Significance
The secretion of papain-like cysteine proteases (PLCPs) is an important component of the immune response across the plant kingdom. Here we show that immune protease Rcr3, a secreted PLCP of tomato, is activated by secreted subtilisins, which are common serine proteases in plants. Subtilase P69B activates proRcr3 by cleaving after aspartates in the junction between the autoinhibitory prodomain and the protease domain of the Rcr3 precursor, thereby activating Rcr3. Subtilases of a different subfamily facilitate proRcr3 processing in a tobacco relative, indicating that this proteolytic cascade might be common in plants. Thus, pathogens that secrete subtilisin inhibitors may indirectly prevent the activation of immune proteases.
Keywords: apoplast, proteolytic cascade, immunity, subtilase, Solanaceae
Abstract
Proteolytic cascades regulate immunity and development in animals, but these cascades in plants have not yet been reported. Here we report that the extracellular immune protease Rcr3 of tomato is activated by P69B and other subtilases (SBTs), revealing a proteolytic cascade regulating extracellular immunity in solanaceous plants. Rcr3 is a secreted papain-like Cys protease (PLCP) of tomato that acts both in basal resistance against late blight disease (Phytophthora infestans) and in gene-for-gene resistance against the fungal pathogen Cladosporium fulvum (syn. Passalora fulva). Despite the prevalent model that Rcr3-like proteases can activate themselves at low pH, we found that catalytically inactive proRcr3 mutant precursors are still processed into mature mRcr3 isoforms. ProRcr3 is processed by secreted P69B and other Asp-selective SBTs in solanaceous plants, providing robust immunity through SBT redundancy. The apoplastic effector EPI1 of P. infestans can block Rcr3 activation by inhibiting SBTs, suggesting that this effector promotes virulence indirectly by preventing the activation of Rcr3(-like) immune proteases. Rcr3 activation in Nicotiana benthamiana requires a SBT from a different subfamily, indicating that extracellular proteolytic cascades have evolved convergently in solanaceous plants or are very ancient in the plant kingdom. The frequent incidence of Asp residues in the cleavage region of Rcr3-like proteases in solanaceous plants indicates that activation of immune proteases by SBTs is a general mechanism, illuminating a proteolytic cascade that provides robust apoplastic immunity.
Rcr3 is a secreted papain-like cysteine protease (PLCP) of tomato that acts as a coreceptor for the Cf-2 resistance protein to detect Avr2, a secreted apoplastic effector of the fungal pathogen Cladosporium fulvum (syn. Passalora fulva) (1, 2). Recognition of Avr2 by Cf-2 results in a localized programmed cell death known as the hypersensitive response (HR). This recognition event is consistent with the classical gene-for-gene interaction, which states that resistance genes in plants confer recognition of specific avirulence effector genes in the pathogen, here Cf-2 and Avr2, respectively. Cf-2 encodes a receptor-like protein with extracellular leucine-rich repeats (3), while Avr2 is a small, secreted cysteine-rich protein (4). Avr2 binds and inhibits Rcr3, and in the current model, this Avr2-Rcr3 complex is recognized by Cf-2 (5) (Fig. 1A). Accordingly, the absence of Rcr3 (e.g., in rcr3-3 null mutant tomato) causes a loss of Avr2 recognition and susceptibility to C. fulvum producing Avr2 (1). The role of Rcr3 in immunity against C. fulvum is dependent on Cf-2 because in the absence of Cf-2, Rcr3 does not contribute to immunity against C. fulvum (6).
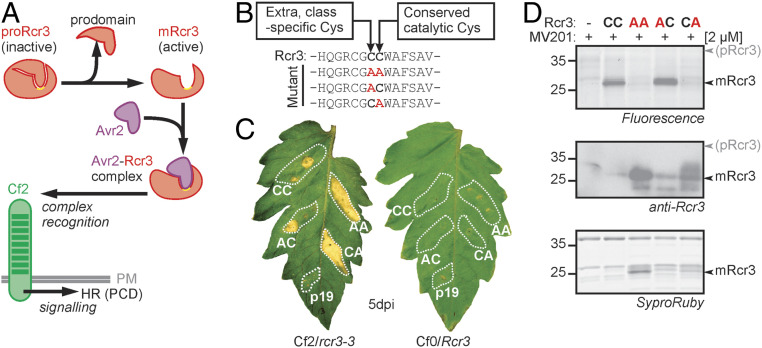
Fig. 1.
Catalytic Rcr3 mutants trigger HR and are processed into mature Rcr3 (mRcr3). (A) Proposed mechanism of Avr2 perception by Rcr3 and Cf-2. The prodomain of Rcr3 is removed to create a binding site for Avr2 and to form a complex that is recognized by the receptor-like protein Cf-2 in the plasma membrane (PM), resulting in HR, a form of programmed cell death. (B) Substitutions of two Cys residues in the active site Rcr3. Rcr3 carries a class-specific Cys residue directly preceding the catalytic Cys residue (CC; bold). All three combinations of Cys-to-Ala substitutions were generated. All Rcr3 constructs were driven by a 35S promoter and cloned into the transfer DNA of a binary plasmid. (C) Catalytic Rcr3 mutants can still trigger HR. AF isolated from agroinfiltrated N. benthamiana plants expressing (mutant) Rcr3 or p19 was coinjected with 300 nM purified Avr2 into leaflets of MM-Cf2/rcr3-3 and MM-Cf0/Rcr3 tomato plants, and pictures were taken at 5 d postinjection (dpi). (D) Catalytic Rcr3 mutants are still processed. AF isolated from agroinfiltrated N. benthamiana plants expressing (mutant) Rcr3 was labeled with 2 µM MV201 for 3.5 h. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by fluorescence scanning (Top), Western blot with an anti-Rcr3 antibody (Middle), and Sypro Ruby staining (Bottom). Rcr3 mutants lacking the catalytic Cys are inactive but accumulate as mature proteins.
The inhibition of Rcr3 by Avr2 indicates that this protease is an important component of the extracellular immune system of tomato. Indeed, Rcr3 is also inhibited by three additional pathogen-derived inhibitors: cystatin-like EpiC1 and EpiC2B from the oomycete potato blight pathogen Phytophthora infestans (7–9), GrVap1 of the potato cyst nematode Globodera rostochiensis (10), and chagasin-like Cip1 from Pseudomonas syringae (11). Further support for the importance of immune PLCPs comes from the observation that the EpiC1 ortholog of Phytophthora mirabilis has evolved genetic adaptations that are crucial for inhibiting Rcr3-like proteases of its host Mirabilis jalapa (12).
The relevance of apoplastic PLCPs in immunity (13) is also apparent in other plant species. The bacterial Huanglongbing pathogen of citrus, for instance, secretes effector SDE1 to suppress citrus PLCPs (14), whereas the maize smut fungus Ustilago maydis secretes effector Pit2 to inhibit maize PLCPs (15, 16). Thus, the emerging picture is that probably most apoplast-colonizing plant pathogens produce inhibitors to suppress defense-related PLCPs secreted by their host plants.
The role of secreted PLCPs in immunity has been demonstrated for Rcr3 and other tomato PLCPs. Depletion of Rcr3 from tomato increases susceptibility to P. infestans (8) independent of Cf-2 (6). Likewise, depletion of Pip1, a paralog of Rcr3, causes hypersusceptibility to bacterial, fungal, and oomycete pathogens (6), and silencing of PLCP CP14 in Nicotiana benthamiana increases the susceptibility to P. infestans (9, 17).
Here we tested whether the catalytic activity of Rcr3 is required for its role in Avr2 perception. Based on the described pH-dependent self-activation mechanism of PLCPs, we hypothesized that Rcr3 mutants lacking the catalytic cysteine would be unable to activate themselves (Fig. 1A). When delivered into an acidic environment, the prodomain of PLCPs unfolds, and the proprotease activates itself by cleaving between the prodomain and the protease domain (18–20). Since Avr2 is thought to inhibit Rcr3 by interacting with the substrate-binding groove (21), the Rcr3 prodomain would prohibit binding of Avr2 to proRcr3. Therefore, catalytically inactive proRcr3 should fail to remove its prodomain and interact with Avr2 to trigger HR. However, during this study, we found that catalytically inactive Rcr3 is still processed and is able to bind Avr2 and trigger HR. Our further studies revealed that proRcr3 is processed by a class of apoplastic serine proteases called subtilases (SBTs). This class includes P69B, also known as pathogenesis-related 7 (PR7), an abundant immune-related SBT in the apoplast of tomato (22). P69B can indeed activate proRcr3, leading to the activation of immune proteases in plants. Interestingly, P69B and other SBTs are inhibited by EPI1, a Kazal-like SBT inhibitor effector produced by P. infestans, indicating that this pathogen may prevent activation of induced immune PLCPs by inhibiting the upstream protease. Our work reveals that a redundant proteolytic cascade in solanaceous plants activates immune proteases to provide robust apoplastic immunity.
Results
Inactive Rcr3 Triggers HR and Accumulates as Mature Protease.
To test whether the catalytic Cys is required for HR, we generated a C154A substitution mutant of Rcr3, Rcr3CA (Fig. 1B). The catalytic Cys in Rcr3 is preceded by an additional Cys residue, as is common in this subfamily of SAG12-like proteases (23). To exclude the possibility that this residue acts redundantly with the catalytic residue, we also generated a double-substitution mutant (Rcr3AA: C153A, C154A) and included a single-substitution mutant of this residue (Rcr3AC: C153A). These three substitution mutants and wild-type (WT) Rcr3 were transiently expressed in N. benthamiana by agroinfiltration. Apoplastic fluid (AF) was isolated, mixed with purified Avr2, and injected into leaflets of Money Maker tomato plants carrying Cf-2 but lacking Rcr3 (Cf2/rcr3-3 mutant plants) (1) or lacking Cf-2 (Cf0/Rcr3 plants). Importantly, all four proteins triggered HR in leaves of Cf2/rcr3 plants but not in leaves of Cf0/Rcr3 plants (Fig. 1C), demonstrating that the two Cys residues are not required for triggering HR.
To detect which of these four proteins are active proteases, we incubated AF containing these proteins with MV201, a fluorescent activity-based probe for PLCPs (23). MV201 labeling was detected for both the WT Rcr3CC and the Rcr3AC mutant (Fig. 1 D, Top), indicating that these proteins are active proteases. In contrast, no labeling was observed for Rcr3AA and Rcr3CA mutants (Fig. 1D), confirming that the C154 residue is a catalytic residue, and that C153 is not essential for protease activity.
To demonstrate that inactive proteases also accumulate in the AF, we performed Western blot analysis on AF using polyclonal antibodies raised against the Rcr3 protein (Materials and Methods). All four Rcr3 proteins were detected (Fig. 1 D, Middle). The inactive Rcr3AA and Rcr3CA proteins accumulated to higher levels compared with the active Rcr3CC and Rcr3AC proteins (Fig. 1D), and were even detected on protein gels stained with Sypro Ruby (Fig. 1 D, Bottom). The greater accumulation of the inactive Rcr3AA and Rcr3CA proteins correlated with their ability to induce a stronger HR (Fig. 1C) and was likely caused by the absence of the self-degradation that occurs with active Rcr3.
Unexpectedly, the inactive Rcr3AA and Rcr3CA mutants accumulated not as the 40-kDa proprotease (proRcr3), but as a 25-kDa mature protease (mRcr3), similar to active Rcr3CC and Rcr3AC (Fig. 1D). Therefore, this experiment indicates that proteases other than Rcr3 can remove the prodomain from proRcr3 and produce (active) mRcr3.
Rcr3 Is Activated in Tomato AF by Another Protease.
To further investigate processing of proRcr3, we produced both WT (CC) and double-mutant (AA) proRcr3 with a C-terminal His-tag in Pichia pastoris (Fig. 2A). Both proteins were purified on nickel-nitrilotriacetic acid (NTA) columns and accumulated as a 40-kDa protein, consistent with being unprocessed proRcr3 (Fig. 2B). However, when incubated with AF from tomato, both proRcr3 proteins were quickly converted into mRcr3 (Fig. 2B). The presence of Rcr3 in tomato AF was not required for this conversion, as it also occurred with AF from plants lacking Rcr3 (Cf2/rcr3-3 plants; Fig. 2B). Time course experiments showed that conversion is completed within 15 min, irrespective of the presence of endogenous Rcr3 in AF (Fig. 2C). These experiments show that AF of tomato contains a protease that cleaves proRcr3 to produce mRcr3.
Fig. 2.
Rcr3 is activated in tomato AF by another protease. (A) Constructs used for the expression of proRcr3 in P. pastoris. pJK209 encodes proRcr3AA, which carries two Cys-to-Ala substitutions in the active site. Both proteins contain a yeast alpha-factor signal peptide for secretion into the medium and a C-terminal His-tag for purification. (B) proRcr3 is processed in tomato AF irrespective of its intrinsic activity. Purified WT and mutant (AA) Rcr3 was incubated for 15 min with AF isolated from WT and rcr3-3 (rcr3) MM-Cf2 tomato plants and analyzed by Western blot using the anti-Rcr3 antibody. (C) proRcr3 conversion in AF is complete in 15 min. Signals were quantified from Western blots with ImageJ. Error bars represent the SD of three replicates.
ProRcr3 Is Processed by Apoplastic Ser Proteases.
To characterize the proteases responsible for proRcr3 cleavage, we preincubated AF of tomato with various protease inhibitors and then added purified proRcr3. ProRcr3 processing was blocked by Ser protease inhibitor PMSF (phenyl-methyl-sulfonyl-fluoride) and Asp protease inhibitor pepstatin A, whereas Ser protease inhibitor DCI (3,4-dichloro-isocoumarin) had an intermediate effect, and no effect was detected for Cys protease inhibitor E-64 and metalloprotease inhibitor phenanthroline (Fig. 3A). However, we noticed that pepstatin A also caused protein precipitation in the AF. At lower pepstatin A concentrations, no protein precipitation was observed, and proRcr3 processing was not inhibited (SI Appendix, Fig. S1).
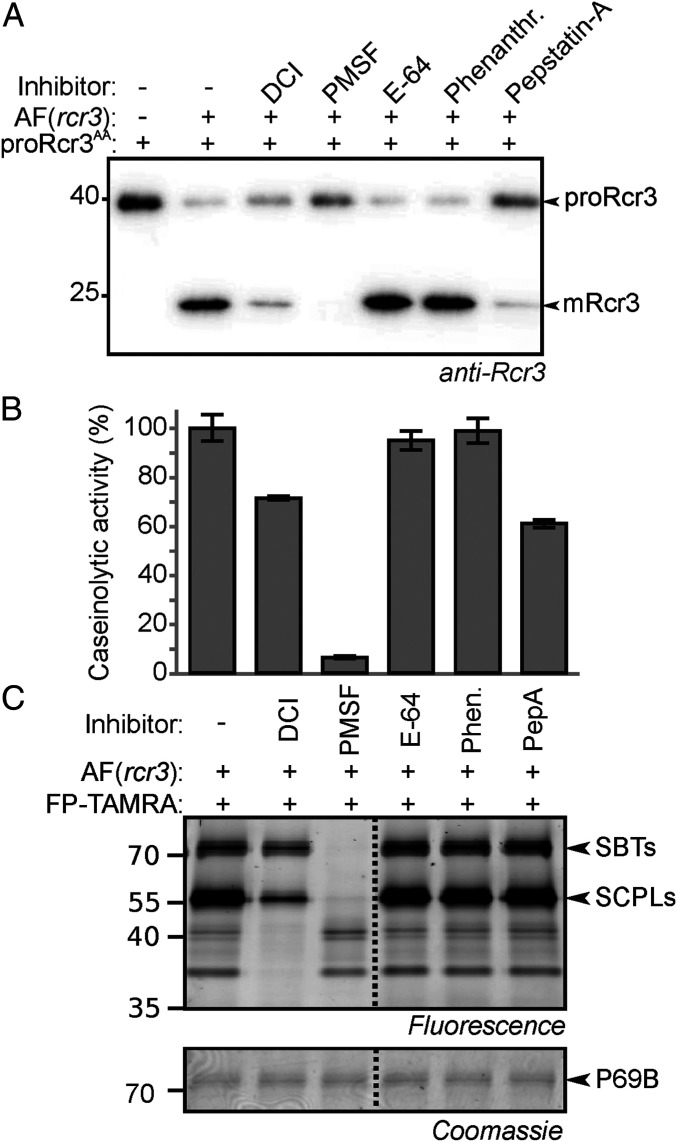
Fig. 3.
proRcr3 is processed by apoplastic Ser proteases. (A) proRcr3 processing is blocked by PMSF. Tomato AFs were preincubated with 1 mM inhibitors for 1 h and then incubated with purified Rcr3AA for 15 min and analyzed by Western blot using anti-Rcr3 antibodies. (B) PMSF strongly reduces proteolytic activity in AF. The proteolytic activity of AF used in A was measured with the general protease substrate FITC-casein in the presence of 1 mM inhibitors. Released fluorescence was measured at 490 nm after a 5-min incubation. (C) PMSF blocks activity-based labeling of SBTs and SCPLs. Tomato AF was preincubated with 1 mM inhibitors as in A and then labeled with 0.2 µM FP-TAMRA for 1 h. Proteins were separated by SDS-PAGE and analyzed by fluorescence scanning.
To monitor proteases that are blocked by the inhibitors, we used the general protease substrate casein-fluorescein isothiocyanate (FITC), which releases fluorescent fragments when processed (24). Incubation of AF with casein-FITC revealed that caseinolytic activity was reduced by DCI and pepstatin A and almost abolished by PMSF (Fig. 3B), indicating that Ser proteases are major contributors to the proteolytic activity in tomato AF.
To detect proteases inhibited by PMSF and DCI, we monitored Ser hydrolase activities using a fluorescent fluorophosphonate probe (FP-TAMRA) (25). PMSF blocked FP labeling of 70- and 55-kDa proteins (Fig. 3C), previously identified as SBTs and Ser carboxypeptidase-like (SCPL) proteins, respectively (26). DCI partially suppressed the labeling of these two signals and blocked the labeling of proteins that migrated at a lower molecular weight (Fig. 3C). The suppression of labeling of SBTs and SCPLs by PMSF and DCI correlated with the suppression of proRcr3 processing by these inhibitors, suggesting that SBTs or SCPLs may process proRcr3. However, SCPLs are carboxypeptidases and acyltransferases and have no known endopeptidase activity (27). In contrast, SBTs are endopeptidases (28) that could facilitate proRcr3 processing.
EPI1 Interferes with proRcr3 Processing by Inhibiting SBTs.
To test whether SBTs could mediate proRcr3 processing, we took advantage of the Kazal-like SBT inhibitor EPI1 of the oomycete potato blight pathogen P. infestans. EPI1 inhibits P69B, an abundant defense-induced SBT in the tomato apoplast (29). EPI1 also inhibits other SBTs and has been used to overcome functional redundancy of the SBT family in regulating floral organ abscission and the maturation of CLEL signaling peptides in Arabidopsis (30, 31) and haustorium development in the parasitic plant Phtheirospermum japonicum (32). Preincubation of tomato AF with purified EPI1 prevented FP-TAMRA labeling of the 70-kDa SBTs but not of the 55-kDa SCPLs (Fig. 4A), indicating that EPI1 inhibits SBTs but not SCPLs. Importantly, preincubation of tomato AF with EPI1 followed by incubation with proRcr3 completely blocked proRcr3 processing (Fig. 4B), indicating that SBTs are responsible for proRcr3 cleavage.
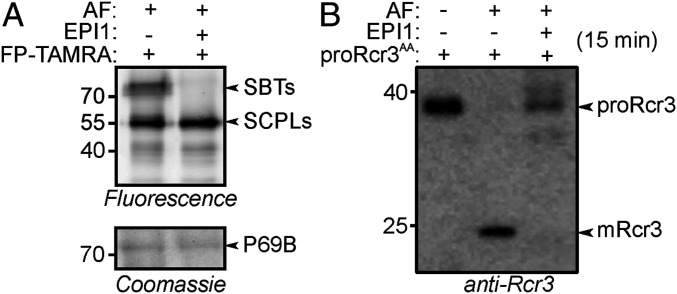
Fig. 4.
SBT inhibition by P. infestans effector EPI1 blocks Rcr3AA maturation. (A) EPI1 blocks SBT labeling in tomato AF. AF from Cf2/rcr3 tomato plants was incubated with and without 17 µM purified EPI1 for 1 h and then labeled with 0.2 µM FP for 1 h. Proteins were separated by SDS-PAGE and analyzed by fluorescence scanning and Coomassie staining. (B) EPI1 blocks Rcr3 maturation. AF from Cf2/rcr3 tomato plants was incubated with 17 µM purified EPI1 for 1 h and then incubated with purified proRcr3AA. Samples were taken after 15 min and analyzed by Western blot using the anti-Rcr3 antibody.
Tomato SBT P69B Can Cleave proRcr3.
To test whether P69B could process Rcr3, we transiently expressed a C-terminally His-tagged P69B by agroinfiltration of N. benthamiana. Labeling of AF isolated from agroinfiltrated plants with FP-TAMRA displayed a strong 70-kDa signal (Fig. 5A), indicating that P69B-His is an active Ser protease. A strong 70-kDa signal was also detected by Coomassie staining (Fig. 5A), indicating that P69B-His accumulates to high levels in AF of agroinfiltrated plants.
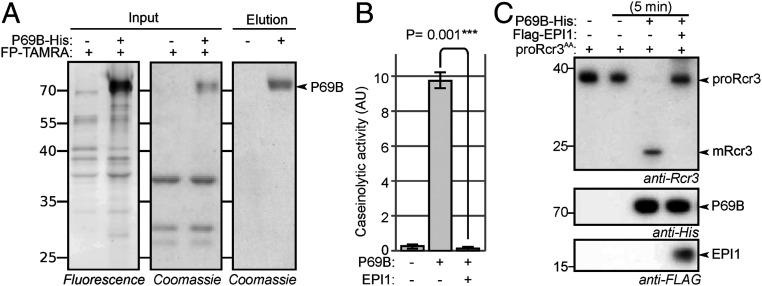
Fig. 5.
Tomato SBT P69B cleaves proRcr3. (A) P69B expression and purification. AF from plants transiently coexpressing P69B-His with p19 or only p19 (control) was isolated at 5 dpi and labeled with FP-TAMRA. Fluorescently labeled proteins were detected from protein gels by fluorescence scanning (Left). Proteins were stained with Coomassie (Middle). His-tagged proteins were purified from AF of p19 and P69B-His expressing leaves on nickel-NTA and detected from protein gels using Coomassie staining (Right). (B) Purified P69B is active and can be inhibited by EPI1. Purified P69B and the control were incubated with FITC-casein, and casein processing was measured at 5 min. Error bars represent SD of three replicates. P values were calculated using Student’s t test. (C) Purified P69B converts purified proRcr3 into mRcr3 in vitro. Purified P69B was preincubated with and without EPI1 for 45 min and then incubated with purified proRcr3 for 5 min. Rcr3, P69B, and EPI1 were detected by Western blot analysis using the respective antibodies.
The caseinolytic activity in AF was strongly increased on transient P69B-His expression (SI Appendix, Fig. S2A), indicating that P69B is an active protease. Incubation of AF from agroinfiltrated plants with proRcr3 showed that the generation of mRcr3 from proRcr3 was faster in the presence of P69B (SI Appendix, Fig. S2B), indicating that P69B can cleave proRcr3. In AF of agroinfiltrated plants that do not produce P69B, proRcr3 processing was reduced but still occurred (SI Appendix, Fig. S2B).
To demonstrate that P69B itself can cleave proRcr3 directly, we purified P69B-His from AF of agroinfiltrated plants (Fig. 5A). This purified P69B had high caseinolytic activity, and this activity was blocked with EPI1 (Fig. 5B). This P69B cleaved purified proRcr3, and also this activity was blocked with EPI1 (Fig. 5C and SI Appendix, Fig. S3). Thus, purified proRcr3 is a substrate for purified P69B in vitro.
Asp Residues Are Required for Cleavage of proRcr3 by P69B.
Based on the alignment with mature papain and other mature PLCPs, we identified a putative cleavage region of 10 residues (NDLSDDYMPS; Fig. 6A). To create uncleavable proRcr3 mutants, we generated a synthetic library in which nine residues in this cleavage region (all except Pro) were randomly substituted into 19 residues: alanine (Ala) and two additional residues in case the Ala substitution could not be achieved with a single nucleotide substitution. This library theoretically contains 49,152 possible amino acid substitution combinations (Fig. 6A).
Fig. 6.
Asp residues in proRcr3 are required for cleavage by P69B. (A) Random mutagenesis of the cleavage region in proRcr3. ProRcr3AA was synthesized with specific degenerate codons in the region encoding the putative cleavage site in proRcr3. All possible amino acid substitutions are shown in red. There are 49,152 possible combinations of these substitutions and original residues. (B) All tested random mutants accumulate as mRcr3 on agroinfiltration. A total of 315 random mutants were transiently expressed in N. benthamiana by agroinfiltration, and total extract was isolated and analyzed by Western blot using the anti-Rcr3 antibody. Shown are sequences of seven random clones, indicating that the mutagenesis was successful. (C) Mutant proRcr33K;AA carries three Asp-into-Lys substitutions in the putative cleavage region of proRcr3AA. (D) Purified proRcr33K;AA is no longer cleaved by purified P69B in vitro. (E) Processing of purified proRcr33K;AA is slower when incubated with AF of tomato.
This substitution library was cloned into a binary vector and expressed transiently by agroinfiltration. We tested 315 different clones by Western blot analysis of total extracts from agroinfiltrated leaves and found that all mutants still accumulated as mRcr3 (Fig. 6B and SI Appendix, Fig. S4). Sequencing of seven random clones indicated that the mutagenesis was successful, as the intended substitutions were found in various combinations (Fig. 6B). This experiment shows that all proRcr3 proteins are cleaved in this region irrespective of the sequence, indicating that different proteases of N. benthamiana may act on this region.
Since proRcr3 is normally processed in tomato, presumably by P69B, we used a different strategy to demonstrate that Rcr3 is cleaved by P69B in the cleavage region. We focused on substituting the three Asp residues contained in proRcr3 in this region (Fig. 6A). We focused on Asp residues for several reasons: 1) Asp residues are common in this region for Rcr3 homologs in solanaceous plants (SI Appendix, Fig. S5); 2) the closest clustering homolog of P69B (P69A) prefers P1 = Asp (33); and 3) EPI1 carries an Asp residue in its recognition loop, corresponding to the P1 position. Indeed, the fluorogenic caspase-1 substrate Ac-YVAD-AMC (carrying P1 = Asp) is cleaved after the Asp residue by P69B (SI Appendix, Fig. S6).
To further investigate the Asp specificity by P69B, we synthesized five YVAX-ACC tetrapeptides with X = Asp(D), Asn (N), Glu(E), Ala(A), and Phe(F). C-terminal processing would release fluorescent ACC [7-amino-4-carbamoylmethylcoumarin (34)]. The presence of P69B in AF of N. benthamiana significantly increases fluorescence on incubation with YVAD-ACC, whereas no strong fluorescence was released on incubation with YVAN-ACC, YVAE-ACC, YVAA-ACC, or YVAF-ACC (SI Appendix, Fig. S7). These data indicate that P69B can distinguish Asp(D) from related Asn(N) or Glu(E) residues, implying that P69B is an Asp-specific protease at apoplastic pH.
Consequently, we substituted the three Asp residues in proRcr3AA into Lys residues (Fig. 6C) and produced this mutant (proRcr33K;AA) in P. pastoris. When expressed in yeast, some of the proRcr33K;AA was processed into a mRcr3 isoform that had a slightly lower molecular weight than mRcr3AA cleaved in planta (mRcr3*; SI Appendix, Fig. S8), indicating that the substitutions had created cleavage sites for one or more yeast protease(s). Since this mRcr3* was smaller than the mRcr3 obtained in plants, we nevertheless could test processing of the remaining proRcr33K;AA.
Incubation of proRcr3 with purified P69B revealed that, in contrast to rapid processing of proRcr3AA, proRcr33K;AA was no longer cleaved by P69B (Fig. 6D). When incubated with tomato AF, proRcr33K;AA processing was significantly reduced compared with proRcr3AA (Fig. 6E), indicating that the Asp residues are important for efficient processing in the tomato apoplast. These data demonstrate that although purified P69B can no longer cleave proRcr33K;AA, other apoplastic proteases still process the mutant cleavage region, albeit at a reduced rate.
ProRcr3 Processing Is Reduced in asP69B Plants.
To test whether P69B is the protease that activates Rcr3 in tomato, we generated two independent transgenic antisense P69B (asP69B) tomato lines (16-32 and 16-34) (16–34) in the Money Maker (MM-Cf0) background. Since both lines behaved similarly in our assays, only 16-32 is shown as representative. The asP69B plants did not have a major growth phenotype compared with the nontransgenic (NT) control (Fig. 7A). Besides reduced transcript levels of P69B, asP69B plants also had reduced transcript levels of P69A, -C, -D, -G, -H, and -I (SI Appendix, Fig. S9), consistent with their >85% shared nucleotide identity with P69B used for silencing (SI Appendix, Fig. S10). Labeling of isolated AF with FP-TAMRA revealed that the 70-kDa signal was no longer present in asP69B plants (Fig. 7B), and also the main 70-kDa signal detected by Coomassie staining was strongly reduced in AF from asP69B plants (Fig. 7B), consistent with P69B and its cosilenced paralogs. Besides the 70-kDa signal, we detected two weaker signals in AF of both NT and asP69B plants (Fig. 7B, red inset). Labeling of these signals was blocked by EPI1 (Fig. 7B, red inset), indicating that these signals originate from EPI1-sensitive SBTs other than P69B or its cosilenced paralogs. These signals may originate from P69E and P69F, which are not silenced in asP69B plants (SI Appendix, Fig. S9).
Fig. 7.
P69B depletion reduces proRcr3AA conversion. (A) asP69B plants have no macroscopic developmental phenotypes. NT MM-Cf0 tomato plants were transformed with an antisense P69B (asP69B) construct. The photographs are of 6-wk-old NT and asP69B (line 16-32) plants. (B) SBT labeling is depleted in AF of asP69B plants. AF of NT and asP69B transgenic MM-Cf0 tomato plants was labeled with 0.2 µM FP, and proteins were analyzed from protein gels by fluorescence scanning and Coomassie staining. (Red Inset) The same image with increased contrast and intensity. (C) Reduced proRcr3 processing in AF of asP69B plants. AF of NT and asP69B tomato plants was incubated with purified proRcr3AA for 15 min, and samples were analyzed by Western blot with the anti-Rcr3 antibody.
When incubated with purified proRcr3AA, AF from asP69B plants showed a slower rate of conversion compared with AF from NT control plants (Fig. 7C), indicating that P69B is required for efficient proRcr3 processing. However, Rcr3 was still converted in AF of asP69B plants, whereas EPI1 inhibited Rcr3 processing in tomato AF (Fig. 4B), indicating that P69B silencing is incomplete, or that other apoplastic EPI1-sensitive SBTs are mediating proRcr3 processing in asP69B lines.
We next investigated the accumulation of endogenous Rcr3 in the asP69B plants. Western blot analysis of AF with anti-Rcr3 antibodies showed that Rcr3 accumulated to a higher level in asP69B plants, but endogenous Rcr3 was still processed into mRcr3 (SI Appendix, Fig. S11A). The seemingly normal processing of endogenous Rcr3 may have been caused by incomplete P69B silencing and/or the activity of other SBTs over the long incubation times with endogenous proRcr3. We also frequently observed increased levels of PR proteins PR2 and PR3 in AF of untreated asP69B plants (SI Appendix, Fig. S11B). These data indicate that the asP69B plants were stressed, causing increased levels of PR proteins, including Rcr3. Although the processing speed of exogenous proRcr3 was reduced in AF from these plants, the accumulation of endogenous proRcr3 was slow enough to allow for processing into mRcr3.
Rcr3 Is Processed by (a) Distantly Related SBT(s) in N. benthamiana.
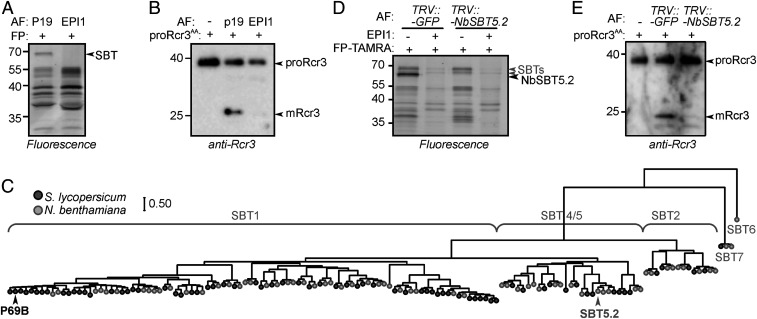
We had noticed earlier that catalytically inactive proRcr3 was still processed into mRcr3 on agroinfiltration of N. benthamiana (Figs. 1D and 5C and SI Appendix, Fig. S2), indicating that N. benthamiana also contains proteases that cleave proRcr3. Labeling of AF of agroinfiltrated N. benthamiana with FP-TAMRA resulted in 70-kDa signals that were suppressed on transient expression of EPI1 (Fig. 8A), indicating that these signals are SBTs.
Fig. 8.
Rcr3 is matured by a distantly related SBT in N. benthamiana. (A) Transient expression of EPI1 suppresses labeling of endogenous SBTs in N. benthamiana AF. AF from agroinfiltrated N. benthamiana plants transiently (co)expressing silencing inhibitor p19 with and without EPI1 was labeled with FP, and labeled proteins were detected from protein gel by fluorescence scanning. (B) EPI1 blocks processing of proRcr3 in AF of N. benthamiana. AF from N. benthamiana leaves transiently expressing silencing inhibitor p19 with and without EPI1 was incubated with purified proRcr3AA for 15 min and analyzed by Western blot using the anti-Rcr3 antibody. (C) The main SBTs in AF of tomato and N. benthamiana AF are distantly related. The phylogenetic tree of tomato and N. benthamiana SBTs was generated using the maximum likelihood method. SI Appendix, Fig. S12 presents the detailed phylogenetic tree. (D) The main 70-kDa SBT signal is depleted in TRV::NbSBT5.2 plants. AF isolated from TRV::GFP and TRV::NbSBT5.2 plants was preincubated with and without purified EPI1 and labeled with 0.2 µM FP-TAMRA. Labeled proteins were detected from protein gel by fluorescence scanning. (E) NbSBT5.2 contributes to proRcr3 processing. AF isolated from TRV::GFP and TRV::NbSBT5.2 plants was incubated with purified proRcr3AA for 15 min and then analyzed by Western blot using the anti-Rcr3 antibody.
Incubation of purified proRcr3AA with AF from agroinfiltrated leaves triggered mRcr3 accumulation, but this was blocked when the leaf also expressed EPI1 (Fig. 8B). This indicates that SBTs in AF of N. benthamiana cleave proRcr3. The N. benthamiana genome has two P69B homologs (Fig. 8C), but these proteases are not abundant in the AF of agroinfiltrated plants (35) (Fig. 8C). We previously identified 28 SBTs from AF of agroinfiltrated leaves and found that NbD038072.1 caused the most abundant signal in proteomics experiments (36). We termed this protease NbSBT5.2, because of its highest sequence identity with Arabidopsis SBT5.2 (37). Notably, NbSBT5.2 belongs to a very different subfamily than P69B, which is a member of the SBT1 subfamily (Fig. 8C and SI Appendix, Fig. S12), indicating that different SBTs dominate the apoplast in different solanaceous plant species.
Because of its relative abundance, we depleted NbSBT5.2 by virus-induced gene silencing (VIGS) to investigate its role in proRcr3 processing. Plants infected with tobacco rattle virus (TRV) carrying a fragment of the NbSBT5.2-encoding gene (TRV::NbSBT5.2 plants) do not grow differently from TRV::GFP control plants (SI Appendix, Fig. S13). Labeling of AF isolated from these plants with FP-TAMRA showed that the main 70-kDa signal in TRV::GFP control plants was absent in TRV::NbSBT5.2 plants (Fig. 8D), indicating that this signal represents the active NbSBT5.2 SBT. Some weaker fluorescent FP-labeled signals in the 70-kDa region remained in AF of TRV::NbSBT5.2 plants. Labeling of these proteins was blocked on preincubation with EPI1 (Fig. 8D), indicating that they are SBTs other than NbSBT5.2. Importantly, processing of purified proRcr3 was suppressed in AF of TRV::NbSBT5.2 plants but present in AF of TRV::GFP plants (Fig. 8E), indicating that NbSBT5.2 is required for proRcr3 processing in N. benthamiana.
Discussion
Apoplastic proteases have emerged as major components of plant immunity against various pathogens (13). One such immune protease is the tomato PLCP Rcr3, which contributes to defenses against fungal and oomycete pathogens. Here we discovered that Rcr3 is activated by P69B and other secreted subtilisins. ProRcr3 is processed by P69B in vitro, and P69B depletion or its inhibition by the P. infestans effector EPI1 prevents proRcr3 processing in vitro. Aspartate residues in the cleavage region of proRcr3 are required for processing, consistent with the ability of P69B to cleave after Asp residues. This discovery shows that immune protease Rcr3 is activated by subtilases, a mechanism that might occur across the plant kingdom.
Proteolytic cascades involve different proteases that consecutively activate one another (38). Several proteolytic cascades have been described for metazoans (39). Blood coagulation, apoptosis, and the development of dorsal-ventral polarity in insects are all regulated through proteolytic cascades (40–42). These proteolytic cascades offer irreversible, fast, nontranscriptional regulatory pathways that can quickly propagate and amplify a signal. Although proteases are equally numerous in plants, proteolytic cascades have not yet been reported in this kingdom, but there are several indications that they probably occur (38). Our results demonstrate that immune protease Rcr3 is released from its inactive proRcr3 precursor by P69B and other SBTs, describing a robust extracellular proteolytic cascade acting in plant immunity in solanaceous plants. Interestingly, P69B itself can be degraded by matrix metalloproteases in tomato, adding an additional layer of proteolytic regulation (43).
The P69B/Rcr3 proteolytic cascade intertwines with the signaling cascade involving Avr2, Rcr3, and Cf-2, leading to HR. At this stage, it seems unlikely that proRcr3 processing is a regulatory step during HR, because only low levels of mRcr3 are required for Avr2 perception, and enough P69B and other SBTs are present in the apoplast of unchallenged plants to facilitate mRcr3 release. However, it is interesting to note that Rcr3 does not require catalytic activity to act in Avr2 recognition, consistent with the decoy hypothesis (44). In contrast, the role of Rcr3 in basal defense against P. infestans (6, 8) is likely dependent on its proteolytic activity, but this remains to be demonstrated.
Activation of Immune Proteases in Solanaceous Plants.
Asp-specific SBTs are common in tomato (33) and likely in other solanaceous plants as well. We discovered that proRcr3 is also processed in N. benthamiana, and that processing was absent when NbSBT5.2 was depleted by silencing. Surprisingly, although P69B and NbSBT5.2 are the dominant active SBTs in the apoplast of tomato and N. benthamiana, respectively, they represent distinct subfamilies, SBT1 and SBT5, respectively. These data show that proRcr3 can be activated by different apoplastic SBTs and indicate that networks of proteolytic cascades may have evolved convergently in solanaceous plants or are ancient and diverged during speciation.
Asp residues occur consistently in the cleavage region of immune proteases Rcr3, Pip1, and C14 in solanaceous plants. The cleavage region in Rcr3 homologs of tobacco, pepper, potato, and tomato contains the consensus kINDLsDDdm, whereas the consensus for this region in Pip1-like proteases in these plants is KYDSvTevPx (Asp residues underlined, conserved residues capitalized; SI Appendix, Fig. S5). Asp residues are also conserved in PLCP subfamily containing tomato C14 (SDRyLPKVGd). PLCP subfamilies that are not defense-induced, such as subfamilies containing Pfp1 and Rfp1 (6), do not have conserved Asp residues in this region (SI Appendix, Fig. S5). These data suggest that Asp-specific SBTs are responsible for the activation of extracellular immune PLCPs in solanaceous plant species.
P. infestans Uses Multiple Strategies to Suppress Immune Proteases.
Kazal-type protease inhibitors are common and highly diversified in oomycetes, and at least two P. infestans-secreted proteins, EPI1 and EPI10, have been proposed to function as effectors that inhibit the SBT P69B (29, 45). These effectors carry P1 = Asp, indicating that they have evolved to suppress SBTs that cleave after Asp residues (saspases) (29). Our data indicate that Asp-specific SBTs may activate immune proteases Rcr3, Pip1, and C14 in solanaceous plants, all of which contribute to the basal defense against P. infestans (6, 8, 9, 17). This leads to the intriguing hypothesis that EPI1 prevents the activation of immune PLCPs during infection. By inhibiting P69B and other SBTs, EPI1 interferes with the activation of Rcr3, Pip1, and C14 delivered into the apoplast during defense responses as part of the PR protein arsenal. Interestingly, P1 = Asp is present in roughly one-half of oomycete Kazal-like inhibitors, whereas it is uncommon in Kazal inhibitors of animals and other eukaryotes (29). This raises the possibility that inhibition of Asp-specific SBTs might be a widespread virulence function in plant pathogenic oomycetes.
P. infestans is already known to suppress immune PLCPs through two mechanisms. P. infestans secretes cystatin-like inhibitors EpiC1 and EpiC2B that target Rcr3, Pip1, and C14 (7–9). In addition, the host-translocated RXLR effector AVRblb2 prevents the secretion of C14 (17). We now hypothesize a third mechanism, in which P. infestans EPI1 prevents activation of PLCPs by inhibiting the upstream protease. Thus, although under unchallenged conditions there could be enough mRcr3 to act in the perception of Avr2, a role for immune PLCP activation by P69B will be more important during defense responses, when large quantities of P69B and immune PLCPs are delivered into the apoplast.
Redundancy in Proteolytic Cascades: An Experimental Challenge and Biological Virtue?
Besides P69B, additional SBTs can also activate proRcr3, because proRcr3 is still processed in plants silenced for P69B. Nevertheless, this processing can be inhibited by EPI1. The tomato genome encodes for 82 SBTs, at least 12 of which have Asp-specificity and are known as phytaspases (33). We have detected active SBTs other than P69B in the apoplast of P69B-silenced plants that are inhibited by EPI1. These data indicate that the activation of immune protease Rcr3 is regulated by several SBTs that act redundantly. This prediction is consistent with the fact that distantly related NbSBT5.2 of N. benthamiana is also involved in proRcr3 processing, implying that more distantly related SBTs of tomato can process proRcr3.
Mutant proRcr3 lacking Asp residues in the cleavage region is no longer processed by P69B but is still processed in AF of tomato. In addition, all random cleavage site mutants of proRcr3 are still processed on expression in N. benthamiana. Furthermore, substitution of the Asp residues in proRcr3 creates a new cleavage site for a protease in the yeast-based expression system. These data indicate that removal of P69B cleavage site creates cleavage sites for other proteases of tomato, N. benthamiana, and yeast. This suggests that cleavage occurs in an exposed loop that is sensitive to multiple proteases, irrespective of the substrate sequence. However, we did note that processing of proRcr3 lacking Asp residues is significantly slower, indicating that these residues are required for efficient activation of proRcr3.
The redundancy in extracellular protein processing is an experimental challenge for research. We found redundancy among Asp-specific SBTs and among proteases acting on different sequences in the same exposed loop of a protein. Thus, both reverse genetics and mutagenesis of the putative cleavage site to generate an uncleavable substrate are limited approaches to demonstrating the relevance of cleavage. The use of EPI1 to block many unrelated SBTs simultaneously has been instrumental in our research and other investigations. EPI1 also has been used to overcome the redundancy of SBTs processing precursors of IDA and CLEL peptides, with regulate organ abscission and gravitropic responses in Arabidopsis (30, 31), and SBTs involved in haustorium development in the parasitic plant P. japonicum (32). Future research strategies will benefit from the use of protease inhibitors like EPI1 in this “effector genetics” strategy. For instance, the tomato cystatin SlCYS8 can inhibit numerous PLCPs and can be used to overcome redundancy within this family (36). EPI1, SlCYS8, and additional protease inhibitors (46) can also be used in combination to block multiple protease families. Clever use of these inhibitors with subcellular targeting signals and inducible or tissue-specific promoters will prove instrumental to future protease research in plants.
The redundancy in extracellular protein processing is probably also a biological virtue. Genetic redundancy enhances the robustness of biological systems and generally results in a network architecture of biological processes (47). Indeed, the observation that multiple SBTs converge on a single immune protease is reminiscent of the networks of immune receptors and signaling components that have been described in plants (48, 49). Redundancy also provides robustness to the extracellular processing that might be an essential defense mechanism against pathogens colonizing the apoplast. This would predict that pathogens are under pressure to produce multiple broad-range protease inhibitors. For instance, P. infestans simultaneously produces Kazal-like EPI1 and EPI10 to target multiple SBTs and cystatin-like EpiC to inhibit multiple PLCPs (7, 9, 29). Our work suggests that these and other apoplastic pathogens will produce an arsenal of protease inhibitors to overcome the redundant proteolytic network of the host apoplast.
Materials and Methods
All generated materials (seeds, plasmids, and antibodies) are available on request. Plasmids have also been deposited to AddGene. Detailed descriptions of the experimental methods are provided in SI Appendix. Tomato cultivar Money Maker (MM) isogenic lines MM-Cf0, MM-Cf2, and MM-Cf2/rcr3-3 (2) and N. benthamiana were grown in a growth chamber at 22 °C with 16 h light per day. MM-Cf2 was transformed with antisense P69B (asP69B) as described in SI Appendix, sections 1 and 7. Molecular cloning procedures are detailed in SI Appendix, section 1, including used plasmids and primers summarized in SI Appendix, Tables S1 and S2, respectively. Purification of Rcr3-His produced in P. pastoris, Flag-EPI1 and Avr2 from Escherichia coli, and P69B-His from agroinfiltrated leaves are detailed in SI Appendix, section 2. The synthesis of fluorogenic substrates YVAX-ACC and their analytical data are described in SI Appendix, section 3. Protease assays with ProRcr3, casein-FITC and YVAX-ACC are detailed in SI Appendix, section 4. Phylogenetic analysis of SBTs is described in SI Appendix, section 5, and agroinfiltration, VIGS, AF isolation, and HR assays are detailed in SI Appendix, section 6. Other biochemical methods, including labeling of Ser hydrolases with FP-TAMRA, Coomassie staining, Western blot, and the anti-Rcr3 antibody, are detailed in SI Appendix, section 8.
Data Availability.
All data supporting the findings of this study are contained in the main text and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by European Research Council Consolidator Grant 616449, “GreenProteases”; Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/S003193/1, “Pip1S”; the Clarendon Fund, the BBSRC Interdisciplinary Doctoral Training Partnership, and the Marie Skłodowska-Curie European Training Network, H2020-MSCA-ITN, PROTECTA.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921101117/-/DCSupplemental.
References
- 1.Dixon M. S., Golstein C., Thomas C. M., van Der Biezen E. A., Jones J. D. G., Genetic complexity of pathogen perception by plants: The example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. U.S.A. 97, 8807–8814 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krüger J. et al., A tomato cysteine protease required for Cf-2–dependent disease resistance and suppression of autonecrosis. Science 296, 744–747 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Dixon M. S. et al., The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Luderer R., Takken F. L., de Wit P. J. G. M., Joosten M. H. A. J., Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45, 875–884 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Rooney H. C. et al., Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308, 1783–1786 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Ilyas M. et al., Functional divergence of two secreted immune proteases of tomato. Curr. Biol. 25, 2300–2306 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Tian M. et al., A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143, 364–377 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J. et al., Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. U.S.A. 106, 1654–1659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaschani F. et al., An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozano-Torres J. L. et al., Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. U.S.A. 109, 10119–10124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shindo T. et al., Screen of non-annotated small secreted proteins of Pseudomonas syringae reveals a virulence factor that inhibits tomato immune proteases. PLoS Pathog. 12, e1005874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong S. et al., Effector specialization in a lineage of the Irish potato famine pathogen. Science 343, 552–555 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Misas-Villamil J. C., van der Hoorn R. A. L., Doehlemann G., Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 212, 902–907 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Clark K. et al., An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 9, 1718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller A. N., Ziemann S., Treitschke S., Aßmann D., Doehlemann G., Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9, e1003177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misas Villamil J. C. et al., A fungal substrate mimicking molecule suppresses plant immunity via an inter-kingdom conserved motif. Nat. Commun. 10, 1576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozkurt T. O. et al., Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U.S.A. 108, 20832–20837 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S., Choudhury D., Aich P., Dattagupta J. K., Biswas S., The structure of a thermostable mutant of pro-papain reveals its activation mechanism. Acta Crystallogr. D Biol. Crystallogr. 68, 1591–1603 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Groves M. R. et al., The prosequence of procaricain forms an alpha-helical domain that prevents access to the substrate-binding cleft. Structure 4, 1193–1203 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Verma S., Dixit R., Pandey K. C., Cysteine Proteases: Modes of activation and future prospects as pharmacological targets. Front. Pharmacol. 7, 107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hörger A. C., van der Hoorn R. A. L., The structural basis of specific protease-inhibitor interactions at the plant-pathogen interface. Curr. Opin. Struct. Biol. 23, 842–850 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Jordá L., Coego A., Conejero V., Vera P., A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J. Biol. Chem. 274, 2360–2365 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Richau K. H. et al., Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 158, 1583–1599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Twining S. S., Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 143, 30–34 (1984). [DOI] [PubMed] [Google Scholar]
- 25.Liu Y., Patricelli M. P., Cravatt B. F., Activity-based protein profiling: The serine hydrolases. Proc. Natl. Acad. Sci. U.S.A. 96, 14694–14699 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sueldo D. et al., Dynamic hydrolase activities precede hypersensitive tissue collapse in tomato seedlings. New Phytol. 203, 913–925 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Milkowski C., Strack D., Serine carboxypeptidase-like acyltransferases. Phytochemistry 65, 517–524 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Schaller A. et al., From structure to function—A family portrait of plant subtilases. New Phytol. 218, 901–915 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Tian M., Huitema E., Da Cunha L., Torto-Alalibo T., Kamoun S., A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 279, 26370–26377 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Schardon K. et al., Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Stührwohldt N. et al., The biogenesis of CLEL peptides involves several processing events in consecutive compartments of the secretory pathway. eLife 9, e55580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa S., et al. , Subtilase activity in the intrusive cells mediates haustorium maturation in parasitic plants. 10.1101/2020.03.30.015149 (30 March 2020). [DOI] [PMC free article] [PubMed]
- 33.Reichardt S. et al., The tomato subtilase family includes several cell death-related proteinases with caspase specificity. Sci. Rep. 8, 10531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris J. L. et al., Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. U.S.A. 97, 7754–7759 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grosse-Holz F. et al., The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol. J. 16, 1068–1084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jutras P. V. et al., Activity-based proteomics reveals nine target proteases for the recombinant protein-stabilizing inhibitor SlCYS8 in Nicotiana benthamiana. Plant Biotechnol. J. 17, 1670–1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serrano I. et al., A non canonical subtilase attenuates the transcriptional activation of defence responses in Arabidopsis thaliana. eLife 5, e19755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulus J. K., Van der Hoorn R. A. L., Do proteolytic cascades exist in plants? J. Exp. Bot. 70, 1997–2002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krem M. M., Di Cera E., Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem. Sci. 27, 67–74 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Amara U. et al., . “Interaction between the coagulation and complement system” in Current Topics in Complement II, Lambris J. D., Ed. (Springer, 2008), pp. 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho Y. S., Stevens L. M., Sieverman K. J., Nguyen J., Stein D., A ventrally localized protease in the Drosophila egg controls embryo dorsoventral polarity. Curr. Biol. 22, 1013–1018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slee E. A. et al., Ordering the cytochrome c-initiated caspase cascade: Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144, 281–292 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmermann D. et al., Cell death control by matrix metalloproteinases. Plant Physiol. 171, 1456–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Hoorn R. A. L., Kamoun S., From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian M., Benedetti B., Kamoun S., A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 138, 1785–1793 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grosse-Holz F. et al., Three unrelated protease inhibitors enhance accumulation of pharmaceutical recombinant proteins in Nicotiana benthamiana. Plant Biotechnol. J. 16, 1797–1810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowak M. A., Boerlijst M. C., Cooke J., Smith J. M., Evolution of genetic redundancy. Nature 388, 167–171 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Wu C. H. et al., NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U.S.A. 114, 8113–8118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C. H., Derevnina L., Kamoun S., Receptor networks underpin plant immunity. Science 360, 1300–1301 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are contained in the main text and SI Appendix.