Abstract
To understand membrane protein biogenesis, we need to explore folding within a bilayer context. Here we describe a single-molecule force microscopy technique that monitors the folding of helical membrane proteins in vesicle and bicelle environments. After completely unfolding the protein at high force, we lower the force to initiate folding while transmembrane helices are aligned in zigzag manners within the bilayer, thereby imposing minimal constraints on folding. We used the approach to characterize the folding pathways of the Escherichia coli rhomboid protease GlpG and the human β2-adrenergic receptor. Despite their evolutionary distance, both proteins fold in a strict N-to-C-terminal fashion, accruing structures in units of helical hairpins. These common features suggest that integral helical membrane proteins have evolved to maximize their fitness with co-translational folding.
One Sentence Summary
Single molecule experiments reveal that translation, insertion and folding can be coupled during membrane protein biogenesis.
Tens of thousands of mutations associated with diseases are thought to affect membrane protein folding and trafficking (1). The biogenesis of most helix bundle membrane proteins has been divided conceptually into two stages (2, 3). First, co-translational insertion of the hydrophobic protein into the membrane occurs through the Sec translocon pathway (4, 5), thereby establishing much of the transmembrane helical structure and initial topology. Second, the protein completes folding to its final tertiary structure. The two stages, however, are not necessarily cleanly separable (6–9). Studying folding mechanisms of membrane proteins by single-molecule force spectroscopy has been challenging and limited mostly to observing unfolding (10–13), because folding intermediates are usually invisible at the lower forces where folding occurs on a practical time scale. Here, we employ physicochemical conditions that strongly favor folding, thereby enabling the observation of folding at forces high enough to achieve 1 nm resolution.
To develop an experimental method that can be generally applied to the observation of the folding pathways of polytopic membrane proteins, we build on a single-molecule approach we have developed using magnetic tweezers (MT) (Fig. 1A) (14, 15). We link DNA handles to the N- and C-termini of the protein using a SpyTag/SpyCatcher attachment system (16). The handles are in turn attached to a magnetic bead and a polymer-coated glass surface, respectively. The target membrane protein is embedded in bicelles that provide a lipid bilayer-like environment (Fig. 1A). While applying pN- to tens of pN-scale force to the magnetic bead, we record the vertical position of the bead relative to a reference bead stuck on the surface (referred to as the extension value) (fig. S1).
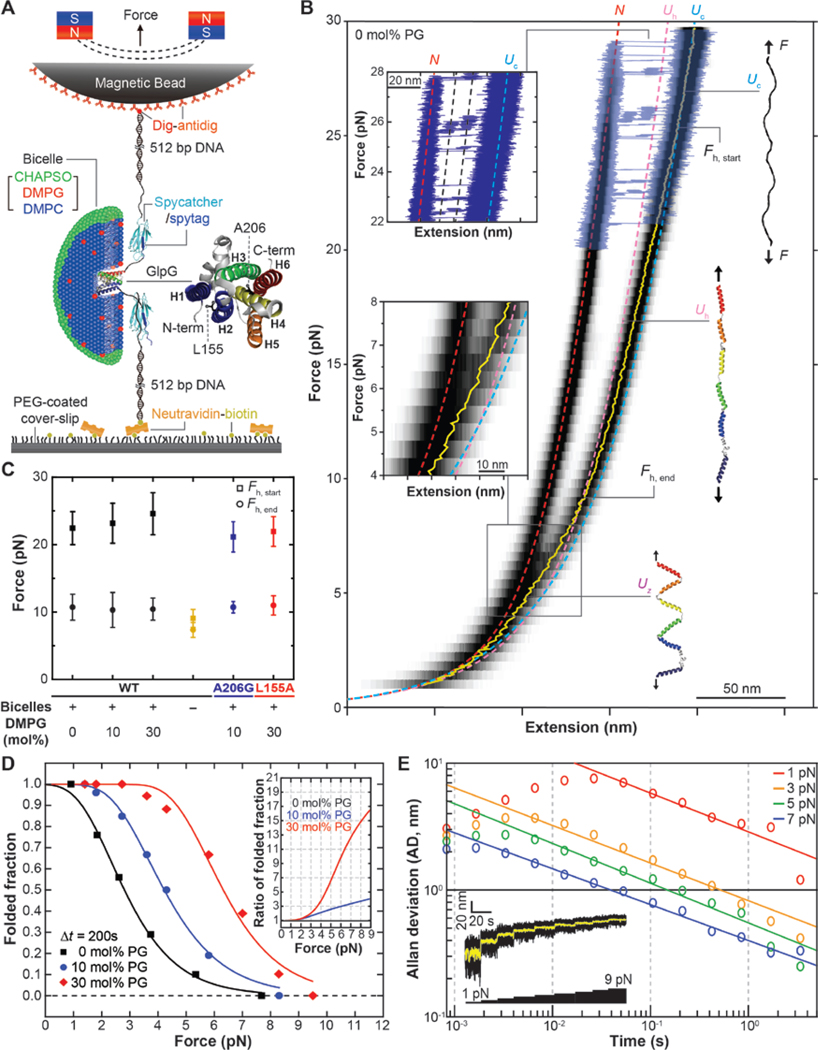
Fig. 1. Physicochemical search for refolding conditions of polytopic helical membrane proteins.
(A) Schematic of single-molecule MT folding experiment for a single GlpG protein reconstituted in a bicelle. (B) FEC of single GlpG proteins averaged over 28 cycles of mechanical stretching and relaxation (black heat map). To show individual unfolding events, representative raw traces are overlaid above 20 pN tension (blue traces). Yellow trace shows the mean extension value in the relaxation phase. Fh,start and Fh,end indicates the force levels at which the coil-to-helix transition starts and ends. Theoretical FECs for the N, Uc and Uh states are shown as red, light blue and pink dashed lines, respectively. Upper inset: a close-up view of the unfolding events. Lower inset: a magnified view of the FEC between 8 and 4 pN force. (C) Average Fh,start and Fh,end values determined under different refolding conditions. n = 34, 42, 58 for the 0, 10 and 30 mol% DMPG cases. n = 16 for the no bicelle case. n = 6, 10 for the A206G and L155A single-point GlpG mutant cases. All error bars represent mean ± s.d. (D) Refolding probability determined using a simple force jump experiment (see fig. S3) at different applied force levels (n = 125, 147, 111 for the 0, 10 and 30 mol% PG cases, respectively). Inset shows the refolding probability normalized to the 0 mol% PG case. (E) Allan deviation of the magnetic bead fluctuation at different force levels. Inset: Representative trace showing the Brownian fluctuation of a magnetic bead at different force levels (black trace: raw data at 1.2 kHz sampling, yellow trace: a median-filtered data with a 5 Hz window).
We applied this method to study the folding of Escherichia coli rhomboid protease GlpG. Figure 1B shows force-extension curves (FECs) averaged over multiple cycles of mechanical stretching and relaxation for single GlpG proteins. At high force levels above 20 pN, the single GlpGs show cooperative unfolding of six transmembrane (TM) helices to an unstructured polypeptide (referred to as the Unfolded Coil or Uc state) while exhibiting two unfolding intermediates as we described previously (Fig. 1B, upper inset) (14, 15). When relaxing the mechanical tension, we detect a gradual transition in FEC from the theoretical curve for Uc to a more compact state that is dependent on the presence of bicelles (Fig. 1, B and C and fig. S2). We designate the new state as Unfolded Helical (Uh) because it fits the FEC expected for a state in which all α-helical structures are restored for the TM helices and linkers. But the protein remains fully stretched along the pulling direction (Fig. 1B). Below the Uc to Uh transition, the FEC shifts to a yet more compact state (Fig. 1B, bottom inset), which we call the Uz state (unfolded zig-zag state). As discussed below, the Uz state appears to consist of bilayer inserted, but weakly interacting TM helices, arranged in a zigzag-like fashion. Finally, at low forces, GlpG finds its folded, native conformation (referred to as the N state) (Fig. 1B). Because formation of the native state could only be achieved at low tension below 2 pN, Brownian motions of the magnetic bead precludes observation of any detailed intermediates during the refolding process under these conditions.
In hopes of seeing folding at higher forces where extension measurements can be more precise, we screened for more favorable folding conditions using a simple force jump experiment (fig. S3) (10, 17, 18). We find that addition of 1,2-dimyristoyl-sn-glycero-3-phosphorylglycerol (DMPG) is effective in enhancing GlpG refolding. When we add 30 mol% DMPG lipids in the bicelle phase, the refolding probability after a waiting time of 200 s at 5 pN tension increases by a factor of 7 (Fig. 1D). The FECs obtained with and without 30 mol% DMPG lipids almost exactly overlap with one another, preserving the coil-to-helix transition as well as the formation of the Uz state (fig. S2 and S4). These observations suggest that the addition of negatively charged lipids does not fundamentally alter the folding pathway, but selectively enhances refolding commencing from the Uz state.
With the ability to observe folding at higher forces, we tested the potential for achieving high resolution by examining the Brownian motion of magnetic beads in the bicelle phase. With high-speed tracking at 1.2 kHz and the force above 5 pN, we obtain an Allan deviation (i.e., uncertainty in our tracking) of less than 1 nm when median filtered at 5 Hz (200 ms), corresponding to a resolution of a few amino acids (Fig. 1E). Also, we observe folding with a reasonable probability up to 8 pN tension, the force where the Uz state starts to form (Fig. 1D).
On the basis of these observations, we developed a force-application protocol to monitor the folding process of single GlpG proteins (Fig. 2). We first induce full unraveling of GlpG to the Uc state by applying a high mechanical tension above 20 pN, and then make a force jump to a low force level between 5 and 8 pN (Fig. 2A). The force jump takes a finite time of ~300 ms, during which single GlpGs relax to the Uz state (Fig. 2B, right). We experimentally confirmed that the force jump indeed reaches the same extension state as that reached through slow gradual force relaxation at –1 pN/s (Fig. 2B, left). When maintained at the low force level, the magnetic bead begins to show complex up-and-down movements, finally culminating in a compact N state (Fig. 2A, 5 pN phases). Achievement of the N state is verified by observing the extension expected for the N state after jumping the tension back to ~20 pN. By repeating our designed mechanical cycle, we can observe folding of single GlpG molecules multiple times.
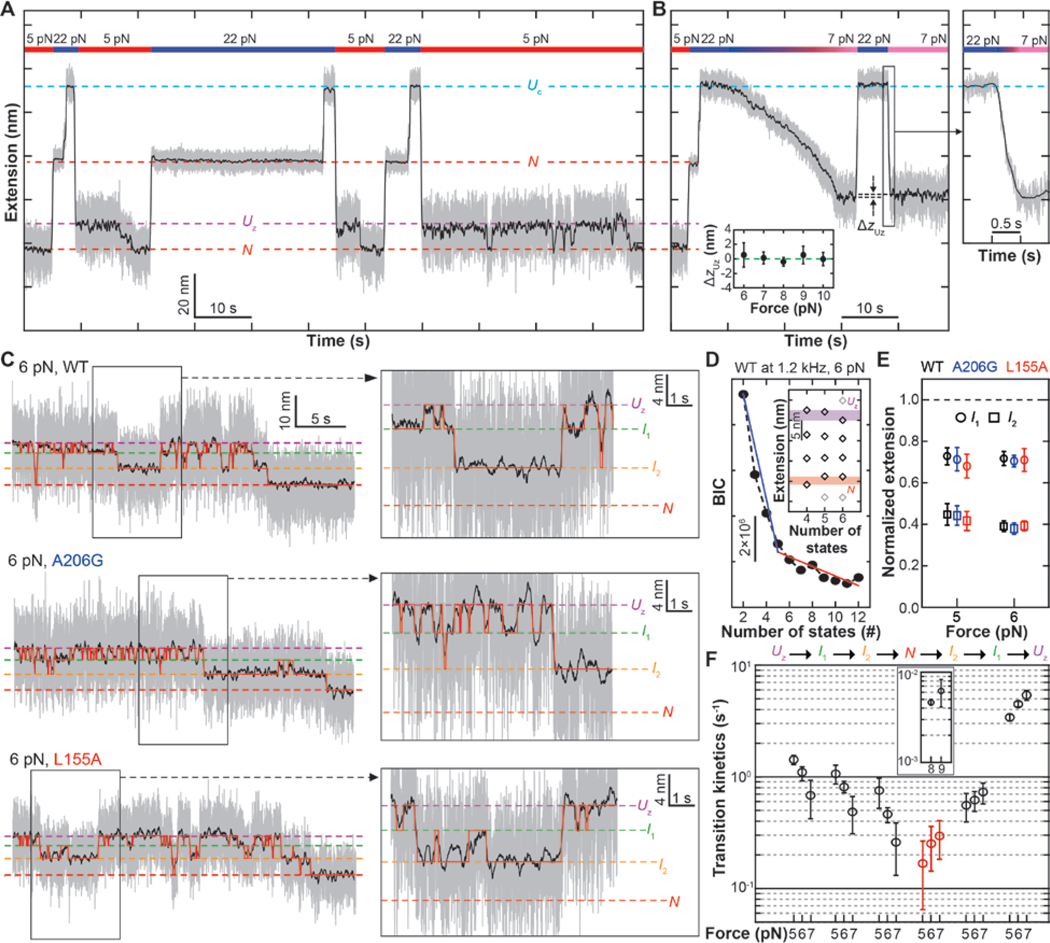
Fig. 2. Direct observation of single GlpG folding.
(A) Designed mechanical cycle for inducing refolding of single GlpG proteins. The gray and black traces are 1.2-kHz raw data and 5 Hz median-filtered data, respectively. (B) Representative time-resolved traces comparing the extensions following slow force relaxation and force jump. Lower inset shows the extension difference (ΔzUz) at indicated force levels. Right inset shows a closed-up trace of the force jump that takes ~300 ms. (C) Representative folding traces for WT, A206G and L155A GlpG under 6 pN tension. Right insets show close-up traces exhibiting reversible transitions among the Uz, I1, and I2 states. (D) BIC values for indicated number of states. n = 20, 21 and 11 for the WT, A206G and L155A cases, respectively. (E) Positions of the intermediate states in the normalized extension space (at 6 pN, n is same as (D); at 5 pN, n = 21, 24 and 14 for WT, A206G and L155A, respectively). Error bars represent s.e.m. (F) Transition kinetics between the neighboring states at indicated force levels. In N-to-I2 transition, both slow (inset, black) and fast (red) rates are displayed. Error bars represent s.e.m.
With the reduced Brownian motion of the magnetic bead in the 5–8 pN force window, we are able to see distinct conformational changes during refolding when the extension traces are median filtered down to 5 Hz (Fig. 2C, black traces). Application of hidden Markov modeling (HMM) and Bayesian information criteria (BIC) to the time-resolved extension traces indicates that the data are best fit by a total of four states: two intermediate states (referred to as I1 and I2) in addition to the Uz and N states (Fig. 2, C to E and fig. S5) (19).
The magnetic beads show many upward (i.e., local unfolding) and downward movements (local folding) before reaching the native N state extension. Thus, the resultant time-resolved traces report reversible intermediate folding/unfolding events—equilibrium processes from which we can directly reconstruct the folding energy landscape. The local folding and unfolding processes pass through the same I1 and I2 intermediate states, justifying a one-dimensional representation of the energy landscape (20, 21). Our HMM analysis indicates that while rates connecting non-neighboring states are negligible, the transitions connecting the neighboring states are well described by single rates falling in a narrow region between 0.1 to 10 s−1 (Fig. 2F and fig. S6). An exception is the N-to-I2 transition that shows two different rates. One N-to-I2 transition is relatively fast with an average dwell time in N of only ~5 s, indicating incomplete refolding (fig. S6 and Fig. 2F, red symbols). The other subset of the N states has higher stability, requiring higher forces (~ 8 pN) to show unfolding within our observation time, and presumably corresponds to a correctly folded state (fig. S7 and Fig. 2F, inset).
We next examined whether we could extend our MT folding experiment to true bilayers, by reconstituting GlpG in vesicles produced through slow detergent removal (Fig. 3A and fig. S8). For the vesicle-reconstituted GlpGs, we observe cooperative unraveling of six TM helices to the Uc state, similar to what is observed for the bicelle-reconstituted GlpGs (Fig. 3B, 1st stretching cycle). We also observe the expected coil-to-helix transition during the relaxation phase (Fig. 3C) as well as complete refolding, albeit with low probability (Fig. 3B, 6th stretching). Since there are no free vesicles, these observations suggest that single GlpGs remain bound to the vesicle membranes after their unraveling to unstructured polypeptides. Nevertheless, many of the vesicle-reconstituted GlpGs fail to refold (e.g., Fig. 3B, 2nd and 5th stretching). The refolding probability at 1 pN is only ~15 % with a 200 s wait time, in contrast with a refolding probability approaching 100 % seen for the bicelle-reconstituted GlpGs. We also find that the FECs of vesicle-reconstituted GlpGs persistently follow the Uh curve and fail to form the loosely stretched Uz state (Fig. 3C, lower inset).
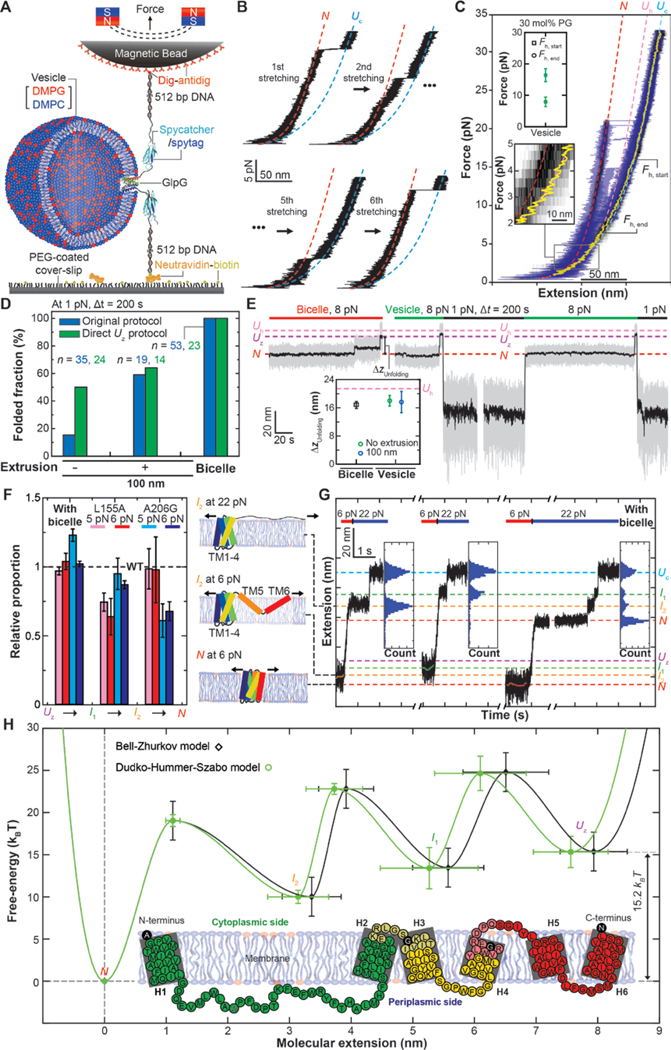
Fig. 3. Characterization of folding properties for vesicle- and bicelle-reconstituted single GlpGs.
(A) Schematic of single-molecule MT folding experiments for GlpGs reconstituted in vesicle membranes. (B) Representative FECs showing successive stretching cycles applied to a single GlpG protein in a vesicle membrane. (C) FEC of vesicle-reconstituted single GlpG proteins averaged over 5 cycles of mechanical stretching and relaxation (black heat map). To show individual unfolding events, representative raw traces during stretching are overlaid (blue traces). Other definitions are the same as in Fig. 1B. Upper inset shows the average Fh,start and Fh,end values. Lower inset: a magnified view of the FEC between 5 and 2 pN force. (D) Refolding probability determined under different membrane conditions. n is the number of trials. (E) Representative traces of the folding protocol that directly induces the Uz state. Inset shows the extension changes (ΔzUnfolding) during unfolding at 8 pN under indicated membrane conditions. Pink dotted line is the expected extension change when reaching the Uh state. (F) Normalized transition rates determined for the A206G and L155A single-point mutants relative to the WT case (dashed line). Right illustrations show an anticipated conformational status of GlpG in each indicated state. Error bars represent s.e.m (G) Representative force-jump experiments applied for the intermediate states. Each inset shows the distribution of extension values recorded during high-force unfolding. Estimated extensions for individual states are shown as the dashed lines. (H) Folding energy landscapes of a single GlpG protein along the molecular extension reconstructed based on the Bell-Zhrukov (black trace) and the Dudko-Hummer-Szabo models (green trace). Inset: detailed structural segments in the folding pathways of GlpG. Each structurally segment is denoted in distinct colors. Black-colored amino acids correspond to the boundaries of the intermediate states. Faint colors around the boundaries represent the measurement errors (s.d.).
We hypothesized that polypeptide insertion into bilayers may be more difficult in vesicles compared to bicelles, which would explain the barrier to folding and the block to formation of the Uz state if the Uz state was indeed membrane inserted. To explore this possibility, we tested whether decreasing vesicle size would enhance folding because increased bilayer curvature may allow more facile insertion of the TM helices into the membrane (22). Indeed, when we decrease the diameter of reconstitution vesicles to 100 nm by extrusion, the refolding probability at 1 pN increases to ~60 % (Fig. 3D and fig. S8).
We next attempted to avoid membrane extraction and directly access the Uz state by applying a moderate force of 8 pN, a tension at which the Uz state was seen to form in FEC (referred to as the direct-Uz protocol). We tested the feasibility of this protocol first with the bicelle-reconstituted GlpGs, and found that application of 8 pN indeed directly induces the Uz state (Fig. 3E, left). Subsequent lowering of the force leads to complete refolding in bicelles (fig. S9). When we apply 8 pN to the vesicle-reconstituted GlpGs, the resultant unfolding step is almost identical to that expected for the Uz state (Fig. 3E, right and inset). When we subsequently induce refolding by lowering the force to 1 pN, the refolding probability in vesicles increases relative to refolding from the Uc state (~50 % versus ~15%) (Fig. 3, D and E), consistent with the possibility that this 8 pN unfolding selectively disrupts the tertiary structure while decreasing exposure of the TM helices to outside of the lipid bilayer, thereby reducing the need to reinsert TM helices during refolding.
Our analysis of the end-to-end distance of the Uz state further suggests that the penetration depths of TM helices in the Uz state might not be enough to completely reach the other side of the lipid bilayer (fig. S10 and Fig. 3F, right). If so, tertiary structure formation would be intimately coupled with the membrane insertion (Fig. 3F, right; compare I2 and N at 6 pN). To determine whether membrane insertion or tertiary structure formation dominate the energy barriers, we examined the local folding/unfolding kinetics of the two single-point GlpG mutants in the bicelle phase (23). The N-terminal (L155A on TM helix 2) and C-terminal (A206G on TM helix 4) mutants selectively slow down the I1-to-I2 and the I2-to-N transitions, respectively (Fig. 3F, left and fig. S11). These observations suggest that the tertiary structure formation makes a major contribution to the observed energy barriers. Moreover, these mutant data are consistent with GlpG folding occurring in a unidirectional manner from the N- to C-terminus.
To map the partially folded structures in I1 and I2, we made force jumps to ~20 pN while the protein sampled either I1 or I2 in the course of refolding (Fig. 3G). Surprisingly, the extensions after the force jump exactly coincide with the two intermediates of the high-force unfolding (Fig. 3G and Fig. 1B, upper inset), which indicates that the low-force folding/unfolding and the high-force unfolding intermediates share the same partially folded structures, albeit with different levels of stretching in the unfolded regions (Fig. 3F, right; compare I2 at 6 pN with I2 at 22 pN). We therefore used the extension difference between I2 and N at 22 pN to estimate that I2 is positioned at the C-terminus of TM helix 4 (Fig. 3H, inset). Likewise, we used the extension difference between Uc and I1 at 22 pN to estimate that I1 is positioned after the TM helix 2. Combined with the mutant data above, we conclude that GlpG folds in an N-to-C direction, largely in units of helical hairpins.
Based on the structural assignments made above, we examined one more GlpG mutant in which two hydrophobic residues in the long linker region between TM helix 1 and 2 are mutated to negatively charged residues (L121E/F133E) (Fig. 3H, inset and fig. S12B). Although such mutations reportedly increase the energy barrier for membrane insertion and flip-flop (24, 25), we did not detect any sign of slowing down in the transition between Uz and I1 (fig. S12). This data supports again our conclusion that the TM helices have made their initial membrane integration as the Uz state forms. In particular, because of the many polar and charged residues in the long linker, we suspect that TM helices 1 and 2 of GlpG are inserted more deeply than other TM helices in the zigzag-aligned Uz state.
Finally, we constructed one-dimensional energy landscapes for the reversible folding/unfolding process of single GlpG proteins using the Bell (26) and the Dudko-Hummer-Szabo models (27). Both models indicate a free energy difference (ΔG0) of 15.2 kBT between the native state (N) and the zigzag Uz state albeit with slightly different intermediate positions (Fig. 3H). As expected, this ΔG0 value is slightly larger than the estimates from previous ensemble measurements under less favorable folding conditions (7.1 to 13.9 kBT) (23, 28, 29). When we apply the Crooks fluctuation theorem to the FECs as shown in Fig. 1B, we obtain an estimate of ~115 kBT for a free energy difference between the N and the Uc states (fig. S13 and Table. S1), almost 8-fold larger than 15.2 kBT estimated between the N and Uz states. We attribute this larger free energy difference to additional processes imposed on the high-force unfolding, such as pulling TM helices out of the membrane and disruption of secondary structures. These observations attest to the fundamental difference between the energy barriers seen during high-force unfolding and the low-force folding/unfolding processes. At lower forces, we can explore rearrangements of intact TM helices that occur largely within the lipid bilayer, more closely reflecting the process expected for second stage folding (2).
Using the experimental methods established with GlpG, we next sought to observe the folding process of a single human β2-adrenergic receptor (β2AR), which belongs to the G protein coupled receptor (GPCR) family (Fig. 4A). We first examined the FEC and again observed a large mechanical hysteresis in the unfolding and refolding of β2AR (Fig. 4B). Because β2AR has an odd number of TM helices, the DNA handles are pulling on opposite sides of the bilayer and we note the possibility that after the cooperative unraveling, some part of β2AR may reside within the lipid bilayer (most likely the first TM helix; see fig. S14). During the relaxation phase, we observed the coil-to-helix transition in nearly the same force range as that observed for GlpG. Moreover, below 8 pN, the FEC of β2AR became shorter than the Uh extension consistent with the formation of a zigzag-aligned Uz state (Fig. 4B and fig. S15).
Fig. 4. Direct observation of the complete folding pathway of human β2AR.
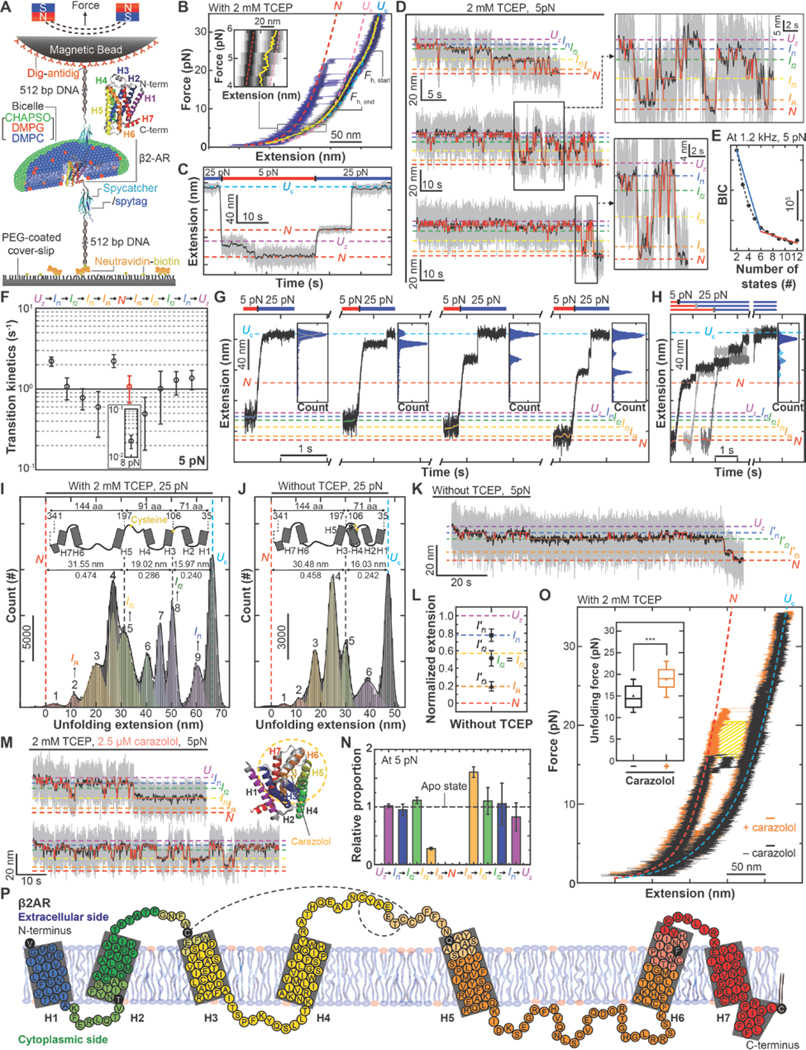
(A) Schematic diagram of single-molecule MT folding experiment for β2AR. (B) FEC of single β2AR proteins averaged over 6 cycles of mechanical stretching and relaxation (black heat map). To show individual unfolding events, representative raw traces during stretching are overlaid (blue traces). Other definitions are the same as in Fig. 1B. Inset: an enlarged view of the FEC between 6 and 4 pN force. (C) Designed mechanical cycle for inducing refolding of human β2AR at low force levels. (D) Representative time-resolved traces for β2AR folding under 5 pN tension (with 2 mM TCEP). Right insets show close-up trajectories. Red traces show the transitions between four intermediates identified the HMM. (E) BIC values for indicated numbers of states (n = 18). (F) Transition kinetics between the neighboring states at 5 pN. In N-to-If4 transition, both slow (inset, black) and fast (red) rates are displayed. Error bars represent s.e.m. (G, H) Representative traces for the force jump experiments applied to individual folding intermediates (G) and the native state (H). Each inset shows an extension distribution during high-force unfolding. (I, J) Extension distribution during high-force unfolding initiated from the native N state. (n = 29, 13 for the cases with 2 mM TCEP and without TCEP, respectively). The peaks indicate the fit centers of multiple Gaussian functions (colored for each function). Upper insets show structural diagrams of β2AR to guide mapping onto the structure. (K) Representative β2AR folding trace at 5 pN with no TCEP (n = 10). HMM analysis finds three intermediate states (If1´, If2´, If3´). (L) Normalized extensions for β2AR folding intermediates under no TCEP condition. Dashed lines are anticipated extensions for the intermediates under 2 mM TCEP condition. Error bars are s.d. (M) Representative folding traces for β2AR at 5 pN in the presence of 2.5 μM carazolol (with 2 mM TCEP). Right inset shows the structure of carazolol-bound human β2AR. Yellow circle indicates interaction regions between carazolol and β2AR. (N) Normalized rates determined for carazolol-bound β2AR relative to the apo β2AR case (dashed line). (O) Representative FECs of β2AR showing high-force cooperative unfolding in the presence (orange) and absence (black) of 2.5 μM carazolol (with 2 mM TCEP). N and Uc are defined the same as in (B). Inset: distributions of unfolding forces with (orange, n = 26) and without (black, n = 35) carazolol. Error bars are s.d. (P) Detailed structural segments in the folding pathways of β2AR. Each structurally segment is denoted in distinct colors. Other notations are same as inset in Fig. 3H.
To observe the folding process of human β2AR, we used the original folding protocol starting from the Uc state (Fig 4C). We first induced mechanical unraveling of a single β2AR Uc state by applying 25 pN tension, and then induced the Uz state through force quenching to 5 pN. We re-confirmed that the force quenching within 300 ms yielded the same Uz state as that obtained through slow force relaxation (fig. S16). With the mechanical tension kept at 5 pN, the magnetic bead shows complex up-and-down movements, ending in a compact N state (confirmed to be the native state through re-application of high force) (Fig. 4C).
By applying the HMM and BIC analyses to the time-resolved extension traces of the magnetic beads, we identify six major states (thus four intermediates) in the folding process of human β2AR (Fig. 4, D and E). Both local folding and unfolding processes share these four intermediate states (referred to as If1, If2, If3 and If4), indicative of the one-dimensionality of the folding energy landscape. The transition rates connecting the neighboring states fall between 10−1 and 10 s−1 while all other rates are negligibly small (Fig. 4F and fig. S17). As was the case for GlpG, we observed two groups of the N states: one with a lower stability (Fig. 4F, red symbol) and the other reflecting a correctly folded structure (Fig. 4F, inset).
To measure the number of amino acids unfolded in the structures of the four intermediates, we applied the force-jump technique to each intermediate observed during the low-force folding/unfolding processes. We find that all the four intermediates correspond to distinct extension states at 25 pN, reflecting a direct connection between the low-force folding/unfolding and the high-force unfolding intermediates (Fig. 4G). We thus sought to use the extension states of the high-force unfolding intermediates to infer the partially folded structures in individual low-force intermediates. The distribution of intermediate extension states during high force unfolding clearly revealed a total of nine peaks (Fig. 4, H and I).
To map the unfolded structures on the high-force unfolding intermediates, we took advantage of the fact that in the native structure of human β2AR, there is one conserved disulfide bond formed between C106 and C191, which locks TM helices 3 and 4 and the extracellular linker 2 (ECL2) into one structural unit (Fig. 4, I and J, upper insets). We reasoned that with removal of the reducing agent tris(2-carboxyethyl)phosphine (TCEP), the high-force unfolding intermediates related to the region linked by the disulfide bond might disappear from the extension distribution. Indeed, in the absence of TCEP, the first five peaks (peaks 1 to 5) and the last two peaks (peaks 9 and Uc) are essentially preserved, but the three peaks in the middle (peaks 6 to 8) selectively disappear (Fig. 4, I and J). We further note that in the absence of TCEP, the extension change spanning the first five peaks is distinctly larger than that spanning the last two peaks by a factor of 1.98. This value closely matches the 2.02 ratio of the number of amino acids placed C–terminal to the disulfide bond (144 a.a.) to that N-terminal to the disulfide bond (71 a.a.) (Fig. 4I). Thus, our observations point to a hypothesis that the first five peaks correspond to unfolding from C-terminus to TM helix 5. In the absence of TCEP, unfolding from ECL2 to TM helix 3 is prevented by the disulfide, so that the last two peaks correspond to unfolding of the two N-terminal helices and ECL1. By aligning the unfolding traces in Figure 4G and 4H with one another, we find that folding intermediates If1, If2, If3 and If4 correspond to the high force unfolding intermediate peaks 9, 8, 5 and 2, respectively (Fig. 4I and fig. S18). Together, our data suggested the human β2AR shows unidirectional folding from the N- to the C-terminus.
To test the validity of our structural assignment, we monitored the folding process at 5 pN in the absence of TCEP. The HMM and BIC analyses indicate a reduction in the number of folding intermediates to three (Fig. 4K and fig. S19). The positions of these three folding intermediates (If1′, If2′ and If3′) matched well with those expected when If2 and If3 are merged (Fig. 4L), reaffirming that the transition from If2 to If3 corresponds to folding of TM helices 3 and 4 and ECL2.
We also examined the effect of carazolol, a partial inverse agonist of human β2ARs, on the 5-pN folding process. While the presence of 2.5 μM carazolol did not change the positions of the four intermediates in the extension space, it markedly inhibited any transition beyond If3 (Fig. 4M and fig. S19). This inhibition was highly selective because the transition Uz to If3 remained minimally affected (Fig. 4N), suggesting that single human β2ARs folds normally up to ECL2, but fails to fold TM helices 5 and 6 onto the growing structure in the presence of carazolol. When we examined unfolding by force ramping, carazolol increases the forces at which unfolding occurs by 4.5 pN on average (Fig. 4O, inset), indicating that additional work of more than 50 kBT is required to induce the unraveling of single β2ARs in the presence of carazolol (Fig. 4O, shaded area). Thus, our observations suggest distinct effects of carazolol on human β2AR folding and unfolding. Carazolol inhibits the addition of TM helices 5 and 6 during folding, perhaps being loosely located in the incomplete ligand binding pocket formed by TM helices 1–4 and sterically interfering with incoming TM helices 5 and 6. In the presence of excess carazolol, it is also possible that carazolol is already bound to TM helices 5 and 6 as carazolol makes an extended aromatic network with the residues on TM helices 5 and 6. However, once folded, carazolol binding dramatically stabilizes the tertiary structure as expected (30, 31).
The identified folding pathway of the human β2AR reveals several interesting features (Fig. 4P). The first intermediate If1 corresponds to association between the first TM helix and the following linker helix, completing insertion of this nascent structure with respect to the residing membrane structure. The second TM helix folds on to this structure to form the first helical hairpin, completing intermediate If2. The next folding step involves the addition of TM helices 3 and 4 as well as ECL2 (forming If3). We note that the positions of If2 and If3 closely map to the cysteine residues of C106 and C191, thereby potentially consolidating the formed tertiary structure via disulfide bonding. The transition from If3 to If4 involves formation of the helical hairpin consisting of the TM helices 5 and 6. This folding step is found to be markedly inhibited in the presence of carazolol. The last step from If4 to N involves addition of the TM helix 7 and the C-terminal membrane-associated helix onto the structure, completing the known structure of human β2AR (30, 31). Although our experimental data consistently support the folding pathway delineated above, we cannot rule out the possibility that an alternative folding pathway exists in physiological milieu. We also note that the folding pathway presented here is a coarse-grained one down to a 5 Hz sampling rate. Enhancing the bandwidth of our methods would reveal a more complex and dynamic nature of the polytopic membrane protein folding (12). Finally, we note the possibility that the strategy of using disulfide bonds to map the four folding intermediates of β2AR can be extended to other membrane proteins.
Although E. coli GlpG and human β2AR are at an enormous evolutionary distance, both integral membrane proteins accrue structure largely in units of helical hairpins, with a unidirectional N-to-C folding as a single, predominant pathway out of a countless number of permutations in the possible folding pathways. Unidirectional N-to-C folding is consistent with several prior studies (28, 32–36), and would permit the nascent N-terminal chain to commence folding without needing to wait for the more C-terminal TM helices to be translated, thereby reducing the risk of generating misfolded structures. Thus, the folding processes of integral membrane proteins may be evolutionarily selected and tailored to fit with co-translational folding.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Creative Research Initiative Program (Center for Single-Molecule Systems Biology to T.-Y.Y.; NRF-2011–0018352), the Bio Medical Technology Development Program (NRF-2018M3A9E2023523 to T.-Y.Y.), the Basic Science Research Program (NRF-2016R1A6A3A03007871 to D.M.) and a NRF grant (NRF-2016R1A2B4013488 to H.-J.C.), all funded by the National Research Foundation of South Korea. This work was also supported by the National Institutes of Health grant (R01GM063919 to J.U.B.).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References and Notes
- 1.Sanders CR, Myers JK, Disease-related misassembly of membrane proteins. Annu. Rev. Biophys. Biomol. Struct 33, 25–51 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Popot J-L, Engelman DM, Membrane protein folding and oligomerization: the two-stage model. Biochemistry 29, 4031–4037 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Bowie JU, Solving the membrane protein folding problem. Nature 438, 581 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Mori H, Ito K, The Sec protein-translocation pathway. Trends Microbiol. 9, 494–500 (2001). [DOI] [PubMed] [Google Scholar]
- 5.White SH, von Heijne G, How translocons select transmembrane helices. Annu. Rev. Biophys 37, 23–42 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Cymer F, Von Heijne G, White SH, Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol 427, 999–1022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodall NB, Yin Y, Bowie JU, Dual-topology insertion of a dual-topology membrane protein. Nat. commun 6, 8099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodall NB, Hadley S, Yin Y, Bowie JU, Complete topology inversion can be part of normal membrane protein biogenesis. Protein Sci. 26, 824–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowhan W, Vitrac H, Bogdanov M, Lipid-assisted membrane protein folding and topogenesis. Protein J. 38, 274–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedrov A, Janovjak H, Ziegler C, Kuhlbrandt W, Muller DJ, Observing folding pathways and kinetics of a single sodium-proton antiporter from Escherichia coli. J. Mol. Biol 355, 2–8 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Serdiuk T. et al. , YidC assists the stepwise and stochastic folding of membrane proteins. Nat. Chem. Biol 12, 911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Siewny MG, Edwards DT, Sanders AW, Perkins TT, Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science 355, 945–950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu P. et al. , Accurate computational design of multipass transmembrane proteins. Science 359, 1042–1046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min D, Jefferson RE, Bowie JU, Yoon T-Y, Mapping the energy landscape for second-stage folding of a single membrane protein. Nat. Chem. Biol 11, 981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min D, Arbing MA, Jefferson RE, Bowie JU, A simple DNA handle attachment method for single molecule mechanical manipulation experiments. Protein Sci. 25, 1535–1544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zakeri B. et al. , Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oesterhelt F. et al. , Unfolding pathways of individual bacteriorhodopsins. Science 288, 143–146 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Fernandez JM, Li H, Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science 303, 1674–1678 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Lee T-H, Extracting kinetics information from single-molecule fluorescence resonance energy transfer data using hidden Markov models. J. Phys. Chem. B 113, 11535–11542 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neupane K, Manuel AP, Woodside MT, Protein folding trajectories can be described quantitatively by one-dimensional diffusion over measured energy landscapes. Nat. Phys 12, 700 (2016). [Google Scholar]
- 21.Neupane K. et al. , Direct observation of transition paths during the folding of proteins and nucleic acids. Science 352, 239–242 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Hatzakis NS et al. , How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol 5, 835 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Baker RP, Urban S, Architectural and thermodynamic principles underlying intramembrane protease function. Nat. Chem. Biol 8, 759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Lehn RC, Zhang B, Miller TF III, Regulation of multispanning membrane protein topology via post-translational annealing. Elife 4, e08697 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seurig M, Ek M, Von Heijne G, Fluman N. J. b., Dynamic membrane topology in an unassembled membrane protein. bioRxiv, 548537 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Bell GI, Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978). [DOI] [PubMed] [Google Scholar]
- 27.Dudko OK, Hummer G, Szabo A, Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys. Rev. Lett 96, 108101 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Paslawski W. et al. , Cooperative folding of a polytopic α-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. Proc. Natl. Acad. Sci. U.S.A 112, 7978–7983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo R. et al. , Steric trapping reveals a cooperativity network in the intramembrane protease GlpG. Nat. Chem. Biol 12, 353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen SG et al. , Crystal structure of the human β 2 adrenergic G-protein-coupled receptor. Nature 450, 383 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Cherezov V. et al. , High-resolution crystal structure of an engineered human β2-adrenergic G protein–coupled receptor. Science 318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curnow P, Booth PJ, The transition state for integral membrane protein folding. Proc. Natl. Acad. Sci. U.S.A 106, 773–778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min D. et al. , Unfolding of a ClC chloride transporter retains memory of its evolutionary history. Nat. Chem. Biol 14, 489–496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ismail N, Hedman R, Schiller N, Von Heijne G, A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat. Struct. Mol. Biol 19, 1018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cymer F, von Heijne G, Cotranslational folding of membrane proteins probed by arrest-peptide–mediated force measurements. Proc. Natl. Acad. Sci. U.S.A 110, 14640–14645 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson OB et al. , Cotranslational folding of spectrin domains via partially structured states. Nat. Struct. Mol. Biol 24, 221 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Li L, Fierer JO, Rapoport TA, Howarth M, Structural analysis and optimization of the covalent association between SpyCatcher and a peptide Tag. J. Mol. Biol 426, 309–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song H, Orr A, Duan M, Merz AJ, Wickner W, Sec17/Sec18 act twice, enhancing membrane fusion and then disassembling cis-SNARE complexes. Elife 6, e26646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon T-Y, Okumus B, Zhang F, Shin Y-K, Ha T, Multiple intermediates in SNARE-induced membrane fusion. Proc. Natl. Acad. Sci. U.S.A 103, 19731–19736 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arenas-Guerrero P. et al. , Determination of the size distribution of non-spherical nanoparticles by electric birefringence-based methods. Sci. Rep 8, 9502 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shon MJ, Kim H, Yoon T-Y, Focused clamping of a single neuronal SNARE complex by complexin under high mechanical tension. Nat. commun 9, 3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shon MJ, Rah S-H, Yoon T-Y, Submicrometer elasticity of double-stranded DNA revealed by precision force-extension measurements with magnetic tweezers. Sci. Adv 5, eaav1697 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrosyan R, Improved approximations for some polymer extension models. Rheol. Acta 56, 21–26 (2017). [Google Scholar]
- 44.Seol Y, Li J, Nelson PC, Perkins TT, Betterton M, Elasticity of short DNA molecules: theory and experiment for contour lengths of 0.6–7 μm. Biophys. J 93, 4360–4373 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar A, Caamano S, Fernandez JM, The mechanical fingerprint of a parallel polyprotein dimer. Biophys. J 92, L36–L38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gebhardt JCM, Bornschlögl T, Rief M, Full distance-resolved folding energy landscape of one single protein molecule. Proc. Natl. Acad. Sci. U.S.A 107, 2013–2018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kessler DA, Rabin Y, Distribution functions for filaments under tension. J. Chem. Phys 121, 1155–1164 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Van Vliet CM, Handel PH, A new transform theorem for stochastic processes with special application to counting statistics. Physica A 113, 261–276 (1982). [Google Scholar]
- 49.Lansdorp BM, Saleh OA, Power spectrum and Allan variance methods for calibrating single-molecule video-tracking instruments. Rev. Sci. Instrum 83, 025115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dulin D. et al. , High spatiotemporal-resolution magnetic tweezers: calibration and applications for DNA dynamics. Biophys. J 109, 2113–2125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Jiao J, Rebane AA, Hidden Markov modeling with detailed balance and its application to single protein folding. Biophys. J 111, 2110–2124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crooks GE, Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys. Rev. E 60, 2721 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Hummer G, Szabo A, Free energy reconstruction from nonequilibrium single-molecule pulling experiments. Proc. Natl. Acad. Sci. U.S.A 98, 3658–3661 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liphardt J, Dumont S, Smith SB, Tinoco I, Bustamante C, Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski’s equality. Science 296, 1832–1835 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Collin D. et al. , Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature 437, 231 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bennett CH, Efficient estimates of free energy differences from Monte Carlo data. J. Comp. Phys 22, 245–268 (1976). [Google Scholar]
- 57.Crooks GE, Path-ensemble averages in systems driven far from equilibrium. Phys. Rev. E 61, 2361–2366 (2000). [Google Scholar]
- 58.Yang WY, Gruebele M, Folding at the speed limit. Nature 423, 193 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Kubelka J, Hofrichter J, Eaton WA, The protein folding ‘speed limit’. Curr. Opin. Struct. Biol 14, 76–88 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Chung HS, Louis JM, Eaton WA, Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc. Natl. Acad. Sci. U.S.A 106, 11837–11844 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dimova R, Pouligny B, Dietrich C, Pretransitional effects in dimyristoylphosphatidylcholine vesicle membranes: optical dynamometry study. Biophys. J 79, 340–356 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shkulipa S, den Otter WK, Briels WJ, Surface viscosity, diffusion, and intermonolayer friction: simulating sheared amphiphilic bilayers. Biophys. J 89, 823–829 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cecconi C, Shank EA, Bustamante C, Marqusee S, Direct observation of the three-state folding of a single protein molecule. Science 309, 2057–2060 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Dudko OK, Graham TG, Best RB, Locating the barrier for folding of single molecules under an external force. Phys. Rev. Lett 107, 208301 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Dudko O. k., A transformation for the mechanical fingerprints of complex biomolecular interactions. Proc. Natl. Acad. Sci. U.S.A 110, 16432–16437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.