Significance
Enhancer of Zeste Homolog 2 (EZH2) is the catalytic subunit of Polycomb Repressive Complex 2, mediating trimethylation of histone H3K27, a hallmark of gene silencing. Paradoxically, accumulating evidence indicates that EZH2 also dictates a unique gene activation program to promote cancer development. The molecular mechanism underlying the functional conversion of EZH2 from a gene repressor to an activator is unclear. In the current study, we report an unexpected finding that EZH2 contains a hidden, partially disordered transactivation domain (TAD), which directly binds to the transcriptional coactivator p300 and activates gene expression in cancer cells. Structural analysis suggests a “lock-and-release” mechanism of the functional control of the EZH2 TAD by cancer-specific EZH2 phosphorylation.
Keywords: EZH2, gene activation, transcriptional activation domain, phosphorylation, p300
Abstract
Enhancer of Zeste Homolog 2 (EZH2) is the catalytic subunit of Polycomb Repressive Complex 2 (PRC2), which minimally requires two other subunits, EED and SUZ12, for enzymatic activity. EZH2 has been traditionally known to mediate histone H3K27 trimethylation, a hallmark of silent chromatin. Emerging evidence indicates that EZH2 also activates gene expression in cancer cells in a context distinct from canonical PRC2. The molecular mechanism underlying the functional conversion of EZH2 from a gene repressor to an activator is unclear. Here, we show that EZH2 harbors a hidden, partially disordered transactivation domain (TAD) capable of interacting with components of active transcription machinery, mimicking archetypal acidic activators. The EZH2 TAD comprises the SRM (Stimulation-Responsive Motif) and SANT1 (SWI3, ADA2, N-CoR, and TFIIIB 1) regions that are normally involved in H3K27 methylation. The crystal structure of an EZH2−EED binary complex indicates that the EZH2 TAD mediates protein oligomerization in a noncanonical PRC2 context and is entirely sequestered. The EZH2 TAD can be unlocked by cancer-specific EZH2 phosphorylation events to undergo structural transitions that may enable subsequent transcriptional coactivator binding. The EZH2 TAD directly interacts with the transcriptional coactivator and histone acetyltransferase p300 and activates gene expression in a p300-dependent manner in cells. The corresponding TAD may also account for the gene activation function of EZH1, the paralog of EZH2. Distinct kinase signaling pathways that are known to abnormally convert EZH2 into a gene activator in cancer cells can now be understood in a common structural context of the EZH2 TAD.
EZH2 (Enhancer of Zeste Homolog 2) and its paralog, EZH1, are the catalytic subunits of Polycomb Repressive Complex 2 (PRC2), which maintains silent chromatin via trimethylation of histone H3 lysine 27 (H3K27me3). EED (Embryonic Ectoderm Development) and SUZ12 (SUppressor of Zeste 12), two other subunits of PRC2, are indispensable for the enzymatic activity of EZH2 (1). Recent crystal structures of a minimally active PRC2 containing EZH2, EED, and the VEFS [VRN2, EMF2, FIS2, and Su(z)12) domain of SUZ12 (SUZ12(VEFS)] from both thermophilic fungi Chaetomium thermophilum and humans indicate that EZH2 adopts an overall modular structural design: Multiple structural domains form intra- and intersubunit interactions that confer gene repression function of EZH2 in the context of PRC2 (2–4). In particular, the SRM (Stimulation-Responsive Motif) of EZH2 is folded as an α-helix to bind H3K27me3 and transmits the stimulating signal to the catalytic SET (Su(var)3-9, Enhancer-of-zeste and Trithorax) domain of EZH2 through an allosteric mechanism (2, 3). In addition, the SANT1 (SWI3, ADA2, N-CoR, and TFIIIB 1) domain of EZH2 that immediately follows the SRM interacts with the SBD (SANT1-Binding Domain) from the N terminus of EZH2, forming an intramolecular α-helix bundle structure (2–4). The SBD−SANT1 region contacts DNA when PRC2 engages with a dinucleosome (5); an overlapping region of EZH2 is also involved in the binding of G-quadruplex RNA (6, 7).
Paradoxically, EZH2 is also linked to gene activation. EZH2 is overexpressed in a wide spectrum of cancers (8). EZH2 has also been repeatedly shown to stimulate gene expression in cancer cells (9–15). In particular, EZH2 is overphosphorylated on residue serine 21 in some cancer types by kinase AKT, and EZH2-S21 phosphorylation promotes formation of non-PRC2 complexes of EZH2 that occupy active chromatin in some prostate cancer cells (9, 16). These EZH2-stimulated loci are devoid of SUZ12, EED, and the repressive H3K27me3 histone mark but are enriched for RNA polymerase II (pol II) and the active H3K4me3 histone mark (9). In addition, phosphorylated EZH2-S21 (EZH2-S21p) has also been found in glioblastoma multiforme stem-like cells to promote tumorigenicity through STAT3 signaling (17). In another notable example, JAK3 kinase converts EZH2 into a gene activator via phosphorylation of residue tyrosine 244, up-regulating genes involved in proliferation, stemness, and invasiveness (18). In line with the noncanonical function of EZH2, enzymatic inhibitors of JAK3 but not of EZH2 confer inhibition of NKTL cell growth (18). EZH1 is known to mediate distinctive cellular function, such as repression of hematopoietic multipotency (19). EZH1 has also been shown to promote transcription during development (20, 21), echoing the noncanonical gene activation function of EZH2 in cancer cells.
EZH2 has been indicated in previous literature here and there to interact with components of active transcription machinery, which partially explains the conundrum of the cell context-dependent gene regulation function of EZH2. For example, EZH2 physically binds to estrogen receptor α (ERα), β-catenin, and Mediator to transactivate c-Myc and cyclin D1 promoters in MCF-7 ER-positive breast cancer cells (14). Another study indicates that EZH2 also interacts with TRIM28 and chromatin remodeler SWI/SNF complex to promote gene expression in MCF-7 cells (22). In contrast, EZH2 forms a complex with RelA/RelB to confer nuclear factor κB target gene activation in ER-negative breast cancer cells (10). Furthermore, PRC2-independent interaction between EZH2 and PAF (PCNA-associated factor) mediates activation of β-catenin target genes in colon cancer cells (13).
Despite these recent advances, it remains conceptually challenging to understand how EZH2 carries out two opposing functions in cancer cells and how EZH2 may be converted from a gene repressor to an activator. Answers to this question may promise novel therapeutics against EZH2-dependent cancers. Here, we report the unexpected finding that both EZH1 and EZH2 contain a partially disordered region that resembles a classical acidic transcriptional activation domain (TAD). The EZH2 TAD is structurally locked in both PRC2 and non-PRC2 contexts, mainly by the binding of the SBD of EZH2. The interaction between the TAD and the SBD of EZH2 can also mediate protein oligomerization, independent of an active PRC2. Cancer-specific EZH2 phosphorylation may release the EZH2 TAD to activate gene expression by, at least in part, recruiting the transcriptional coactivator p300. Our work provides a structural mechanism that may connect gene repression and activation functions of EZH2.
Results
EZH2 Contains a Partially Disordered Acidic TAD.
As a starting point and similar to previous data (23, 24), we showed that a GAL4DBD-HA tagged full-length EZH2 (EZH2-FL) efficiently repressed the expression of a luciferase reporter gene under control of a “6XGAL4UAS-Thymidine Kinase (TK) promoter” cassette. Reporter gene repression by EZH2 was dependent on GAL4DBD-mediated promoter targeting, as removal of the GAL4DBD tag abolished gene repression (SI Appendix, Fig. S1).
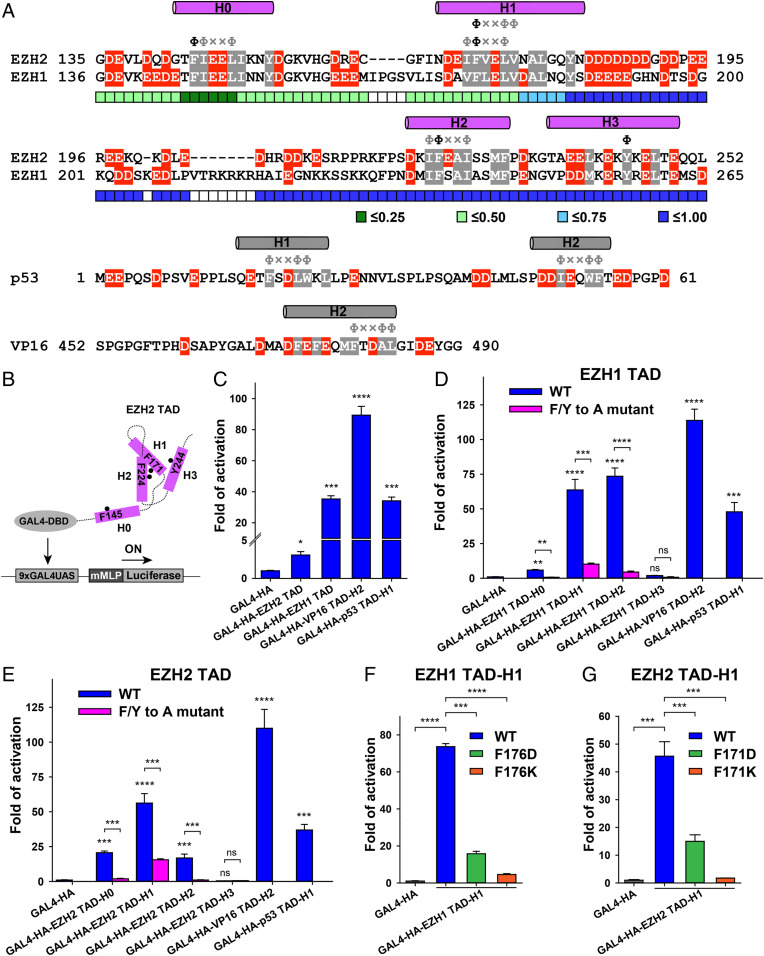
How may EZH2 become an activator? A close examination of protein sequence and structure revealed that the SANT1 domain of both EZH1 and EZH2 contains a cluster of three acidic α-helices (designated as H1, H2, and H3) (Fig. 1A). The SRM of EZH2 (designated as H0) that immediately precedes the SANT1 domain adopts a similar α-helical structure and was also included in our analysis (Fig. 1A). The SBD−SRM−SANT1 module of EZH2 displays a large degree of structural flexibility in PRC2, becoming partially or completely disordered in crystal structures under certain circumstances (SI Appendix, Fig. S2 A–D). This observation confirms the dynamic nature of this module, suggesting they are not structurally confined to “PRC2-only” functions. In addition, the SRM and SANT1 are predicted to be partially disordered based on the protein sequence (SI Appendix, Fig. S3 and also discussed below) (25). Intrinsically disordered polypeptides are known to frequently serve as components of cell signaling and gene regulation machineries (26).
Fig. 1.
The TADs of EZH1 and EZH2. (A) The TADs of EZH2 and EZH1 are aligned. Helix H0 of the EZH2(SRM) and helices H1, H2, and H3 of the EZH2(SANT1) are drawn above the sequence alignment. Hydrophobic residues are highlighted in gray and negatively charged acidic residues in red. Selected Φ-x-x-Φ-Φ and Φ-Φ-x-x-Φ motifs in helices H0, H1, and H2 are indicated by gray symbols, with the aromatic residues mutated in the reporter assay colored in black. Residue Y244 on helix H3 not belonging to the consensus motifs is also indicated by Φ. Throughout the paper, “4A” indicates mutants containing a quadruple alanine mutation on residues F145, F171, F224, and Y244 of EZH2 (or the equivalent of EZH1), and “2A” indicates mutants containing a double alanine mutation on residues F171 and F224 of EZH2. The p53 TAD (helices H1 and H2) and the second part of the VP16 TAD (helix H2) are annotated similarly for comparison. The partially disordered region of the EZH2 TAD was predicted using the VSL2 predictor in PONDR. The PONDR score is color-coded, with scores greater than 0.5 indicating intrinsically disordered residues. (B) Schematic of a GAL4-based reporter gene activation assay. (C) Luciferase gene activation by the TADs of EZH1 and EZH2. The TADs of p53 and VP16 were positive controls. The P values were derived from a two-tailed Student’s t test: ns (not significant), P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. (D and E) Gene activation by individual helices H0, H1, H2, and H3. The critical role of the characteristic aromatic residues of the EZH1 TAD (residues F146, F176, and F237) (D) and of the EZH2 TAD (residues F145, F171, and F224) (E) within the Φ-x-x-Φ-Φ or Φ-Φ-x-x-Φ consensus motif in gene activation was tested by alanine mutation. Residue Y257 of the EZH1 TAD (D) and residue Y244 of the EZH2 TAD (E) on helix H3 were also mutated and tested as a negative control. (F and G) Gene activation by helix H1 with an aspartate or lysine mutation. Residue F176 of the EZH1 TAD (F) and residue F171 of the EZH2 TAD (G) were mutated to a charged aspartate or lysine residue and tested for the gene activation activity.
Surprisingly, the acidic helices H0, H1, and H2 harbor the characteristic Φ-x-x-Φ-Φ and/or Φ-Φ-x-x-Φ motifs (Φ represents hydrophobic residues and x represents any residues), mimicking the TADs of the archetypal acidic activators, such as the tumor suppressor p53 and human herpes simplex virus (HSV) protein VP16 (Fig. 1A) (27, 28). In contrast, helix H3 lacks such motifs (Fig. 1A). The TADs of both p53 and VP16 contain two acidic regions (designated as H1 and H2); H1 and H2 of the p53 TAD as well as H2 of the VP16 TAD are also known to adopt α-helical structures (Fig. 1A) (27, 28). Accordingly, we proposed that the SRM−SANT1 region might be broadly considered as the TADs of EZH1 and EZH2 on the protein sequence level (Fig. 1A).
To test the gene activation activity of the newly identified TADs of EZH1 and EZH2 in cells, we first used a reporter gene assay, in which the luciferase gene was placed downstream of a tandem 9XGAL4UAS DNA sequence followed by a minimal adenovirus major late promoter (mMLP) (Fig. 1B). Compared to the GAL4DBD-HA vector control, the GAL4DBD-HA–tagged EZH1 TAD and, to a lesser extent, EZH2 TAD, activated luciferase gene expression in HEK293T cells, similar to gene activation by the TADs of p53 and VP16 in the same context (Fig. 1C and SI Appendix, Fig. S4). In addition, the individual acidic helices—H0, H1, and H2 but not H3—from the TADs of EZH1 and EZH2 were adequate to activate the reporter gene (Fig. 1 D and E and SI Appendix, Fig. S5).
Alanine mutation of a single phenylalanine residue in the EZH1 TAD (residue F146, F176, or F237) and the EZH2 TAD (residue F145, F171, or F224) within the Φ-x-x-Φ-Φ or Φ-Φ-x-x-Φ consensus motif diminished gene activation by the individual helix H0, H1, or H2, correspondingly (Fig. 1 A, D, and E and SI Appendix, Fig. S5). In addition, replacing residue EZH1-F176 or EZH2-F171 on helix H1 with a charged aspartate or lysine residue also greatly reduced gene activation (Fig. 1 F and G and SI Appendix, Fig. S5), highlighting the specificity of the reporter gene activation assay. As expected, alanine mutation of residue EZH1-Y257 or EZH2-Y244 on helix H3 did not show any effect (Fig. 1 A, D, and E and SI Appendix, Fig. S5). Similarly, gene activation by the TADs of p53 and VP16 was long known to require individual bulky hydrophobic residues from the consensus motifs to mediate direct interactions with multiple components of active transcription machineries—usually transcription factors or transcriptional coactivators (27–30). Collectively, these data support the notion that the TADs of EZH1 and EZH2 may function as bona fide TADs in cells.
The EZH2 TAD Is Structurally Locked.
As shown by the previous crystal structures of an active PRC2, the EZH2 TAD is structurally locked, not available for binding active transcription machinery: Specifically, 1) the SRM (i.e., helix H0) binds to H3K27me3 and the SET domain simultaneously to stimulate the enzymatic activity of EZH2 (SI Appendix, Fig. S2B) (3, 4) and 2) the SANT1 (i.e., helices H1, H2, and H3) of EZH2 forms an intramolecular complex with the SBD of EZH2 (SI Appendix, Fig. S2 B and C) (3, 4).
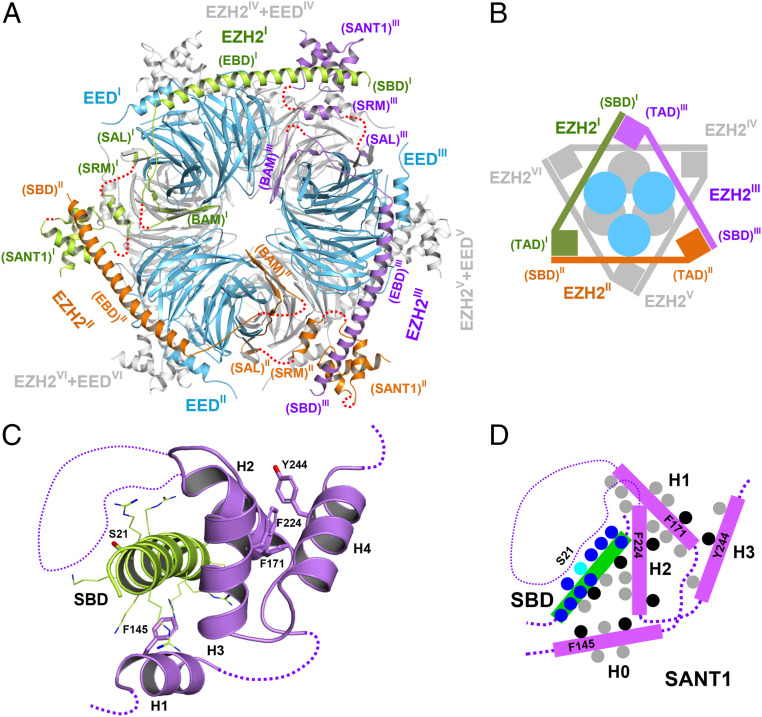
In fact, EZH2 is thought to activate gene expression independent of a canonical, enzymatically active PRC2 that normally confers gene repression (9, 18). However, the structural information of the EZH2 TAD in a noncanonical context is lacking; it is unclear whether it may change structure and become accessible for gene activation in this context. In an attempt to answer this question, we determined the crystal structure of a minimal EZH2−EED complex (SI Appendix, Table S1). The structure shows that the EZH2 TAD is largely involved in protein oligomerization, a role not previously observed in an active PRC2 (Fig. 2 A and B).
Fig. 2.
Crystal structure of an EZH2−EED complex. (A) Top view of the structure of the binary complex, containing EZH2 (1 to 254, residues 183 to 219 replaced by GSSG) and EED (residues 78 to 441). Three copies of the EZH2−EED binary complex form a trimeric structure largely through the intermolecular interaction between the SBD and the TAD of EZH2; two copies of the trimeric complexes stack face to face to form a hexameric complex. The trimer facing inward is shown in gray. Three EZH2 molecules from the trimer facing outward are shown in lime, orange, and magenta, respectively, with all of the three bound EED molecules shown in blue. The SBD, EBD, BAM, SAL, SRM, and SANT1 domains from each EZH2 molecule are labeled and numbered by superscripts. (B) Cartoon illustration of A. Protein oligomerization is largely mediated by the EZH2 TAD, including the SRM and SANT1, and the SBD of EZH2 in a “head-to-tail” mode. (C) Close-up view of the intermolecular α-helix bundle structure formed by the TAD and SBD of EZH2. The complex is shown in cartoon. Side chains of residues F145, F171, F224, and Y244 from the TAD and of residue S21 from the SBD are highlighted in sticks and labeled. Positively charged residues from the SBD are also shown in sticks. Dotted lines indicate disordered loops. (D) Cartoon illustration of C. Hydrophobic residues of the binary complex are represented by gray discs, except that aromatic residues are shown as black discs. Residue S21 from the SBD is shown as a cyan disc and the positively charged residues from the SBD are indicated by blue discs.
Overall, six functional domains of EZH2 are present in the current structure, including the SBD, the EBD (EED-Binding Domain), the BAM (β-Addition Motif), the SAL (SET Activation Loop), the SRM, and the SANT1 (Fig. 2 A and B) (2–4). The EZH2−EED binary complex adopts a hexameric structure in the following way: The TAD, including the SRM and SANT1, of one EZH2 molecule is bound by the SBD of another EZH2 molecule such that a trimeric structure is formed (Fig. 2 A and B); EZH2 molecules in two EZH2−EED trimers exchange the SAL pairwise to mediate new β-addition interactions with respective EED molecules to form a dimer of trimers (SI Appendix, Fig. S6). A series of oligomerization states of the EZH2−EED binary complex were formerly noted biochemically (31), and the current crystal structure captures one such state. Moreover, formation of the EZH2−EED trimer appears to largely depend on the intermolecular interactions between neighboring EZH2 molecules, indicating that EZH2 alone may also self-associate using a similar structural mechanism.
A direct comparison of the current structure to the structure of an active PRC2 reveals three major differences (SI Appendix, Figs. S6 and S7) (3). First, the SAL of EZH2 that is cofolded with the SET and required for the enzymatic activity of EZH2 in PRC2 becomes a binding partner of EED in the current structure, contributing to protein oligomerization (SI Appendix, Figs. S6 and S7 A and B). Second, the SBD and SANT1 of EZH2 in the current structure are engaged in intermolecular interactions but not intramolecular interactions as in canonical PRC2, mediating protein oligomerization (Fig. 2 A and B), although the mode of interaction per se is almost identical in the two contexts (SI Appendix, Fig. S7 A and C). Third, the SRM is loosely associated with the SBD−SANT1 complex, forming a bundle of five α-helices that is unique to the current structure (Fig. 2 C andD and SI Appendix, Fig. S7C and S8).
In particular, the conserved phenylalanine residues from the SRM and SANT1 of EZH2 that are important for transactivation are mostly buried in the hydrophobic core of the helix bundle in the current structure (Fig. 2 C and D). Considering these results, we concluded that the EZH2 TAD is largely sequestered in this noncanonical PRC2 context and that, different from the TADs of p53 and VP16, the EZH2 TAD in its native form is likely unable to mediate gene activation.
EZH2 Phosphorylation Unlocks the EZH2 TAD.
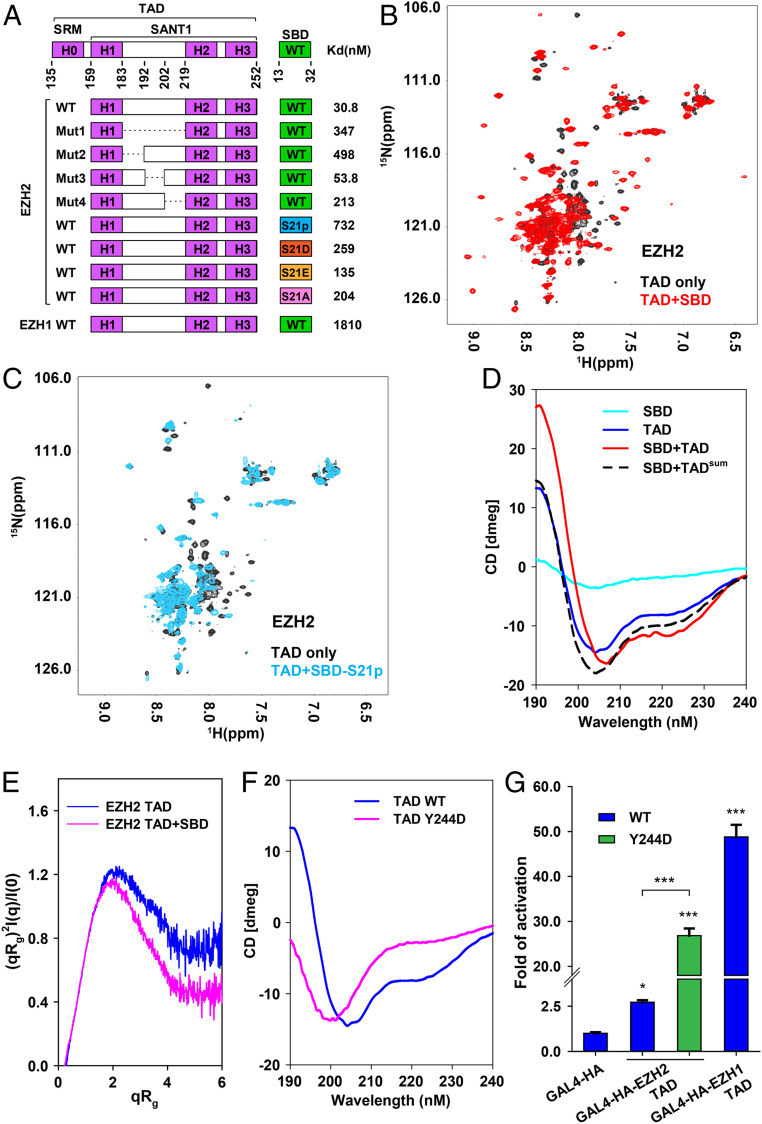
How can the locked gene activation activity of the EZH2 TAD be released in cells? EZH2 phosphorylation in the SBD by kinase AKT provides a possible mechanism (9, 16). We first used isothermal titration calorimetry (ITC) to monitor complex formation and disruption. The binding affinity of the SBD−SANT1 interaction is 30.8 nM (dissociation constant, Kd) (Fig. 3A and SI Appendix, Fig. S9). A long linker region (residues 183 to 219) between helices H1 and H2 of the SANT1 was absent in the crystal structure, and we found that replacement of this overall negatively charged linker with a GSSG sequence weakened the SBD−SANT1 interaction by over 10-fold (Figs. 1A and 3A and SI Appendix, Fig. S10). In addition, variation of three subregions of the linker caused distinct binding defects (Figs. 1A and 3A and SI Appendix, Fig. S10). Notably, substitution of a (GGS)3G sequence for the first 10 residues of the linker, 9 of which are aspartate, was sufficient to reduce SBD binding by over 15-fold (Figs. 1A and 3A and SI Appendix, Fig. S10), suggesting that this subregion may contact the SBD directly. In agreement with this possibility, the amphiphilic SBD helix that is spatially proximal to the linker in the SBD−SANT1 complex is highly positively charged (Fig. 2 C and D). Interestingly, the high density of acidic residues within the classical TADs was previously suggested to confer a kinetic advantage for attracting basic transcription factors or transcriptional coactivators over a large distance during gene activation (28, 32).
Fig. 3.
Conformational control of the EZH2 TAD function. (A) Summary of the binding affinities between the the EZH2(SANT1) (residues 159 to 254, wild type [WT] and variants) and the EZH2(SBD) peptide (residues 13 to 32, WT and variants) as measured by ITC. The binding affinity between the EZH1(SANT1) (residues 160 to 267, WT) and the EZH1(SBD) peptide (residues 13 t 32, WT) is also indicated. Mut1, 2, 3, and 4 contained residue replacement mutations of the linker between helices H1 and H2: Residues 183 to 219 were replaced by GSSG in Mut 1; residues 183 to 192 were replaced by (GGS)3G in Mut 2; residues 193 to 202 were replaced by (GGS)3G in Mut 3; residues 203 to 219 were replaced by (GGS)5GG in Mut 4. (B) Superimposed 1H-15N HSQC spectra of the 15N-labeled EZH2 TAD (TAD only, black, residues 135 to 254) and the EZH2 TAD mixed with unlabeled WT EZH2(SBD) peptide (SBD, red). (C) Superimposed 1H-15N HSQC spectra of the 15N-labeled EZH2 TAD (TAD only, black) and the EZH2 TAD mixed with EZH2-S21 phosphorylated EZH2(SBD) peptide (SBD-S21p, cyan). (D) Far-UV CD spectra of the SBD of EZH2, the unbound EZH2 TAD, the sum of the unmixed components and the SBD-bound EZH2 TAD. (E) Dimensionless Kratky plots of the unbound EZH2 TAD and the SBD-bound EZH2 TAD. (F) Far-UV CD spectra of the EZH2 TAD-WT and -Y244D. The EZH2 TAD-WT is the same as the unbound EZH2 TAD in D. (G) GAL4-based reporter gene activation assay in HEK293T cells. The EZH2 TAD WT and Y244D mutant were compared for the activation activity. GAL4-HA and GAL4-HA-EZH1 TAD are the negative and positive controls, respectively. The P values were derived from two-tailed Student’s t test: *P ≤ 0.05; ***P ≤ 0.001.
EZH2-S21, located in the middle of the SBD of EZH2, is overphosphorylated by kinase AKT in some cancer cells to promote gene activation (9, 16). Remarkably, we found that EZH2-S21p weakened the SBD−SANT1 complex by over 20-fold, which may unlock the EZH2 TAD (Fig. 3A and SI Appendix, Fig. S11). The EZH2-S21D and EZH2-S21E phosphomimetic mutants also displayed a binding defect, although to a lesser extent (Fig. 3A and SI Appendix, Fig. S11). EZH2-S21p partially neutralizes the local positive charges in the SBD, which may directly impair SBD binding to the negatively charged linker region of the EZH2 TAD, as discussed above (Fig. 2 C and D and also see the analysis above). Interestingly, an EZH2-S21A mutation of the SBD also reduced the binding affinity (Fig. 3A and SI Appendix, Fig. S11), suggesting that residue EZH2-S21, like the surrounding positively charged residues in the SBD, may also interact with the linker region of the SANT1 not observed in the crystal structure (dotted curve in Fig. 2 C and D). The SBD of EZH1 displayed a binding affinity of only 1.8 μM toward the SANT1 of EZH1 (Fig. 3A and SI Appendix, Fig. S12), implicating that the EZH1 TAD may naturally be more accessible for the binding of active transcription machineries compared to the EZH2 TAD.
To analyze the structural changes associated with binding of the SBD to the TAD of EZH2, we used NMR spectroscopy. The 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of the EZH2 TAD exhibited poor dispersion and spectral overlap (Fig. 3B and SI Appendix, Fig. S13A, black contours), which is characteristic of polypeptides that are unstructured or lack a stable tertiary structure (33). Consistently, at least the linker region of about 40 residues between helices H1 and H2 within the EZH2 TAD contains sequences of low complexity and is predicted to be intrinsically disordered (Fig. 1A). We note that better dispersion was observed in the 1H-15N HSQC spectrum of a slightly shorter fragment of EZH2 (residues 141 to 251 instead of residues 135 to 254, present in our fragment) (34), which indicated that at least part of this shorter fragment adopts a stable tertiary structure. Hence, the additional residues in our fragment appear to destabilize this structure. Addition of the SBD of EZH2 caused dramatic spectral changes in the 1H-15N HSQC spectrum of the EZH2 TAD, and we observed the appearance of multiple well-resolved cross-peaks that can be associated to the formation of a defined structure upon binding to the SBD (Fig. 3B and SI Appendix, Fig. S13B; compare black and red contours). In contrast, the SBD peptide harboring the S21p modification caused less-dramatic changes and did not induce the appearance of the well-resolved cross-peaks observed for the TAD−SBD complex (Fig. 3C and SI Appendix, Fig. S13C; compare black and blue contours). These results show that SBD-S21p can still bind to the EZH2 TAD but is unable to stabilize its structure like the unphosphorylated SBD does. Phosphorylation of EZH2-S21 may therefore function to structurally unlock the EZH2 TAD, which is otherwise sequestered by the SBD.
We next collected far-ultraviolet (UV) circular dichroism (CD) spectra to examine the secondary structure content of the EZH2 TAD (Fig. 3D). Crystal structure analysis suggested that when bound to the SBD ∼30 to 40% of the EZH2 TAD adopts α-helical structures, with the rest being largely disordered (Fig. 1A). The CD spectrum of the unbound EZH2 TAD possessed two minima at 205 nm and 222 nm, agreeing with a mixture of random coils with a typical minimum at below 200 nm and α-helices with minima at 208 nm and 222 nm (Fig. 3D). Compared to the sum of the unmixed components, the CD spectrum of the TAD−SBD complex displayed an enhanced negative ellipticity at the 222 nm minimum, a shift of the other minimum from 205 nm to 208 nm, and an increase of the ellipticity at 190 nm, all of which indicate a gain of α-helical conformation upon complex formation (Fig. 3D) (35). We note that the increase in helical structure is modest, which is consistent with the observation that only a limited number of new well-dispersed cross-peaks arising from formation of a defined structure were observed in the 1H-15N HSQC spectrum acquired in the presence of the SBD (Fig. 3B). Kratky plot analysis in small-angle X-ray scattering (SAXS) experiments also allowed us to qualitatively assess disordered states of the EZH2 TAD (36). The normalized dimensionless Kratky plot of the unbound EZH2 TAD was less converged to the baseline compared to the SBD-bound state (Fig. 3E and SI Appendix, Fig. S14), indicating that the unbound EZH2 TAD is less compact and has more disordered regions (36).
EZH2-S21 phosphorylation by AKT is not the only mechanism that may unlock the EZH2 TAD. Kinase JAK3 was also shown to convert EZH2 from a gene repressor into an activator by phosphorylating residue EZH2-Y244 in NKTL cells (18). Notably, based on the crystal structure, phosphorylated EZH2-Y244 (EZH2-Y244p) on helix H3 may also weaken SBD binding by disrupting the hydrophobic core of the α-helix bundle structure (Fig. 2 C and D). In addition, according to the CD analysis, an EZH2-Y244D phosphomimetic mutation made the EZH2 TAD much more disordered, with an almost complete loss of the 222-nm minimum and a shift of the other minimum to 200 nm (Fig. 3F). Correspondingly, the EZH2-Y244D mutation greatly enhanced gene activation by the EZH2 TAD in the reporter gene assay (Fig. 3G and SI Appendix, Fig. S15), suggesting that the partial folding of the EZH2 TAD stabilized by residue EZH2-Y244 on helix H3 may impose a second layer of conformational control of the gene activation activity, in addition to SBD binding, whereas helix H3 per se is not directly involved in gene activation.
These results show that the gene activation function of the EZH2 TAD is normally sequestered by SBD binding and partial protein folding. Cancer-specific phosphorylation of EZH2 by distinct kinases, AKT and JAK3, can remove these locking mechanisms that likely exist independent of other PRC2 subunits, to support a “lock-and-release” model for the functional conversion of EZH2.
The EZH2 TAD Interacts with the Transcriptional Coactivator p300 Directly.
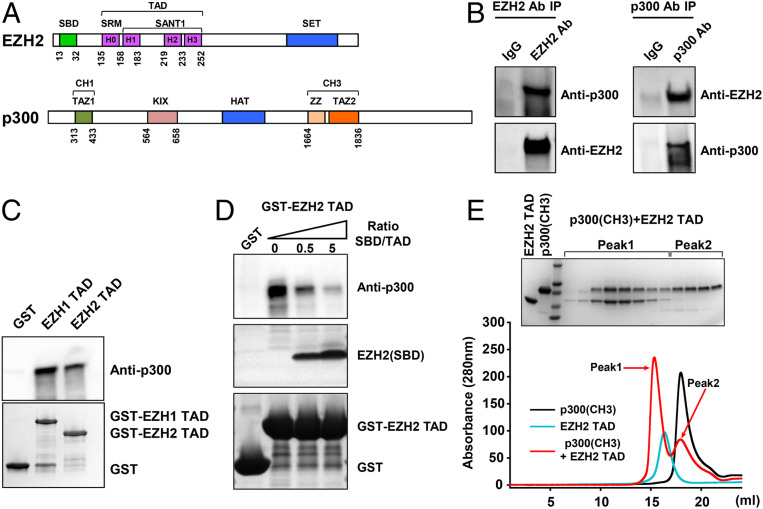
Acidic activators are known to activate gene expression by binding to transcription factors and transcriptional coactivators. In particular, the gene activation function of the p53 TAD depends on the interaction with the transcriptional coactivator p300/CBP (37, 38) (Fig. 4A). Here, we found that EZH2 was also bound by p300 in the nuclear extracts of 22Rv1 prostate cancer cells (Fig. 4B), in which EZH2 was previously shown to function as an activator (9, 15). A large-scale nuclear extract preparation from 1 × 109 cells was necessary to capture the interaction between EZH2 and p300 in a reciprocal coimmunoprecipitation (Co-IP) experiment (Fig. 4B), indicative of substoichiometric or weak binding in vivo. In addition, the TADs of EZH1 and EZH2 were sufficient to pull down p300 from HeLa nuclear extracts (Fig. 4C).
Fig. 4.
Direct interaction between EZH2 and p300. (A) Schematic domain organization of EZH2 and p300. The SBD, SRM, SANT1, and SET are illustrated in EZH2. The CH1/TAZ1, KIX, HAT, and CH3/TAZ2 are illustrated in p300. (B) Reciprocal Co-IP between EZH2 and p300 in 22Rv1 nuclear extracts. (C) Binding of the recombinant TADs of EZH1 and EZH2 to p300 from HeLa nuclear extracts. (D) Binding of the EZH2 TAD to p300 competitively inhibited by the EZH2(SBD) peptide. (E) Formation of a stoichiometric complex between the EZH2 TAD and the CH3/TAZ2 domain of p300 on a SEC column. Peak 1 and Peak 2 indicate the binary complex and the excessive unbound p300(CH3), respectively.
We next sought to biochemically dissect the interaction using purified recombinant proteins. The EZH2 TAD formed a binary complex with full-length p300 in a GST pull-down assay, and the SBD of EZH2 disrupted this complex in a dose-dependent manner through direct competition (Fig. 4D), in agreement with the “lock-and-release” mechanism that promotes coactivator binding. Furthermore, we showed that the TADs of EZH1 and EZH2 were efficiently bound to the CH1/TAZ1, KIX, and CH3/TAZ2 domains of p300 (SI Appendix, Fig. S16) and that a quadruple alanine mutation of residues F145, F171, F224, and Y244 of the EZH2 TAD (EZH2-TAD-4A) greatly reduced CH3/TAZ2 binding (SI Appendix, Fig. S17), underlining the functional similarity to the p53 TAD that also directly interacted with the same set of p300 domains (30).
Notably, the EZH2 TAD formed a stoichiometric complex with the CH3/TAZ2 domain of p300 on a size-exclusion chromatography (SEC) column (Fig. 4E), and this stoichiometric complex was subjected to disruption by the SBD of EZH2 (SI Appendix, Fig. S18). Furthermore, the p53 TAD was found to use two helices to bind the CH3/TAZ2 domain of CBP (27); likewise, a core fragment spanning from helix H1 to H2 of the EZH2 TAD (EZH2-TAD-core, residues 159 to 232) was also sufficient for the stoichiometric binding to the CH3/TAZ2 domain of p300 (SI Appendix, Fig. S19A). As expected, a double alanine mutation of residues F171 and F224 (EZH2-TAD-core-2A) destabilized the stoichiometric complex, confirming the binding specificity (SI Appendix, Fig. S19B). In another SEC run, the CH3/TAZ2 domain of p300 did not coelute with a SANT1 construct that contains helices H1, H2, and H3 with the long linker region (residues 183 to 219) replaced by a GSSG sequence as in the crystal structure, indicating a critical role of this negatively charged linker in coactivator binding (SI Appendix, Fig. S19C).
The EZH2 TAD-Mediated Gene Activation Depends on p300.
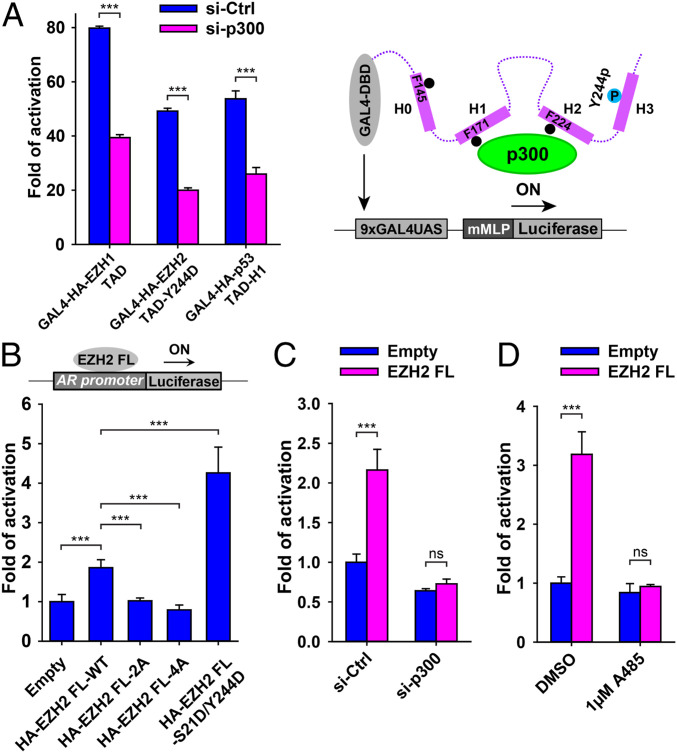
The histone acetyltransferase activity of p300 is known to mediate H3K27 acetylation (H3K27ac) during gene activation in cells. A recent genome-wide study reported extensive correlation between EZH2 occupancy and active H3K27ac histone mark on chromatin during gene activation in LNCaP prostate cancer cells, in which EZH2 knockdown decreased expression of the Prostate-Specific Antigen (PSA) gene (15). We obtained the same result and further showed that p300 knockdown also reduced PSA gene expression (SI Appendix, Fig. S20), supporting a functional correlation between EZH2 and p300. As a negative control, SUZ12 knockdown did not change gene expression (SI Appendix, Fig. S20). Moreover, p300 knockdown in HEK293T cells impaired the GAL4-dependent reporter gene activation by the EZH1 TAD, the EZH2 TAD harboring the potentiating EZH2-Y244D mutation, and the p53 TAD as a positive control (Fig. 5A and SI Appendix, Fig. S21).
Fig. 5.
Dependence of EZH2-mediated gene activation on p300. (A) Reporter gene activation assay in HEK293T cells in the absence and presence of p300 knockdown. The p53 TAD was used as a positive control. Schematic of the reporter gene activation assay for the case of the EZH2 TAD-Y244D is drawn. The P values were derived from two-tailed Student’s t test: ns, P > 0.05; ***P ≤ 0.001. (B) AR promoter-based reporter gene assays in HEK293T cells using full-length EZH2 constructs. EZH2-FL-4A and EZH2-FL-2A containing loss-of-function mutations and EZH2-FL-S21D/Y244D containing gain-of-function mutations were compared to EZH2-FL-WT. Empty vector served as a negative control. (C and D) AR promoter-based reporter gene assays in 22Rv1 prostate cancer cells using full-length EZH2 constructs. Effects of p300 knockdown (C) or chemical inhibition of the histone acetyltransferase activity of p300 (D) on the EZH2-mediated gene activation were assayed.
EZH2-FL was shown to occupy the androgen receptor (AR) promoter in both LNCaP and HEK293T cells and activate an AR promoter-based reporter gene in HEK293T cells (15). Using a similar reporter gene assay in HEK293T cells, we found that gene activation by EZH2-FL was considerably enhanced in the presence of the EZH2-S21D/Y244D double mutation, which was designed to mimic EZH2 phosphorylation by both AKT and JAK3 to unlock the EZH2 TAD (Fig. 5B and SI Appendix, Fig. S22). In contrast, EZH2-FL containing alanine mutation of residues F145, F171, F224, and Y244 (EZH2-FL-4A) or of residues F171 and F224 (EZH2-FL-2A) abolished reporter gene activation (Fig. 5B and SI Appendix, Fig. S22), possibly due to compromised coactivator binding as biochemically shown for the binding of p300 to EZH2-TAD-4A and EZH2-TAD-core-2A above (SI Appendix, Figs. S17 and S19).
EZH2-FL also activated the AR promoter-based reporter gene in 22Rv1 prostate cancer cells (SI Appendix, Fig. S23). Pretreatment of 22Rv1 cells with the PI3K-AKT kinase inhibitor LY294002 that was known to abolish EZH2-S21 phosphorylation eliminated reporter gene activation by EZH2-FL (SI Appendix, Fig. S23) (16). EZH2-FL containing an S21A mutation was unable to activate the reporter gene; in contrast, EZH2-FL with an S21D phosphomimetic mutation activated the reporter gene, whether or not 22Rv1 cells were pretreated with the kinase inhibitor (SI Appendix, Fig. S23), agreeing with the hypothesis that AKT-dependent EZH2-S21 phosphorylation promotes the gene activation function of EZH2 (9). As another control, the EZH2-FL-Y244D phosphomimetic mutation that unlocks the EZH2 TAD also constitutively activated the reporter gene irrelevant to the PI3K-AKT kinase inhibitor (SI Appendix, Fig. S23). In addition, both p300 knockdown and chemical inhibition of the histone acetyltransferase activity of p300 by A485 resulted in loss of the AR promoter-based reporter gene activation by EZH2-FL in 22Rv1 cells (Fig. 5 C and D and SI Appendix, Fig. S24), highlighting a critical role of p300 in EZH2-mediated gene activation in cancer cells.
Discussion
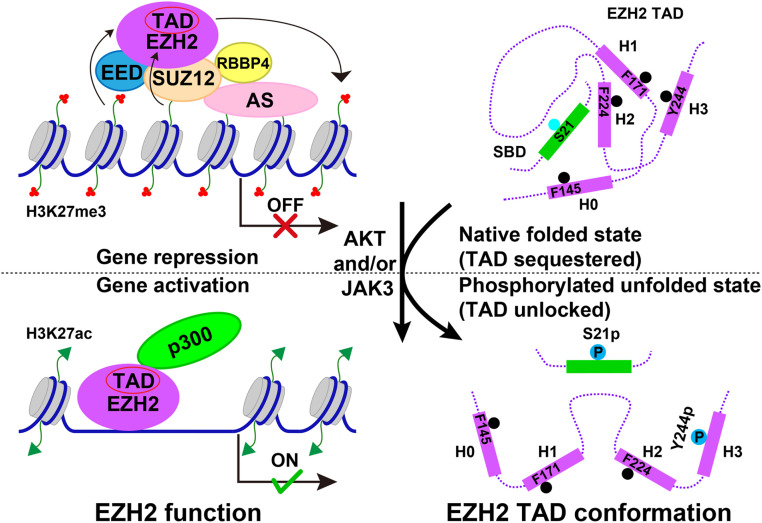
Accumulating evidence indicates that EZH2 plays a critical role in cancer development by activating gene expression independent of PRC2, which contradicts the canonical role of EZH2 in gene repression through PRC2-dependent H3K27 methylation (9–15). In the current study, we discovered based on sequence similarity, structural comparison, and gene expression analysis that EZH2 contains a partially disordered TAD that resembles the prototypical acidic TADs from p53 and VP16 in gene activation (27, 28). The TADs of EZH1 and EZH2 consist of the SRM (helix H0) and the SANT1 (helices H1, H2, and H3) (Fig. 6) and were sufficient to specifically activate reporter gene expression, when artificially recruited to the target promoter through GAL4-DNA binding. Consensus motifs necessary for gene activation by acidic activators are also present in the TADs of EZH1 and EZH2 (Fig. 6) (27, 28).
Fig. 6.
A simplified model of gene activation by EZH2. Gene repression and activation by EZH2 are illustrated on the left. EZH2 is recruited to chromatin together with other core subunits, EED, SUZ12, and RBBP4, at least in part by accessory subunits (AS) to mediate H3K27me3 in the context of PRC2. EZH2 can also be recruited to active chromatin independent of PRC2 through unknown mechanisms, where it interacts with components of active transcription machineries, such as the transcriptional coactivator p300. On the right EZH2 phosphorylation by AKT and JAK3 in cancer cells triggers structural transitions of the EZH2 TAD (helices H0, H1, H2, and H3), unlocking the otherwise sequestered gene activation activity. The core fragment of the EZH2 TAD becomes accessible to transcription factors and transcriptional coactivators.
Unlike the TADs of p53 and VP16, the EZH2 TAD is structurally locked in an α-helix bundle structure by the SBD of EZH2, unable to interact with transcription factors or transcriptional coactivators (Fig. 6). Based on additional binding and biophysical data, we showed that the EZH2 TAD is partially disordered and that SBD binding changes the conformation of the EZH2 TAD, making it more compact and ordered. Correspondingly, the locked gene activation activity of the EZH2 TAD may be released by at least two cancer pathways (Fig. 6). First, EZH2-S21 phosphorylation by AKT in the SBD of EZH2 weakens SBD binding to the EZH2 TAD (9, 16). Second, although helix H3 of the EZH2 TAD does not mediate gene activation directly, phosphorylation of EZH2-Y244 in helix H3 by JAK3 may not only impair SBD binding but also disrupt the intrinsic partial folding of the EZH2 TAD, exposing its core fragment for gene activation (18). The mechanisms of the functional conversion of EZH2 by distinct kinases now appear to converge in the same structural context of the EZH2 TAD.
Notably, we observed the stoichiometric binding between the EZH2 TAD and the CH3/TAZ2 domain of the transcriptional coactivator and histone acetyltransferase p300. Indeed, the core fragment of the EZH2 TAD spanning from helix H1 to H2 was adequate for stable binding to p300. Specific mutations of the gene activation consensus motifs within the EZH2 TAD that impeded p300 binding in vitro also hampered EZH2-mediated gene activation in vivo, highlighting the specificity of our gene expression assays.
A number of accessory subunits, including AEBP2 (Adipocyte Enhancer-Binding Protein 2), JARID2 (Jumonji/AT-Rich Interaction Domain Containing 2), and PCL (PolyComb-Like) proteins, are believed to facilitate chromatin targeting of PRC2 in cells, to allow EZH2-mediated, H3K27me3-dependent gene repression (Fig. 6) (39). When artificially targeted by GAL4-DNA binding, EZH2-FL repressed a constitutively active TK promoter, whereas the EZH2 TAD construct activated an mMLP promoter; gene activation was even more robust with EZH2-TAD-Y244D, in which the gene activation activity is completely unlocked. In addition, when overexpressed and abnormally phosphorylated in cancer cells, EZH2-FL may bind to transcriptional coactivators and form non-PRC2 complexes on certain promoters, like the AR and Cyclin D1 promoters, to activate gene expression without requiring the GAL4-based artificial chromatin recruitment (9, 14–16, 18). Apparently, whether EZH2 functions as a repressor or activator is likely promoter and cell context-dependent, and EZH2 phosphorylation may promote the activator function by structurally unlocking the TAD of EZH2.
Although EZH2 contains the TAD, it appears to lack specific DNA-binding activity; how EZH2 as an activator is recruited to chromatin in cells remains unclear. Although we have been mostly focused on p300 in this study, the TAD of EZH2 may bind to other transcription factors or coactivators in different cells, similar to the TADs of classical acidic activators. In addition, the binding affinity between endogenous EZH2 and p300 in nuclear extracts is markedly reduced compared to that between the EZH2 TAD and the CH3/TAZ2 domain of p300 in vitro, implicating that additional cellular factors may regulate formation of EZH2-containing gene activation complexes in vivo. That being said, it is not impossible that proper chromatin contexts, not recapitulated in nuclear extracts, may also be necessary for assembly of these noncanonical complexes of EZH2.
Materials and Methods
Cell Culture.
Mammalian cell lines used in this study are human embryonic kidney cell line HEK293T, HeLa, and prostate cancer cell lines LNCaP and 22Rv1. HEK293T, HeLa, LNCaP, and 22Rv1 were obtained from American Type Culture Collection (ATCC). HEK293T and HeLa were grown and maintained in Dulbecco’s modified Eagle’s medium (D5796; Sigma) supplemented with 10% fetal bovine serum (FBS) (F2442; Sigma) and 1× penicillin–streptomycin (P0781; Sigma). LNCaP and 22Rv1 cells were cultured in RPMI1640 medium (R8758; Sigma) containing with 10% FBS and 1× penicillin–streptomycin.
Plasmids.
Luciferase plasmid pGL4.35 containing a 9XGAL4UAS-Adenovirus mMLP-driven luciferase gene was purchased from Promega. DNA sequence of HSV thymidine kinase promoter (HSV-TK promoter) was amplified from pGL4.54 (Promega). A 6XGAL4UAS was added N-terminal to HSV-TK promoter and was then cloned into pGL4.35 backbone by replacing the original DNA cassette 9XGAL4UAS-mMLP using Gibson Assembly Master Mix (NEB). This generated reporter vector G6-TK-luc. AR promoter-based reporter plasmid AR-luc (fragment +1.1 kb-2.5 kb) was a gift from Jindan Yu, Northwestern University, Evanston, IL (15). Plasmid pCMVβ constitutively expressing β-galactosidase was used for normalization (40).
In the GAL4UAS-mMLP–based reporter assay, a DNA fragment encoding GAL4-HA was fused to target genes using PCR amplification. The fused constructs were then cloned into pCS2+ vector by using EcoRI and XhoI sites. In the cyclin D1 promoter-driven and the AR-promoter driven reporter gene assays, constructs encoding HA-tagged target genes were cloned into pCS2+ vector by using EcoRI and XhoI sites. Detailed information of target genes is provided in SI Appendix, Table S2.
For preparation of the TADs of EZH1 and EZH2 with or without GST tag, the DNA sequence encoding either the human EZH2 TAD (residues 135 to 254) or the human EZH1 TAD (residues 136 to 267) was cloned into pGEX 4T-1 (GE Healthcare) vector by using BamH1 and XhoI sites. The same strategy was used for the preparation of the SANT1 domains of EZH1 (residues 160 to 267) and EZH2 (residues 159 to 254) for ITC assays. Genes encoding domains of human p300, including CH1/TAZ1 (residues 313 to 433), KIX (residues 564 to 658), and CH3/TAZ2 (residues 1664 to 1836), were also cloned into the pGEX-4T-1 vector by using BamH1 and XhoI sites.
For preparation of human EZH2–EED binary complex for crystallization, the truncated human EED gene (residues 78 to 441) was ligated into pGEX 4T-1 vector by using BamH1 and XhoI sites. Human EZH2 (residues 1 to 254 with residues 183 to 219 replaced by GSSG) was tagged with an N-terminal His6-SUMO tag and was cloned into pET28 vector (EMD Biosciences) by using BamH1 and EcoRI sites.
Constructs harboring mutations were generated with the site-directed mutagenesis strategy following the manufacture’s instruction (Agilent). All plasmids were verified by Sanger DNA sequencing.
Luciferase Reporter Assay.
In the GAL4UAS-mMLP–based reporter assay, HEK293T cells were plated at a density of ∼0.35 × 106 cells per well in six-well plates and allowed to grow for 20 h prior to reporter transfection. For assays with small interfering RNA (siRNA) or inhibitor treatment, cells were treated by p300 siRNA (50 nM si-p300; Sigma) or p300 inhibitor (1 μM A485; Tocris Bioscience) (41) for another 20 h prior to the reporter transfection. Reporter plasmid pGL4.35 of 250 ng was cotransfected with 500 ng pCS2+ plasmid expressing GAL4-HA tagged EZH2 WT/mutant and 100 ng pCMVβ vector using Xtrememe Gene 9 DNA (Roche). Twenty-four hours after transfection, cells were harvested, suspended, and lysed in 1× Reporter Lysis Buffer (Promega). The luciferase activity was measured using Luciferase Assay System kit (Promega) following the manufacturer’s instructions. The β-Gal activity was measured using β-Galactosidase Enzyme Assay System (Promega). The β-Gal normalized luciferase measurements were plotted in Prism7. Each experiment was performed in triplicate using cells from three different wells. Protein expression was detected by Western blot using anti-HA antibody (3724; Cell Signaling). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a loading control and detected with anti-GAPDH antibody (MA5-15738; Invitrogen). The GAL4-TK promoter-based reporter assay was performed in the same way except that pGL4.35 was replaced by the reporter vector G6-TK-luc.
In the AR promoter-based reporter assay, 22Rv1 cells were plated at a density of ∼0.5 × 106 cells per well in six-well plates and allowed to grow for 20 h prior to reporter transfection. For assays with inhibitor treatment, cells were treated by PI3K-Akt inhibitor (20 μM LY294002; Cayman) (16) for another 20 h prior to reporter transfection. Reporter plasmid AR-luc of 250 ng was cotransfected with 500 ng pCS2+ plasmid expressing HA-tagged EZH2 WT/mutant and 100 ng pCMVβ vector using Xtrememe Gene 9 DNA (Roche). Cells were harvested 24 h after transfection and assay measurements were performed the same as mentioned above.
Protein Expression and Purification.
To prepare proteins of the EZH2 TAD, the EZH1 TAD or p300 domains, the gene-harboring pGEX-4T-1 plasmid was transformed into Escherichia coli Rosetta2 (DE3) cells (EMD Biosciences). Cells were grown in Luria–Bertani (LB) medium at 37 °C until optical density at 600 nm (OD600nm) reached 0.8. The culture was then induced with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) overnight at 18 °C. Cells were harvested and resuspended in 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 2 mM dithiothreitol (DTT). After lysis by sonication and clarification by ultracentrifugation, glutathione agarose beads (Thermo Scientific) was added to the supernatant and incubated at 4 °C for 2 h. The beads were captured and washed thoroughly. The bound GST-tagged proteins were eluted with the cell lysis buffer supplemented with 20 mM glutathione. Alternatively, target proteins were released and eluted by Thrombin protease (Enzyme Research) cleavage at 4 °C overnight. Eluted proteins were concentrated and loaded to Superdex S200 SEC column (GE Healthcare) equilibrated with 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 2 mM DTT. The purified proteins were then concentrated, aliquoted, and stored at −80 °C until further use.
To prepare the EZH2–EED binary complex, two plasmids, pGEX-4T-1 harboring EED (78 to 441) and pET28 harboring His6-SUMO-tagged EZH2 (1 to 254, residues 183 to 219 replaced by GSSG) were cotransformed into E. coli Rosetta2 (DE3) competent cells (EMD Biosciences). Cells were grown in LB medium containing ampicillin (100 μg/mL), kanamycin (50 μg/mL), and chloramphenicol (25 μg/mL) at 37 °C until OD600nm reached 0.8. The culture was induced with 0.5 mM IPTG overnight at 18 °C. Cells were harvested and resuspended in 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 2 mM DTT. After lysis by sonication and clarification by ultracentrifugation, the supernatant was mixed with glutathione agarose beads (Thermo Scientific) preequilibrated in lysis buffer. The binding was performed at 4 °C for 2 h. The beads was captured and washed extensively with lysis buffer. Homemade SUMO protease (Ulp1) was added to the resin and incubated at 4 °C for at least 2 h. The resin was washed again thoroughly. The bound binary complex was eluted by Thrombin protease cleavage at 4 °C overnight. After SEC by Superdex S200 column equilibrated with 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 2 mM DTT, the peak fractions containing the binary complex were concentrated to about 30 mg/mL and stored at −80 °C.
SAXS.
SAXS data for the EZH2 TAD and the EZH2 TAD bound to a synthesized SBD peptide were collected at SIBYLS beamline 12.3.1 at the Advanced Light Source (ALS) (42). Each sample was collected at three concentrations: 1.5, 3.0, and 6.0 mg/mL in a buffer composed of 20 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, and 2 mM DTT. A series of exposures, in equal subsecond time slices, were taken. The scattering intensity I(q) was measured for q values (q = 4πsinθ/λ, where 2θ is the scattering angle) ranging from 0.01 to 0.4 Å−1. The data were integrated, scaled, and buffer-subtracted to get the standard scattering curves. Multiple curves with different concentrations and different exposure times were scaled and averaged in PRIMUS from ATSAS package (43). The initial I(0) and Rg values were analyzed by the Guinier plot analysis in PRIMUS (44). Data for the 1.5 mg/mL sample was used for the Kratky plot analysis. The normalized dimensionless Kratky plot was drawn as (qRg)2I(q)/I(0) vs. qRg.
CD.
CD spectra were measured using a J-815 Circular Dichroism Spectrometer. Proteins were first buffer-exchanged by using Superdex S200 SEC column (GE Healthcare) equilibrated with 10 mM NaHPO4-Na2HPO4, pH 7.0, and 40 mM NaF. The measurements were performed at room temperature using a quartz glass cell at a protein concentration of 0.1 to 0.5 mg/mL. The spectra were measured at a wavelength range of 190 to 240 nm and recorded five scans each. Data analysis and secondary structure estimation were performed with the CONTINLL algorithm implemented in the DichroWeb server (45–48).
Crystallization and Structure Determination of the Human EZH2–EED Binary Complex.
Crystal screening was performed by the sitting-drop vapor diffusion method. The crystals were refined by the hanging-drop vapor diffusion method, by mixing 1 μL of the protein solution at 10∼20 mg/mL and 1 μL of the reservoir solution containing 0.8 to 1 M NaK tartrate, 0.2 M Li2SO4, and 0.1 M Tris⋅HCl, pH 7.0, and equilibrating over 500 μL of the reservoir solution. Crystals were harvested within 1 wk, cryoprotected by mother liquor with 20% glycerol, and flash-frozen in liquid nitrogen. Diffraction data used for structure determination was collected at Advanced Photo Source beamline 19ID. The crystal belongs to space group P2221 with cell dimensions of a = 181.6 Å, b = 114.7 Å, c = 131.5 Å, α = β = γ = 90° and contains three binary complexes per asymmetry unit. The actual biological complex contains six binary complexes from two neighboring asymmetric units. Datasets were indexed, integrated, and scaled using HKL2000 (49) and further processed with CCP4 suite programs (50). The structure was solved by molecular replacement using the coordinates of an EED structure (Protein Data Bank ID code 2QXV) as a searching model (51). Molecular replacement was performed using Phaser (52). Further model building and iterative refinement were carried out using Coot (53), PHENIX (54), Refmac (55), and autoBUSTER (56). The final model was obtained by using TLS (translation, libration, and screw-motion) refinement with autoBUSTER (56). Molecular graphics were generated using PyMOL (57). Statistics for data collection and structure refinement are summarized in SI Appendix, Table S1.
NMR Spectroscopy.
The 15N-labeled EZH2 TAD and EZH1 TAD proteins were prepared similarly as the unlabeled proteins, except that the E. coli BL21 (DE3) cells were grown in M9 medium containing glucose (0.2% M/V) and 15N ammonium chloride (1 g/L) (Cambridge Isotope Laboratories). Before NMR data acquisition, the purified 15N-labeled proteins were loaded onto Superdex S200 column equilibrated with 50 mM NaH2PO4-Na2HPO4 (pH 6.5), 50 mM NaCl, and 1 mM DTT and concentrated to a concentration of ∼0.5 mM. The 15N-labeled EZH2 TAD protein (∼0.05 mM) was used for NMR data acquisition in the absence or presence the corresponding synthesized SBD peptide (∼0.15 mM). All NMR experiments were performed at 25 °C on an Agilent DD2 800-MHz spectrometer equipped with a cold probe. 1H-15N HSQC spectra were acquired with 50 μM samples of uniformly 15N-labeled EZH2 TAD in the absence or presence of 60 μM SBD or SBD-S21p peptide. NMRPipe (58) was used for NMR data processing and NMRViewJ (59) for data display and analysis.
ITC.
The purified SANT1 domains of EZH1 and EZH2 were dialyzed into a buffer containing 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 2 mM DTT. Synthetic peptides were resuspended in the same buffer. Concentrations for proteins were measured using absorbance at 280 nm. Prior to titration, the sample solutions were centrifuged at 18,000 × g at 4 °C for 10 min to remove any debris and air bubbles. The calorimetric titrations were performed on a MicroCal iTC200 instrument (Microcal Inc.) at 20 °C. The reactants (50 µM protein) were placed in the 300-µL sample chamber and the peptides (500 µM) were added using the syringe with 20 successive additions of 2 µL (with an initial injection of 0.5 µL), spaced 120 s apart. Data were analyzed using programs NITPIC (60) and SEDPHAT (61), and the dissociation constant (Kd) and binding stoichiometry (N) were calculated by curve fitting.
Protein Interaction Pull-Down Assay.
HeLa nuclear extracts were initially used to analyze the interaction of GST-EZH2 TAD or GST-EZH1 TAD with p300. The nuclear extracts were prepared as described previously (62) and dialyzed into binding buffer containing 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 2 mM DTT, 10% glycerol, and 0.1% Nonidet P-40 overnight at 4 °C before use. In the binding assay, the purified GST-tagged EZH2 TAD or EZH1 TAD of ∼50 µg was first bound to ∼20 µL glutathione agarose beads equilibrated with binding buffer for 1 h at 4 °C. After several washes, the dialyzed HeLa nuclear extracts of ∼10 mg were applied per binding assay and incubated for 4 h at 4 °C. The beads were then washed thoroughly with binding buffer and boiled at 95 °C for 5 min in 1× sample loading dye. The bound p300 was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE), transferred to poly(vinylidene difluoride) (PVDF) membrane, and detected by Western blot with anti-p300 primary antibody (ab14984; Abcam).
The whole-cell lysates of HEK293T cells overexpressing myc-p300 protein were used to analyze the competition the SBD of EZH2 on p300 binding to GST-EZH2 TAD. Briefly, HEK293T cells were transfected using polyethylenimine by the pCMV-myc-p300 vector, a gift from Laura Banaszynski, University of Texas Southwestern Medical Center, Dallas, TX. Cells were harvested after 48 h and lysed in a buffer containing 50 mM Tris⋅HCl, pH 8.0, 250 mM NaCl, 2 mM DTT, and 1% Triton X-100 in the presence of protease inhibitors for 30 min at 4 °C. Similar to the binding assay with HeLa nuclear extracts, the purified GST-tagged EZH2 TAD of ∼50 µg was bound to 20 µL glutathione agarose beads equilibrated with the same binding buffer for 1 h at 4 °C in the presence of different concentrations of a synthetic SBD peptide. After several washes, ∼10 mg of cell lysates containing overexpressed myc-p300 was added into the system and incubated for 4 h at 4 °C. The beads then washed thoroughly and boiled at 95 °C for 5 min in 1× sample loading dye. The bound p300 was separated by SDS/PAGE, transferred to PVDF membrane, and detected by Western blot with an anti-p300 primary antibody (ab14984; Abcam).
Co-IP Assay.
Nuclear extracts of 22Rv1 cells were used in the Co-IP assay to analyze the binding of endogenous EZH2 with endogenous p300. Briefly, 100 µg of anti-EZH2 (5246; Cell Signaling) or anti-p300 (ab14984; Abcam) antibody was coupled to cyanogen bromide-activated-Sepharose 4B beads (C9142; Sigma) and used for the Co-IP assay. IgG Sepharose 6 beads (GE Healthcare) were used as a negative control. Nuclear extracts of 22Rv1 cells were prepared as described previously (62). After dialysis against a buffer containing 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 2 mM DTT, 10% glycerol, and 0.1% Nonidet P-40 overnight at 4 °C, nuclear extracts of ∼50 mg were used and incubated with antibody coupled beads for 4 h at 4 °C. The antibody beads were washed thoroughly. The bound fractions were eluted using 100 mM glycine, pH 2.5, and concentrated using standard trichloroacetic acid precipitation. The pelleted samples were resuspended and boiled at 95 °C for 5 min in 1× sample loading dye. The bound proteins were separated by SDS/PAGE, transferred to PVDF membrane, and detected by Western blot with either anti-EZH2 antibody (5246; Cell Signaling) or anti-p300 primary antibody (ab14984; Abcam).
Gene Expression Analysis by RT-PCR.
LNCaP cells were used for endogenous gene expression analysis. Cells were seeded at a density of 0.3 × 106 cells per well in six-well plates. Lipofectamine RNAimax (Life Technology) was used for siRNA duplexes transfection following the manufacturer’s instructions. Briefly, 100 pmol siRNA duplex and 5 μL Lipofectamine RNAimax were diluted separately in Opti-MEM, mixed, and used for each transfection. After 48 h, cells were harvested in 1 mL TRIzol Reagent (Invitrogen). Total RNA was then isolated following the standard protocol. Reverse transcription was performed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). The synthesized complementary DNA was subjected to real-time qPCR (SYBR Green PCR Master Mix; Applied Biosystems) using the gene-specific primers as reported previously (15). Relative messenger RNA levels were shown from three independent replicates using GAPDH as the internal control. The control siRNA MISSION siRNA Universal Negative Control #1 (si-Ctrl) and siRNA duplexes targeting EZH2 (si-EZH2_CDS), SUZ12 (si-SUZ12), and p300 (si-p300) were purchased from Sigma. The sequences of each siRNA and the PCR primers are listed in SI Appendix, Table S3.
Data Availability.
Materials are available upon request. Coordinates and structure factors of the crystal structure reported in this study have been deposited in the Protein Data Bank under the accession number 6U4Y.
Supplementary Material
Acknowledgments
We thank Dr. Jindan Yu from Northwestern University for providing the AR promoter-containing reporter gene plasmids and for the discussion of gene expression analysis in prostate cancer cells. We thank Dr. Laura Banaszynski from University of Texas (UT) Southwestern Medical Center for providing the plasmid for transient expression of p300. This research was supported by Welch Foundation research grant I-1790 to X.L. and I-1304 to J.R. and Cancer Prevention and Research Institute of Texas research grant R1119, Rita Allen Foundation research grant, UT Southwestern Medical Center Endowed Scholar fund, and NIH grants GM114576, GM121662, and GM136308 to X.L. L.J. was supported by American Heart Association postdoctoral fellowship 16POST30700004. X.L. is a W. W. Caruth, Jr. Scholar in Biomedical Research. This research also received support from the Cecil H. and Ida Green Center Training Program in Reproductive Biology Sciences Research. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US DOE under contract DE-AC02-05CH11231. Use of the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, is supported by the US DOE, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH, National Institute of General Medical Sciences (NIGMS) (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. SAXS data were collected at SIBYLS beamline 12.3.1 at the ALS. SAXS data collection at SIBYLS is funded through the DOE Biological and Environmental Research Integrated Diffraction Analysis Technologies program and NIGMS grant P30 GM124169-01, ALS-ENABLE.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors of the crystal structure reported in this study have been deposited in the Protein Data Bank, http://www.wwpdb.org (PDB ID code 6U4Y).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914866117/-/DCSupplemental.
References
- 1.Cao R., Zhang Y., SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15, 57–67 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Jiao L., Liu X., Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Justin N., et al. , Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 7, 11316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooun A., et al. , Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun. 7, 11384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poepsel S., Kasinath V., Nogales E., Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 25, 154–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long Y., et al. , Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. eLife 6, e31558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q., et al. , RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat. Struct. Mol. Biol. 26, 237–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K. H., Roberts C. W., Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu K., et al. , EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S. T., et al. , Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell 43, 798–810 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez M. E., et al. , EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 3098–3103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan J., et al. , EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 121, 4512–4520 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Jung H. Y., et al. , PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Mol. Cell 52, 193–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi B., et al. , Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 27, 5105–5119 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J., et al. , Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 25, 2808–2820.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha T. L., et al. , Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Kim E., et al. , Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23, 839–852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J., et al. , EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood 128, 948–958 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Vo L. T., et al. , Regulation of embryonic haematopoietic multipotency by EZH1. Nature 553, 506–510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J., et al. , Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell 57, 304–316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mousavi K., Zare H., Wang A. H., Sartorelli V., Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol. Cell 45, 255–262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J., et al. , TRIM28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene 36, 2991–3001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen K. H., et al. , A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Margueron R., et al. , Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng K., Radivojac P., Vucetic S., Dunker A. K., Obradovic Z., Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 7, 208 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krois A. S., Ferreon J. C., Martinez-Yamout M. A., Dyson H. J., Wright P. E., Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein. Proc. Natl. Acad. Sci. U.S.A. 113, E1853–E1862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uesugi M., Nyanguile O., Lu H., Levine A. J., Verdine G. L., Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 277, 1310–1313 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Sullivan S. M., et al. , Mutational analysis of a transcriptional activation region of the VP16 protein of herpes simplex virus. Nucleic Acids Res. 26, 4487–4496 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teufel D. P., Freund S. M., Bycroft M., Fersht A. R., Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl. Acad. Sci. U.S.A. 104, 7009–7014 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L., Murat P., Matak-Vinkovic D., Murrell A., Balasubramanian S., Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry 52, 9519–9527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dyson H. J., Wright P. E., Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Yao J., Dyson H. J., Wright P. E., Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins. FEBS Lett. 419, 285–289 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Weaver T. M., et al. , The EZH2 SANT1 domain is a histone reader providing sensitivity to the modification state of the H4 tail. Sci. Rep. 9, 987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenfield N. J., Circular dichroism analysis for protein-protein interactions. Methods Mol. Biol. 261, 55–78 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Kikhney A. G., Svergun D. I., A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 589, 2570–2577 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Avantaggiati M. L., et al. , Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89, 1175–1184 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Gu W., Shi X. L., Roeder R. G., Synergistic activation of transcription by CBP and p53. Nature 387, 819–823 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Schuettengruber B., Bourbon H. M., Di Croce L., Cavalli G., Genome regulation by polycomb and Trithorax: 70 years and counting. Cell 171, 34–57 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Kim M. Y., Woo E. M., Chong Y. T., Homenko D. R., Kraus W. L., Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol. Endocrinol. 20, 1479–1493 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lasko L. M., et al. , Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Classen S., et al. , Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 46, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franke D., et al. , ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 50, 1212–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003). [Google Scholar]
- 45.Whitmore L., Wallace B. A., DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Provencher S. W., Glöckner J., Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 (1981). [DOI] [PubMed] [Google Scholar]
- 47.van Stokkum I. H., Spoelder H. J., Bloemendal M., van Grondelle R., Groen F. C., Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 191, 110–118 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Sreerama N., Woody R. W., Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han Z., et al. , Structural basis of EZH2 recognition by EED. Structure 15, 1306–1315 (2007). [DOI] [PubMed] [Google Scholar]
- 52.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Bricogne G., et al. , BUSTER Version 2.10.1 (Global Phasing Ltd., Cambridge, UK, 2011).
- 57.Schrodinger, LLC , The PyMOL Molecular Graphics System, Version 1.3r1 (Schrodinger, LLC, 2010).
- 58.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Johnson B. A., Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Keller S., et al. , High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Houtman J. C., et al. , Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: Application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carey M. F., Peterson C. L., Smale S. T.. Dignam and Roeder nuclear extract preparation. Cold Spring Harb. Protoc. 2009, pdb.prot5330 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials are available upon request. Coordinates and structure factors of the crystal structure reported in this study have been deposited in the Protein Data Bank under the accession number 6U4Y.