Significance
RAS oncoproteins have long been considered among the most elusive drug targets in cancer research. At issue is the lack of accessible drug-binding sites and the extreme affinity for GTP. Covalent inhibitors against the KRAS G12C mutant have shown early clinical promise; however, targeting the other oncogenic RAS mutants across three RAS isoforms has proven challenging. Inhibition of activated wild-type RAS in the absence of canonical RAS mutations is also highly desirable in certain tumors. Here, we demonstrate delivery of an extremely potent pan-RAS and RAP1-cleaving enzyme in therapeutic quantities to specific receptor-bearing cells in vitro and in vivo. We aim to advance this approach to engineer the first targeted pan-RAS inhibitor for cancer therapy.
Keywords: RAS, RRSP, xenograft, cancer, recombinant toxins
Abstract
Despite nearly four decades of effort, broad inhibition of oncogenic RAS using small-molecule approaches has proven to be a major challenge. Here we describe the development of a pan-RAS biologic inhibitor composed of the RAS-RAP1–specific endopeptidase fused to the protein delivery machinery of diphtheria toxin. We show that this engineered chimeric toxin irreversibly cleaves and inactivates intracellular RAS at low picomolar concentrations terminating downstream signaling in receptor-bearing cells. Furthermore, we demonstrate in vivo target engagement and reduction of tumor burden in three mouse xenograft models driven by either wild-type or mutant RAS. Intracellular delivery of a potent anti-RAS biologic through a receptor-mediated mechanism represents a promising approach to developing RAS therapeutics against a broad array of cancers.
More than one-third of all human cancers harbor activating mutations in RAS oncogenes. Among the major isoforms, KRAS is the most frequently mutated oncogene, found in nearly 25% of malignancies and 85% of RAS-driven cancers (1–3). Notably, three of the four deadliest cancers (pancreatic, colorectal, and lung) exhibit a high frequency of KRAS mutations (2, 3). Moreover, NRAS and HRAS are also known oncogenic drivers in other neoplasms (2). Activating point mutations in RAS genes impair the intrinsic capacity of RAS proteins to hydrolyze GTP, thus locking them in a constitutively activated GTP-bound state. This leads to constitutive activation of downstream transduction-signaling networks, such as the RAF/MEK/ERK (MAPK) axis, which drives survival and uncontrolled proliferation (4–6). Even in the absence of gain-of-function mutations, RAS genes still play a major role in tumorigenesis due to hyperactivation of RAS-signaling pathways via overexpression of upstream receptor tyrosine kinases (RTKs) and/or amplification of wild-type RAS, such as in head and neck squamous cell carcinoma (7), esophageal and gastric cancers (8), ovarian adenocarcinoma (9), and triple-negative breast cancer (TNBC) (10, 11).
Due to their major role in a wide spectrum of cancers, RAS proteins have become a primary target for drug discovery, and extensive effort has been directed to the development of selective RAS inhibitors (12). However, the high affinity for GTP and the absence of drug-accessible binding pockets have complicated efforts for decades, earning RAS the moniker “undruggable” (12–14). Nevertheless, recent success has been achieved by selectively targeting KRAS G12C with small molecules that covalently bind to Cys12 in the KRAS Switch-II pocket (15–17), and clinical trials are underway to validate their effectiveness (18–20). However, these pharmacophores are specific for KRAS G12C and cannot be expanded to other mutants. Furthermore, KRASG12C mutations account for only ∼11% of all KRAS mutations in cancer (21), detected mainly in lung (14%), colorectal (5%), and pancreatic (1 to 3%) cancers (13, 15). Therefore, there remains an urgent need for a broadly applicable pan-RAS inhibitor for use against all RAS-driven tumors, either mutation-dependent or -independent.
Recently, we discovered a RAS/RAP1 specific endopeptidase (RRSP) from Vibrio vulnificus that site-specifically cleaves RAS and its close homolog RAP1 between residues Y32 and D33 within the Switch I, a region crucial to RAS-mediated signal transduction (22). RRSP is highly specific for RAS and RAP1 and does not cleave other closely related GTPases (23). Importantly, RRSP cleaves all three major RAS isoforms, as well as oncogenic RAS with mutations at position 12, 13, and 61 (22). RRSP also targets both active (GTP-bound) and inactive (GDP-bound) RAS, resulting in destruction of the entire cellular RAS pool (23). By proteolytically cleaving the Switch I loop, RRSP prevents RAS from undergoing GDP-GTP exchange and binding the downstream effector kinase RAF, ultimately terminating ERK signaling in cells (24).
Assessing the therapeutic potential of RRSP, however, is precluded by the fact that RRSP is a 56-kDa domain of a larger protein toxin that alone does not readily diffuse across biological membranes. Recently, we demonstrated that the translocation machinery of diphtheria toxin (DT) can be engineered to deliver a broad diversity of passenger proteins into target cells (25, 26). In this study, we exploit the receptor-targeting and membrane translocation properties of DT to deliver RRSP into targeted cancer cells.
We created an engineered chimeric toxin that uses the DT translocation system and show that RRSP potently and irreversibly destroys RAS. We demonstrate the anticancer properties of this engineered chimeric toxin both in vitro and in vivo, providing proof-of-concept for the therapeutic development of RRSP as a pan-RAS inhibitor.
Results
Engineered Diphtheria Toxin Binding and Translocation Domain DTB Efficiently Delivers RRSP into Cells.
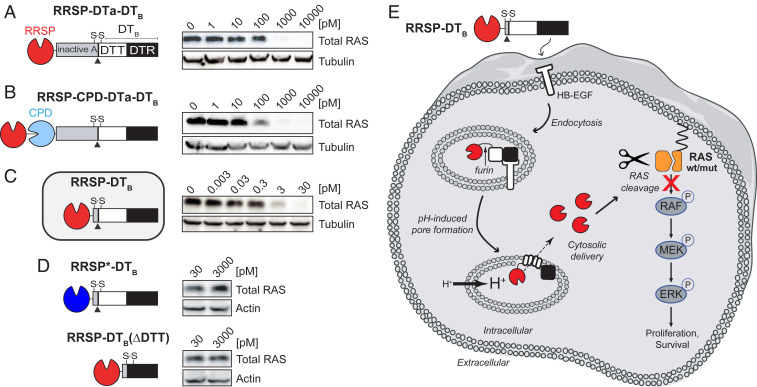
DT consists of a catalytically active A fragment (DTA) and a B fragment (DTB) that includes both a receptor-binding domain (DTR) and a translocation domain (DTT). DTB alone can bind its surface receptor, the heparin-binding epidermal growth factor-like growth factor (HB-EGF), and transfer a wide range of protein cargos to the cell cytosol via a receptor-mediated endocytic mechanism (25). The introduction of two amino acid substitutions (K51E and E148K) into DTA resulted in a nontoxic, catalytically inactive A fragment, herein referred to as DTa. To examine RRSP intracellular delivery, we expressed RRSP fused at the amino terminus of DTa with an intervening (G4S)2 linker, a design previously used to deliver other protein cargos (25, 26). Western blotting with the RAS10 pan-RAS antibody showed that treatment with RRSP-(G4S)2-DTa-DTB resulted in depletion of RAS in HCT-116 (KRASG13D) cells at 1 nM (Fig. 1A and SI Appendix, Fig. S1A).
Fig. 1.
Engineered diphtheria toxin delivery machinery enables intracellular delivery of RRSP. Schematic of chimeric fusions of RRSP to (A) the amino terminus of nontoxic, full-length diphtheria toxin (RRSP-DTa-DTT-DTR), (B) to autoprocessing cysteine protease domain (CPD) of MARTX toxin from V. vulnificus (RRSP-CPD-DTa-DTT-DTR), and (C) to DTT-DTR without the DTa domain (RRSP-DTT-DTR or RRSP-DTB) and corresponding immunoblots against total RAS of lysates prepared from HCT-116 cells treated with RRSP-DT variants for 24 h. (D) Immunoblots showing that the catalytically dead RRSP*-DTB mutant and the translocation-deficient control (RRSP-DTB[ΔDTT]) do not affect RAS levels in HCT-116 cells. (E) Schematic diagram illustrating transport and intracellular trafficking of RRSP fused to the translocation T and receptor R binding domains of diphtheria toxin fragment B (DTB) via a receptor-mediated endocytic mechanism. Once in the cytosol, RRSP cleaves RAS resulting in down-regulation of the MAPK/ERK-signaling cascade. Empty cell and vesicles images credit: Servier Medical Art (https://smart.servier.com), which is licensed under CC BY 3.0.
An intrinsic feature of the first-generation design is that RRSP is codelivered with the DTa domain. To generate chimeras that would deliver RRSP alone into cells without DTa, we added the autoprocessing cysteine protease domain (CPD) from V. vulnificus MARTX toxin that autocleaves and thereby releases its toxin effectors (i.e., RRSP) upon binding inositol hexakisphosphate in the host cytosol (27). When added to cells, RRSP-CPD-DTa-DTB improved RAS cleavage (partial inactivation at 100 pM) (Fig. 1B and SI Appendix, Fig. S1B). In the third iteration design, we removed DTa entirely and appended RRSP via a (G4S)2 linker to the native release machinery of DT that includes DTT and DTR (DTB) and named it RRSP-DTB. Indeed, cells treated with this much smaller protein showed complete loss of detectable RAS at concentrations as low as 3 pM (Fig. 1C and SI Appendix, Fig. S1C). Control proteins with the catalytically inactive mutant RRSPH4030A (RRSP*-DTB) or with the DTT domain removed (RRSP-DTB[ΔDTT]) failed to deplete RAS from cells (Fig. 1D). The uptake of RRSP-DTB, translocation to cytosol, and subsequent RAS cleavage are summarized in Fig. 1E. Altogether, these results demonstrate that DTB is highly effective for intracellular translocation of RRSP across membranes to target RAS within cells. This final chimeric recombinant toxin was purified on a large scale, enabling the experimental investigation of the anticancer potential of RRSP (SI Appendix, Fig. S2 A–H).
RAS Processing by RRSP Strongly Affects Viability and Proliferation of TNBC Cancer Cells with Activated Wild-Type RAS.
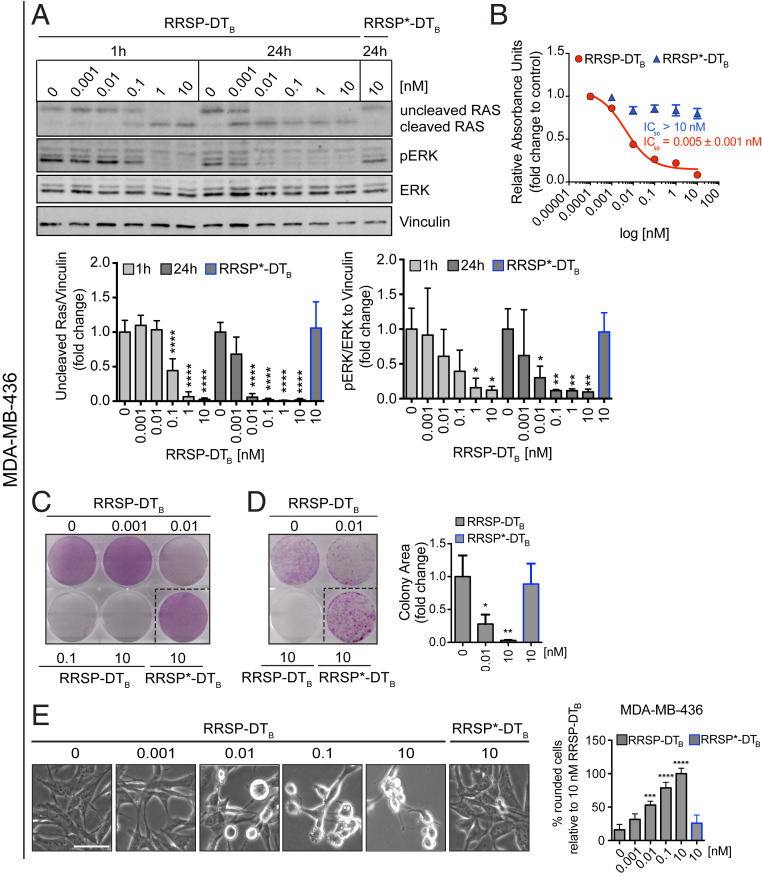
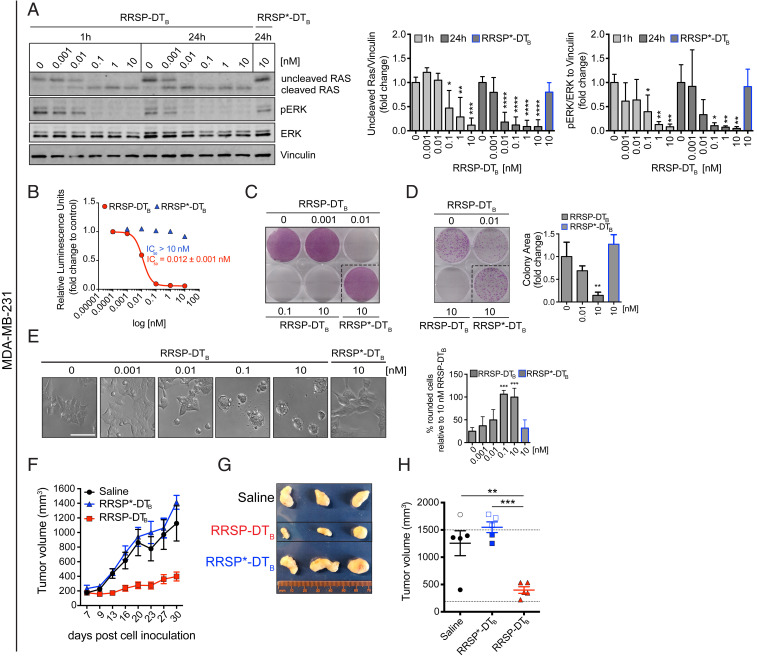
We first investigated the anticancer potential of RRSP-DTB in TNBC, a devastating disease and the last frontier in breast cancer research (28). Although only ∼5% of breast cancers harbor RAS mutations (10), RAS proteins can be pathologically activated in TNBC via up-regulation of RTKs and/or amplification of wild-type RAS (10, 29, 30). The basal-like MDA-MB-436 cell line features overexpression of KRASWT and hyperactivation of ERK signaling (31). Protein levels of HB-EGF, the DT receptor, were assessed by Western blotting in MDA-MB-436 cells and additional cell lines used in this study as a correlation parameter between HB-EGF expression and cytotoxicity (SI Appendix, Fig. S3A). HB-EGF–expressing MDA-MB-436 cells were treated with increasing concentrations of RRSP-DTB for 1 and 24 h, and levels of intracellular RAS and phosphorylated ERK1/2 (pERK) were measured by Western blot. Treatment with RRSP-DTB resulted in complete cleavage of RAS at 10 nM after 1 h, while 0.01 nM RRSP-DTB was sufficient to completely cleave RAS after 24 h. At all concentrations, levels of pERK were reduced in lockstep with RAS cleavage (Fig. 2A). While pERK levels were markedly reduced after 24 h, effects on cell viability reached a maximum at 72 h (IC50 = 0.005 ± 0.001 nM) (Fig. 2B and SI Appendix, Fig. S3B). In addition, crystal violet staining showed cell loss starting at 0.01 nM after a 72-h exposure to RRSP-DTB (Fig. 2C and SI Appendix, Fig. S3C), while a colony formation assay showed that RRSP-DTB had a significant effect on cell proliferation at 0.01 nM and completely prevented colony formation at 10 nM over 10 d (Fig. 2D). Phenotypically, RRSP-DTB caused significant cell rounding in MDA-MB-436 cells at 0.01 nM (Fig. 2E). Overall, RRSP-DTB causes highly potent cytotoxic effects via the efficient ablation of RAS signaling in a KRASWT cell line.
Fig. 2.
RAS processing by RRSP leads to reduced viability and proliferation of a triple-negative KRASWT breast cancer cell line. (A) Representative Western blot and densitometric analysis of uncleaved RAS and pERK levels in MDA-MB-436 KRASWT cells treated with increasing doses of RRSP-DTB for 1 and 24 h. The catalytically inactive RRSP*-DTB mutant was used as a negative control at 10 nM and vinculin as a gel loading control. Results are expressed as means ± SD of four independent experiments (*P < 0.05, **P < 0.01, ****P < 0.0001 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test; n = 4). (B) Fitted dose–response curve of RRSP-DTB in MDA-MB-436 cells following 72 h of treatment. Results are expressed as mean ± SEM (n = 3). (C) Representative images of crystal violet staining of MDA-MB-436 cells treated with RRSP-DTB or RRSP*-DTB as indicated for 72 h. (D) Representative images and quantitative analysis of crystal violet-stained colonies from MDA-MB-436 cells pretreated with RRSP-DTB and RRSP*-DTB at the indicated concentrations for 72 h and replated at 2,500 cells/well to form colonies over 10 d. Results are expressed as means ± SD of three independent experiments (*P < 0.05, **P < 0.01 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test, n = 3). (E) Bright-field images of MDA-MB-436 cells treated with RRSP-DTB and RRSP*-DTB at the indicated doses for 48 h and corresponding cell rounding quantification (***P < 0.001, ****P < 0.0001 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test; n = 3). (Scale bar, 50 µM.)
RRSP Halts Tumor Growth of a KRASWT TNBC Xenograft.
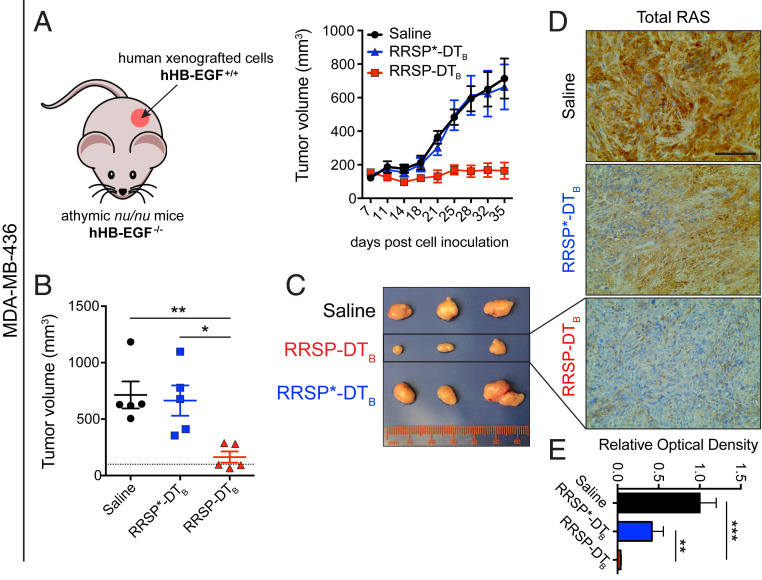
Retaining the native receptor targeting of DTB to translocate RRSP is advantageous as a test system for mouse xenografts since DT is at least 1,000-fold less potent in cells expressing murine HB-EGF than in cells expressing human HB-EGF (32–34). Indeed, RRSP-DTB treatment did not affect the viability of mouse embryonic fibroblasts (MEFs) (SI Appendix, Fig. S4 A and B). A maximum tolerated dose study determined that 0.1 mg/kg administered intraperitoneally (i.p.) every day (q.d.) was well tolerated, while dosage at or above 0.5 mg/kg caused weight loss (SI Appendix, Fig. S4 C and D). In the MDA-MB-436 xenograft, intraperitoneal administration of RRSP-DTB at 0.1 mg/kg every other day (weekends excluded) for 4 wk halted tumor growth (Fig. 3 A and B). Importantly, three of five mice treated with RRSP-DTB showed tumor regression by the end of the treatment schedule (Fig. 3B). Most importantly, catalytically inactive RRSP*-DTB did not affect tumor growth compared to vehicle (saline) control, demonstrating that tumor regression was due specifically to RRSP-mediated RAS-processing activity. Indeed, RRSP-DTB dramatically reduced total RAS immunoreactivity in residual tumors, confirming effective RAS cleavage in vivo (Fig. 3 D and E and SI Appendix, Fig. S5A). Additional experimental groups using slight variations of the dosing strategy corroborated these findings (SI Appendix, Fig. S6 A–F). Since ERK phosphorylation was limited to the outer rim of control MDA-MB-436 tumors excised from mice at 4 wk (SI Appendix, Figs. S5B and S6G), we assessed pERK levels at an earlier time point when the smaller size of large control tumors is more amenable to pERK detection. In tumors harvested at 2 wk, we observed staining throughout the sections with high basal pERK levels in both saline and RRSP*-DTB–treated tumors and with significant reduction in pERK in the RRSP-DTB treatment group (SI Appendix, Fig. S5C). These results demonstrate that RRSP-DTB effectively engages and ablates RAS in vivo, resulting in decreased ERK activation and tumor regression.
Fig. 3.
RRSP halts tumor growth in a KRASWT TNBC xenograft in vivo. (A) Tumor growth curve of vehicle, RRSP-DTB and RRSP*-DTB–treated athymic nu/nu female mice bearing MDA-MB-436–derived tumors. Mice received 0.1 mg/kg of RRSP-DTB and 0.1 mg/kg of RRSP*-DTB every other day (weekends excluded) via i.p. injection. (B). Column scatter plots showing individual tumor volumes at the end of the treatment schedule for MDA-MB-436 xenografts. Horizontal dotted line indicates the average tumor volume (baseline) on the first day of treatment (140 mm3). Data are means ± SEM (n = 5 mice per group; *P < 0.05, **P < 0.01). (C) Representative images of MDA-MB-436 tumors at the experimental endpoint. (D) Representative images of immunoreactivity to total RAS in sections from MDA-MB-436 tumors and (E) corresponding quantification of DAB (3,3′-diaminobenzidine) optical density (**P < 0.01, ***P < 0.001, one-way ANOVA followed by Tukey’s multiple comparison test, n = 3). (Scale bar = 100 µM.)
Assessment of Relative Susceptibility to RRSP-DTB Using the NCI-60 Panel.
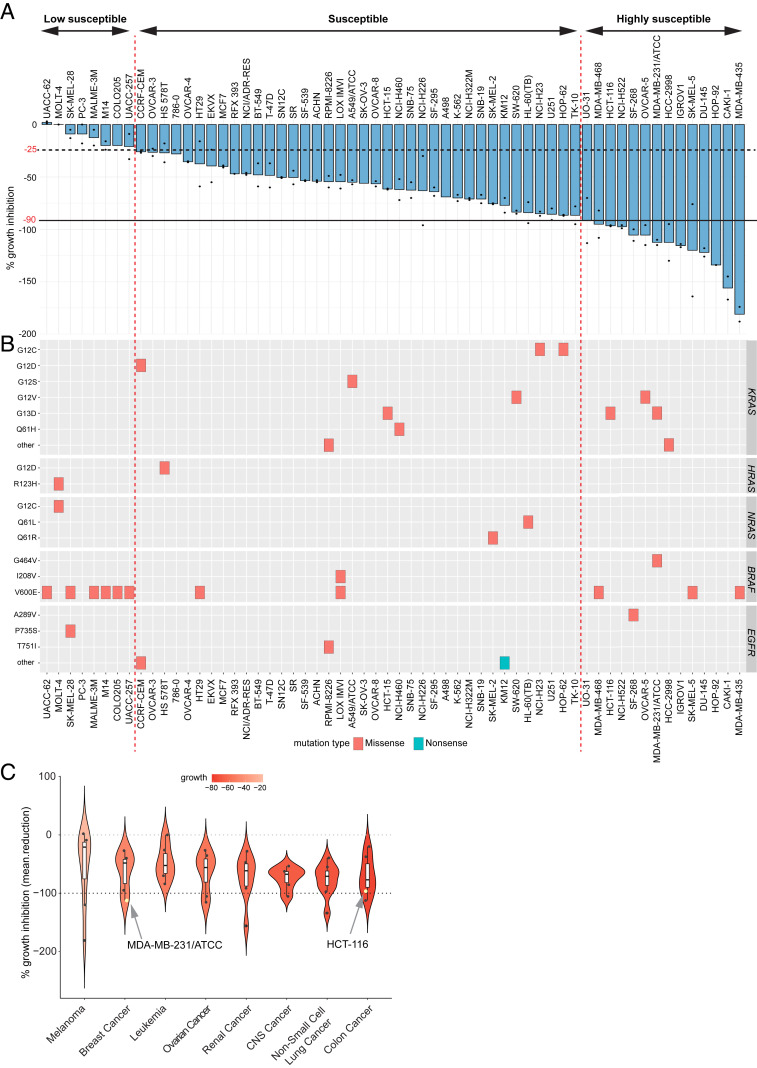
We next screened RRSP-DTB against the National Cancer Institute NCI-60 human tumor cell line panel composed of 60 cell lines representing 9 different cancer types with various genetic backgrounds, including RAS mutations. Growth inhibition caused by RRSP-DTB was measured by sulforhodamine B assay after 48 h and reported in Fig. 4A for the highest dose employed (13.5 nM). Fourteen cell lines were classified as “highly susceptible” to RRSP-DTB as they showed growth inhibition greater than or equal to 90%. Thirty-eight cell lines showed varying degrees of growth inhibition from 25 to 90% and were designated as “susceptible” to RRSP-DTB (Fig. 4A).
Fig. 4.
Comprehensive analysis of the effect of RRSP on the NCI-60 cancer cell line panel. (A) Bar plot showing the percentage of growth inhibition of RRSP-DTB in the NCI-60 panel. Cancer cell lines were ranked in descending order based on their growth inhibition percentage value. The presence of two dots on the bars indicates that two replicates were performed per each cell line, and bars represent means. No dots indicate that only one replicate was available. (B) Spectrum of missense and nonsense mutations in KRAS, HRAS, NRAS, BRAF, and EGFR genes in the 60 cell lines of the panel. (C) Violin plot showing the median percentage of growth inhibition of cell lines treated with RRSP-DTB and grouped per tumor type in descending order. Only tumor types that had at least five cell lines per group were plotted on the graph.
Generally, cell lines that carry KRAS missense mutations were among the most responsive to RRSP-DTB, while cell lines with mutations in BRAF (especially BRAFV600E) tended to be less responsive (Fig. 4B). Mutations in HRAS, NRAS, or EGFR were not associated with a response pattern to RRSP-DTB treatment (Fig. 4B). Notably, analysis of copy number alterations from exome data available for 53 of 60 National Cancer Institute (NCI) cell lines showed amplification of KRAS and NRAS, as well as deletions in EGFR, in the cell lines most sensitive to RRSP-DTB (SI Appendix, Fig. S7 B and C). Furthermore, colon cancer cell lines as a group were the most sensitive to RRSP-DTB overall, followed by nonsmall cell lung cancer lines (Fig. 4C).
Eight cell lines in the NCI-60 screen showed <25% growth inhibition compared to mock-treated and were categorized as “less susceptible.” In this group, growth of UACC-62 and MOLT-4 cells was not affected by RRSP-DTB. One requirement for RRSP-DTB cytotoxicity is expression of the DT receptor HB-EGF on the cell surface. Available HBEGF gene expression data (SI Appendix, Fig. S7A) showed that, while the correlation between RRSP-DTB sensitivity and HBEGF expression is not linear, many of the cell lines that responded to RRSP-DTB had higher expression of HBEGF. In addition, the TNBC Hs578T cell line (HRASG12D), categorized as less sensitive in the NCI-60 screen, was confirmed to have lower expression of HB-EGF protein and a moderate reduction in cell viability after 72 h, although RAS cleavage and ERK dephosphorylation were detected at earlier time points (SI Appendix, Figs. S3A and S8 A–D). Overall, results from the NCI-60 screen indicate that most tumor types were sensitive to RRSP-DTB, and cell lines with genomic abnormalities in RAS genes were markedly sensitive to RRSP-DTB treatment.
RRSP Reduces Tumor Burden in a Mutant KRAS TNBC Xenograft Model.
The highly sensitive basal-like MDA-MB-231 TNBC cell line in the NCI-60 screen (Fig. 4A) has a KRASG13D mutation, is a KRAS-dependent cell line (31, 35), and was among the first cell lines characterized as sensitive to RRSP (22) (Fig. 4A). Consistent with these findings, treatment of MDA-MB-231 cells with the engineered chimeric toxin RRSP-DTB cleaved RAS and reduced pERK levels to a similar extent as in MDA-MB-436 cells (Fig. 5A). Also similar to MDA-MB-436 cells, cell viability (IC50 = 0.012 ± 0.001 nM; Fig. 5 B and C and SI Appendix, Fig. S9 A and B) and cell proliferation (Fig. 5D) were strongly affected, and RRSP-DTB induced significant cell rounding in MDA-MB-231 cells starting at 0.1 nM (Fig. 5E). As this cell line has a more rapid doubling time than MDA-MB-436 cells (36), mice with MDA-MB-231 xenografts were treated daily (5 d ON/2 d OFF) (Fig. 5F). After 4 wk, the RRSP-DTB treatment group had markedly smaller tumors than saline and RRSP*-DTB controls (Fig. 5 G and H). Moreover, the RRSP-DTB–treated tumors had a more focal/patchier staining pattern for RAS and pERK than tumors from saline- and RRSP*-DTB–treated mice, which instead exhibited a more diffuse staining pattern (SI Appendix, Fig. S9 C and D). Indeed, MDA-MB-231 tumors (Fig. 5G) were pale in color compared to the MDA-MB-436 tumors (Fig. 3C) consistent with reports that MDA-MB-231 tumors exhibit low vascularization (37). This supports that MDA-MB-231 tumors in our study may be poorly vascularized, and tumor size reduction may be predominantly due to RRSP-DTB diffusion from the tumor periphery, resulting in the partial cleavage of RAS in the center of residual tumors. Even still, RRSP-DTB was highly effective in targeting TNBC MDA-MB-231 tumors resulting in a significant inhibition of tumor growth, thus confirming that RRSP-DTB is effective against TNBC.
Fig. 5.
RRSP inhibits cell viability, proliferation, and tumor growth in a KRASG13D-dependent TNBC xenograft in vivo. (A) Representative Western blot and densitometric analysis of uncleaved RAS and pERK levels in MDA-MB-231 KRASG13D cells treated with increasing doses of RRSP-DTB and with 10 nM of RRSP*-DTB. Results are expressed as means ± SD of four independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test; n = 4). (B) Fitted dose–response curve of RRSP-DTB in MDA-MB-231 cells following 72 h of treatment. Results are expressed as means ± SEM (n = 3). (C) Representative images of crystal violet staining of MDA-MB-231 cells treated with RRSP-DTB or RRSP*-DTB as indicated for 72 h. (D) Representative images and quantitative analysis of crystal violet-stained colonies from MDA-MB-231 cells pretreated with RRSP-DTB and RRSP*-DTB at the indicated concentrations for 72 h and replated at 2,500 cells/well to form colonies over 10 d. Results are expressed as means ± SD of three independent experiments (**P < 0.01 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test; n = 3). (E) Bright-field images of MDA-MB-231 cells treated with RRSP-DTB and RRSP*-DTB at the indicated concentrations for 48 h and corresponding cell rounding quantification (***P < 0.001 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test; n = 3). (Scale bar, 50 µM.) (F) Tumor growth curve of vehicle, RRSP-DTB, and RRSP*-DTB–treated athymic nu/nu female mice bearing MDA-MB-231–derived tumors. Mice received 0.1 mg/kg of RRSP-DTB and 0.1 mg/kg of RRSP*-DTB every day (weekends excluded). (G) Representative images of tumors at the experimental end point. (H) Column scatter plots showing individual tumor volumes at the end of the treatment schedule. Horizontal dotted line indicates the average tumor volume (baseline) on the first day of treatment (194 mm3). Empty points indicate tumors from mice that were killed earlier because they exceeded the 1,500-mm3 threshold. Data are means ± SEM (n = 5 mice per group). Statistical analysis between vehicle and treatment groups was performed using one-way ANOVA followed by Tukey’s multiple comparison test (**P < 0.01, ***P < 0.001).
RRSP-DTB Inhibits Cell Viability of Colorectal Cancer Cells in Two-Dimensional Monolayers and Three-Dimensional Spheroids.
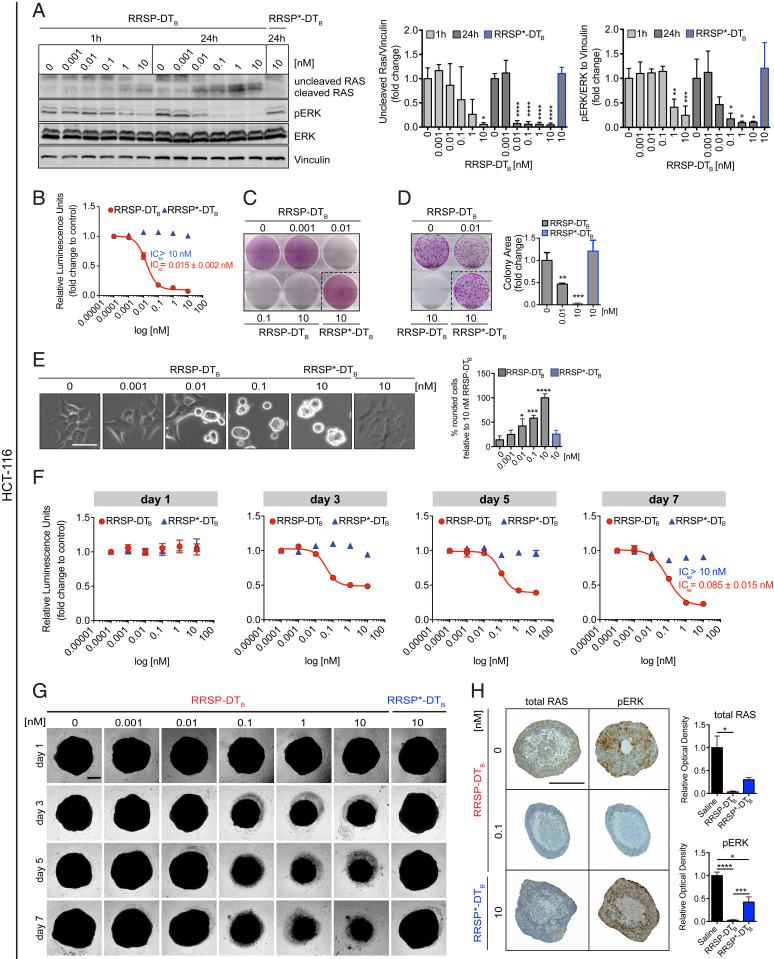
KRAS mutations are found in ∼50% of colorectal carcinomas (CRC), the third leading cause of cancer-related deaths, representing a major target population for anti-RAS therapy (38). Colon cancer cell lines were the most susceptible to RRSP-DTB in the NCI-60 screen. Here, we examined the anticancer potential of RRSP-DTB in the CRC cell line HCT-116 harboring a KRASG13D mutation. Treatment of cells with RRSP-DTB at 10 pM led to RAS processing and reduced pERK levels at 10 pM after 24 h (Fig. 6A) and strongly decreased the viability of HCT-116 cells after 72 h (IC50 = 0.0015 ± 0.002 nM) (Fig. 6B and SI Appendix, Fig. S10A). Crystal violet staining showed remarkable cell loss after treatment with RRSP-DTB as low as 10 pM (Fig. 6C and SI Appendix, Fig. S10B). Clonogenic assays showed a significant reduction in HCT-116 colonies following treatment with 10 pM of RRSP-DTB and almost no colonies were found at 10 nM RRSP-DTB (Fig. 6D) demonstrating complete loss of cell proliferation. These data agree with the RRSP-DTB–induced cell rounding phenotype observed with this and other sensitive cell lines (Fig. 6E).
Fig. 6.
Effect of RRSP on viability and proliferation of colorectal HCT-116 KRASG13D cells in 2D and 3D cellular models. (A) Representative Western blot and densitometric analysis of uncleaved RAS and phosphorylated ERK in HCT-116 cells treated with increasing doses of RRSP-DTB for 1 and 24 h. Catalytically inactive RRSP*-DTB was used as the negative control at 10 nM. Results are expressed as means ± SD of three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test; n = 3). (B) Fitted dose–response curve of RRSP-DTB in HCT-116 cells following 72 h of treatment. (C) Representative images of crystal violet staining of HCT-116 cells treated with RRSP-DTB or RRSP*-DTB as indicated for 72 h. (D) Representative images and quantitative analysis of crystal violet-stained colonies from HCT-116 cells pretreated with RRSP-DTB or RRSP*-DTB at the indicated concentrations for 72 h and reseeded at 2,500 cells/well to form colonies over 10 d. Results are expressed as means ± SD of three independent experiments (**P < 0.01, ***P < 0.001 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test, n = 3). (E) Bright-field images of HCT-116 cells treated with RRSP-DTB and RRSP*-DTB at the indicated concentrations for 48 h and corresponding cell rounding quantification (*P < 0.05, ***P < 0.001, ****P < 0.0001 versus corresponding control 0 nM; one-way ANOVA followed by Dunnett’s multiple comparison test; n = 3). (F) Fitted dose–response curves in HCT-116 spheroids following treatment with RRSP-DTB and RRSP*-DTB at the indicated time and concentration. Results are expressed as means ± SEM (n = 4). (G) Representative bright-field images of HCT-116 spheroids treated at the indicated time and concentrations with RRSP-DTB and RRSP*-DTB and quantitative analysis of spheroids’ volume. (Scale bar, 200 µm.) (H) Representative images of immunoreactivity to total RAS and pERK in sections from HCT-116 spheroids and corresponding quantification of DAB optical density via color deconvolution (*P < 0.05, ***P < 0.001, ****P < 0.0001; one-way ANOVA followed by Tukey’s multiple comparison test; n = 3). (Scale bar, 400 µM.)
Unlike two-dimensional (2D) cell monolayers, three-dimensional (3D) spheroids can recapitulate the architectural, microenvironmental, and functional features of in vivo tumors, while retaining reproducibility and easy-to-use properties (39, 40). Effects on cell viability of spheroids from HCT-116 cells were observed after 3 d, with maximum effect observed after 7 d of treatment (IC50 = 0.085 ± 0.015 nM) (Fig. 6F). Bright-field images and quantitative analysis showed a dose- and time-dependent reduction in spheroid size following RRSP-DTB treatment (Fig. 6G and SI Appendix, Fig. S10C). Immunostaining of spheroid sections treated with 0.1 nM RRSP-DTB for 3 d showed that intracellular RAS was essentially absent and pERK undetectable (Fig. 6H). Collectively, these results demonstrate that RRSP-DTB–dependent RAS ablation and subsequent loss of pERK is highly cytotoxic to CRC HCT-116 (KRASG13D) cells in 2D monolayers and 3D spheroids.
RRSP-DTB Exhibits Antitumor Activity in a Colorectal Cancer Xenograft Model.
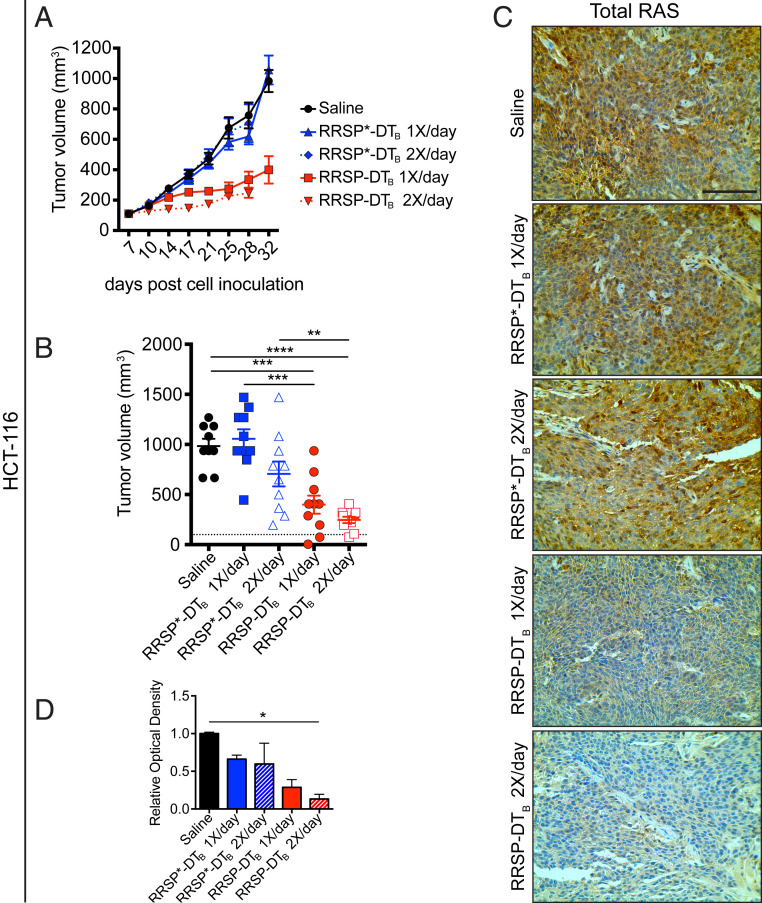
Since HCT-116 cells are fast-growing cells (doubling time: ≤20 h) that generate fast-growing tumors in vivo (41), we administered 0.1 mg/kg of RRSP-DTB to mice on a q.d. (1×/day) or twice per day (2×/day, b.i.d.) schedule (weekends excluded) for 4 wk. Both dosing schedules resulted in significant tumor size reduction, and tumor regression was observed in 2/10 mice in both RRSP-DTB treatment groups (Fig. 7 A and B). The residual tumors showed that RRSP-DTB effectively cleaved RAS, although only the b.i.d. dosing group achieved statistical significance (Fig. 7 C and D and SI Appendix, Fig. S11A). Quantitative analysis of pERK showed that the b.i.d. dosing group had a 2.5-fold reduction in pERK levels relative to controls and that some tumors displayed focal staining patterns (SI Appendix, Fig. S11B). Overall, these data show that RRSP-DTB exhibited strong antitumor activity in a CRC xenograft model via irreversible inactivation of RAS.
Fig. 7.
RRSP slows tumor growth in a CRC xenograft in vivo. (A) Tumor growth curve of vehicle, RRSP-DTB– and RRSP*-DTB–treated athymic nu/nu female mice bearing HCT-116–derived tumors. Mice received 0.1 mg/kg of RRSP-DTB and 0.1 mg/kg of RRSP*-DTB every day (1×/day) or twice per day (2×/day) with weekends excluded. (B) Column scatter plots showing HCT-116 individual tumor volumes at the end of the treatment schedule. Horizontal dotted lines indicate the average tumor volume (baseline) on the first day of treatment (100 mm3). Data are means ± SEM. In all groups, n = 10 mice. In the saline group, one mouse was killed on day 28 due to a too large tumor. Statistical analysis between vehicle and treatment groups was performed using one-way ANOVA followed by Tukey’s multiple comparison test (**P < 0.01, ***P < 0.001, ****P < 0.0001). (C) Representative IHC (immunohistochemistry) images of immunoreactivity to total RAS in sections from HCT-116 tumors and (D) corresponding quantification of DAB optical density (*P < 0.05, one-way ANOVA followed by Tukey’s multiple comparison test, n = 3). (Scale bar, 100 µM.)
Discussion
Almost four decades ago, the discovery of RAS as the first human oncogene changed our understanding of cancer. Despite tremendous effort, the three RAS isoforms (KRAS, NRAS, and HRAS) have been called “undruggable,” and no direct therapies are currently in clinical use (1, 12–14). Nevertheless, promising results for small molecules that irreversibly bind the G12C mutant form of KRAS have led to ongoing phase I clinical trials to evaluate the efficacy and safety profile of AMG510, MRTX849, and ARS3248 (18–20, 42, 43). However, the KRASG12C mutation is found in only a subpopulation of cancers, limiting the applicability of these compounds. Furthermore, while RAS oncoproteins remain the main oncogenic drivers in RAS-addicted tumors, several studies have pinpointed the tumorigenic role of wild-type RAS proteins (44). Indeed, amplification of wild-type RAS genes or activation of wild-type RAS proteins via acute growth factor stimulation have been shown to sustain growth of multiple tumor types (7–10). Moreover, it has been previously reported that depletion of mutant RAS in heterozygous RAS cells can lead to overactivation of EGFR/RAS signaling from the remaining wild-type RAS (45). Therefore, there is an urgent need for broadly applicable pan-RAS inhibitors that target not only the most common RAS mutants, but also wild-type RAS proteins aberrantly overactivated by mutation-independent mechanisms.
Here, we describe an engineered chimeric toxin composed of an endopeptidase from V. vulnificus that is highly specific for RAS and RAP1 and the protein translocation machinery of DT. This fusion protein mediates the endocytosis and cytosolic delivery of RRSP exclusively into HB-EGF receptor-bearing cells. Of note, HB-EGF is highly expressed in several human cancers, including gastric, ovarian, and TNBC, and the nontoxic diphtheria toxoid CRM197 showed antitumor effects on TNBC xenografts (46). As such, we predicted that TNBC could be highly susceptible to RRSP-DTB. We show that picomolar amounts of RRSP-DTB completely cleaved RAS proteins in these cells, eliminating RAS-dependent ERK phosphorylation. This resulted in a very potent effect on MDA-MB-436 cell viability (IC50 = 5 pM). Similarly, RRSP-DTB treatment led to a large reduction in tumor burden in MDA-MB-436 xenografts, with all mice showing significantly smaller tumors than control groups and 60% of mice showing tumor regression. Target engagement was confirmed by immunohistochemistry analysis of tumor sections showing complete depletion of RAS in residual tumors. We also observed low pERK staining in small dissected RRSP-DTB–treated tumors, although pERK levels in these specimens were not significantly different from control tumors that showed limited detection of pERK after 4 wk. However, assessment of resected tumors from a shorter 14-d xenograft study revealed that RRSP-DTB significantly reduced levels of pERK. This effect was masked at 4 wk likely due to technical factors. Specifically, a previous study reported that in large (>1 cm) glioma specimens, immunoreactivity of pERK was limited to the tumor periphery and was very low in the tumor core, as we also observed. This study suggested that, due to time-dependent penetration of the formalin fixative, pERK state is lost in the deeper tissue cores by the time the fixative has permeated (47).
The study of TNBC was extended then to demonstrate the susceptibility of another TNBC cell line, MDA-MB-231. This cell line, which harbors the oncogenic mutations KRASG13D and BRAFG464V, is characterized as KRAS-dependent (35) and is highly sensitive to RRSP (22). In xenograft studies using MDA-MB-231, RRSP-DTB induced a significant reduction in tumor size. Interestingly, the residual tumors did not show significant reduction in RAS or pERK, but instead revealed a patchy or focal distribution. Thus, the true reduction in RAS detection may be masked by the low vascularization of MDA-MB-231 tumors such that RRSP-DTB reduces overall tumor size by diffusion from the tumor periphery rather than infiltrating equally throughout the tumor center, as occurred with the MDA-MB-436 xenografts.
We finally investigated the effects of RRSP-DTB treatment on the CRC cell line HCT-116 (KRASG13D). As with the other cell lines tested, RRSP-DTB treatment resulted in complete RAS cleavage and pERK reduction in the low-picomolar range with a corresponding reduction in cell viability. Similar results were obtained in HCT-116 spheroids, including complete loss of RAS and pERK immunoreactivity. Notably, depletion of RAS and reduction in pERK were also observed in spheroids treated with the catalytically inactive mutant RRSP*-DTB, but at a 10-fold higher dose. Previous binding studies showed that RRSP* retains its ability to bind recombinant KRAS, especially at high KRAS:RRSP* molar ratios (24). Therefore, although RRSP*-DTB does not cleave RAS, it might impact its activity when used at high doses and after longer exposure both in vitro and in vivo.
As with the other tumor models, RRSP-DTB treatment resulted in significant reduction in tumor size in an HCT-116 xenograft study. Both q.d. and b.i.d. dosing strategies showed similar median reductions in tumor size; however, the b.i.d. arm showed less variability represented by a small interquartile range. Effective target engagement was demonstrated by a reduction in total RAS levels in residual RRSP-DTB–treated tumors. We did not observe statistically significant differences between controls and RRSP-DTB–treated tumors when analyzing pERK immunoreactivity. However, as noted above, large control tumors also showed poor immunostaining for pERK, and thus low values for control may have caused the lack of statistical significance in the small sample size when data were normalized. Even still, we observed reduced tumor size in all treatment groups and a decrease in pERK in the b.i.d. treatment group.
In order to assess the broader applicability of RRSP-DTB and have a better understanding of its efficacy as an anticancer agent, RRSP-DTB was tested against the NCI-60 cell line panel. Results showed that RRSP-DTB inhibited cell growth in a wide variety of cell lines. Cell lines with missense mutations or amplifications in RAS genes were enriched in the highly responsive group. Moreover, although the expression levels of the DT receptor HB-EGF play an important role in determining RRSP-DTB susceptibility, some cancer cell lines that express less HB-EGF were still sensitive to RRSP-DTB, suggesting differential susceptibility of tumor cells to RAS/RAP1 inhibition. Thus, RRSP-DTB has the potential to be broadly applicable against many types of cancers, including both wild-type and mutant RAS tumors. In addition, cancer cell lines from colon and lung are particularly sensitive to RRSP-DTB. However, several tumor cell lines with no genetic defects in RAS or the MAPK pathway remain very sensitive to RRSP-DTB.
In total, this study provides solid proof-of-concept that the effective, receptor-mediated, intracellular delivery of a potent anti-RAS biologic represents a promising approach for the development of RAS-targeted therapeutics. We contend that the ability of RRSP to directly and irreversibly inactivate both wild-type and mutant RAS proteins represents an attractive mechanism compared to the current approach of targeting RAS mutants individually. However, a pan-RAS inhibitor is expected to induce dose-limiting toxicity due to the critical importance of RAS signaling in noncancerous tissues. The ability of DT to deliver cargos exclusively to receptor-bearing cells provides a solution to this toxicity. While HB-EGF is up-regulated in various tumor types, it is also widely expressed in humans and may not represent the ideal receptor for tumor-targeting. In order to restrict the delivery of RRSP to tumor cells, the DT-based delivery platform described here can be retargeted to various cell types by replacing the receptor-binding domain of DT with other binding moieties, such as antibody fragments or ligands. For example, the recombinant immunotoxins Ontak and Tagraxofusp comprise wild-type DTA with DTR replaced by interleukin 2 (IL-2) or interleukin 3 (IL-3), respectively (48, 49). These drugs specifically target cells expressing the IL-2 receptor (IL-2R) or IL-3 receptor (IL-3R) and rely on the membrane-translocating ability of DT to deliver the DTA domain into the cell cytosol, where it terminates protein synthesis leading to cell death. These immunotoxins are extremely potent and are indicated for treatment of cutaneous T cell lymphoma (Ontak) and blastic plasmacytoid dendritic-cell neoplasm (Tagraxofusp) (48, 49). RRSP-DTB can be retargeted in the same fashion, and RRSP-DTT-IL2 is able to efficiently cleave RAS in both MOLT-4 and Jurkat cell lines which express IL-2R, but not in CFPAC-I cells, which do not (SI Appendix, Fig. S12). In fact, MOLT-4 was among the least responsive cell lines in the NCI-60 screen, suggesting that even cancers currently in the low-sensitivity group would be much more sensitive to RRSP-DTB following advanced engineering to target the chimeric toxin to alternative receptors appropriated for that cell type. Further study into engineered chimeric toxins, such as RRSP-DTB, could usher in the next generation of anticancer biologics.
Materials and Methods
Plasmids Design, Protein Purification, and Endotoxin Removal.
Detailed methods for plasmid design, purification of RRSP-DTB and RRSP*-DTB, cell culture, chemical, cell-based assays, and animal studies can be found in SI Appendix. All animal studies were approved by the Northwestern University Institutional Animal Care and Use Committee (IACUC).
Data and Materials Availability.
All data are available in this paper or SI Appendix. Plasmids for expression of RRSP-DT are available with materials transfer agreement from R.A.M., Hospital for Sick Children.
Supplementary Material
Acknowledgments
We thank the Robert H. Lurie Comprehensive Cancer Center Pathology Core Facility and B. Shmaltuyeva, B. Frederick, and Dr. D. Gursel for assistance with immunohistochemical staining; A. Dean and C. Nordloh for assistance with xenografts; S. Son for performing validation of the pan-RAS antibody; Dr. Damiano Fantini for bioinformatics assistance; and the Frederick National Laboratory for Cancer Research (FNLCR) for providing the RAS-less KRASWT MEF cells and the anti-RAS 4E8 hybridoma cells. This work was funded by the Lynn Sage Cancer Research Foundation; Northwestern University Clinical and Translational Sciences Institute (which is supported by NIH/National Center for Advancing Translational Sciences Award UL1TR001422); the Northwestern Medicine Catalyst Fund; and the Robert H. Lurie Comprehensive Cancer Research Center (to K.J.F.S.). D.R.G. is supported by NIH/NCI Grants R01CA152601, R01CA152799, R01CA168292, and R01CA214025, the Avon Breast Cancer Foundation, the Zell Family Foundation, the Chicago Biomedical Consortium, the Searle Funds at The Chicago Community Trust, and Cancer Prevention & Research Institute of Texas Grant RR200112. Additional support from the SickKids Proof-of-Principal Funding and the Canadian Institutes of Health Research Grant 366017 (to R.A.M.). M.B. was supported by a PanCan/FNLCR Fellowship. G.L.B. was supported by a SickKids Restracomp Fellowship.
Footnotes
Competing interest statement: M.B. and K.J.F.S. are authors of a pending patent on use of RRSP as a therapeutic for cancer (WO2016019379A1). K.J.F.S. is author of a published patent on use of cysteine protease domain for autoprocessing of proteins (US20100137563A1). R.A.M. and G.L.B. are authors of a published patent on the use of diphtheria toxin for protein delivery and of a pending patent on RRSP-DTB as a RAS-directed therapeutic (US20180080033A1 and WO2019104433A1, respectively). K.J.F.S. has a significant financial interest in Situ Biosciences LLC, which conducts contract research unrelated to this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000312117/-/DCSupplemental.
Change History
December 30, 2021: The Acknowledgments have been updated.
References
- 1.Cox A. D., Der C. J., Ras history: The saga continues. Small GTPases 1, 2–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hobbs G. A., Der C. J., Rossman K. L., RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prior I. A., Lewis P. D., Mattos C., A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos J. L., Rehmann H., Wittinghofer A., GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129, 865–877 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Schubbert S., Shannon K., Bollag G., Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Downward J., Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Hoa M., Davis S. L., Ames S. J., Spanjaard R. A., Amplification of wild-type K-ras promotes growth of head and neck squamous cell carcinoma. Cancer Res. 62, 7154–7156 (2002). [PubMed] [Google Scholar]
- 8.Dulak A. M.et al., Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 72, 4383–4393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross J. S.et al., Comprehensive genomic profiling of epithelial ovarian cancer by next generation sequencing-based diagnostic assay reveals new routes to targeted therapies. Gynecol. Oncol. 130, 554–559 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Eckert L. B.et al., Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res. 64, 4585–4592 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Network , Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papke B., Der C. J., Drugging RAS: Know the enemy. Science 355, 1158–1163 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCormick F., KRAS as a therapeutic target. Clin. Cancer Res. 21, 1797–1801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janes M. R.et al., Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Ostrem J. M., Peters U., Sos M. L., Wells J. A., Shokat K. M., K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patricelli M. P.et al., Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 6, 316–329 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Amgen (2019) A phase 1/2, study Evaluating the Safety, Tolerability, PK, and Efficacy of AMG 510 in Subjects With Solid Tumors With a Specific KRAS Mutation. https://clinicaltrials.gov/ct2/show/NCT03600883. Accessed 3 January 2020.
- 19.Janssen Research & Development (2019) First-in-Human Study of JNJ-74699157 in Participants With Tumors Harboring the KRAS G12C Mutation. https://clinicaltrials.gov/ct2/show/NCT04006301. Accessed 3 January 2020.
- 20.Mirati Therapeutics Inc . (2019) Phase 1/2 Study of MRTX849 in Patients With Cancer Having a KRAS G12C Mutation. https://clinicaltrials.gov/ct2/show/NCT03785249. Accessed 3 January 2020.
- 21.O’Bryan J. P., Pharmacological targeting of RAS: Recent success with direct inhibitors. Pharmacol. Res. 139, 503–511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antic I., Biancucci M., Zhu Y., Gius D. R., Satchell K. J. F., Site-specific processing of Ras and Rap1 Switch I by a MARTX toxin effector domain. Nat. Commun. 6, 7396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biancucci M.et al., Substrate recognition of MARTX Ras/Rap1-specific endopeptidase. Biochemistry 56, 2747–2757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biancucci M.et al., The bacterial Ras/Rap1 site-specific endopeptidase RRSP cleaves Ras through an atypical mechanism to disrupt Ras-ERK signaling. Sci. Signal. 11, eaat8335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auger A.et al., Efficient delivery of structurally diverse protein cargo into mammalian cells by a bacterial toxin. Mol. Pharm. 12, 2962–2971 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Park M.et al., Intracellular delivery of human purine nucleoside phosphorylase by engineered diphtheria toxin rescues function in target cells. Mol. Pharm. 15, 5217–5226 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Egerer M., Satchell K. J., Inositol hexakisphosphate-induced autoprocessing of large bacterial protein toxins. PLoS Pathog. 6, e1000942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzger-Filho O.et al., Dissecting the heterogeneity of triple-negative breast cancer. J. Clin. Oncol. 30, 1879–1887 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Adeyinka A.et al., Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin. Cancer Res. 8, 1747–1753 (2002). [PubMed] [Google Scholar]
- 30.Giltnane J. M., Balko J. M., Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov. Med. 17, 275–283 (2014). [PubMed] [Google Scholar]
- 31.Mokhlis H. A.et al., The modulatory role of MicroRNA-873 in the progression of KRAS-driven cancers. Mol. Ther. Nucleic Acids 14, 301–317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cha J. H., Chang M. Y., Richardson J. A., Eidels L., Transgenic mice expressing the diphtheria toxin receptor are sensitive to the toxin. Mol. Microbiol. 49, 235–240 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Mitamura T., Higashiyama S., Taniguchi N., Klagsbrun M., Mekada E., Diphtheria toxin binds to the epidermal growth factor (EGF)-like domain of human heparin-binding EGF-like growth factor/diphtheria toxin receptor and inhibits specifically its mitogenic activity. J. Biol. Chem. 270, 1015–1019 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Palmiter R., Interrogation by toxin. Nat. Biotechnol. 19, 731–732 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Scholl C.et al., Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821–834 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Hassan S., Esch A., Liby T., Gray J. W., Heiser L. M., Pathway-enriched gene signature associated with 53BP1 response to PARP inhibition in triple-negative breast cancer. Mol. Cancer Ther. 16, 2892–2901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fleming J. M., Miller T. C., Meyer M. J., Ginsburg E., Vonderhaar B. K., Local regulation of human breast xenograft models. J. Cell. Physiol. 224, 795–806 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferlay J.et al., Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Ferreira L. P., Gaspar V. M., Mano J. F., Design of spherically structured 3D in vitro tumor models: Advances and prospects. Acta Biomater. 75, 11–34 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Vidimar V.et al., The AKT/BCL-2 Axis mediates survival of uterine leiomyoma in a novel 3D spheroid model. Endocrinology 159, 1453–1462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmed D.et al., Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2, e71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canon J.et al., The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Hallin J.et al., The KRASG12C inhibitor, MRTX849, provides insight toward therapeutic susceptibility of KRAS mutant cancers in mouse models and patients. Cancer Discov. 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou B., Der C. J., Cox A. D., The role of wild type RAS isoforms in cancer. Semin. Cell Dev. Biol. 58, 60–69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young A., Lou D., McCormick F., Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 3, 112–123 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Nam S. O.et al., Anti-tumor effect of intravenous administration of CRM197 for triple-negative breast cancer therapy. Anticancer Res. 36, 3651–3657 (2016). [PubMed] [Google Scholar]
- 47.Mandell J. W., Phosphorylation state-specific antibodies: Applications in investigative and diagnostic pathology. Am. J. Pathol. 163, 1687–1698 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jen E. Y.et al., FDA approval summary: Tagraxofusp-erzs for treatment of blastic plasmacytoid dendritic cell neoplasm. Clin. Cancer Res. 26, 532–536 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Prince H. M.et al., Phase III placebo-controlled trial of denileukin diftitox for patients with cutaneous T-cell lymphoma. J. Clin. Oncol. 28, 1870–1877 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.