Significance
Adiponectin, a member of the CTRP family of secreted proteins produced by adipose tissue, is thought to function as an endocrine hormone. An unusual feature of adiponectin is that it is present in high concentrations in circulation and thus may have noncanonical endocrine functions. Here we show that adiponectin as well as several other CTRPs and C1q family proteins bind anionic phospholipids, suggesting that their mode of action is opsonization of cells with specific plasma membrane phospholipid compositions. These findings suggest a new perspective on CTRP family function.
Keywords: adiponectin, adipokine, opsonin, lipid binding
Abstract
Adiponectin (Acrp30) is an adipokine associated with protection from cardiovascular disease, insulin resistance, and inflammation. Although its effects are conventionally attributed to binding Adipor1/2 and T-cadherin, its abundance in circulation, role in ceramide metabolism, and homology to C1q suggest an overlooked role as a lipid-binding protein, possibly generalizable to other C1q/TNF-related proteins (CTRPs) and C1q family members. To investigate this, adiponectin, representative family members, and variants were expressed in Expi293 cells and tested for binding to lipids in liposomes using density centrifugation. Binding to physiological lipids were also analyzed using gradient ultracentrifugation, liquid chromatography-mass spectrometry, and shotgun lipidomics. Interestingly, adiponectin selectively bound several anionic phospholipids and sphingolipids, including phosphatidylserine, ceramide-1-phosphate, glucosylceramide, and sulfatide, via the C1q domain in an oligomerization-dependent fashion. Binding to lipids was observed in liposomes, low-density lipoproteins, cell membranes, and plasma. Other CTRPs and C1q family members (Cbln1, CTRP1, CTRP5, and CTRP13) also bound similar lipids. These findings suggest that adiponectin and CTRPs function not only as hormones, but also as lipid opsonins, as may other C1q family proteins.
Since the discovery of adiponectin as a metabolic hormone secreted by adipocytes, many studies have described the protective effects of adiponectin against cardiovascular disease, insulin resistance, and inflammation in sepsis and metabolic disease (1–8). Many of these discoveries were complemented by studies on the putative adiponectin receptors Adipor1 and Adipor2 (1, 9), which are thought to mediate the myriad effects of adiponectin by inducing 5′ AMP-activated protein kinase (AMPK) signaling, peroxisome proliferator-activated receptor-α transcriptional programs, and receptor-intrinsic ceramidase activity (10). This ability of adiponectin to reduce tissue ceramide levels has been posited to explain its insulin-sensitizing and anti-inflammatory properties (11–13). Adiponectin also has been shown to bind a separate glycosylphosphatidylinositol-linked receptor, T-cadherin (14), which has recently been shown to similarly reduce tissue ceramide levels by triggering the release of exosomes (15). Together, these findings suggest that adiponectin ameliorates metabolic disease by altering tissue lipids, especially ceramides, via receptor binding and activation of downstream intracellular processes. Closely related C1q/TNF-related proteins (CTRPs) have been found to similarly affect metabolism (16, 17).
However, several features of adiponectin biology are not readily consistent with the prevailing view that adiponectin functions solely as a classical hormone. First, adiponectin circulates in blood at micromolar levels (18), in the range of carrier proteins such as insulin-like growth factor-binding proteins and retinol-binding proteins. Other hormones, such as insulin and leptin, circulate in the nanomolar range. Another unexplained feature of adiponectin is its existence as multimers and cleaved forms, which appear to have different physiological effects. Adiponectin spontaneously forms trimers, which assemble into dimers of trimers and 4- and 5-mers of trimers, respectively referred to as low molecular weight (LMW), medium molecular weight (MMW), and high molecular weight (HMW) complexes. Adiponectin can also be cleaved by serum elastases and thrombin between the globular C1q-like head domain and the collagenous tail domain, forming globular adiponectin (gAd). While gAd appears to bind AdipoR1/2 and activate AMPK more strongly than full-length protein (19, 20), levels of HMW multimers are more strongly associated with insulin sensitivity (21, 22), have a longer half-life in circulation (18), and are selectively bound by T-cadherin over globular/trimeric forms (14). The basis of these differences is difficult to explain with the current receptor-ligand paradigm of adiponectin. Finally, several studies have shown an important protective role of adiponectin in clearing apoptotic cells and preventing the development of autoimmunity, supposedly mediated in part by binding to calreticulin (23, 24). How this nonmetabolic role of adiponectin relates to its metabolic role is also unclear.
Interestingly, HMW adiponectin appears almost identical to C1q (25), with which adiponectin and CTRPs share extensive structural and sequence homology (16, 26, 27). While these similarities have been noted, the possibility that these proteins may have similar functions has not been explored. C1q is best known in innate immunity as an opsonin and initiator of the classical complement cascade. Unlike adiponectin, which forms homotrimers, C1q is a heterotrimer of three distinct subunits (C1qA, B, and C), which together bind diverse ligands including the FC domain of various immunoglobulins, C-reactive protein, β-amyloid, carbohydrates, and anionic phospholipids like lipopolysaccharide, cardiolipin, and phosphatidylserine (27). Adiponectin, CTRPs, and C1q family proteins also resemble collectins (e.g., surfactant proteins, mannose-binding lectins) and ficolins, which, like C1q, bind various phospholipids and carbohydrates (28–30).
Given adiponectin’s structural similarity to C1q, involvement in apoptotic cell clearance, unexplained high circulating levels, and relation with tissue ceramides, along with the known effects of ectopic lipid deposition on insulin resistance and metabolic disease (31, 32), we hypothesized that adiponectin may bind and help clear unwanted lipids. Using biochemical approaches, we find that adiponectin binds a range of anionic phospholipids and sphingolipids, including phosphatidylserine, sulfatide, glucosylceramide, and ceramide-1-phosphate, suggesting a modified model of adiponectin function in which its activity as a lipid opsonin promotes lipid clearance. We also find that this property extends to multiple CTRPs and C1q family members, which may influence our understanding of their biology as well.
Results
Adiponectin Binds Multiple Anionic Phospholipids and Sphingolipids.
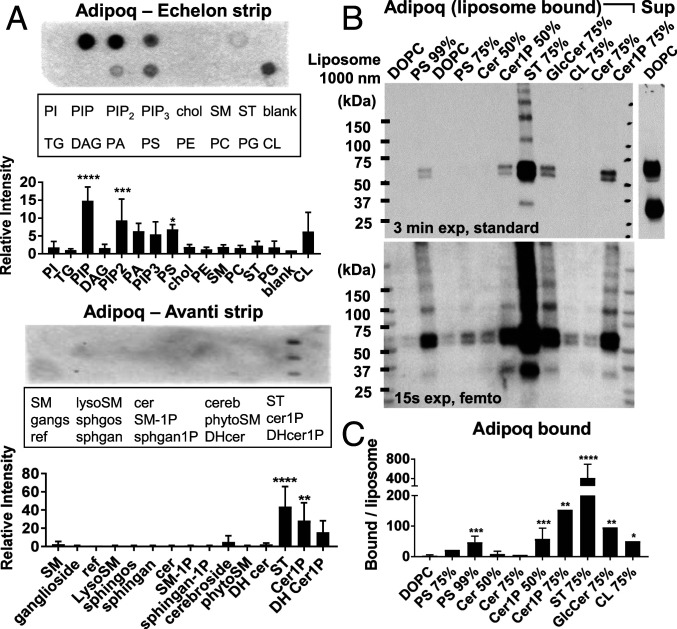
For initial screens, recombinant murine adiponectin cloned with a C-terminal V5-6xHis tag was expressed in Expi293 cells, harvested in cell supernatant, and tested for binding to lipids on nitrocellulose strips using dot blot assays. Remarkably, adiponectin showed binding to a variety of anionic phospholipids and sphingolipids, including phosphatidic acid, phosphatidylserine (PS), phosphatidylinositol phosphate (PIP), phosphatidylinositol 4,5-bisphosphate (PIP2), phosphatidylinositol (3,4,5)-trisphosphate (PIP3), cardiolipin (CL), sulfatide (ST), ceramide-1-phosphate (Cer1P), and dihydroceramide-1-phosphate (DHCer1P) (Fig. 1A). Binding to Cer1P is especially interesting given the relationship of adiponectin with ceramide metabolism (10, 11). Little or no binding was observed to triglycerides, diacylglycerol, cholesterol, neutral sphingolipids and phospholipids—including ceramide and phosphatidylcholine—or single-tailed sphingosines. Lipid binding was lost on deletion of the C1q domain, suggesting a specific protein–lipid interaction. Binding patterns were also identical between unpurified supernatant and adiponectin isolated by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography (SI Appendix, Fig. S1), suggesting direct lipid binding. Purified protein was used for all subsequent experiments.
Fig. 1.
Adiponectin binds multiple anionic phospholipids and sphingolipids. (A) Binding of recombinant murine adiponectin to lipids on commercial lipid strips by dot blot, quantified at right. Density was normalized to a blank or reference spot. n ≥ 3 experiments. (B) Binding of adiponectin to liposome lipids in pulldown assay. Adiponectin in pellet (liposome-bound) and in supernatant (Sup) are shown. The percentage of target lipids in liposomes is indicated above each lane. Two exposures are shown to highlight strong and weak bands. (C) Densiometric quantification of adiponectin V5 signal in liposome pulldown Western blots over multiple experiments. Band densities are normalized to liposome rhodamine fluorescence and averaged over n ≥ 5 experiments. Error bars represent SD from the mean. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Because dot plots are prone to false-positive results from nonphysiological interactions of proteins with randomly oriented desiccated lipids, binding of adiponectin to a subset of lipid targets was validated using a liposome pulldown assay, in which lipids are presented to proteins in a more physiological orientation and environment. In brief, sucrose-loaded liposomes with varying amounts of target lipid in the membrane were blocked, incubated with protein at 37 °C, washed, isolated by density centrifugation, and analyzed for bound protein. Plain DOPC liposomes were used as a negative control. By this method, adiponectin binding was validated to PS, Cer1P, glucosylceramide (GlcCer), CL, and ST, to which binding was remarkably strong (Fig. 1B and SI Appendix, Fig. S2). Of note, a large excess of recombinant protein remained unbound in the supernatant irrespective of liposome type, despite differences in protein binding. Detectable liposome binding also required a high concentration of target lipids: >50% for PS and Cer1P and >10% lipids for ST (SI Appendix, Fig. S3). Binding to dot blot lipids furthermore diminished with decreasing tail length (SI Appendix, Fig. S4A). Interestingly, despite apparent coordination of Ca2+ ions by adiponectin (33), calcium removal or chelation did not affect lipid binding to strips or liposomes (SI Appendix, Fig. S4 B and C).
Lipid Binding Occurs via the C1q Domain of Adiponectin and Is Affected by Mutation of R134.
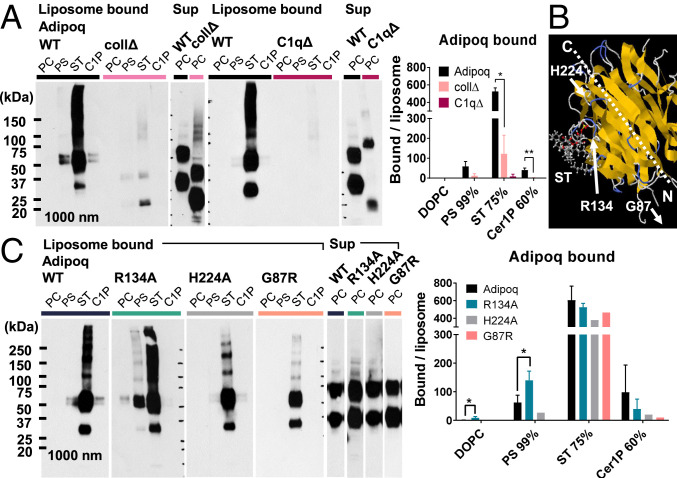
The loss of adiponectin lipid binding in dot blots on deletion of the C1q domain suggests that it mediates the observed lipid binding. Additional experiments with “headless” C1q domain-deleted (C1qΔ) and “tailless” collagenous domain-deleted (collΔ) adiponectin mutants corroborate this idea, showing that the C1q domain is both necessary and sufficient for lipid binding to PS, ST, and Cer1P in dot plots and liposomes (Fig. 2A and SI Appendix, Figs. S1 and S5). However, lipid binding in liposomes was much weaker for collΔ than for WT adiponectin, suggesting that isolated C1q domains bind lipids less avidly than full-length protein.
Fig. 2.
Lipid-binding properties of adiponectin are mediated by the C1q domain and altered by R134A mutation. (A) Binding of WT, collΔ, and C1qΔ adiponectin to liposome lipids by pulldown assay. Liposome ligands are labeled above each lane: PC, 99% DOPC; PS, 99% PS; ST, 75% ST; C1P, 60% Cer1P. Excess unbound protein (Sup) is shown for comparison. Normalized band density is averaged over n ≥ 2 experiments. (B) Position of adiponectin residues selected for site-directed mutagenesis relative to a potential ST-binding site predicted by SwissDock on a single-chain trimer of recombinant globular human adiponectin (Protein Data Bank ID 4DOU) (33, 35). (C) Binding of WT adiponectin and point mutants to liposome lipids, quantified over n ≥ 2 experiments. Error bars represent SD from the mean. **P < 0.01; *P < 0.05.
Given these data and the preference of adiponectin for anionic phospholipids and sphingolipids, we hypothesized that positive residues in the adiponectin C1q head domain may be involved in ligand selectivity. Since previous work implicated the R111 residue of human C1qC (R139 in mouse) in interactions with the serine headgroup of PS (34), we investigated an analogous arginine (R134) in murine adiponectin conserved in humans. This residue also interacted with the negative headgroups of ST and Cer1P when docked onto a model of the adiponectin C1q domain (Protein Data Bank ID 4DOU) (33) using SwissDock (35), yielding conformations with estimated ΔG = −10.24 kcal/mol and −8.75 kcal/mol, respectively. Interestingly, site-directed mutagenesis of the R134 residue to alanine did not abolish lipid binding; however, it did increase the relative affinity of adiponectin to PS and decrease binding to Cer1P (Fig. 2C). In addition, a G87R mutation that is outside the C1q domain but has a weak clinical association with diabetes (21) marginally decreased binding of adiponectin to PS and Cer1P, and an H224A mutation targeting the tip of the C1q protein had no effect on lipid binding.
Oligomerization of Adiponectin Is Required for Lipid Binding.
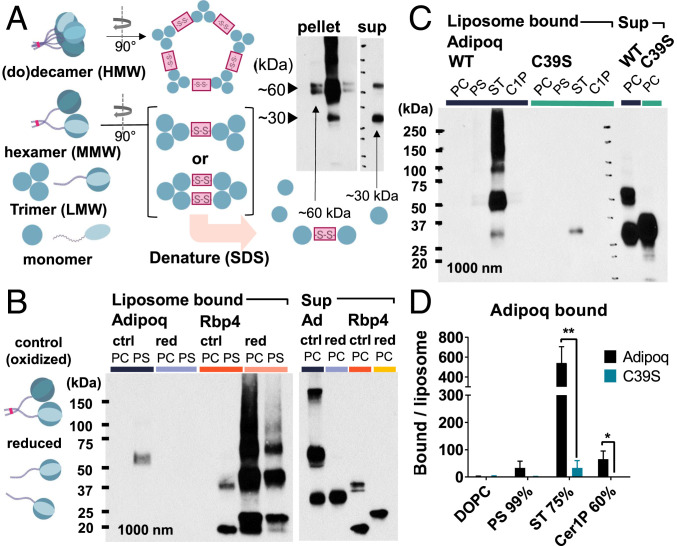
In the adiponectin bands in the pulldown assays, we noted a predominance of the upper 60-kDa band in liposome-bound protein vs. an equivalent or flipped distribution of the 30-kDa and 60-kDa bands in the unbound supernatant (Fig. 3A). As adiponectin monomers run around 30 kDa, the 60-kDa band, which collapses to 30 kDa in reducing conditions, was thought to represent a disulfide-bonded adiponectin dimer. Since adiponectin trimers are held together by hydrophobic interactions (36), we surmised that the dimers must be formed by intertrimer bonds at the N-terminal cysteine in HMW and MMW oligomers, analogous to those seen in C1q complexes (37). Thus, the predominance of the dimer band in liposome-bound fractions may reflect preferential lipid binding by HMW adiponectin oligomers, which is corroborated by the observed differences in lipid binding between WT and collΔ adiponectin and the distinct physiological effects of adiponectin oligomers. Gel filtration also showed evidence of HMW, MMW, and LMW adiponectin complexes in the isolated adiponectin used in the foregoing assays (SI Appendix, Fig. S6).
Fig. 3.
Oligomerization of adiponectin via disulfide bonds is required for lipid binding. (A) Possible intertrimer disulfide bond orientations in adiponectin multimers giving rise to ∼60-kDa and ∼30-kDa bands on nonreducing denaturing SDS-polyacrylamide gel electrophoresis Western blots, which disrupts intratrimer hydrophobic interactions. (B) Liposome binding of adiponectin and RBP4 in normal (oxidized) or reducing conditions. (C and D) Binding of WT and C39S adiponectin to liposome lipids. PC, 99% DOPC; PS, 99% PS; ST, 75% ST; C1P, 60% Cer1P. Excess unbound protein (Sup) is shown at the right. Band density quantified in D normalized to total liposome pellet fluorescence and expressed as mean and SD over n = 3 experiments. **P < 0.01; *P < 0.05.
To assess the contribution of adiponectin oligomerization to liposome lipid binding, we tested the effect of reducing the disulfide bonds by β-mercaptoethanol. Retinol-binding protein 4 (RBP4), a monomeric protein with some binding to PS, was used to control for nonspecific effects of reducing conditions on lipid binding. Consistent with our hypothesis, reduction of adiponectin disulfide bonds abrogated adiponectin binding to PS liposomes and eliminated the 60-kDa and higher MW adiponectin bands (Fig. 3B and SI Appendix, Fig. S7). Conversely, RBP4 showed increased binding to PC and PS liposomes in the reduced state, suggesting that reducing conditions do not indiscriminately reduce lipid binding. To confirm the effect of adiponectin oligomerization on lipid binding in native conditions, site-directed mutagenesis was used to introduce an adiponectin C39S mutation that had been previously shown to abrogate oligomerization (20). As with reduced protein, C39S adiponectin did not form a 60-kDa band, confirming that it arises from oligomerization, and had vastly diminished binding to target liposomes (Fig. 3 C and D). Light binding with ST was still seen, however, indicating that binding was not completely lost.
Adiponectin Binds Lipids in Low-Density Lipoprotein, Cell Membranes, and Plasma.
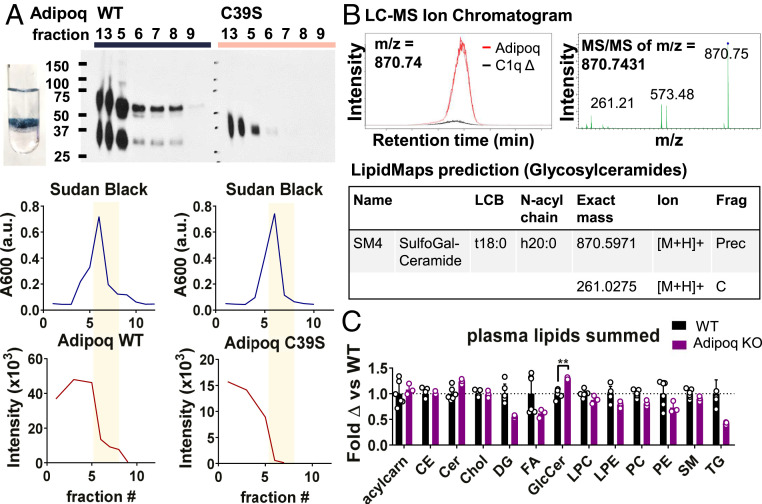
Our experiments thus far investigated binding of adiponectin to selected target lipids at high concentrations in liposomes. While this demonstrates adiponectin lipid binding in physiological conditions, we wondered whether adiponectin also binds physiological substrates with more complex lipid composition and less abundant targets, and also what might be “natural” ligands of adiponectin in the body. Since its discovery, adiponectin has been associated with cardiovascular protection (3) and is known to bind atherosclerotic lesions (38, 39). Given these observations and the apparent lipid-binding property of adiponectin, we tested binding of WT and C39S adiponectin to low-density lipoprotein (LDL) particles. Using an LDL flotation assay, in which blocked LDL is incubated with adiponectin, isolated by gradient flotation, and then assessed for bound protein, we observed a substantial amount of WT adiponectin in the LDL layer that was lost in reduced protein and the C39S mutant, suggesting that adiponectin binds to LDL via LDL lipids (Fig. 4A and SI Appendix, Fig. S8). As in the liposome pulldown assays, most protein remained unbound in the lower fractions.
Fig. 4.
Adiponectin binds physiological lipid substrates. (A) Oligomerization-dependent binding of adiponectin to LDL lipids. (Top) WT or C39S adiponectin in iodixanol gradient fractions in LDL flotation assay detected by Western blot analysis. The photograph shows an LDL band in the gradient after ultracentrifugation. (Bottom) Quantification of Sudan black (Upper) and adiponectin (Lower) bands in fractions. Yellow bars highlight LDL fractions. Data are representative of n = 3 experiments. (B) Adiponectin isolated from cell culture is associated with ST. (Left) LC-MS peak of ligand bound to adiponectin but not C1qΔ or RBP4 controls. (Right) MS/MS of m/z = 870.74. (Bottom) LipidMaps prediction matching the observed fragmentation pattern. (C) Shotgun lipidomics of WT and Adipoq KO mouse plasma. Peak intensity reads, normalized to that of each internal standard, are summed over all identified lipids within a category and graphed as fold change over the WT average. Error bars represent SD from the mean. Open circles are values from individual mice. **P < 0.01; *P < 0.05.
We also tested whether adiponectin binds target lipids in cell membranes using an untargeted liquid chromatography-mass spectrometry (LC-MS) analysis of all lipids associated with adiponectin simply on isolation from transfected cell supernatant. Because Expi293 medium lacks serum or protein additives, any bound lipids presumably are derived from the surrounding cells, potentially from plasma membrane, exosomes, or other membranous debris. Because lipids frequently adsorb nonspecifically to proteins, leading to a high rate of false-positives, we performed LC-MS with three protein isolates: WT adiponectin, C1qΔ adiponectin, and RBP4. Only significant peaks specific to WT adiponectin in both WT-C1qΔ and WT-RBP4 comparisons were considered. Among the peaks meeting this criterion, 11 had ≥2.5-fold enrichment in WT adiponectin in both comparisons, P < 0.01, q < 0.01, and peak maximum intensity ≥20,000 (SI Appendix, Fig. S9 and Table S3 and Dataset S1). While many peaks could not be mapped to a known compound using the XCMS-MRM platform (40), two were mapped to triglyceride or PC, one was mapped to PIP2, one was mapped to a glucosaminyldiphosphoenol, and one was mapped to galactosylceramide or sphingomyelin. The last peak (m/z = 870.7) was further fragmented by MS/MS, given its potential similarity to adiponectin lipid ligands in liposomes, and searched against METLIN MS/MS and LipidMaps libraries (41, 42) (Fig. 4B). Of all possible hits, only one hit obtained using the LipidMaps glycosylceramide prediction tool accounted for two observed fragment peaks, mapping the m/z = 870.7 and 261.2 peaks to the precursor and C-fragment (glycosidic bond) ions of sulfogalactosyl-ceramide (also known as ST). This corroborated the liposome lipid-binding data and demonstrated that adiponectin binds ST in cell membranes.
Given that adiponectin binds physiological lipid substrates, we investigated whether differences in lipid levels could be observed in vivo in WT vs. adiponectin (Adipoq) KO mice, especially in plasma, where adiponectin circulates in high concentrations. To do this, plasma of WT and KO mice were subjected to shotgun lipidomics analysis. While most detected lipids were not adiponectin ligands and were not significantly different between the WT and KO mice, GlcCer species, which bind adiponectin in liposome pulldowns (Fig. 1B and SI Appendix, Fig. S10), were significantly elevated in KO plasma (Fig. 4C and SI Appendix, Fig. S11 and Dataset S2), suggesting that adiponectin lipid binding in vivo reduces levels of lipid ligands. Of note, sulfatides were not included in the shotgun lipidomics panel.
Adiponectin Binds to Dead Cells but Not in a Lipid-Specific Manner.
Since adiponectin was seen to bind PS in liposomes, we tested whether adiponectin also binds PS in necrotic or apoptotic cells using flow cytometry. For this, purified WT adiponectin and C39S adiponectin were used to stain a mixed-viability population of Expi293 cells costained with Annexin V and propidium iodide (PI). While recombinant WT adiponectin was able to clearly bind to dead cells (Annexin V+, PI+) and a fraction of apoptotic cells (Annexin V+, PI−), the same binding pattern was observed with C39S adiponectin, seen earlier to lack PS binding (SI Appendix, Fig. S12). This suggests that adiponectin may bind additional nonlipid ligands exposed on dead cells, such as calreticulin.
Adiponectin Does Not Have Intermembrane Lipid Transfer Activity.
As a lipid-binding protein, adiponectin may function primarily by binding, like an opsonin, or have additional lipid extraction or transfer activity. While adiponectin lacks an obvious hydrophobic pocket that might mediate lipid transfer, we formally assessed the possibility using a lipid transfer assay based on loss of quenching by Forster resonance energy transfer, where transfer of fluorescently labeled lipid ligands from quenched donor liposomes to unlabeled acceptor liposomes is monitored by the appearance of fluorescence over time. From these experiments, adiponectin was found to have no lipid transfer activity for ceramide, Cer1P, or PS (SI Appendix, Fig. S13). Adiponectin also appeared unable to extract or transfer ST, although high spontaneous lipid transfer between donor and acceptor liposomes confounded this assessment.
CTRPs and C1q Family Members Bind a Similar Set of Anionic Phospholipids and Sphingolipids.
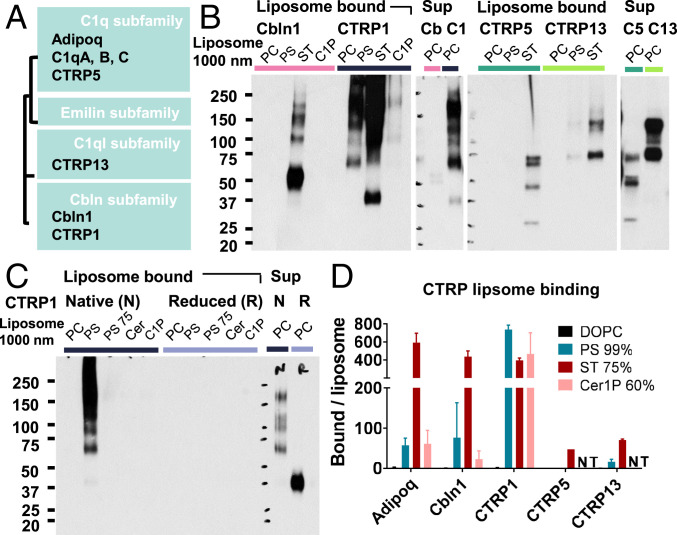
Adiponectin, C1q, and CTRPs belong to the larger C1q family, which consists of C1q, emilin/multimerin, C1q-like, and cerebellin subfamilies (43, 44). Because these proteins share similar structures with conservation of the C1q domain, differing primarily in the N-terminal tail, we wondered whether lipid binding also occurs in CTRPs and C1q family proteins. To test this, liposome pulldown experiments were performed with Expi293-expressed isolates of V5-6xHis–tagged murine Cbln1, CTRP1 (C1qtnf1), CTRP5 (C1qtnf5), and CTRP13 (C1ql3) (Fig. 5A and SI Appendix, Fig. S14). CTRP1, an adipokine that affects glucose and lipid homeostasis (45), and CTRP5, a retinal protein involved in age-related macular degeneration that also inhibits insulin signaling (46, 47), belong to the C1q subfamily with C1q and adiponectin. Cbln1 is a cerebellin that acts as a synaptic organizer (48, 49), and CTRP13 is a C1q-like subfamily protein expressed in brain cortex and retina that regulates synaptogenesis and activates AMPK (50, 51). Expression levels of each protein in Expi293 cells differed quite dramatically, which limited the availability of CTRP5 and CTRP13 to test Cer1P binding. Nonetheless, all CTRPs and C1q family proteins tested showed binding to PS, Cer1P, and ST that, as with adiponectin, depended on oligomerization mediated by disulfide bonds (Fig. 5B and C and SI Appendix, Fig. S15). Qualitatively, affinities of each protein to various lipids were similar to those of adiponectin, but some variability was observed among family members. CTRP1 had stronger binding to PS relative to ST, reminiscent of the R134A adiponectin mutant. Cbln1 also appeared to have a higher affinity for ST, as evidenced by the strong binding to ST liposomes despite lower levels of protein in the supernatant.
Fig. 5.
Other CTRPs and C1q family proteins also bind lipids in liposomes with similar but distinct affinity patterns. (A) Simplified tree diagram showing the relationships between CTRPs and C1q family members cloned for this study. (B) Binding of Cbln1, CTRP1, CTRP5, and CTRP13 to liposome lipids. PC, 99% DOPC; PS, 99% PS; PS 75, 75% PS; ST, 75% ST; C1P, 60% Cer1P; Cer, 60% ceramide. (C) Loss of lipid binding of CTRP1 to target liposome lipids in reducing conditions. (D) Quantification of CTRP lipid binding in liposome pulldown blots. Band density is normalized to liposome rhodamine fluorescence. NT, not tested. Bars represent mean and SD over n ≥ 2 experiments.
In sequence alignments of human and mouse adiponectin and C1qC, Cbln1, CTRP1, CTRP5, and CTRP13, differences in lipid ligand affinity approximately correlated with differences in basic residues in the first C-terminal–directed solvent-exposed loop of each protein’s C1q domain (SI Appendix, Fig. S16). Specifically, the mAdipoq R134 residue, conserved across human and mouse adiponectin and C1qC (green triangle) and partially conserved in Cbln1, CTRP5, and CTRP13, is lost in CTRP1, making the C1 loop of CTRP1 electrostatically similar to the R134A adiponectin mutant and explaining their similar lipid affinity patterns. Additional basic residues further upstream in the C1 segment likely maintain their overall preference for anionic lipids, however. Because the solvent-exposed loops are less critical for structural integrity of the C1q domain, the variability there likely contributed to the specificity of different C1q family proteins for various C-terminal ligands and other special properties; for instance, the (Y/F)FGGWP sequence in the C3 loop of CTRP5 forms a unique hydrophobic patch (52). Of note, however, all family members show strong conservation of the mAdipoq C39 residue (blue triangle), suggesting that oligomerization is a key unifying functional feature.
Discussion
In this study, we have demonstrated that adiponectin selectively binds anionic phospholipids and sphingolipids, including PS, Cer1P, GlcCer, and ST, in synthetic liposomes, LDL, biological cell membranes, and plasma. The C1q domain was necessary and sufficient for binding, but protein oligomerization via disulfide bonds at C39 in the collagenous domain was also required. Properties of the amino acids in the C-terminal solvent-exposed loops also modulated the selectivity for lipid ligands, as an R134A mutation altered the relative affinity of adiponectin for Cer1P and PS. More generally, we found that other representative CTRPs and C1q family members also bind to similar lipid ligands in a likewise oligomerization-dependent manner, and that natural variability in the first C-terminal solvent-exposed loop corresponds to different lipid affinity patterns.
The ability of adiponectin to bind anionic phospholipids and sphingolipids is consistent with reports of C1q binding to CL and PS (27, 34), as well as with the known carbohydrate- and phospholipid-binding properties of collectins and ficolins (28–30). In C1q, collectins, and ficolins, collectively referred to as “defense collagens,” ligand binding mediates opsonization of microbes. By analogy, lipid binding in adiponectin may facilitate opsonization of extracellular lipids like LDL and other membranous debris enriched in target anionic phospholipids and sphingolipids. While adiponectin-mediated lipid uptake was not directly shown (given confounding C1q secretion by phagocytes), it is likely, given the stable binding of adiponectin to target liposomes through multiple washes, lack of lipid transfer activity, and putative orientation of bound adiponectin with C1q heads toward target lipids and collagen tails outward, available for recognition by phagocyte collagen and scavenger receptors. It is also supported by the accumulation of GlcCer in Adipoq KO vs. WT plasma, as well as the previously reported evidence that adiponectin and C1q promote apoptotic cell uptake (24).
Of note, the requirement for high ligand density and oligomerization for lipid binding by adiponectin and CTRP/C1q proteins suggests that individual protein–lipid interactions are weak, and that multiple interactions are required for sufficient avidity. This may actually serve to regulate the extent and location of lipid binding by adiponectin and C1q proteins. In most membranes, target lipid abundance is low (∼3 to 10% for PS and ST and <0.1% for Cer1P in some membranes) (53, 54). Thus, binding may occur only at patches of high-density target lipids assembled in lipoproteins or membrane rafts or selectively exposed during cell activation or apoptosis. The common chemical properties of the observed target lipids also may mark lipids to be opsonized; PS is known to mark apoptotic cells, CL is associated with necrosis, and ST is released from damaged myelin and senescent fibroblasts (55, 56). Indeed, the macrophage scavenger receptor Trem2 binds a similar set of anionic and zwitterionic “damage-associated” lipids, including PS and ST, mediating amyloid accumulation in brain and lipid homeostasis in adipose tissue (55, 57).
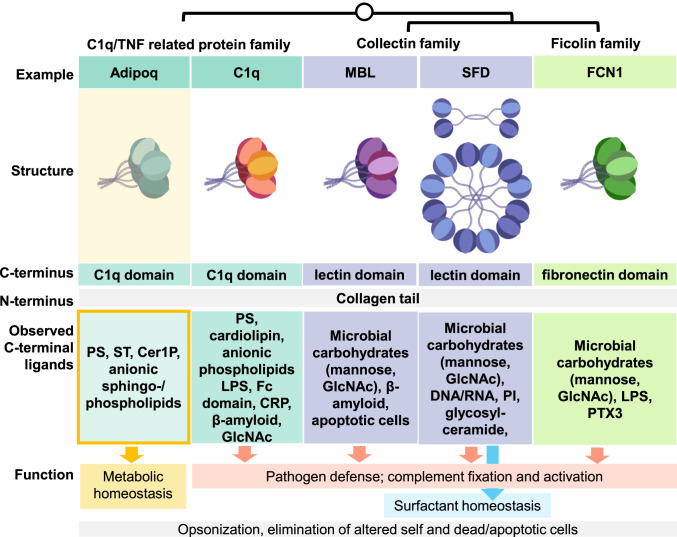
While opsonization of lipids followed by phagocytosis is likely a common function of C1q, adiponectin, CTRPs, collectins, and ficolins, each of these proteins may also have specific functions mediated by different downstream binding partners (Fig. 6). For instance, C1q, mannose-binding lectin, and ficolins fix complement via C1r/s or MASP serine proteases (58). Adiponectin does not appear to activate complement in the native state (59) but instead exerts metabolic effects, potentially by engaging Adipor1/2 and promoting uptake and recycling of lipid cargo via Appl- and Cav3-mediated endocytosis and receptor-activated ceramidase activity. Although the enzymatic activity of the Adipor1/2 receptors has been characterized for ceramides, other phospholipids and sphingolipids could be similarly hydrolyzed if able to enter the active site. For lipids with large or anionic headgroups, this could be facilitated by coupled phosphatase or glycosidase activity on endocytosis; however, this assumes that lipid binding does not impede receptor activation and that adiponectin actually binds Adipor1/2, which is generally accepted but has been debated (60). This and other specific functions may be key in differentiating the biological roles of adiponectin and CTRPs, which have roles in metabolism (61), from other C1q proteins. The specific functions may also compete; for instance, adiponectin lipid binding may reduce the substrates available to bind C1q or other complement fixing proteins, which may promote inflammation if a critical amount of C3b is deposited (62).
Fig. 6.
Binding of adiponectin to anionic sphingolipids and phospholipids fits known structural and ligand-binding features of C1q, collectins, and ficolins but fills a functional niche in metabolism. Similarities and differences between proteins in terms of oligomeric structure, C- and N-terminal domains, C-terminal binding targets, and currently understood function are shown. Possible shared features, like opsonization, are in gray; possible unique features are in colored boxes. Adipoq, adiponectin; MBL, mannose-binding lectin; SFD, surfactant protein D; FCN1, ficolin 1.
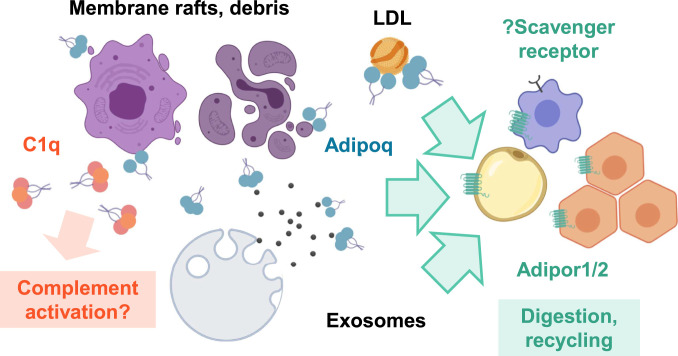
Thus, our working model is that adiponectin HMW/MMW oligomers bind anionic phospholipids and sphingolipids enriched in LDL, lipid rafts, or other membranous debris and mediate their delivery to phagocytes recognizing the collagenous domain or Adipor1/2-expressing cells for recycling by receptor-intrinsic hydrolysis, removing those lipids from vessels and extracellular tissue spaces where they can cause insulin resistance, inflammation, and/or inappropriate complement fixation (Fig. 7). LMW/globular adiponectin may complement this opsonization activity by promoting receptor activation without lipid delivery, priming the metabolic state of the receiving tissue for lipid oxidation. This model is consistent with the known metabolic effects of adiponectin and suggests a more complete mechanism for achieving those effects, for instance, by targeting ectopic lipids to macrophages and AdipoR1/2-expressing cells while activating AMPK to prevent insulin resistance, or by clearing LDL while directly limiting inflammation in macrophages to prevent atherosclerosis (3, 63). Our findings also explain the association of HMW vs. LMW adiponectin with protection against type 2 diabetes and metabolic syndrome (21, 22), as well as the observed induction of and binding to exosomes by adiponectin (15, 64). Furthermore, they have interesting implications for the mechanism of action of other CTRPs, which circulate at 100- to 1,000-fold lower levels than adiponectin but still may have important local effects, and the contribution of lipid binding to the function of other C1q family proteins. Continuing research in this area will be instrumental in building a clearer understanding of extracellular lipid handling and its role in organismal health and disease.
Fig. 7.
Adiponectin lipid binding may promote lipid clearance and limit complement-mediated inflammation. Adiponectin, C1q, and CTRP/C1q family proteins bind similar target lipids, including PS, Cer1P, GlcCer, and ST potentially found in lipoproteins, exosomes, and membranous debris. Unlike C1q, however, adiponectin does not generally fix complement and may mediate clearance by binding AdipoR1/2. The collagenous N-terminal domain may also permit uptake via macrophage scavenger receptors. Thus, adiponectin may promote clearance and recycling of lipid debris and also dampen activation of complement by competing for overlapping lipid targets.
Materials and Methods
Additional information on the materials and methods used in this study available in SI Appendix.
Protein Expression and Isolation.
cDNAs for murine adiponectin, CTRPs, and C1q family proteins were cloned into pcDNA 3.1 vectors with a C-terminal V5 6xHis tag and expressed using the Expi293 mammalian cell expression system. On day 5 posttransfection, protein was harvested, filtered, and isolated using Ni-NTA affinity purification in HNC isolation buffer (25 mM Hepes, 150 mM NaCl, and 1 mM CaCl2, pH 7.4). Protein was stored at 4 °C until use in experiments.
Dot Blot.
Lipids spotted on nitrocellulose strips were obtained commercially from Echelon Biosciences and Avanti Polar Lipids or made in-house with lipids purchased from Avanti Polar Lipids. Blots were blocked with 3% fatty acid-free BSA in TBST, incubated with protein overnight, washed, probed with rabbit anti-6×His primary antibody followed by goat anti-rabbit peroxidase-conjugated secondary antibody, and then developed using enhanced chemiluminescence substrate.
Liposome Pulldown Assay.
Here 1,000-nm sucrose-containing liposomes of varying lipid composition with 1% rhodamine-PE for visualization were synthesized and then incubated at 0.5 mM with 10 μg/mL isolated recombinant protein in the presence of 10 mg/mL fatty acid-free BSA block in HNC buffer at 37 °C for 1 h. For use in experiments, reduced protein was prepared by adding β-mercaptoethanol to a final concentration of 2% to recombinant protein before addition to lipids. After binding, liposomes were washed twice with HNC (with centrifugation at 15,000 rpm for 15 min) and then lysed with Triton X-100 (final concentration 1%) in a black opaque 96-well plate. Rhodamine fluorescence was measured at 553/627 nm, and samples were denatured with 5× nonreducing sodium dodecyl sulfate (SDS) sample buffer, boiled, and analyzed for V5 by Western blot analysis. For each protein in each experiment, liposome-bound samples were run alongside excess unbound protein in supernatant, diluted 1:20, for comparison.
LDL Flotation Assay.
Human LDL (final concentration 1 mg/mL) was incubated with 10 μg/mL isolated recombinant protein in the presence of 10 mg/mL fatty acid-free BSA block and 1 mg/mL Sudan black in HNC buffer at 37 °C for 1 h. Reduced protein was prepared as described above. After incubation, the 50-μL LDL-protein mixture was added to 280 μL of OptiPrep iodixanol solution (STEMCELL Technologies), then overlaid with an iodixanol-HNC gradient (1×, 70%; 2×, 36%; 2×, 0% OptiPrep). Pairs of gradients were then ultracentrifuged at 50,000 rpm for 20 h at 4 °C, and fractions were collected by tube puncture. Absorbance at 600 nm was measured for each fraction, then selected fractions were collected into 5× nonreducing SDS sample buffer and analyzed for V5 by Western blot analysis.
Protein-Bound Lipid Analysis by LC-MS and MS/MS.
Expressed proteins concentrated to 80 to 100 μL were capped under N2 atmosphere and then snap- frozen. Subsequent LC-MS and MS/MS analyses were performed by the Scripps Center for Metabolomics and Mass Spectrometry. Samples were extracted with 50:50 methanol:acetonitrile and subjected to LC-MS (Bruker Impact II Q-TOF coupled to an Agilent 1200 LC stack). Positive ions were analyzed using XCMS (65). MS/MS peaks were searched against the METLIN and LipidMaps databases, including the glycerophospholipid search tool (www.lipidmaps.org/tools/ms/glycosylcer_gen.html).
Plasma Collection and Lipidomics.
Whole blood was harvested by retro-orbital collection from 8- to 10-wk-old male WT C57B6/J (The Jackson Laboratory; stock no. 000644) and Adipoq KO mice (B6;129-Adipoqtm1Chan/J; The Jackson Laboratory; stock no. 008195) housed in a specific pathogen-free facility with a regulated 12-h light/dark cycle and ad libitum access to standard chow and water. Plasma was isolated using BD lithium-heparin coated plasma separator tubes and stored at −20 °C. Lipidomics was performed by the West Coast Metabolomics Center at University of California Davis. Sample extraction was performed in methyl tertiary-butyl ether with the addition of internal standards, followed by ultra-high-performance liquid chromatography on a Waters CSH column, interfaced to a quadrupole time-of-flight mass spectrometer. Data were collected in positive and negative ion modes and analyzed using MassHunter (Agilent).
Quantification and Statistical Analysis.
Films and images were quantified using FIJI. For liposome pulldown assays, band densities were normalized to total pellet rhodamine fluorescence. Means, SDs, and statistical significance were calculated using GraphPad Prism. Student’s t test (two-tailed) was used for single comparisons between groups, and ANOVA with Dunnett’s correction for multiple testing was used when multiple comparisons were made with the same control. P values <0.05 were considered significant.
Data Availability Statement.
All data necessary for replication of these experiments are provided in the main text and SI Appendix, including lipidomics datasets in SI Appendix and Datasets S1 and S2. No original code was written for analysis of the data; algorithms and tools used for lipidomics analysis have been published previously as referenced.
Supplementary Material
Acknowledgments
We thank Daniel Okin and all other current and former members of the R.M. laboratory for discussions. Special thanks also to the De Camilli lab, Cresswell laboratory, and the Yale Department of Immunobiology for their contributions of ideas, comments, tools, and reagents. This paper is based on a dissertation by J.J.Y. submitted to fulfill in part the requirements for a doctoral degree at Yale University. Work in the R.M. laboratory was supported by the HHMI, the Blavatnik Family Foundation, and the NIH (Grant 1R01 AI144152-01). J.J.Y. is supported by the Yale School of Medicine Medical Scientist Training Program (GM007205) and in part by NIH Training Grant T32AI07019 (Yale Interdisciplinary Immunobiology Training Program). X.B. is supported by a Human Frontier Science Program Long-Term Fellowship. J.L. is supported by a Jane Coffin Childs Fellowship and a Human Frontiers Science Program Long-Term Fellowship.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922270117/-/DCSupplemental.
References
- 1.Kadowaki T. et al., Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Invest. 116, 1784–1792 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z. V., Scherer P. E., Adiponectin, the past two decades. J. Mol. Cell Biol. 8, 93–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto Y. et al., Adiponectin reduces atherosclerosis in apolipoprotein E-deficient mice. Circulation 106, 2767–2770 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Lindberg S. et al., Adiponectin, type 2 diabetes, and cardiovascular risk. Eur. J. Prev. Cardiol. 22, 276–283 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Konter J. M. et al., Adiponectin attenuates lipopolysaccharide-induced acute lung injury through suppression of endothelial cell activation. J. Immunol. 188, 854–863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao L. et al., Adiponectin cardioprotection after myocardial ischemia/reperfusion involves the reduction of oxidative/nitrative stress. Circulation 115, 1408–1416 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Vachharajani V. et al., Adiponectin-deficiency exaggerates sepsis-induced microvascular dysfunction in the mouse brain. Obesity (Silver Spring) 20, 498–504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Buechler N. L., Yoza B. K., McCall C. E., Vachharajani V., Adiponectin treatment attenuates inflammatory response during early sepsis in obese mice. J. Inflamm. Res. 9, 167–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamauchi T. et al., Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat. Med. 13, 332–339 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Vasiliauskaité-Brooks I. et al., Structural insights into adiponectin receptors suggest ceramidase activity. Nature 544, 120–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holland W. L. L. et al., An FGF21-adiponectin-ceramide axis controls energy expenditure and insulin action in mice. Cell Metab. 17, 790–797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia J. Y. et al., Targeted induction of ceramide degradation leads to improved systemic metabolism and reduced hepatic steatosis. Cell Metab. 22, 266–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y. et al., Adiponectin inhibits tumor necrosis factor-α-induced vascular inflammatory response via caveolin-mediated ceramidase recruitment and activation. Circ. Res. 114, 792–805 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hug C. et al., T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc. Natl. Acad. Sci. U.S.A. 101, 10308–10313 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obata Y. et al., Adiponectin/T-cadherin system enhances exosome biogenesis and decreases cellular ceramides by exosomal release. JCI Insight 3, e99680 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong G. W., Wang J., Hug C., Tsao T.-S., Lodish H. F., A family of Acrp30/adiponectin structural and functional paralogs. Proc. Natl. Acad. Sci. U.S.A. 101, 10302–10307 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong G. W. et al., Molecular, biochemical, and functional characterizations of C1q/TNF family members: Adipose-tissue-selective expression patterns, regulation by PPAR-γ agonist, cysteine-mediated oligomerizations, combinatorial associations, and metabolic functions. Biochem. J. 416, 161–177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halberg N. et al., Systemic fate of the adipocyte-derived factor adiponectin. Diabetes 58, 1961–1970 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamauchi T. et al., Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423, 762–769 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Pajvani U. B. et al., Structure-function studies of the adipocyte-secreted hormone Acrp30/adiponectin. Implications for metabolic regulation and bioactivity. J. Biol. Chem. 278, 9073–9085 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Waki H. et al., Impaired multimerization of human adiponectin mutants associated with diabetes. Molecular structure and multimer formation of adiponectin. J. Biol. Chem. 278, 40352–40363 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Liu Y. et al., Total and high molecular weight but not trimeric or hexameric forms of adiponectin correlate with markers of the metabolic syndrome and liver injury in Thai subjects. J. Clin. Endocrinol. Metab. 92, 4313–4318 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Takemura Y. et al., Adiponectin modulates inflammatory reactions via calreticulin receptor-dependent clearance of early apoptotic bodies. J. Clin. Invest. 117, 375–386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galvan M. D., Hulsebus H., Heitker T., Zeng E., Bohlson S. S., Complement protein C1q and adiponectin stimulate Mer tyrosine kinase-dependent engulfment of apoptotic cells through a shared pathway. J. Innate Immun. 6, 780–792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radjainia M., Wang Y., Mitra A. K., Structural polymorphism of oligomeric adiponectin visualized by electron microscopy. J. Mol. Biol. 381, 419–430 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Ressl S. et al., Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure 23, 688–699 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Kishore U. et al., C1q and tumor necrosis factor superfamily: Modularity and versatility. Trends Immunol. 25, 551–561 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Wesener D. A., Dugan A., Kiessling L. L., Recognition of microbial glycans by soluble human lectins. Curr. Opin. Struct. Biol. 44, 168–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endo Y., Matsushita M., Fujita T., The role of ficolins in the lectin pathway of innate immunity. Int. J. Biochem. Cell Biol. 43, 705–712 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Seaton B. A. et al., Review: Structural determinants of pattern recognition by lung collectins. Innate Immun. 16, 143–150 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Grundy S. M., Overnutrition, ectopic lipid and the metabolic syndrome. J. Investig. Med. 64, 1082–1086 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Chaurasia B., Summers S. A., Ceramides—Lipotoxic inducers of metabolic disorders. Trends Endocrinol. Metab. 26, 538–550 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Min X. et al., Crystal structure of a single-chain trimer of human adiponectin globular domain. FEBS Lett. 586, 912–917 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Païdassi H. et al., C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol. 180, 2329–2338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grosdidier A., Zoete V., Michielin O., SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270-7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapiro L., Scherer P. E., The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8, 335–338 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Lu J., Kishore U., C1 complex: An adaptable proteolytic module for complement and non-complement functions. Front. Immunol. 8, 592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almer G. et al., Globular domain of adiponectin: Promising target molecule for detection of atherosclerotic lesions. Biologics 5, 95–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori T. et al., Ultrastructural localization of adiponectin protein in vasculature of normal and atherosclerotic mice. Sci. Rep. 4, 4895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domingo-Almenara X. et al., XCMS-MRM and METLIN-MRM: A cloud library and public resource for targeted analysis of small molecules. Nat. Methods 15, 681–684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guijas C. et al., METLIN: A technology platform for identifying knowns and unknowns. Anal. Chem. 90, 3156–3164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sud M. et al., LMSD: LIPID MAPS structure database. Nucleic Acids Res. 35, D527–D532 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tom Tang Y. et al., The complete complement of C1q-domain-containing proteins in Homo sapiens. Genomics 86, 100–111 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Matsuda K., Synapse organization and modulation via C1q family proteins and their receptors in the central nervous system. Neurosci. Res. 116, 46–53 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez S. et al., Loss of CTRP1 disrupts glucose and lipid homeostasis. Am. J. Physiol. Endocrinol. Metab. 311, E678–E697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chavali V. R. M., Sommer J. R., Petters R. M., Ayyagari R., Identification of a promoter for the human C1Q-tumor necrosis factor-related protein-5 gene associated with late-onset retinal degeneration. Invest. Ophthalmol. Vis. Sci. 51, 5499–5507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lei X. et al., Loss of CTRP5 improves insulin action and hepatic steatosis. Am. J. Physiol. Endocrinol. Metab. 310, E1036–E1052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirai H. et al., Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat. Neurosci. 8, 1534–1541 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Elegheert J. et al., Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinelli D. C. et al., Expression of C1ql3 in discrete neuronal populations controls efferent synapse numbers and diverse behaviors. Neuron 91, 1034–1051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei Z., Peterson J. M., Wong G. W., Metabolic regulation by C1q/TNF-related protein-13 (CTRP13): Activation OF AMP-activated protein kinase and suppression of fatty acid-induced JNK signaling. J. Biol. Chem. 286, 15652–15665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu X., Palczewski K., Crystal structure of the globular domain of C1QTNF5: Implications for late-onset retinal macular degeneration. J. Struct. Biol. 180, 439–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norton W. T., Poduslo S. E., Myelination in rat brain: Changes in myelin composition during brain maturation. J. Neurochem. 21, 759–773 (1973). [DOI] [PubMed] [Google Scholar]
- 54.Shaner R. L. et al., Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50, 1692–1707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y. et al., TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buratta S. et al., Extracellular vesicles released by fibroblasts undergoing H-Ras-induced senescence show changes in lipid profile. PLoS One 12, e0188840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaitin D. A. et al., Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. Cell 178, 686–698.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallis R., Mitchell D. A., Schmid R., Schwaeble W. J., Keeble A. H., Paths reunited: Initiation of the classical and lectin pathways of complement activation. Immunobiology 215, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peake P., Shen Y., Factor H binds to the N-terminus of adiponectin and modulates complement activation. Biochem. Biophys. Res. Commun. 397, 361–366 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Kita S., Fukuda S., Maeda N., Shimomura I., Native adiponectin in serum binds to mammalian cells expressing T-cadherin, but not AdipoRs or calreticulin. eLife 8, e48675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seldin M. M., Tan S. Y., Wong G. W., Metabolic function of the CTRP family of hormones. Rev. Endocr. Metab. Disord. 15, 111–123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giles B. M., Boackle S. A., Linking complement and anti-dsDNA antibodies in the pathogenesis of systemic lupus erythematosus. Immunol. Res. 55, 10–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Folco E. J., Rocha V. Z., López-Ilasaca M., Libby P., Adiponectin inhibits pro-inflammatory signaling in human macrophages independent of interleukin-10. J. Biol. Chem. 284, 25569–25575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phoonsawat W., Aoki-Yoshida A., Tsuruta T., Sonoyama K., Adiponectin is partially associated with exosomes in mouse serum. Biochem. Biophys. Res. Commun. 448, 261–266 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Tautenhahn R., Patti G. J., Rinehart D., Siuzdak G., XCMS online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary for replication of these experiments are provided in the main text and SI Appendix, including lipidomics datasets in SI Appendix and Datasets S1 and S2. No original code was written for analysis of the data; algorithms and tools used for lipidomics analysis have been published previously as referenced.