Significance
Flagella are used by many pathogenic bacteria not only for motility but also for adhesion to host cells and to other bacteria in biofilm formation. Since thousands of copies of flagellin, the protein that forms the flagellar filament, are exposed on the surface of bacteria, they are a target for immune surveillance by hosts. Some bacteria important to human health, such as Campylobacter jejuni and Helicobacter pylori, have managed to evade the innate immune recognition of their flagellin. We show, with an atomic structure of the Campylobacter jejuni flagellar filament, how mutations that destabilize the filament in one region recognized by vertebrates have been compensated for by new contacts not seen in other flagellar filaments that would stabilize these filaments.
Keywords: cryo-EM, bacterial flagella, innate immunity
Abstract
Vertebrates, from zebra fish to humans, have an innate immune recognition of many bacterial flagellins. This involves a conserved eight-amino acid epitope in flagellin recognized by the Toll-like receptor 5 (TLR5). Several important human pathogens, such as Helicobacter pylori and Campylobacter jejuni, have escaped TLR5 activation by mutations in this epitope. When such mutations were introduced into Salmonella flagellin, motility was abolished. It was previously argued, using very low-resolution cryoelectron microscopy (cryo-EM), that C. jejuni accommodated these mutations by forming filaments with 7 protofilaments, rather than the 11 found in other bacteria. We have now determined the atomic structure of the C. jejuni G508A flagellar filament from a 3.5-Å-resolution cryo-EM reconstruction, and show that it has 11 protofilaments. The residues in the C. jejuni TLR5 epitope have reduced contacts with the adjacent subunit compared to other bacterial flagellar filament structures. The weakening of the subunit−subunit interface introduced by the mutations in the TLR5 epitope is compensated for by extensive interactions between the outer domains of the flagellin subunits. In other bacteria, these outer domains can be nearly absent or removed without affecting motility. Furthermore, we provide evidence for the stabilization of these outer domain interactions through glycosylation of key residues. These results explain the essential role of glycosylation in C. jejuni motility, and show how the outer domains have evolved to play a role not previously found in other bacteria.
Innate immunity, as opposed to acquired immunity, involves a genetically preprogrammed potential for the recognition of pathogens that exists before an organism ever encounters the threat. Toll-like receptor 5 (TLR5) has been shown to recognize a highly conserved epitope in bacterial flagellin as part of this innate immunity (1), and various experiments have shown that this epitope is crucial for the assembly of bacterial flagellar filaments (2–5). Since this conserved epitope is recognized by vertebrates from zebrafish to humans, the assumption has been that this epitope in many pathogenic bacteria has not diverged in more than 350 million years, and that TLR5 has evolved to recognize a region that is functionally constrained (3, 4). However, it is apparent that some bacteria, such as the α and ε Proteobacteria, have tolerated sequence changes within this epitope and escape TLR5 activation. Since such important human pathogens as Helicobacter pylori and Campylobacter jejuni are members of the ε Proteobacteria, it is of great interest to understand how they evolved to avoid TLR5 activation.
A number of studies have provided a very detailed picture of the interactions of bacterial flagellin with TLR5. Mutational analysis initially mapped the epitope to residues 79 to 117 and 408 to 439 of Salmonella typhimurium flagellin (5). A subsequent study showed that the evasion of TLR5 activation by H. pylori and C. jejuni was due to amino acid substitutions in the region 89 to 96. A crystal structure of a fragment of flagellin bound to TLR5 showed the structural basis for this recognition (3). Bacterial flagellins have two largely coiled-coil domains that are called D0 and D1, and these are highly conserved across all bacteria. These domains assemble to form the core of the flagellar filament. In contrast, there are outer, globular domains that are highly variable. In S. typhimurium these are D2 and D3 (6), while, in Bacillus subtilis, the outer domains do not exist (7). The TLR5−flagellin crystal structure showed that the main interaction with TLR5 involved three helices in flagellin D1, one of which included the previously described 89 to 96 epitope. A subsequent crystallographic and mutational analysis extended these results (2) and showed that three residues in B. subtilis flagellin formed a “hot spot” that was the largest determinant of this interaction: R89, L93, and E114. These three residues are quite conserved among the bacterial flagellins that activate TLR5, while, in C. jejuni, the corresponding residues are T91, K95, and D115.
During the dark ages of cryoelectron microscopy (cryo-EM), a very low-resolution reconstruction of C. jejuni flagellar filaments containing the G508A mutation suggested that C. jejuni was able to accommodate the mutations in the TLR5 89 to 96 epitope by having a dramatically different quaternary structure, with 7 protofilaments rather than the 11 seen in all other bacterial flagellar filaments that had been studied (8). The G508A mutation was used, as it led to straight flagellar filaments, rather than the supercoiled filaments that exist in the wild type (WT). The use of straight filaments (9) had been crucial to the structural studies of Salmonella flagellar filaments (6, 10) which have served as a model system for understanding flagellar structure.
The resolution revolution in cryo-EM (11–13) has led to the ability to reach near-atomic resolution almost routinely for many specimens. At less than near-atomic resolution, it is possible for ambiguities in helical symmetry to exist (14), and we now understand that, for some specimens, ambiguities can exist at even 5-Å resolution. We have reinvestigated the structure of the C. jejuni flagellar filaments using a direct electron detector, and have generated a cryo-EM reconstruction at 3.5-Å resolution. There are clearly 11 protofilaments in this structure, not 7, and the resolution is sufficient to model glycans into the density for the outer domains. We show that the amino acid changes in the TLR5 epitope would destabilize the filament, and stabilization of the filament occurs by large interactions among the outer domains rather than by simply compensatory mutations at the site on a neighboring subunit facing the TLR5 epitope. We further show the role of glycosylation in the interactions between neighboring outer domains, and thus explain why glycosylation is necessary for filament formation in C. jejuni (15). In contrast, the outer domains in Salmonella do not interact with each other (6, 10), and mutations have been found lacking most or all of the outer domains that assembled flagellar filaments and were motile (16). As previously stated, in the B. subtilis WT filament, the outer domains are almost completely absent (7). While the outer domains in H. pylori have diverged significantly from C. jejuni, we show that a similar mechanism of stabilization is likely to occur in H. pylori.
Results
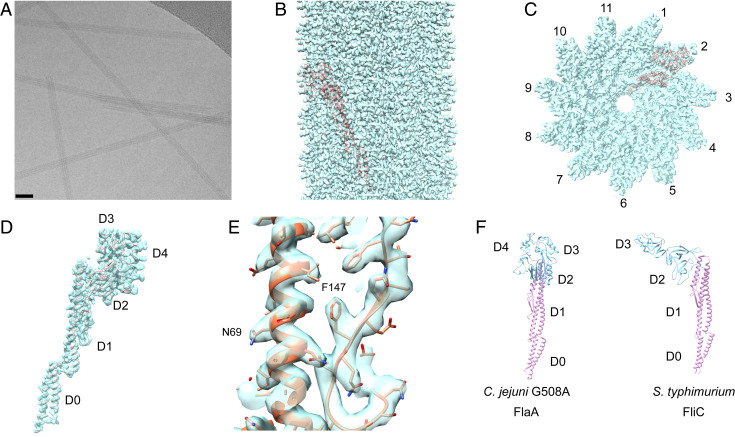
We used C. jejuni strain 81176, where the minor flagellin FlaB was knocked out, and the WT flagellin was replaced by the G508A mutant (8). The C. jejuni G508A flagellar filaments were imaged by cryo-EM (Fig. 1A) and were revealed, as expected, to be straight filaments. Using Iterative Helical Real Space Reconstruction (IHRSR) (17), the helical structure of the C. jejuni filaments was solved. Analysis of the averaged power spectrum from thousands of images of the flagellar filament revealed several possible symmetries arising from either 7, 9, or 11 protofilaments. Trying each of these, we were able to get a high-resolution structure (Fig. 1 B–E) for the C. jejuni flagellar filaments showing that it has 11 protofilaments (Fig. 1C). The correctly indexed power spectrum is shown in SI Appendix, Fig. S1A. Using Relion (18), we were able to reach a resolution of 3.5 Å as determined by the “gold-standard” map:map Fourier shell correlation (FSC) plot (SI Appendix, Fig. S1B).
Fig. 1.
Cryo-EM structure of the C. jejuni G508A flagellar filament. (A) Cryoelectron micrograph of the flagellar filaments. (Scale bar, 40 nm.) (B) Side view of the 3.5-Å resolution C. jejuni G508A density map. (C) A top view of the density map shows 11 protofilaments. A model of a single flagellin subunit (orange) is fit into the map. (D) The model of a single C. jejuni G508A flagellin fit into its corresponding electron density. (E) A blowup showing the quality of the map around residues 50 to 160. (F) Comparison of the C. jejuni and S. typhimurium flagellins. The inner core domains (D0 and D1) are in pink, and are shown in the same orientation. The outer domains for C. jejuni (D2, D3, and D4) and S. typhimurium (D2 and D3) are shown in cyan.
The filaments have a canonical L-type flagellar symmetry with an axial rise of 4.8 Å and a twist of 65.32°. This means that the protofilaments supercoil in a left-handed manner, due to their being very slightly more than 11 subunits in every two turns of the right-handed 1-start helix. A model of a single C. jejuni flagellin subunit fit into its corresponding electron density is shown in Fig. 1D. Using the model:map FSC with a threshold of 0.5, a resolution of 3.5 Å was obtained (SI Appendix, Fig. S1C), consistent with the map:map FSC results from Relion. The flagellin subunit, as shown in Fig. 1D, can be subdivided into five structural domains. These are the two conserved inner core domains D0 and D1, and three domains on the outside of the filaments. These outer domains are referred to as D2, D3, and D4 and are shown in greater detail in SI Appendix, Fig. S2 A and B. A close-up of the atomic model for D1 fit to the map is shown in Fig. 1E. While domains D0 and D1 of the C. jejuni flagellin look the same as those of all other existing bacterial flagellin structures, the outer domains are ∼80 amino acids larger than those of the next largest solved flagellin structure, from S. typhimurium (6). A comparison of the C. jejuni and S. typhimurium flagellins (Fig. 1F) shows the conservation of D0 and D1 (pink) and the absence of detectable homology in the outer domains (blue). Thus, domains D2 and D3 in C. jejuni have no homology with domains D2 and D3 in S. typhimurium.
The Dali server (19) was used to search for structural homologs of each individual C. jejuni FlaA outer domain. Domains D2 and D3 are, in fact, structural homologs of each other (SI Appendix, Fig. S2B), and searching for homologs to D2 and D3 yielded the same top hits but in different order. Interestingly, all of these top hit homologs are domains from various flagella-related proteins (SI Appendix, Fig. S2C). Two of the top hits for D2 of C. jejuni were the outer domain from the Pseudomonas aeruginosa A-type flagellin FliC (20) and filament cap protein, FliD, from Bdellovibrio bacteriovorus (21). Some of C. jejuni FlaA D3′s top hits were FliD from H. pylori (22) and D4 from the C. jejuni flagellar hook protein FlgE (23). The top Dali server hits for C. jejuni FlaA D2 and D3 are shown in SI Appendix, Table S1, along with the corresponding Z score.
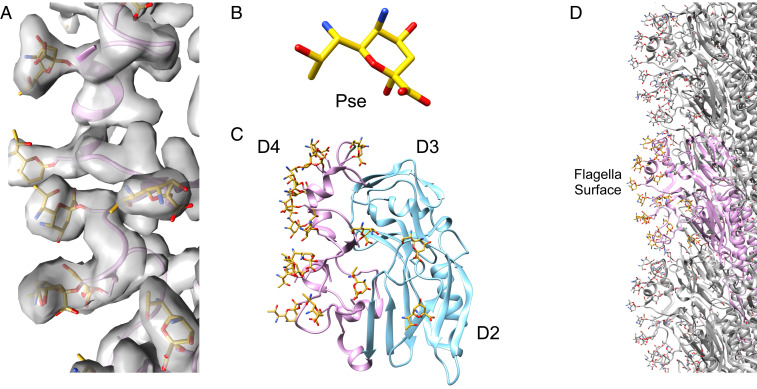
During the flagellin model building process, large extra densities were observed in the map contiguous with serine and threonine residues. These densities were all quite uniform in size and are indicative of O-linked glycosylation. FlaA from C. jejuni strain 81176 has been shown to be glycosylated entirely with pseudaminic acid (Pse) and its derivatives (24). Fig. 2A shows that these glycans bound to their serine or threonine residues provide an excellent fit to the map. Due to the inability to distinguish between the slightly different-in-size Pse derivatives at ∼3.5-Å resolution, only the basic Pse glycan (Fig. 2B) was modeled at each site. Density corresponding to glycosylation was found for 17 different residues on the outer domain. These residues are listed in SI Appendix, Fig. S3A and shown in the sequence in SI Appendix, Fig. S3B. The majority of these residues (15 of the 17) are in D4 (Fig. 2C). As might be expected, all of the glycosylation is present on the surface of the flagellar filament (Fig. 2D).
Fig. 2.
Extensive glycosylation of the C. jejuni outer domains with Pse glycans. (A) The density map (transparent gray) with the fit of Pse glycans (gold). (B) The atomic model of a single Pse glycan. (C) Flagellin outer domains are shown in pink (D4) and blue (D2, D3), with its glycan moieties shown in gold. (D) The surface-exposed region of a single 11-start protofilament.
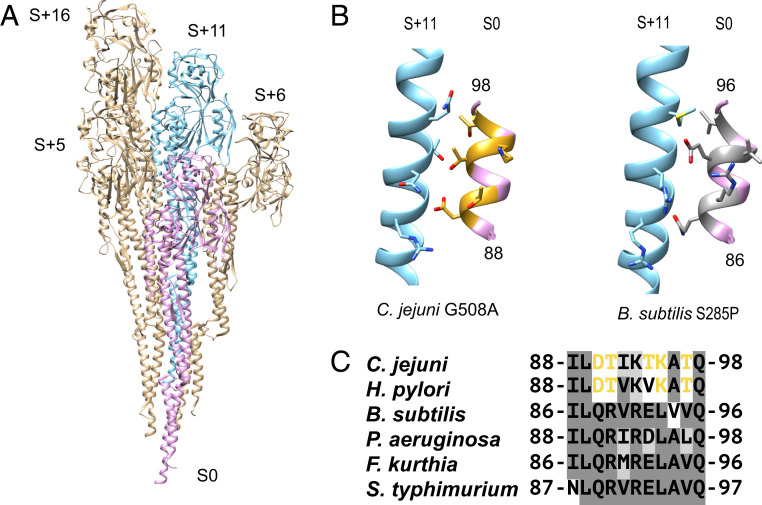
It was previously proposed (8) that changes in the TLR5 epitope that would destabilize the flagellar filament led to a different packing with only seven protofilaments. Since we now know that is wrong, the key question is, How does the C. jejuni accommodate mutations in D1 which are known to abrogate filament formation in Salmonella FliC? We considered amino acids 88 to 98 in C. jejuni which include four more amino acids than those focused on by Andersen-Nissen et al. (4). First, we examined how these residues interacted with neighboring flagellin subunits. Fig. 3A shows all of the flagellin subunits (S+5, S+6, S+11, S+16) that make contacts with a reference subunit (S0, in pink). The most prominent interactions the Campylobacter TLR5 epitope makes are with the S+11 subunit, in blue. Fig. 3B shows a comparison of the TLR5 epitope interactions along the 11-start helix between C. jejuni and B. subtilis, which has a similar TLR5 epitope to S. typhimurium. In the C. jejuni model, amino acids that are significantly different from the ones conserved across most bacteria are colored gold. In B. subtilis, these corresponding residues are colored gray. Fig. 3C shows a sequence alignment of the TLR5 epitope in C. jejuni and H. pylori against several flagellins whose filament structure has been solved. In C. jejuni, D90, T91, T94, and T97 are at the interface of the S0 subunit with S+11. In B. subtilis, the corresponding residues are Q88, R89, E92, and V95.
Fig. 3.
Interactions of the TLR5 recognition site (C. jejuni residues 88 to 98) with the next 11-start flagellin subunit (S+11). (A) Each subunit in the filament contacts four other subunits. The subunits that contact a single flagellin (S0) are S+5, S+6, S+11, and S+16. (B) Comparison of the contacts between the TLR5 site (pink) and the next 11-start subunit, S+11 (blue), in C. jejuni (Left) and B. subtilis (Right). Residues that are not identical between the two TLR5 sites are shown in gold for C. jejuni and dark grey for B. subtilis. (C) Alignment of the TLR5 sequences from C. jejuni flagellin with those of five flagellins from different species whose high-resolution structures have been determined by cryo-EM.
Using PDBePISA (25), we calculated the interfacial surface area between the TLR5 epitope of the S0 subunit and the entire S+11 subunit (S0-TLR5/S+11) of several flagellins from different bacteria. The interfacial surface areas between the entire S0 and S+11 subunits were also calculated (Table 1). The S0-TLR5/S+11 interface is much smaller in C. jejuni than in the other flagellar filaments. Strikingly, the interface between the entire C. jejuni S0/S+11 subunit interface was greater than in any other flagellar filament. These results suggest that, in C. jejuni, the weakened interactions between the TLR5 interface and the adjacent subunit are compensated for by interactions between the subunits not found in the other structures. Most of this arises from interactions of the outer domains.
Table 1.
Comparison of the interactions between the TLR5 recognition sequence and the next 11-start flagellin across different flagellar filament structures, with the PDB ID code
| Bacteria, PDB ID code | TLR5 site 11-start interactions interfacial surface area, Å2 | 11-Start full model interfacial area, Å2 |
| C. jejuni, G508A 6X80 | 300.6 (AA 88 to 98) | 2,869.9 |
| B. subtilis, S285P 5WJY | 410.8 (AA 86 to 96) | 1,881.5 |
| B. subtilis, N226Y 5WJT | 381 (AA 86 to 96) | 1,858.9 |
| P. aeruginosa, A443V 5WK5 | 369.5 (AA 88 to 98) | 1,845 |
| F. kurthia, 6T17 | 371.4 (AA 86 to 96) | 1,964.9 |
| S. typhymurium, FljB A461V 6JY0 | 385.8 (AA 87 to 97) | 1,787.3 |
AA, amino acid.
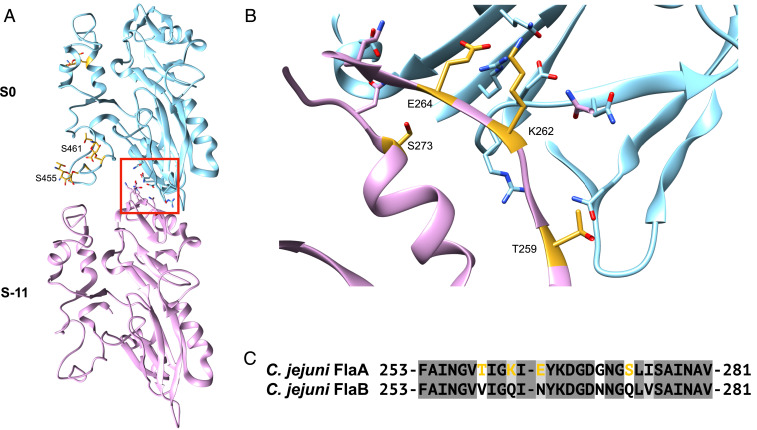
The outer domains of the C. jejuni flagellin strongly interact with the outer domains of adjacent subunits on the same protofilament. Fig. 4A shows the interactions of two adjacent flagellin outer domains along the same 11-start protofilament. Previously, it has been shown that when any one of the glycosylation sites S426, S455, or S461 are mutated to alanine, C. jejuni 81 to 176 is unable to assemble full-length flagellar filaments and exhibits greatly reduced motility (15). In our filament model, two of these glycosylated serines (S455 and S461) are directly at the 11-start interface between the D4 domains of both subunits, and are indicated (Fig. 4A). Extensive contacts are made between D3 of S−11 and D1 and D2 of S0. These are highlighted by the red box and shown in greater detail in Fig. 4B. When FlaA has been deleteriously mutated or deleted, C. jejuni has been known to form very short less-stable filaments using only FlaB, and the bacteria exhibit greatly reduced motility (26). There is 95% sequence identity between the two flagellins, and some sequence differences between FlaA and FlaB occur in D3 (Fig. 4C); however, there are some in D0 and D1 as well. The gold colored residues in Fig. 4B correspond to residues in D3 at the 11-start outer domain interface that are significantly different between FlaA and FlaB. Thus, the instability of the FlaB filaments may be directly due to the loss of interactions involving D3.
Fig. 4.
Outer domain interactions between adjacent subunits along the same 11-start protofilament. (A) Two neighboring subunits with the glycans attached to S455 and S461 in domain D4 are shown in gold. The 11-start interaction site between D2 in one subunit and D3 in an adjacent subunit is boxed in red. (B) Close-up of this D2/D3 11-start interface. Residues at the interface between the two subunits in the FlaA filament that are significantly different in the C. jejuni FlaB sequence are in gold. Sequence alignment of C. jejuni FlaA and FlaB at residues 253-281 which are in domain D3 and contact domains D1 and D2 of the adjacent outer domain. FlaA residues colored gold are the same ones indicated in B.
Discussion
The structure of the C. jejuni flagellar filaments provides clear evidence for how ε Proteobacteria evolved to evade detection by TLR5. The amino acids in the C. jejuni TLR5 epitope display a dramatically weakened ability to interact with the adjacent subunit on the same protofilament. However, the entire C. jejuni flagellin has much stronger interactions with its neighboring 11-start subunit. These are the result of outer domain interactions among the adjacent subunits that are entirely absent in all other existing flagellar filament structures. The two outer domains (D2 and D3) that make these contacts are actually structural homologs of not only each other but of domains found in various flagellar structural proteins across different species of bacteria (SI Appendix, Figs. S2 and S4).
Two of the structural homologs are the flagellar hook protein FlgE domain D4 and the hook−filament junction protein FlgK D4, both from C. jejuni. SI Appendix, Fig. S4 shows how both FlaA D3 and FlgE D4 have conserved interactions along the 11-start interface and a few conserved residues. The homology between these domains is likely due to FlaA, FlgE, and FlgK sharing a common ancestor protein which functioned as the protein subunit for the filament before the specialized hook evolved. The conservation of a few key residues in the 11-start interface in both the C. jejuni FlaA D3 and FlgE D4 suggests that there are similar stabilizing interactions among the outer domains of the hook, hook junction protein, and flagellar filament. The role of homologs playing a role at filament ends and in filament nucleation has been seen for both actin and tubulin. In the case of actin, we know that Arps (actin-related proteins), which are structural homologs of actin, such as Arp2/3, play an important role in actin filament nucleation and branching (27). In the case of tubulin, gamma-tubulin, a homolog of alpha- and beta-tubulin, plays a key role in nucleating microtubules (28).
The glycosylation of Ser426, Ser455, and Ser-461 has been previously shown to be critical for filament formation and motility (15). The glycans attached to Ser455 and Ser461 in the 11-start interface are part of the extensive interactions of the flagellin outer domains. Ser426 is not at the interface with any neighboring flagellins and may play a role in the stabilization of the outer domains. Many of the other serine/threonine residues that are glycosylated in our structure have been shown to be critically important for autoagglutination, which is correlated with C. jejuni virulence (15, 29). It was previously observed that Ser207 in D2, Ser409 in D4, and Thr482 in D1 were glycosylated. We detect no significant glycosylation in our structure at these sites. This is likely due to either disorder of the glycans or a heterogeneity in glycosylation, such that the glycans are attached at these residues to only some subunits. For the glycosylated residues that we do see clearly (Fig. 2A), the glycans must be both highly ordered and stoichiometrically attached to these residues in almost every subunit. Most of the glycosylation was not seen in the cryo-EM map of an extensively glycosylated archaeal Type 4 pilus (30). The glycosylation we see at Thr469 has not previously been reported.
All ε Proteobacteria possess an alanine at A493, the position corresponding to I411 in S. typhimurium. When I411 is mutated to alanine in S. typhimurium, TLR5 activation is reduced and motility is completely lost (5). Follow-up studies showed that, when S. typhimurium FliC I411A amino acids Arg58 and Gly59 were mutated to the corresponding H. pylori residues (Ser59 and Ser-60), motility and filament formation were restored, but the TLR5 activation was still greatly reduced (4). These results were mapped to the structure of the S. typhimurium flagellar filament to show that these three residues were predicted to be at the interface between two subunits along the same protofilament. We show (SI Appendix, Fig. S5) that the predictions about the H. pylori and C. jejuni flagellin interactions were correct and that A493 in C. jejuni would be in the same position as S. typhimurium I411. C. jejuni has amino acids A59 and T60 instead of the two serine side chains in H. pylori at these positions.
While we have only determined the structure of the C. jejuni flagellar filament, our results give insights into the structure of the H. pylori filament, and it is certainly likely that extensive outer domain interactions in H. pylori also stabilize that flagellar filament. SI Appendix, Fig. S6A shows a comparison between the observed secondary structure of the outer domains of C. jejuni FlaA and the predicted (31) secondary structure of H. pylori FlaB. The predicted H. pylori structure closely resembles the secondary structure observed, except for a few places, most notably at the N terminus of the outer domains. As a control, the predicted C. jejuni secondary structure agrees quite well with the observed secondary structure (SI Appendix, Fig. S6B). Strikingly, H. pylori appears to lack most of D4, but the expectation would be that the extensive interactions observed in C. jejuni between D2 in one subunit and D3 in an adjacent subunit would be preserved in H. pylori.
Our results reemphasize two points. First, at low resolution, it may never be possible to determine the correct helical symmetry (14) for a filament or tube. Second, although the bacterial flagellum has been a model for Creationists of “irreducible complexity” showing that the flagellum could never have evolved (32), it actually serves as a beautiful system for studying evolution (33). In response to the innate immune surveillance that vertebrates developed to recognize pathogenic bacteria, ε Proteobacteria such as C. jejuni and H. pylori compensated for mutations in the TLR5 recognition site that destabilize the flagellar filament by developing outer domain interactions that stabilize these filaments. We expect that future studies of bacterial flagella will further elucidate the evolutionary divergence of extant flagellar systems.
Methods
Preparation of C. jejuni G508A Flagellar Filaments.
C. jejuni cells were obtained from a Mueller Hinton broth culture incubated at 37 °C in a microaerobic atmosphere (5% O2, 10% CO2, 85% N2). The culture was spun, and the pellet was resuspended in a low-salt buffer (20 mM Tris, 50 mM sodium chloride, pH 7.4). The bacterial suspension was subjected to bead beating, 6 × 30 s. Cell debris was collected by centrifugation, resuspended in the low-salt buffer, and centrifuged at 45,000 rpm for 1 h. The pellet was washed in the low-salt buffer and centrifuged at 45,000 rpm for one additional hour. After the spin was complete, the pellet was resuspended in the low-salt buffer, and the sample was stored at 4 °C until processing.
Cryo-EM Imaging.
The sample was imaged at the Molecular Electron Microscopy Core (MEMC) at the University of Virginia on a Titan Krios equipped with a K3 direct electron detector with a nominal magnification of 81,000× (1.08 Å per pixel). Movies of each area imaged contained 40 frames with a total dose of 51 (e−/Å2).
Image Processing.
Motion correction on the images was done using Motioncorr2 (34). Motion correction was done using both the full frames and then frames 2 to 17 (∼20 e−/Å2) using dose weighting. CTFfind3 (35) was used to calculate the contrast transfer function (CTF) for each image, and images with poor CTFs were discarded. Flagellar filaments were boxed using the e2helixboxer program from the Eman2 software package (36).
Helical Reconstruction.
Initial helical reconstruction on CTF-multiplied dose-weighted images was done using the IHRSR implemented in Spider (17). The power spectra of hundreds of nonoverlapping images of flagellar filaments were calculated, and, from this, an averaged power spectrum was created. Indexing this power spectrum revealed the possible helical symmetries of the C. jejuni G508A flagellar filaments (17, 37). The most likely symmetries were ones with 7, 9, and 11 protofilaments. Using IHRSR, we tried each of the possible symmetries, and the canonical flagellar symmetry with 11 protofilaments was determined to be correct, given that we were able to clearly see the pitch and hand of the α-helices and some bulky side chains were visible.
The 11-protofilament volume from IHRSR in Spider was used as the input volume for Refine3d in Relion (18, 38). With ∼120,000 images (we started with 280,000 before Class2d) and Refine3d followed by CTFRefine, Particle Polishing, and Postprocessing (38), we were able to reach a resolution of 3.5 Å using the “gold-standard” map:map FSC.
Model Building.
Using Chimera (39), the L-type flagellin from B. subtilis S285P, Protein Data Bank (PDB) ID code 5WJY (7), was fit into the D0/D1 region of the C. jejuni G508A density map. Using Coot (40), the amino acids in D0/D1 of the starting model were manually replaced with the corresponding ones from C. jejuni G508A and fit to the map. The outer domain region was then built manually, starting with a Cα trace. The structure of the Pse glycan was generated from PubChem Compound ID 101137651. A single C. jejuni flagellin and its corresponding map density served as inputs into RosettaCM (41) which rebuilt the model with better geometries and secondary structure. The side chain placement and secondary structure of the model were further refined in Coot and Phenix (42). Finally, a full filament model was generated with Rosetta scripts (43) and real-space refined in Phenix. The final model refinement statistics (Table 2) were validated with MolProbity (44).
Table 2.
CryoEM imaging and model refinement statistics for the C. jejuni G508A flagellar filament
| Data collection | Parameter value |
| Magnification | 81,000 |
| Voltage (kV) | 300 |
| electron exposure (e-Å2) | 20 |
| Defocus range (um) | 1.25 to 2.25 |
| Pixel size (Å) | 1.08 |
| Image processing | |
| Helical rise (Å) | 4.8 |
| Helical twist (°) | 65.32 |
| Initial number of particles | 276,180 |
| Final number of particles | 116,959 |
| Map sharpening B factor (Å2) | −90 |
| Model:map FSC (0.50) | 3.5 |
| Gold-standard” map:map FSC (0.143) | 3.5 |
| Model composition (single flagellin) | |
| Nonhydrogen atoms | 4431 |
| Number of amino acid residues | 574 |
| Number of ligands | 17 |
| B factors (Å2) | |
| Protein | 76.03 |
| Ligand | 106.41 |
| Root-mean-square deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (deg) | 0.628 |
| Validation | |
| MolProbity score | 1.89 |
| Clashscore | 7.08 |
| Rotamer outliers (%) | 0 |
| Protein geometry | |
| Ramachandran favored (%) | 91.58 |
| Ramachandran allowed (%) | 9.24 |
| Ramachandran outliers (%) | 0.17 |
| Model PDB ID | 6X80 |
| Density map EMDB ID | EMD-22088 |
Sequence Alignments.
All sequence alignments in this study were done in Jalview 2 (45) using the Clustal Omega alignment (46). Amino acids in a particular flagellin that were identical to the majority of the amino acids at the same position in other flagellins were shaded in dark gray (Figs. 3C and 4C). Similar amino acids were shaded in light gray (Figs. 3C and 4C). Amino acids with no similarity were not shaded. The alignment in SI Appendix, Fig. S4A was done according to a Blosum62 scoring matrix.
Structural Analysis of C. jejuni G508A.
Structural homologs to the C. jejuni FlaA outer domains D2, D3, and D4 were identified with the Dali server (19, 47). All secondary structure predictions were made with JPred (31). Interfacial analyses were made using the PDBePISA server (25). For interfacial analysis, at least one structure from each of the various bacteria whose flagellar filaments have been studied was chosen. These were S. typhimurium (48), Firmicute kurthia (49), P. aeruginosa (7), and B. subtilis (7).
Data Availability.
The atomic coordinates have been deposited at the PDB under accession number 6X80. The density map has been deposited at the Electron Microscopy Data Bank under accession number EMD-22088.
Supplementary Material
Acknowledgments
Cryo-EM imaging of was done at the MEMC facility at the University of Virginia, which is supported by the School of Medicine. Within the Core, the Titan Krios (Grant SIG S10-RR025067) and K3/GIF (Grant U24-GM116790) were purchased, in part, with the designated NIH grants. This work was supported by NIH Grant R35GM122510 (E.H.E.). M.A.B.K. was supported by NIH Grant T32 GM080186 and the University of Virginia Robert R. Wagoner Fellowship. This work was also supported by US Navy Work Unit (Program 6000.RAD1.DA3.A0308). The views expressed in this article reflect the results of research conducted by the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, or the US Government. F.P. is a federal employee of the US Government. This work was prepared as part of his official duties. Title 17 U.S.C. 105 provides that copyright protection under this title is not available for any work of the US Government. Title 17 U.S.C. 101 defines a US Government work as work prepared by a military service member or employee of the US Government as part of that person’s official duties.
Footnotes
The authors declare no competing interest.
Data deposition: The atomic coordinates have been deposited at the Protein Data Bank under accession number 6X80. The density map has been deposited at the Electron Microscopy Data Bank under accession number EMD-22088.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010996117/-/DCSupplemental.
References
- 1.Hayashi F. et al., The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Song W. S., Jeon Y. J., Namgung B., Hong M., Yoon S. I., A conserved TLR5 binding and activation hot spot on flagellin. Sci. Rep. 7, 40878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon S. I. et al., Structural basis of TLR5-flagellin recognition and signaling. Science 335, 859–864 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen-Nissen E. et al., Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 9247–9252 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith K. D. et al., Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4, 1247–1253 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Yonekura K., Maki-Yonekura S., Namba K., Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Wang F. et al., A structural model of flagellar filament switching across multiple bacterial species. Nat. Commun. 8, 960 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galkin V. E. et al., Divergence of quaternary structures among bacterial flagellar filaments. Science 320, 382–385 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Hyman H. C., Trachtenberg S., Point mutations that lock Salmonella typhimurium flagellar filaments in the straight right-handed and left-handed forms and their relation to filament superhelicity. J. Mol. Biol. 220, 79–88 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Maki-Yonekura S., Yonekura K., Namba K., Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat. Struct. Mol. Biol. 17, 417–422 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Egelman E. H., The current revolution in cryo-EM. Biophys. J. 110, 1008–1012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai X. C., McMullan G., Scheres S. H., How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Kühlbrandt W., Cryo-EM enters a new era. eLife 3, e03678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egelman E. H., Ambiguities in helical reconstruction. eLife 3, e04969 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewing C. P., Andreishcheva E., Guerry P., Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191, 7086–7093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshioka K., Aizawa S., Yamaguchi S., Flagellar filament structure and cell motility of Salmonella typhimurium mutants lacking part of the outer domain of flagellin. J. Bacteriol. 177, 1090–1093 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egelman E. H., A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 18.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm L., Rosenström P., Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song W. S., Yoon S. I., Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament. Biochem. Biophys. Res. Commun. 444, 109–115 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Cho S. Y., Song W. S., Yoon S. I., Crystal structure of the flagellar cap protein FliD from Bdellovibrio bacteriovorus. Biochem. Biophys. Res. Commun. 519, 652–658 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Cho S. Y. et al., Structural analysis of the flagellar capping protein FliD from Helicobacter pylori. Biochem. Biophys. Res. Commun. 514, 98–104 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Matsunami H., Barker C. S., Yoon Y. H., Wolf M., Samatey F. A., Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat. Commun. 7, 13425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thibault P. et al., Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276, 34862–34870 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Wassenaar T. M., Bleumink-Pluym N. M., van der Zeijst B. A., Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10, 2055–2061 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoddard P. R., Williams T. A., Garner E., Baum B., Evolution of polymer formation within the actin superfamily. Mol. Biol. Cell 28, 2461–2469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kollman J. M., Merdes A., Mourey L., Agard D. A., Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misawa N., Blaser M. J., Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect. Immun. 68, 6168–6175 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F. et al., An extensively glycosylated archaeal pilus survives extreme conditions. Nat. Microbiol. 4, 1401–1410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drozdetskiy A., Cole C., Procter J., Barton G. J., JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dembski W. A., No Free Lunch: Why Specified Complexity Cannot Be Purchased without Intelligence, (Rowman & Littlefield, 2001). [Google Scholar]
- 33.Egelman E. H., Reducing irreducible complexity: Divergence of quaternary structure and function in macromolecular assemblies. Curr. Opin. Cell Biol. 22, 68–74 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Zheng S. Q. et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mindell J. A., Grigorieff N., Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Tang G. et al., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Egelman E. H., Reconstruction of helical filaments and tubes. Methods Enzymol. 482, 167–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zivanov J. et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen E. F. et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Song Y. et al., High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das R., Baker D., Macromolecular modeling with rosetta. Annu. Rev. Biochem. 77, 363–382 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Williams C. J. et al., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F., Higgins D. G., Clustal omega. Curr. Protoc. Bioinformatics 48, 3 13 11-3 13 16 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Holm L., Sander C., Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi T. et al., Structural and functional comparison of Salmonella flagellar filaments composed of FljB and FliC. Biomolecules 10, 246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blum T. B., Filippidou S., Fatton M., Junier P., Abrahams J. P., The wild-type flagellar filament of the Firmicute Kurthia at 2.8 Å resolution in vivo. Sci. Rep. 9, 14948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates have been deposited at the PDB under accession number 6X80. The density map has been deposited at the Electron Microscopy Data Bank under accession number EMD-22088.