Significance
MicroRNAs (miRNAs) are indispensable regulators of gene expression. Hyponastic Leaves 1 (HYL1) is a core component of the microprocessor that processes primary miRNAs (pri-miRNAs) into mature miRNAs. Here we report a noncanonical function of HYL1 in protecting pri-miRNAs from unwanted nuclear exosome activities during processing. Our observations also suggest that HYL1 may contribute differently to the Dicer-like 1 (DCL1)-mediated maturation of loop-to-base– and base-to-loop–processed pri-miRNAs. This study thus integrates the RNA surveillance system to the miRNA biogenesis pathway and sheds light on the multifaced roles of HYL1 in miRNA biogenesis.
Keywords: HYL1, miRNA, exosome, HEN2, SOP1
Abstract
Biogenesis of plant microRNAs (miRNAs) takes place in nuclear dicing bodies (D-bodies), where the ribonulease III-type enzyme Dicer-like 1 (DCL1) processes primary transcripts of miRNAs (pri-miRNAs) into miRNA/miRNA* (*, passenger strand) duplexes from either base-to-loop or loop-to-base directions. Hyponastic Leaves 1 (HYL1), a double-stranded RNA-binding protein, is crucial for efficient and accurate processing. However, whether HYL1 has additional function remains unknown. Here, we report that HYL1 plays a noncanonical role in protecting pri-miRNAs from nuclear exosome attack in addition to ensuring processing. Loss of functions in SOP1 or HEN2, two cofactors of the nucleoplasmic exosome, significantly suppressed the morphological phenotypes of hyl1-2. Remarkably, mature miRNAs generated from loop-to-base processing were partially but preferentially restored in the hyl1 sop1 and hyl1 hen2 double mutants. Accordingly, loop-to-base–processed pri-miRNAs accumulated to higher levels in double mutants. In addition, dysfunction of HEN2, but not of SOP1, in hyl1-2 resulted in overaccumulation of many base-to-loop–processed pri-miRNAs, with most of their respective miRNAs unaffected. In summary, our findings reveal an antagonistic action of exosome in pri-miRNA biogenesis and uncover dual roles of HYL1 in stabilizing and processing of pri-miRNAs.
MicroRNAs (miRNAs) are a class of endogenously expressed small noncoding RNAs that play crucial roles in diverse biological processes, including reproduction, cell fate determination and transitions, organ formation and development, as well as responses to environmental disturbances (1). Primary transcripts of MIRNA genes (pri-miRNAs) are folded into stem-loop structures, which can be recognized and subsequently processed by an RNase III-type endonuclease Dicer-like 1 (DCL1) with the help of the double-stranded RNA-binding protein Hyponastic Leaves 1 (HYL1) and the zinc finger protein Serrate (SE) (2–8). Plant pri-miRNAs are more variable in the foldback length (from ∼50 nt to over 500 nt) than their animal counterparts (∼70 nt) (9). Consequently, dozens of plant miRNAs have been shown to be matured through a loop-to-base processing mode, rather than the canonical base-to-loop processing (10, 11).
In the canonical base-to-loop processing mode, the initial cut position is defined by an internal loop followed by a 15- to 17-bp structured lower stem (11–14). However, such features are absent in pri-miRNAs that are processed from loop to base (10). Instead, a structured terminal region that contains a short terminal loop and a 15- to 17-bp stem is utilized for first cut registration (10, 11). Moreover, pri-miRNAs with long stems can undergo sequential base-to-loop or loop-to-base processing, which generate both miRNAs and additional siRNAs (10, 15, 16).
Processing of pri-miRNAs occurs in subnuclear speckles termed dicing bodies (D-bodies). Core members of the processing machinery in the D-body include DCL1, HYL1, and SE (17–19). HYL1 is the most important chaperone of DCL1, and its function in miRNA processing is thought to be similar to that of Drosha’s chaperone DGCR8 and Dicer’s chaperone TRBP (20). While knockout of DCL1 causes embryo lethality, loss-of-function mutations in HYL1 are still viable, suggesting that HYL1 is not essential (21, 22). Previous studies indicated that HYL1 forms a homodimer, probably binds to the miRNA/miRNA* (*, passenger strand) region of precursor RNAs, and improves both efficiency and accuracy of DCL1-mediated processing (7, 23–25). Nevertheless, its detailed mechanism of action and whether HYL1 has additional biological functions remains unclear.
Exosomes are 3′ to 5′ exoribonucleolytic machineries that play key roles in many RNA metabolic processes such as rRNA maturation, cryptic unstable transcripts (CUTs) surveillance, and aberrant RNA clearance (26). Depending on their subcellular location, exosomes are categorized into cytoplasmic and nuclear exosomes. The latter is further divided into nucleolar and nucleoplasmic exosomes. Exosomes recognize and degrade diverse RNA substrates via interacting with different cofactors. In yeast, RNA helicases SKI2 and MTR4 are key cofactors of the cytoplasmic exosome and the nuclear exosome, respectively (27). In addition to SKI2, there are two MTR4 homologs in plants, namely MTR4 and HEN2. Recent studies have shown that MTR4 and HEN2 are cofactors of the nucleolar exosome and nucleoplasmic exosome, respectively. While MTR4 is mainly involved in the maturation of rRNAs and clearance of processing byproducts, HEN2 is responsible for the degradation of various nucleoplasmic RNAs including snoRNA precursors, partially spliced RNAs, and notably a few miRNA precursors (28). Genetic screen for morphological suppressors of the PAS2 splicing-site mutant pas2 (sop) led to the identification of three suppressors, sop1, -2, and -3 (29). Among them, sop2 is a weak allele of RRP4 and sop3 bears a mutation in the HEN2 gene. SOP1 contains five tandem repeats of the CCCH-type zinc finger domain at its C terminus, which may be required for RNA binding. SOP1 and HEN2 colocalize and participate in the degradation of a subset of nucleoplasmic exosome substrates (29).
In this study, we conducted a genetic screen of hyl1-2 suppressors. Consistent with previous studies (30, 31), we identified a series of DCL1 allelic mutations. Interestingly, we characterized a non-DCL1 mutation that partially suppressed the hyl1-2 phenotypes (named suppressor of hyl1, 43 [shy43]). We mapped shy43 to the SOP1 locus. Similar to sop1, the HEN2 mutation also suppressed the hyl1-2 phenotype. Small RNA sequencing (sRNA-seq) analysis revealed that miRNAs generated from loop-to-base processing are preferentially restored in hyl1 sop1 and hyl1 hen2 mutants relative to those in hyl1-2. Accordingly, their pri-miRNAs accumulated to higher levels in hyl1 sop1 and hyl1 hen2. Intriguingly, HEN2 also targets base-to-loop–processed pri-miRNAs, with their mature miRNA levels largely unaffected. Thus, HYL1 plays dual roles in promoting pri-miRNAs processing and guarding against nuclear exosome attack.
Results
Genetic Screen of hyl1-2 Suppressors Identifies Additional Alleles of DCL1 and SOP1.
Previously, two laboratories conducted genetic screens in the hyl1 background and identified several dominant DCL1 alleles that can restore the processing defects of miRNAs and consequently suppress the hyl1-2 phenotype to varying degrees. Interestingly, all characterized amino acid substitutions were either in the helicase ATP binding or the RNaseIIIa domain of DCL1, which likely enhanced DCL1 processing efficiency (30, 31). Using the same strategy but with a more extensive level of screening, we isolated a series of putative suppressors of hyl1 (shy), from which we identified a total of eight DCL1 alleles via targeted sequencing of the DCL1 genomic region. Among them, three were identical to previously characterized ones, and the other five were designated dcl1-25 to dcl1-29, to be consecutive with numbering in a previous report (31) (SI Appendix, Fig. S1). Not surprisingly, all these alleles harbored amino acid substitutions in either the helicase ATP binding or the RNaseIIIa domains (SI Appendix, Fig. S1). Of note, an A166T substitution at N terminus was detected in the shy28-2/hyl1-2 dcl1-26 mutant in addition to the V392M mutation at helicase ATP binding domain, although its relative contribution to DCL1 function remains to be investigated.
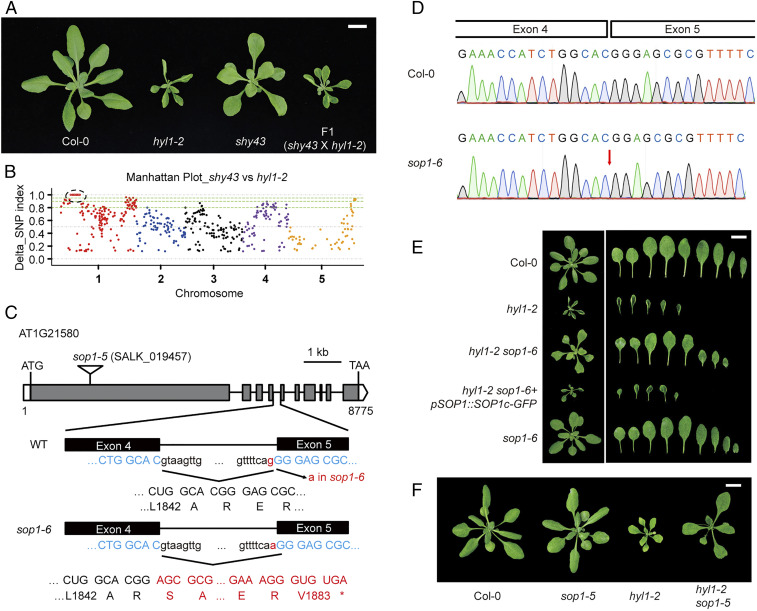
Fortunately, we also isolated a new suppressor (dubbed shy43) with no mutation detected in DCL1 (Fig. 1A). As compared with hyl1-2, shy43 had larger leaf blades with reduced curvature, increased leaf emergence rate, and partially restored fertility (Fig. 1A and SI Appendix, Fig. S2 A and B). F1 plants from a backcross of shy43 with hyl1-2 were morphologically identical to hyl1-2 (Fig. 1A), and F2 progenies segregated in a 3:1 ratio (303 hyl1-like: 92 shy43-like; χ2 = 0.615, P > 0.1), implying that the suppressed phenotype is conferred by a single, recessive mutation. To map the causal mutation, we selected and pooled ∼80 F2 individuals showing the shy43-like phenotype and performed whole genome resequencing. A pooled sample of 50 hyl1-2 plants was sequenced in parallel as a control. The causal mutation was mapped to the left arm of chromosome 1 (Fig. 1B). Point mutations introduced by ethyl methanesulfonate (EMS) served as molecular markers for fine mapping. With this approach, a G-A point mutation disrupting the splicing acceptor site of the fourth intron in the SOP1 gene (designated sop1-6) was determined, causing abnormal splicing and consequently a 1-bp frame shift and pretermination of the gene (Fig. 1 C and D and SI Appendix, Fig. S2C). Transformation of a genomic fragment of SOP1 into shy43 restored the morphology to the hyl1-2 level (Fig. 1E). Moreover, we obtained a T-DNA insertion mutant of SOP1 (sop1-5, SALK_019457) (SI Appendix, Fig. S2 D and E), and crossed it to hyl1-2. The hyl1-2 sop1-5 double mutant exhibited significantly recovered phenotype (Fig. 1F). Taken together, our results demonstrated that SOP1 is a genetic suppressor of hyl1-2. Thus, we designated the shy43 mutant hyl1-2 sop1-6.
Fig. 1.
SOP1 mutations suppress the morphological defects of hyl1-2. (A) Vegetative phenotype of shy43, hyl1-2, and F1 plants from a cross between shy43 and hyl1-2. Aerial parts of 4-wk-old plants were photographed. (Scale bar, 1 cm.) (B) Manhattan plot showing the delta single nucleotide polymorphism (SNP) index between the shy43-like F2 pool and the hyl1-2 pool. SNP index was calculated by MutMAP (51). The causal gene was mapped to the left arm of Chr. 1, as highlighted by the black dotted circle. (C) Schematic structure of the SOP1 gene, the sop1-5 T-DNA insertion, and the sop1-6/sop1shy43 mutation. The sop1-6 mutation and the resultant amino acid changes are highlighted in red. Open box, 5′ or 3′ untranslated regions; gray box, exons; solid line, introns; *, stop codon. (D) Sanger sequencing of the SOP1 cDNA from the Col-0 and sop1-6 plants. In sop1-6, the G-to-A mutation at acceptor site of the fourth intron of SOP1 resulted in a 1-bp shift of the splicing acceptor site, and consequently led to one guanine deletion. (E) Phenotype of different mutants and transgenic plants. Four-week-old plants and their rosette leaves of the indicated genotypes are shown. (Scale bar, 1 cm.) (F) sop1-5 rescues the morphological defects of hyl1-2. Four-week-old plants are photographed. (Scale bar, 1 cm.)
sop1-6 Partially and Selectively Rescues the Functions of a Subset of miRNAs in hyl1-2.
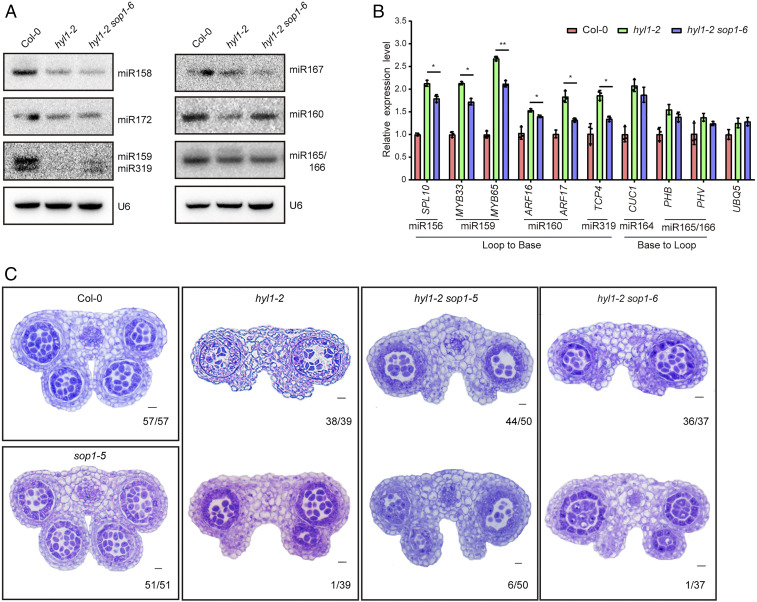
HYL1 is a core factor in miRNA biogenesis and its malfunction resulted in drastic reduction of miRNA abundance (3, 4). We hypothesized that the suppressed phenotypes in hyl1-2 sop1-6 are likely due to either global or selective recovery of miRNA expression. As HYL1 functions in both vegetative and reproductive development, we performed small RNA sequencing (sRNA-seq) analysis with both seedling and inflorescence samples from Col-0, hyl1-2, and hyl1-2 sop1-6 (Datasets S1 and S2). The results showed that miRNA abundance globally plummeted in hyl1-2 at both developmental stages. Surprisingly, the overall expression pattern of miRNAs in hyl1-2 sop1-6 was similar to that in hyl1-2, suggesting that SOP1 only affects a subset of miRNAs (SI Appendix, Fig. S2 F and G). Since hyl1-2 sop1-6 is a suppressor of hyl1-2, we focused on miRNAs with elevated expression. We found that some miRNAs related to development (e.g., miR156, miR159, miR162, and miR319) showed up-regulation in at least one of the tissue samples (fold change ≥1.5, P < 0.05) (Datasets S1 and S2). Consistently, Northern blot analysis revealed that miR159, miR160, and miR319, but not other tested miRNAs, were preferentially restored in hyl1-2 sop1-6 (Fig. 2A). qPCR analysis showed that target genes of miR156, miR159, miR160, and miR319, but not those of miR164 and miR165/166, were elevated in hyl1-2 and partially receded in hyl1-2 sop1-6 (Fig. 2B). Partial restoration of miR156, miR159, miR160, and miR319 functions in hyl1-2 sop1-6 was in accordance with previous results that the developmental defects of hyl1 could be largely explained by the lack of specific miRNAs (i.e., miR156, miR160, miR165/166, and miR319) (32–34).
Fig. 2.
sop1-6 rescues a subset of miRNAs in hyl1-2. (A) Northern blot analysis of miRNA expression in Col-0, hyl1-2, and hyl1-2 sop1-6. U6 served as loading control. (B) qPCR analysis of the expression levels of miRNA target genes. ACTIN2 serves as an internal control. Another housekeeping gene UBQ5 was analyzed in parallel. Data are means of three biological replicates and error bars denote SD. Each dot point is the average value of three technical replicates. *P < 0.05; **P < 0.01 (paired t test). Inflorescence tissues of mixed stages were used in A and B. (C) Anther microsporangia phenotypes in Col-0 and different mutants. Numbers in the Lower Right corner of each panel indicate the number of anthers with shown phenotype vs. the total number of examined anthers.
Dysfunction of HYL1 also affected anther development, with the loss of two inner microsporangia. This defect was largely due to the lack of miR165/166 and consequently ectopic overexpression of their target genes in hyl1 (34). However, our data showed that both miR165/166 reduction and target genes overaccumulation in hyl1-2 were not rescued by SOP1 mutation (Fig. 2 A and B). We thus checked the microsporangia development in the hyl1-2, hyl1-2 sop1-6, and hyl1-2 sop1-5 mutants. Indeed, in sharp contrast to having four well-developed anther microsporangia in wild-type and sop1-5 stamens, most stamens in the hyl1-2 (97.4%, 38/39), hyl1-2 sop1-6 (97.3%, 36/37), and hyl1-2 sop1-5 (88%, 44/50) mutants had only two outer anther microsporangia, with the rest possessing an additional inner microsporangium (Fig. 2C). Taken together, these data suggested that SOP1 only affects a subset of miRNAs in the absence of HYL1.
Genetic and Biochemical Interactions between Nuclear Exosome and Core miRNA Pathway Components.
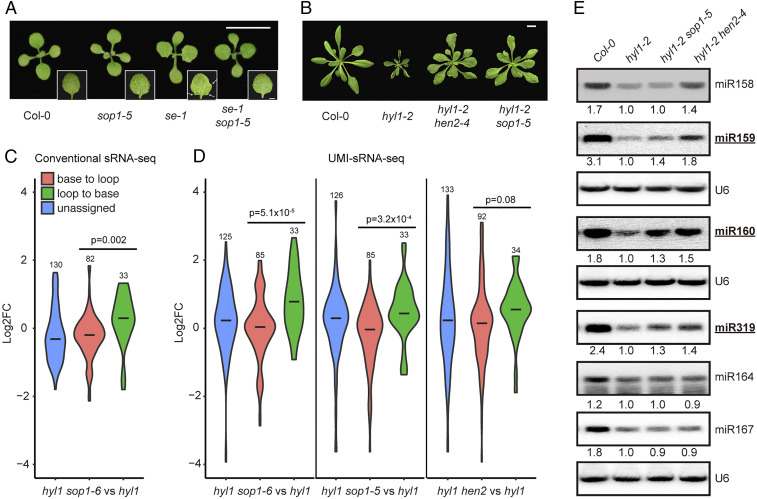
We next asked whether SOP1 genetically interacts with other miRNA pathway genes or its activity is specific to the HYL1 mutation. For this purpose, we introduced sop1-5 into mutant alleles of DCL1 (dcl1-7), SE (se-1), and AGO1 (ago1-27) via genetic crossing. In sharp contrast to its capability to rescue the hyl1-2 phenotypes, sop1-5 had a nearly negligible effect on both the morphological phenotypes and mature miRNA levels of all tested mutants (SI Appendix, Fig. S3 A–D). We only observed the disappearance of leaf serration in the first pair of true leaves in se-1 sop1-5, which is evident in the se-1 single mutant (Fig. 3A). This implied that SE may also counteract with SOP1 activity, but to a lesser extent than HYL1, or due to the use of a weak allele of se mutant.
Fig. 3.
Loop-to-base–processed miRNAs are preferentially restored in hyl1 sop1 and hyl1 hen2 mutants. (A) Twelve-day-old seedlings of Col-0, se-1, sop1-5, and se-1 sop1-5. White arrows indicate serrated leaf margins. (Scale bar, 1 cm.) (B) Twenty-five-day-old plants of Col-0, hyl1-2, hyl1-2 hen2-4, and hyl1-2 sop1-5. (Scale bar, 1 cm.) (C and D) Violin plots showing differential expression of miRNAs with different maturation ways in respective double mutants as compared with those in the hyl1-2 single mutant. Statistical differences between loop-to-base– and base-to-loop–processed miRNAs are calculated using two-tailed t test and shown as P values. Numbers on top of each plot indicate the numbers of miRNAs analyzed. See SI Appendix, Fig. S4 A and E for comparisons between mutants and wild type (i.e., Col-0). (E) Northern blot analysis of miRNA expression in different genotypes. Inflorescence tissues of mixed stages were used. Numerals indicate relative abundance of respective miRNAs relative to those in hyl1-2. U6 snRNA is a loading control. miRNAs generated from loop-to-base processing direction are bold and underlined.
SOP1 was recently characterized as a new cofactor of the nucleoplasmic exosome, which colocalizes with HEN2, and is involved in the degradation of a subset of HEN2 substrates (29). To test whether SOP1 modifies miRNA biogenesis through its role as an exosome component, we constructed the hyl1-2 hen2-4 double mutant. The hyl1-2 hen2-4 mutant phenotypically resembled hyl1-2 sop1 during vegetative growth, but had significantly better fertility (Fig. 3B and SI Appendix, Fig. S2 A and B). Of note, neither SOP1 nor HEN2 single loss-of-function mutations caused visible developmental defects under normal growth conditions (SI Appendix, Figs. S2 A and B and S3 E and F).
Since SOP1 and HEN2 colocalize in nuclear speckles, a pattern similar to the dicing bodies, we thus asked whether there were any spatial overlaps and/or physical interactions between the nucleoplasmic exosome speckles and the dicing bodies. Although the pSOP1::SOP1-GFP constructs fully complemented the hyl1-2 sop1-6 phenotype, no SOP1-GFP signals were detected either by Western blot or by confocal microscopy, likely due to weak expression and/or large proteins (the fusion protein has an estimated size of larger than 260 kDa). Fortunately, we were able to detect robust HEN2-GFP signals from the p35S::HEN2-GFP transgenes (29). To test whether HEN2 is connected to the dicing body, we performed an in situ immunofluorescence assay using an anti-GFP antibody for HEN2-GFP and anti-HYL1 antibody for endogenous HYL1. The results suggested that there was no evident colocalization between HEN2 and HYL1 speckles (SI Appendix, Fig. S3G). We also performed an in vivo coimmunoprecipitation assay between HEN2-GFP and endogenous HYL1 and SE. The results showed that HEN2-GFP interacted with SE but not HYL1 (SI Appendix, Fig. S3H). The interaction between HEN2-GFP and DCL1 remained unknown because we were unable to detect HEN2-GFP signals in young floral buds where DCL1 could only be detected using our anti-DCL1 antibody.
Loop-to-Base–Processed miRNAs Are Partially but Preferentially Restored in hyl1 sop1 and hyl1 hen2 Suppressor Mutants.
We noticed a potential enrichment of loop-to-base–processed miRNAs among the ones up-regulated in hyl1-2 sop1-6. To further assess this, we classified miRNAs into three groups based on published literature (i.e., loop-to-base, base-to-loop, and unassigned) and reanalyzed our small RNA-seq data (10, 11). Seedling samples were not analyzed due to relatively low expression of miRNAs; consequently, an insufficient number of miRNAs could be used for statistical analysis. Indeed, miRNAs processed from loop to base showed a clear trend toward up-regulation. By contrast, base-to-loop–processed miRNAs had a median value close to but lower than zero, indicating that SOP1 preferentially affected the expression of loop-to-base–processed miRNAs (Fig. 3C and SI Appendix, Fig. S4A).
To further strengthen our conclusion, we added the hyl1-2 sop1-5 and hyl1-2 hen2-4 samples into analysis and repeated the experiment. Moreover, we employed the unique molecular identifier (UMI)-based method during library construction, which eliminates PCR bias during library amplification (35). Clustering analysis revealed that all hyl1-2-containing mutants separated from Col-0, further suggesting that sop1 and hen2 only partially modify the microRNAome (SI Appendix, Fig. S4B). Notably, hyl1-2 hen2-4 was relatively distant from hyl1-2 sop1-6, hyl1-2 sop1-5, and hyl1-2 (SI Appendix, Fig. S4B). Consistently, we detected more differentially expressed miRNAs in hyl1-2 hen2-4 than in hyl1-2 sop1 as compared with those in hyl1-2 (SI Appendix, Fig. S4 C and D and Dataset S3). This observation strongly suggested that HEN2 may play a broader role in miRNA regulation than SOP1, which is consistent with previous results that SOP1 contributes to a subset of HEN2 substrates (29). Nevertheless, all double mutants preferentially restored the expression of loop-to-base–processed miRNAs, though the effect is less significant in hyl1-2 hen2-4, presumably due to its broader effect on base-to-loop–processed pri-miRNAs (Fig. 3D and SI Appendix, Fig. S4E). Three classical loop-to-base–processed miRNAs (miR319, miR159, and miR160) and three base-to-loop–processed miRNAs (miR158, miR164, and miR167) were chosen for Northern blot validation. As shown in Fig. 2E, all three loop-to-base–processed miRNAs showed higher expression levels in hyl1-2 sop1-5 and hyl1-2 hen2-4, albeit to varying degrees, relative to those in hyl1-2. By contrast, among base-to-loop–processed miRNAs, only miR158 was up-regulated in hyl1-2 hen2-4. Moreover, neither SOP1 nor HEN2 mutation rescued the DCL1-processing imprecision defects caused by HYL1 dysfunction (SI Appendix, Fig. S4F).
HYL1 Protects Pri-miRNAs from Nuclear Exosome-Mediated Decay.
Nucleoplasmic exosomes are involved in the surveillance and degradation of many types of nuclear RNAs including some pre-mRNAs (36). We first examined whether the expression of known miRNA biogenesis pathway genes was disturbed. Ten miRNA pathway genes, including DCL1, SE, AGO1, TGH, and DDL, were chosen for analysis. DCL1 expression was elevated when HYL1 was compromised, consistent with its role as a target gene of miR162 and miR838 (SI Appendix, Fig. S5) (37, 38). Nevertheless, all tested genes showed comparable expression levels between hyl1-2 sop1-6 and hyl1-2, except for DCL1, which was moderately down-regulated in hyl1-2 sop1-6, likely due to the elevation of miR162 expression (SI Appendix, Fig. S5).
When the exosome activity was compromised, several miRNA precursors and/or processing intermediates were overaccumulated, although their respective mature miRNA levels were seemingly unaffected (39, 40). We thus investigated the extent to which SOP1 and HEN2 affect miRNA precursor accumulation in the hyl1 background. Considering that miRNA precursors usually have relatively shorter sizes than mRNAs, we performed deeper fragmentation of our RNA samples and selected 40- to 100-nt cDNA insertions during library construction (SI Appendix, Fig. S6A). RNA-seq data obtained from two biological replicates of inflorescence tissues from Col-0, sop1-5, hen2-4, hyl1-2, hyl1-2 sop1-5, and hyl1-2 hen2-4 were used for differential gene expression analysis (Dataset S4).
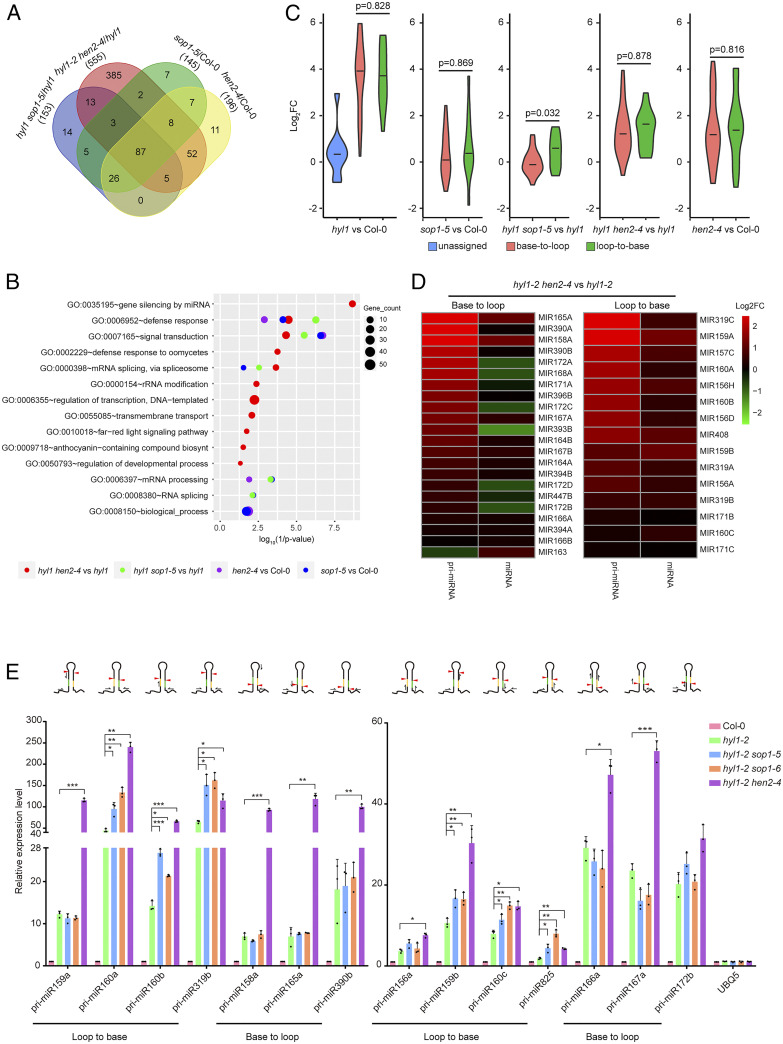
Known SOP1-, HEN2-, and HYL1-dependent transcripts [i.e., up-regulated transcripts in sop1 and hen2 based on previous studies (29, 40) and pri-miRNAs] were firstly evaluated as quality controls. Known SOP1-dependent transcripts and pri-miRNAs were significantly up-regulated in the sop1-5 and hyl1-2 mutants, respectively (SI Appendix, Fig. S6 B and C and Datasets S5 and S6). hen2-4, but not hyl1-2, also overaccumulated SOP1-dependent transcripts. On the other hand, hen2-4, but not sop1, caused moderate overaccumulation of pri-miRNAs (SI Appendix, Fig. S6 B and C). Dozens of mRNAs have been characterized as putative HEN2-dependent transcripts. However, they were of low confidence due to few overlaps with core exosome substrates and low reproducibility (40). Indeed, we didn’t see global up-regulation of these mRNAs in hen2-4 (SI Appendix, Fig. S6D and Dataset S7). By contrast, HEN2-dependent noncoding RNAs, including snoRNAs were remarkably up-regulated in the hen2-4 and hyl1-2 hen2-4 samples (SI Appendix, Fig. S6E and Dataset S8). We next characterized significant up-regulated genes in four different comparison groups (i.e., sop1-5 vs. Col-0, hen2-4 vs. Col-0, hyl1-2 sop1-5 vs. hyl1-2, and hyl1-2 hen2-4 vs. hyl1-2) (Dataset S9). A Venn diagram revealed that a total of 87 genes were up-regulated in all four groups, with most up-regulated genes in sop1-5 (83%, 121/145) and hen2-4 (77.6%, 152/196) also up-regulated in hyl1-2 sop1-5 and hyl1-2 hen2-4, respectively (Fig. 4A and SI Appendix, Fig. S6F). More impressively, 88.3% (128/145) of up-regulated genes in sop1-5 overlapped with hen2-4 (Fig. 4A and SI Appendix, Fig. S6F). Interestingly, we found that hyl1-2 hen2-4 had the largest number of up-regulated genes, with 69% (385/555) of them unique (Fig. 4A). Remarkably, gene ontology (GO) analysis on up-regulated genes revealed a significant enrichment of miRNA-mediated gene silencing (all were pri-miRNAs) in hyl1-2 hen2-4 (vs. hyl1-2), but not in hen2-4 (vs. Col-0) (Fig. 4B), indicating that pri-miRNAs are more sensitive to nucleoplasmic exosome attack in the hyl1 mutant background.
Fig. 4.
Transcriptome analysis and accumulation of pri-miRNAs in different mutants. (A) Venn diagram showing the overlap of significantly up-regulated genes among different groups. Numbers in parentheses indicate the total number of up-regulated genes in the corresponding comparison group. (B) GO analysis (biological process [BP]) of 403 genes specifically up-regulated in hyl1-2 hen2-4 vs. hyl1-2 but not in hen2-4 vs. Col-0. (C) Violin plots showing differential expression of pri-miRNAs with different maturation ways in different mutants. Statistical differences between loop-to-base– and base-to-loop–processed pri-miRNAs are calculated using two-tailed t test and shown as P values. See also SI Appendix, Fig. S6G for comparisons between additional mutants and wild type. (D) Heatmap showing correlation between pri-miRNA overaccumulation and mature miRNA expression in hyl1-2 hen2-4 vs. hyl1-2. See SI Appendix, Fig. S6 H and I for comparisons of hyl1 sop1 vs. hyl1 and mutants vs. Col-0, respectively. (E) qPCR analysis of the expression levels of pri-miRNAs. The expression changes are normalized to those of ACTIN2, with expression levels in Col-0 set to 1. Data are means of three biological replicates and error bars denote SD. Each dot point is the average value of three technical replicates. *P < 0.05; **P < 0.01; ***P < 0.001 (paired t test). The Top shows schematic structures of pri-miRNAs and positions of primers relative to first cut sites. miRNA and miRNA* are highlighed in yellow and green, respectively. Red arrow heads depict first cut sites. Black half arrows indicate relative positions of primers used for qPCR analysis (not in scale).
While most of loop-to-base and base-to-loop pri-miRNAs were dramatically up-regulated when HYL1 is compromised, many unassigned pri-miRNAs were largely unresponsive (Fig. 4C and SI Appendix, Fig. S6G). We noticed that all of these unresponsive pri-miRNAs are from the ones that are numbered after 400, and have not been rigorously tested, suggesting that a substantial proportion of these loci may not be bona fide MIRNA genes. We next compared the expression changes between loop-to-base– and base-to-loop–processed pri-miRNAs in hyl1-2 sop1-5 and hyl1-2 hen2-4. As expected, loop-to-base–processed pri-miRNAs were preferentially up-regulated in hyl1-2 sop1-5 relative to those in hyl1-2 (Fig. 4C). Strikingly, both types of pri-miRNAs were up-regulated to very high levels in hyl1-2 hen2-4 (Fig. 4C). This result indicated that SOP1 may have more of a specific role in the degradation of loop-to-base–processed miRNA precursors than HEN2. To further validate a direct role of SOP1 in pri-miRNA degradation, we compared the decaying rate of pri-miRNAs among Col-0, hyl1-2, and hyl1-2 sop1-5. The result showed that five out of six tested pri-miRNAs were more stable in hyl1-2 as compared with Col-0, consistent with compromised DCL1 processing in hyl1-2. All three loop-to-base–processed pri-miRNAs (i.e., pri-miR159b, pri-miR160a, and pri-miR319b), but not base-to-loop ones (i.e., pri-miR167a, pri-miR172a, and pri-miR172b), decayed more slowly in hyl1-2 sop1-5 relative to those in hyl1-2 (SI Appendix, Fig. S7).
Overaccumulation of base-to-loop–processed pri-miRNAs but seemingly not mature miRNAs in hyl1-2 hen2-4 promoted us to check correlation between expression changes in pri-miRNAs and their corresponding miRNAs (Fig. 4D and SI Appendix, Fig. S6 H and I). A good correlation was only detected in the loop-to-base group. In sharp contrast, expression of base-to-loop–processed miRNAs in hyl1-2 hen2-4, however, was largely dissociated from pri-miRNA elevation (Fig. 4D), suggesting that processing of pri-miRNAs from base-to-loop by DCL1 remains stagnant in hyl1-2 hen2-4. Consistent with previous results, mutating SOP1 increased loop-to-base–, but not base-to-loop–processed pri-miRNAs and mature miRNAs in the hyl1 background (SI Appendix, Fig. S6 H and I). Fourteen pri-miRNAs exhibiting different response patterns were selected for qPCR validation. The result showed high concordance between RNA-seq and qPCR results (Fig. 4E). Since most of our primers (11/14) span first DCL1 cutting sites and thus recognize pri-miRNAs only (Fig. 4E), we concluded that the nucleoplasmic exosome mainly targets pri-miRNAs in hyl1, although we cannot exclude the possibility that it can also target processing intermediates for degradation, as proposed previously (39).
Discussion
In summary, we uncover a noncanonical role for HYL1 in antagonizing nucleoplasmic exosome activities. In the presence of HYL1, the nucleoplasmic exosome mainly serves as a surveillance machinery to eliminate processing byproducts or aborted processing intermediates (39, 40). In the absence of HYL1, however, pri-miRNAs and/or processing intermediates overaccumulated and/or were changed in their secondary structures, theoretically leaving them more vulnerable to exosome attack. Interestingly, we found that a subset of pri-miRNAs, especially loop-to-base–processed ones, were preferentially targeted by SOP1 (Fig. 4C). It is not known how such selectivity is achieved. Considering SOP1 is a putative RNA-binding protein, it will be interesting to test whether SOP1 directly and selectively binds to loop-to-base–processed miRNA precursors. The exosome usually requires RNA-binding adaptors for substrate recognition (27). It is tempting to speculate that SOP1 may serve as an RNA-binding adaptor to recruit the exosome to loop-to-base–processed pri-miRNAs. On the other hand, we found that HEN2 interacts with SE and indiscriminately targets loop-to-base– and base-to-loop–processed pri-miRNAs (SI Appendix, Fig. S3H and Fig. 4 C and D), suggesting that the nucleoplamic exosome itself has no preference on pri-miRNAs with either processing direction. Although the biological significance of HEN2 and SE interaction remains to be further studied, it is possible that SE may serve as a scaffold protein for the recruitment of the HEN2-containing exosome to the pri-miRNA substrates. Our data also imply the existence of yet uncharacterized adaptor proteins with functions parallel to SOP1 in base-to-loop–processed pri-miRNA recognition.
Although HEN2 targets base-to-loop–processed pri-miRNAs, only a few of their corresponding mature miRNAs were rescued in the hyl1-2 hen2-4 mutants (Figs. 3 D and E and 4 C and D), suggesting that higher pri-miRNA levels alone do not guarantee more miRNA production. A previous report showed that three base-to-loop–processed pri-miRNAs (i.e., pri-miR163, pri-miR164b, and pri-miR166a), but not their respective pre-miRNAs, were overaccumulated in hyl1-2, suggesting that pri-miRNA to pre-miRNA processing is more severely impeded than pre-miRNA to mature miRNA in hyl1-2 (8). Moreover, HYL1 has been proposed to play distinct roles on different types of substrates, either by promoting the correct loading of DCL1 onto its substrate RNA (e.g., pri-miR166b, a base-to-loop–processed miRNA) to initiate the first cut, or by activating the second cut through triggering a helicase-dependent conformational change in DCL1 (e.g., pri-miR156a, a loop-to-base–processed miRNA) (31). If this were the case, more loop-to-base–processed intermediates would be present in hyl1 mutants, which are also favored substrates of exosomes. Compromised nucleoplasmic exosome function in the hyl1 mutant background presumably increases the chance of the second cut by DCL1, thereby generating more mature miRNA/miRNA*s. Clearly, careful followup investigations are required to test this intriguing hypothesis. In addition to exosome mediated 3′ to 5′ decay, XRN2/3-dependent 5′ to 3′ exoribonucleases have also been shown to degrade pri-miRNA processing byproducts (41). It thus will be interesting to test the dynamic interaction and coordination between nuclear RNA surveillance machinery and pri-miRNA biogenesis machinery.
Materials and Methods
Plant Materials and Growth Conditions.
EMS mutagenesis was performed as previously described (42). Genotypes used in this study were Col-0, hyl1-2 (SALK_064863), se-1 (CS3257) (43), dcl1-7 (CS6953) (22), ago1-27 (44), sop1-5 (SALK_019457), sop1shy43/sop1-6 (this work), and hen2-4 (SALK_091606C). Double mutants were created by genetic crosses, and F2 plants were genotyped for homozygotes. sop1-6 was obtained from an F3 generation of a cross between hyl1-2 sop1-6 and Col-0. Transgenic plants were generated by the floral dip method (45). All plants were grown on soil or 1/2 Murashige and Skoog medium at 22 ± 2 °C under long-day conditions (16 h light of ∼120 µmol m−2 s−1/8 h dark).
Genetic Mapping.
Genetic mapping was conducted as previously described (46). dCAPS (short for derived cleaved amplified polymorphic sequence) markers (listed in Dataset S10), designed based on single point mutations generated by EMS, were used for fine mapping.
Small RNA Northern Blot.
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific,15596018). For small RNA Northern blot assay, low molecular weight RNA was enriched and separated on a 16% denatured polyacrylamide gel electrophoresis gel. Small RNA Northern blot was conducted as previously described (47). 32P-end–labeled probes were used for Northern blot (see Dataset S10 for probe sequences). Radioactive signals were detected using a Typhoon scanner (GE Healthcare, FLA-9000) and quantified with ImageQuant (Molecular Dynamics).
Anther Anatomy Using Semithin Section.
Inflorescences of Col-0 and mutant plants were collected and fixed immediately in the FAA solution (38% formaldehyde: acetic acid: 50% ethanol = 1:1:18), and then embedded in Technovit 7100 resin (Kulzer Technique, https://www.kulzer-technik.com/en_kt/kt/home_15/startseite_7.aspx#) as described previously (48). Samples were sectioned to a thickness of 1.0 μm with a motorized RM2265 rotary microtome (Leica, https://www.leica-microsystems.com/) and stained for 2 min with 0.005% Toluidine Blue O (Sangon Biotech, 92-31-9) before being photographed under the Axio Scope A1 microscope (Carl Zeiss).
Small RNA Deep Sequencing.
Conventional small RNA library construction and deep sequencing analyses were performed as described previously (49). For UMI-sRNA-seq, a 10-bp UMI-labeled primer was used for first strand synthesis. Small RNA libraries were sequenced with single-end 50 bp (SE50) using the HisEq. 2500 platform at Novagene (for conventional sRNA-seq) or BGI (for UMI-sRNA-seq). Data pretreatment and quality-control analysis were conducted according to our previously established pipeline (46). Differential expression analysis was conducted using DEGseq (v1.39.0) with a modified miRNA annotation file, which merges miRNAs with identical sequences (46). miRNA processing imprecision was calculated as described previously (31). Annotation of loop-to-base and base-to-loop miRNAs was based on a previous study (10).
RNA Sequencing.
Total RNA was extracted from inflorescence tissues of different genotypes. Sequencing libraries were constructed using KAPA mRNA HyperPrep kit with a modified protocol. In brief, mRNA was captured by magnetic oligo-dT beads (Kapa Biosystems) and fragmented into short sizes by incubating at 94 °C for 15 min (KAPA mRNA HyperPrep Kit, Cat. No. KR1352). After second strand synthesis and A-tailing, cDNA fragments larger than 150 bp were discarded by right side selection (i.e., larger fragments bound to beads are discarded and smaller fragments in the supernatant are kept) using 1× volume of Ampure beads (Beckman Coulter). Two additional right-side selections using 1.8× volume of Ampure beads were conducted after adaptor ligation and library amplification, respectively. Library DNAs ranging from 160 to 220 bp (corresponding to insert size 40 ∼ 100 bp) were recovered from polyacrylamide gel and run at pair end 150 bp (PE150) using the Illumina Hiseq X-Ten platform (Genenergy Inc.). Differential gene expression analysis was conducted using DESeq2 v1.12.3 (50). Only pri-miRNAs with fragments per kilobase of transcript, per million fragments sequenced (FPKM) >10 in at least one sample were analyzed.
Data Availability.
Raw high-throughput sequencing data have been deposited to the NCBI Sequence Read Archive database with the following project identification nos. PRJNA563051 (genome resequencing), PRJNA563021 (conventional sRNA-seq, seedlings), PRJNA563026 (conventional sRNA-seq, inflorescences), PRJNA563040 (UMI-sRNA-seq), and PRJNA563043 (RNA-seq). All other data and associated protocols are available in the manuscript or in SI Appendix.
Plasmid Construction, Immunofluorescence, Coimmunoprecipitation, qPCR, and RNA Decay.
Plasmid construction, immunofluorescence, coimmunoprecipitation, qPCR, and RNA decay analysis are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Jirong Huang for sharing the sop1-5 and hen2-4 mutant seeds and to Dominique Gagliardi for the p35S::HEN2-GFP transgenic lines. This work was supported by grants from the National Natural Science Foundation of China (31970275, 91740101, and 31622009 to G.R. and 31771480 to N.J.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw high-throughput sequencing data have been deposited to the National Center for Biotechnology Information (NCBI) Sequence Read Archive database with the following project identification nos. PRJNA563051 (genome resequencing), PRJNA563021 (conventional sRNA-seq, seedlings), PRJNA563026 (conventional sRNA-seq, inflorescences), PRJNA563040 (UMI-sRNA-seq), and PRJNA563043 (RNA-seq).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007203117/-/DCSupplemental.
References
- 1.Li S., Castillo-González C., Yu B., Zhang X., The functions of plant small RNAs in development and in stress responses. Plant J. 90, 654–670 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P., MicroRNAs in plants. Genes Dev. 16, 1616–1626 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han M. H., Goud S., Song L., Fedoroff N., The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U.S.A. 101, 1093–1098 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez F., Gasciolli V., Crété P., Vaucheret H., The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Lobbes D., Rallapalli G., Schmidt D. D., Martin C., Clarke J., SERRATE: A new player on the plant microRNA scene. EMBO Rep. 7, 1052–1058 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L., Liu Z., Lu F., Dong A., Huang H., SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47, 841–850 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Dong Z., Han M. H., Fedoroff N., The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 9970–9975 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurihara Y., Takashi Y., Watanabe Y., The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Z., Khanna K., Ruan S., Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 21, 790–797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bologna N. G. et al., Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 23, 1675–1689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moro B. et al., Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res. 46, 10709–10723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateos J. L., Bologna N. G., Chorostecki U., Palatnik J. F., Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 20, 49–54 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Song L., Axtell M. J., Fedoroff N. V., RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20, 37–41 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Werner S., Wollmann H., Schneeberger K., Weigel D., Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 20, 42–48 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Addo-Quaye C. et al., Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15, 2112–2121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bologna N. G., Mateos J. L., Bresso E. G., Palatnik J. F., A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28, 3646–3656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang Y., Spector D. L., Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17, 818–823 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujioka Y., Utsumi M., Ohba Y., Watanabe Y., Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 48, 1243–1253 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Song L., Han M. H., Lesicka J., Fedoroff N., Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. U.S.A. 104, 5437–5442 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren G., Yu B., Critical roles of RNA-binding proteins in miRNA biogenesis in Arabidopsis. RNA Biol. 9, 1424–1428 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Lu C., Fedoroff N., A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12, 2351–2366 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauer S. E., Jacobsen S. E., Meinke D. W., Ray A., DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Liu Q. et al., Complementation of HYPONASTIC LEAVES1 by double-strand RNA-binding domains of DICER-LIKE1 in nuclear dicing bodies. Plant Physiol. 163, 108–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X. et al., Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res. 42, 12224–12236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S. W. et al., Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18, 594–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilchert C., Wittmann S., Vasiljeva L., The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 17, 227–239 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Schmid M., Jensen T. H., The exosome: A multipurpose RNA-decay machine. Trends Biochem. Sci. 33, 501–510 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Lange H., Sement F. M., Gagliardi D., MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 68, 51–63 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Hématy K. et al., The zinc-finger protein SOP1 is required for a subset of the nuclear exosome functions in Arabidopsis. PLoS Genet. 12, e1005817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tagami Y., Motose H., Watanabe Y., A dominant mutation in DCL1 suppresses the hyl1 mutant phenotype by promoting the processing of miRNA. RNA 15, 450–458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C., Axtell M. J., Fedoroff N. V., The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol. 159, 748–758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Jia L., Wang H., He Y., HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 62, 4367–4381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Yang X., Wu F., He Y., HYL1 controls the miR156-mediated juvenile phase of vegetative growth. J. Exp. Bot. 63, 2787–2798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lian H., Li X., Liu Z., He Y., HYL1 is required for establishment of stamen architecture with four microsporangia in Arabidopsis. J. Exp. Bot. 64, 3397–3410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kivioja T. et al., Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Houseley J., LaCava J., Tollervey D., RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7, 529–539 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Rajagopalan R., Vaucheret H., Trejo J., Bartel D. P., A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Z., Kasschau K. D., Carrington J. C., Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA. Curr. Biol. 13, 784–789 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Chekanova J. A. et al., Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131, 1340–1353 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Lange H. et al., The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 10, e1004564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gy I. et al., Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X. et al., Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genet. 11, e1005091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prigge M. J., Wagner D. R., The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1279 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morel J. B. et al., Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629–639 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Mei J., Jiang N., Ren G., The F-box protein HAWAIIAN SKIRT is required for mimicry target induced microRNA degradation in Arabidopsis. J. Integr. Plant Biol. 61, 1121–1127 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Ren G. et al., Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 12817–12821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang F., Zhang Z., Jin Y., Ma H., Cell biological analyses of anther morphogenesis and pollen viability in Arabidopsis and rice. Methods Mol. Biol. 1110, 203–216 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Wang J. et al., Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana. Nucleic Acids Res. 47, 7886–7900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abe A. et al., Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw high-throughput sequencing data have been deposited to the NCBI Sequence Read Archive database with the following project identification nos. PRJNA563051 (genome resequencing), PRJNA563021 (conventional sRNA-seq, seedlings), PRJNA563026 (conventional sRNA-seq, inflorescences), PRJNA563040 (UMI-sRNA-seq), and PRJNA563043 (RNA-seq). All other data and associated protocols are available in the manuscript or in SI Appendix.