SUMMARY
Mitogen-activated protein kinase kinase kinase kinases (MAP4Ks) constitute a mammalian STE20-like serine/threonine kinase subfamily. Recent studies provide substantial evidence for MAP4K family kinases in the Hippo pathway regulation, suggesting a broad role of MAP4Ks in human physiology and diseases. However, a comprehensive analysis of the regulators and effectors for this key kinase family has not been fully achieved. Using a proteomic approach, we define the protein-protein interaction network for human MAP4K family kinases and reveal diverse cellular signaling events involving this important kinase family. Through it, we identify a STRIPAK complex component, STRN4, as a generic binding partner for MAP4Ks and a key regulator of the Hippo pathway in endometrial cancer development. Taken together, the results of our study not only generate a rich resource for further characterizing human MAP4K family kinases in numerous biological processes but also dissect the STRIPAK-mediated regulation of MAP4Ks in the Hippo pathway.
Graphical Abstract
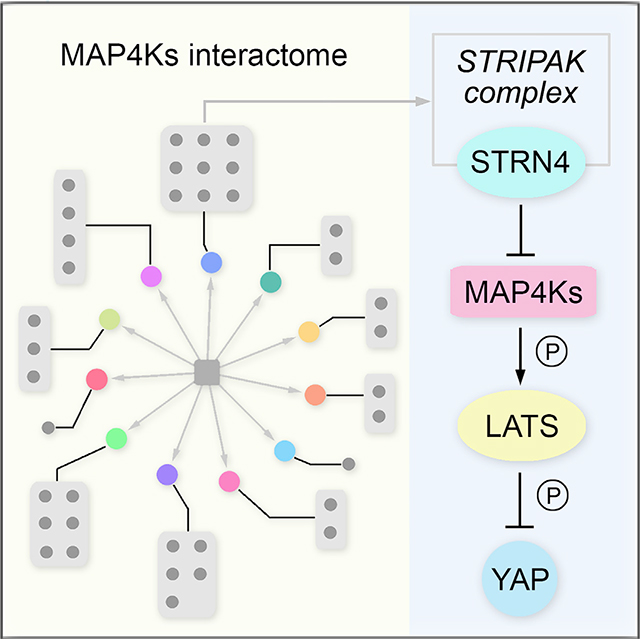
In Brief
Seo et al. conduct a proteomic analysis to define the protein-protein interaction network for the human MAP4K family kinases. Through functional validation, they identify a STRIPAK complex component, STRN4, as a common binding partner for MAP4Ks and a key regulator of the Hippo pathway in endometrial cancer development.
INTRODUCTION
Mitogen-activated protein kinase kinase kinase kinases (MAP4Ks) belong to the mammalian STE20-like serine/threonine kinase family, which are known to play critical roles in diverse cellular functions (Dan et al., 2001; Kyriakis and Avruch, 2012). This kinase family contains seven members, namely, hematopoietic progenitor kinase 1 (HPK1/MAP4K1), germinal center kinase (GCK/MAP4K2), germinal center kinase-like kinase (GLK/MAP4K3), HPK/GCK-like kinase (HGK/MAP4K4), the kinase homologous to SPS1/STE20 (KHS/MAP4K5), misshapen-like kinase 1 (MINK1/MAP4K6), and TRAF2 and NCK interacting kinase (TNIK/MAP4K7), which share a similar protein structure with an N-terminal kinase domain, proline-rich motifs, and a C-terminal citron-homology domain (CNH) (Chuang et al., 2016).
Once activated, MAP4Ks exert numerous cellular effects on cell proliferation, apoptosis, cell migration, and autophagy, mostly through the control of c-Jun N-terminal kinase (JNK) and MAPK kinase cascades (Kyriakis and Avruch, 2012; Miller et al., 2019). In addition, MAP4Ks play important roles in immunity and inflammation-related signaling events, such as the nuclear factor κB (NF-κB) pathway, tumor necrosis factor alpha (TNF-α) pathway, T cell receptor signaling, and B cell receptor signaling (Chuang and Tan, 2019; Chuang et al., 2016). Notably, recent findings in both Drosophila and mammals revealed MAP4Ks as key components in the Hippo tumor suppressor pathway (Li et al., 2014, 2015, 2018; Meng et al., 2015; Zheng et al., 2015), which largely enriched our understanding of this key kinase family in tissue homeostasis and cancer.
The Hippo pathway, which was identified and elucidated in Drosophila, has been established as a conserved signaling pathway in organ/organism size control by restricting cell proliferation and promoting apoptosis (Halder and Johnson, 2011; Piccolo et al., 2014; Yu et al., 2015; Zheng and Pan, 2019). In the mammalian Hippo pathway, MAP4K1/2/3/4/6/7 function in parallel to another two STE20-like kinases, MST1/2, to phosphorylate and activate two ACG serine/threonine kinases, LATS1/2. Upon phosphorylation, LATS1/2, together with their adaptor proteins MOB1A/B, in turn phosphorylate the downstream effectors YAP and TAZ. Phosphorylated YAP/TAZ can be recognized by 14-3-3 proteins, retained in the cytoplasm, and eventually degraded in proteasome by a β-TRCP E3 ligase complex. When the Hippo pathway is inhibited, un-phosphorylated YAP/TAZ can be released into the nucleus, where they form a complex with TEAD1–4 transcription factors and regulate the transcription of genes involved in cell proliferation and survival. In Drosophila, MAP4K1/2/3/5 orthologous protein Happyhour and MAP4K4/6/7 orthologous protein Misshapen are also able to directly phosphorylate Warts (LATS1/2 in mammals) independently of Hippo (MST1/2 in mammals) (Li et al., 2014, 2015, 2018; Meng et al., 2015; Zheng et al., 2015), highlighting the evolutionarily conserved roles of MAP4Ks in the Hippo pathway.
Although MAP4K family members have been implicated to act in multiple physiological and pathological processes related to human health, including glucose metabolism (Bouzakri and Zierath, 2007), lipogenesis (Danai et al., 2013), protein synthesis (Resnik-Docampo and de Celis, 2011), inflammation (Chuang et al., 2016), cell adhesion (Yue et al., 2014), and tumor metastasis (Loftus et al., 2013), the underlying mechanisms are still largely unknown. Moreover, how MAP4Ks are regulated and whether they have additional important cellular functions remain to be elucidated. Therefore, a comprehensive analysis of the MAP4K-centered protein interaction landscape will not only provide a chance to uncover regulators and targets for this key kinase family but also help to identify functional players for the known MAP4K-associated cellular events, such as Hippo signaling.
In this study, we took advantage of our previously established tandem affinity purification coupled with mass spectrometry (TAP-MS) platform (Li et al., 2016, 2017; Wang et al., 2014) to define the human MAP4K-based protein-protein interaction (PPI) network. Through it, we connected the MAP4K family kinases with various biological processes, identified potentially universal regulators for this key kinase family, and discovered interacting proteins for MAP4K family members that may exert their cellular functions. In addition, we applied our MAP4K-based PPI network to their associated signaling events and uncovered STRN4 (also named zinedin) as an important regulator of the Hippo pathway by modulating the MAP4K-LATS1/2 kinase cascade. STRN4 is a component of the striatin-interacting phosphatase and kinase (STRIPAK) complex that has been shown to function upstream of the Hippo pathway in both Drosophila and mammals (Bae et al., 2017; Chen et al., 2019; Shi et al., 2016; Zheng et al., 2017). Here, our study demonstrated an adaptor role of STRN4, which recruited STRIPAK to inhibit MAP4Ks and subsequently activated YAP/TAZ. Taken together, our proteomic analysis of the MAP4K-associated PPI network provides a rich resource for the further exploration of this key kinase family in various signaling events and biological processes.
RESULTS
Mapping the MAP4K PPI Network
To achieve a comprehensive understanding of the MAP4K PPI network, we established HEK293T cells stably expressing each MAP4K family member fused with SFB triple tags (S protein tag-FLAG tag-SBP tag) through lentiviral infection and puromycin selection (Figures 1A and 1B). These MAP4Ks stable cells were then subjected to two independent TAP experiments. The associated proteins within each isolated protein complex were further examined using MS. A complete list of the peptides and proteins identified in this study can be found in Tables S1 and S2, respectively. We also created HEK293A cells stably expressing SFB-tagged MAP4K kinases to determine their sub-cellular localization (Figures 1B and 1C), allowing us to further validate their binding partners with known cellular localization.
Figure 1. Proteomic Analysis of MAP4K Protein Interaction Network.
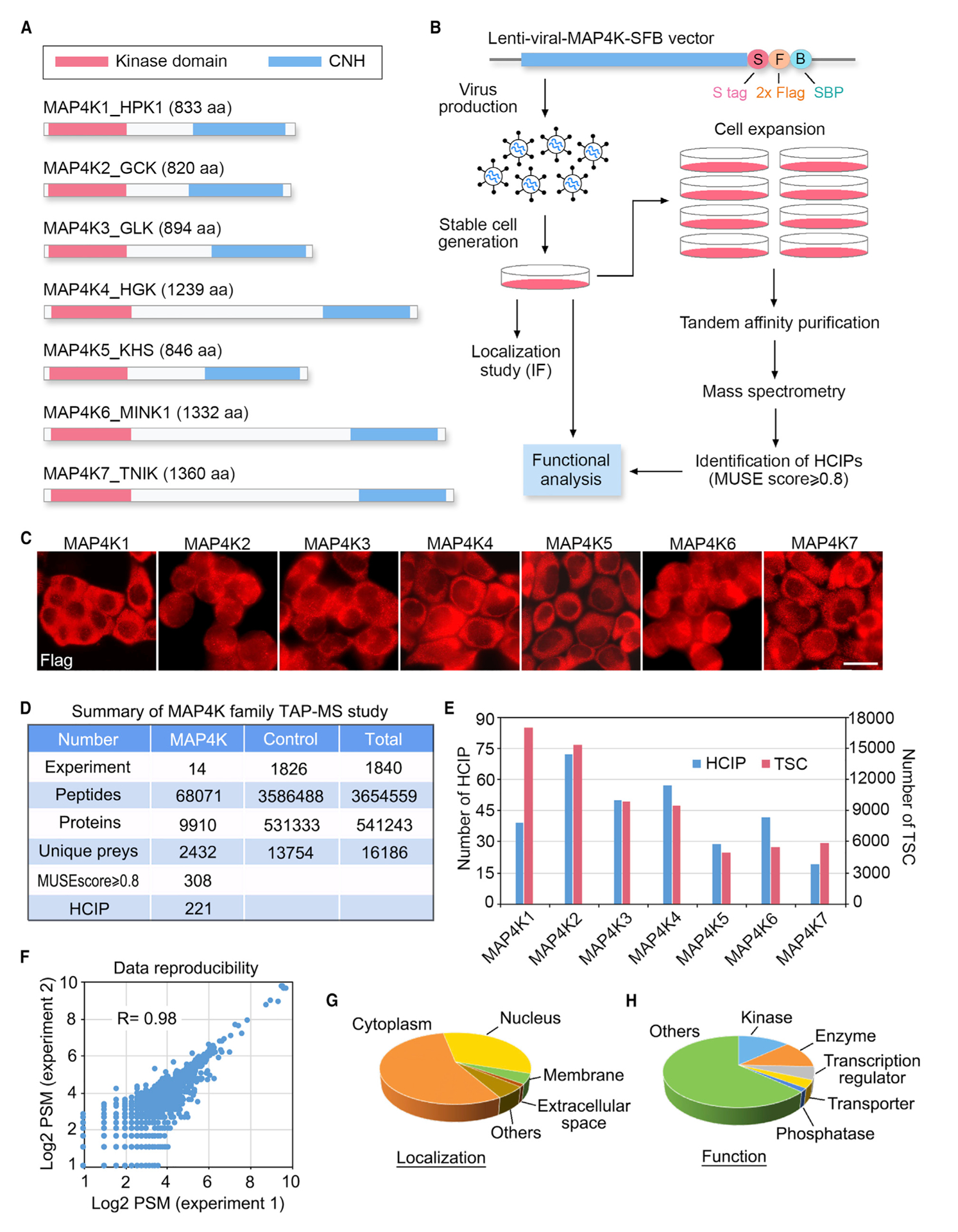
(A) Schematic illustration of the protein domains for the MAP4K family kinases.
(B) Schematic illustration of the major steps used in TAP-MS analysis of human MAP4K-associated protein complexes. MAP4Ks were constructed into a C-terminal SFB-tag-fused lentiviral vector. HEK293T cells stably expressing each bait protein were generated by lentiviral infection and puromycin selection. The purified protein complexes were identified by MS analysis, and final interacting proteins were generated using the MUSE statistical model.
(C) Localization of MAP4Ks in the HEK293A cells stably expressing each MAP4K family member. Immunofluorescent staining was performed by using FLAG antibody. Scale bar, 20 μm.
(D) Summary of the MAP4K TAP-MS analysis. Experiment information and total numbers of peptides and proteins identified in the MS analysis are shown. A MUSE score of ≥0.8 was used as the cutoff to identify HCIPs.
(E) The total spectral counts (TSCs) and corresponding numbers of HCIPs for MAP4Ks are shown.
(F) Data reproducibility between two biological TAP-MS experiments is evaluated using the number of peptide spectrum matches (PSMs).
(G and H) GO annotations of the identified MAP4Ks-HCIPs. Cellular localization (G) and cellular functions (H) of the MAP4Ks-HCIPs are shown. See also Tables S1, S2, S3, and S4.
To refine the MAP4K proteomic data, we used the MUSE algorithm (Li et al., 2016) to assign quality scores to each identified PPI. To achieve this, a group of 1,826 unrelated TAP-MS experiments (1,806 experiments using stably overexpressed SFB-tagged proteins and 20 experiments using empty vector) performed under the similar experimental conditions were included for the comparative analysis (Figures 1B and 1D). We considered any interaction with a MUSE score of least 0.8 and raw spectra counts greater than 1 as a high confidence interacting protein (HCIP) (Figures 1D and 1E). Through it, a total of 221 HCIPs among the 2,432 preys were identified for the MAP4K kinase family (Figure 1D; Tables S3). Both the HCIP number and total spectral count (TSC) number for each MAP4K family member were summarized in Figure 1E. We also confirmed our data reproducibility by comparing the two biological TAP-MS repeats for MAP4Ks (R = 0.98) (Figure 1F), underscoring the reliability of our dataset as generated here.
Gene Ontology (GO) analysis of these identified MAP4K HCIPs suggested that they were widely distributed in the cells with different sub-cellular localization (Figure 1G; Table S3) and were involved in diverse cellular functions (Figure 1H; Table S4). Because the stable cells collected for the TAP-MS analysis were grown under normal culture conditions, the identified HCIPs for each MAP4K family member should be considered as the basal-state interactome of the human MAP4K kinase family.
Overview of the MAP4K Family Protein Interaction Landscape
Given the structural similarity and redundant functions for MAP4K family kinases, first, we compared their associated HCIPs through the prey-oriented hierarchical clustering analysis. As shown in Figure 2, although many HCIPs were identified only for individual MAP4K, some common subsets were revealed between different MAP4K family members. For example, MAP4K1 and MAP4K5 both interacted with several adaptor proteins involved in signaling transduction, including CRK (cluster 1), CRKL (cluster 2), NCK2 (cluster 2), and GRB2 (cluster 2) (Figure 2), which were consistent with the previous reports (Huttlin et al., 2017; Ling et al., 1999; Shi et al., 2000); MAP4K2 and MAP4K3 both interacted with ribosomal proteins MRPL11 and RPL36 and two nuclear proteins LIN28B and SNRPB2 (cluster 3) (Figure 2), suggesting their potentially redundant roles in the related biological processes; MAP4K4/6/7 shared several HCIPs that belonged to the STRIPAK complex (clusters 4 and 5), such as STRN4, FGFR1OP2, STRIP1, STRN3, CTTNBP2NL, STRN, and MOB4, (Figure 2), which is consistent with the previous studies (Hein et al., 2015; Huttlin et al., 2017).
Figure 2. Hierarchical Clustering Analysis of the MAP4K-HCIPs.
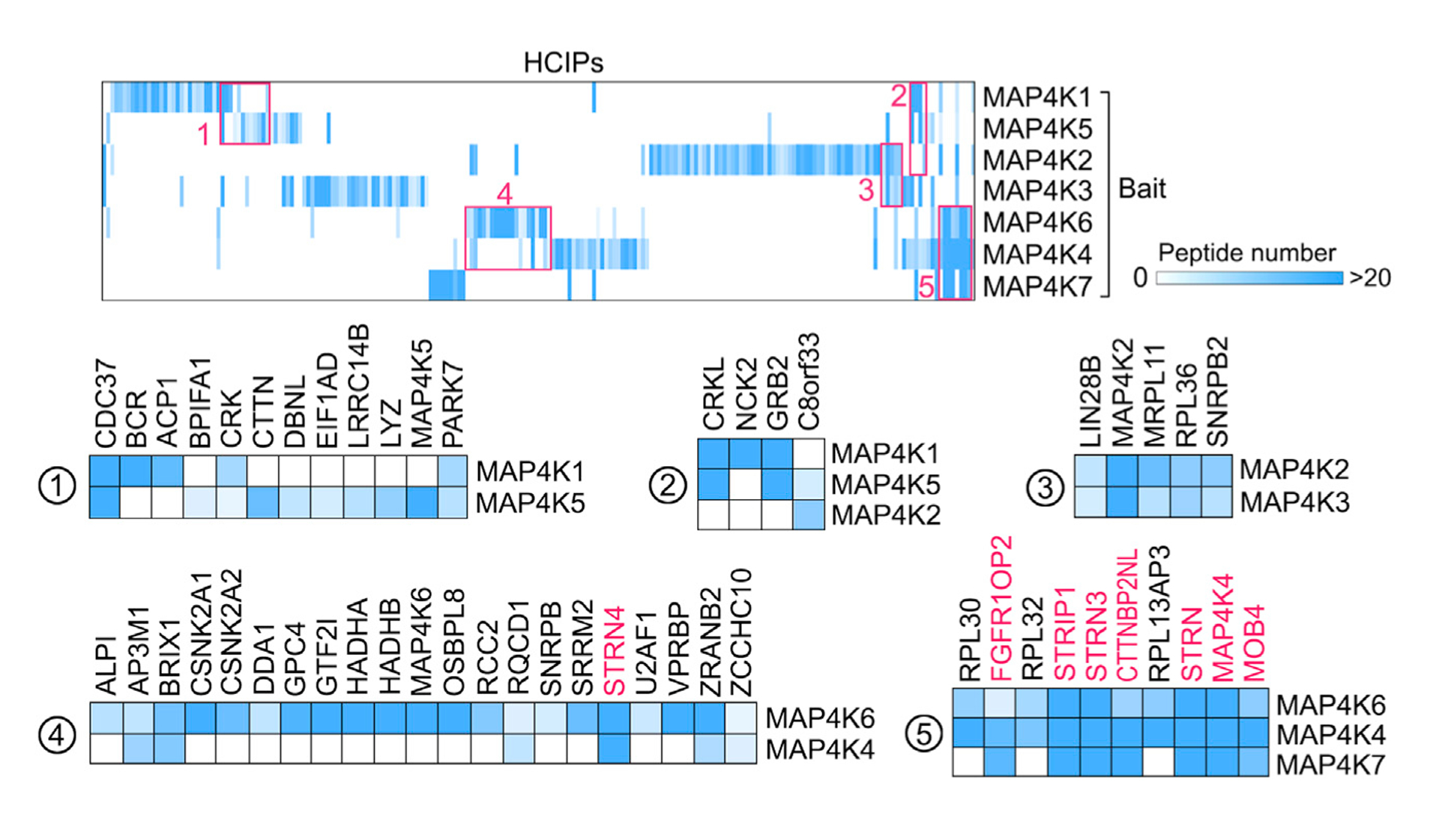
A heatmap was generated from hierarchical clustering of 221 HCIPs for MAP4K family kinases. Five prominent HCIP clusters were manually selected and enlarged below. The color of squares in the heatmap indicates the identified HCIP peptide number for each MAP4K protein. The shared STRIPAK complex components are shown in red. See also Table S3.
Interestingly, our clustering analysis also revealed a frequent homo/hetero-dimer formation among the MAP4K family members, such as MAP4K2/MAP4K2 (cluster 3), MAP4K2/MAP4K3 (cluster 3), MAP4K6/MAP4K6 (cluster 4), MAP4K4/MAP4K6 (cluster 5), MAP4K4/MAP4K4 (cluster 5), and MAP4K4/MAP4K7 (cluster 5). Actually, it has been evident that some STE20-like kinases (e.g., MST1, MST2, and MAP4K3) can exert auto-phosphorylation through dimerization (Glantschnig et al., 2002; Jin et al., 2012; Yan et al., 2010). Our findings further indicated that such a regulatory or activating mechanism could widely exist among MAP4K family members.
Given the relatively individual HCIP subsets identified for each MAP4K family member (Figure 2), next, we organized the MAP4K PPI network individually and functionally by clustering the HCIPs based on their biological functions (Figure 3). Consistent with our GO analysis (Figure 1H), MAP4K HCIPs were functionally distributed in various biological processes but still showed some enrichments, such as transcription regulation, ribosomal biogenesis, signal transduction, protein folding (chaperone proteins), cell shape (cell junction/cytoskeleton regulation), phosphorylation (kinases and phosphatases), metabolism, protein stability (E3 ligases and de-ubiquitin enzymes), cell cycle, and vesicle transport (Figure 3). Such a comparative interaction network organization highlighted a group of individual HCIPs for each MAP4K family member, implicating their particular roles in the related biological processes. For example, MAP4K2 specifically interacted with GABARAP/L1/L2 proteins (Figures 3, 4A, and 4B), suggesting its potential role in autophagy (Schaaf et al., 2016); MAP4K3/5 may function in endocytosis by forming a complex with SH3GL proteins (Figures 3, 4A, and 4C; Chan Wah Hak et al., 2018); and MAP4K5 could play a role in the CTTN-mediated lamellipodia formation and cell migration (Figure 3, 4A, and 4D; He et al., 2015). Moreover, this function-based interaction network also pointed to several cellular events shared by different MAP4K kinases, such as ribosome biogenesis (MAP4K1/2/3/4/5/6), adaptor-mediated signaling transduction (MAP4K1/3/4/5/7), AP3-coated vesicle transport (MAP4K2/4/6), and STRIPAK-complex-dependent protein de-phosphorylation (MAP4K4/6/7). Collectively, these results show that MAP4K family members could share common upstream regulators and display both redundant and specific functions within different cellular events.
Figure 3. Integrated Interaction Map of the Human MAP4K Protein Interaction Network.
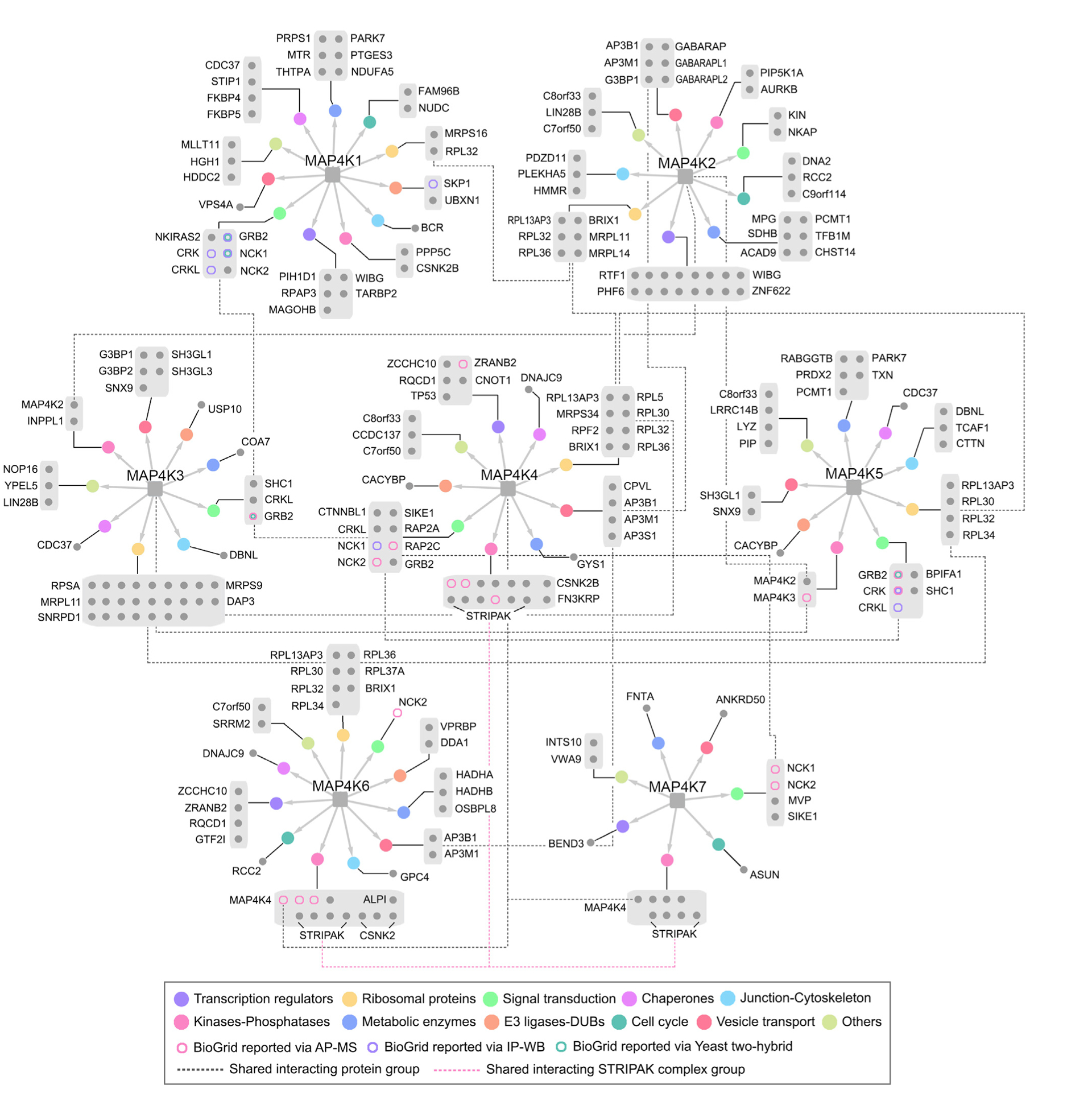
As for each MAP4K family member, HCIPs are grouped based on their cellular functions, as indicated by GO analysis and literature search. The different circle colors indicate different cellular functions. The shared HCIP groups are connected by dot-lines. The identified HCIPs are compared to the known MAP4K-binding proteins reported in the BioGrid database and are indicated in different colors. See also Tables S3 and S4.
Figure 4. Validation of MAP4K Protein Interaction Network.
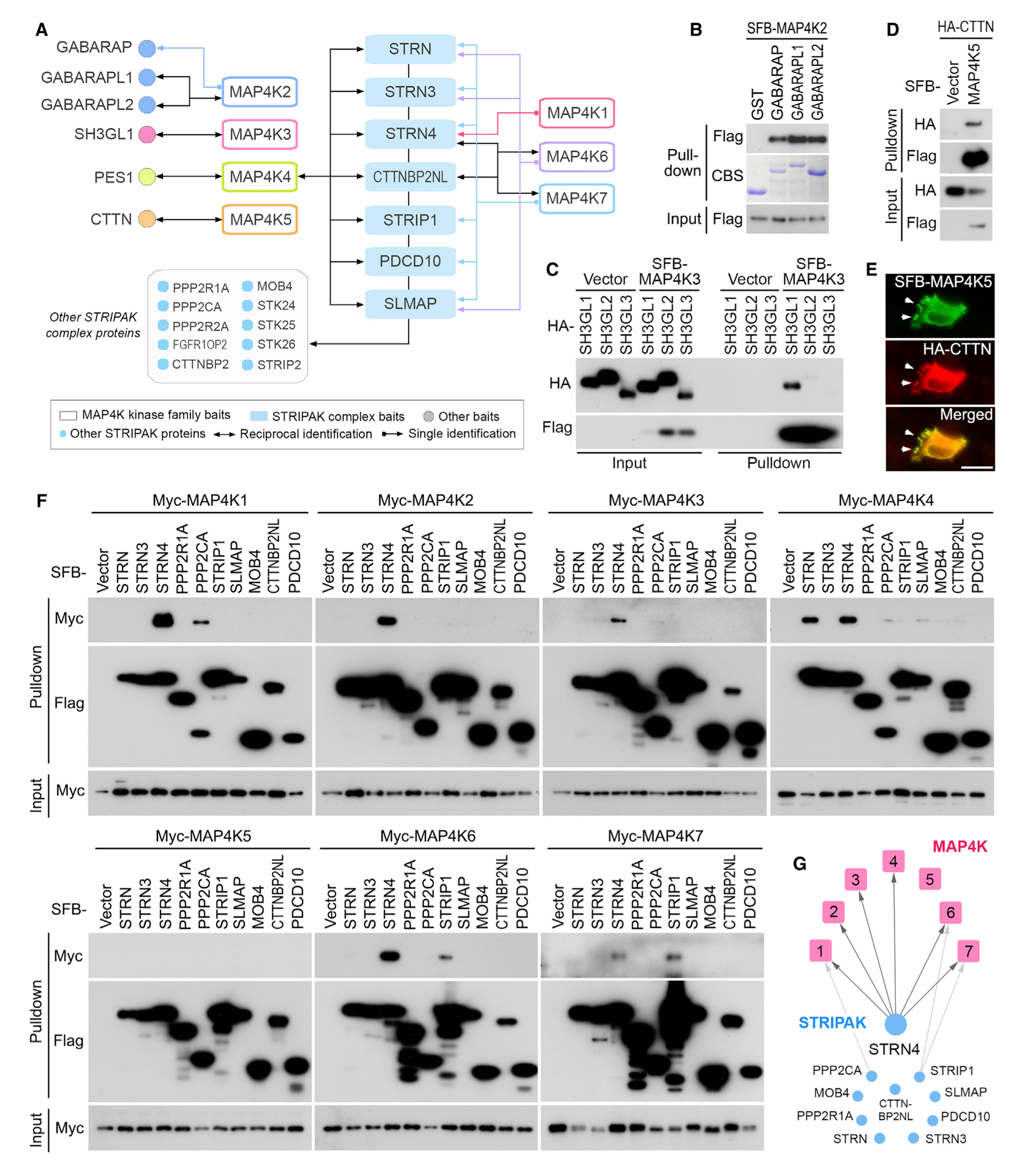
(A) A summary of reciprocal TAP-MS analyses for the selected MAP4K-HCIPs.
(B) Validation of the interaction between MAP4K2 and GABARAP family proteins. The bacterially purified GST-GABARAP family proteins were subjected to the pull-down assay. CBS, Coomassie blue staining.
(C) Validation of the interaction between MAP4K3 and SH3GL1. HEK293T cells were transfected with the indicated constructs and subjected to the pull-down assay.
(D and E) Validation of the interaction between MAP4K5 and CTTN. HEK293T cells were transfected with the indicated constructs and subjected to the pull-down assay (D). The co-localization between MAP4K5 and CTTN in lamellipodia was indicated by arrows (E). Scale bar, 20 μm.
(F and G) Validation of the interaction between MAP4K family kinases and the STRIPAK complex components. HEK293T cells were transfected with the indicated constructs and subjected to the pull-down assay (F). The pull-down experiment results were summarized (G).
To validate the MAP4K PPI network, we performed reciprocal TAP-MS experiments for 13 selected HCIPs, including the STRIPAK complex components (STRN, STRN3, STRN4, CTTNBP2NL, STRIP1, PDCD10, and SLMAP) and several preys identified for each MAP4K family member (GABARAP, GABAR-APL1, GABARAPL2, SH3GL1, PES1, and CTTN) (Figure 4A). As for the tested STRIPAK complex components, they all reciprocally identified MAP4K4 in their TAP-MS experiments, whereas only STRN4 and CTTNBP2NL reciprocally uncovered MAP4K6 and MAP4K7 (Figure 4A). As for the six TAP-MS experiments with the above selected HCIPs as baits, they all identified their corresponding MAP4Ks except for GABARAP (Figure 4A). Moreover, we experimentally validated the interaction for these protein complexes. As shown in Figure 4B, all the bacterially purified GABARAP family proteins can pull down SFB-tagged MAP4K2. Moreover, SH3GL1 dominantly associated with MAP4K3, as compared with SH3GL2 and SH3GL3 (Figure 4C). Consistently, MAP4K5 and CTTN formed as a protein complex (Figure 4D) and were co-localized in the cell lamellipodia (Figure 4E). Taken together, these validation studies not only confirmed our current MAP4K PPI network but also pinpointed several MAP4K-associated protein complexes whose functional significance deserves further investigation.
STRIPAK Complex Component STRN4 Is a Shared Binding Partner for MAP4Ks
Because our MAP4Ks PPI network strikingly pointed to a connection between MAP4Ks and the STRIPAK complex (Figures 2, 3, and 4A), we further examined the interaction between MAP4Ks and the STRIPAK complex components. Interestingly, only few STRIPAK complex components interacted with MAP4Ks, where STRN4 is the only shared STRIPAK component among MAP4K family members (Figures 4F and 4G). As for MAP4K family kinases, only MAP4K5 failed to bind any tested STRIPAK complex components (Figures 4F and 4G). These data suggest STRN4 as a special component of the STRIPAK complex in mediating the regulation of MAP4Ks by STRIPAK.
STRN4 Inhibits the Hippo Pathway
Studies in both Drosophila and human cells have shown that STRIPAK negatively regulated the Hippo pathway by inhibiting Hippo or MST1/2 through the STRIPAK complex component SLMAP (Bae et al., 2017; Zheng et al., 2017). Because MAP4Ks also directly phosphorylate LATS1/2, we hypothesized that STRN4 could play a similar role as SLMAP in regard to regulating MAP4Ks. To test this hypothesis, we first determined the binding specificity between STRN4 and MAP4Ks in the Hippo pathway. As shown in Figure 5A, STRN4 specifically associated with MAP4K4 among the tested Hippo pathway components, whereas SLMAP majorly formed a complex with MST1/2 and their adaptor SAV1. In addition, we hardly detected the interaction between SLMAP and MAP4K family kinases (Figures 4F and 4G). Taken together, these results suggest that STRN4 could play a critical role in the Hippo pathway by mediating the STRI-PAK-induced MAP4Ks inhibition.
Figure 5. STRN4 Negatively Regulates the Hippo Pathway.
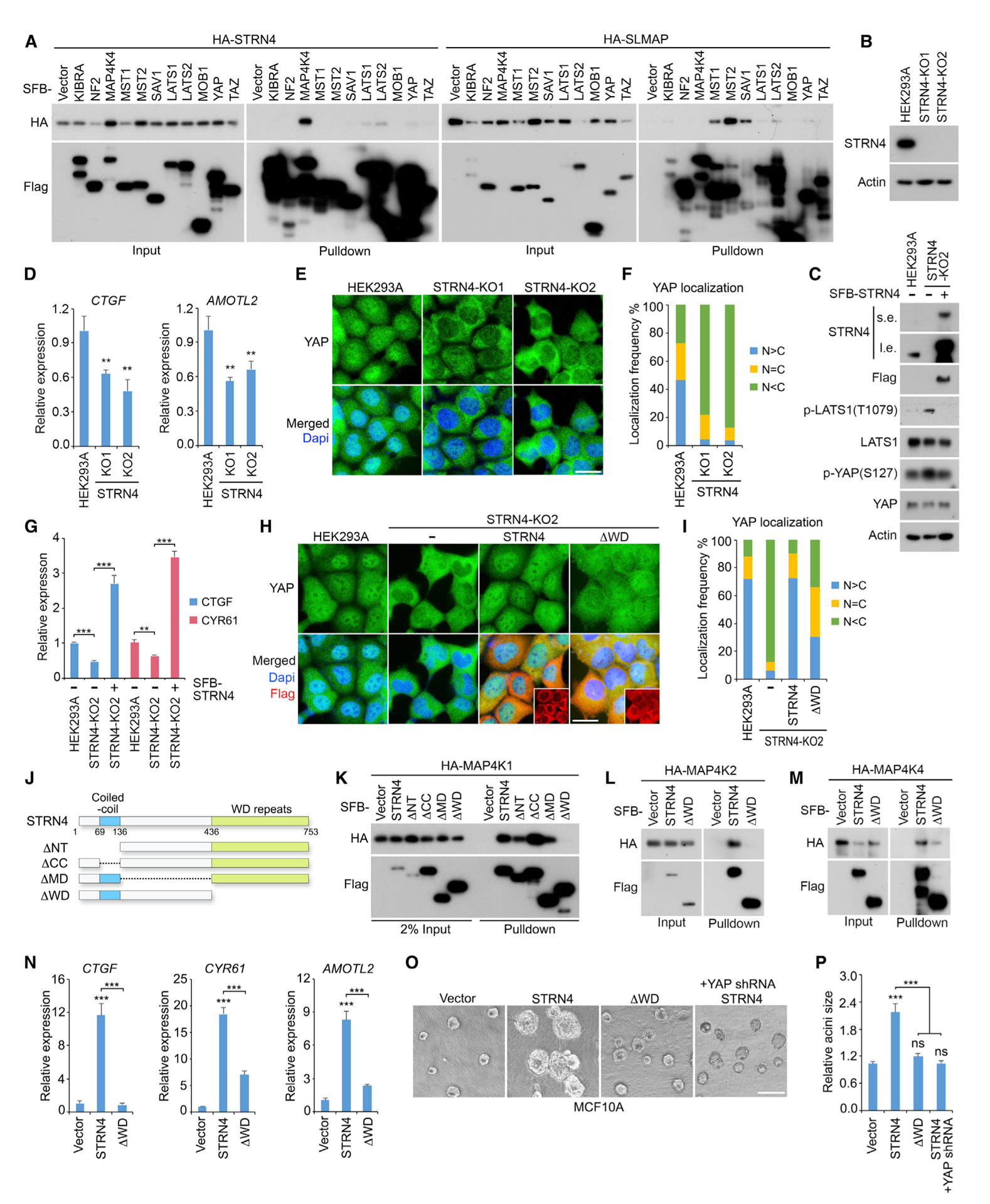
(A) Binding specificity for STRN4 and SLMAP are respectively examined within the Hippo pathway components. HEK293T cells were transfected with the indicated constructs and subjected to the pull-down assay.
(B–F) Depletion of STRN4 inactivates YAP. Loss of STRN4 (B) increased the phosphorylation of LATS1 and YAP (C), which can be rescued by re-expressing STRN4 in the knockout cells. Loss of STRN4 suppressed YAP downstream gene transcription (mean ± SD, n = 3 biological replicates) (D) and induced YAP cytoplasmic translocation (E). STRN4 knockout cells (~200 cells in total) were randomly selected and quantified for YAP localization (F). Western blotting was performed with the indicated antibodies. **p < 0.01 (Student’s t test). Scale bar, 20 μm.
(G–I) Reconstituting STRN4 rescues YAP activity in the STRN4 knockout cells. SFB-tagged STRN4 was re-expressed in the STRN4 knockout cells, where YAP downstream gene transcription (mean ± SD, n = 3 biological replicates) (G) and YAP cellular localization were examined (H and I). FLAG-positive cells (~200 cells in total) were randomly selected and quantified for YAP localization (I). **p < 0.01, ***p < 0.001 (Student’s t test). ΔWD, STRN4 WD-repeat-deletion mutant. Scale bar, 20 μm.
(J–M) The WD-repeat region is required for the association between STRN4 and MAP4Ks. A series of STRN4 truncation and deletion mutants used in this study were illustrated (J). HEK293T cells were transfected with the indicated constructs and subjected to the pull-down assay for MAP4K1 (K), MAP4K2 (L), and MAP4K4 (M).
(N–P) Overexpression of STRN4 but not its ΔWD mutant induces YAP oncogenic activity in MCF10A cells. The indicated MCF10A stable cells were subjected to examination of YAP downstream gene transcription (mean ± SD, n = 3 biological replicates) (N) and acini formation. The representative acini were shown (O) and the relative acini size was quantified (~50 acini were randomly selected in different views) (mean ± SD, n = 3 biological replicates) (P). ***p < 0.001 (Student’s t test). ns, no significance. Scale bar, 200 μm.
Indeed, a loss of STRN4 (Figure 5B) enhanced MAP4K kinase activity (Figure S1) and induced the phosphorylation of LATS1 at T1079 and YAP at S127 (Figure 5C), which can be rescued by re-expressing STRN4 in the knockout cells (Figures S1 and 5C). Consistently, STRN4 deficiency significantly inhibited the transcription of YAP downstream genes (Figure 5D) and promoted YAP’s cytoplasmic localization (Figures 5E and 5F). Re-expressing STRN4 largely rescued YAP’s transcriptional activity (Figure 5G) and nuclear localization (Figures 5H and 5I) in the STRN4 knockout cells. These results demonstrate that STRN4 is a negative regulator of the Hippo pathway.
Next, we examined whether STRN4 regulates YAP through MAP4Ks. To achieve this, we generated a series of STRN4 domain deletion/truncation mutants (Figure 5J) and found that its C-terminal WD (tryptophan-aspartic acid) repeat region was required for the association between STRN4 and MAP4K family members (Figures 5K–5M). In contrast to wild-type STRN4, its WD-repeat-region-deletion mutant (ΔWD) only partially rescued YAP nuclear localization in the STRN4-deficient cells (Figures 5H and 5I), indicating that the association with MAP4Ks was required for STRN4-mediated YAP activation.
This hypothesis was further confirmed in an immortalized normal mammary epithelial cell line, MCF10A. Loss of the WD repeat region largely attenuated the ability of STRN4 to induce the transcription of YAP downstream genes CTGF, CYR61, and AMOTL2 (Figure 5N) and the MCF10A acini growth in Matrigel (Figures 5M and 5N). Moreover, YAP depletion significantly reduced the size of the STRN4-overexpressed MCF10A acini (Figures 5O and 5P). Taken together, these data suggest that STRN4 could promote oncogenic transformation by activating YAP, where its association with MAP4Ks is required.
STRN4 Is Highly Expressed in Endometrial Cancer and Correlated with YAP Activation
By analyzing The Cancer Genome Atlas (TCGA) database, we found that STRN4 is frequently altered in multiple human cancers and highly amplified in endometrial cancer (Figure 6A). Consistently, STRN4 showed a relatively abundant expression in most of the tested endometrial cancer cell lines (Figure 6B). In contrast, STRN4 only had a low or moderate expression in MCF10A and an immortalized endometrial stromal cell line, MAD11 (Figure 6B).
Figure 6. STRN4 Is a Potential Oncoprotein in Human Endometrial Cancer.
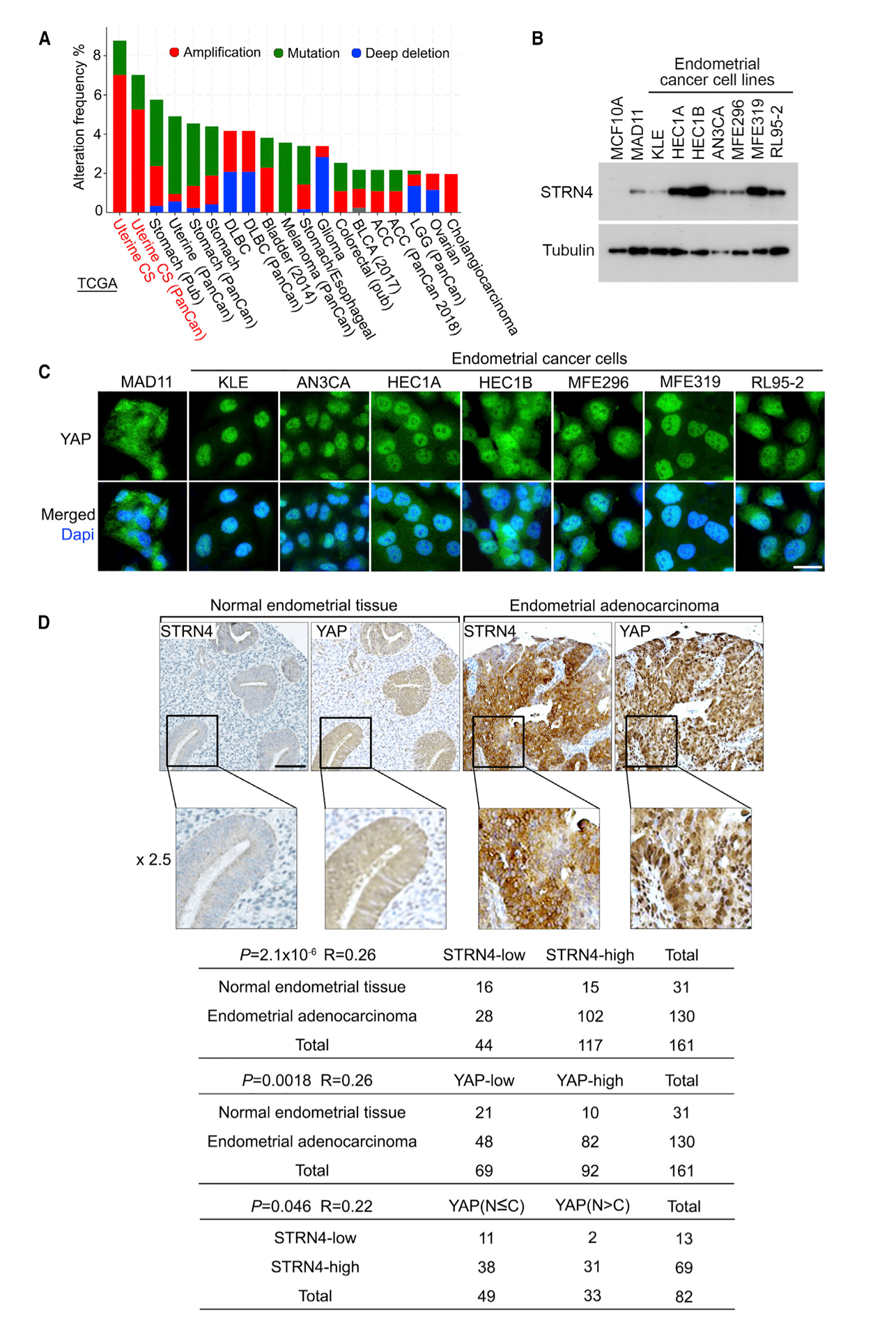
(A) STRN4 is highly expressed in human endometrial cancer. The alteration frequency of STRN4 is analyzed through the cBioportal database (https://www.cbioportal.org).
(B) STRN4 expression is examined in a panel of endometrial cancer cell lines by western blotting.
(C) YAP cellular localization is examined in a panel of endometrial cancer cell lines by immunofluorescent staining. Scale bar, 20 μm.
(D) Immunohistochemical staining of STRN4 and YAP were performed in endometrial cancer tissue microarrays, where the indicated regions in the box were shown 2.5 times enlarged. Brown staining indicates positive immunoreactivity. Correlation analyses between STRN4 and YAP in human normal endometrial tissue and endometrial adenocarcinoma samples are shown as tables. Statistical significance was determined by chi-square test. R, correlation coefficient; N, nuclear localization; C, cytoplasmic localization. Scale bar, 100 μm.
Given the positive role of STRN4 in activating YAP (Figure 5) and the reported oncogenic functions of YAP in endometrial cancer (Dasari et al., 2017; Tsujiura et al., 2014; Wang et al., 2017, 2019), we also explored the pathological relevance between STRN4 and YAP in endometrial cancer development. First, we examined the YAP localization in the same panel of endometrial cancer cell lines as described above (Figure 6B). As shown in Figure 6C, YAP majorly localized in the nucleus of the tested endometrial cancer cell lines with a high expression of STRN4 (Figure 5B), whereas YAP showed increased cytoplasmic localization in MAD11 cells (Figure 5B). Although a low expression of STRN4 was observed in KLE cells, we were still able to detect YAP nuclear enrichment there, suggesting the complicated regulation of YAP in endometrial cancer cells.
Moreover, we further tested this hypothesis through an endometrial tissue microarray analysis. Consistently, upregulation of STRN4 was observed in 78.5% (102 of 130) of endometrial adenocarcinoma tissue samples, whereas this ratio was decreased to 48.4% (15 of 31) for the normal endometrial tissue samples (Figure 6D). Moreover, a positive correlation between high STRN4 expression and YAP nuclear enrichment was found in the endometrial adenocarcinoma tissue samples with YAP high expression (p = 0.046, R = 0.22), where 45% (31 of 69) of the tested endometrial adenocarcinoma tissue samples with high STRN4 expression had high nuclear enrichment of YAP (Figure 6D). Notably, there were still 15.4% (2 of 13) of the total tested patient specimens showing low expression of STRN4 but YAP nuclear enrichment (Figure 6D). These results suggest that upregulation of STRN4 may contribute to YAP activation in a substantial fraction of endometrial adenocarcinoma, whereas additional mechanisms may exist to account for the YAP activation in other tumors.
Collectively, these results not only unveiled a likely mechanism that is required for YAP activation in endometrial cancer but also suggested STRN4 as a potential oncogenic player in endometrial cancer development.
DISCUSSION
In this study, we comprehensively defined the protein interaction landscape for the MAP4K family kinases and identified over 200 HCIPs, which greatly expanded our knowledge of this group of kinases in diverse signaling events and cellular functions.
Because MAP4Ks have similar protein structures and evolutionary origins, members of this family of kinases were found to share a group of binding partners despite their individual PPI network (Figures 2 and 3), providing us an opportunity to further explore both generic and specific functions/regulations for MAP4K family members. Indeed, our validation studies not only revealed several specific MAP4K-associated complexes, such as MAP4K2-GABARAP/L1/L2, MAP4K3-SH3GL1, and MAP4K5-CTTN (Figure 4), but also discovered STRIPAK complex component STRN4 as a universal binding partner for MAP4Ks (Figure 4).
STRIPAK is a striatin-centered multicomponent complex known for its crucial roles in regulating protein phosphorylation events under various biological processes (Hwang and Pallas, 2014; Shi et al., 2016). Although STRIPAK and MAP4Ks have been both connected with Hippo signaling in different organisms, their architectural interplay and functional regulation have not been elucidated at the same details. Interestingly, our proteomic analysis uncovered STRN4 as a key STRIPAK component involved in Hippo pathway regulation (Figure 5). The STRN4-based regulatory mechanism is similar to the one involved in the SLMAP-mediated regulation of MST1/2 by STRIPAK. These findings together place STRIPAK upstream of both MST1/2 and MAP4Ks in the Hippo pathway to control their activities through SLMAP and STRN4, respectively. Given the redundant roles between MST1/2 and MAP4Ks in regulating LATS1/2 kinases, this model highlighted a critical role of STRIPAK in regulating the Hippo pathway. Indeed, a recent study showed that STRIPAK can integrate several Hippo upstream signaling events to control the Hippo pathway and its downstream YAP activity (Chen et al., 2019).
Given the critical roles of STRN4 and SLMAP in mediating the STRIPAK-dependent Hippo pathway inhibition, modulating the expression of these two adaptor proteins are expected to affect YAP/TAZ activities. Indeed, our data also demonstrated STRN4 as an activator of YAP and a potential oncogene in driving cell transformation (Figure 5). Notably, our current studies were mostly performed in HEK293A and MCF10A cells, where MST1/2 kinases were found dispensable for LATS1/2 activation (Kim et al., 2011; Meng et al., 2015) and MAP4Ks are likely to play a dominant role for it, allowing us to characterize the role of STRN4 in the Hippo pathway regulation (Figure 5). Moreover, although our data uncovered an “adaptor” role of STRN4 in mediating the STRIPAK complex to bind and inhibit MAP4Ks, it should be noted that the STRN4-independent recruitment for several MAP4K members may also exist (Figure 4F), indicating a complex regulation of MAP4Ks by the STRIPAK complex.
In this study, we also examined the pathological relevance between STRN4 and YAP in human cancers, where STRN4 was found highly expressed in endometrial cancer and positively correlated with YAP activation. YAP is known to be highly expressed and activated in many types of human cancers; however, the genetic mutation of the Hippo pathway core components is rare (Yu et al., 2015). This discrepancy indicates that additional oncogenic events may account for YAP activation in tumorigenesis. Actually, YAP has been shown to be highly expressed and activated in endometrial cancer (Figure 6D; Dasari et al., 2017; Tsujiura et al., 2014; Wang et al., 2017, 2019), but its upstream oncogenic alteration has not been fully elucidated. Here, our findings suggest that upregulation of STRN4 could be at least one of these oncogenic events required for YAP activation in endometrial cancer. Of course, it is not surprising that not all of those cancers would work through the STRN4-YAP axis.
In summary, our proteomic study of the MAP4K-centered PPI network not only reveals a number of cellular functions/regulations for the MAP4K-related signaling events but also generates a comprehensive resource for further characterization of key kinase families in various biological processes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wenqi Wang (wenqiw6@uci.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The proteomic data have been deposited in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2013) with the project identifier PXD016931. The detailed project information is as follows:
Project Name: Human MAP4K proteins TAP-LC-MSMS Project accession: PXD016931
Project https://doi.org/10.6019/PXD016931
Sequence information about all the oligos used in this study can be found in Table S5.
Original Western Blot data have been deposited to Mendeley Data at: http://dx.doi.org/10.17632/r7sdn4kzx4.1
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HEK293T (a female cell line, ATCC: CRL-3216) and MCF10A (a female cell line, ATCC: CRL-10317) cells were purchased from ATCC and kindly provided by Dr. Junjie Chen (MD Anderson Cancer Center). HEK293A (a female cell line, ThermoFisher: R70507) cells were purchased from Thermo Fisher and kindly provided by Dr. Jae-Il Park (MD Anderson Cancer Center). MAD11 (a female cell line), KLE (a female cell line), AN3CA (a female cell line), MFE296 (a female cell line), MFE319 (a female cell line) and RL95–2 (a female cell line) endometrial cancer cells were kindly provided by Drs. Kyle Hoehn (University of New South Wales), Doug Marchion (Moffitt Cancer Center) and Hui Li (University of Virginia). HEC1A (a female cell line, ATCC: HTB-112) and HEC1B (a female cell line, ATCC: HTB-113) endometrial cancer cells were purchased from the Characterized Cell Line Core Facility at MD Anderson Cancer Center.
HEK293A cells were maintained in Dulbecco’s modified essential medium (DMEM) supplemented with 10% bovine growth serum supplemented calf at 37°C in 5% CO2 (v/v). MCF10A cells were maintained in DMEM/F12 medium supplemented with 5% horse serum, 200 ng/mL epidermal growth factor, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin and 10 mg/mL insulin at 37°C in 5% CO2 (v/v). HEK293T and MAD11 cells were maintained in DMEM supplemented with 10% fetal bovine serum at 37°C in 5% CO2 (v/v). KLE and RL95–2 cells were maintained in DMEM/F12 with 10% fetal bovine serum at 37°C in 5% CO2 (v/v). HEC1A and HEC1B cells were cultured in McCoy’s 5A medium with L-glutamine supplemented with 10% fetal bovine serum at 37°C in 5% CO2 (v/v). AN3CA, MFE296 and MFE319 cells were grown in minimum essential medium (MEM) supplemented with 10% fetal bovine serum at 37°C in 5% CO2 (v/v). All the culture media contain 1% penicillin and streptomycin. Plasmid transfection was performed using a polyethylenimine reagent.
Patient tissues
The endometrial cancer tissue arrays were purchased from US Biomax, Inc. According to the Declaration of Specimen Collection provided by US Biomax, each specimen collected from any clinic was consented by both hospital and individual.
METHOD DETAILS
Antibodies and chemicals
For western blotting, anti-Flag (M2) (F3165–5MG, 1:5000 dilution), anti-α-tubulin (T6199–200UL, 1:5000 dilution) and anti-β-actin (A5441–100UL, 1:5000 dilution) monoclonal antibodies were obtained from Sigma-Aldrich. An anti-Myc (sc-40, 1:500 dilution) antibody was purchased from Santa Cruz Biotechnology. An anti-hemagglutinin (HA) monoclonal antibody (MMS-101P, 1:3000 dilution) was obtained from Covance. An anti-STRN4 (A304–573A-T, 1:1000 dilution) was obtained from Bethyl laboratories. Anti-phospho-YAP (S127) (4911S, 1:1000 dilution), anti-phospho-LATS1 (T1079) (8654S, 1:1000 dilution) and anti-LATS1 (3477S, 1:1000 dilution) antibodies were purchased from Cell Signaling Technology. The YAP polyclonal antibody was raised against a GST-YAP fusion protein and the antisera were affinity-purified using an AminoLink Plus Immobilization and Purification Kit (Pierce) (Wang et al., 2011).
For immunostaining, an anti-YAP (sc-101199, 1:200 dilution) monoclonal antibody was purchased from Santa Cruz Biotechnology. An anti-HA polyclonal antibody (3724S, 1:3000 dilution) was obtained from Cell Signaling Technology.
For immunohistochemical staining, an anti-YAP (14074S, 1:20 dilution) monoclonal antibody was purchased from Cell Signaling Technology. An anti-STRN4 (A304–573A-T, 1:20 dilution) was obtained from Bethyl laboratories.
Constructs and viruses
Plasmids encoding the indicated genes were obtained from the Human ORFeome V5.1 library or purchased from Dharmacon and Harvard Plasmid DNA Resource Cor. All constructs were generated via polymerase chain reaction (PCR) and sub-cloned into a pDONOR201 vector using Gateway Technology (Invitrogen) as the entry clones. For tandem affinity purification (TAP), all entry clones were subsequently recombined into a lentiviral gateway-compatible destination vector for the expression of C-terminal SFB-tagged fusion proteins. Gateway-compatible destination vectors with the indicated SFB tag, HA tag, Myc tag, GST tag and MBP tag were used to express various fusion proteins. PCR-mediated site-directed mutagenesis was used to generate deletion or truncation mutations for STRN4: deletion of N-terminus (ΔNT), missing residues 1~135; deletion of coiled-coil domain (ΔCC), missing residues 69~136; deletion of middle region (ΔMD), missing residues 137~435; and deletion of WD repeats (ΔWD), missing residues 437~753. YAP shRNA was obtained from Addgene (#27369) (Zhao et al., 2008).
All lentiviral supernatants were generated by transient transfection of HEK293T cells with the helper plasmids pSPAX2 and pMD2G (kindly provided by Dr. Zhou Songyang, Baylor College of Medicine) and harvested 48 hours later. Supernatants were passed through a 0.45-μm filter and used to infect cells with the addition of 8 μg/mL hexadimethrine bromide (Polybrene) (Sigma-Aldrich).
Purification of SFB-tagged MAP4K complexes
HEK293T cells stably expressing SFB-tagged MAP4K proteins were generated by culturing in medium containing 2 μg/mL puromycin and validated by immunostaining and western blotting as described previously (Wang et al., 2014). For tandem affinity purification, the HEK293T stable cells were lysed in NETN buffer with protease and phosphatase inhibitors at 4 C for 20 minutes. The crude lysates were centrifuged at 14,000 rpm for 15 minutes at 4°C. The supernatants were incubated with streptavidin-conjugated beads (GE Healthcare) for 1 hour at 4°C. The beads were then washed 3 times with NETN buffer, and bound proteins were eluted with NETN buffer containing 2 mg/mL biotin (Sigma-Aldrich) for 2 hours at 4°C. The elutes were incubated with S protein beads (Novagen) for 1 hour. The beads were washed three times with NETN buffer and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis. Each sample was run into the separation gel for a short distance, so that the whole bands could be excised as one sample for in-gel trypsin digestion and MS analysis.
Mass spectrometry analysis
The mass spectrometry was performed by the Taplin Mass Spectrometry Facility (Harvard Medical School) as described previously (Li et al., 2016; Wang et al., 2014). Briefly, the excised gel bands described above were cut into approximately 1-mm3 pieces. The gel pieces were then subjected to in-gel trypsin digestion (Shevchenko et al., 1996) and dried. Samples were reconstituted in 5 μL of high-performance liquid chromatography (HPLC) solvent A (2.5% acetonitrile, 0.1% formic acid). The facility packed a nanoscale reverse-phase HPLC capillary column by packing 5-μm C18 spherical silica beads (Thermo Fisher Scientific) into a fused silica capillary (100 μm inner diameter × ~20 cm length) with a flame-drawn tip. After the column was equilibrated, each sample was loaded onto the column via a Famos autosampler (LC Packings). A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid).
As the peptides eluted, they were subjected to electrospray ionization and then entered into an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). The peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein databases with the fragmentation pattern acquired by the software program SEQUEST (ver. 28) (Thermo Fisher Scientific). Enzyme specificity was set to partially tryptic with 2 missed cleavages. Modifications included carboxyamidomethyl (cysteines, fixed) and oxidation (methionine, variable). Mass tolerance was set to 5 ppm for precursor ions and 0.5 Da for fragment ions. The database searched was UniProt. Spectral matches were filtered to contain a false discovery rate of less than 1% at the peptide level using the target-decoy method (Elias and Gygi, 2007), and the protein inference was considered followed the general rules (Nesvizhskii and Aebersold, 2005), with manual annotation based on experiences applied when necessary. This same principle was used for isoforms when they were present in the database. The longest isoform was reported as the match.
Bioinformatic analysis
As for the TAP-MS data analysis, a group of unrelated TAP-MS experiments (1,806 experiments using stably expressed TAP-tagged protein baits and 20 experiments using empty vector baits) were included as a control group. Using the “sensitivity1/2-(1-specificity)1/2” measurement as determined by the BioGrid and CRAPome databases (Chatr-Aryamontri et al., 2017; Mellacheruvu et al., 2013), we considered any interaction with a MUSE score of at least 0.8 and raw spectra count greater than 1 to be a high-confident interacting protein (HCIP) (Li et al., 2016, 2017; Vargas et al., 2020). The overall HCIP reproducibility rate was close to 80%, which increased when the cutoff peptide number increased. The MAP4Ks interactome was enriched in signaling pathways, diseases & disorders and molecular & cellular functions using the HCIP sets (Table S4). The P values were estimated using the Knowledge Base provided by Ingenuity Pathway software (Ingenuity Systems, https://www.ingenuity.com/), which contains findings and annotations from multiple sources including the Gene Ontology database, KEGG pathway database, and Panther pathway database. Only statistically significant correlations (p < 0.05) are shown. The −log10 (P value) for each function with related HCIPs is listed.
Immunofluorescent staining
Immunofluorescent staining was performed as described previously (Wang et al., 2008). Briefly, cells cultured on coverslips were fixed with 4% paraformaldehyde for 10 minutes at room temperature and then extracted with 0.5% Triton X-100 solution for 5 minutes. After blocking with Tris-buffered saline with Tween 20 containing 1% bovine serum albumin, the cells were incubated with the indicated primary antibodies for 1 hour at room temperature. After that, the cells were washed and incubated with fluorescein isothiocyanate- or rhodamine-conjugated secondary antibodies for 1 hour. Cells were counterstained with 100 ng/mL 4′,6-diami-dino-2-phenylindole (DAPI) for 2 minutes to visualize nuclear DNA. The coverslips were mounted onto glass slides with an anti-fade solution and visualized under a Nikon Ti2-E inverted microscope.
Gene inactivation by CRISPR/Cas9 system
To generate the STRN4 knockout cells, five distinct single-guide RNAs (sgRNA) were designed by CHOPCHOP website (https://chopchop.cbu.uib.no/), cloned into lentiGuide-Puro vector (Addgene plasmid # 52963) (Sanjana et al., 2014) and transfected into HEK293A cells with lentiCas9-Blast construct (Addgene plasmid # 52962) (Sanjana et al., 2014). The next day, cells were selected with puromycin (2 μg/mL) for two days and sub-cloned to form single colonies. Knockout cell clones were screened by western blotting to verify the loss of STRN4 expression.
The oligo sequence information of sgRNAs used for knockout cell generation is listed in the Table S5.
RNA extraction, reverse transcription and real-time PCR
RNA samples were extracted with TRIzol reagent (Invitrogen). Reverse transcription assay was performed using the Script Reverse Transcription Supermix Kit (Bio-Rad) according to the manufacturer’s instructions. Real-time PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems). For quantification of gene expression, the 2−ΔΔCt method was used. GAPDH expression was used for normalization. The sequence information of q-PCR primers used for gene expression analysis is listed in the Table S5.
In vitro kinase assay
SFB-MAP4K2 and SFB-MAP4K6 were expressed in the indicated HEK293A cells for 48 hours. The kinases were then pulled down by S protein beads, washed three times in washing buffer (40 mM HEPES, 250 mM NaCl), and subjected to the kinase assay in the presence of cold ATP (500 μM) and 2 μg bacterially-purified MBP-LATS1-C3 (Han et al., 2018). The reaction mixture was incubated at 30°C for 30 min, terminated with 2x SDS loading buffer and subjected to SDS–PAGE. Phosphorylation of LATS1-hydrophobic motif (HM) in the LATS1-C3 region were determined using anti-phospho-LATS1 (T1079).
3D culture of MCF10A cells for acini formation
MCF10A acini formation assay was performed as previously described (Wang et al., 2012). Briefly, the indicated MCF10A cells (5 × 103) were grown in growth factor-reduced BD Matrigel matrix (BD Biosciences) within the eight-well chamber slide system (Fisher Scientific). The cultured acini were analyzed after 4 days of growth in Matrigel.
Immunohistochemical analysis
The endometrial cancer tissue arrays (T091 and EMC1502) were purchased from US Biomax, deparaffinized and rehydrated. The antigens were retrieved by applying Unmask Solution (Vector Laboratories) in a steamer for 40 min. To block endogenous peroxidase activity, the sections were treated with 3% hydrogen peroxide for 30 min. After 1 hour of pre-incubation in 10% goat serum to prevent non-specific staining, the samples were incubated with an antibody at 4°C overnight. The sections were incubated with SignalStain Boost detection reagent at room temperature for 30 min. Color was developed with SignalStain 3,3′-diaminobenzidine chromogen-diluted solution (all reagents were obtained from Cell Signaling Technology). Sections were counterstained with Mayer hematoxylin. To quantify the results, a total score of protein expression was calculated from both the percentage of immunopositive cells and immunostaining intensity. High and low protein expressions were defined using the mean score of all samples as a cutoff point. Pearson chi-square analysis test was used for statistical analysis of the correlation of STRN4 with tissue type (normal versus cancer) and the correlation between STRN4 and YAP.
QUANTIFICATION AND STATISTICAL ANALYSIS
Each experiment was repeated twice or more, unless otherwise noted. There were no samples excluded for the analyses in this study. The Student’s t test was used to analyze the differences between groups. Data were analyzed by Student’s t test or Pearson chi-square analysis. SD was used for error estimation. A P value < 0.05 was considered statistically significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | ||
|---|---|---|---|---|
| Antibodies | ||||
| Anti-Myc | Santa Cruz Biotechnology | sc-40; RRID:AB_627268 | ||
| Anti-Flag (M2) | Sigma-Aldrich | F3165–5MG; RRID:AB_259529 | ||
| Anti-HA | Cell Signaling Technology | 3724S; RRID:AB_1549585 | ||
| Anti-hemagglutinin (HA) | Covance | MMS-101P; RRID:AB_439687 | ||
| Anti-LATS1 | Cell Signaling Technology | 3477S; RRID:AB_2133513 | ||
| Anti-phospho-LATS1 (T1079) | Cell Signaling Technology | 8654S; RRID:AB_10971635 | ||
| Anti-phospho-YAP (S127) | Cell Signaling Technology | 4911S; RRID:AB_2218913 | ||
| Anti-STRN4 | Bethyl laboratories | A304–573A-T; RRID:AB_2782004 | ||
| Anti-YAP | Wang et al., 2011 | N/A | ||
| Anti-YAP | Santa Cruz Biotechnology | sc-101199; RRID:AB_1131430 | ||
| Anti-YAP | Cell Signaling Technology | 14074S; RRID:AB_2650491 | ||
| Anti-α-tubulin | Sigma-Aldrich | T6199–200UL; RRID:AB_477583 | ||
| Anti-β-actin | Sigma-Aldrich | A5441–100UL; RRID:AB_476744 | ||
| Bacterial and Virus Strains | ||||
| DH5α competent E. coli | Laboratory of Junjie Chen | N/A | ||
| BL21 competent E. coli | Laboratory of Junjie Chen | N/A | ||
| DB3.1 competent E. coli | Laboratory of Junjie Chen | N/A | ||
| Biological Samples | ||||
| Endometrial cancer tissue arrays | US Biomax, Inc | T091 and EMC1502 | ||
| Chemicals, Peptides, and Recombinant Proteins | ||||
| Hexadimethrine bromide (Polybrene) | Sigma Aldrich | Cat# H9268 | ||
| 4’,6-diamidino-2-phenylindole (DAPI) | Sigma Aldrich | Cat# D9542 | ||
| TRIzol | Thermo Fisher | Cat# 15596026 | ||
| Power SYBR Green PCR master mix | Thermo Fisher | Cat# 4367659 | ||
| Streptavidin-conjugated beads | GE Healthcare | Cat# 17-5113-01 | ||
| Biotin | Sigma Aldrich | Cat# B4639 | ||
| S protein beads | Novagen | Cat# 69704 | ||
| Growth factor-reduced BD Matrigel matrix | BD Biosciences | Cat# 354230 | ||
| Polyethylenimine | VWR | Cat# 87001–912 | ||
| Critical Commercial Assays | ||||
| Gateway BP Clonase Enzyme Mix for BP assay | Thermo Fisher | Cat# 11789021 | ||
| Gateway LR Clonase Enzyme Mix for LR assay | Thermo Fisher | Cat# 11791043 | ||
| Script Reverse Transcription Supermix Kit for reverse transcription assay | Bio Rad | Cat# 1708841 | ||
| Deposited Data | ||||
| Proteomic data | ProteomeXchange Consortium database | Project identifier: PXD016931 | ||
| Experimental Models: Cell Lines | ||||
| HEK293T | ATCC, MD Anderson Cancer Center (Junjie Chen Lab) | N/A | ||
| MCF10A | ATCC, MD Anderson Cancer Center (Junjie Chen Lab) | N/A | ||
| HEK293A | Thermo Fisher and MD Anderson Cancer Center (Jae-Il Park Lab) | N/A | ||
| MDA11, KLE, AN3CA, MFE296, MFE319, RL95–2 | University of New South Wales (Kyle Hoehn Lab), Moffitt Cancer Center (Doug Marchion Lab) and University of Virginia (Hui Li Lab) | N/A | ||
| HEC1A, HEC1B | MD Anderson Cancer Center (Characterized Cell Line Core Facility) | N/A | ||
| Oligonucleotides | ||||
| STRN4 guide RNAs, see Table S5 | This paper | N/A | ||
| qPCR primers, see Table S5 | This paper | N/A | ||
| Recombinant DNA | ||||
| Lenti Guide-Puro vector | Sanjana et al., 2014 | Addgene #52963 | ||
| Lenti Cas9-Blast construct | Sanjana et al., 2014 | Addgene #52962 | ||
| pLKO-YAP shRNA | Zhao et al., 2008 | Addgene #27369 | ||
| Lenti-MAP4K1-SFB | This paper | N/A | ||
| Lenti-MAP4K2-SFB | This paper | N/A | ||
| Lenti-MAP4K3-SFB | This paper | N/A | ||
| Lenti-MAP4K4-SFB | This paper | N/A | ||
| Lenti-MAP4K5-SFB | This paper | N/A | ||
| Lenti-MAP4K6-SFB | This paper | N/A | ||
| Lenti-MAP4K7-SFB | This paper | N/A | ||
| SFB-STRN4-Full length (1–753 amino acids) | This paper | N/A | ||
| SFB-STRN4-ΔNT (136–753 amino acids) | This paper | N/A | ||
| SFB-STRN4-ΔCC (1–68, 137–753 amino acids) | This paper | N/A | ||
| SFB-STRN4-ΔMD (1–136, 436–753 amino acids) | This paper | N/A | ||
| SFB-STRN4-DWD (1–435 amino acids) | This paper | N/A | ||
| MBP-LATS1-C3 | Han et al., 2018 | N/A | ||
| Software and Algorithms | ||||
| Optimized CRISPR Design | CHOPCHOP website | http://chopchop.cbu.uib.no/ | ||
| ImageJ | ImageJ Software | RRID: SCR_003070 | ||
| Ingenuity Pathway software | Ingenuity Systems | https://www.ingenuity.com/ | ||
Highlights.
Proteomic analysis defines the human MAP4K family interactome
The STRIPAK complex component STRN4 interacts with and inhibits MAP4Ks
STRN4 is a positive regulator of YAP
Elevated STRN4 expression is associated with YAP activation in endometrial cancer
ACKNOWLEDGMENTS
W.W. thanks Dr. Junjie Chen (MD Anderson Cancer Center) for his mentoring and help with the MAP4K TAP-MS analyses. We thank Dr. Ross Tomaino (Taplin Mass Spectrometry Facility, Harvard Medical School) for the mass spectrometry analysis. We also thank Drs. Kyle Hoehn (University of New South Wales), Doug Marchion (Moffitt Cancer Center), and Hui Li (University of Virginia) for providing the endometrial cancer cell lines. This work was supported in part by a NIH grant (GM126048) and an American Cancer Society Research Scholar grant (RSG-18-009-01-CCG) to W.W. R.E.V. is supported by a NIH Initiative for Maximizing Student Development (IMSD) Fellowship (GM055246). Research reported in this publication was also supported in part by a NIH-NCI grant (P30CA062203) and the UC Irvine Chao Family Comprehensive Cancer Center using UCI Anti-Cancer Challenge funds. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Chao Family Comprehensive Cancer Center.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107860.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Bae SJ, Ni L, Osinski A, Tomchick DR, Brautigam CA, and Luo X (2017). SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK. eLife 6, e30278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzakri K, and Zierath JR (2007). MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-alpha-induced insulin resistance. J. Biol. Chem 282, 7783–7789. [DOI] [PubMed] [Google Scholar]
- Chan Wah Hak L, Khan S, Di Meglio I, Law AL, Lucken-Ardjomande Häsler S, Quintaneiro LM, Ferreira APA, Krause M, McMahon HT, and Boucrot E (2018). FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat. Cell Biol 20, 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O’Donnell L, Oster S, Theesfeld C, Sellam A, et al. (2017). The Bio-GRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Xie R, Meng Z, Ma S, and Guan KL (2019). STRIPAK integrates upstream signals to initiate the Hippo kinase cascade. Nat. Cell Biol 21, 1565–1577. [DOI] [PubMed] [Google Scholar]
- Chuang HC, and Tan TH (2019). MAP4K Family Kinases and DUSP Family Phosphatases in T-Cell Signaling and Systemic Lupus Erythematosus. Cells 8, 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HC, Wang X, and Tan TH (2016). MAP4K Family Kinases in Immunity and Inflammation. Adv. Immunol 129, 277–314. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, and Kusumi A (2001). The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220–230. [DOI] [PubMed] [Google Scholar]
- Danai LV, Guilherme A, Guntur KV, Straubhaar J, Nicoloro SM, and Czech MP (2013). Map4k4 suppresses Srebp-1 and adipocyte lipogenesis independent of JNK signaling. J. Lipid Res 54, 2697–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari VR, Mazack V, Feng W, Nash J, Carey DJ, and Gogoi R (2017). Verteporfin exhibits YAP-independent anti-proliferative and cytotoxic effects in endometrial cancer cells. Oncotarget 8, 28628–28640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, and Gygi SP (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. [DOI] [PubMed] [Google Scholar]
- Glantschnig H, Rodan GA, and Reszka AA (2002). Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J. Biol. Chem 277, 42987–42996. [DOI] [PubMed] [Google Scholar]
- Halder G, and Johnson RL (2011). Hippo signaling: growth control and beyond. Development 138, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Qi R, Zhou JJ, Ta AP, Yang B, Nakaoka HJ, Seo G, Guan KL, Luo R, and Wang W (2018). Regulation of the Hippo Pathway by Phosphatidic Acid-Mediated Lipid-Protein Interaction. Mol. Cell 72, 328–340.e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Ren Y, Wu B, Decourt B, Lee AC, Taylor A, and Suter DM (2015). Src and cortactin promote lamellipodia protrusion and filopodia formation and stability in growth cones. Mol. Biol. Cell 26, 3229–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, and Pallas DC (2014). STRIPAK complexes: structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol 47, 118–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Dong L, Lu Y, Wu W, Hao Q, Zhou Z, Jiang J, Zhao Y, and Zhang L (2012). Dimerization and cytoplasmic localization regulate Hippo kinase signaling activity in organ size control. J. Biol. Chem 287, 5784–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NG, Koh E, Chen X, and Gumbiner BM (2011). E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA 108, 11930–11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis JM, and Avruch J (2012). Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol. Rev 92, 689–737. [DOI] [PubMed] [Google Scholar]
- Li Q, Li S, Mana-Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, Nie Y, Kaneko S, Yao X, Chen X, et al. (2014). The conserved misshapenwarts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev. Cell 31, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Cho YS, Yue T, Ip YT, and Jiang J (2015). Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling. Cell Discov. 1, 15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tran KM, Aziz KE, Sorokin AV, Chen J, and Wang W (2016). Defining the Protein-Protein Interaction Network of the Human Protein Tyrosine Phosphatase Family. Mol. Cell. Proteomics 15, 3030–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Han H, Zhou MT, Yang B, Ta AP, Li N, Chen J, and Wang W (2017). Proteomic Analysis of the Human Tankyrase Protein Interaction Network Reveals Its Role in Pexophagy. Cell Rep. 20, 737–749. [DOI] [PubMed] [Google Scholar]
- Li Q, Nirala NK, Nie Y, Chen HJ, Ostroff G, Mao J, Wang Q, Xu L, and Ip YT (2018). Ingestion of Food Particles Regulates the Mechanosensing Misshapen-Yorkie Pathway in Drosophila Intestinal Growth. Dev. Cell 45, 433–449.e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P, Yao Z, Meyer CF, Wang XS, Oehrl W, Feller SM, and Tan TH (1999). Interaction of hematopoietic progenitor kinase 1 with adapter proteins Crk and CrkL leads to synergistic activation of c-Jun N-terminal kinase. Mol. Cell. Biol 19, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus JC, Yang Z, Kloss J, Dhruv H, Tran NL, and Riggs DL (2013). A Novel Interaction between Pyk2 and MAP4K4 Is Integrated with Glioma Cell Migration. J. Signal Transduct 2013, 956580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. (2013). The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S, et al. (2015). MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun 6, 8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CJ, Lou HJ, Simpson C, van de Kooij B, Ha BH, Fisher OS, Pirman NL, Boggon TJ, Rinehart J, Yaffe MB, et al. (2019). Comprehensive profiling of the STE20 kinase family defines features essential for selective substrate targeting and signaling output. PLoS Biol. 17, e2006540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, and Aebersold R (2005). Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, and Cordenonsi M (2014). The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev 94, 1287–1312. [DOI] [PubMed] [Google Scholar]
- Resnik-Docampo M, and de Celis JF (2011). MAP4K3 is a component of the TORC1 signalling complex that modulates cell growth and viability in Drosophila melanogaster. PLoS One 6, e14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf MB, Keulers TG, Vooijs MA, and Rouschop KM (2016). LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J. 30, 3961–3978. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, and Mann M (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shi CS, Tuscano J, and Kehrl JH (2000). Adaptor proteins CRK and CRKL associate with the serine/threonine protein kinase GCKR promoting GCKR and SAPK activation. Blood 95, 776–782. [PubMed] [Google Scholar]
- Shi Z, Jiao S, and Zhou Z (2016). STRIPAK complexes in cell signaling and cancer. Oncogene 35, 4549–4557. [DOI] [PubMed] [Google Scholar]
- Tsujiura M, Mazack V, Sudol M, Kaspar HG, Nash J, Carey DJ, and Gogoi R (2014). Yes-associated protein (YAP) modulates oncogenic features and radiation sensitivity in endometrial cancer. PLoS One 9, e100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas RE, Duong VT, Han H, Ta AP, Chen Y, Zhao S, Yang B, Seo G, Chuc K, Oh S, et al. (2020). Elucidation of WW domain ligand binding specificities in the Hippo pathway reveals STXBP4 as YAP inhibitor. EMBO J. 39, e102406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. (2013). The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen L, Ding Y, Jin J, and Liao K (2008). Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J. Cell Sci 121, 1334–1343. [DOI] [PubMed] [Google Scholar]
- Wang W, Huang J, and Chen J (2011). Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem 286, 4364–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park JI, and Chen J (2012). PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 26, 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li X, Huang J, Feng L, Dolinta KG, and Chen J (2014). Defining the protein-protein interaction network of the human hippo pathway. Mol. Cell. Proteomics 13, 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Gu C, Jeong KJ, Zhang D, Guo W, Lu Y, Ju Z, Panupinthu N, Yang JY, Gagea MM, et al. (2017). YAP/TAZ-Mediated Upregulation of GAB2 Leads to Increased Sensitivity to Growth Factor-Induced Activation of the PI3K Pathway. Cancer Res. 77, 1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Song T, Zhou S, and Kong X (2019). YAP promotes the malignancy of endometrial cancer cells via regulation of IL-6 and IL-11. Mol. Med 25, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, and Lamb RF (2010). PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol. Cell 37, 633–642. [DOI] [PubMed] [Google Scholar]
- Yu FX, Zhao B, and Guan KL (2015). Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 163, 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Xie M, Gou X, Lee P, Schneider MD, and Wu X (2014). Microtubules regulate focal adhesion dynamics through MAP4K4. Dev. Cell 31, 572–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, and Pan D (2019). The Hippo Signaling Pathway in Development and Disease. Dev. Cell 50, 264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang W, Liu B, Deng H, Uster E, and Pan D (2015). Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev. Cell 34, 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Liu B, Wang L, Lei H, Pulgar Prieto KD, and Pan D (2017). Homeostatic Control of Hpo/MST Kinase Activity through Autophosphorylation-Dependent Recruitment of the STRIPAK PP2A Phosphatase Complex. Cell Rep. 21, 3612–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomic data have been deposited in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2013) with the project identifier PXD016931. The detailed project information is as follows:
Project Name: Human MAP4K proteins TAP-LC-MSMS Project accession: PXD016931
Project https://doi.org/10.6019/PXD016931
Sequence information about all the oligos used in this study can be found in Table S5.
Original Western Blot data have been deposited to Mendeley Data at: http://dx.doi.org/10.17632/r7sdn4kzx4.1


