Significance
Aneuploid cells experience significant cellular stress; however, the transcriptional consequences of aneuploidy remain highly debated. In aneuploid budding yeast, two opposing gene-expression patterns have been reported: the “environmental stress response” (ESR) and the “common aneuploidy gene-expression” (CAGE) signature, in which many ESR genes are oppositely regulated. Here we resolve this controversy. We show that the ESR represents a common response to aneuploidy and explain why the CAGE signature does not. Our data further suggest that activation of the ESR has profound consequences on the cellular physiology of aneuploid cells, leading to ribosome loss. Thus, our work provides critical insights into the coordination between cell division and macromolecule biosynthesis and a potential explanation for why aneuploid cells are less dense.
Keywords: aneuploidy, ESR, CAGE, ribosome loss
Abstract
Aneuploidy, a condition characterized by whole chromosome gains and losses, is often associated with significant cellular stress and decreased fitness. However, how cells respond to the aneuploid state has remained controversial. In aneuploid budding yeast, two opposing gene-expression patterns have been reported: the “environmental stress response” (ESR) and the “common aneuploidy gene-expression” (CAGE) signature, in which many ESR genes are oppositely regulated. Here, we investigate this controversy. We show that the CAGE signature is not an aneuploidy-specific gene-expression signature but the result of normalizing the gene-expression profile of actively proliferating aneuploid cells to that of euploid cells grown into stationary phase. Because growth into stationary phase is among the strongest inducers of the ESR, the ESR in aneuploid cells was masked when stationary phase euploid cells were used for normalization in transcriptomic studies. When exponentially growing euploid cells are used in gene-expression comparisons with aneuploid cells, the CAGE signature is no longer evident in aneuploid cells. Instead, aneuploid cells exhibit the ESR. We further show that the ESR causes selective ribosome loss in aneuploid cells, providing an explanation for the decreased cellular density of aneuploid cells. We conclude that aneuploid budding yeast cells mount the ESR, rather than the CAGE signature, in response to aneuploidy-induced cellular stresses, resulting in selective ribosome loss. We propose that the ESR serves two purposes in aneuploid cells: protecting cells from aneuploidy-induced cellular stresses and preventing excessive cellular enlargement during slowed cell cycles by down-regulating translation capacity.
Dividing cells rely on multiple complex mechanisms to correctly segregate their chromosomes and create euploid progeny. When chromosome missegregation occurs, daughter cells can acquire an incorrect number of chromosomes that is not a complete multiple of the haploid genome, a condition termed aneuploidy. Aneuploidy can occur naturally; for example, 17% of wild budding yeast isolates harbor aneuploidies and are thought to have evolved mechanisms to tolerate these aneuploid karyotypes (1, 2). In most cases, however, aneuploidy is highly detrimental, especially in multicellular animals (3).
Various models have been developed to study aneuploidy in Saccharomyces cerevisiae. Their analyses led to the conclusions that aneuploidy affects a wide range of cellular processes, such as protein homeostasis, metabolism, and cell wall integrity, and results in an overall decrease in cellular fitness (3, 4). However, how aneuploidy affects gene expression has remained controversial. While it is clear that gene-expression scales with gene copy number in aneuploid cells, there is not yet a consensus on whether aneuploidy elicits a global transcriptional response in yeast and what this response may be.
We previously described that haploid aneuploid yeast cells harboring only one additional chromosome (henceforth disomic yeast strains) experience an environmental stress response (ESR) (5). The ESR is a transcriptional signature observed in response to nearly every type of exogenous stress, including hyperosmotic, heat shock, oxidative and reductive stress, and nutrient limitation. These conditions cause the coordinated up-regulation of ∼300 genes, also known as the “induced (i)ESR” and down-regulation of ∼600 genes, also known as the “repressed (r)ESR” (6, 7). Genes that are up-regulated compensate for various stressors and encode chaperones, amino acid transporters, and proteins involved in increasing endocytosis and proteasome activity. Down-regulated genes encode factors critical for transcription and translation, among them are genes encoding ribosomal proteins and proteins involved in ribosome biogenesis (5, 6). The ESR is not only observed in response to stress but also in cells that grow slowly or cells that are cell cycle arrested (4, 5, 8). Indeed, the strength of the ESR, that is the degree to which iESR genes are up-regulated and rESR genes are down-regulated, correlates remarkably well with growth rate, suggesting that this transcriptional signature is primarily determined by proliferation rate (4).
A recent study by Tsai et al. (9) reported that yeast cell populations harboring heterogenous aneuploidies do not exhibit the ESR. Instead, these aneuploid populations were described to exhibit a transcriptional response, termed the “common aneuploidy gene-expression” (CAGE) response. In the CAGE response, the genes that are up-regulated in the ESR are down-regulated, and those that are down-regulated in the ESR are up-regulated. The authors further found that the CAGE response bears similarity to a hypoosmotic shock gene-expression pattern, which was proposed to counter a decrease in cytoplasmic density observed in aneuploid cells (9, 10).
We report here the reanalysis of the gene-expression data generated by Tsai et al. (9) as well as replication of their experimental approach. These analyses showed that the CAGE gene-expression signature described by Tsai et al. (9) is an artifact caused by normalizing the gene expression of actively dividing aneuploid cells to that of euploid control cells that had grown to stationary phase. Growth into stationary phase is among the strongest inducers of the ESR (6). Thus, when Tsai et al. (9) compared the gene-expression pattern of euploid stationary phase cells to that of aneuploid cells that, due to their poor proliferation, had not yet reached stationary phase, the ESR caused by aneuploidy was obscured. We find that when exponentially growing euploid cells were used in gene-expression comparisons with aneuploid cells, the CAGE signature of aneuploid cells is no longer evident. Instead, aneuploid cell populations are found to exhibit the ESR, confirming previous reports (5). Using strains harboring multiple aneuploidies, we further show that the ESR causes a selective loss of ribosomes in aneuploid cells, providing a potential explanation for decreased cellular density previously reported to occur in response to chromosome gains and losses. We conclude that aneuploid budding yeast cells mount the ESR in response to aneuploidy-induced cellular stresses that results in ribosome loss.
Results
Exponentially Growing Haploid Cells Exhibit a Transcriptional Response, Previously Described to Be Unique to Aneuploid Cells.
A recent study (9) reported the absence of the ESR in populations of yeast cells harboring different, random aneuploid karyotypes. Instead, it was reported that these heterogeneous aneuploid yeast populations exhibit the CAGE signature. In this study, Tsai et al. (9) developed two protocols to generate heterogeneous populations of aneuploid cells, taking advantage of the fact that sporulation of triploid cells results in high levels of aneuploid progeny. In the first method, Tsai et al. (9) dissected spores obtained from triploid cells, grew individual aneuploid spores into colonies, pooled these colonies, and analyzed the gene-expression pattern of these cells ( SI Appendix, Fig. S1A ). We will refer to these aneuploid populations as “aneuploid populations obtained from tetrads.” Euploid haploid cells obtained from sporulating diploid cells and handled in the same manner as aneuploid cells served as the control (henceforth “euploid populations obtained from tetrads”). In the second protocol, Tsai et al. (9) sporulated triploid cells and then selected viable MATa aneuploid colonies by selecting for histidine prototrophy brought about by HIS5 expressed from the MATa-specific STE2 promoter ( SI Appendix, Fig. S1A ). We will refer to these aneuploid populations as “aneuploid populations obtained from MATa selection.” Again, euploid haploid cells obtained by sporulating diploid cells and MATa selected served as the euploid control (henceforth “euploid populations obtained from MATa selection”). Gene-expression analysis of these cell populations led to the identification of an expression signature Tsai et al. (9) termed CAGE response. This gene-expression signature resembles a hypoosmotic stress response and is essentially oppositely regulated to the ESR; 59.8% of genes up-regulated in the CAGE response are down-regulated in the ESR, while 13.2% of CAGE down-regulated genes are up-regulated in the ESR (9).
Having previously identified the ESR in yeast strains harboring defined aneuploidies (4, 5), we wished to determine why pooled aneuploid populations did not exhibit the ESR but instead the CAGE gene-expression signature. To this end, we reanalyzed the gene-expression data reported by Tsai et al. (9) by individually processing the samples rather than normalizing the aneuploid cell populations to the euploid control populations.
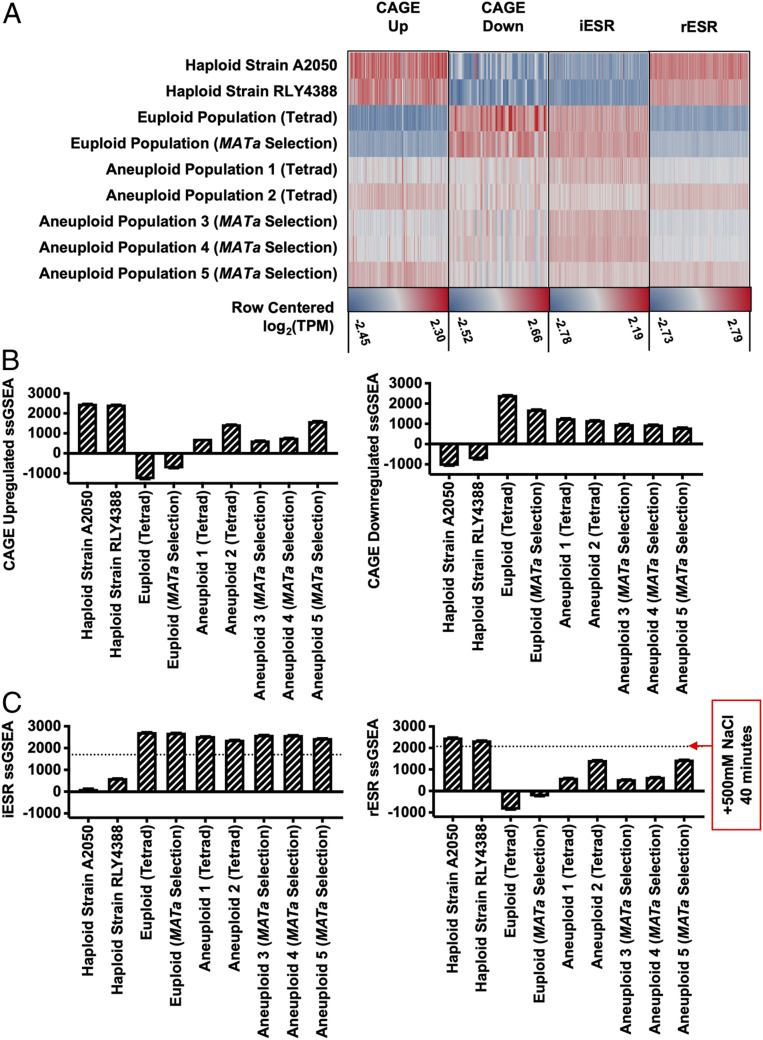
Among the RNA-sequencing (RNA-Seq) datasets deposited by Tsai et al. (9) was one termed “haploid control” that was obtained from a haploid strain RLY4388 grown in test tubes on roller drums (accession nos. GSM2886452 and GSM2886453) that the authors did not analyze. Using the RNA-Seq by expectation maximization (RSEM) processing method, we calculated the raw transcripts per million (TPM) values for the aneuploid and euploid cell populations as well as strain RLY4388, then log2 transformed these values with a +1 offset to avoid negative expression values, and created row-centered heatmaps for genes up-regulated and down-regulated in both the CAGE and ESR gene-expression signature (Fig. 1A ). As previously reported, we observed the CAGE gene-expression signature in the pooled aneuploid populations. Unexpectedly, however, strain RLY4388 exhibited the strongest CAGE gene-expression signature, and the pooled euploid populations, used as normalization controls by Tsai et al. (9), exhibited the strongest ESR gene-expression signature (Fig. 1A ).
Fig. 1.
Reanalysis of published aneuploid transcription data from Tsai et al. (9). Transcription data of haploid strain RLY4388 and euploid and aneuploid cell populations obtained from tetrad dissection (tetrad) or MATa selection (MATa selection) were reanalyzed with the RSEM processing method [Tsai et al. (9), accession no. GSE107997]. Raw TPM values were calculated for euploid cell populations, aneuploid cell populations, the haploid strain RLY4388 and exponentially growing haploid strain A2050. (A) Row centered log2(TPM) values for each gene-expression set (CAGE up-regulated, CAGE down-regulated, iESR, and rESR). Each gene set was row centered individually and has a separate maximum (red) and minimum (blue) value, noted underneath. (B) CAGE up-regulated and down-regulated ssGSEA projection values for the haploid strains A2050 and RLY4388, and euploid and aneuploid cell populations (tetrad and MATa selection). (C) iESR and rESR ssGSEA projection values for the haploid strains A2050 and RLY4388, and euploid and aneuploid cell populations (tetrad and MATa selection). The horizontal lines represent the iESR and rESR ssGSEA projection values for W303 wild-type cells (A2587) treated with 500 mM NaCl for 40 min, a positive control for the ESR induction. Error bars represent SD from the mean of technical replicates.
Given that the haploid strain RLY4388 exhibited the strongest CAGE gene-expression signature, it was of interest to determine the growth state of these cells. According to Tsai et al. (9), this strain was grown in regular test tubes, but the OD(600 nm) at which it was harvested was not recorded. To determine in which growth phase haploid strain RLY4388 was when harvested, we compared its gene-expression profile to that of an exponentially growing haploid strain of the same genetic background (S288C) from our laboratory (A2050) ( SI Appendix, Table S1). We observed a strong correlation between the genes expressed in both strains, indicating that haploid strain RLY4388 was in exponential phase when harvested ( SI Appendix, Fig. S2; Pearson, R 2 = 0.91, P < 0.001).
Tsai et al. (9) discovered the CAGE response and the absence of the ESR in aneuploid cell populations by normalizing the gene expression of aneuploid cell populations to euploid control cell populations (ref. 9 and SI Appendix, Fig. S3 A and B ). Given that our analysis of their raw data showed that the euploid control populations exhibited a strong ESR, we used the gene-expression dataset obtained from the haploid strain RLY4388 to normalize the gene-expression data from aneuploid populations instead of normalizing the data to that of euploid control populations. When compared to the gene-expression data generated from strain RLY4388, aneuploid populations obtained from tetrad dissection and MATa selection exhibited the ESR, and the CAGE signature was no longer evident ( SI Appendix, Fig. S3 C and D ). When we measured the differential gene expression between euploid populations and strain RLY4388, it was also apparent that the euploid populations (tetrad and MATa selection) were experiencing the ESR ( SI Appendix, Fig. S3 E and F ).
Given that the choice of euploid control (euploid populations versus a haploid wild-type strain RLY4388) made such a large difference in the experimental outcome, we decided to employ a data analysis method that does not depend on normalization. Single-sample gene set enrichment analysis (ssGSEA) generates a single projection value for a set of genes within a sample. These values can then be compared between samples in order to measure how the gene-expression distribution of that gene set changes across an experiment, i.e., overall increased or decreased expression of gene sets across samples (11). Using this approach, we confirmed that aneuploid cell populations exhibited the CAGE signature, while the euploid control cell populations did not (Fig. 1B ). However, the samples with the strongest CAGE signature, thought to be a characteristic of aneuploidy, were obtained from exponentially growing haploid strain A2050 and strain RLY4388 (Fig. 1B ).
ssGSEA analysis of the ESR in aneuploid and euploid cell populations revealed equally unanticipated results. As expected, the exponentially growing haploid strain A2050 and strain RLY4388 did not exhibit the ESR (Fig. 1C ). Consistent with our previous observations in disomic yeast strains (5), aneuploid cell populations showed the ESR, but the euploid control populations exhibited the ESR even more strongly (Fig. 1C ). The degree to which the ESR was induced in these euploid control populations was greater than in exponentially growing wild-type cells (A2587) treated with 500 mM NaCl for 40 min. We conclude that the euploid control populations analyzed by Tsai et al. (9) exhibit the strongest ESR signature, indicating that they experienced significant exogenous stress.
Stationary Phase Cells Exhibit the Environmental Stress Response.
It was curious that the euploid control populations generated by Tsai et al. (9) strongly exhibited the ESR. To determine the cause of this robust ESR, we repeated their tetrad dissection protocol to obtain euploid and aneuploid cell populations, employing the strains used by Tsai et al. (9) after detailed consultation with the authors. We dissected 200 and 770 tetrads obtained from diploid and triploid cells, respectively. Spore viability for the euploid strain was 97.3% and, as expected, significantly lower for triploid strains (40.2%) because many aneuploid strains are inviable. We then followed the protocol developed by Tsai et al. (9) and grew colonies obtained from viable spores in individual wells of a 96-deep well plate for 14 to 16 h in 200 μL YEPD medium at 25 °C ( SI Appendix, Fig. S1A ) Thereafter, we added 300 μL YEPD medium to cultures and grew them for an additional 5 h at 25 °C. The euploid and aneuploid cultures were then separately pooled to create heterogeneous euploid and aneuploid cell populations. Using this growth protocol, pooled euploid populations had reached an OD(600 nm) of 8.54. As expected, owing to aneuploid cells having significant proliferation defects, pooled aneuploid populations reached an OD(600 nm) of only 2.62.
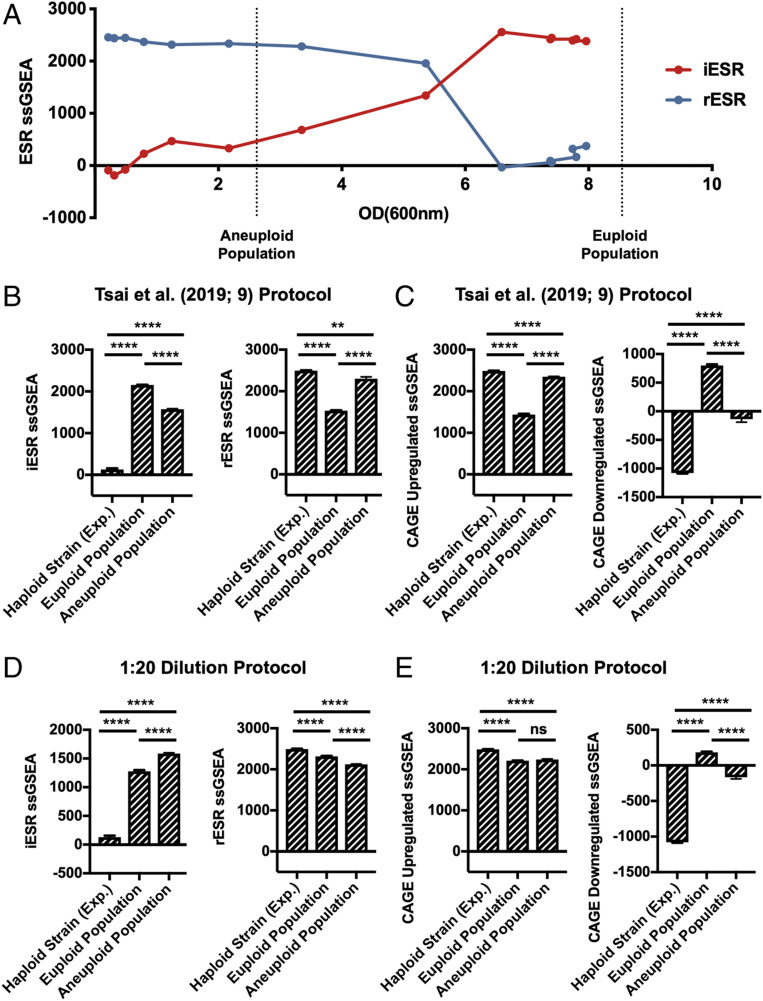
The high OD(600 nm) values reached by the euploid population provided a potential explanation for why they exhibited a strong ESR. As cultures approach stationary phase, cells experience starvation, which is among the strongest inducers of the ESR (6). To test this hypothesis, we determined at which OD(600 nm) S288C wild-type haploid cells activate the ESR. We grew cells into stationary phase in YEPD medium and measured rESR and iESR gene expression over time (Fig. 2A ). iESR gene induction was observed at around an OD(600 nm) of 3.5, determined by an increase in iESR ssGSEA projection values; rESR gene expression began to decline dramatically at an OD(600 nm) of 5.5. These results provided a potential explanation for why euploid control populations analyzed by Tsai et al. (9) exhibited such a strong ESR but aneuploid populations did not. The aneuploid cells had not yet reached an OD(600 nm) value where starvation-induced ESR induction occurs.
Fig. 2.
Effects of culture density on ESR strength in aneuploid cell populations. (A) iESR (red) and rESR (blue) ssGSEA projection values were determined at the indicated OD(600 nm) for S288C wild-type haploid cells (A2050) grown in YEPD over 28 h. Vertical lines represent the OD(600 nm) values of pooled euploid and aneuploid cell populations generated by tetrad dissection. Error bars represent SD from the mean of technical replicates. (B and C) Tetrads of sporulated S288C diploid and triploid cells (A40877 and A40878) were dissected to produce heterogeneous haploid and aneuploid cell populations, respectively. A total of 144 individual haploid colonies and 432 aneuploid colonies were inoculated and grown overnight in 200 μL YEPD. The next morning 300 μL YEPD were added to cultures and grown for an additional 5 h. Individual euploid and aneuploid cultures were then pooled and their transcriptomes analyzed. An exponentially growing haploid strain (A2050) was included as a control. Gene-expression data were analyzed by calculating ssGSEA projection values for the (B) iESR and rESR and (C) CAGE up-regulated and down-regulated genes. Error bars represent SD from the mean of technical replicates; one-way two-tailed ANOVA test with multiple comparisons and Bonferroni correction, P < 0.0001 (****), P = 0.0021 (**). For additional statistical analysis see SI Appendix, Fig. S4. (D and E) Tetrads of sporulated S288C diploid and triploid cells (A40877 and A40878) were dissected to produce heterogeneous haploid and aneuploid cell populations, respectively. A total of 144 individual haploid colonies and 432 aneuploid colonies were inoculated and grown overnight in 200 μL YEPD. The next morning cultures were diluted 1:20 and grown for an additional 5 h. Colonies were then pooled, further diluted to approximately OD(600 nm) = 0.3, and grown for 2 additional hours. Transcriptomes of pooled euploid and aneuploid populations and an exponentially growing haploid strain (A2050) were analyzed with RNA-Seq, and ssGSEA projection values were calculated for (D) iESR and rESR and (E) CAGE up-regulated and down-regulated genes. Error bars represent SD from the mean of technical replicates; one-way two-tailed ANOVA test with multiple comparisons and Bonferroni correction, P < 0.0001 (****), P = 0.1234 (ns, no statistical significance). For additional statistical analysis see SI Appendix, Fig. S4.
To confirm this conclusion, we analyzed the gene-expression profile in our euploid and aneuploid cell populations grown using the protocol employed by Tsai et al. (9). Euploid control populations exhibited a stronger ESR than aneuploid cell populations (Fig. 2B and SI Appendix, Fig. S4). Consistent with the idea that the CAGE signature is essentially the opposite of the ESR, aneuploid strains exhibited a stronger CAGE signature than euploid strains (Fig. 2C and SI Appendix, Fig. S4).
We also included an exponentially growing haploid wild-type strain (A2050) in our analysis. As expected, this strain did not exhibit the ESR (Fig. 2B and SI Appendix, Fig. S4) but instead showed the strongest CAGE response among all of the cultures analyzed (Fig. 2C and SI Appendix, Fig. S4). Together, these data indicate that the CAGE response is not an aneuploidy-specific gene-expression signature but the result of differences in proliferation rates between aneuploid and euploid cell populations. In the growth protocol employed by Tsai et al. (9), euploid cells had reached stationary phase, which causes a very strong ESR. In contrast, aneuploid cells had not. Because the ESR of aneuploid cells is weaker than that of stationary phase euploid cells, normalization of the aneuploid gene-expression profile to that of stationary euploid cells led to the incorrect conclusion that aneuploid cells exhibited a transcriptional signature opposite of the ESR. This conclusion predicts that when growth into stationary phase is avoided, aneuploid cell populations ought to exhibit the ESR stronger than euploid control populations.
Aneuploid Cell Populations Exhibit the ESR.
To determine whether growth of the control euploid population into stationary phase precluded the identification of the ESR in aneuploid cell populations, we repeated the protocol developed by Tsai et al. (9) to generate euploid and aneuploid cell populations. However, instead of diluting cultures 1:2 fold after 14 to 16 h of growth in 200 μL of YEPD, we diluted cultures 1:20 fold ( SI Appendix, Fig. S1B ; henceforth 1:20 dilution protocol). This prevented either culture from reaching stationary phase, and the final OD(600 nm) of pooled euploid and aneuploid populations was 0.29 and 0.3, respectively.
Gene-expression analysis of these cultures resulted in a strikingly different outcome compared to that obtained from cells where euploid control populations had reached stationary phase. Aneuploid populations exhibited a stronger ESR than euploid control populations (Fig. 2D and SI Appendix, Fig. S4; iESR P < 0.0001, rESR P < 0.0001). It is, however, noteworthy that euploid control populations also exhibited the ESR, although not as strong as aneuploid populations, when compared to an exponentially growing haploid strain (Fig. 2D and SI Appendix, Fig. S4). This is likely due to the fact that euploid cell populations grown in deep wells experience nutrient limitation. Aeration is poorer and proliferation rate slower in deep well plates compared to in a vigorously shaking flask.
Analysis of the CAGE signature revealed that the exponentially growing haploid strain A2050 expressed CAGE genes much more strongly than either the euploid (CAGE up-regulated P < 0.0001, CAGE down-regulated P < 0.0001) or aneuploid cell populations (Fig. 2E and SI Appendix, Fig. S4; CAGE up-regulated P < 0.0001, CAGE down-regulated P < 0.0001). We note that the aneuploid population showed a slightly greater decrease in expression of the down-regulated CAGE response than euploid control populations (Fig. 2E ). While this difference is statistically significant (P < 0.0001), it is likely biologically irrelevant, given the dramatically higher down-regulation of CAGE genes in the exponentially growing haploid strain. We conclude that aneuploid cell populations exhibit the ESR and that the previously reported aneuploidy-specific CAGE signature is most prominent in an exponentially growing haploid strain.
Degree of Aneuploidy Correlates with ESR Strength in Complex Aneuploid Strains.
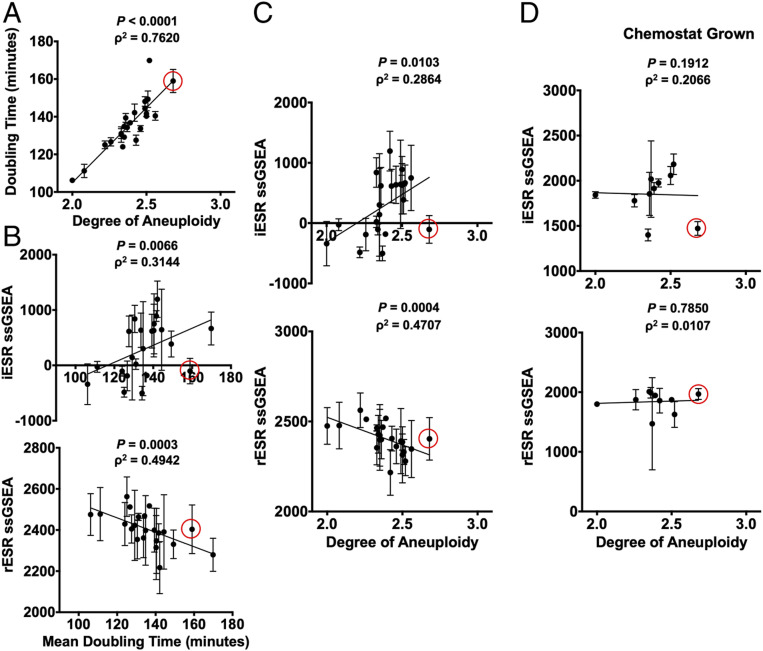
Previous results from our laboratory indicated that yeast strains harboring an additional chromosome (disomes) activate the ESR, and our results shown here demonstrate that heterogeneous aneuploid populations do too (5). We next wished to determine whether this gene-expression signature is also present in yeast strains harboring multiple specific aneuploidies. Pavelka et al. (12) created a large number of yeast strains carrying multiple aneuploidies by sporulating a pentaploid strain (12). Strains obtained from such spores harbor multiple aneuploidies ranging in genome content between 2N and 3N ( SI Appendix, Table S2 and ref. 12). Because the strength of the ESR is largely defined by proliferation rate (4, 8), we first measured doubling times of these complex aneuploid strains to ask whether proliferation rate was correlated with degree of aneuploidy also in strains harboring multiple aneuploidies. We calculated degree of aneuploidy as the fraction of base pairs in the aneuploid strain vis à vis a haploid euploid control strain. We found that the proliferation defect of aneuploid strains correlated remarkably well with their degree of aneuploidy (Fig. 3A ; Spearman, ρ2 = 0.7620, P < 0.0001).
Fig. 3.
Complex aneuploid yeast strains exhibit the ESR. (A–C) Aneuploid yeast strains harboring aneuploidies ranging from 2N to 3N were grown to log phase in YEPD. For each strain, degree of aneuploidy was calculated as the fraction of base pairs in the aneuploid strain/base pairs in a haploid control strain. Doubling times were calculated from growth curves generated by measuring OD(600 nm) in 20-min intervals over 5 h in a plate reader. (A) Correlation between doubling time and degree of aneuploidy (Spearman, ρ2 = 0.7620, P < 0.0001). Transcriptomes of the complex aneuploid strains were analyzed by RNA-Seq, and ssGSEA projection values were calculated for iESR and rESR genes. Correlations between iESR ssGSEA projections and mean doubling time (Spearman, ρ2 = 0.3144, P = 0.0066) and rESR ssGSEA projections and mean doubling time (Spearman, ρ2 = 0.4942, P = 0.0003) are shown in B. Correlations between iESR ssGSEA projections and degree of aneuploidy (Spearman, ρ2 = 0.2864, P = 0.0103) and rESR ssGSEA projections and degree of aneuploidy (Spearman, ρ2 = 0.4707, P = 0.0004) are shown in C. Error bars represent SD from the mean. (D) Select complex aneuploid strains were grown in a phosphate-limiting chemostat until steady state was reached. Transcriptomes of harvested cells were analyzed by RNA-Seq. ssGSEA projection values were calculated for iESR and rESR genes. Correlations between iESR ssGSEA projections and degree of aneuploidy (Spearman, ρ2 = 0.1912, P = 0.2066) and rESR ssGSEA projections and degree of aneuploidy (Spearman, ρ2 = 0.0107, P = 0.7850) are shown. Error bars represent SD from the mean of experimental replicates. The data point circled in red represents a complex aneuploid strain that does not mount the ESR.
Gene-expression analysis of these complex aneuploid strains further revealed a strong correlation between mean doubling time and ESR strength (Fig. 3B ; Spearman, iESR ρ2 = 0.3144, P = 0.0066; Spearman, rESR ρ2 = 0.4942, P = 0.0003) as well as between degree of aneuploidy and ESR strength (Fig. 3C ; Spearman, iESR ρ2 = 0.2864, P = 0.0103; Spearman, rESR ρ2 = 0.4707, P = 0.0004). It is worth noting that a few aneuploid strains were not able to mount the ESR, despite their slowed proliferation (i.e., strain A22 from Pavelka et al.) (12) (circled in red in Fig. 3). The strains that were unable to activate the ESR all harbored gains of chromosomes 2, 7, 11, 15, and 16. This observation suggests that some specific gene combinations prevent activation of the ESR despite slow proliferation. It will be interesting to determine the mechanism of this ESR suppression.
Finally, we probed for the existence of the CAGE signature in these complex aneuploid strains. The up-regulated CAGE signature did not correlate with degree of aneuploidy ( SI Appendix, Fig. S5A ; Spearman, up-regulated CAGE ρ2 = 0.0679, P = 0.2416). We observed a correlation between the down-regulated CAGE signature and degree of aneuploidy, but it was opposite to what would be expected if it were determined by degree of aneuploidy. Increased degree of aneuploidy correlated with increased expression of CAGE down-regulated genes ( SI Appendix, Fig. S5A ; Spearman, down-regulated CAGE ρ2 = 0.2368, P = 0.0217). We conclude that the CAGE signature is not a common aneuploidy gene-expression signature among complex aneuploid strains, but the ESR is.
Proliferation Rate Determines ESR Strength.
The strong correlation between ESR strength and doubling times in complex aneuploid strains suggested that proliferation rate was the primary determinant of ESR strength. To directly test this possibility, we examined whether equalizing proliferation rate among complex aneuploid strains and euploid control strains affected the correlation between ESR strength and degree of aneuploidy by culturing cells in a phosphate-limited chemostat (4, 13). When proliferation rate was equalized in this manner, the ESR gene-expression signature was no longer evident in aneuploid strains (Fig. 3D ; Spearman, iESR ρ2 = 0.1912, P = 0.2066; Spearman, rESR ρ2 = 0.0107, P = 0.7850). We also probed for the existence of the CAGE signature in complex aneuploid strains grown under phosphate-limiting conditions. We observed no correlation between degree of aneuploidy and genes up-regulated in the CAGE response and the opposite correlation as would have been expected for the down-regulated genes of the CAGE signature ( SI Appendix, Fig. S5B ; Spearman, up-regulated CAGE ρ2 = 0.0030, P = 0.8916; Spearman, down-regulated CAGE ρ2 = 0.4364, P = 0.0.0438). We conclude that when proliferation is equally slow in euploid and aneuploid cells, the ESR caused by aneuploidy is no longer evident. This suggests that in both euploid and aneuploid cells, proliferation rate is the primary determinant of ESR strength.
ESR Induction in Aneuploid Cells Causes Ribosome Loss.
Tsai et al. (9) reported the CAGE gene-expression signature as aneuploidy specific and most similar to the hypoosmotic stress response. This similarity is not surprising, given that both the CAGE signature and the hypoosmotic stress response are essentially oppositely regulated to the ESR. Given our findings that heterogeneous aneuploid populations do not exhibit a CAGE signature, we next determined whether aneuploid cells indeed experience hypoosmotic stress that was proposed to occur in response to a decrease in cytoplasmic density in aneuploid cells (9).
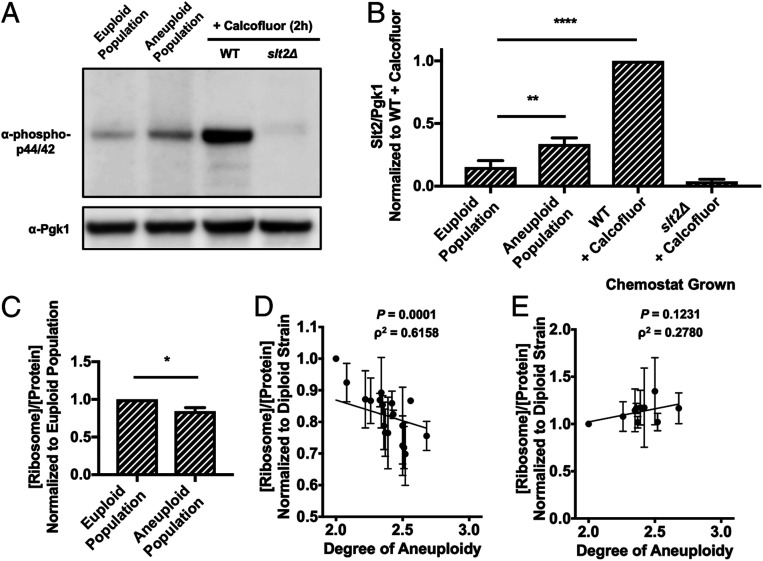
To assess induction of the hypoosmolarity stress pathway, we probed activation of the hypoosmolarity pathway MAP kinase Slt2 using a phospho-specific antibody that recognizes active phospho-Slt2 (14). Aneuploid cell populations showed a 2.22-fold increase in mean Slt2 phosphorylation compared to euploid control populations (Fig. 4 A and B ). The activation in the aneuploid cell populations was subtle compared to cell wall stress induced by prolonged Calcofluor White treatment (6.59-fold increase over euploid cell population). This difference in induction was not due to acute versus chronic induction of the hypoosmolarity pathway because we treated cells with Calcofluor White for 2 h before analyzing the phosphorylation state of Slt2. The subtle activation of the hypoosmolarity pathway in aneuploid strains suggested that either all aneuploidies cause weak activation of this stress pathway or that only a subset of aneuploid cells activates the pathway. We favor the latter possibility because we previously showed that not all disomies cause cell wall defects in yeast (15).
Fig. 4.
ESR induction causes ribosome loss in aneuploid strains. (A and B) Euploid and aneuploid cell populations were grown in YEPD with the 1:20 dilution protocol, and Slt2 Thr202/Tyr204 phosphorylation was determined. Wild-type euploid (A2050) and slt2Δ cells (A41265) treated with 5 μg/mL Calcofluor White for 2 h served as positive and negative controls, respectively, in immunoblots (A). Pgk1 served as a loading control. Quantifications of Slt2 Thr202/Tyr204 phosphorylation are shown in B. Slt2/Pgk1 values were normalized to the wild-type cells treated with Calcofluor White. Error bars represent SD from the mean of experimental replicates; one-way ANOVA test with multiple comparisons and Bonferroni correction, P < 0.0001 (****), P = 0.0021 (**). All other comparisons between samples had a significant difference of P < 0.0001 (****) with the exception of the euploid populations and slt2Δ + Calcofluor, which had a significant difference of P = 0.0288. (C) The fraction of ribosome in total protein extracts ([ribosome]/[protein]) was determined in euploid and aneuploid cell populations grown with the 1:20 dilution protocol. [ribosome]/[protein] in aneuploid cell populations was normalized to that in euploid cell populations. Error bars represent SD from the mean of technical replicates; unpaired two-tailed t test test, P = 0.0332 (*). (D) Aneuploid yeast strains harboring aneuploidies ranging from 2N to 3N were grown to log phase in YEPD and the fraction of ribosomes in total protein extracts ([ribosome]/[protein]) was determined. Correlation between [ribosome]/[protein] and degree of aneuploidy (ρ2 = 0.6158, P = 0.0001, Spearman) is shown. The calculated values were normalized to the [ribosome]/[protein] of a diploid control. (E) Aneuploid yeast strains harboring aneuploidies ranging from 2N to 3N were grown in a phosphate-limited chemostat and the fraction of ribosome in total protein extracts ([ribosome]/[protein]) was determined. Correlation between [ribosome]/[protein] and degree of aneuploidy (ρ2 = 0.2780, P = 0.1231, Spearman) is shown. The calculated values were normalized to the [ribosome]/[protein] of a diploid control. Error bars represent SD from the mean of experimental replicates.
The subtle activation of the hypoosmolarity pathway in aneuploid cell populations was hard to reconcile with the comparatively dramatic effects on cytoplasmic density reported to occur in aneuploid cell populations (9). Our observation that aneuploid cell populations exhibit the ESR provided an alternative hypothesis. A recent study by Delarue et al. (16) showed that the major determinant of cytoplasm density is the ribosomal fraction within a cell’s proteome. Given that aneuploid cell populations exhibit the ESR, which is characterized by the down-regulation of ribosomal protein and ribosome biogenesis gene expression, we asked whether aneuploid cell populations harbor fewer ribosomes than euploid control populations.
To address this question, we isolated ribosomes from aneuploid and euploid populations grown using the 1:20 dilution protocol ( SI Appendix, Fig. S1B ). For this, we used a protocol that was adapted from methods developed to purify 80S assembled ribosomes (17 –19). We previously examined the purity of this ribosome preparation in ref. 20. To ensure that this preparation consisted largely of 80S assembled ribosomes, we quantified the Coomassie staining pattern of our ribosome preparation published in Brennan et al. (20) and compared it with that obtained by Munoz et al. (18), who published the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) banding pattern of purified 40S and 60S subunits ( SI Appendix, Fig. S6 A and B ). This analysis confirmed that the method we employed to isolate ribosomes enriches for assembled ribosomes. Ribosome preparations were then subjected to absorbance measurements at 260 nm to determine ribosome content. To assess the sensitivity of this quantification method, we compared protein and ribosome content between a wild-type haploid strain (A2587) and a wild-type diploid strain (A33821) ( SI Appendix, Table S1). Diploid cells are twice as large as haploid cells and are thus expected to have twice as many proteins and ribosomes. Our measurements showed this to be the case ( SI Appendix, Fig. S6 C and D ), indicating that our method was highly sensitive.
Having established a method to determine a cell’s ribosome content, we next compared the ribosome content between aneuploid and euploid populations. This analysis revealed that ribosomes made up a significantly smaller fraction of total protein in aneuploid cell populations than in euploid cell populations (Fig. 4C ). Our analysis of complex aneuploid strains confirmed these results. Ribosome content inversely correlated with degree of aneuploidy (Fig. 4D ; Spearman, ρ2 = 0.6158, P = 0.0001), which is consistent with ESR strength correlating with degree of aneuploidy. This correlation suggested that the ESR triggered ribosome loss and hence loss of cytoplasmic density in aneuploid cells. If true, we would predict that equalizing proliferation rates among aneuploid and euploid cells should eliminate this correlation. This is what we observed. When we grew euploid and complex aneuploid strains in continuous culture under phosphate-limiting conditions, all strains not only exhibited similar ESR strengths, but ribosome content was no longer inversely correlated with degree of aneuploidy (Fig. 4E ; Spearman, ρ2 = 0.2780, P = 0.1231). We propose that the decrease in cytoplasmic density observed in aneuploid cells is caused by an ESR-induced loss of ribosomes.
Discussion
Whether aneuploidy elicits a stereotypic transcriptional response in yeast and what this response may be has been controversial. The analysis of a series of disomic yeast strains harboring an additional copy of one of yeast’s 16 chromosomes showed that these strains exhibit the generic ESR. However, other reports found this not to be the case. The analysis of five complex aneuploid yeast strains showed that only three out of these five strains exhibited the ESR (12). Most recently, Tsai et al. (9) reported that heterogeneous aneuploid cell populations also lack the ESR. We reevaluated these reports to find that when a large number of complex aneuploid strains is analyzed and when gene-expression profiles of mixed aneuploid cell populations are normalized to euploid populations that are in the same proliferation state as aneuploid populations, the ESR is evident. Together, these results indicate that aneuploid yeast strains exhibit the ESR. We consider this result not surprising, given that the ESR is largely a consequence of slowed cell division (4, 8) and that aneuploidy causes proliferation defects (5). We further note that the ESR-like transcriptional signature is also observed in aneuploid mammalian cells (4). Together, these results indicate that the ESR and relatives of this signature are a pervasive response to aneuploidy.
In wild isolates of yeast that are naturally aneuploid, the ESR is less prevalent (1, 2). This observation indicates that under selective pressure, aneuploidies evolve and adaptation to aneuploidy-induced cellular stresses occurs such that proliferation rate is less affected by a particular chromosome gain or loss. Indeed, an aneuploidy tolerating natural gene variant has recently been described (2). Based on the analysis of two wild yeast isolates disomic for either chromosome 8 or 12, Hose et al. (2) proposed that the ESR in aneuploid cells is strongly influenced by the type of SSD1 allele that a strain carries. Specifically, the ESR observed in aneuploid strains of the W303 background was attributed to the fact that W303 harbors a truncation allele of SSD1. The complex aneuploid strains and the aneuploid cell populations employed in our study are derivatives of the S288C strain background, which harbors a full-length SSD1 allele. The finding that ESR strength correlates with degree of aneuploidy in these aneuploid S288C derivatives indicates that the ESR is not defined by SSD1 allele identity but degree of aneuploidy. It is nevertheless possible that specific SSD1 alleles and other genomic alterations exist that suppress the ESR gene signature. We found that strains harboring a gain of chromosomes 2, 7, 11, 15, and 16 do not exhibit the ESR despite proliferating slowly. Understanding why these chromosome combinations suppress the ESR will be interesting. We speculate that growth-promoting pathways known to negatively regulate the ESR, such as the PKA and TOR pathways, are hyperactive in these strains.
Our data indicate that the previously described aneuploidy-specific CAGE gene-expression signature is an artifact caused by normalizing the gene expression of actively dividing aneuploid cells to that of euploid control cells that had grown to stationary phase. We show that the ESR in the euploid control populations that had grown into stationary phase was stronger than the ESR observed in aneuploid cell populations that, due to their poor proliferation, were still actively proliferating. Thus, when euploid stationary phase cells were used for normalization, aneuploid cells exhibited the CAGE signature in which many ESR genes are oppositely regulated. As such, it is not surprising that the CAGE signature is most similar to the previously described hypoosmolarity gene-expression signature. Gasch et al. (6) identified two stresses that do not result in activation of the ESR: cold shock and hypoosmotic shock. Under these stresses, the ESR is oppositely regulated. In particular the rESR, which encompasses genes encoding transcription and translation factors and ribosomal proteins are up-regulated rather than down-regulated (6). While it is not clear why ribosome production must be up-regulated during cold shock, we understand why this occurs during hypoosmotic shock. During hypoosmotic shock, water uptake increases (10). To avoid cytoplasm dilution during this water influx, production of ribosomes, which are the main determinants of cytoplasm density (16), must be up-regulated or at least prevented from being down-regulated. Further analysis of these exceptional stress conditions, during which the rESR is not down-regulated, will provide key insights into regulation of this central stress and slow proliferation response.
Under most, if not all stress conditions, cell proliferation is slowed or halted, and the ESR signature is evident with the exceptions noted above. Down-regulation of the rESR generally correlates much better with degree of slow proliferation than induction of the iESR. This is not surprising. The vast majority of the genes that are part of the rESR are involved in transcription and translation. Cell growth, or cellular enlargement, needs to be attenuated during any stress that causes a slowing or halting of cell division to prevent cells from growing too large. When growth continues unabated during cell cycle arrest, DNA becomes limiting, causing numerous defects including impaired cell proliferation, cell signaling, and gene expression (21, 22). We speculate that a role of the rESR is to attenuate macromolecule biosynthesis and hence cell growth during cell cycle arrest. In contrast, the iESR may be aimed at alleviating cellular stress, which requires expression of genes unique to specific stresses rather than control of biomass production.
Our results suggest that repression of rESR genes in response to aneuploidy has profound consequences on cellular proteome composition. It leads to a significant drop in the contribution of ribosomes to the cell’s total protein. This not only leads to down-regulation of translational capacity but, because ribosomes are the key determinant of cytoplasmic density (16), is likely the major cause of loss in cellular density previously reported to occur in aneuploid cells (9). We propose that activation of the ESR in aneuploid cells serves two purposes. It protects cells from cellular stresses caused by an unbalanced genome and prevents excessive cellular enlargement during their slowed cell cycles. Understanding how slowed proliferation leads to activation of the ESR will provide critical insights into the coordination between cell division and macromolecule biosynthesis.
Materials and Methods
Dataset Processing and Single-Sample Gene Set Enrichment Analysis.
Raw RNA-Seq data were obtained as described below or through download from Tsai et al. (9) with gene accession no. GSE107997. Reads were aligned to a S. cerevisiae transcriptome with STAR version 2.5.3a (23) and gene expression was quantified with RSEM version 1.3.0 (24). TPM values were offset by +1, log2 transformed, and used to prepare gene cluster text files for ssGSEA (version 7.7; refs. 25 and 26) obtained from the Indian University public GenePattern server (27). ssGSEA projections were prepared for the ESR originally described by Gasch et al. (6) CAGE signatures identified in Tsai et al. (9). Sort order has a subtle effect on ssGSEA.
Differential Gene-Expression Analysis.
Raw RNA-Seq data were obtained through download from Tsai et al. (9) with gene accession no. GSE107997. Integer count values derived from RSEM processing were used as input to differential expression analysis with DESeq2 (version 1.24.0; ref. 28) using normal log fold change shrinkage. Expression data from aneuploid cell populations generated by tetrad dissection were pooled to create “aneuploid populations (tetrad).” Expression data from aneuploid populations obtained from MATa selection were pooled to create “aneuploid populations (MATa selection).” These populations, as well as both the euploid populations obtained from tetrad dissection and MATa selection, were compared to the exponentially growing haploid control. Differential expression data were visualized using TIBCO Spotfire Analyst version 7.11.1.
Stationary-Phase Growth Timecourse.
S288C wild-type cells (A2050) were inoculated into 25 mL YEPD and grown overnight at 25 °C. After ∼12 to 14 h of growth, cells were diluted to OD(600 nm) = 0.1 with YEPD + 2% glucose and grown for an additional 4 h at 25 °C. Once the OD(600 nm) reached approximately OD(600 nm) = 0.3, samples for transcriptomics were taken and OD(600 nm) was measured every 2 h for 28 h. Optical density was measured at 600 nm with a spectrophotometer.
Heterogeneous Euploid and Aneuploid Population Generation Using the Tsai et al. (9) Protocol.
pRS315-STE2pr-spHIS5 S288C diploids (A40877) and triploids (A40878) ( SI Appendix, Table S1) were grown overnight in SD −Leu medium and subsequently sporulated in “super sporulation medium” (1% potassium acetate and 0.02% raffinose) from Tsai et al. (9). Sporulated tetrads were then dissected. [Note in the publication by Tsai et al. (9), sporulated tetrads were also MATa selected through histidine prototrophy. We did not generate cell populations in this manner.] Individual colonies were grown for 14 to 16 h in 200 μL of YEPD + 2% glucose in a 96-deep well plate. A total of 300 μL of YEPD was then added to cultures. The cultures were grown for 5 additional hours, pooled, and samples for RNA-Seq and [ribosome]/[protein] content measurements were taken.
Heterogeneous Euploid and Aneuploid Population Generation Using the 1:20 Dilution Protocol.
pRS315-STE2pr-spHIS5 S288C diploids (A40877) and triploids (A40878) ( SI Appendix, Table S1) were grown overnight in SD −Leu medium and subsequently sporulated in the super sporulation media mentioned above. Sporulated tetrads were then tetrad dissected. Individual colonies obtained from spores were grown for 14 to 16 h in 200 μL of YEPD in a 96-deep well plate. Cultures were then diluted 1:20 in YEPD + 2% glucose, grown for another 5 h, then pooled, and diluted to approximately OD(600 nm) = 0.3. The pooled aneuploid populations were grown for an additional 2 h, and samples for RNA-Seq, [ribosome]/[protein] content measurements, and Slt2 phosphorylation state analysis were taken.
ssGSEA Bootstrapping Analysis.
Four groups of 1,000 random gene sets were generated. Each group was the same size as one of the following gene sets: iESR (283 genes), rESR (585 genes), CAGE up-regulated (169 genes), and CAGE down-regulated (53 genes). For the exponentially growing haploid strain A2050 and euploid and aneuploid populations (grown using the 1:20 dilution protocol), ssGSEA projections were prepared for the 4,000 total gene sets. For each gene set, a one-way two-tailed ANOVA test with multiple comparisons and Bonferroni correction (P value multiplied by 3) was computed for ssGSEA values of euploid population vs. aneuploid population, euploid population vs. haploid strain, and aneuploid population vs. haploid strain. In the end, 1,000 P values were generated for each of the three comparisons, for each of the four differently sized groups of 1,000 random gene sets. P values from the randomly generated gene sets were then compared to the P value of the corresponding gene set and samples used. The overall significance of this bootstrapping analysis was determined by dividing the number of randomly generated comparisons which had a P value smaller than the original P value by 1,000. For example, of the 1,000 randomly generated gene sets of 283 genes comparing euploid vs. aneuploid populations, three comparisons had a P value smaller than the P value generated for the iESR gene set (which contains 283 genes) in these strains. The bootstrapping P values were not multiple-test corrected.
Growth Conditions for Complex Aneuploid Strains.
Complex aneuploid strains were generated by Pavelka et al. (12) ( SI Appendix, Table S2). Complex aneuploid strains were grown on YEPD + 2% glucose plates for 2 d at 25 °C. Colonies were inoculated overnight in 25 mL YEPD + 2% glucose and grown at 25 °C. After 12 to 14 h of growth at 25 °C, cells were diluted to approximately OD(600 nm) = 0.1 and grown for an additional 4 h at 25 °C. Cells were then harvested for RNA-Seq and [ribosome]/[protein] content measurements.
Doubling Time Measurements.
Doubling time of complex aneuploid strains were measured in a 96-well format in YEPD + 2% glucose at 25 °C. OD(600 nm) values were taken in 20-min intervals over 5 h, and doubling time was calculated from the growth curves generated.
Growth in Phosphate-Limiting Chemostats.
Selected complex aneuploid strains were grown on YEPD + 2% glucose plate for 2 d at 30 °C. Strains were inoculated in phosphate-limited media (29) and grown overnight. Once chemostats were set up and filled, phosphate-limited media in the chemostat was inoculated with 2 mL of overnight culture, and cells were allowed to grow for 30 to 36 h although some strains required 48+ hours of growth. The dilution pumps were then turned on at a dilution rate of 0.11 ± 0.01 chemostat volumes per hour. The chemostat was sampled daily to measure effluent volume, hemocytometer counts, and OD(600 nm) measurements. When growth in the chemostat had reached a steady state, defined by less than 5% change from the previous day’s measurements, samples were harvested for RNA-Seq and [ribosome]/[protein] content.
RNA-Seq.
Three- to 5-mL samples of culture were taken, spun down at 3,000 rpm for 5 min, washed with 1 mL diethylpyrocarbonate-treated water, and transferred to a 2-mL RNase-free screw-cap tube. Samples were spun again at 8,000 rpm for 3 min, and supernatant was aspirated. Cells were snap frozen with liquid nitrogen and stored at −80 °C. RNA samples were prepared with RNeasy mini kit from Qiagen and treated with DNase on-column treatment (RNase-free) from Qiagen. Purified RNA was used in two different library preparation methods. In experiments with complex aneuploid strains, total RNA was sequenced using Illumina Truseq followed by Nextera or Roche KAPA. In all other experiments, total RNA was sequenced using Illumina Truseq followed by Roche KAPA. All sequencing was done using an Illumina HiSeq2000.
[Ribosome]/[Protein] Content Measurements.
A total of 1 mL of 1:100 diluted samples of culture were used to determine cell number in the culture using a Beckman Multisizer 3 Coulter Counter ([cell]). Concurrently, 50-mL samples of culture were taken, spun down at 3,000 rpm for 5 min, frozen with liquid nitrogen, and stored at −80 °C. Cells were resuspended in 30 mL lysis buffer (20 mM Hepes pH 7.4, 100 mM potassium acetate, 2 mM magnesium acetate, 3 mM dithiothreitol (DTT), and ethylenediaminetetraacetic acid-free protease inhibitor [Roche, 11836170001]), and 0.5 mg/mL zymolyase was added. Resuspended cells were lysed twice with a French press. Lysed samples were then spun at 19,000 rpm for 20 min at 4 °C to remove cell debris. Protein concentration of the cell lysate ([protein]) was measured in triplicate by Bradford assay.
A total of 10 mL cell lysate was overlaid onto 15-mL prechilled (4 °C) sucrose solution (30% sucrose, 20 mM Hepes pH 7.4, 500 mM potassium acetate, 2 mM magnesium acetate, and 3 mM DTT), which was then spun at 50,000 rpm for 4 h at 4 °C to purify 80S ribosomes (17 –19). Solution above assembled ribosome pellet was poured from the tube after spin, and the tubes were dried upside down for 10 min to remove excess liquid. Pellets were resuspended with 1 mL lysis buffer, and absorbance at 260 nm was measured with a Nanodrop to obtain concentration of purified assembled ribosomes ([ribosome]).
Calcofluor Treatment.
S288C wild-type (A2050) and slt2Δ (A41265) cells were inoculated into 25 mL YEPD + 2% glucose, and grown at 25 °C for 12 to 14 h. Cells were diluted to approximately OD(600 nm) = 0.1, grown for an additional 4 h, and treated with 5 μg/mL Calcofluor White for 2 h. Samples were harvested for Western blot analysis.
Western Blot and Quantification.
One OD(600 nm) unit of sample was trichloroacetic acid (TCA) precipitated. A total of 20 μL of each sample was run on a NuPAGE 4 to 12% Bis-Tris protein gel from Invitrogen and then transferred to polyvinylidene difluoride membrane from EMD Millipore. Phospho-p44/42 MAPK (Thr202/Tyr204) antibody (1:1,000; Cell Signaling Technology, 9101) was used to detect phosphorylated Slt2. Pgk1 (Pgk1 antibody, 1:4,000; Thermo Fisher, 22C5D8) was used as a loading control. Immunoblots were incubated with horseradish peroxidase-conjugated secondary antibodies and ECL Western blotting detection reagents from Amersham and then scanned on an ImageQuant LAS4000.
Signals were quantified on an ImageQuant LAS4000 and integrated densities of bands were quantified using ImageJ. Three separate immunoblots were quantified and normalized to wild-type cells treated with Calcofluor. To quantify ribosomal proteins ( SI Appendix, Fig. S6) we mimicked a densitometer measurement. A line was drawn in the middle of the lane of the Coomassie-stained gels from Munoz et al. (18) and Brennan et al. (20). The pixel intensity, measured as gray values, along this line was then quantified using ImageJ and normalized to the maximum pixel intensity value. For the 40S + 60S measurements, pixel intensity measurements of the 40S subunit and the 60S subunit were averaged prior to normalization. Quantifications were recorded starting directly above the slowest migrating ribosomal band and ending below the fastest migrating band. This distance was set to 1 arbitrary unit.
Data Availability.
The data discussed in the paper are included in the figures and SI Appendix . RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE146791).
Supplementary Material
Acknowledgments
Thank you to Summer Morrill, Xiaoxue Zhao, and David Waterman for comments and the Massachusetts Institute of Technology (MIT) BioMicroCenter for RNA-Seq. This work was supported by NIH grant R35 GM118066 to A.A., who is an investigator of the Howard Hughes Medical Institute, the Paul F. Glenn Center for Biology of Aging Research at MIT and the Ludwig Center at MIT. A.K. was supported in part by National Human Genome Research Institute grant T32 HG-00035. The research of M.J.D. was supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute and by National Institute of General Medical Sciences grant P41 GM103533. M.J.D. is a Senior Fellow in the Genetic Networks program at the Canadian Institute for Advanced Research.
Footnotes
Editor J.S.W. and authors A.T., A.S., C.A.W., and A.A. are affiliated with Massachusetts Institute of Technology. They are not current collaborators. The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE146791).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005648117/-/DCSupplemental.
Change History
May 18, 2022: A competing interests footnote has been added; please see accompanying Correction for details.
References
- 1. Peter J.et al., Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hose J.et al., The genetic basis of aneuploidy tolerance in wild yeast. eLife 9, e52063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chunduri N. K., Storchová Z., The diverse consequences of aneuploidy. Nat. Cell Biol. 21, 54–62 (2019). [DOI] [PubMed] [Google Scholar]
- 4. Sheltzer J. M., Torres E. M., Dunham M. J., Amon A., Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. U.S.A. 109, 12644–12649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Torres E. M.et al., Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317, 916–924 (2007). [DOI] [PubMed] [Google Scholar]
- 6. Gasch A. P.et al., Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho Y. H., Shishkova E., Hose J., Coon J. J., Gasch A. P., Decoupling yeast cell division and stress defense implicates mRNA repression in translational reallocation during stress. Curr. Biol. 28, 2673–2680.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brauer M. J.et al., Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 19, 352–367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai H. J.et al., Hypo-osmotic-like stress underlies general cellular defects of aneuploidy. Nature 570, 117–121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altenburg T., Goldenbogen B., Uhlendorf J., Klipp E., Osmolyte homeostasis controls single-cell growth rate and maximum cell size of Saccharomyces cerevisiae . NPJ Syst. Biol. Appl. 5, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tarca A. L., Bhatti G., Romero R., A comparison of gene set analysis methods in terms of sensitivity, prioritization and specificity. PLoS One 8, e79217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavelka N.et al., Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunham M. J., Kerr E. O., Miller A. W., Payen C., Chemostat culture for yeast physiology and experimental evolution. Cold Spring Harb. Protoc. 2017, pdb.top077610 (2017). [DOI] [PubMed] [Google Scholar]
- 14. Hahn J. S., Thiele D. J., Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase. J. Biol. Chem. 277, 21278–21284 (2002). [DOI] [PubMed] [Google Scholar]
- 15. Dodgson S. E.et al., Chromosome-specific and global effects of aneuploidy in Saccharomyces cerevisiae. Genetics 202, 1395–1409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delarue M.et al., mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivera M. C., Maguire B., Lake J. A., Isolation of ribosomes and polysomes. Cold Spring Harb. Protoc. 2015, 293–299 (2015). [DOI] [PubMed] [Google Scholar]
- 18. Munoz A. M.et al., Active yeast ribosome preparation using monolithic anion exchange chromatography. RNA Biol. 14, 188–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Algire M. A.et al., Development and characterization of a reconstituted yeast translation initiation system. RNA 8, 382–397 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brennan C. M.et al., Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells. Genes Dev. 33, 1031–1047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neurohr G. E.et al., Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 176, 1083–1097.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neurohr G. E., Amon A., Relevance and regulation of cell density. Trends Cell Biol. 30, 213–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dobin A.et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian A.et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barbie D. A.et al., Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reich M.et al., GenePattern 2.0. Nat. Genet. 38, 500–501 (2006). [DOI] [PubMed] [Google Scholar]
- 28. Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dunham M., Mitchell E., Dunham Lab Chemostat Manual, (Dunham Lab, University of Washington, 2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in the paper are included in the figures and SI Appendix . RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE146791).