Abstract
Targeted tandem mass spectrometry (LC–MS/MS) has been extremely useful for profiling small molecules extracted from biological sources, such as cells, bodily fluids and tissues. Here, we present a protocol for analysing incorporation of the non-radioactive stable isotopes carbon-13 (13C) and nitrogen-15 (15N) into polar metabolites in central carbon metabolism and related pathways. Our platform utilizes selected reaction monitoring (SRM) with polarity switching and amide hydrophilic interaction liquid chromatography (HILIC) to capture transitions for carbon and nitrogen incorporation into selected metabolites using a hybrid triple quadrupole (QQQ) mass spectrometer. This protocol represents an extension of a previously published protocol for targeted metabolomics of unlabeled species and has been used extensively in tracing the metabolism of nutrients such as 13C-labeled glucose, 13C-glutamine and 15N-glutamine in a variety of biological settings (e.g., cell culture experiments and in vivo mouse labelling via i.p. injection). SRM signals are integrated to produce an array of peak areas for each labelling form that serve as the output for further analysis. The processed data are then used to obtain the degree and distribution of labelling of the targeted molecules (termed fluxomics). Each method can be customized on the basis of known unlabeled Q1/Q3 SRM transitions and adjusted to account for the corresponding 13C or 15N incorporation. The entire procedure takes ~6–7 h for a single sample from experimental labelling and metabolite extraction to peak integration.
Introduction
There has been rapidly growing interest in and application of metabolomics technologies over the past decade due to advancements in mass spectrometry (MS) technology and the realization that central carbon metabolism is reprogrammed in a variety of diseases, including cancer and diabetes. Targeted MS is very powerful because it can enhance the sensitivity of detection of compounds of interest in the face of enormous molecular background. High-resolution MS has been used to track specific compounds and their 13C incorporation through subsets of central carbon metabolism pathways1-6. Previous work in other laboratories has included tracing 13C-labeled glucose into central carbon metabolism pathways and amino acids in microbes using gas chromatography–mass spectrometry (GC–MS)7-9. In addition, nuclear magnetic resonance (NMR) has also been used to track 13C-labeled carbons in targeted molecules in microorganisms10, and software such as SUMOFLUX, 13C-FLUX211,12, X13CMS or Omix for GC–MS and LC–MS data13,14 has been used for tracking 13C fluxes. Here, we show a targeted tandem mass spectrometry (LC–MS/MS)-based approach for analysing 13C or 15N incorporation into polar metabolites. This protocol represents an extension of a previously published protocol for targeted steady-state metabolomics of unlabeled species that now includes specific methods for tracking the fate of 13C and 15N from glucose and glutamine15. Our platform utilizes SRM with polarity switching and amide HILIC on a hybrid triple quadrupole MS to capture transitions reflecting the incorporation of heavy atoms. This technology is fast, reproducible and robust for studying large numbers of research and clinically stable isotope-labeled metabolite samples. The platform can be customized to target any forms of heavy carbon or nitrogen incorporation into metabolites, provided that the Q1 precursor ion and Q3 fragment ion are adjusted to account for the labeled species. The benefit of using SRM versus untargeted high-resolution LC–MS/MS approaches is improved sensitivity and molecule coverage for specific molecules and pathways of interest. This technique isolates specific unlabeled (12C) and 13C (or 15N) isotopomers away from the molecular background, thus improving signal-to-noise ratio and dynamic range.
In this protocol, we address dynamic changes in metabolism in response to drug treatments or nutrient changes. Therefore,, the 13C/15N labelling sources are added minutes to hours prior to harvesting cells in order to probe how central carbon metabolism and related pathways are altered in response to a stimulus or to probe how tumour cells utilize metabolism compared to healthy cells. We have used this targeted triple quadrupole technology to track 13C or 15N metabolic flux across central carbon metabolism in >30 articles to date, with many in major peer-reviewed journals, including Science, Cell, and Nature.16-27. Noteworthy examples discussed in more detail in the Anticipated results section include the application of this technology to discern the metabolic reprogramming of the tricarboxylic acid (TCA) cycle in pancreatic cancer16,20,26 and the regulation of pyrimidine and purine synthesis by the mammalian target of rapamycin (mTOR)17,18. The entire Procedure does not add a substantial amount of time to our previously published protocol (Yuan et. al.)15 and can be used in cell culture to study the intra- and extracellular metabolome, for in vivo tissue samples, and for bodily fluids from isotopically labeled animals.
For some SRM transitions that involve labeled versions of metabolites, it is possible that the inherent biological sample contains an unlabeled and unrelated molecule that may mimic the labeled SRM transition, resulting in a false-positive signal, known as an isobaric peak. This situation Usually, reveals itself when an unlabeled sample is analysed using a 13C/15N tracer method. In some cases, the signal can be baseline-subtracted from the labeled sample on the basis of the unlabeled control. In other cases, if the Q1 precursor ion is altered by only 1 mass unit in the 13C or 15N flux tracer method, but the Q3 fragment ion does not change (i.e., phosphate loss), there can be a small degree of signal bleed due to peak width isolation.
This protocol focuses on targeted LC–MS/MS-based flux from the most common 13C- or 15N-labeled energy sources, such as glucose and glutamine. The number and location of labeled carbon or nitrogen atoms per molecule are dependent upon the specific flux questions that need to be addressed in the experimental setup. Common labelling patterns for flux experiments utilize either fully labeled glucose (13C6-U-glucose) or fully labeled glutamine (13C5-U-glutamine), as well as two-carbon-labeled glucose (13C1,2-glucose) and one-nitrogen-labeled glutamine in either the amide or amine positions (15N1-amide-glutamine or 15N1-amine-glutamine). Targeting 13C or 15N atom incorporation yields various isotopologs for each metabolite, and these data can be used to obtain flux information (Fig. 1). We have used this platform for analysing stable isotope tracer flux in a variety of cell types with a variety of labeled energy sources16-25.
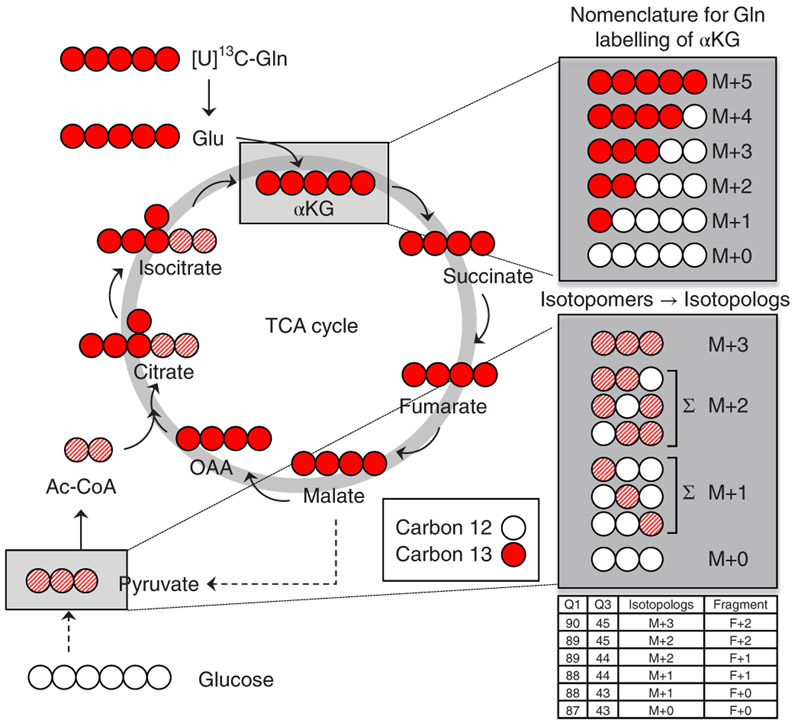
Fig. 1 ∣. Isotopomer depiction of 13C-glutamine incorporation into the TCA cycle.
Schematic example of uniformly 13C-labeled glutamine ([U]13C-Gln) metabolism in the TCA cycle. Labeled carbons are depicted in red; unlabeled carbons are white. Gln carbon incorporation into alpha-ketoglutarate (αKG) during the first turn of the TCA cycle results in fully labeled αKG (M+5). Successive turns contribute other carbon sources to the TCA cycle, resulting in partial labelling. The full spectrum of labeled positions is denoted in the box at the top right. Gln-labeled malate can leave the mitochondria, where it is converted into pyruvate. This can equilibrate with glucose-derived pyruvate and the resulting mixed label is depicted as red hash marks. The mixed labelling can exist in several forms. Mixed labelling within a pyruvate isotopolog (e.g., M+2) can exist as three possible isotopomers. Using the SRM protocol, each isotopomer is identified individually and the ion currents are summed to obtain the labelling data for the isotopolog. M+, mass plus.
Experimental design
When performing metabolic flux experiments, it is important to also include unlabeled (12C or 14N) control samples. These are used to eliminate signals in 13C SRM targets that may be due to unrelated molecules that coincidentally have the same parent mass, fragment ion mass and elution time of a labeled molecule. The unlabeled control also provides a true reference value for the peak area intensity for each targeted molecule because the signal is not distributed among isotopologs. We also suggest that one includes three (or more) biological replicates per experimental condition in order to calculate the statistical significance of signal changes across samples. We also advise injection of a water or methanol blank sample between unique sample sets in order to eliminate the possibility of signal carryover from, e.g., the column or the injector. Although this is rare, it can occur if metabolite samples contain a very large quantity of material (e.g., >4–5 million cells, >400 μL of serum or plasma, or >15 mg of tissue).
Materials
▲ CRITICAL The Reagents and Equipment lists are similar to those in Yuan et al.15 (https://www.nature.com/articles/nprot.2012.024).
Biological materials
Cultured cells: We recommend using ≥1–2 million cultured cells at 70–80% confluence, ~5–10 mg of solid tissue (tumours, organs) or ~50–200 μL of bodily fluids (serum, urine, plasma, cerebrospinal fluid (CSF)) for performing both 80% (vol/vol) methanol and methyl tert-butyl ether (MTBE) extractions. ! CAUTION If stable cell lines are to be used in your research, they should be regularly checked to ensure that they are authentic and that they are not infected with mycoplasma.
Animals: Any animals (e.g., mice) can be used for in vivo 13C/15N labelling via i.p. or tail-vein injections, or oral gavage. ! CAUTION All experiments involving animals must be approved by the relevant animal care and use committee and adhere to local and national regulations.
Reagents
▲ CRITICAL Alternative 13C and 15N molecules can be purchased for customized studies.
d-Glucose-13C6 (≥99% atom) (Cambridge Isotope Laboratories, cat. no. CLM-1396 (250-mg to 2-g quantities); MilliporeSigma, cat. no. 389374 (250-mg to 25-g quantities))
d-Glucose-13C1 (98% atom) (Cambridge Isotope Laboratories, cat. no. CLM-420-PK (250-mg to 10-g quantities); MilliporeSigma, cat. no. 297046 (250-mg to 10-g quantities))
l-Glutamine-13C5 (98% atom) (Cambridge Isotope Laboratories, cat. no. CLM-1822-H (100- to 500-mg quantities); MilliporeSigma, cat. no. 605166 (100- to 500-mg quantities))
l-Glutamine-15N2 (98% atom) (Cambridge Isotope Laboratories, cat. no. NLM-1328 (250-mg quantity); MilliporeSigma, cat. no. 490032 (100-mg quantity))
l-Glutamine-(amide-15N) (98% atom) (Cambridge Isotope Laboratories, cat. no. NLM-557-PK (500-mg to 1-g quantities); MilliporeSigma, cat. no. 490024 (100-mg quantity))
l-Glutamine-(amine-15N) (98% atom) (Cambridge Isotope Laboratories, cat. no. NLM-1016-PK (100-mg to 1-g quantities); MilliporeSigma, cat. no. 486809 (100-mg quantity))
Methyl tert-butyl ether (MTBE; anhydrous 99.8%; MilliporeSigma, cat. no. 306975-1L)
LC/MS-grade water (Fisher Scientific, cat. no. MWX00011)
LC/MS-grade acetonitrile (Fisher Scientific, cat. no. AC61514-0025)
HPLC-grade methanol (Fisher Scientific, cat. no. AC61009-0040)
Ammonium hydroxide, 25% (wt/vol) solution, puriss grade (Fisher Scientific, cat. no. 1336-21-6)
Ammonium acetate crystals, puriss grade (Fisher Scientific, cat. no. 09688)
Equipment
Mass spectrometer: We advise using a triple quadrupole–style mass spectrometer that is capable of positive/negative polarity switching, such as a 5500 QTRAP or 6500 QTRAP (AB/SCIEX) or a TSQ Quantum or TSQ Quantiva (Thermo Fisher Scientific)
HPLC system: For chromatography, an HPLC system capable of flow rates up to 1 mL/min is also needed, such as a Prominence UFLC (Shimadzu) or a 1200 HPLC (Agilent Technologies)
SpeedVac concentrator or lyophilizer for drying metabolite supernatants to a pellet (Thermo Fisher Scientific or other vendor)
1.5-mL polypropylene microcentrifuge tubes, (Fisher Scientific, cat. no. 05-408-129)
2.0-mL polypropylene microcentrifuge tubes, (Fisher Scientific, cat. no. 02-682-558)
12 x 32-mm polypropylene autosampler vials with caps (National Scientific, cat. no. C4000-87)
Small pestle or tissue grinder for use with 1.5- and 2.0-mL microcentrifuge tubes
Software
Analyst v.1.6.2 (https://sciex.com/products/software/analyst-software); used for acquisition and for setting up SRM transition methods for QTRAP instrumentation
MultiQuant v.3.0 or higher (https://sciex.com/products/software/multiquant-software); used for peak area integration of the Q3 signals
Microsoft Excel 2010 or later (https://products.office.com/en-US/compare-all-microsoft-office-products-b?tab=1&OCID=AID737190_SEM_IpiPmp7w&lnkd=Google_O365SMB_Mixed&gclid=EAIaIQobChMIr6PSkemV3wIVCl8NCh0sNQwFEAAYASAAEgKdsfD_BwE); used for data analysis
MetaboAnalyst v.4.0 software (http://www.MetaboAnalyst.ca); used for data analysis (e.g., normalization, heat maps, clustergrams, pathway enrichment)
SUMOFLUX (https://omictools.com/sumoflux-tool), Omix Visualization (https://www.omix-visualization.com/index.jsp#sthash.UXgEqfAI.dpbs), or 13C-FLUX2 (https://www.13cflux.net/13cflux2/); used for calculating 13C flux in known pathways
Cytoscape v.3.5.1 or later (https://cytoscape.org/); used for visualization of data and pathway information.
Reagent setup
Medium preparation
It is recommended that the representative base medium (e.g., DMEM or RPMI) be purchased without the nutrient being supplemented. For example, to trace 13C6-glucose, glucose-free base medium is preferred, and is supplemented with dialyzed serum (e.g., 10% (vol/vol)) per standard cell growth medium formulations. In doing so, the glucose-derived signal arising from the labeled molecule is enriched. The concentration of the label should be added at the typical final concentration of the non-labeled nutrient. In this example, 25 mM 13C6-glucose can be used in place of 25 mM glucose (4.5 g/L for DMEM), or 4 mM 13C5-glutamine can be used in place of 4 mM glutamine (for DMEM).
80% (vol/vol) methanol
To prepare the 80% (vol/vol) methanol solution for metabolite extraction, add 40 mL of HPLC-grade methanol to a 50-mL polypropylene conical tube, followed by 10 mL of LC/MS-grade water, then cap and gently shake to mix. Store in a −80 °C freezer for 4–6 h for further use. Note that methanol can be replaced or combined with acetonitrile to an 80% total organic concentration for metabolite extractions, although methanol is preferred for intracellular metabolites and most applications
HPLC buffer A
HPLC buffer A is (pH 9.0: 95% (vol/vol) water, 5% (vol/vol) acetonitrile, 20 mM ammonium hydroxide, 20 mM ammonium acetate). To prepare HPLC buffer A, add 1.54 g of ammonium acetate to a 1-L HPLC bottle, and then add 950 μL of LC/MS-grade water and gently stir by rotating the bottle. Next, add 2.8 mL of 25% (wt/vol) ammonium hydroxide solution, followed by 50 mL of HPLC-grade acetonitrile. Cap the bottle and mix by gently shaking. Check the pH value to ensure it is at 9.0.
HPLC buffer B
HPLC buffer B is 100% acetonitrile. To prepare HPLC buffer B, add 1 L of LC/MS-grade acetonitrile to a 1-L HPLC bottle and cap the bottle.
Equipment setup
QTRAP mass spectrometer with polarity switching
Use the following settings for the QTRAP mass spectrometer: +4,800 V positive/−4,500 V negative; 475 °C; curtain N2 gas set to 25; high collision energy; ion source gas 1 and 2 set to 35; declustering potential +93 positive/−93 negative; entrance potential +10 positive/−10 negative; collision cell exit potential +10 positive/−10 negative.
HPLC system
The buffers, column and gradient conditions can be reproduced from those in Yuan et al.15; these include a HILIC (4.6-mm column, Amide XBridge, Waters) with ammonium hydroxide buffers at pH 9.0 over an ~26-min targeted acquisition. Buffer A is 95% (vol/vol) water, 5% (vol/vol) acetonitrile, 20 mM ammonium hydroxide, and 20 mM ammonium acetate; buffer B is 100% (vol/vol) acetonitrile; gradient conditions are as follows.
| Time (min) | %B | Flow rate (μL/min) |
|---|---|---|
| 0.0 | 85 | 400 |
| 3.5 | 32 | 400 |
| 12.0 | 2 | 400 |
| 16.5 | 2 | 400 |
| 17.0 | 85 | 400 |
| 25.5 | 85 | 400 |
Software
Several of these tools, including Cytoscape and Omix Visualization, require the user to first calculate 13C/12C flux ratios in Excel before mapping over known or custom metabolic pathways for flux visualization.
Procedure
Metabolite extraction
-
Metabolites can be extracted from a variety of different biological sources; in some cases, there are multiple options for extraction. Although the traditional approach uses 80% (vol/vol) methanol for extraction15, one can use MTBE/methanol and collect the middle polar layer for polar metabolomics analysis28,29. The benefit of MTBE extraction is the ability to collect labeled nonpolar lipids (top layer) for labeled lipid flux analysis.
This section also includes the addition of the isotopically labeled reagent. The nature of the experiment should be used to determine the duration of exposure to the label. To gain an understanding of label incorporation at steady state, overnight labelling (~12–16 h) is typically sufficient. For flux analysis, the time series should be determined experimentally for each biological system. To track flux into a pathway using a 13C or 15N isotope label, cells can be prepared for a time course analysis. For example, to track glucose incorporation into glycolysis, time points on the order of minutes are suggested16. To track glutamine carbon incorporation into the TCA cycle, time points spanning ~4 h are recommended20. However, if one wishes to observe acute labelling effects in response to kinase inhibitor drugs, we advise priming the cells with the drug and then adding your labelling source (e.g., glucose or glutamine), followed by harvesting the cells at the time frames for the label indicated above30.
To extract polar metabolites from adherent cells using 80 %(vol/vol) MeOH/H2O, use option A; to extract polar metabolites from suspension cells, use option B; to extract metabolites from adherent cells using the three-phase system with MTBE, use option C; to extract metabolites from suspension cells using the three-phase system with MTBE, use option D; to extract polar metabolites from tissues, use option E; to extract metabolites from tissues with the three-phase system, use option F; to extract metabolites from liquid samples with MeOH, use option G; and to extract metabolites from liquid samples with MTBE, use option H.- Adherent cell sample preparation: polar metabolite extraction from isotopically labeled cells ● Timing ~2–3 h
-
Seed the cells on a 10-cm2 cell culture plate with unlabeled medium. The cells are grown to 75–80% confluence in log-phase in the cell culture plate.▲ CRITICAL STEP Prepare an extra plate or well from which proteins will be later collected; this will serve as a loading control.▲ CRITICAL STEP Recommendations for cell number per replicate range from ~105 to 106, depending on the cell type. As an example, cancer cells tend to be much larger than lymphocytes, and using ~10× of the latter is recommended. We also recommend a minimum of three biological replicates, although four is preferred.
- After the cells have reached the desired confluence, remove the unlabeled medium, wash the cells using ~3 mL of 1× PBS and aspirate the wash volume.
-
Replace the medium with an equal volume of the 13C/15N isotopic tracer–labeled medium (e.g., 25 mM 13C6 glucose or 4 mM 13C5 glutamine). For example, 10 mL of medium is used for cells in a 10-cm2 dish, and 2 mL is used for cells in a well of a 6-well dish. Incubate the cells with labeled medium depending on the nature of the experimental inquiry, as described above.▲ CRITICAL STEP For some experiments, including drug treatments, pre-treatment of the cells with the small-molecule drug is preferred in order to induce an acute effect on the cells (<2 h) before the 13C/15N labelling procedure.
-
When the desired time point has been reached, remove the cells from the incubator and aspirate the medium completely. This can be performed on a benchtop (i.e., non-sterile).▲ CRITICAL STEP Collect the proteins from the loading control well described in Step 1A (i), using standard lysate preparation procedures. Calculate protein concentration and apply this for metabolite loading in Step 1A(xi).
- Place the plates on dry ice and add 4 mL of 80% (vol/vol) methanol (cooled to −80 °C) for 10 cm2 dishes or 1 mL/well of a 6-well dish.
- Incubate the plates at −80 °C for 20 min.
- Scrape the entirety of the contents of the plate or well and transfer to a 15-mL conical tube.
- (viii) Vortex for 1 min at 4–8 °C, and then incubate for 30 min at −80 °C.
- Centrifuge at >4,000g for 10 min (at 4–8 °C) to precipitate the insoluble material.
- Transfer the supernatant to a new 15-mL conical tube.
-
Make aliquots of the supernatant that account for the protein concentration of the experimental samples (see the CRITICAL STEP note below) in 1.5-mL microcentrifuge tubes. Typically this is on the order of ~1 mL.▲ CRITICAL STEP The protein concentration of the protein extract from the additional biological replicate isolated in Step 1A(iv) should now be calculated. This can be done by BCA or Bradford assay. This will serve as a scaling factor for normalization because sample source protein amounts scale with metabolite amounts. MS is a quantitative analytical procedure, meaning that the amount of sample loaded onto the instrument will be proportional to the output. Normalization of sample loading is especially important in instances in which an experimental variable may affect the growth/proliferation of one group.■ PAUSE POINT The samples can be stored at −80 °C for up to 6 months, although they should be used within 4–8 weeks for optimal reliability.
-
SpeedVac/lyophilize to a pellet in 1.5-mL microcentrifuge tubes, using no heat. A nitrogen EVAP system can also be used to dry the samples.■ PAUSE POINT The samples can be stored at −80 °C for <6 months.
-
- Suspension cell sample preparation ● Timing ~2–3 h
- Transfer the medium with the cells to a 15-mL conical tube, and spin at 750g for 5 min at 4 °C.
- Completely remove the medium, add 4 mL of 80% (vol/vol) methanol (−80 °C), vortex to stir the cells and incubate for 20 min at −80 °C.
- Continue as described in Step 1A(vii–xii).
-
Extraction of metabolites from adherent cells using MTBE ● Timing ~2–3 h▲ CRITICAL One can use MTBE to collect the middle polar layer for metabolomics analysis4,12,13,29. The benefit of MTBE extraction is that it provides the additional ability to collect labeled nonpolar lipids (top layer) for labeled lipid flux analysis.
- Perform Step 1A(i) and (ii).
- Harvest the cells from the plates at room temperature (RT; ~23 °C) and transfer to a 12-mL glass vial.
- Spin for 5 min at 1,000g (microcentrifuge, RT) and remove the supernatant.
- Add 200 μL of 1× PBS to the vial.
- Add 1.5 mL of HPLC-grade methanol and vortex for 1 min.
- Add 5 mL of MTBE and rock for 1 h at RT.
- Add 1.2 mL of water and vortex again for 1 min.
- (viii) Spin for 10 min at 1,000g (microcentrifuge, RT).
- Collect the upper MTBE liquid phase containing the nonpolar lipids.
- Re-extract the lower liquid phase with two volumes of MTBE/methanol/water (10:3:2.5, vol/vol/vol) by repeating Step 1C(v–ix).
-
Dry the combined MTBE phases in a SpeedVac or under a nitrogen stream.■ PAUSE POINT Dried metabolite samples can be stored at −80 °C for <6 months.
- MTBE extraction for suspension cell sample preparation ● Timing ~2–3 h
- Transfer the medium with the cells to a 15-mL conical tube, and spin at 750g for 5 min at 4 °C.
- Completely remove the medium, and proceed as described in Step 1C(iv–xi).
-
Metabolite extraction from tissues, organs and tumours of isotopically labeled animals ● Timing ~2–3 h▲ CRITICAL The procedure below will provide information about how, and the specific pathways into which, a labeled nutrient incorporates in vivo. For example, using 13C6-glucose, one can determine if the cells in a tissue/organ/tumour incorporate glucose label into various metabolic pathways. This method relies on a single i.p. bolus injection of an isotopically labeled metabolite and is limited to assessing the degree of incorporation. Accordingly, one tissue may experience a different exposure than another tissue, or looking across experimental animals, a tumour in one animal may experience more labelling than another. To accurately compare the degree of labelling, steady-state incorporation of the label must be achieved and experimentally validated31. The extraction and analysis method described below can also be applied to tissues that have been labeled to steady state. Finally, care should be taken when interpreting data from in vivo–labeled animals because the nutrients are readily processed by the liver and then re-centre circulation, where the tissue/organ/tumour in question can access the processed nutrients. The steps below use the single-bolus glucose labelling strategy to assess the degree of incorporation, as an example.
- For glucose, inject mice i.p. with a 13C6-glucose solution (2 g/kg in PBS). Numerous other protocols exist for in vivo labelling studies, including tail-vein injection and jugular-vein injection, plus or minus the application of an osmotic pump31-34. To assess label incorporation, we advise the use of i.p. injection, as it is the simplest of the above-noted protocols. (Labeled glutamine can be applied similarly with a 10 mg/kg 13C5-glutamine solution in PBS35.)
-
Use a glucometer to monitor blood glucose at 15- to 30-min intervals, and sacrifice the mice when blood glucose levels begin to decline36. Harvest the tissues/organs/tumours for metabolite extraction, as follows.Metabolite sample preparation: polar metabolite extraction from isotopically labeled samples
- Weigh the tissue/organ/tumour on a sensitive analytical balance to ±0.1 mg. Record the weight. This will be used later when normalizing sample loading.
-
Add the tissue/organ/tumour to a 1.5-mL or 2.0-mL round-bottom microcentrifuge tube containing 1 mL of 80% (vol/vol) HPLC-grade methanol (cooled to −80 °C) and a small steel ball bearing. ~10–20 mg of tissue/organ/tumour can be readily processed in this manner.▲ CRITICAL STEP Conical-bottom microcentrifuge tubes may crack using this protocol, resulting in loss of contents. Round-bottom microcentrifuge tubes are highly recommended.
- Place the tubes into a TissueRupter (Qiagen) or similar device. Apply the tissue-ruption program for 30 s pulses at 26 Hz, cooling on dry ice between cycles, until the tissue/organ/tumour is fully dissociated.
- Centrifuge at 14,000g (or the highest speed) for 10 min using a refrigerated centrifuge (4–8 °C).
- Transfer the supernatant to a new 1.5-mL microcentrifuge tube.
-
Repeat the centrifugation and supernatant transfer.▲ CRITICAL STEP Ball bearings can be washed and reused.■ PAUSE POINT Supernatants can be stored at −80 °C for up to 6 months.
-
Aliquot the equivalent of 10 mg of supernatant into a new 1.5-mL microcentrifuge tube and dry using SpeedVac/lyophilization or nitrogen gas.■ PAUSE POINT Dried, labeled metabolite samples can be stored at −80°C for <1 year.
- MTBE extraction of tissues
- Add 200 μL of 1× PBS and disrupt the tissue, as in Step 1E(v).
- Perform the MTBE extraction in the same way as you would for adherent cells; i.e., follow Step 1C(v–xi).
- Extraction from bodily fluids (serum/urine/CSF/cell culture medium)
- Obtain ~100–250 μL of the fluid.
- Centrifuge at 14,000g for 10 min at 4–8 °C to remove contaminating cells.
- Transfer the supernatant to a new 1.5-mL microcentrifuge tube.
- Add enough methanol (cooled to −80 °C) to the supernatant to make a final 80% (vol/vol) methanol solution.
- Gently shake to mix and incubate for 20 min at −80 °C.
-
Follow Step 1A(viii–xii) as in the adherent cell extraction procedure.■ PAUSE POINT Dried metabolite samples can be stored at −80 °C for <1 year.
- MTBE extraction of liquid (blood/urine/CSF) samples
- For liquid samples of 200–250 μL, follow Step 1C(v–xi).
- For liquid samples of <200 μL, add enough 1× PBS buffer to obtain a 200-μL volume; then follow Step 1C(v–xi).
LC–MS/MS acquisition
-
2
Set up the MS method as follows: modify the Q1/Q3 SRM transition list to accommodate the 13C or 15N labeled versions of each metabolite for both the increase of C or N atom mass for the precursor ion (Q1) and the fragment ion (Q3) in each polarity mode. In many cases, one will create all possible options for 13C or 15N incorporation for isotopomer/isotopolog analysis. Representative methods for glucose and glutamine tracing are provided as Supplementary Datasets. For example, to measure 13C-glucose incorporation into glucose 6-phosphate (G6P), the mass of the deprotonated Q1 precursor ion is 259 Da (C6H12O9P) in negative ion mode and that of the Q3 fragment ion is 199 Da (C6H12O6). Both the Q1 and the Q3 ions contain six carbon atoms (product ion, loss of PO3−). So, if one wants to analyse fully 13C labeled G6P (referred to as mass plus 6, M+6), 6 Da must be added to the Q1 and Q3 transitions to form 265/205. In addition, if one chooses to analyse the isotopologs of G6P, a transition for each carbon atom needs to be made, i.e., 259/199 (M+0, unlabeled), 260/200 (M+1), 261/201 (M+2), 262/202 (M+3), 263/203 (M+4) and 264/204 (M+5).
-
3
Inject ~5 μL of each metabolite sample from the sample list via the autosampler after resuspension in ~20–25 μL of LC–MS-grade water, using a targeted acquisition method defined in the Analyst software in Step 2.
-
4
Using analysis and peak integration software such as MultiQuant with SCIEX instrumentation, look for 13C/15N peaks in the mass spectra in addition to the unlabeled 12C peaks.
▲ CRITICAL STEP When creating SRM transition lists for the QTRAP 5500, it is important to keep the total number of SRM transitions <300 (~150 per polarity mode) because scheduled SRM is not used. By doing so, it ensures that the combined total cycle time will be <~1.5 s, resulting in at least 10 data points per peak for a standard 30-min LC–MS/MS run time per sample with amide HILIC. Of note, each isotopomer transition counts against the total ~300 transitions targeted per run, and thus, inclusion of more isotopomers comes at the expense of increased total metabolite coverage. Since the scan speed is higher on the QTRAP 6500, the total number of SRM transitions can be increased to ~350–375 without scheduling.
▲ CRITICAL STEP For some metabolites, multiple isotopomer patterns are possible for a single isotopolog, depending upon which carbon atoms are labeled. In this case, the ion current for each isotopomer for a given isotopolog is summed to obtain the total ion current for the individual isotopolog. The only substantial change to the original method from 2012 is the application of the new set of SRM transitions for the 13C labeled carbon atoms or 15N labeled nitrogen atoms. Supplementary Datasets 1-5 contain five different methods for SRM-based tracer flux profiling. Supplementary Dataset 1 is for 13C isotopomer analysis focusing on glycolysis, the TCA cycle, the pentose phosphate pathway (PPP) and nucleotides; Supplementary Dataset 2 is for 13C fully labeled metabolite analysis from glucose; Supplementary Dataset 3 is for 13C fully labeled metabolite analysis from glutamine; Supplementary Dataset 4 is for 15N labelling from glutamine (amide) and Supplementary Dataset 5 is for 15N labelling from glutamine in the amine position. These Supplementary Datasets contain the necessary information needed for creating customized SRM methods in both positive and negative ion modes and can be downloaded and incorporated into Analyst (SCIEX) software or modified for other vendor software.
▲ CRITICAL STEP In addition to using the protein concentration from a parallel sample for cells, the wet weight for tissues/organs/tumours, or the volume for bodily fluids for normalization (see above), one can also use the median or average peak area values from each sample in resulting datasets to normalize instrument loading across sample conditions.
-
5
As an alternative to Step 4, it is possible to run high-resolution LC–MS/MS using a mass spectrometer capable of high mass accuracy in both MS and MS/MS mode with polarity switching, e.g., a QExactive Orbitrap in data-dependent acquisition (DDA) mode. However, this approach does not involve setting Q1/Q3 transitions. If this approach is chosen, reconstruct the parent ion and fragment ion peak areas from the full fragmentation profile.
? TROUBLESHOOTING
Troubleshooting
Metabolite peaks are uniformly higher/lower in some samples.
-
Loading inconsistency can be alleviated by careful attention to protein concentration (for cells in culture), wet weight (for tissues/organs/tumours) and volume (for biological fluids) across replicates/samples. Unfortunately,, once run, inconsistent loading is not readily corrected by computational approaches. It is recommended that the lyophilized sample volume be recalculated and the samples be re-prepared and re-run.
The mass spectrometer is not showing any Q1/Q3 transition peaks above baseline for 13C isotopomer transitions.
This would indicate either that not enough metabolite content is on the HPLC column during the acquisition or, more likely, that the flux of 13C atoms into specific Q1/Q3 transitions is below detectable levels. Check the levels of HPLC buffers, and collision and source gases, and that the column or tubing is not leaking.
Check that the instrument is well-calibrated in both positive and negative ion modes.
Check whether the parent ion can be detected in the unlabeled sample.
Check how many cells or how much tissue was used. It is possible that more sample is required (equivalent of >2 million cells).
-
The ion source area has a tendency to get very dirty under the conditions described in this protocol, hence, it will be necessary to clean the ion source more frequently.
The Q1/Q3 transitions for 13C isotopomers are displaying peaks at a different retention time from that of the 12C unlabeled version.
-
The peaks at altered retention times are not the same molecule and cannot be integrated as such. 13C or 15N stable isotopes are chemically identical to the unlabeled forms, and there should be no shift in retention time.
The metabolite peaks either are not being retained or are eluting very late on the HPLC column.
Check that the HPLC buffer concentrations and pH are correct; and make fresh solutions if necessary.
Check that the HPLC A and B pumps are properly purged to remove air.
-
If the buffers are good and the pumps are properly purged and the problem persists, it may indicate that either your pre-column filter is clogged or the HPLC column has been contaminated or has undergone passivation. First, try changing the pre-column filter, and if this does not work, change the analytical column. After making any changes to the pre-column or analytical column, it is important to run a series of standards of known concentration for the metabolites of interest, to re-establish their retention times.
Some metabolites are never detected, no matter what changes are done or what concentrations are injected.
It is possible that the metabolite Q1/Q3 values are not optimal or are incorrect, or that the collision energy selected is not ideal for the selected fragmentation event.
The particular metabolite of interest may be present only at very low concentrations in your sample. It may also degrade quickly, either in vivo or after extraction. For example, -CoA derivatives degrade quickly and sulfhydryl-bearing molecules are readily oxidized, which make these metabolites more difficult to detect than many other metabolites.
It is also possible that methanol and MTBE are not ideal solvents for the metabolite extraction, or that the chromatography used is not suitable for the compound of interest.
Timing
Step 1, from tissue/cells/fluid collection to sample injection of metabolite extracts: ~4.5–6 h, mainly depending upon the drying speed of the SpeedVac/lyophilizer
Steps 2 and 3, total LC–MS/MS run time: 25.5 min (16.5-min MS acquisition) from injection to injection
Step 4, peak integration and verification of raw data: ~1.0 h per dataset; data analysis using various bioinformatics software tools: variable, depending on the degree of interpretation, ~1.0–2.5 h per dataset
Anticipated results
The targeted SRM LC–MS/MS method should be selected on the basis of the biological question (Fig. 2). We have included methods for the profiling of >140 fully labeled metabolites and isotopolog analysis of up to 40–50 unique metabolites (Supplementary Datasets 1-5). The fully labeled methods target the unlabeled and fully labeled metabolites, whereas the isotopolog method will reveal all incremental forms of 13C or 15N labelling (Fig. 1). The output will be a peak area array of all detected metabolite forms from each tested sample in a .txt or Excel format similar to the output from an unlabeled profiling experiment. We advise that pathways of interest be plotted using Excel and more global bioinformatics analyses, including heat maps and PCA, which can be obtained using free online MetaboAnalyst 4.0 software (https://www.metaboanalyst.ca/). One can also run untargeted metabolomics of isotope-labeled samples with high-resolution tandem MS; however, the data will need to be interpreted manually or 13C/15N labelling software must be used37.
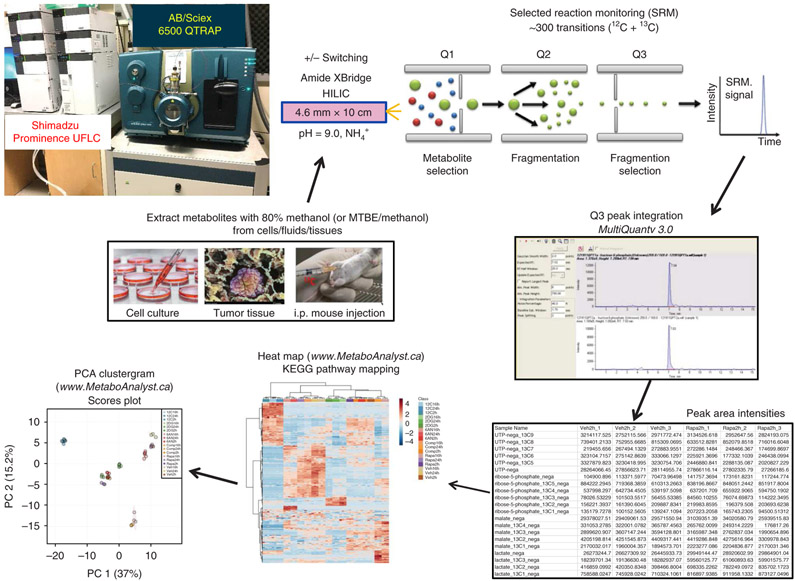
Fig. 2 ∣. Targeted 13C metabolomics workflow.
Platform for targeted SRM for 13C-labeled polar metabolites, from metabolite extraction to LC–MS/MS through peak integration and data analysis. PCA, principal component analysis.
Figure 2 shows the platform that is used for both unlabeled (12C or 14N) and labeled (13C or 15N) metabolites. To achieve a minimum of 10 data points per peak, the total SRM transition list is kept below ~300 transitions because scheduled SRM is not used. After the Q3 signals are integrated, an array of peak areas are available, representing each labelling form, which serve as the output for further analysis.
In Fig. 1, an example of 13C5-labeled glutamine (also often referred to as uniformly labeled [U]13C) incorporation into the TCA cycle is presented schematically. Alpha-ketoglutarate (αKG) is used in this example to illustrate isotopolog-labelling nomenclature. Labelling of pyruvate is used to show how isotopomer-labelling patterns can be obtained and consolidated to find the isotopolog distribution. The white circles indicate carbon-12 (12C) atoms from glucose via glycolysis, and the red circles indicate 13C atoms from glutamine via the TCA cycle. The circles containing white and red stripes indicate carbon atoms that can have contribution from 12C via glucose/glycolysis or 13C via glutamine/TCA cycle. The data in Fig. 1 show that pyruvate and acetyl-coA contain carbon atoms from both 12C and 13C energy sources. Further, two of the six carbon atoms on citrate and isocitrate are labeled from either glycolysis (12C-glucose) or the TCA cycle (13C-glutamine) (hashed circles), whereas four carbon atoms in citrate/isocitrate are derived from oxidative 13C-glutamine metabolism in the TCA cycle. In the example in Fig. 1, αKG is shown with all five fully labeled with 13C from glutamine.
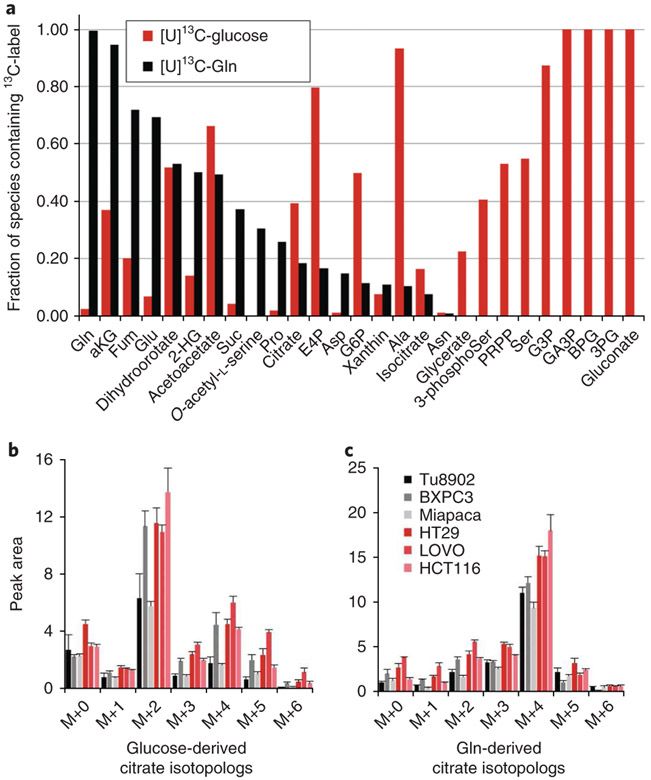
In Fig. 3 (top), this concept is applied in pancreatic cancer cells by tracing either U13C-glucose or U13C-glutamine (in parallel experiments) to analyse 13C label incorporation across a spectrum of metabolites in central carbon metabolism. In Fig. 3b, isotopolog analysis is performed for citrate.
Fig. 3 ∣. Fractional labelling of central carbon metabolism in cancer cell lines with 13C-glucose and 13C-glutamine.
a, Bar plot of the fraction of 13C incorporated into a spectrum of metabolites from central carbon metabolism for both 13C6-glucose and 13C5-glutamine in murine pancreatic cancer cells. Glucose and glutamine labelling data are generated from parallel samples. b,c, The sums of all isotopologs containing label are presented as single bars. b, Isotopolog analysis of 13C6-glucose. c, 13C5-Gln incorporation into citrate across a panel of pancreatic cancer (shades of black) and colon cancer (shades of red) cell lines. Data are presented as ion current for samples normalized for loading by protein concentration in parallel samples. Gln, glutamine; Glu, glucose.
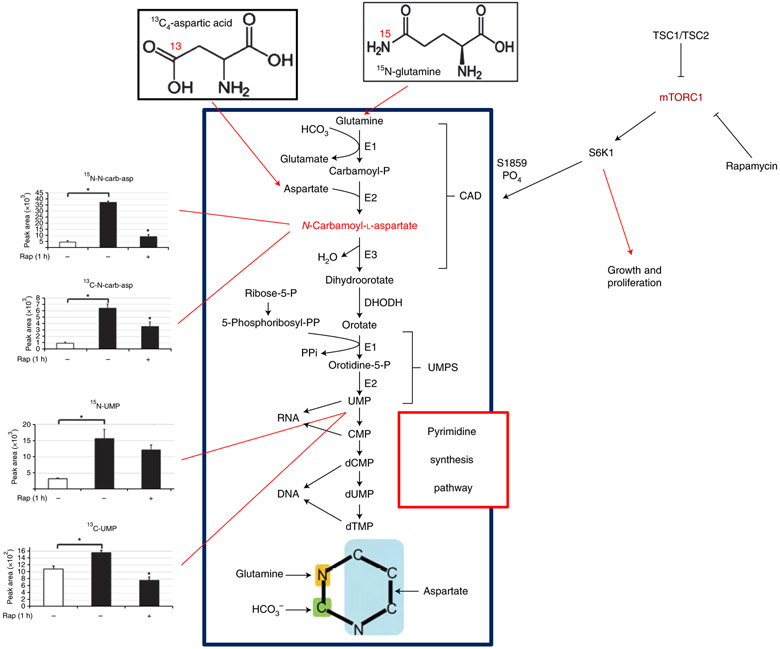
In addition to glucose and glutamine, it is possible to label cells with any molecule that contains heavy atoms involved in the metabolic pathways of interest. For example, using labelling of 15N-glutamine in the amide position, or using 13C4 aspartic acid, we found that mTORC1 increased cellular levels of N-carbamoyl-aspartate flux through the de novo pyrimidine synthesis pathway (Fig. 4)17. This was validated using 15N- and 13C-labeled N-carbamoyl-aspartate as well as 15N- and 13C-labeled uridine monophosphate (UMP). Furthermore, we also found that treatment with the TOR inhibitor rapamycin, a specific inhibitor of mTORC1 signalling, reduces the 13C/15N flux from labeled glutamine and aspartic acid to N-carbamoyl-aspartate and UMP, which suggests that the pyrimidine synthesis pathway is under the control of mTORC1. The data in Fig. 4 show that multiple labelling sources can be used to interrogate isotopomer flux through a pathway in order to determine the source of carbon or nitrogen flow from a specific molecule.
Fig. 4 ∣. Isotope labelling of de novo pyrimidine biosynthesis.
13C and 15N flux into nucleotides and precursors of the pyrimidine synthesis pathway from glutamine and aspartic acid in TSC2−/− cells are under mTORC1 control via CAD protein phosphorylation. In the graphs at left, white bars represent TSC2+/+ cells and black bars represent TSC2−/− cells. UMPS, uridine monophosphate synthetase. Parts of this figure reprinted with permission from Ben-Sahra et. al. 17, American Association for the Advancement of Science.
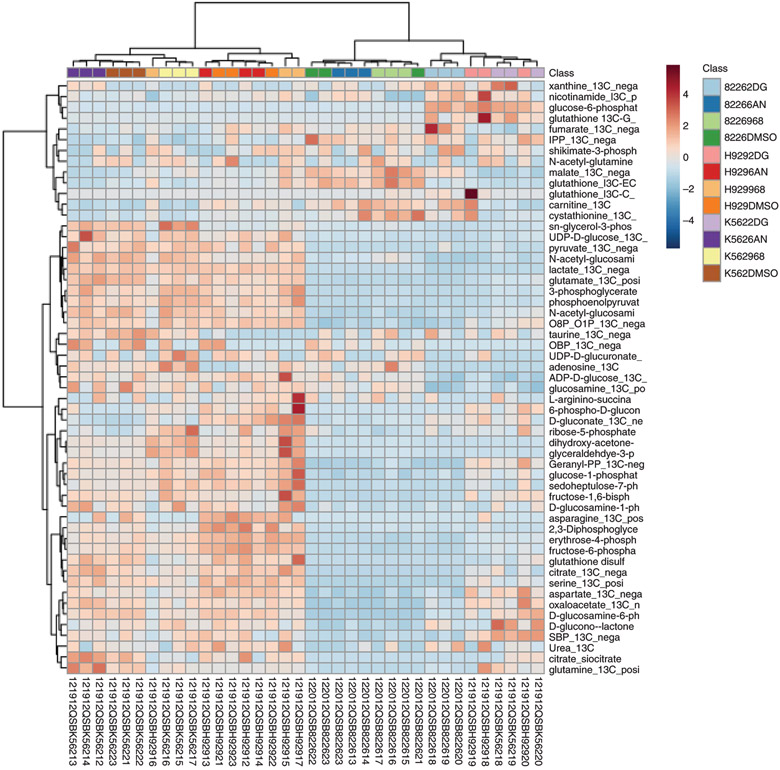
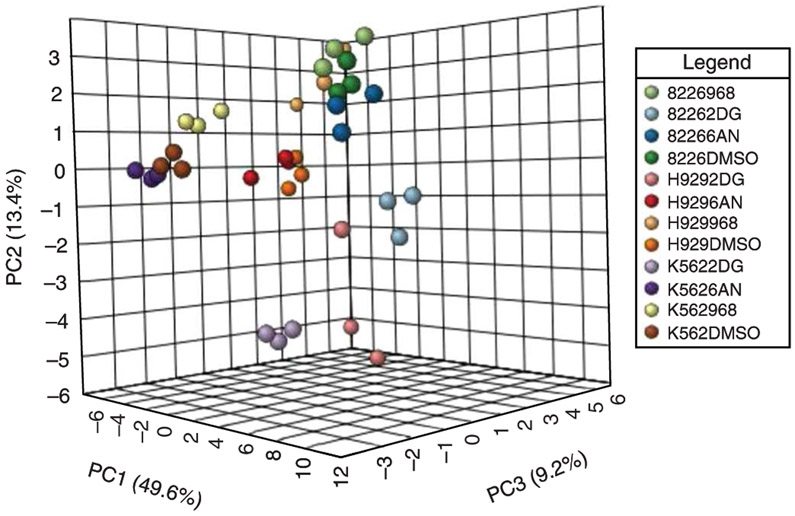
Beyond the targeted studies outlined in Figs. 1, 3, and 4, one can also analyse the stable isotope flux into various metabolites in a more global manner by using a method that targets both the unlabeled and fully labeled (all heavy labeled carbon or nitrogen atoms) versions of the metabolites of interest. Using H929 multiple myeloma cells labeled with 13C6-glucose and treated with drugs that block metabolic pathways, we targeted >125 unique metabolites. Figure 5 shows a Ward clustering heat map in 13C6-glucose-treated H929 and RPMI 8226 multiple myeloma cells and K562 chronic myeloid leukaemia cells that had been exposed to the metabolic inhibitors 2-deoxyglucose (2-DG, glycolysis inhibitor), 6-aminonicotinamide (6-AN, pentose phosphate pathway inhibitor) or compound 968 (glutaminase inhibitor)30,38 as compared to a vehicle control (DMSO). The cells were treated for 2 h with drugs and 13C label was applied at the time of drug treatment. The data show distinct effects across each cell line as well as with each drug treatment displaying the resolution that can be obtained by targeting both the unlabeled and fully labeled versions of central carbon metabolite intermediates. The data analysed in Fig. 5 can also be presented as a PCA clustergram (Fig. 6), which demonstrates the reproducibility across the experiment. Further, such an analysis can be useful for revealing features that give rise to distinct clusters. The metabolic effects of multiple myeloma cells as well as their relationship to proteomics, lipidomics and phosphoproteomics, was established for H929 cells recently30.
Fig. 5 ∣. Clustering of unlabeled (12C) metabolites and labeled (13C) isotopomers from targeted LC–MS/MS data.
Heat map illustration of both labeled (13C) and unlabeled (12C) metabolites from 13C6-glucose-labeled RPMI 8226 and H929 myeloma cells and K562 CML cells with metabolic inhibitors 2-deoxyglucose, 6-aminonicotinamide and compound 968 for the 55 most significant features using MetaboAnalyst software.
Fig. 6 ∣. Clustergrams of labeled cancer cells.
PCA clustergram for triplicate analyses of 13C6-glucose-labeled H929, RPMI 8226 and K562 cells treated with metabolic inhibitor treatments from MetaboAnalyst software.
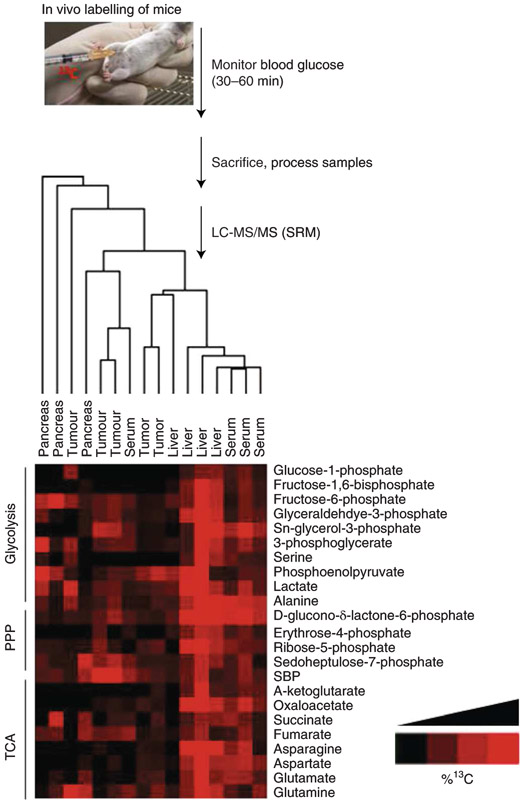
In addition to flux profiling of cells in culture, we have also traced metabolism in vivo by labelling mice via i.p. injection of a 10% (wt/vol) U-13C6-glucose solution prior to sacrificing mice for analysis of tissue and tumour labelling. As noted in the methods description above, this strategy is useful for determining the profile of nutrients that contribute to a given metabolic pathway. However, it cannot be used to compare the degree of labelling across tissues or animals. For such analyses, we recommend efforts to achieve steady-state label incorporation, as have been described31. Representative data for the degree of labelling and variability among animals are presented for tissues and tumours in Fig. 7. Furthermore, to achieve enrichment of glucose incorporation, we found that while monitoring blood glucose levels, and in particular, while waiting for glucose levels to fall, use of a hand-held metre is able to enhance the tissue/tumour enrichment. The optimal time after injection to maximize uptake into tissues before sacrifice and resection is approximately 30–60 min. The data show the highest level of 13C labelling enrichment in the liver and serum (Fig. 7). We have also explored other in vivo mouse systems, including triple-negative breast tumours and analysed the central carbon metabolite pathways, including glycolysis, the TCA cycle and the PPP following treatment with various kinase inhibition36.
Fig. 7 ∣. In vivo 13C/15N labelling for flux analysis.
Flowchart of protocol for in vivo 13C6-glucose mouse labelling by i.p. injection followed by targeted LC–MS/MS via SRM. PPP, pentose phosphate pathway; TCA, tricarboxylic acid.
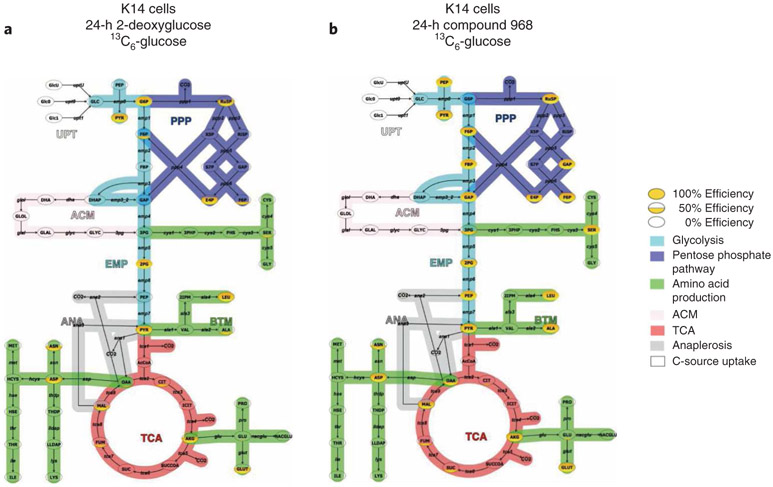
One can also map the 13C/12C flux ratio data onto known metabolic pathways after data manipulation in Excel using programs such as Omix Visualization. Figure 8 shows K14 triple-negative breast cancer cells that contain Brca1−/− and P53−/− mutations28. Cells were labeled overnight with 13C6-glucose and treated with 2-DG or compound 968 for 24 h. The solid yellow-filled metabolite bubbles represent a high percentage of 13C flux through the metabolic intermediate; whereas a partially filled or empty bubble represents less 13C flux through that molecule. The data show that 2-DG markedly reduces the flow of 13C from glucose (flux) through glycolysis and PPP (Fig. 8a) and that compound 968 does not inhibit 13C glucose flux through glycolysis and may compensate for TCA cycle inhibition by increasing flux through glycolysis and PPP in K14 cells (Fig. 8b). Note that 13C flux is not observed in high abundance in the TCA cycle from either drug treatment because 13C6-labeled glucose was used in this experiment rather than glutamine, which feeds the TCA cycle via glutamic acid. One can also customize one’s own pathway maps in order to customize one’s flux observations.
Fig. 8 ∣. Visualization of metabolic flux analyses with Omix Visualization.
a,b, Pathway map and flux of metabolite intermediates through glycolysis, TCA cycle and pentose phosphate pathway from K14 cells labeled with 13C6-glucose and treated for 24 h with the glycolysis inhibitor 2-deoxyglucose (2-DG) (a) and the TCA cycle inhibitor compound 968 (b).
Conclusion
These data demonstrate that a variety of 13C/15N isotopic labelling reagents (e.g., 13C-glucose, 13C-glutamine, 15N-glutamine) can be used in a variety of sample sources from cell lines to in vivo tumours and can be used with not only glucose and glutamine, but also with many metabolizable nutrients. The targeted isotopic metabolic fluxomic platform presented here can trace the fate of central carbon metabolites as well as their dynamic metabolic flux patterns in biological systems. We also showed that the platform can be applied to both in vitro cell/tissue/biofluid experiments and in vivo biological systems.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
This research was supported by grants from the National Institutes of Health (5P01CA120964 to J.M.A. and B.D.M.; 5R35CA197459 to B.D.M. and J.M.A.; and 5P30CA006516 to J.M.A.) and the BIDMC Research Capital Fund for funding the mass spectrometry instrumentation. C.A.L. was supported by a Pancreatic Cancer Action Network/AACR Pathway to Leadership award (13-70-25-LYSS), a Dale F. Frey Award for Breakthrough Scientists from the Damon Runyon Cancer Research Foundation (DFS-09-14), a Junior Scholar Award from The V Foundation for Cancer Research (V2016-009), a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research (SKF-16-005), and a 2017 AACR NextGen Grant for Transformative Cancer Research (17-20-01-LYSS). D.M.K was supported by a University of Michigan Program in Chemical Biology Graduate Assistance in Areas of National Need (GAANN) award. We thank L.C. Cantley (Weill-Cornell MC) and G.M. Wulf (BIDMC/HMS) for helpful discussions and the donation of K14 cells.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-018-0102-x.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jorda J et al. Quantitative metabolomics and instationary 13C-metabolic flux analysis reveals impact of recombinant protein production on trehalose and energy metabolism in Pichia pastoris. Metabolites 4, 281–299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metallo CM, Walther JL & Stephanopoulos G Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J. Biotechnol 144, 167–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nissim I et al. Effects of a glucokinase activator on hepatic intermediary metabolism: study with 13C-isotopomer-based metabolomics. Biochem. J 444, 537–551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zamboni N & Sauer U Novel biological insights through metabolomics and 13C-flux analysis. Curr. Opin. Microbiol 12, 553–558 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Wills J, Edwards-Hicks J & Finch AJ AssayR: a simple mass spectrometry software tool for targeted metabolic and stable isotope tracer analyses. Anal. Chem 89, 9616–9619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamboni N, Saghatelian A & Patti GJ Defining the metabolome: size, flux, and regulation. Mol. Cell 58, 699–706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamboni N, Fendt S-M, Rühl M & Sauer U 13C-based metabolic flux analysis. Nat. Protoc 4, 878–892 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Nanchen A, Fuhrer T & Sauer U Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol 358, 177–197 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Crown SB, Long CP & Antoniewicz MR Optimal tracers for parallel labeling experiments and (13)C metabolic flux analysis: a new precision and synergy scoring system. Metab. Eng 38, 10–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiechert W 13C metabolic flux analysis. Metab. Eng 3, 195–206 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Wiechert W, Mollney M, Petersen S & de Graaf AA A universal framework for 13C metabolic flux analysis. Metab. Eng 3, 265–283 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Weitzel M et al. 13CFLUX2--high-performance software suite for (13)C-metabolic flux analysis. Bioinformatics 29, 143–145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X et al. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal. Chem. 86, 1632–1639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noh K, Droste P & Wiechert W Visual workflows for 13C-metabolic flux analysis. Bioinformatics 31, 346–354 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Yuan M, Breitkopf SB, Yang X & Asara JM A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc 7, 872–881 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying H et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Sahra I, Howell JJ, Asara JM & Manning BD Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM & Manning BD mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shyh-Chang N et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son J et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolay BN et al. Loss of RBF1 changes glutamine catabolism. Genes Dev. 27, 182–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox AG et al. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat. Cell Biol 18, 886–896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locasale JW et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet 43, 869–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu H et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164, 433–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lien EC et al. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat. Cell Biol 18, 572–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sousa CM et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viale A et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breitkopf SB, Taveira MO, Yuan M, Wulf GM & Asara JM Serial-omics of P53−/−, Brca1−/− mouse breast tumor and normal mammary gland. Sci. Rep 7, 14503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitkopf SB et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 13, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breitkopf SB, Yuan M, Helenius KP, Lyssiotis CA & Asara JM Triomics analysis of imatinib-treated myeloma cells connects kinase inhibition to RNA processing and decreased lipid biosynthesis. Anal. Chem 87, 10995–11006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hensley CT et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson SM et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane AN, Yan J & Fan TW 13C Tracer studies of metabolism in mouse tumor xenografts. Bio Protoc. 5, e1650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mashimo T et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159, 1603–1614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glick GD et al. Anaplerotic metabolism of alloreactive T cells provides a metabolic approach to treat graft-versus-host disease. J. Pharmacol. Exp. Ther 351, 298–307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juvekar A et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc. Natl. Acad. Sci. USA 113, E4338–E4347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Yuan M, Wulf GM & Asara JM in Proceedings from the ASMS Conference on Mass Spectrometry and Allied Topics (San Diego, CA, 2018). [Google Scholar]
- 38.Wang JB et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18, 207–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.