Abstract
Nitrogen (N) is one of the most important macronutrients for plant growth and development. However, the concentration and distribution of N varies in soil due to a variety of environmental factors. In response, higher plants have evolved a developmentally flexible root system to efficiently take up N under N-limited conditions. Over the past decade, significant progress has been made in understanding this form of plant ‘root-foraging’ behavior, which is controlled by both a local and a long-distance systemic nitrate signaling pathway. In this review, we focus on the key components of nitrate perception, signaling, and transduction and its role in lateral root development. We also highlight recent findings on the molecular mechanisms of the nitrate systemic signaling pathway, including small signaling peptides involved in long-distance shoot–root communication. Furthermore, we summarize the transcription factor networks responsible for nitrate-dependent lateral root and root hair development.
Keywords: Lateral root, local signaling, long-distance communication, nitrate signaling, root foraging, root hair, systemic signaling
This review provides an update on the molecular mechanisms involved in the long-distance shoot–root communication in systemic nitrate signaling, and outlines the transcription factor network responsible for nitrate regulation of lateral root and root hair growth.
Introduction
Nitrogen (N) is one of the primary mineral nutrients for plant growth and development, as well as one of the main components of commercial fertilizers. N applications need to be repeated in order to maintain N availability and soil fertility (Mallory et al., 2010). Nitrate (NO3–) and ammonium (NH4+) are the main mineral forms of N that plants utilize from their external environment, but the fluctuating environment and the intrinsic complexity of soils cause numerous reactions, transformations, and N losses that generate tremendous variation in soil N distribution (Zhu et al., 2011; Ye et al., 2015; Berhe and Torn, 2017; Pandey et al., 2019). The highly plastic plant root system is able to respond developmentally to the N nutrient signal, enabling exploration and colonization into N-rich patches of soil. However, crops are only able to use 30–50% of the applied N in general. Therefore, an understanding of the mechanism of nitrate-regulated lateral root (LR) and root hair development may lead to increased nitrogen use efficiency (NUE) in crops (Bowles et al., 2018).
The plant root system is not only regulated by the ‘N signal’, which represents the autonomous response to local nitrate availability (Remans et al., 2006), but it is also affected by the so-called ‘systemic N signal’ pathway, which selectively promotes colonization of plant roots in N-rich patches (Bellegarde et al., 2017; Ohkubo et al., 2017). Recent studies demonstrated that nitrate-dependent root foraging is controlled by complex interactions between nitrate perception, signaling, and the systemic regulatory pathways. This review will summarize our understanding of these interactions and provide an outline of shoot–root communication in nitrate systemic regulation. In addition, we describe current progress in discovering the genetic network for nitrate-dependent LR and root hair regulation.
Lateral root and root hair development response to external nitrate
The root systems of higher plants have evolved to be highly plastic, capable of recognizing and colonizing fertile soils; a phenomenon known as ‘root foraging’ (Drew et al., 1973; Motte and Beeckman, 2019). In general, root foraging is triggered by an uneven distribution of nutrients in the environment. For example, a localized concentration of nitrate, as compared with uniform nitrate levels, promotes more significant root development changes in plants (Zhang and Forde, 2000). Such a localized external nitrate treatment could stimulate LR development in many plant species, including maize, barley, rice, and Arabidopsis (Xu et al., 2012; Forde, 2014). In homogenous nitrate conditions, N deficiency (<1 mM) represses LR development, and excessive N supply (>10 mM) exerts a systemic inhibitory effect on LR development (Zhang et al., 1999, 2007). It is interesting that the post-embryonic LRs rather than the embryonic primary root or seminal root tend to show more sensitivity and plasticity in response to external nitrate signals in both dicots and monocots, despite the substantial differences in monocot and dicot root system architecture (Forde, 2014; Tian et al., 2014; Steffens and Rasmussen, 2016).
In a typical dicot tap root system, such as Arabidopsis, the embryonic primary root grows vertically downward, and smaller post-embryonic LRs arise from the sides (Petricka et al., 2012). In monocot plants, such as rice, wheat, and maize, their fibrous root systems contain embryonic primary and seminal roots, as well as post-embryonic adventitious roots (also called nodal roots, brace roots, or crown roots) and LRs (Coudert et al., 2010; Hochholdinger et al., 2018). Despite the major divergence of root system architecture between monocots and dicots, the formation of LRs seems to be fairly conserved at the anatomical level (Motte and Beeckman, 2019). LR development begins from founder cells in the pericycle tissue; these founder cells divide to form an LR primordium, and finally the developing LR primordium emerges from the epidermis and becomes the newly formed LR (Peret et al., 2009). The subsequent elongation of LRs is regulated by both external and internal stimuli (Fig. 1A) (Zhang et al., 1999; Motte and Beeckman, 2019).
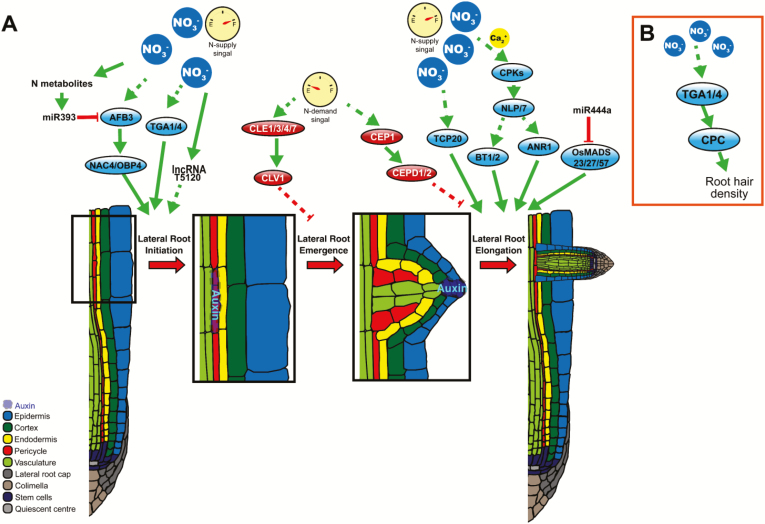
Fig. 1.
Schematic presentation of the genetic network regulating LR and root hair development in plants. (A) Schematic presentation of the genetic network regulating LR development: LR initiation, LR emergence, and LR elongation. (B) Schematic presentation of the genetic network regulating root hair development. Blue ovals indicate positive regulators. Red ovals indicate negative regulators. Green arrows indicate the positive regulation, and red lines indicate negative regulation.
By utilizing a segmented agar plate technique or split-root system, the local nitrate effect on LR initiation, emergence, and elongation has been studied. In non-legume dicot plants, such as Arabidopsis thaliana and tomato, the local nitrate treatment mainly affects the elongation of LRs, and has little or no effect on LR numbers (Zhang et al., 1999; Linkohr et al., 2002; Lu et al., 2010; Forde, 2014), except that a local nitrate deprivation signal does suppress LR initiation in Arabidopsis (Linkohr et al., 2002). In legume dicots, such as Medicago truncatula and Phaseolus vulgaris, nitrate not only promotes growth of LRs but also affects root nodule formation (Guo et al., 2007; Ruffel et al., 2008). On the other hand, in monocots (such as cereals), the local nitrate effect displays a more complicated behavior as LRs originate from different types of roots. For example, both the initiation and elongation of LRs could be induced by local nitrate supply in barley and wheat (Hackett, 1972; Drew et al., 1973). However, in 7- or 14-day-old rice seedling, local nitrate supply only affected elongation of LRs on primary and seminal roots, and LR number was not changed (Wang et al., 2002). In 7-day-old maize seedlings, only elongation of LRs on primary roots is induced (P. Yu et al., 2014), but older maize seedlings show alterations in the density and length of LRs on crown roots by local nitrate treatment (in ‘t Zandt et al., 2015). Thus, the LRs of various cereal species respond differently to nitrate, implying that both species-specific and developmental stage-specific effects are involved. Further research into these differing mechanisms will be essential for better understanding of the nitrate-triggered LR-foraging phenomenon.
Root hairs are specialized epidermal cells that participate in water and nutrient uptake. Surprisingly, there are only few reports about the effect of nitrate availability on root hair development (Foehse and Jungk, 1983; Robinson and Rorison, 1987; Vatter et al., 2015). In tomato, spinach, and rape, the density and length of root hairs were reported to be negatively correlated with homogenous nitrate availability (Foehse and Jungk, 1983). In four grass species, the root hair density and length were also reported to decrease as the nitrate concentration increases in nutrient solution (Robinson and Rorison, 1987). These data indicate that, in a homogenous nitrate environment, root hair development is gradually inhibited by a systemic effect similar to the effect on LR development. Interestingly, in a recent report, root hair development in Arabidopsis was found to respond to local nitrate availability (Alvarez et al., 2014; Vatter et al., 2015). In Arabidopsis, root hair density generally increased with local nitrate concentration in different ecotypes (Vatter et al., 2015). These results indicate the root hair development, like LR development, is under the control of both local and systemic nitrate signaling effects.
Regulation of lateral roots and root hair development by a nitrate-responsive regulatory network
Nitrate serves not only as an N source but also as a signaling molecule in plant nutritional response (Wang et al., 2007). In Arabidopsis, the nitrate signaling pathways have been studied intensively. External nitrate is initially perceived by the dual-affinity transceptor AtNRT1.1/CHL1/AtNPF6.3 (AT1G12110) (Tsay et al., 1993; Liu et al., 1999). After perception, the signal is further transduced by a calcium-dependent pathway and a calcium-independent pathway (Riveras et al., 2015; Zhang et al., 2019). In the calcium-dependent pathway, calcium acts as a secondary messenger for the nitrate signal. Its accumulation in the cytoplasm and nucleus accompanies the nuclear translocation of CALCIUM-SENSOR PROTEIN KINASES (CPKs), such as AtCPK10, AtCPK30, and AtCPK32 (Liu et al., 2017). The nuclear translocation of AtCPK10/30/32 transduces the signal into the regulatory networks in the nucleus, triggers the primary nitrate response, and further influences the development of the root system (Liu et al., 2017). In the calcium-independent pathway, the expression of AUXIN SIGNALING F-BOX3 (AtAFB3) is triggered by intracellular nitrate, and this regulates downstream genes such as NAC3/OBP4 that influence primary root and LR development (Vidal et al., 2010, 2013; Riveras et al., 2015).
Although nitrate perception and primary nitrate response is a rapid and local process that takes a few minutes, the signal needs to be further transmitted to initiate the physiological and developmental response (Wang et al., 2003; Hu et al., 2009). These kinds of response to nitrate, such as the regulation of root system development, are much slower, usually needing hours or days. The transcription factors involved in nitrate-dependent LR development collaborate with both local and systematic signals, and these transcription factors are discussed below (Fig. 1A, B).
The nitrate–CPK–NLP signaling plays a central role in nitrate-responsive lateral root regulation
Ca2+ is an universal second messenger of diverse signaling pathways, involved in biotic and abiotic stresses (Knight et al., 1996; Boudsocq et al., 2010). By introducing aequorin reporter lines, the levels of calcium were observed to fluctuate in cytoplasm in response to nitrate availability. In addition, this process occurs downstream of AtNRT1.1, which indicates that Ca2+ can serve as a second messenger in nitrate signaling (Riveras et al., 2015). On the other hand, application with a phospholipase C (PLC) inhibitor (U73122), calcium chelator EGTA, or the calcium channel blocker La3+ could also repress the nitrate-induced expression of NIR and NIA genes in Arabidopsis, maize, and barley (Sakakibara et al., 1997; Sueyoshi et al., 1999; Riveras et al., 2015). These data demonstrated that the PLC activity and Ca2+ play a crucial role in nitrate signaling. In the Arabidopsis chemically engineered mutant icpk, mutation of CPK10, CPK30, and CPK32 disrupts not only the nitrate-specific stimulation of LR initiation and elongation, but also the nuclear location and phosphorylation of NIN-like proteins (NLPs; Liu et al., 2017). Further studies show that CPK–NLP signaling controls both the primary nitrate response and nitrate-responsive root regulation (Castaings et al., 2009; Liu et al., 2017). In Arabidopsis, AtNLP7 was reported to be capable of binding to the promotor region of several nitrate-responsive transcription factors, such as ANR1 and BT1/2 (Marchive et al., 2013). In legume species, NRSYM1/LjNLP4 in Lotus japonicus and MtNLP1/4 in M. truncatula were shown to be involved in nitrate-dependent nodule formation (Lin et al., 2018; Nishida et al., 2018). Combined together, the nitrate–CPK–NLP signaling pathway may be genetically conserved in the nitrate response regulatory network in most plants, thereby influencing LR development and nodule formation (Liu et al., 2017; Mu and Luo, 2019).
miR393/AFB3 module involved in systemic regulation of lateral root initiation
In the Arabidopsis calcium-independent nitrate signaling pathway, elevated intracellular nitrate induces expression of AFB3 in root tips and the pericycle area (Vidal et al., 2010; Riveras et al., 2015). Mutation of AFB3 leads to shorter primary roots and lower LR density in the nitrate-rich side of a split-root system, which indicates that AFB3 is involved in regulating nitrate-dependent LR initiation and primary root development (Vidal et al., 2010). Further studies revealed that AFB3 acts upstream of NAC3 and OBP4 by up-regulating the expression of NAC3 and OBP4 to further control LR initiation under auxin signaling (Vidal et al., 2013). In response to a systemic signal, the AFB3 module could be feedback regulated by AtmiR393, an miRNA induced by products of nitrate assimilation (Vidal et al., 2010). Working together, AtmiR393/AFB3, NAC3, and OBP4 are involved in the crosstalk between nitrate signaling and auxin signaling to regulate LR initiation (Fig. 1A).
The role of AGL17-Like MADS-box transcription factor in nitrate-dependent regulation of lateral root elongation
In Arabidopsis, the AGL17-Like gene AGL44/AtANR1 was first reported as a MADS-box transcription factor, involved in promoting LR elongation in response to localized nitrate treatment (Zhang and Forde, 1998; Gan et al., 2012). AtANR1 is expressed in LR primordia and the primary root apex, it promotes meristematic activity, and it was induced by nitrate signal downstream of AtNRT1.1 (Remans et al., 2006). Overexpression of AtANR1 significantly increased the LR number and length in Arabidopsis (Gan et al., 2012). In rice, there are five AGL17-Like homologs (OsMADS23, 25, 27, 57, and 61), and three of them (OsMADS25, 27, and 57) were responsive to nitrate signal (C.Y. Yu et al., 2014). OsMAD25 and 27 exert an inhibitory effect on primary root development, but they promote the elongation of LRs in an auxin-dependent manner (Yu et al., 2015; Chen et al., 2018; Zhang et al., 2018). Additionally, the overexpression of OsMAD25 and 27 enhances salinity tolerance in rice via modulation of the ABA signaling pathway (Chen et al., 2018; Xu et al., 2018). The monocot-specific miR444 affects post-transcription inhibition of OsMADS genes, because overexpression of OsmiR444 reduced the accumulation of OsMADS23, 27, and 57 which in turn decreased LR elongation in a nitrate-dependent manner (Yan et al., 2014). Additionally, the AGL17-Like homolog in chrysanthemum, CmANR1, had also been reported to positively modulate both adventious root and LR development, which occurs by directly regulating the auxin transport gene CmPIN2 (Sun et al., 2018). Despite the evolutionary divergence between these species, the role of ANR1-related genes seems to be conserved for regulating nitrate-dependent LR elongation in plants (Fig. 1A).
BT1/2 is involved in systemic regulation of lateral root elongation
In Arabidopsis, the Bric-a-Brac/Tramtrack/Broad (BTB) and TAZ DOMAIN PROTEIN 2 (BT2) was first found as an activator of telomerase in mature leaves, and it also plays a crucial role in gametophyte development (Ren et al., 2007). Further studies have demonstrated that the expression of BT2 is under the control of light, nutrients, hormones, and stresses, suggesting that BT2 is a key element in integrating multiple signaling networks (Mandadi et al., 2009). Using systems biology and bioinformatic tools, BT2 has been predicted to be the potential regulator in the NUE network (Araus et al., 2016). BT1 is the closest homolog of BT2, and expression of both BT1 and BT2 is nitrate inducible (Sato et al., 2017). Under low nitrate availability conditions, the bt1/bt2 double mutant exhibited an extended LR phenotype as compared with wild-type plants (Araus et al., 2016). However under high nitrate conditions, the bt1-1 bt2-4 double mutant showed shorter LRs in comparison with wild-type plants (Sato et al., 2017). These results suggest that BT1 and BT2 are required not only for LR elongation under high nitrate conditions, but also for the inhibition of LR elongation under low nitrate availability. The distinct roles played by BT1 and BT2 under different nitrate conditions implicates them in systemic regulation of LR development (Fig. 1A).
TCP20 interacts with NLP6/7 to control systemic regulation of LR elongation
In Arabidopsis, by studying the nitrate-responsive cis-element (NRE) region of NRT2.1 and NIA1 using the yeast one-hybrid assay, the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1-20 (AtTCP20) was identified as binding the promotor of NRT1.1, NRT2.1, and NIA1 (Guan et al., 2014). Mutation of AtTCP20 causes defective nitrate foraging root phenotypes in split-root systems, indicating that AtTCP20 plays a key role in the systemic nitrate response (Guan et al., 2014). Further studies show the AtTCP20 physically interacts with AtNLP6/7, and subcellular localizations of TCP20/NLP6/7 complexes depend on nitrate availability (Guan et al., 2017). The intranuclear interaction between AtTCP20 and AtNLP6&7 represses expression of the cell cycle marker gene CYCB1;1 in roots under N starvation (Guan et al., 2017). In other species, such as chrysanthemum, the overexpression of CmTCP20 influences auxin accumulation and promotes LR development (Fan et al., 2019a, b). Further studies show that CmTCP20 also interacts with CmARF8, binding to the proximal site in the promoter region of CmCYCB1;1 to positively regulate the cell cycle in the root (Fan et al., 2019b). These results indicate that TCP20 has two distinct functions in root development under the control of local and systemic signaling, and may be genetically conserved in plants (Fig. 1A).
TGA1 and TGA4 respond to local nitrate and regulate LR development and root hair initiation
By integrative network bioinformatics, TGA1 and TGA4 were identified as potential regulatory factors of nitrate response in Arabidopsis (Alvarez et al., 2014). Expression of both TGA1 and TGA4 is induced downstream of AtNRT1.1 with external nitrate application (Alvarez et al., 2014). ChIP assays revealed that TGA1 could directly bind to the promoter of AtNRT2.1/2.2 and promote the expression of NRT2.1/2.2 (Alvarez et al., 2014). Mutations of both TGA1 and TGA4 inhibit LR initiation, emergence, root hair initiation, and primary root length (Alvarez et al., 2014; Canales et al., 2017). These results indicate that the TGA1/4 genes regulate LR and root hair initiation by acting downstream of AtNRT1.1 but upstream of AtNRT2.1/2.2 (Fig. 1A) (Alvarez et al., 2014). However, only a few studies revealed that root hair development also responds to nitrate levels. Nitrate was reported to play a key role in controlling root hair initiation on developing roots (Canales et al., 2014, 2017; Vatter et al., 2015). The meta-transcriptomics analysis demonstrated that a set of co-expressed genes involved in root hair development also respond to external nitrate (Canales et al., 2014). Similar to LR development, root hair density of the NRT1.1-related mutant displayed a significant reduction in comparison with wild-type plants, suggesting that root hair development is also controlled by NRT1.1-based nitrate signaling (Vatter et al., 2015; Canales et al., 2017). Further experiments revealed that external nitrate treatment could increase root hair density in Arabidopsis. The increased root hair number is mainly due to a reduction in the longitudinal cell length of trichoblasts, and this process requires NRT1.1, TGA1/4, and CPC (Canales et al., 2017). The ChIP assay demonstrated that TGA1 could directly bind to the –1839 to –1831 region of the CPC promoter, which promotes CPC expression (Canales et al., 2017). These results suggest that a TGA1/CPC module responds to an NRT1.1-mediated external nitrate signal to control root hair density and nitrate uptake efficiency in Arabidopsis (Fig. 1B).
A CLE–CLV1 module involved in systemic lateral root inhibition
The CLAVATA3 (CLV3)/ENDOSPERM SURROUNDING REGION (ESR)-related (CLE) family were first identified as extracellular peptides that interact with CLAVATA1 (CLV1), a type XI leucine-rich repeat receptor-like kinase (LRR-RLK), to control cell proliferation and differentiation in the shoot apical meristem in Arabidopsis (Fiers et al., 2007; Butenko et al., 2009). Among 32 CLE genes in Arabidopsis, the expression of four CLE genes (CLE1, 3, 4, and 7) was found to be induced in a dose-dependent manner in roots under nitrate deficit conditions (Araya et al., 2014; Goad et al., 2017). Promoter-driven green fluorescent protein (GFP) analyses indicate that CLE1, 2, 3, 4, and 7 are predominantly expressed in root pericycle cells, and overexpression of any of these genes is sufficient to inhibit LR development (Araya et al., 2014). On the other hand, among mutants of the nine type XI LRR-RLK genes, only the clv mutant displayed a significant extension of LRs under N deficit conditions (Araya et al., 2014). Overexpression of CLE3 leads to significant LR inhibition, but not in the clv1-4 mutant background, which indicates that CLE-induced LR inhibition requires CLV1 (Araya et al., 2014). CLV1 expression is located in phloem companion cells, separate from pericycle cells expressing CLE3, implying that the CLE–CLV1 module participates in intercellular signaling for the systemic N response. The CLV3/ESR (CLE) protein family is found in at least 19 species across monocots and dicots (Oelkers et al., 2008). In addition to data in Arabidopsis, the CLEs were reported to be involved in root nodulation of legumes and root development in wheat (Mortier et al., 2010; Nishida et al., 2018; Li et al., 2019). The above evidence implies that CLE function in nitrate-dependent root regulation may be conserved in other species.
Long non-coding RNA T5120 regulates nitrate-dependent lateral root initiation
In Arabidopsis, TCONS_00005120(T5120) was identified as a novel long non-coding RNA (lncRNA) induced by external nitrate, using high-throughput strand-specific RNA-seq (Liu et al., 2019). The expression of T5120 was very low in all tested organs, and it was strongly induced by nitrate in a time- and concentration-dependent manner (Liu et al., 2019). In an NR-null mutant (nia1 nia2), the expression of T5120 could still be specifically induced by external nitrate, meaning that T5120 responds to nitrate itself but not its reduction products (Liu et al., 2019). Further experiments demonstrated that AtNLP7 directly binds to the NRE-like motif of the T5120 promoter and positively regulates T5120 transcription (Liu et al., 2019). The lncRNA T5120 acts downstream of AtNLP7 and AtNRT1.1 and promotes nitrate assimilation and LR initiation in Arabidopsis (Liu et al., 2019). The lncRNAs have been demonstrated to play multiple roles in plant development, but given that lncRNAs are highly variable during evolution, the functions of lncRNAs in nitrate-dependent processes need to be further investigated in other species (Ulitsky, 2016). Thus, screening potential lncRNAs involved in nitrate-dependent root development could be a practical tool for improved NUE in crops.
Distance communication involved in nitrate-dependent root regulation
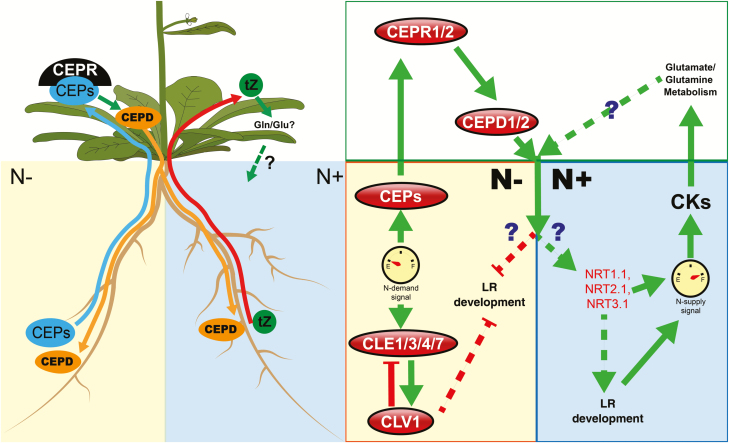
The root system represents the only subterranean portion of the plant, and recent studies show that the complicated root-foraging response depends on long-distance root–shoot–root communication (Ruffel et al., 2011; de Bang et al., 2017; McCleery et al., 2017; Ohkubo et al., 2017; Roy, 2018). These studies suggest that N demand signals are generated from the N-deprived side of the root system, and are transmitted from root to shoot (Ohkubo et al., 2017). On the other hand, N supply signals are generated from the N-rich side of the root system and are also transmitted from root to shoot (Ruffel et al., 2011). The shoot perceives these N demand versus N supply signals and, in turn, generates corresponding signals that are transmitted to each side of the root system to modulate root development, NRT2.1 expression, and nitrate uptake (Fig. 2).
Fig. 2.
Schematic presentation of distance communication in nitrate systemic signaling in plants. Left: distance communication in nitrate systemic signaling. The blue arrow indicates the ascending N-demanding signal, the red arrow indicates the ascending N supply signal, yellow arrows indicate the descending signal, and green arrows indicate positive regulation. Right: schematic presentation of the genetic network involved in nitrate systemic signaling. A green oulined square represents the shoots. The orange outlined square represents the N-deprived side of of root. The blue outlined square represents the N-rich side of the root. Green arrows indicate positive regulation, and red lines indicate negative regulation.
A CEP1–CEPR1/2–CEPD1/2 small peptide hormone pathway is required for N demand signaling
The C-TERMINALLY ENCODED PEPTIDE genes were discovered using an in silico approach. They are post-translationally modified, secreted peptide hormones that respond to N demand signaling, and they act through their receptors to control nodulation and root architecture in plants (Taleski et al., 2018). In Arabidopsis, C-terminally encoded peptide 1 (AtCEP1) was first reported as a small peptide hormone expressed in LR primordia to function in inhibition of root growth (Ohyama et al., 2008). In silico analyses confirmed a total of 15 CEP genes in the Arabidopsis genome, including seven (AtCEP1/3/5/6/7/8/9) up-regulated when roots suffer from N starvation conditions. These small peptides are synthesized in the stele of LRs (Roberts et al., 2013; Tabata et al., 2014), and then loaded into the xylem vessels and further transported from root to shoot as an ascending systemic N demand signal to leaves (Tabata et al., 2014). In vascular tissues of the leaf, AtCEP1 is perceived by an LRR-RK, named CEP receptor 1/2 (AtCEPR1/2) (Tabata et al., 2014). Mutating both AtCPER1 and AtCEPR2 not only caused growth retardation in N-rich medium with N deficiency symptoms, but also impaired the systemic N demand signaling response (Tabata et al., 2014). When shoots perceived the CEP1 signal from roots by AtCEPR1 and AtCEPR2, the non-secreted small signaling peptides AtCEPD1 and AtCEPD2 were induced and then translocated to both N-rich and N-deprived sides of the root. However, only the expression of NRT2.1 in the N-rich side of LRs was induced (Ohkubo et al., 2017). These results suggest that the descending AtCEPD1/2 signal needs to cooperate with local signals on both sides (Ohkubo et al., 2017). On the other hand, the cepd1,2 double mutant produces longer LRs, which indicates that the descending AtCEPD1/2 signal might be involved in systemic inhibition of LR elongation in the nitrate-deprived side (Fig. 2) (Ohkubo et al., 2017).
In other species, the CEPs were also reported to be involved in nitrate-dependent root development. In M. truncatula, the expression of MtCEP1/2 was up-regulated under low nitrate conditions, inhibited LR initiation and emergence, and promoted nodule formation (Imin et al., 2013). In Oryza sativa, the expression level of OsCEP6.1 was induced by low nitrate availability. The overexpression of OsCEP6.1 and application of synthesized OsCEP6.1 significantly reduced root growth (Sui et al., 2016). In addition, phylogenetic analyses indicated that CEP genes exist in most angiosperm plants, which means that CEP-mediated pathways may be conserved during plant evolution (Ogilvie et al., 2014). Together, these studies suggested that CEP peptide hormones play important roles in orchestrating N demand signaling, root nodulation, and LR development in plants.
Root–shoot communication of cytokinin involved in N supply systemic signaling
Among phytohormones, cytokinin (CK) has been considered to be closely linked to N signaling (Miyawaki et al., 2004; Sakakibara et al., 2006). Previous studies demonstrated that the CK-dependent systemic N demand signaling controls LR initiation in Arabidopsis (Ruffel et al., 2016). Recently, by introducing abcg14 and the isopentenyl transferase ipt3 ipt5 ipt7 triple mutant to the typical split-root system, CK was confirmed to play an essential role in root to shoot communication of systemic N demand signaling (Ruffel et al., 2011; Poitout et al., 2018). Compared with wild-type plants, the ipt3 ipt5 ipt7 triple mutant failed to maintain a proper primary response in the split-root system, which means that biosynthesis of CK is required for N systemic signaling under heterogeneous nitrate conditions (Poitout et al., 2018). Similarly, the abcg14 mutant, which cannot translocate CKs from root to shoot, also displayed an impaired systemic N response phenotype (Poitout et al., 2018). A UHPLC-MS analyses of various active forms of CK in shoots and roots revealed that trans-zeatin (tZ) accumulation in shoots is required for the systemic N response in roots (Poitout et al., 2018). Collectively, the biosynthesis of CKs was triggered by N-rich conditions in roots, then the tZ-type CK was translocated to the shoot by ABCG14 as an ascending signal, causing accumulation of active tZ in shoots that controls the N supply signaling in roots (Fig. 1) (Takei et al., 2004; Ko et al., 2014; Zhang et al., 2014; Poitout et al., 2018). Transcriptomic analyses indicate that the translocated active tZ in shoots results in modified expression of glutamate and glutamine biosynthesis (Fig. 2) (Poitout et al., 2018). This further confirms the importance of CK-dependent root–shoot communication in nitrate systemic signaling, and leads to a model in which CK-dependent root–shoot communication affects glutamate/glutamine metabolism in shoots, emphasizing the role of CK in nitrate systemic signaling.
Concluding remarks and future perspectives
Over the past several years, an increasing number of players involved in local and systemic nitrate signaling have been discovered. The emerging story of nitrate-dependent systemic regulation places particular emphasis on root–shoot–root distance communication (Ohkubo et al., 2017; Poitout et al., 2018). After external nitrate signals are perceived by roots, a signal is generated that travels from the root to the shoot. In turn, the shoots produce a descending signal responsible for the root-foraging phenomenon. However, we still lack an understanding of how the descending shoot–root signals act differently on each side of split-root systems (Ohkubo et al., 2017). Furthermore, the nitrate-dependent LR development is clearly under control of both local and systemic nitrate signaling pathways. Since the systemic signaling is a new and emerging field of nitrate signaling, we have highlighted some of the key components responsible for root foraging in both local and systemic signaling pathways. For example, the CEPD1/2 descending signal is distributed equally, but it causes different phenotypes under different nitrate conditions (Ohkubo et al., 2017). Further, the transcription factors TGA1/4, TCP20, and BT1/BT2 were shown to have essential roles in systemic nitrate signaling, but we still have little mechanistic understanding of the crosstalk between the local and systemic nitrate signaling pathways (Guan et al., 2014; Araus et al., 2016; Sato et al., 2017). There is a need to use integrated systems biology approaches and functional genomics tools to dissect the complex regulatory networks behind these processes.
At the same time, many regulators have been identified that influence LR development in the nitrate-dependent signaling pathway in model plants, such as Arabidopsis and rice (Vidal et al., 2010; Guan et al., 2014; Wang et al., 2018). The LR initiation determines the number of LRs that branch from the primary root, while the elongation of LRs enhances nitrate uptake (Zhan and Lynch, 2015). Moreover, root hair development is also regulated by nitrate (Vatter et al., 2015). As shown in Fig. 1, there are multiple regulators responsible for the same process, implicating that these regulatory mechanisms are redundant, and in some cases antagonistic to each other. Thus, the genetic relationship between the various regulators still remains elusive and requires the use of an array of molecular and genetic tools to decipher. The nitrate-dependent regulation of LR and root hair development may share a common intrinsic mechanism (such as NLPs, AGL14-like MADS-box transcription factors, and TCP20), but some of the regulators (such as OsmiR444 and lncRNA T5120) are species specific. Considering that most of these regulator effects on nitrate-dependent LR and root hair development have only been reported in Arabidopsis, more species need to be tested for these regulators in order to dissect the specificity or generality of the regulatory pathways. Although there is increased understanding of nitrate signaling in grasses, it will be important to further investigate any potential new regulatory mechanisms in more species. Progress in understanding nitrate signaling and regulation mechanisms in monocot crops will probably contribute to the development of sustainable and energy-efficient agriculture.
Acknowledgements
The research was supported by The National Key R & D Program of China [2016YFD0100701], the National Natural Science Foundation of China [grant nos 31570183, 31529001, and 31370215], and the China Postdoctoral Science Foundation [grant no. 2018M632476]. We thank Professor Hao Yu for critical reading of the manuscript.
Glossary
Abbreviations:
- ABCG
ATP-binding cassette subfamily G
- AFB
auxin signaling F-box
- ANR1
Arabidopsis nitrate regulated 1
- BT2
Bric-a-brac/Tramtrack/Broad (BTB) and TAZ domain protein 2
- CBL
calcineurin B-like protein
- CEP
C-terminally encoded peptide
- CEPD
CEP downstream
- CEPR
CEP receptor
- CHL
chlorate
- CK
cytokinin
- CLE
CLAVATA3/endosperm surrounding region (ESR)-related
- CLV
CLAVATA
- CPC
CAPRICE
- CPK
calcium sensor protein kinase
- GFP
green fluorescent protein
- IPT
isopentenyl transferase
- LR
lateral root
- LRR-RLK
leucine-rich repeat receptor-like kinase
- MADS
MINICHROMOSOME MAINTENANCE1/AGAMOUS/DEFICIENS/SERUM RESPONSE FACTOR
- NAC
NAM/ATAF/CUC
- NBIP1
NRT1.1B-interacting protein 1
- NIA
nitrate reductase
- NIN
NODULE INCEPTION
- NIR
nitrite reductase
- NLP
NIN-like protein
- NPF
nitrate transporter 1/peptide transporter family
- NRT
nitrate transporter
- NUE
nitrogen use efficiency
- OBP
OCS element-binding factor (OBF)-binding protein
- TCP
TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR
- TGACG
sequence-specific binding protein
- tZ
trans-zeatin
References
- Alvarez JM, Riveras E, Vidal EA, et al. 2014. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal 80, 1–13. [DOI] [PubMed] [Google Scholar]
- Araus V, Vidal EA, Puelma T, Alamos S, Mieulet D, Guiderdoni E, Gutiérrez RA. 2016. Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiology 171, 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H. 2014. CLE–CLAVATA1 peptide–receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proceedings of the National Academy of Sciences, USA 111, 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellegarde F, Gojon A, Martin A. 2017. Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. Journal of Experimental Botany 68, 2553–2565. [DOI] [PubMed] [Google Scholar]
- Berhe AA, Torn MS. 2017. Erosional redistribution of topsoil controls soil nitrogen dynamics. Biogeochemistry 132, 37–54. [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. 2010. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles TM, Atallah SS, Campbell EE, Gaudin ACM, Wieder WR, Grandy AS. 2018. Addressing agricultural nitrogen losses in a changing climate. Nature Sustainability 1, 399–408. [Google Scholar]
- Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. 2009. Plant peptides in signalling: looking for new partners. Trends in Plant Science 14, 255–263. [DOI] [PubMed] [Google Scholar]
- Canales J, Contreras-López O, Álvarez JM, Gutiérrez RA. 2017. Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. The Plant Journal 92, 305–316. [DOI] [PubMed] [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA. 2014. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Frontiers in Plant Science 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, et al. 2009. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. The Plant Journal 57, 426–435. [DOI] [PubMed] [Google Scholar]
- Chen H, Xu N, Wu Q, Yu B, Chu Y, Li X, Huang J, Jin L. 2018. OsMADS27 regulates the root development in a NO3–-dependent manner and modulates the salt tolerance in rice (Oryza sativa L.). Plant Science 277, 20–32. [DOI] [PubMed] [Google Scholar]
- Coudert Y, Périn C, Courtois B, Khong NG, Gantet P. 2010. Genetic control of root development in rice, the model cereal. Trends in Plant Science 15, 219–226. [DOI] [PubMed] [Google Scholar]
- de Bang TC, Lay KS, Scheible WR, Takahashi H. 2017. Small peptide signaling pathways modulating macronutrient utilization in plants. Current Opinion in Plant Biology 39, 31–39. [DOI] [PubMed] [Google Scholar]
- Drew MC, Saker LR, Ashley TW. 1973. Nutrient supply and growth of seminal root system in barley. 1. Effect of nitrate concentration on growth of axes and laterals. Journal of Experimental Botany 24, 1189–1202. [Google Scholar]
- Fan HM, Liu BW, Ma FF, Sun X, Zheng CS. 2019a Proteomic profiling of root system development proteins in chrysanthemum overexpressing the CmTCP20 gene. Plant Science 287, 110175. [DOI] [PubMed] [Google Scholar]
- Fan HM, Sun CH, Wen LZ, Liu BW, Ren H, Sun X, Ma FF, Zheng CS. 2019b CmTCP20 plays a key role in nitrate and auxin signaling-regulated lateral root development in chrysanthemum. Plant & Cell Physiology 60, 1581–1594. [DOI] [PubMed] [Google Scholar]
- Fiers M, Ku KL, Liu CM. 2007. CLE peptide ligands and their roles in establishing meristems. Current Opinion in Plant Biology 10, 39–43. [DOI] [PubMed] [Google Scholar]
- Foehse D, Jungk A. 1983. Influence of phosphate and nitrate supply on root hair formation of rape, spinach and tomato plants. Plant and Soil 74, 359–368. [Google Scholar]
- Forde BG. 2014. Nitrogen signalling pathways shaping root system architecture: an update. Current Opinion in Plant Biology 21, 30–36. [DOI] [PubMed] [Google Scholar]
- Gan Y, Bernreiter A, Filleur S, Abram B, Forde BG. 2012. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant & Cell Physiology 53, 1003–1016. [DOI] [PubMed] [Google Scholar]
- Goad DM, Zhu C, Kellogg EA. 2017. Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytologist 216, 605–616. [DOI] [PubMed] [Google Scholar]
- Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM. 2017. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proceedings of the National Academy of Sciences, USA 114, 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Wang R, Nacry P, Breton G, Kay SA, Pruneda-Paz JL, Davani A, Crawford NM. 2014. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proceedings of the National Academy of Sciences, USA 111, 15267–15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SW, Shen QR, Brueck H. 2007. Effects of local nitrogen supply on water uptake of bean plants in a split root system. Journal of Integrative Plant Biology 49, 472–480. [Google Scholar]
- Hackett C. 1972. Method of applying nutrients locally to roots under controlled conditions, and some morphological effects of locally applied nitrate on branching of wheat roots. Australian Journal of Biological Sciences 25, 1169–1180. [Google Scholar]
- Hochholdinger F, Yu P, Marcon C. 2018. Genetic control of root system development in maize. Trends in Plant Science 23, 79–88. [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. 2009. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. The Plant Journal 57, 264–278. [DOI] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA. 2013. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. Journal of Experimental Botany 64, 5395–5409. [DOI] [PubMed] [Google Scholar]
- in ‘t Zandt D, Le Marie C, Kirchgessner N, Visser EJW, Hund A. 2015. High-resolution quantification of root dynamics in split-nutrient rhizoslides reveals rapid and strong proliferation of maize roots in response to local high nitrogen. Journal of Experimental Botany 66, 5507–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. 1996. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. The Plant Cell 8, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D, Kang J, Kiba T, et al. 2014. Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proceedings of the National Academy of Sciences, USA 111, 7150–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu D, Xia Y, Li Z, Niu N, Ma S, Wang J, Song Y, Zhang G. 2019. Identification and functional analysis of the CLAVATA3/EMBRYO SURROUNDING REGION (CLE) gene family in wheat. International Journal of Molecular Sciences 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Li X, Luo Z, Mysore KS, Wen J, Xie F. 2018. NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nature Plants 4, 942–952. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. The Plant Journal 29, 751–760. [DOI] [PubMed] [Google Scholar]
- Liu F, Xu Y, Chang K, Li S, Liu Z, Qi S, Jia J, Zhang M, Crawford NM, Wang Y. 2019. The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis. New Phytologist 224, 117–131. [DOI] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. 1999. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. The Plant Cell 11, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Niu Y, Konishi M, et al. 2017. Discovery of nitrate–CPK–NLP signalling in central nutrient-growth networks. Nature 545, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Li Q, Xu X, Zhu Y, Dong C, Shen Q. 2010. Responses of tomato seedling to different nitrogen forms supplied by either homogenous or localized culture. Journal of Nanjing Agricultural University 33, 43–49. [Google Scholar]
- Mallory EB, Griffin TS, Porter GA. 2010. Seasonal nitrogen availability from current and past applications of manure. Nutrient Cycling in Agroecosystems 88, 351–360. [Google Scholar]
- Mandadi KK, Misra A, Ren S, McKnight TD. 2009. BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiology 150, 1930–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A. 2013. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nature Communications 4, 1713. [DOI] [PubMed] [Google Scholar]
- McCleery WT, Mohd-Radzman NA, Grieneisen VA. 2017. Root branching plasticity: collective decision-making results from local and global signalling. Current Opinion in Cell Biology 44, 51–58. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. 2004. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. The Plant Journal 37, 128–138. [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S. 2010. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology 153, 222–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motte H, Beeckman T. 2019. The evolution of root branching: increasing the level of plasticity. Journal of Experimental Botany 70, 785–793. [DOI] [PubMed] [Google Scholar]
- Mu X, Luo J. 2019. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cellular and Molecular Life Sciences 76, 3753–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Tanaka S, Handa Y, et al. 2018. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nature Communications 9, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. 2008. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie HA, Imin N, Djordjevic MA. 2014. Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes. BMC Genomics 15, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa-Ohnishi M, Matsubayashi Y. 2017. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nature Plants 3, 17029. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. The Plant Journal 55, 152–160. [DOI] [PubMed] [Google Scholar]
- Pandey A, Suter H, He JZ, Hu HW, Chen DL. 2019. Dissimilatory nitrate reduction to ammonium dominates nitrate reduction in long-term low nitrogen fertilized rice paddies. Soil Biology & Biochemistry 131, 149–156. [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science 14, 399–408. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. 2012. Control of Arabidopsis root development. Annual Review of Plant Biology 63, 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitout A, Crabos A, Petřík I, Novák O, Krouk G, Lacombe B, Ruffel S. 2018. Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-zeatin in shoots. The Plant Cell 30, 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. 2006. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proceedings of the National Academy of Sciences, USA 103, 19206–19211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Mandadi KK, Boedeker AL, Rathore KS, McKnight TD. 2007. Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. The Plant Cell 19, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA. 2015. The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiology 169, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I, Smith S, De Rybel B, Van Den Broeke J, Smet W, De Cokere S, Mispelaere M, De Smet I, Beeckman T. 2013. The CEP family in land plants: evolutionary analyses, expression studies, and role in Arabidopsis shoot development. Journal of Experimental Botany 64, 5371–5381. [DOI] [PubMed] [Google Scholar]
- Robinson D, Rorison IH. 1987. Root hairs and plant-growth at low nitrogen availabilities. New Phytologist 107, 681–693. [Google Scholar]
- Roy S. 2018. Nitrate Ahoy! Shoot cytokinin signals integrate growth responses with nitrogen availability. The Plant Cell 30, 1169–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Freixes S, Balzergue S, et al. 2008. Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula. Plant Physiology 146, 2020–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. 2011. Nitrogen economics of root foraging: transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences, USA 108, 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Poitout A, Krouk G, Coruzzi GM, Lacombe B. 2016. Long-distance nitrate signaling displays cytokinin dependent and independent branches. Journal of Integrative Plant Biology 58, 226–229. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kobayashi K, Deji A, Sugiyama T. 1997. Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant & Cell Physiology 38, 837–843. [Google Scholar]
- Sakakibara H, Takei K, Hirose N. 2006. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends in Plant Science 11, 440–448. [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Konishi M, Yoshioka N, Sasaki Y, Maeda H, Ishida T, Kato Y, Yamaguchi J, Yanagisawa S. 2017. Direct transcriptional activation of BT genes by NLP transcription factors is a key component of the nitrate response in Arabidopsis. Biochemical and Biophysical Research Communications 483, 380–386. [DOI] [PubMed] [Google Scholar]
- Steffens B, Rasmussen A. 2016. The physiology of adventitious roots. Plant Physiology 170, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi K, Mitsuyama T, Sugimoto T, Kleinhofs A, Warner RL, Oji Y. 1999. Effects of inhibitors for signaling components on the expression of the genes for nitrate reductase and nitrite reductase in excised barley leaves. Soil Science and Plant Nutrition 45, 1015–1019. [Google Scholar]
- Sui ZP, Wang TY, Li HJ, Zhang M, YY Li, Xu RB, Xing GF, Ni ZF, Xin MM. 2016. Overexpression of peptide-encoding OsCEP6.1 results in pleiotropic effects on growth in rice (O. sativa). Frontiers in Plant Science 7, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CH, Yu JQ, Duan X, Wang JH, Zhang QY, Gu KD, Hu DG, Zheng CS. 2018. The MADS transcription factor CmANR1 positively modulates root system development by directly regulating CmPIN2 in chrysanthemum. Horticulture Research 5, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. 2014. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. 2004. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant & Cell Physiology 45, 1053–1062. [DOI] [PubMed] [Google Scholar]
- Taleski M, Imin N, Djordjevic MA. 2018. CEP peptide hormones: key players in orchestrating nitrogen-demand signalling, root nodulation, and lateral root development. Journal of Experimental Botany 69, 1829–1836. [DOI] [PubMed] [Google Scholar]
- Tian H, De Smet I, Ding Z. 2014. Shaping a root system: regulating lateral versus primary root growth. Trends in Plant Science 19, 426–431. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. 1993. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. [DOI] [PubMed] [Google Scholar]
- Ulitsky I. 2016. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nature Reviews. Genetics 17, 601–614. [DOI] [PubMed] [Google Scholar]
- Vatter T, Neuhäuser B, Stetter M, Ludewig U. 2015. Regulation of length and density of Arabidopsis root hairs by ammonium and nitrate. Journal of Plant Research 128, 839–848. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. 2010. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA. 2013. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proceedings of the National Academy of Sciences, USA 110, 12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. 2003. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiology 132, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Crawford N. 2007. Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiology 145, 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu P, Hu B, Chen QS. 2002. Effects of nitrate on the growth of lateral root and nitrogen absorption in rice. Acta Botanica Sinica 44, 678–683. [Google Scholar]
- Wang YY, Cheng YH, Chen KE, Tsay YF. 2018. Nitrate transport, signaling, and use efficiency. Annual Review of Plant Biology 69, 85–122. [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Xu N, Chu Y, Chen H, Li X, Wu Q, Jin L, Wang G, Huang J. 2018. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genetics 14, e1007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Wang H, Hamera S, Chen X, Fang R. 2014. miR444a has multiple functions in the rice nitrate-signaling pathway. The Plant Journal 78, 44–55. [DOI] [PubMed] [Google Scholar]
- Ye C, Cheng X, Liu W, Zhang Q. 2015. Revegetation impacts soil nitrogen dynamics in the water level fluctuation zone of the Three Gorges Reservoir, China. The Science of the Total Environment 517, 76–85. [DOI] [PubMed] [Google Scholar]
- Yu CY, Liu YH, Zhang AD, Su S, Yan A, Huang L, Ali I, Liu Y, Forde BG, Gan YB. 2015. MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS One 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Su S, Xu YC, Zhao YQ, Yan A, Huang LL, Ali I, Gan YB. 2014. The effects of fluctuations in the nutrient supply on the expression of five members of the AGL17 clade of MADS-box genes in rice. PLoS One 9, e0135196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Li X, Yuan L, Li C. 2014. A novel morphological response of maize (Zea mays) adult roots to heterogeneous nitrate supply revealed by a split-root experiment. Physiologia Plantarum 150, 133–144. [DOI] [PubMed] [Google Scholar]
- Zhan A, Lynch JP. 2015. Reduced frequency of lateral root branching improves N capture from low-N soils in maize. Journal of Experimental Botany 66, 2055–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GP, Xu N, Chen HL, Wang GX, Huang JL. 2018. OsMADS25 regulates root system development via auxin signalling in rice. The Plant Journal 95, 1004–1022. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. 2000. Regulation of Arabidopsis root development by nitrate availability. Journal of Experimental Botany 51, 51–59. [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences, USA 96, 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D. 2007. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. Journal of Experimental Botany 58, 2329–2338. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Forde BG. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang K, Novak O, Wei Z, Gou M, Zhang X, Yu Y, Yang H, Cai Y, Strnad M, Liu CJ. 2014. Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nature Communications 5, 3274. [DOI] [PubMed] [Google Scholar]
- Zhang X, Cui Y, Yu M, Su B, Gong W, Baluška F, Komis G, Šamaj J, Shan X, Lin J. 2019. Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiology 181, 480–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GB, Wang SY, Wang Y, Wang CX, Risgaard-Petersen N, Jetten MSM, Yin CQ. 2011. Anaerobic ammonia oxidation in a fertilized paddy soil. Isme Journal 5, 1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]