This first systematic characterization of the whole wheat NPF gene family contributes to a better understanding of this large gene family in wheat and lays the foundation for further functional analysis.
Keywords: Gene expression, nitrate, nitrogen, NPF gene, phylogeny, wheat (Triticum aestivum)
Abstract
NPF genes encode membrane transporters involved in the transport of a large variety of substrates including nitrate and peptides. The NPF gene family has been described for many plants, but the whole NPF gene family for wheat has not been completely identified. The release of the wheat reference genome has enabled the identification of the entire wheat NPF gene family. A systematic analysis of the whole wheat NPF gene family was performed, including responses of specific gene expression to development and nitrogen supply. A total of 331 NPF genes (113 homoeologous groups) have been identified in wheat. The chromosomal location of the NPF genes is unevenly distributed, with predominant occurrence in the long arms of the chromosomes. The phylogenetic analysis indicated that wheat NPF genes are closely clustered with Arabidopsis, Brachypodium, and rice orthologues, and subdivided into eight subfamilies. The expression profiles of wheat NPF genes were examined using RNA-seq data, and a subset of 44 NPF genes (homoeologous groups) with contrasting expression responses to nitrogen and/or development in different tissues were identified. The systematic identification of gene composition, chromosomal locations, evolutionary relationships, and expression profiles contributes to a better understanding of the roles of the wheat NPF genes and lays the foundation for further functional analysis in wheat.
Introduction
Nitrogen (N) is the key nutrient for plant growth and development (Xu et al., 2012). Nitrate transporters are responsible for nitrate uptake from the environment and for internal transport (Y.-Y. Wang et al., 2018). The four gene families involved in nitrate transport comprise NPF (nitrate transporter 1/peptide family), NRT2 (nitrate transporter 2), CLC (chloride channel). and SLAC/SLAH (slow anion channel-associated homologues), and have been reported in Arabidopsis and rice, and were reviewed recently (O’Brien et al., 2016; Fan et al., 2017; H. Li et al., 2017a; Undurraga et al., 2017; Xuan et al., 2017; Tegeder and Masclaux-Daubresse, 2018; Y-Y. Wang et al., 2018). The NPF family includes low-affinity nitrate and peptide transporters sharing high sequence homology and a conserved structural arrangement (Tsay et al., 2007; Léran et al., 2014). The composition of the NPF gene family is complex, with between 51 (in Capsella rubella) and 139 (in Malus domestica) unique members of the NPF gene family having been identified in 33 fully sequenced plant genomes, subdivided into eight subfamilies numbered NPF1–NPF8 (Léran et al., 2014).
NPF genes have diverse and important functions for nitrogen utilization described in the model plants Arabidopsis and rice (Y.-Y. Wang et al., 2018). AtNPF6.3 (CHL1/AtNRT1.1) encodes a dual-affinity (low and high substrate affinity controlled by protein phosphorylation) nitrate transporter, expressed predominantly in roots, and is regulated by N status (Liu et al., 1999; Liu and Tsay, 2003). In addition to roles in nitrate uptake and nitrate translocation from roots to shoots (Tsay et al., 1993; Léran et al., 2013), AtNPF6.3 acts as a nitrate sensor involved in early nitrate signalling of the primary nitrate response (Ho et al., 2009). Subsequently, many other NPF genes have been characterized in Arabidopsis and rice, and shown to be involved in different nitrate transport steps in the plant during development (Y.-Y. Wang et al., 2018). In non-plant species, NPF-encoded proteins transport di- and tripeptides (Tsay et al., 2007). In Arabidopsis and rice, members of NPF subfamilies 7 and 8 have been identified as plasma membrane-located dipeptide transporters involved in dipeptide uptake in roots or pollen tissues, the control of flowering/seed development, and regulating total N content and plant growth (Chiang et al., 2004; Dietrich et al., 2004; Komarova et al., 2008; Fang et al., 2017). Some NPF homologues show chloride or potassium transport activity. AtNPF2.4 loads chloride into the xylem to enable root to shoot chloride transport, and AtNPF2.5 seems to be involved in cortical chloride efflux in the root (B. Li et al., 2016, 2017). AtNPF7.3 may be responsible for proton-coupled potassium loading into the xylem (H. Li et al., 2017b). Although many studies have been conducted on physiological functions of NPF genes, little systematic analysis of the NPF gene family has been reported, especially in hexaploid wheat (Triticum aestivum L.). Previously, wheat NPF genes have been identified and partly described, including detailed gene expression analysis (Buchner and Hawkesford, 2014; Bajgain et al., 2018). With the aid of the recently released wheat reference genome (International Wheat Genome Sequencing Consortium, 2018), we carried out a detailed analysis of the complete NPF gene family in wheat. This systematic analysis included gene composition, chromosomal locations, and phylogenetic relationships with other plant species including Arabidopsis, Brachypodium, and rice. A nomenclature for wheat NPF genes is proposed. Further detailed analysis of expression profiles of wheat NPF genes was performed using RNA sequencing (RNA-seq) data, as well as using quantitative real-time PCR to investigate responses to nitrogen supply and/or development in different tissues.
Materials and methods
Database mining and identification of NPF genes in wheat
Protein sequences of four Arabidopsis nitrate transporter gene families, NPF (53 members), NRT2 (7 members), CLC (7 members), and SLAC/SLAH (5 members), were queried based on a blast analysis in InterPro (http://www.ebi.ac.uk/interpro/) for protein domain analysis. A local wheat protein database was established based on the wheat genome (IWGSCv1.1) (https://wheat-urgi.versailles.inra.fr; International Wheat Genome Sequencing Consortium, 2018). Wheat NPF gene homologues containing the NPF-specific protein domain of ‘Proton-dependent oligopeptide transporter family’ (IPR000109) HMM profiles (http://www.ebi.ac.uk/interpro/) were identified using HMMER v3.0, with the default parameters and an E-value cut-off of 1e−5. A partial domain of IPR000109 and potential false positives were eliminated manually. Candidate NPF genes were subjected to analysis for integrities of ORF, protein length, reliability of gene prediction, and sequence redundancy. Homoeologous groups (HGs) were defined by both transcript and protein sequences with >90% (Wan et al., 2017) sequence identities originating from homoeologous chromosomes.
Phylogenetic analysis
Protein sequence alignments were carried out using MUSCLE (Edgar, 2004) within Geneious® 10.2.3, with default parameters. A phylogenetic tree based on 113 single wheat NPF homoeologues together with 53 Arabidopsis, 75 Brachypodium, and 81 rice NPF protein sequences was constructed as described in Wan et al. (2017) using PHYML (Guindon et al., 2010) and 100 bootstraps.
Chromosomal localization of the NPF gene family
Physical positions of 331 wheat NPF genes were downloaded from URGI (https://wheat-urgi.versailles.inra.fr), and the gene distribution on chromosomes was drawn with MapChart software (Voorrips, 2002) and modified with annotation. Forward and reverse locations of NPF genes are indicated by ‘+’ and ‘–’, respectively. Duplicated NPF genes were marked with Roman numerals (I, II, III, IV, and V); TaNPF7.6(1B) and TaNPF8.27(7A) each contains two linked partial genes, annotated with an asterisk.
Expression analysis of the NPF gene family from RNA-seq data
RNA-seq data based on developmental time-course analysis of Chinese Spring (Choulet et al., 2014) and different abiotic and biotic stress experiments were downloaded from the Wheat Expression Browser (www.wheat-expression.com;Ramírez-González et al., 2018). The first study represented five organs (roots, leaves, stem, spikes, and grains) at three developmental stages (two biological replicates). The study on stresses included drought, heat, and drought+heat stress (Liu et al., 2015); cold stress (Li et al., 2015); polyethylene glycol (PEG) stress (N/A); spike drought stress (Ramírez-González et al., 2018); phosphate (Pi) starvation-stressed roots/shoots (Oono et al., 2013); spikelets with Fusarium/abscisic acid (ABA)/gibberellin (GA) stress (Buhrow et al., 2016); leaf powdery mildew/stripe rust stress (Zhang et al., 2014); and leaf fungal Magnaporthe oryzae stress (Islam et al., 2016). Expression values of NPF genes as transcripts per million (tpm) were extracted and summed across homoeologues (see Supplementary Table S6A and B at JXB online). The heatmaps (Fig. 3; Supplementary Fig. S3) were constructed by pheatmaps (v1.0.8, Kolde, 2019) and R (v3.5.2) on log2-transformed data tpm+1 for 113 NPF HGs deduced from 331 NPF genes (Supplementary Table S6A, B). Variations of NPF gene expressions under abiotic and biotic stresses were listed for analysis by using a threshold of 3-fold changes (Supplementary Table S7).
Plant material, nitrogen analysis, and RNA extraction
As described previously (Wan et al., 2017), the wheat variety Hereward was grown in field trials in 2015, with 200 kg ha–1 (high) or no (low) N application. Roots at Zadoks 23 (Z23; 2–3 tiller stage) and Z45 (booting stage) were excavated from the soil with a garden fork and washed several times using deionized water. Excess water was removed using a soft tissue, and the roots were immediately immersed in liquid nitrogen. Leaves at Z23, Z45, as well as at 5, 14, and 21 dpa (days post-anthesis), stems at Z45, and at 5, 14, 21, and 28 dpa, flag leaf nodes at 5 and 14 dpa, and whole caryopses at 5, 10, 14, 21, and 28 dpa were harvested, freezer milled (Freezer Mill 6870, Spex SamplePrep, Stanmore, UK), and stored at –80 °C for RNA extraction.
Total RNA was isolated by a modified method (Verwoerd et al., 1989) including additional phenol–chloroform–isoamylalcohol extractions. The N concentration of oven-dried subsamples was measured by the Dumas method using a LECO CN628 Combustion Analyser (LECO Corporation, St Joseph, MI, USA) and is expressed in percentage dry matter.
Reverse transcription–quantitative real-time PCR analysis (RT–qPCR)
First-strand cDNA synthesis was performed using 2 µg of total RNA based on the Invitrogen Superscript III standard protocol. Real-time PCR was performed on an ABI7500 (Applied Biosystems) thermocycler using SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma-Aldrich). The 20 μl reactions contained 1 µl of cDNA and 250 nM of each primer. Primer efficiency was analysed and only primer combinations were used with primer efficiencies between 90% and 110%. Primers of TaNPF2.11(7D), TaNPF5.8(5A), TaNPF5.16(3B), TaNPF7.4(2D), and TaNPF7.7(1B) were designed according to only one single homoeologue (indicated in parentheses) as no proper common primers for all homoeologues were obtained. Primers of TaNPF5.9(1) were designed based on the common sequences of three (TraesCS5A02G485200, TraesCS5B02G498500, and TraesCS5D02G498700) out of six homoeologues. For the remaining NPF genes, partly degenerated primers were designed to cover the expression from all the gene homoeologues. TraesCS4A02G035500/TraesCS4B02G268200/TraesCS4D02G267600 (cell division control protein) was used as the internal control gene for validation in root, leaf, stem, node, and spike (Paolacci et al., 2009), and TraesCS3A02G186600/TraesCS3B02G216100/TraesCS3D02G190500 (proteasome subunit) was used as the internal control gene for validation in grain. All primer sequences are listed in Supplementary Table S10.
For each primer pair, PCR efficiency was calculated in each run from a pool of all available cDNAs by using LinRegPCR software (Ramakers et al., 2003). All time points had three biological replicates. The normalized relative quantity (NRQ) of expression was calculated in relation to the Ct values and the primer efficiency (E) of both the target gene (X) and reference genes (N) as normalized relative expression (NRE) based on Rieu and Powers (2009): NRE=(EX)–Ct, X/(EN)–Ct, N (all NRE results are in Supplementary Table S11).
Heatmap presentations of RT–qPCR results were constructed for each tissue. All the gene expression data were normalized to the mean expression value of the first detected stage (root/leaf, Z23; stem, Z45; node, 5 dpa; spike, Z45; and grain, 5 dpa) including both low and high N treatments. The heatmaps (Figs 4–8; Supplementary Figs S4–S6) were created by pheatmaps (v1.0.8, Kolde, 2019) and R (v3.5.2) on log2-transformed data of normalized data+1.
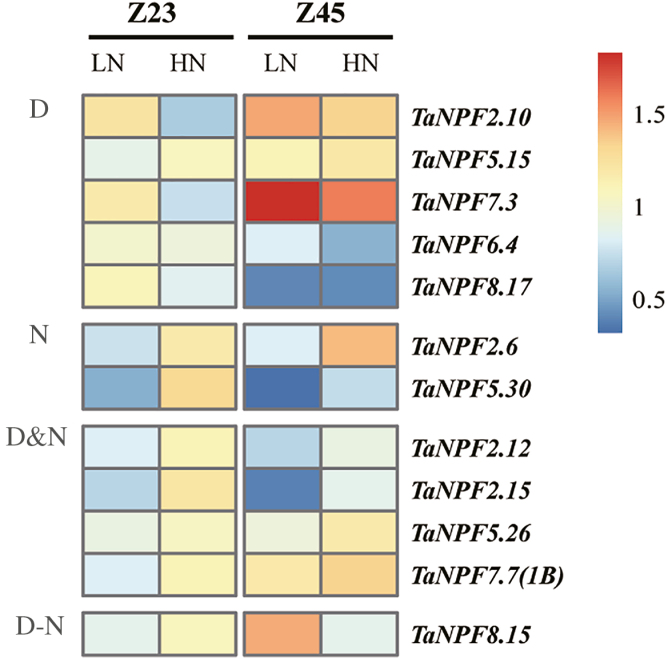
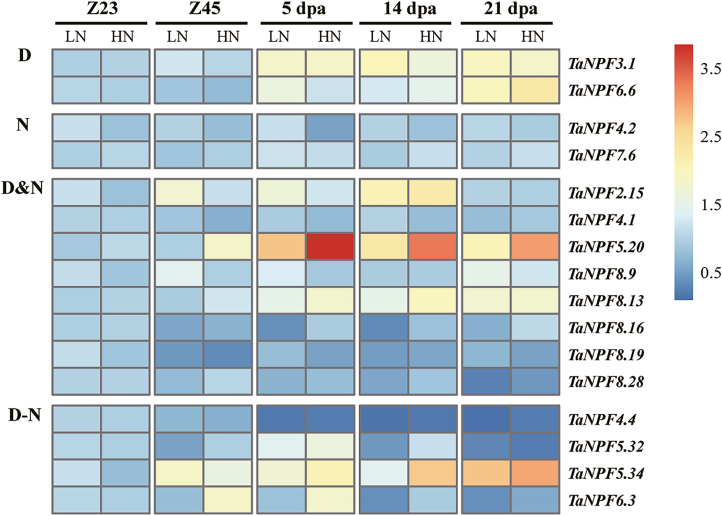
Fig. 4.
Heatmap of NPF expression profiles in root in relation to N fertilization and development by RT–qPCR. Gene expression data were normalized for each gene and are shown as log2-transformed data of normalized data+1. Z23, tillering stage; Z45, booting stage; LN, low nitrogen (0 kg ha–1) application; HN, high nitrogen (200 kg ha–1) application. Groups D, N, D&N, and D–N indicate development effect (D), nitrogen effect (N), development and nitrogen effects (D&N), and interacting effects of development and nitrogen (D–N) on NPF gene expression (n=3 replicates; P<0.05), respectively.
Statistics
The statistical validation of the effects of development (D), nitrogen (N), development and nitrogen (D&N) as well as the interaction of development and nitrogen (D–N) on NPF gene expression was evaluated on log2-transformed NRE and ANOVA using Genstat 18th Edition (VSN International Ltd, UK). Comparisons between relevant means of n=3 replicates were made using the error of the difference (SED) on the residual degrees of freedom from the ANOVA, thus invoking the least significant difference (LSD) at the 5% level of significance (pairs of means different by more than the LSD are statistically significantly different, P<0.05).
Results
Identification of NPF genes in wheat
Protein sequence analysis based on the proton-dependent oligopeptide transporter family (IPR000109) protein domain specific for the NPF family (Supplementary Table S1) and the wheat genome database (IWGSCv1.1) initially identified a total of 365 genes including 331 high confidence (HC) and 34 low confidence (LC) genes (Supplementary Table S2). Among these candidates, 15 LC genes lacked start and/or stop codons and 14 genes (4 HC genes and 10 LC genes) encoded very short proteins (< 200 amino acids) which did not meet the multitransmembrane structure of NPF proteins. Three gene sequences (1 unknown chromosomal location HC gene and 2 LC genes) were partial and redundant with another two HC genes, three LC genes had very low gene prediction scores (<30), and one LC gene had an extra stop codon in the coding sequence (Supplementary Table S3). These 34 genes were removed from the candidate NPF gene cluster, and 331 NPF members including 326 HC and 5 LC genes were classified as wheat NPF genes (Supplementary Table S4).
Homoeologous genes were further defined as those with both transcript and protein sequence similarities >90% to corresponding chromosomes in the different subgenomes (Wan et al., 2017), and the majority of homoeologous genes showed >95% similarity. The 331 NPF genes were subdivided into 113 NPF HGs which contained 73 groups with three, 19 groups with two, seven groups with 4, three groups with 6, one group with 7, and one group with 12 homoeologues, respectively. In addition, there were nine NPF genes without any homoeologues in the wheat genome (Supplementary Table S4).
Phylogenetic analysis of wheat NPF genes
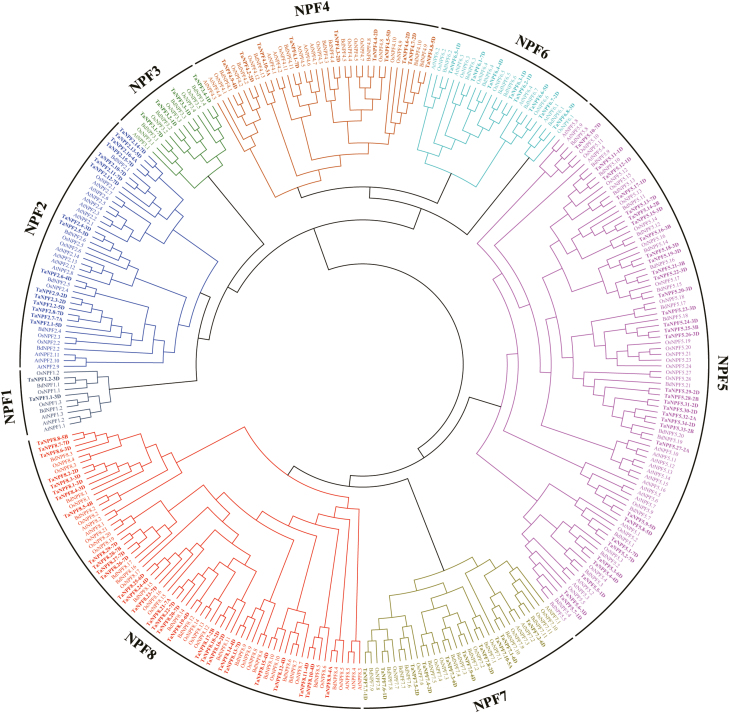
NPF genes have been identified in many plants and are classified into eight subfamilies (Léran et al., 2014). For phylogenetic analysis, classification, and systematic nomenclature of wheat NPF genes, full-length protein sequences of the 113 single homoeologues from each HG were aligned with the orthologues of Arabidopsis, Brachypodium, and rice, and a phylogenetic tree was constructed (Fig. 1). NPF orthologues from Arabidopsis, Brachypodium, and rice located together with individual wheat NPF members in the same clades representing the eight subfamilies (NPF1–NPF8). Among the wheat NPF gene family, the NPF5 subfamily contains the most (34 members) NPF genes, followed by subfamilies NPF8 (29 members), NPF2 (16 members), NPF4 and NPF7 (10 members for each subfamily), NPF6 (8 members), NPF3 (4 members), and NPF1 (2 members) with the least NPF genes. As 16 wheat NPF genes have been previously isolated, numbered, and named (Buchner and Hawkesford, 2014), the other wheat NPF genes were further assigned based on their locations and relationships with orthologues of Arabidopsis, Brachypodium, and rice with the proposed nomenclature (Léran et al., 2014) (Fig. 1; Supplementary Table S4).
Fig. 1.
Phylogenetic analysis of the NPF gene family of wheat. A total of 113 single wheat NPF genes (one from each homeologous group) were aligned with 53 Arabidopsis (A. thaliana), 75 Brachypodium (B. distachyon), and 81 rice (O. sativa) NPF genes using protein sequences. The tree was constructed using CLUSTALW and PHYML programs in Geneious using the Neighbor–Joining method with 100 bootstrap replicates.
Chromosomal locations of wheat NPF genes
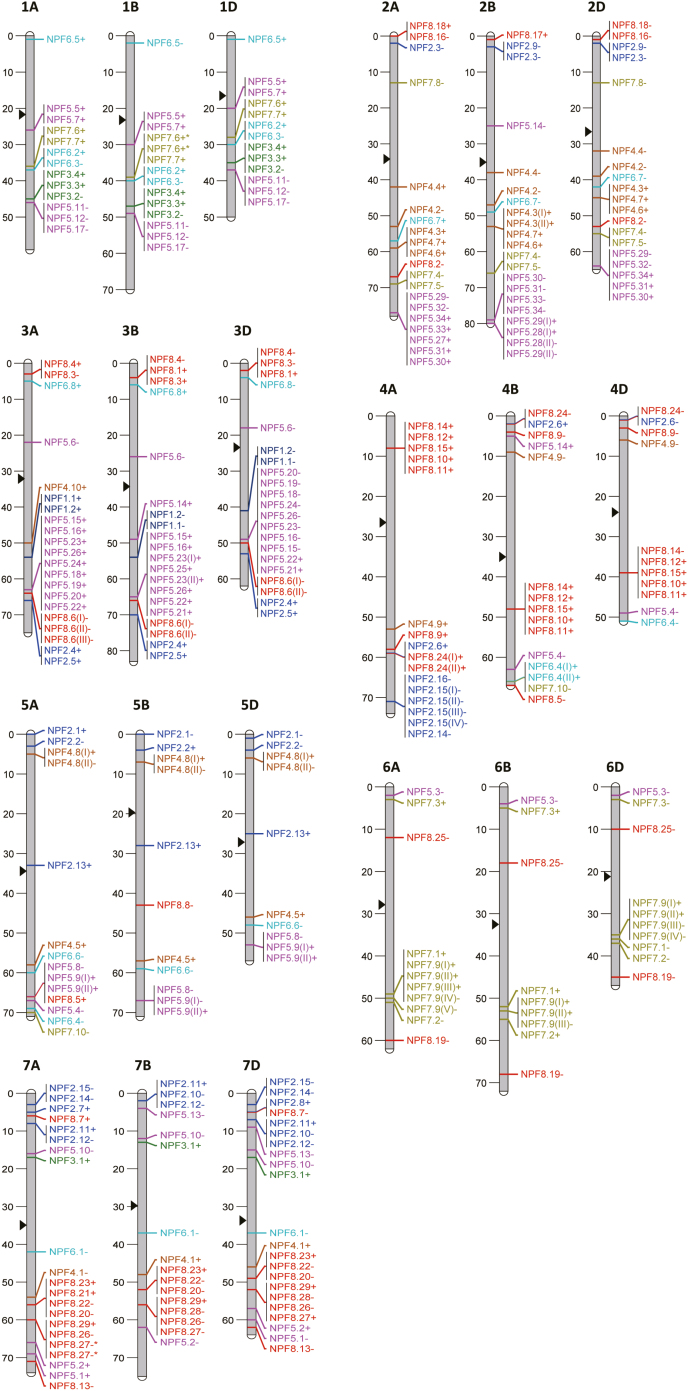
To determine the chromosomal distributions of NPF genes, 328 out of 331 genes were mapped on the wheat chromosomes using physical positions (Fig. 2; Supplementary Table S5). Chromosomes 3, 2, and 7 have the most abundant NPF genes (62, 60, and 59 members, respectively), followed by chromosomes 4, 1, and 5 (42, 40, and 35 members, respectively), while chromosome 6 has the least NPF genes (30 members; Fig. 2). Three NPF genes, TaNPF7.10 (TraesCSU01G130200), TaNPF8.5 (TraesCSU01G115500), and TaNPF8.18 (TraesCSU01G307300LC), are probably located on Chr4D, Chr4D, and Chr2B, respectively, according to the locations of the respective homoeologues (Supplementary Table S4). The majority of NPF genes are located distant from the middle of the chromosomes, and only a few genes are present near to the centromere (Fig. 2). On each chromosome, more NPF genes are located on the long arm compared with the short arm of the chromosome (Fig. 2).
Fig. 2.
Distribution of NPF genes on wheat chromosomes. The centromeres are indicated by arrowheads.
Expression patterns and validation of wheat NPF genes
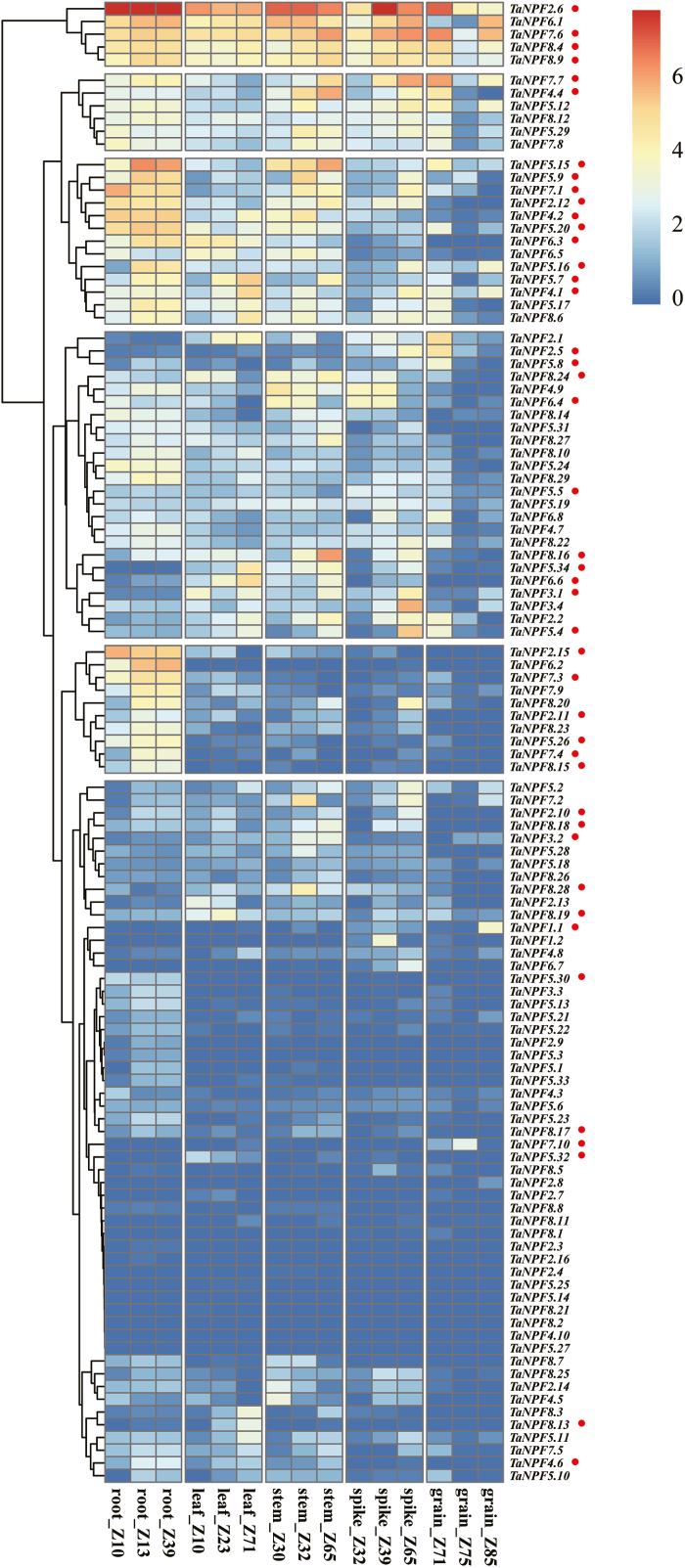
To gain insight into the spatial and temporal expression patterns of NPF genes in wheat, the RNA-seq data sets derived from root, leaf, stem, spike, and grain of the wheat cultivar Chinese Spring (Choulet et al., 2014) were explored. The possible interactions of NPF gene expression in relation to biotic and abiotic stresses were verified by analysis of RNA-seq data sets derived from different stress experiments (Ramírez-González et al., 2018). The spatio-temporal expression profiles of NPF genes were clustered into six groups (Fig. 3) with no specific relationship to the wheat NPF phylogenetic subfamily structure. NPF genes in group I were highly expressed in almost all tissues, with TaNPF2.6 the most abundantly expressed gene. In groups II and III, the majority of the NPF genes were expressed at medium to high levels. In general, the expression of group II NPF genes in stems, spikes, and grains depended on the growth stage, whereas expression in roots was relatively constant. The majority of group III NPF genes had relatively high expression levels in roots, and lower expression in stems and leaves. Most NPF genes in group IV were expressed at intermediate levels in all tissues. Interestingly, all NPF genes in group V were expressed mainly in the roots, indicating root-specific transport functions. Group VI included most of the NPF genes with a less complex tissue specificity pattern and typically low or extremely low expression levels (Fig. 3).
Fig. 3.
Heatmap of spatio-temporal expression profiles of wheat NPF genes based on RNA-seq data (Choulet et al., 2014). Zadoks (Z) developmental stages are shown. Z10, seedling stage; Z13, three leaf stage; Z23, tillering stage; Z30, 1 cm spike; Z32, two nodes detectable; Z39, flag leaf stage; Z65, anthesis; Z71, 2 d post-anthesis; Z75, 14 d post-anthesis; Z85, 30 d post-anthesis. The different colours represent the abundances of the transcripts based on log2-transformed tpm (transcript per million)+1. Red dots marked TaNPF genes selected for RT–qPCR gene expression analysis.
Selected spatio-temporal RNA-seq data were verified by RT–qPCR for eight NPF genes from the six expression groups in roots, leaves, stems, and grain for the wheat cultivar Hereward. The results of the real-time expression analysis are in agreement with the RNA-seq data, except for TaNPF1.1, which had relatively high expression levels in roots in addition to in the grain (Supplementary Figs S1, S2).
The stress-related RNA-seq expression analysis was clustered in relation to the phylogenetic NPF subfamily structure (Supplementary Fig. S3). The majority of the NPF HGs reacted specifically to different stresses. When using a minimum of a 3-fold change as a threshold, 21 out of the 113 HGs did not show any changes of gene expression (Supplementary Table S7). There was no obvious correlation of specific stress-related changes of NPF HG expression in relation to the phylogenetic NPF subfamily structure (Supplementary Fig. S3; Supplementary Table S7). Stress-related up- or down-regulation of members of all NPF subfamilies were seen, apart from leaf heat stress (NPF1), leaf PEG stress (NPF1 and NPF3), spike drought stress (NPF3 and NPF6), phosphate starvation (NPF1, NPF2, NPF4, and NPF8), leaf stripe rust pathogen inoculation (NPF1, NPF3, and NPF6), and leaf M. oryzae inoculation (NPF1, NPF3, NPF4, and NPF6). Gene expression of 38 HGs representing all NPF subfamilies was changed specifically by abiotic stresses, and only eight NPF HGs from NPF2, NPF5, NPF7, and NPF8 subfamilies were changed solely by biotic stress. Leaf heat and drought stress resulted in the strongest response of the whole NPF gene family, where the majority of NPF HGs were down-regulated (heat, 44 members; drought, 25 members; heat+drought, 55 members). The spike response to drought comprised 8 up- and 13 down-regulated NPF HGs, with lower intensity compared with leaves. A larger number of NPF HGs were up-regulated rather than down-regulated in cold-treated shoots and PEG-treated leaves. Biotic-related stresses such as Fusarium/ABA/GA treatments, leaf powdery mildew, and leaf strip rust resulted in an equal number of NPF HGs up- or down-regulated, although for fungal stresses more NPF HGs were down-regulated. Only two members of NPF subfamily 1 were mostly up-regulated under different stresses: TaNPF1.1 responded to cold, Fusarium/ABA, and powdery mildew stress, and TaNPF1.2 in an opposite way for drought in leaves and spikes. The majority of NPF genes reacted to both abiotic and biotic stresses and some were regulated in an opposite way under different stresses. Some NPF genes which were down-regulated by drought/heat stress were up-regulated by cold stress. Some members of NPF subfamily 7 reacted with a strong up-regulation under most biotic stresses in contrast to their reaction to abiotic stresses. The same was seen for some members of NPF subfamilies 5 and 8. A very small number of NPF HGs responded to phosphate starvation in roots and shoot: two NPF subfamily 5 HGs were down-regulated in roots, whereas in shoots up- and down-regulation were seen for four NPF subfamily HGs (Supplementary Table S7).
NPF gene expression profiles under nitrogen treatments
Most NPF genes are linked to N acquisition and utilization, as indicated by their respective substrates. To determine NPF gene expression responses to N supply during development, 44 NPF genes were selected according to their expression patterns from the RNA-seq data (Fig. 3), with the feature of being highly expressed in tissues, or preferred expression in a specific tissue at different expression levels (for details of the selection, see Supplementary Protocol S1). These selected NPF genes represented the eight phylogenetic subfamilies and the six NPF family gene expression groups (Figs 1, 3). After further subselection based on the tissue distribution of the six expression groups (Supplementary Protocol S1), their gene expression patterns were analysed in roots, leaves, stems, nodes, spikes, and developing grains at different growth stages cultivated under high and low N fertilizer treatments in field experiments. Zero N fertilization had a strong influence on plant performance in comparison with 200 kg N ha–1. Grain and straw yields were reduced by 58% and 70%, respectively, with grain and straw N contents reduced by 30% and 76%, respectively. N deficiency was indicated by a drastically reduced grain and straw N accumulation (70% and 78%), with a harvest index slightly higher under the zero N treatment (Table 1). The effects of development, N, and interaction of development and N on gene expression were analysed.
Table 1.
Field performance of wheat cv. Hereward in relation to reduced nitrogen fertilization
| N rate (kg ha–1) | Grain yield (t ha–1, 85% DM) | Straw yield (t ha–1, 85% DM) | Grain % N | Straw % N | Grain N accumulation (kg ha–1) | Straw N accumulation (kg ha–1) | Harvest index |
|---|---|---|---|---|---|---|---|
| 0 | 4.84±0.2 | 2.89±0.5 | 1.33±0.008 | 0.256±0.006 | 54.91±2.9 | 6.21±0.9 | 63.56±3.7 |
| 200 | 11.24±0.12 | 9.42±1.0 | 1.84±0.01 | 0.354±0.01 | 176.13 ±3.3 | 28.54±3.9 | 54.82±2.6 |
Root NPF gene expression
The root plays roles in initial N acquisition. Previously, no NPF root expression of field-grown root samples has been analysed. As root N uptake depends on plant development, root gene expression was analysed at two growth stages, Z23 (2–3 tiller stage) and Z5 (booting stage) (Fig. 4; Supplementary Fig. S4). Expression of the majority of the 28 subselected NPF genes were not affected by development or N supply (Supplementary Fig. S4). However, the expression of seven NPF genes was significantly affected by N supply (Fig. 4). For six genes, transcript levels were decreased by low N (Fig. 4), and four of these NPF genes were also developmentally up- or down-regulated at the two growth stages (Fig. 4). The expression of TaNPF8.15 showed an interaction between N treatment and development, with a significant increase of transcript abundance at the booting stage at low N supply (Fig.4). Five NPF genes were developmentally regulated, with three and two NPF genes significantly up-regulated or down-regulated, respectively, from Z23 to Z45 (Fig. 4).
Leaf NPF gene expression
Depending on the plant growth stage, leaves may be N sinks or N sources, both of which involve N allocation and redistribution. The gene expression patterns of 16 selected NPF genes (Supplementary Protocol S1) were analysed in flag leaves at different growth stages (Z23 and Z45; and 5, 14, and 21 dpa) (Fig. 5). The transcript abundances of 14 of these NPF genes were influenced by the N fertilization and resulted in increased or reduced gene expression with additional or interactive developmental effects in both up and down directions (Fig. 5). For just two NPF genes (Fig. 5), N fertilization had no effect on the expression pattern, but both genes were up-regulated post-anthesis.
Fig. 5.
Heatmap of NPF expression profiles in the flag leaf in relation to development and N fertilization by RT–qPCR. Gene expression data were normalized for each gene and are shown as log2-transformed data of normalized data+1. Z23, tillering stage; Z45, booting stage; dpa, days post-anthesis; LN, low (0 kg ha–1) nitrogen application; HN, high (200 kg ha–1) nitrogen application. Groups D, N, D&N, and D–N indicate development effect (D), nitrogen effect (N), development and nitrogen effects (D&N), and interacting effects of development and nitrogen (D–N) on NPF gene expression (n=3 replicates; P<0.05), respectively.
Stem NPF gene expression
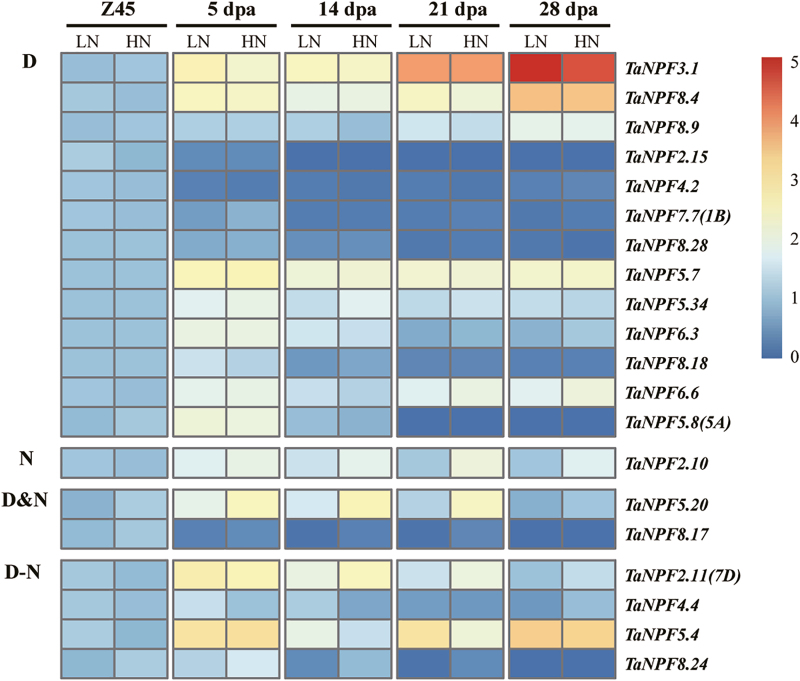
The stem is the conduit for N compound translocation. Gene expression responses to N supply from Z45 to 28 dpa were analysed for 20 subselected NPF genes (Fig. 6). Seven NPF genes were found to be responsive to N supply, mainly by down-regulation under low N (Fig. 6). For TaNPF4.4, low N initiated a post-anthesis increase of transcript which diminished until ripening, whereas TaNPF5.4 was more highly expressed in the post-anthesis period under low N compared with sufficient N. For most of the NPF genes, gene expression in the stem was influenced by development with three typical patterns (Fig. 6): a steady post-anthesis increase or decrease of transcript, or a short increase of expression between Z45 and 5 dpa, followed by steady state or declining levels.
Fig. 6.
Heatmap of NPF expression profiles in the stem in relation to development and N fertilization by RT–qPCR. Gene expression data were normalized for each gene and are shown as log2-transformed data of normalized data+1. Z45, booting stage; dpa, days post-anthesis; LN, low nitrogen (0 kg ha–1) application; HN, high nitrogen (200 kg ha–1) application. Groups D, N, D&N, and D–N indicate development effect (D), nitrogen effect (N), development and nitrogen effects (D&N), and interacting effects of development and nitrogen (D–N) on NPF gene expression (n=3 replicates; P<0.05), respectively.
Node NPF gene expression
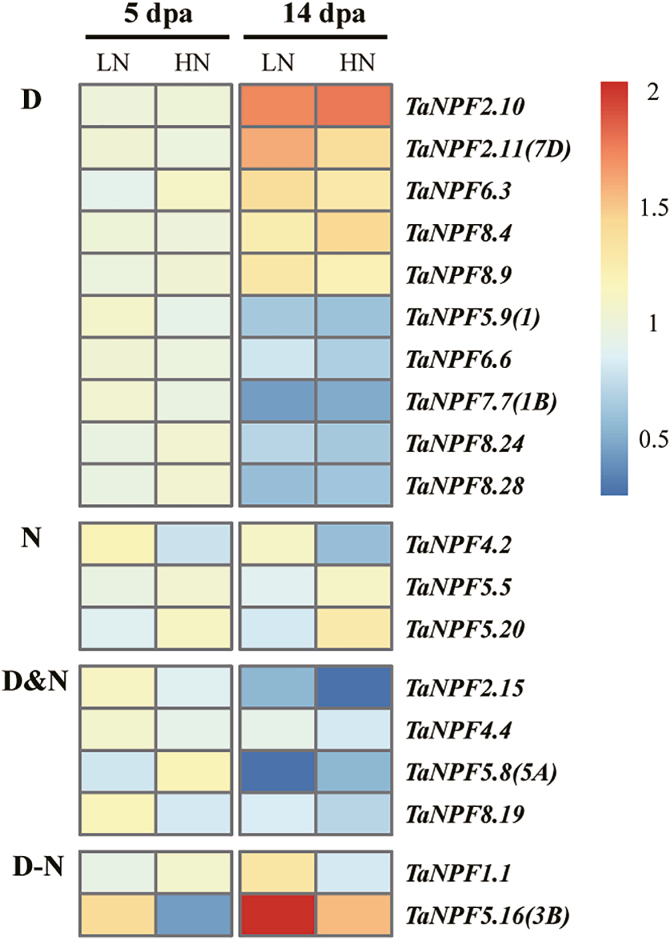
The node is the junction of stem, leaf, and axillary bud/tiller, and possesses well-developed vascular bundles in Poaceae such as rice, barley, and wheat (Yamaji and Ma, 2017). Twenty-seven NPF genes were selected (Supplementary Protocol S1) for analysis of gene expression in the flag leaf node at 5 and 14 dpa (Fig. 7; Supplementary Fig. S5). Expression levels of three of the nine N-responsive NPF genes were not developmentally influenced (Fig. 7). The other NPF genes were additionally developmentally down-regulated between both time points (Fig. 7). Some significant N–development interactions were seen: TaNPF5.16 was up-regulated under both N conditions, whereas for TaNPF1.1 only low N resulted in a significant increase of transcript abundance at 14 dpa (Fig. 7). Ten NPF genes were responsive to node development only, with increased or reduced transcript abundance between 5 and 14 dpa (Fig. 7). Eight NPF genes were not significantly responsive to either N supply or development (Supplementary Fig. S5).
Fig. 7.
Heatmap of NPF expression profiles in the node in relation to development and N fertilization by RT–qPCR. Gene expression data were normalized for each gene and are shown as log2-transformed data of normalized data+1. dpa, days post-anthesis, LN, low nitrogen (0 kg ha–1) application; HN, high nitrogen (200 kg ha–1) application. Groups D, N, D&N, and D–N indicate development effect (D), nitrogen effect (N), development and nitrogen effects (D&N), and interacting effects of development and nitrogen (D–N) on NPF gene expression (n=3 replicates; P<0.05), respectively.
Spike NPF gene expression
Expression levels of 20 selected NPF genes (Supplementary Protocol S1) were analysed in spikes at Z45 stage. As shown in Supplementary Fig. S6, none of the genes showed significant effects of N treatments, indicating consistent expression of NPF genes in the young reproductive organs.
Grain NPF gene expression
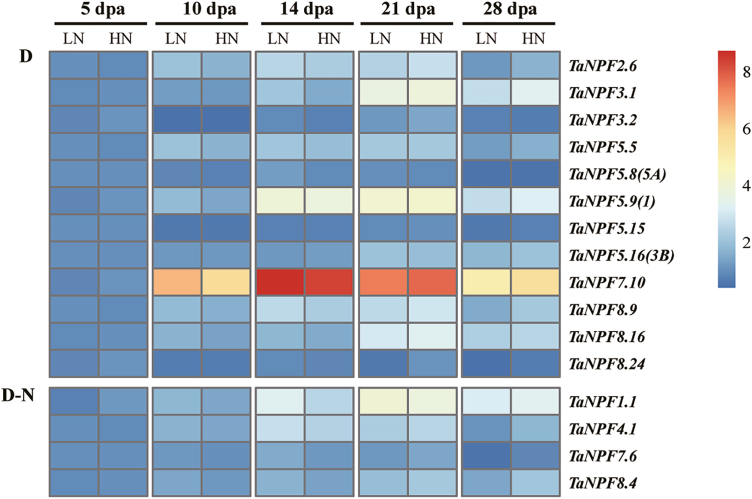
During post-anthesis development, grains are the main sinks for N, originating particularly from redistribution from vegetative organs (leaf, stem, etc.). The expression of 16 NPF genes was analysed in developing grains between 5 and 28 dpa (Fig. 8). Gene expression in the grain of four NPF genes responded to N fertilization with a specific developmental pattern (Fig. 8). During early to mid-grain filling, between 5 and 14/21 dpa, there was an increase of transcript levels of the four NPF genes, with higher expression levels at low N. With further grain development, gene expression of NPF genes decreased rapidly under low N, with no or reduced change under sufficient N supply. This overall post-anthesis developmental pattern was very similar for nearly all 16 NPF genes for both N treatments (Fig. 8).
Fig. 8.
Heatmap of NPF expression profiles in the grain by RT–qPCR. Gene expression data were normalized for each gene and are shown as log2-transformed data of normalized data+1. dpa, days post-anthesis, LN, low nitrogen (0 kg ha–1) application; HN, high nitrogen (200 kg ha–1) application. Groups D, N, D&N, and D–N indicate development effect (D), nitrogen effect (N), development and nitrogen effects (D&N), and interacting effects of development and nitrogen (D–N) on NPF gene expression (n=3 replicates; P<0.05), respectively.
Discussion
Composition of the NPF gene family in wheat
To date, the composition of NPF gene families has been identified for >30 plants (Léran et al., 2014). In wheat, NPF genes were previously described (Buchner and Hawkesford, 2014; Bajgain et al., 2018). The completeness of the newly released wheat genome data (International Wheat Genome Sequencing Consortium, 2018) allowed the identification of 331 non-redundant NPF genes (Supplementary Table S4). Among the wheat NPF gene family, five low confidence (LC) genes were included due to genome sequence gene structure re-validation and confirmed expression signals in RNA-seq data. Recently, a pseudo reference consisting of 500 potential wheat NRT (nitrate transporter) gene sequences was established for wheat seedling RNA-seq expression analysis (Bajgain et al 2018). After re-analysis of the 500 gene IDs based on the TGAC (The Genome Analysis Centre) 2015 assembly annotation in comparison with the current wheat genome IWGSCv1.1 database, we identified just 270 individual NRT genes. Some of the 500 gene IDs mapped as duplicates and not as individual NRT genes. Additionally, some of the provided gene IDs do not represent individual NRT genes or are mapped on different chromosomes indicated by the former TGAC assembly. Some of the genes represent HGs. Finally, 234 genes of the study corresponded to unique NPF genes (Bajgain et al., 2018) (Supplementary Table S8). The remaining 36 NRT genes included 28 clade 5 NRT2 family and eight clade 1 NAR2 family genes (Bajgain et al., 2018) (Supplementary Tables S1, S9). The present analysis identified 97 new NPF genes to complete the whole NPF gene family in wheat, which contains more genes than the other three nitrate transporter gene families, NRT2, CLC, and SLAC/SLAH (Krapp et al., 2014; Fan et al., 2017; H. Li et al., 2017a; Y.-Y. Wang et al., 2018). The chromosomal locations of the wheat NPF genes are unevenly distributed, with high gene abundances in chromosomes 2, 3, and 7 (Fig. 2). This is also the case for members of another large N-related gene family in wheat, the amino acid transporter (AAT) family (Wan et al., 2017). Whereas the AAT genes are mostly located close to the centromere of each chromosome (Wan et al., 2017), the majority of the NPF genes are located distant from the centromere, with a favoured distribution on the long arm of the chromosomes (Fig. 2).
The wheat NPF gene family represents one of the largest NPF gene families among the currently analysed species (Léran et al., 2014). Theoretically, every wheat gene would have three homoeologous genes as the result of allohexaploidization, but many wheat NPF genes have no or only one homoeologous gene, resulting from the side effects of polyploidy, and evolutionary and acclimation processes (Lynch and Force, 2000). The 331 wheat NPF genes were subdivided into 113 HGs (Supplementary Table S4). In addition to 73 (64.6% of the total) groups with three HGs, 19 (16.8%) groups contained two HGs and nine (8.0%) groups were singleton genes without any homoeologues. Gene duplication is a common phenomenon contributing to gene family expansion (Zhang, 2003), and they are commonly detected in wheat interspecific whole-genome analysis (International Wheat Genome Sequencing Consortium, 2014). For the wheat NPF family, seven (6.2%), three (2.7%), one (0.9%), and one (0.9%) HGs contained four, six, seven, and as many as 12 homoeologues, respectively (Supplementary Table S4). The group with 12 homoeologous genes was composed of five, three, and four neighbour-linked duplicated genes located on Chr6A, Chr6B, and Chr6D, respectively. The results revealed that the composition of the wheat NPF gene family is much more complex than that in most other plant species. The complexity of the wheat NPF gene family results mainly from integration of normally three homoeologues and additional gene duplication and/or loss of genes during wheat evolution. This complexity does not influence their phylogeny relationships with orthologues in other plants. The 113 wheat NPF HGs could be classified into eight clades in the phylogenetic tree, similarly to the NPF genes in other plant species (Fig. 1; Supplementary Table S4), providing the basis for a systematic nomenclature of wheat NPF genes (TaNPFX.Y) as proposed, except for the 16 NPF genes already named previously (Buchner and Hawkesford, 2014; Léran et al., 2014). Each clade shows further subtree structure leading to direct and indirect orthologous relationships between the plant species.
Diverse wheat NPF gene expression patterns and responses to abiotic and biotic stresses
In addition to nitrate and peptides, NPF transporters have diverse and mixed substrate specificities including ions (chloride, nitrite), organic compounds (glucosinolates, dimethylarsinate), and hormones (auxin, ABA, jasmonates, and GAs) (Corratgé-Faillie and Lacombe, 2017; Y.-Y. Wang et al., 2018), indicating the broad participation of NPF genes in biological processes involved in adaptation to adverse environmental conditions. Apart from N starvation, some studies have shown specific NPF gene responses to abiotic and biotic stresses (Pike et al., 2014; Taochy et al., 2015; Zhang et al., 2018). The RNA-seq analysis in relation to different abiotic and biotic stresses in wheat illustrates a complex pattern throughout the whole NPF gene family (Supplementary Fig. S3; Supplementary Table S7). These patterns did not show any correlation to the phylogenetic subfamily structure of the wheat NPF gene family. A mixed pattern of up- or down-regulation of NPF genes indicated multiple functions of the NPF family in response to most of the abiotic and biotic stresses. The strongest response was observed for drought/heat stress, for which 61 of the 113 HGs were affected with 55 HGs down-regulated by the stress (Supplementary Table S7). A major consequence of leaf drought stress is an imbalance of carbon and N metabolism due to a decrease in photosynthesis with a strong effect on carbon and N compound translocation (Yang et al., 2019). The wide responses of NPF genes to drought suggest a strong down-regulation of N substrate transport in leaves, reflecting the slow down of N metabolism under drought stress. However, the drought response of NPF genes was not so drastic in spikes (22 NPF genes) with partly opposite up or down gene regulation in response to the stress compared with leaves, indicating a different regulation mechanism in the reproductive organ. Co-application of Fusarium stress and ABA to wheat spikelets resulted in ABA-specific up- or down-regulation of different NPF genes (Supplementary Table S7). Interestingly, most of the ABA-responsive wheat NPF genes were also influenced by heat and/or drought and/or cold stresses. ABA plays an important role in abiotic stress response and tolerance by regulating stomatal response and stress-related gene expression (Chinnusamy et al., 2008). The broad responses of NPF genes to ABA and abiotic stresses suggest that these wheat NPF genes are possibly involved in ABA transport and/or are hormonally regulated. The response of co-application of Fusarium with GA to spikelets induced only three NPF genes, TaNPF3.4, TaNPF5.3, and TaNPF7.10. TaNPF3.4 may play an important role in abiotic/biotic stress responses, as it is also highly up-regulated by co-application of Fusarium with ABA, as well as by heat, drought, and cold stresses (Supplementary Table S7). The only NPF3 orthologue in Arabidopsis, AtNPF3.1, in addition to nitrate/nitrite transport, also transports and is transcriptionally regulated by both GA and ABA (Pike et al., 2014; Tal et al., 2016), suggesting a similar function for the wheat TaNPF3.4. In grapevine and Arabidopsis, NPF3 subfamily genes are induced by leaf powdery mildew infection (Pike et al., 2014), which was not observed for orthologous wheat NPF3 genes (Supplementary Table S7). However, NPF genes of other subfamilies, especially NPF7, showed a strong up-regulation by the fungal stress (Supplementary Table S7). In comparison, NPF5 subfamily genes showed a strong down-regulation by the fungal stress (Supplementary Table S7). In addition, NPF2, NPF5, and NPF8 subfamilies showed more reaction to leaf stripe rust and M. oryzae, compared with other subfamilies (Supplementary Table S7). These results indicated the specific roles of individuals from these subfamilies in fungal stress responses. In general, the stress analysis of wheat NPF family genes revealed a complex regulation with a mix of common or opposite regulation of genes under different abiotic and biotic stresses with additional components of tissue and developmental regulation of the stress response. Together with the diverse and mixed substrate specificities in addition to nitrate and peptides, these results strengthen the hypothesis that NPF members are the basis of the integration of environmental and physiological information linked to the relative availability of nutrients (Corratgé-Faillie and Lacombe, 2017).
Wheat NPF genes are regulated by nitrogen fertilization and/or development
The further validation of NPF gene expression response to N fertilization by RT–qPCR demonstrated the complex expression pattern of the wheat NPF gene family. Expression of a total of 44 pre-selected NPF genes based on phylogeny and the RNA-seq analysis was monitored in root, leaf, stem, node, spike, and grain samples (Figs 4–8; Supplementary Figs S4–S6). This analysis indicated that the expression of NPF genes in wheat showed dynamic variations throughout development and in response to available N supply. In summary, tissue-, development-, and N supply-related expression patterns identified six general expression groups. In roots, expression of 16 out of 28 analysed NPF genes did not change in relation to N fertilization and development (Supplementary Fig. S4), and most had stable expression levels in RNA-seq analysis of root samples (Fig. 3), suggesting a constitutive function in N homeostasis and root growth. The other NPF genes were up-regulated under high N fertilizer and/or by developmental stages (Fig. 4). The up-regulation of NPF genes by N provision in roots probably facilitates N uptake and/or translocation from root to shoot as seen in Arabidopsis and rice (Supplementary Table S12). The developmental up- or down-regulation of NPF genes may be related to the drastically contrasting demands for root N uptake, translocation, and growth between the Z23 stage (2–3 tillers) and Z45 stage (up to 6 tillers). The nodes are the hub for nutrient distribution in graminaceous plants with their complex vascular system (Yamaji and Ma, 2014, 2017). The development- and/or N-regulated expression of as many as 19 out of 27 NPF genes found in nodes suggests a strong participation in transport and regulatory processes of N compound delivery to the reproductive tissues/organs (Fig. 7). The stem mediates transport between root, leaves, and reproductive tissue/organs in varying directions depending on the developmental demand. The expression patterns of NPF genes in the stem from the pre-anthesis vegetative stage to the post-anthesis reproductive stage were mostly characterized by increasing or decreasing transcript levels throughout the period, or peaking at the beginning of grain development (5 dpa), followed by reduction until complete ripening (Fig. 6). These expression patterns indicate that the demand for individual NPF genes in the stem depends on development, and may involve different transport actions/directions which have to ensure N compound delivery for grain development with N accumulation. The expression of NPF genes in leaves also followed the vegetative/reproductive transition. Similarly, to the stem, most NPF gene expression in leaves showed developmental regulation, with increasing, decreasing, or a mixed expression pattern with development (Fig. 5). The analysis revealed that NPF gene expression was more responsive to N supply in flag leaves compared with other organs. With the exception of TaNPF3.1 and TaNPF6.6, all the 14 other NPF genes had increased or decreased expression in response to N fertilization, with partial additional or interactive developmental effects in both up and down directions. Generally, leaves are important N sinks during vegetative growth and convert to N sources during reproductive growth (Tegeder and Masclaux-Daubresse, 2018). The present results indicate participation of NPF genes in leaf N compound allocation and remobilization, respectively, depending on the plant growth stage. In comparison, the expression of NPF genes in reproductive tissues (spike and grain) was not influenced by N supply as much as in vegetative tissues (Fig. 8; Supplementary Fig. S6). With the exception of four NPF genes (TaNPF1.1, TaNPF4.1, TaNPF7.6, and TaNPF8.4) in grains showing interactive regulation by N and development, none of the other NPF genes showed responses to N supply in spikes and grains.
Orthologues and NPF functions
NPF genes play fundamental roles and participate widely in the complex processes of N utilization (Y.-Y. Wang et al., 2018). There are functional data of 38 and 17 characterized NPF genes in Arabidopsis and rice, respectively (Supplementary Table S12). Putative functions in N utilization of the individual wheat NPF genes may be deduced by linking the wheat analysis with the orthologues reported in Arabidopsis and rice. The phylogenetic analysis of the wheat NPFs revealed multiple orthologous relationships to Arabidopsis and rice, with direct orthologous genes present in the same subtree and indirect orthologous relationships present in the same NPF subfamily (Fig. 1). AtNPF6.3 (NRT1.1) was the first identified nitrate transporter, functioning in nitrate uptake in root, nitrate translocation from root to shoot, and as a nitrate transceptor to govern many molecular, physiological, and morphological responses to nitrate (Tsay et al., 1993; Liu et al., 1999; Ho et al., 2009; Léran et al., 2013; Bouguyon et al., 2015; Supplementary Table S12). Two rice orthologues of NRT1.1, OsNPF6.5 and OsNPF6.3, have diverged in subcellular location, and N (nitrate/ammonium) response and utilization, but both showed potential for improving N use efficency (NUE) and yield of rice (Hu et al., 2015; W. Wang et al., 2018; Supplementary Table S12). The two maize orthologues of NRT1.1, ZmNPF6.6 and ZmNPF6.4, showed different substrate preferences and different expression responses to N supply, and were used to improve NUE of maize (Allen et al., 2016; Wen et al., 2017). There are four NRT1.1 orthologues in wheat, which showed varied expression patterns in RNA-seq and/or RT–qPCR analysis (Figs 1, 3; Supplementary Table S4; Buchner et al., 2014). TaNPF6.1 was one of the most highly expressed genes of group I, with constitutive expression in different tissues (Fig. 3). TaNPF6.2 was predominantly expressed in roots, while TaNPF6.3 was mainly expressed in root, leaf, and stem, and was up-regulated by high N supply in leaves but not in stems and nodes (Figs 3, 5–7). TaNPF6.4 showed preferred expression in spike and node, and a lower expression in root and leaf (Fig. 3). The expansion in gene number and variation in expression patterns suggests the divergence of NRT1.1 orthologues in wheat and indicates possible important roles in NUE as previously reported.
Rice OsNPF2.4 mediates not only nitrate acquisition, but also root to shoot nitrate transport and N remobilization from source to sink organs (Xia et al., 2015). OsNPF2.4 (also OsNPF6.5) was discovered by a genome-wide association study (GWAS) on NUE-related agronomic traits (Tang et al., 2019). Wheat TaNPF2.6 is the closest orthologue of OsNPF2.4 and shares high sequence identity (87%) with OsNPF2.4 (Fig. 1; Supplementary Table S12). Interestingly, TaNPF2.6 is the most highly expressed gene among the 113 NPF genes in wheat (Fig. 3 ; Supplementary Fig. S3), and was induced by N supply in roots (Fig. 4). Whether TaNPF2.6 plays the important roles in wheat N utilization as seen for OsNPF2.4 in rice needs to be characterized in the future.
Recently, members of NPF7 subfamily genes, OsNPF7.1–OsNPF7.4 and OsNPF7.7, were reported to be involved in N allocation and shown to have specific roles in regulating tiller number and subsequently the grain yield of rice (Hu et al., 2016; Fang et al., 2017; Huang et al., 2018, 2019; J. Wang et al., 2018; Supplementary Table S12). In wheat, there are 10 NPF7 members (TaNPF7.1–TaNPF7.10) which showed variable expression patterns (Figs 1, 3; Supplementary Table S4). Further detection by RT–qPCR revealed that the expression patterns of some wheat NPF7 genes were responsive to N supply and/or development similarly to the rice NPF7 orthologues. For example, TaNPF7.6 is among the most highly expressed genes in group I (Fig. 3) and was influenced by N supply in leaf and grain (Figs 5, 8). TaNPF7.3 and TaNPF7.10 were mainly expressed in root and grain, respectively (Fig. 3), and the developmental expression patterns were further verified by RT–qPCR.
Among the eight NPF subfamilies of wheat, NPF5 is the largest subfamily and includes as many as 34 members. Combination of the RNA-seq and RT–qPCR analysis suggests a physiological role for some wheat NPF5 genes. TaNPF5.20 was mainly expressed in vegetative tissues, and was up-regulated by N supply in leaves, stems, and nodes, but not in roots. TaNPF5.26 and TaNPF5.30 transcripts were concentrated in roots, although at different expression levels, and showed up-regulation by N supply in roots. In Arabidopsis, three tonoplast-localized NPF5 genes, AtNPF5.11, AtNPF5.12, and AtNPF5.16, have been reported to be involved in vacuolar nitrate efflux and reallocation. TaNPF5.20, TaNPF5.26, and TaNPF5.30 may be involved in nitrate reallocation in different tissues and govern the balance of nitrate between the cytoplasm and vacuole in response to the changeable N supply. Dipeptide transport activity has been verified so far only for Arabidopsis subfamily 8 NPF and one member of subfamily 5, AtNPF5.2 (Supplementary Table S12).
Members of the wheat NPF subfamily 8 have been shown to be strongly developmentally regulated in different tissues, and partly regulated also by N availability. For example, gene expression of TaNPF8.4 and TaNPF8.9 is up-regulated post-anthesis in nodes and stems, in contrast to TaNPF8.19, TaNPF8.24, and TaNPF8.28, which are down-regulated. Different expression patterns of wheat subfamily 8 have also been found in other tissues, suggesting a developmental/tissue-specific and N-dependent regulation of dipeptide distribution within the plant.
As already mentioned, Arabidopsis NPFs of nearly all subfamilies are also involved in plant hormone transport (Supplementary Table S12). The different plant hormones have crucial functions in controlling nearly all aspects of plant growth and development. In addition to the role as a nutrient, nitrate acts as a signal, and N nutrition and plant hormone signalling pathways are closely interconnected (Vega et al., 2019). In Arabidopsis, the auxin transport activity of NPF6.3 regulates N auxin accumulation in lateral roots which prevents lateral root elongation and outgrowth (Krouk et al., 2010). Gene expression of wheat NPF orthologues TaNPF6.2 and TaNPF6.3 is regulated by N availability (Buchner et al., 2014). Two potential orthologous auxin-transporting NPFs may provide wheat with a more sensitive modulation of root system architecture in relation to N availability. In addition to the stress response, ABA plays essential roles in different physiological processes. Loss-of-function mutants of the vascular-located AtNPF4.6 exhibit less sensitivity to ABA during seed germination and seedling growth (Kanno et al., 2012). Four out of seven Arabidopsis NPF subfamily 4 have been identified as ABA transporters (Supplementary Table S12). Additional members of the NPF subfamilies 1, 2, 5, and 8 have been confirmed to be able to transport ABA. Long-distance transport of ABA regulates stomatal activity in relation to water availability (Kanno et al., 2012) as well as promoting tiller bud dormancy in cereals (Luo et al., 2019). Short-distance ABA transport has been shown to influence root growth by accumulation of ABA in the root meristem, as well as root hair growth (Ondzighi-Assoume et al., 2016; Rymen et al., 2017). Transcripts of orthologous wheat NPFs are present in all tissues, partly developmentally and/or nitrogen regulated. Involvement in ABA transport may be possible but needs to be verified.
GAs are involved in many developmental processes such as seed germination, root and shoot elongation, flowering, and fruit patterning. In almost all subfamilies, GA-transporting NPFs are present with different affinities for different active GAs (Chiba et al., 2015). Overexpression of AtNPF3.1 enhances GA3 flux into all cells of the root, and npf3 mutants have impaired hypocotyl elongation and seed germination. The post-anthesis developmental up-regulation of the direct wheat orthologue, TaNPF3.1, in the grain, leaves, and stems suggests more participation in N- than GA-related transport involved in N remobilization and N transport to the grain. In contrast to the single Arabidopsis NPF3, there are three further indirect orthologous subfamily 3 NPFs which may be potentially involved in GA transport. The majority of the orthologous Arabidopsis subfamily 2 NPFs are able to transport GA and jasmonate as well as nitrate (Supplementary Table S12), indicating a potential dual action in relation to the substrate (nitrate and/or hormones), and involvement in hormonal control in relation to nutrition as well as development. Mutations of rice OsNPF2.2 and OsNPF2.4 resulted in severe dwarfism and reduced panicle length (Li et al., 2015; Xia et al., 2015), indicating involvement in GA transport. Different wheat subfamily 2 NPFs are developmentally, N only, or developmentally/N regulated in different tissues, which does not exclude possible activity in GA transport.
Strategies for improvement of crop productivity
Transgenic approaches by overexpression of NPF and other nitrate transporter genes have been successfully used to improve crop productivity and NUE in rice (Hu et al., 2015, Chen et al., 2016; Fan et al., 2016; J. Wang et al., 2018), maize (Allen et al., 2016), and tomato (Fu et al., 2015). Overexpression of OsNPF6.5 (wheat orthologues TaNPF6.2, 6.3), OsNRT2.1 and OsNRT2.3b in rice, and ZmNRT1.1A (wheat orthologue TaNPF6.1) in maize increased grain yield, above-ground biomass, and NUE. Overexpression of OsNPF6.3 (wheat orthologue TaNPF6.1) in rice significantly shortened maturation time by 9–13 d and 10–18 d under low and high N conditions, respectively, with the grain yield per plant being increased by 32–50% (W. Wang et al., 2018). Tiller number and grain yield were also increased in rice by overexpression of four OsNPF7 subfamily genes (Supplementary Table S12). The NPF gene expression profiles in different wheat tissues in this study indicate additional promising candidates. Further approaches are necessary to understand the specific functions of these candidate genes for optimizing productivity and NUE, by manipulation of these NPF genes in future wheat breeding.
Conclusions
In this study, we performed a systematic analysis of the wheat NPF gene family, including gene composition, chromosome locations, and phylogenetic relationships. We carried out detailed RNA-seq and experimental analysis to identify NPF gene expression responses in relation to tissue specificity, abiotic and biotic stresses, and more closely to N supply and/or development in different tissues. Our experimental analysis was based on materials derived from field trial experiments for verification of the putative roles and functions of individual NPF genes in N utilization. The results offer a foundation for future work aimed at both elucidating the molecular mechanisms underlying NPF gene functions in N utilization and optimizing productivity and NUE by manipulation of these NPF genes in wheat.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. RNA-seq expression profiles of selected NPF genes.
Fig. S2. Validation of the expression profiles of selected NPF genes by RT–qPCR analysis.
Fig. S3. Heatmap of NPF expression profiles in relation to different stresses.
Fig. S4. Heatmap of expression profiles of non-regulated NPF genes in roots at growth stages Z23 and Z45 by RT–qPCR.
Fig. S5. Heatmap of post-anthesis expression profiles of non-regulated NPF genes in nodes by RT–qPCR.
Fig. S6. Heatmap of NPF expression profiles in spikes by RT–qPCR.
Table S1. Analysis of Arabidopsis NPF protein domain.
Table S2. Wheat NPF gene candidates containing the IPR000109 protein domain.
Table S3. List of wheat genes excluded from the NPF gene family.
Table S4. Classification of the NPF gene family in wheat.
Table S5. Chromosomal locations of wheat NPF genes.
Tables S6. RNA-seq expression data of Choulet et al. (2014).
Table S7. Fold change (≥3-fold) of NPF gene expression under various abiotic and biotic stresses
Table S8. Comparison of identified wheat NPF genes with Bajgain et al. (2018).
Table S9. Identification of non-NPF NRT genes in Bajgain et al. (2018).
Table S10. Primer sequences used for RT–qPCR expression analysis.
Table S11. Normalized relative expression (NRE) data of RT–qPCR expression analysis.
Table S12. Summary of potential functions of identified NPF genes in Arabidopsis and rice, and their phylogeny orthologues in wheat.
Protocol S1. Detailed explanation about the NPF gene selection for RT–qPCR gene expression analysis
Acknowledgment
This work was carried out at Rothamsted Research and received support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK, as part of the Designing Future Wheat (DFW) project (BB/P016855/1), and by the Defra-sponsored Wheat Genetic Improvement Network project (CH0109). Additional funds were provided by the National Natural Science Foundation of China (31501819) and Jiangsu Agricultural Science and Technology Innovation Fund (CX(18)1001).
References
- Allen S, Guo M, Loussaert D, Rupe M, Wang H. 2016. Enhanced nitrate uptake and nitrate translocation by over-expressing maize functional low-affinity nitrate transporters in transgenic maize. US Patent 20160010101 A1. [Google Scholar]
- Bajgain P, Russell B, Mohammadi M. 2018. Phylogenetic analyses and in-seedling expression of ammonium and nitrate transporters in wheat. Scientific Reports 8, 7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, et al. 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1, 15015. [DOI] [PubMed] [Google Scholar]
- Buchner P, Hawkesford MJ. 2014. Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. Journal of Experimental Botany 65, 5697–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrow LM, Cram D, Tulpan D, Foroud NA, Loewen MC. 2016. Exogenous abscisic acid and gibberellic acid elicit opposing effects on Fusarium graminearum infection in wheat. Phytopathology 106, 986–996. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang Y, Tan Y, Zhang M, Zhu L, Xu G, Fan X. 2016. Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnology Journal 14, 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CS, Stacey G, Tsay YF. 2004. Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. Journal of Biological Chemistry 279, 30150–30157. [DOI] [PubMed] [Google Scholar]
- Chiba Y, Shimizu T, Miyakawa S, Kanno Y, Koshiba T, Kamiya Y, Seo M. 2015. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. Journal of Plant Research 128, 679–686. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Gong Z, Zhu JK. 2008. Abscisic acid-mediated epigenetic processes in plant development and stress responses. Journal of Integrative Plant Biology 50, 1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F, Alberti A, Theil S, et al. 2014. Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721. [DOI] [PubMed] [Google Scholar]
- Corratgé-Faillie C, Lacombe B. 2017. Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. Journal of Experimental Botany 68, 3107–3113. [DOI] [PubMed] [Google Scholar]
- Dietrich D, Hammes U, Thor K, Suter-Grotemeyer M, Flückiger R, Slusarenko AJ, Ward JM, Rentsch D. 2004. AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. The Plant Journal 40, 488–499. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan SC, Lin CS, Hsu PK, Lin SH, Tsay YF. 2009. The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. The Plant Cell 21, 2750–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Feng H, Tan Y, Xu Y, Miao Q, Xu G. 2016. A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. Journal of Integrative Plant Biology 58, 590–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Naz M, Fan X, Xuan W, Miller AJ, Xu G. 2017. Plant nitrate transporters: from gene function to application. Journal of Experimental Botany 68, 2463–2475. [DOI] [PubMed] [Google Scholar]
- Fan X, Tang Z, Tan Y, Zhang Y, Luo B, Yang M, Lian X, Shen Q, Miller AJ, Xu G. 2016. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proceedings of the National Academy of Sciences, USA 113, 7118–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Bai G, Huang W, Wang Z, Wang X, Zhang M. 2017. The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield. Frontiers in Plant Science 8, 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yi H, Bao J, Gong J. 2015. LeNRT2.3 functions in nitrate acquisition and long-distance transport in tomato. FEBS Letters 589, 1072–1079. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hu R, Qiu D, Chen Y, Miller AJ, Fan X, Pan X, Zhang M. 2016. Knock-down of a tonoplast localized low-affinity nitrate transporter OsNPF7.2 affects rice growth under high nitrate supply. Frontiers in Plant Science 7, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang W, Ou S, et al. 2015. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nature Genetics 47, 834–838. [DOI] [PubMed] [Google Scholar]
- Huang W, Bai G, Wang J, Zhu W, Zeng Q, Lu K, Sun S, Fang Z. 2018. Two splicing variants of OsNPF7.7 regulate shoot branching and nitrogen utilization efficiency in rice. Frontiers in Plant Science 9, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Nie H, Feng F, Wang J, Lu K, Fang Z. 2019. Altered expression of OsNPF7.1 and OsNPF7.4 differentially regulates tillering and grain yield in rice. Plant Science 283, 23–31. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- Islam MT, Croll D, Gladieux P, et al. 2016. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biology 14, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. 2012. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proceedings of the National Academy of Sciences, USA 109, 9653–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2019. pheatmap: Pretty Heatmaps. Implementation of heatmaps that offers more control over dimensions and appearance. https://rdrr.io/cran/pheatmap/. [Google Scholar]
- Komarova NY, Thor K, Gubler A, Meier S, Dietrich D, Weichert A, Suter Grotemeyer M, Tegeder M, Rentsch D. 2008. AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiology 148, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince AS, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F. 2014. Nitrate transport and signalling in Arabidopsis. Journal of Experimental Botany 65, 789–798. [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell 18, 927–937. [DOI] [PubMed] [Google Scholar]
- Léran S, Muños S, Brachet C, Tillard P, Gojon A, Lacombe B. 2013. Arabidopsis NRT1.1 is a bidirectional transporter involved in root-to-shoot nitrate translocation. Molecular Plant 6, 1984–1987. [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, et al. 2014. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends in Plant Science 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Li B, Byrt C, Qiu J, et al. 2016. Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis. Plant Physiology 170, 1014–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qiu J, Jayakannan M, et al. 2017. AtNPF2.5 modulates chloride (Cl–) efflux from roots of Arabidopsis thaliana. Frontiers Plant Science 7, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hu B, Chu C. 2017a Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. Journal of Experimental Botany 68, 2477–2488. [DOI] [PubMed] [Google Scholar]
- Li H, Yu M, Du XQ, Wang ZF, Wu WH, Quintero FJ, Jin XH, Li HD, Wang Y. 2017b NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. The Plant Cell 29, 2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zheng Q, Shen W, Cram D, Fowler DB, Wei Y, Zou J. 2015. Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. The Plant Cell 27, 86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ouyang J, Wang YY, Hu R, Xia K, Duan J, Wang Y, Tsay YF, Zhang M. 2015. Disruption of the rice nitrate transporter OsNPF2.2 hinders root-to-shoot nitrate transport and vascular development. Scientific Reports 5, 9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pan X, Guo X, Fan K, Lin W. 2019. Physiological and transcriptome analyses of early leaf senescence for ospls1 mutant rice (Oryza sativa L.) during the grain-filling stage. International Journal of Molecular Science 20, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. 1999. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. The Plant Cell 11, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF. 2003. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. The EMBO Journal 22, 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, Sun Q. 2015. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biology 15, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Takahashi M, Kameoka H, Qin R, Shiga T, Kanno Y, Seo M, Ito M, Xu G, Kyozuka J. 2019. Developmental analysis of the early steps in strigolactone-mediated axillary bud dormancy in rice. The Plant Journal 97, 1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473.10629003 [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA. 2016. Nitrate transport, sensing, and responses in plants. Molecular Plant 9, 837–856. [DOI] [PubMed] [Google Scholar]
- Ondzighi-Assoume CA, Chakraborty S, Harris JM. 2016. Environmental nitrate stimulates abscisic acid accumulation in arabidopsis root tips by releasing it from inactive stores. The Plant Cell 28, 729–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Kobayashi F, Kawahara Y, Yazawa T, Handa H, Itoh T, Matsumoto T. 2013. Characterisation of the wheat (Triticum aestivum L.) transcriptome by de novo assembly for the discovery of phosphate starvation-responsive genes: gene expression in Pi-stressed wheat. BMC Genomics 14, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M. 2009. Identification and validation of reference genes for quantitative RT–PCR normalization in wheat. BMC Molecular Biology 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike S, Gao F, Kim MJ, Kim SH, Schachtman DP, Gassmann W. 2014. Members of the NPF3 transporter subfamily encode pathogen-inducible nitrate/nitrite transporters in grapevine and Arabidopsis. Plant & Cell Physiology 55, 162–170. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Ramírez-González R, Borrill P, Lang D, et al. 2018. The transcriptional landscape of polyploid wheat. Science 361, eaar6089. [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ. 2009. Real-time quantitative RT–PCR: design, calculations, and statistics. The Plant Cell 21, 1031–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Schäfer S, et al. 2017. ABA suppresses root hair growth via the OBP4 transcriptional regulator. Plant Physiology 173, 1750–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal I, Zhang Y, Jørgensen ME, et al. 2016. The Arabidopsis NPF3 protein is a GA transporter. Nature Communications 7, 11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ye J, Yao X, et al. 2019. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nature Communications 10, 5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taochy C, Gaillard I, Ipotesi E, et al. 2015. The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. The Plant Journal 83, 466–479. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C. 2018. Source and sink mechanisms of nitrogen transport and use. New Phytologist 217, 35–53. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. 2007. Nitrate transporters and peptide transporters. FEBS Letters 581, 2290–2300. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. 1993. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. [DOI] [PubMed] [Google Scholar]
- Undurraga SF, Ibarra-Henríquez C, Fredes I, Álvarez JM, Gutiérrez RA. 2017. Nitrate signaling and early responses in Arabidopsis roots. Journal of Experimental Botany 68, 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega A, O’Brien JA, Gutiérrez RA. 2019. Nitrate and hormonal signaling crosstalk for plant growth and development. Current Opinion in Plant Biology 52, 155–163. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. 1989. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Research 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93, 77–78. [DOI] [PubMed] [Google Scholar]
- Wan Y, King R, Mitchell RAC, Hassani-Pak K, Hawkesford MJ. 2017. Spatiotemporal expression patterns of wheat amino acid transporters reveal their putative roles in nitrogen transport and responses to abiotic stress. Scientific Reports 7, 5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lu K, Nie H, Zeng Q, Wu B, Qian J, Fang Z. 2018. Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield. Rice 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu B, Yuan D, et al. 2018. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. The Plant Cell 30, 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-Y, Cheng Y-H, Chen K-E, Tsay Y-F. 2018. Nitrate transport, signaling, and use efficiency. Annual Review of Plant Biology 69, 85–122. [DOI] [PubMed] [Google Scholar]
- Wen Z, Tyerman SD, Dechorgnat J, Ovchinnikova E, Dhugga KS, Kaiser BN. 2017. Maize NPF6 proteins are homologs of arabidopsis CHL1 that are selective for both nitrate and chloride. The Plant Cell 29, 2581–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Fan X, Wei J, Feng H, Qu H, Xie D, Miller AJ, Xu G. 2015. Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. Journal of Experimental Botany 66, 317–331. [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Xuan W, Beeckman T, Xu G. 2017. Plant nitrogen nutrition: sensing and signaling. Current Opinion in Plant Biology 39, 57–65. [DOI] [PubMed] [Google Scholar]
- Yamaji N, Ma JF. 2014. The node, a hub for mineral nutrient distribution in graminaceous plants. Trends in Plant Science 19, 556–563. [DOI] [PubMed] [Google Scholar]
- Yamaji N, Ma JF. 2017. Node-controlled allocation of mineral elements in Poaceae. Current Opinion in Plant Biology 39, 18–24 [DOI] [PubMed] [Google Scholar]
- Yang M, Geng M, Shen P, Chen X, Li Y, Wen X. 2019. Effect of post-silking drought stress on the expression profiles of genes involved in carbon and nitrogen metabolism during leaf senescence in maize (Zea mays L.). Plant Physiology and Biochemistry 135, 304–309. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends in Ecology and Evolution 18, 292–298. [Google Scholar]
- Zhang G-B, Meng S, Gong J-M. 2018. The expected and unexpected roles of nitrate transporters in plant abiotic stress resistance and their regulation. International Journal of Molecular Sciences 19, 3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang Y, Wang C, Liu M, Li H, Fu Y, Wang Y, Nie Y, Liu X, Ji W. 2014. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics 15, 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.