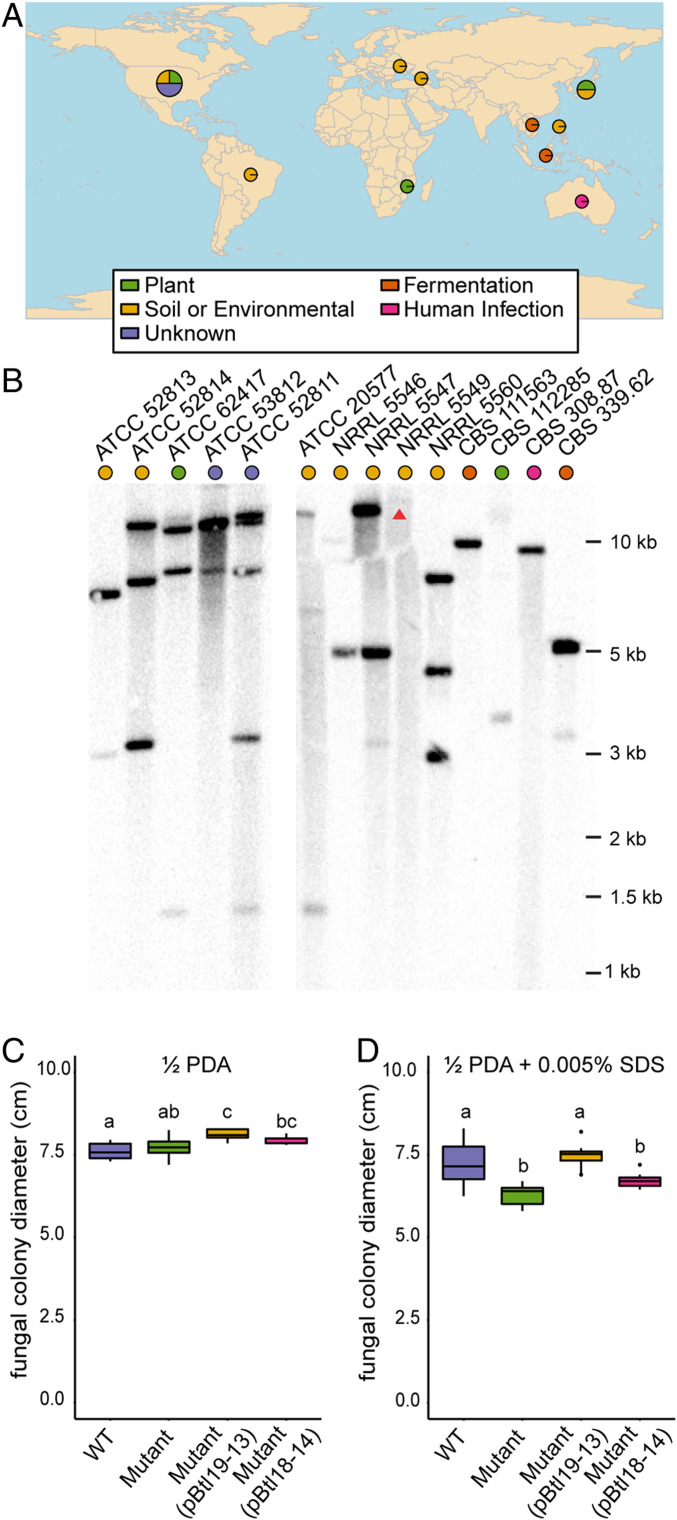
Given that Btl19-13 does not appear to be integral to the formation or maintenance of the symbiosis, but instead appears to enhance the tolerance of the host to cell membrane stress, it was unclear whether all Mycetohabitans spp. could be expected to have btl genes serving the same function. The btl genes of strains B1, B3, and B14 encode distinct arrays of RVDs, which would indicate differences in their targeted BEs (SI Appendix, Table S1). To survey btl genes across Mycetohabitans isolates, we assayed by Southern blot a collection of 14 geographically diverse strains isolated from across the variety of niches that R. microsporus inhabits (Fig. 4A and SI Appendix, Table S4), including B1, B13, and B14. We used the cloned btl19-13 gene of B13 as a probe. The numbers and sizes of bands detected for B1, B13, and B14 matched what was predicted based on the genome sequences (Fig. 4B). While B13 only has one predicted functional btl gene, there is also a small btl gene fragment (435 nucleotides) that we do not believe encodes a functional Btl protein based on its small size, lack of nonrepeat domains, and dissimilar promoter with no predicted HrpB-binding element. Each of the additional 11 strains yielded at least one strongly hybridizing band in the Southern blot, except NRRL 5549, which showed a faint, high-molecular-weight band that is clearer at longer exposures (Fig. 4B and SI Appendix, Fig. S5). A few bands were common to multiple strains, but no two strains shared the same overall banding pattern, suggesting that while btl genes are widely distributed in Mycetohabitans, they and/or their genomic contexts are variable.
Fig. 4.
Presence of btl genes in geographically diverse isolates of Mycetohabitans spp. and the effect of substituting btl18-14 for btl19-13. (A) World map showing the location and substrate from which the R. microsporus hosts of the Mycetohabitans spp. assessed were isolated, generated using rworldmap (39). More information on the strains is available in SI Appendix, Table S3. (B) Southern blots of genomic DNA from each strain, digested with AatII and probed with btl19-13 amplified from B13. Strains are identified by the culture collection accession number of their fungal hosts. ATCC 52813 represents B13, ATCC 52814 B14, and ATCC 62417 B1. The arrow points to a faint high-molecular-weight band in an otherwise empty lane, which can be more clearly seen in SI Appendix, Fig. S5. (C and D) Growth of R. microsporus infected with wild-type B13 (WT), B13∆btl19-13 (mutant), the mutant strain with pBtl19-13, or the mutant strain with pBtl18-14, on half-strength potato dextrose agar without (C) or with (D) 0.005% SDS after 3 or 6 d, respectively, at 28 °C. Data shown represent values from 10 replicate plates each and were analyzed by ANOVA with a post hoc Tukey’s test. The experiment was repeated twice and yielded the same result. In each plot, different lowercase letters above any two groups indicate a significant difference between the means (P < 0.001).
The apparent diversity of btl genes across strains suggests that Btl proteins have differentiated in function. To explore this possibility more directly, we cloned the 18-repeat btl gene from strain B14 (btl18-14), which is the sequenced btl gene encoding the RVD sequence most similar to that of Btl19-13 (SI Appendix, Table S1), and tested whether it would rescue the B13Δbtl19-13 phenotype of reduced tolerance to SDS. It did not (Fig. 4 C and D), indicating that at least between these two strains, Btl proteins are not interchangeable.
Discussion
The T3 translocation of Btl19-13, its nuclear localization, its contribution to host cell membrane stress, and the fact that it alters the host transcriptome support the hypothesis that Btl proteins act as effectors in Mycetohabitans–Rhizopus symbioses. Btl19-13 displays these properties despite largely truncated N- and C-terminal regions and degenerate or cryptic nuclear localization and T3S signals compared to Xanthomonas and Ralstonia TAL effectors. Endosymbiotic bacteria are observed to undergo genome reduction and to accumulate deleterious mutations relative to free-living bacteria due to their smaller population sizes, reduced recombination, and stable environment, which imposes purifying selection on a smaller number of genes (40, 41). It seems plausible that Btl proteins represent degenerate descendants of ancestral TAL effector-like proteins. Indeed, it is striking, given the structural differences, that Btl19-13 retains the ability to be T3-secreted and nuclear-localize. Likewise striking, Btl proteins specifically bind DNA despite the degeneracy of their central repeat sequences relative to TAL effectors. Together these observations are consistent with Btl proteins being under relatively strong selection as effectors that act on host DNA. The properties of Btl proteins contrast with those of a distinct class of structurally degenerate TAL effector-like proteins, TruncTALEs, which are found in some strains of the rice pathogen Xanthomonas oryzae. TruncTALEs lack the ability to bind DNA but block the function of a certain class of plant disease resistance gene, possibly through protein–protein interaction (42).
While the data presented demonstrate that Btl19-13 alters the host transcriptome, the mechanism remains unclear, as we could not detect an effect of Btl19-13 on transcription in an in planta reporter gene assay commonly used for TAL effector studies (e.g., refs. 24, 35). Specifically, Btl19-13 failed to activate a reporter with a confirmed Btl19-13 BE, while a dTALE matching that BE did, thus functionally differentiating Btl proteins and TAL effectors. As Btl19-13 does not have the C-terminal activation domain of TAL effectors, it is perhaps not surprising that it did not up-regulate gene expression in this assay. Btl19-13 also did not measurably reduce dTALE-driven expression by direct competition, nor did it actively repress expression driven by a TAL effector targeting a downstream BE. Btl proteins have been observed to repress transcription of a reporter in Escherichia coli when the promoter is amended with a corresponding BE (17). Therefore, Btl repressor activity might depend on affinity relative to any concurrent activator(s), or on promoter context. We did not assess the affinity of Btl19-13 for the BE relative to dT19-13, nor did we test repression at other locations within the promoter or gene body. It remains to be seen whether Btl proteins act as transcriptional activators or as repressors within Rhizopus, or in fact impact transcription in the host in some other way, by remodeling chromatin, for example. Given the coevolution of the endosymbiont and host, Btl proteins may be adapted to function by partnering with specific Rhizopus proteins or with another codelivered effector not present in the in planta assay. Determining the binding site(s) of Btl19-13 in Rhizopus chromatin in relation to the gene expression changes we observed in our RNAseq experiment, and identifying any interacting proteins, are important future steps toward determining how Btl proteins alter host transcription.
The mechanism by which Btl19-13 increases the tolerance of Rhizopus to detergent is likewise unclear, but could be related to major changes in Rhizopus lipid metabolism observed during infection (43). Reprogramming of lipid metabolism in the fungus has been suggested to help meet the bacterium’s nutritional requirement or favor invasion by the bacterium (43). While our study showed that Btl19-13 is not required for host infection, the protein could confer a quantitative benefit we did not measure. Annotated lipid metabolism genes were not present among the genes differentially expressed in response to Btl19-13, but some of the DE genes might still influence lipid metabolic pathways. It is also possible that the effect of Btl19-13 is highly localized and not adequately captured by RNAseq of whole fungal colonies. Sampling of nuclei adjacent to bacterial cells could overcome this limitation. Furthermore, metabolome or lipidome analysis could reveal changes dependent on Btl19-13 and suggest genomic targets to investigate. Although beyond the scope of the current study, such approaches are an attractive first step toward understanding the connection between the observed influence of Btl19-13 on fungal SDS tolerance and the relevant biological function of Btl19-13. In other fungi, intolerance to SDS has been associated with changes not only in lipid metabolism (44), but also fungal virulence (45) and amino acid metabolism (46). Thus, Btl19-13 could confer a variety of biologically relevant benefits to the fungus, the bacterium, or both.
The hypothesis that Btl proteins are important for the symbiosis is strengthened by the observation that, despite the reduced genomes of Mycetohabitans spp., btl genes are present in all strains we examined in a collection of R. microsporus isolates from diverse geographic locations and substrates. The structural features of Btl19-13 are likely conserved in the other Btl proteins, based on similarity of the sequenced btl genes. However, there are a number of lines of evidence that these proteins do not all have the same functional target: 1) the number of btl genes in each strain varies, 2) the RVD sequences of the sequenced btl genes differ, and 3) B13Δbtl19-13 could not be rescued with btl18-14. Btl proteins in different strains may target host genes with distinct functions or different genes with related functions. Or they may target regions of the same gene(s) that are polymorphic across Rhizopus isolates. In the latter scenario, although Btl19-13 is not required for infection and is thus not a host specificity determinant in the strict sense, Btl proteins may influence patterns of association between specific bacterial and fungal isolates by contributing differentially to the persistence of the symbiosis depending on the host genotype. Further characterization of the sequence diversity of btl genes, comparative genomics of the corresponding host fungal isolates, and analysis of the Btl protein-dependent changes in the host transcriptome across a wide array of isolates will be important future steps toward determining the role(s) Btl proteins and their targets play in bacterial–fungal symbioses.
Methods
Bacteria were isolated from fungi by tissue disruption and filtration, and fungi were cured with antibiotics and reinfected by cocultivation, as described (43). A variety of plasmids (SI Appendix, Table S5) with btl gene derivatives were constructed by PCR, restriction enzyme digests, and mutagenesis. Microscopy of R. microsporus was done with a DeltaVision imaging station, and yeast microscopy was done with a Zeiss 710 microscope. Type III secretion was investigated using a Cya assay, as described (47). The reporter gene assay was done as described (35). For RNAseq, RNA was extracted from day-old cultures of R. microsporus using an RNeasy Plant Mini Kit (Qiagen) and sent to Novogene for library preparation and Illumina sequencing. For amended media experiments, plates of R. microsporus were started with 2 × 2 mm plugs of 2-d-old fungus, and the colony diameter was measured daily. For the Southern blot, bacterial genomic DNA was prepared using the MasterPure Gram Positive DNA Purification Kit (Lucigen) and fragmented by AatII digestion, and the blot was probed with btl19-13. Full details of methods and materials are presented in SI Appendix.
Data Availability.
RNAseq data have been deposited in the Sequence Read Archive of the NCBI (BioProject: PRJNA629487). Isolates are available from culture collections as listed in SI Appendix, Table S4, and all plasmids and novel strains are available upon request.
Supplementary Material
Acknowledgments
The authors thank Megan Feely, Pallavi Singh, and Ashli Gerschutz for laboratory assistance; Melanie Filiatrault for N. benthamiana plants; Alan Collmer and Suma Chakravarthy for Pst strains and plasmids, as well as guidance on the Cya assay; Wojtek Pawlowski for the use of the DeltaVision imaging workstation; Kathie Hodge for the use of the Olympus SZX12 dissecting microscope; Bryan Swingle for use of the plate reader; and the Cornell University Biotechnology Resource Center for the shared Zeiss LSM 710 Confocal Microscope (NIH 1S10RR025502). This work was supported by the US Department of Agriculture National Institute of Food and Agriculture (Predoctoral Fellowship Award 2018-67011-28015 to M.E.C.), the National Science Foundation (Grant IOS-1261004 to T.E.P.), and the NIH (Grant R01GM098861 to A.J.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: RNAseq data have been deposited in the Sequence Read Archive of the National Center for Biotechnology Information (NCBI) under BioProject PRJNA629487.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003857117/-/DCSupplemental.
References
- 1.Arora P., Riyaz-Ul-Hassan S., Endohyphal bacteria; The prokaryotic modulators of host fungal biology. Fungal Biol. Rev. 33, 72–81 (2018). [Google Scholar]
- 2.Estrada-de Los Santos P. et al., Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes (Basel) 9, 389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Partida-Martinez L. P., Hertweck C., Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Lackner G. et al., Global distribution and evolution of a toxinogenic Burkholderia-Rhizopus symbiosis. Appl. Environ. Microbiol. 75, 2982–2986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Partida-Martinez L. P., Monajembashi S., Greulich K.-O., Hertweck C., Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17, 773–777 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Mondo S. J. et al., Bacterial endosymbionts influence host sexuality and reveal reproductive genes of early divergent fungi. Nat. Commun. 8, 1843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He S. Y., Nomura K., Whittam T. S., Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694, 181–206 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Lackner G., Moebius N., Hertweck C., Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 5, 252–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfano J. R., Collmer A., Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Hu Y. et al., A global survey of bacterial type III secretion systems and their effectors. Environ. Microbiol. 19, 3879–3895 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Nazir R., Mazurier S., Yang P., Lemanceau P., van Elsas J. D., The ecological role of type three secretion systems in the interaction of bacteria with fungi in soil and related habitats is diverse and context-dependent. Front. Microbiol. 8, 38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange O. et al., Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain. Nucleic Acids Res. 42, 7436–7449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lackner G., Moebius N., Partida-Martinez L., Hertweck C., Complete genome sequence of Burkholderia rhizoxinica, an Endosymbiont of Rhizopus microsporus. J. Bacteriol. 193, 783–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juillerat A. et al., BurrH: A new modular DNA binding protein for genome engineering. Sci. Rep. 4, 3831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boch J., Bonas U., Lahaye T., TAL effectors—Pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hutin M., Pérez-Quintero A. L., Lopez C., Szurek B., MorTAL Kombat: The story of defense against TAL effectors through loss-of-susceptibility. Front. Plant Sci. 6, 535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lange O. et al., DNA-binding proteins from marine bacteria expand the known sequence diversity of TALE-like repeats. Nucleic Acids Res. 43, 10065–10080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szurek B., Rossier O., Hause G., Bonas U., Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 46, 13–23 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Szurek B., Marois E., Bonas U., Van den Ackerveken G., Eukaryotic features of the Xanthomonas type III effector AvrBs3: Protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 26, 523–534 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Van den Ackerveken G., Marois E., Bonas U., Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87, 1307–1316 (1996). [DOI] [PubMed] [Google Scholar]
- 21.de Lange O. et al., Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 199, 773–786 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Deng D. et al., Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335, 720–723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak A. N.-S., Bradley P., Cernadas R. A., Bogdanove A. J., Stoddard B. L., The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335, 716–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boch J. et al., Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Moscou M. J., Bogdanove A. J., A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Doyle E. L., Stoddard B. L., Voytas D. F., Bogdanove A. J., TAL effectors: Highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 23, 390–398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Lange O., Binder A., Lahaye T., From dead leaf, to new life: TAL effectors as tools for synthetic biology. Plant J. 78, 753–771 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Badel J. L., Shimizu R., Oh H. S., Collmer A., A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant Microbe Interact. 19, 99–111 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Stella S. et al., BuD, a helix-loop-helix DNA-binding domain for genome modification. Acta Crystallogr. D Biol. Crystallogr. 70, 2042–2052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eichinger V. et al., EffectiveDB—Updates and novel features for a better annotation of bacterial secreted proteins and Type III, IV, VI secretion systems. Nucleic Acids Res. 44, D669–D674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenselau S., Bonas U., Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant Microbe Interact. 8, 845–854 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Cunnac S., Boucher C., Genin S., Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186, 2309–2318 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schechter L. M., Roberts K. A., Jamir Y., Alfano J. R., Collmer A., Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186, 543–555 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ladant D., Ullmann A., Bordatella pertussis adenylate cyclase: A toxin with multiple talents. Trends Microbiol. 7, 172–176 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Cernadas R. A. et al., Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 10, e1003972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Römer P. et al., Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 187, 1048–1057 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter M. E., et al. , RNAseq of Rhizopus microsporus with mutant Mycetohabitans. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA629487. Deposited 30 April 2020.
- 39.South A., rworldmap: A new R package for mapping global data. R J. 3, 35–43 (2011). [Google Scholar]
- 40.Moran N. A., Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U.S.A. 93, 2873–2878 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller H. J., The relation of recombination to mutational advance. Mutat. Res. 106, 2–9 (1964). [DOI] [PubMed] [Google Scholar]
- 42.Read A. C. et al., Suppression of Xo1-mediated disease resistance in rice by a truncated, non-DNA-binding TAL effector of Xanthomonas oryzae. Front. Plant Sci. 7, 1516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lastovetsky O. A. et al., Lipid metabolic changes in an early divergent fungus govern the establishment of a mutualistic symbiosis with endobacteria. Proc. Natl. Acad. Sci. U.S.A. 113, 15102–15107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gsell M. et al., A yeast mutant deleted of GPH1 bears defects in lipid metabolism. PLoS One 10, e0136957 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López-Fernández L. et al., Understanding Mucor circinelloides pathogenesis by comparative genomics and phenotypical studies. Virulence 9, 707–720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schroeder L., Ikui A. E., Tryptophan confers resistance to SDS-associated cell membrane stress in Saccharomyces cerevisiae. PLoS One 14, e0199484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakravarthy S., Huot B., Kvitko B. H., . “Effector translocation: Cya reporter assay” in Bacterial Protein Secretion Systems: Methods and Protocols, Journet L., Cascales E., Eds. (Springer, 2017), pp. 473–487. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data have been deposited in the Sequence Read Archive of the NCBI (BioProject: PRJNA629487). Isolates are available from culture collections as listed in SI Appendix, Table S4, and all plasmids and novel strains are available upon request.