Genetic factors determine how local and systemic nitrogen signals shape root system architecture in higher plants.
Keywords: Brassinosteroids, auxin, lateral root development, local signal, nitrate transporter, nitrogen signaling, nutrient efficiency, primary root development, root traits, systemic signal
Abstract
Among all essential mineral elements, nitrogen (N) is required in the largest amounts and thus is often a limiting factor for plant growth. N is taken up by plant roots in the form of water-soluble nitrate, ammonium, and, depending on abundance, low-molecular weight organic N. In soils, the availability and composition of these N forms can vary over space and time, which exposes roots to various local N signals that regulate root system architecture in combination with systemic signals reflecting the N nutritional status of the shoot. Uncovering the molecular mechanisms underlying N-dependent signaling provides great potential to optimize root system architecture for the sake of higher N uptake efficiency in crop breeding. In this review, we summarize prominent signaling mechanisms and their underlying molecular players that derive from external N forms or the internal N nutritional status and modulate root development including root hair formation and gravitropism. We also compare the current state of knowledge of these pathways between Arabidopsis and graminaceous plant species.
Introduction
As a major constituent of biomolecules, nitrogen (N) has a strong impact on plant growth and development, in both natural and agricultural ecosystems. Plant roots preferentially take up the inorganic N forms NO3– and NH4+, but also organic forms, including urea, amino acids, and peptides (Kojima et al., 2007; Komarova et al., 2008; Ganeteg et al., 2017). The abundance of these N forms varies with soil type and climate. In well-aerated, neutral to high pH soils, nitrate is the most abundant N source, whereas ammonium can be a dominant N form in water-logged or low pH soils. In boreal and cold habitats, amino acids can account for a major fraction of the soluble N pool (Rothstein, 2009). The availability of different N forms in soils underlies considerable variation in space and time, as demonstrated by rapid turnover of amino acid-N and large concentration gradients of nitrate varying over one to several orders of magnitude (Lark et al., 2004; Kielland et al., 2007; Rothstein, 2009). Considering the sessile nature of plants, they must adapt continuously to spatiotemporal N fluctuations by sensing available N forms and modulating N transport or metabolism and, in the long run, their root system architecture to sustain plant growth.
Referring to root system architecture as the three-dimensional configuration of the root system, it is determined mainly by four parameters—growth, branching, surface area, and angle (Morris et al., 2017). In dicotyledonous plants, such as Arabidopsis thaliana, the root system is composed of an embryonically formed primary root and individual post-embryonic lateral roots (Osmont et al., 2007). In contrast, graminaceous species, such as maize or rice, form a fibrous root system composed of embryonic seminal roots, including eventually a primary root, as well as post-embryonic shoot-borne nodal or crown roots, which all undergo higher order lateral branching (Hochholdinger et al., 2018). Despite these morphological differences, responses of many architectural traits of the two distinct root systems appear to be conserved at the genetic level (Rogers and Benfey, 2015). Such highly dynamic architectural responses of roots allow plants to optimize spatially defined soil exploration, improving plant performance under challenging N conditions. For instance, plant species forming more root biomass in deeper soil layers also deplete nitrate pools more efficiently (Heuermann et al., 2019), and local proliferation of lateral roots into N-rich soil patches contributes significantly to plant N nutrition (Hodge et al., 1999; Robinson et al., 1999; Ma et al., 2013). Under N-deficient conditions, plants develop a steeper root angle, which promotes deep rooting and facilitates N acquisition from subsoil layers, especially under soil conditions with high potential for nitrate leaching (Trachsel et al., 2013; Saengwilai et al., 2014). These observations imply that the simultaneous genetic improvement of root system architecture (RSA) and its plasticity to nutrient dynamics represents an effective strategy to improve N use efficiency. Here, we review the mechanisms by which external and internal N signals modulate root system architecture and, as far as possible, we compare these adaptive responses between the model species Arabidopsis and graminaceous crop species.
Local N signals modulate lateral root elongation and branching
Under natural conditions, soil is a spatially and temporally heterogeneous growth substrate for plant roots, in which a range of biotic and abiotic factors modulate the amount and distribution of different N forms. Precise proliferation of roots into N-rich soil zones is of crucial importance for plants to maximize soil exploitation for N. Plant strategies allowing an increase in foraging of localized N include the local stimulation of lateral root branching and elongation (Forde, 2014; Giehl et al., 2014; Giehl and von Wirén, 2014; Liu and von Wirén, 2017; Fig. 1).
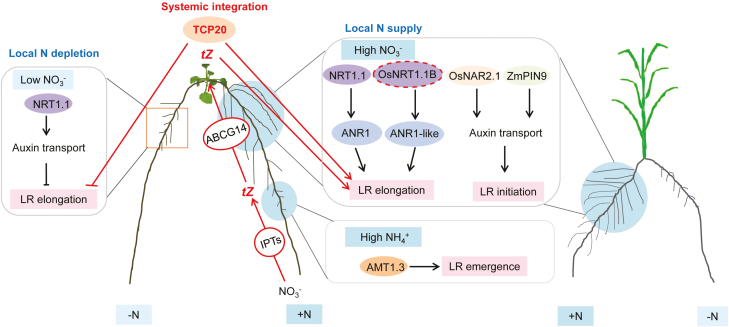
Fig. 1.
Local and systemic signaling involved in lateral root growth in response to local N supply in Arabidopsis and graminaceous species. In Arabidopsis, NRT1.1-dependent auxin removal from the lateral root (LR) primordium prevents LR elongation into N-depleted sites, while local high NO3– promotes LR elongation involving the NRT1.1–ANR1 signaling pathway that is probably conserved in cereals. Local N signaling is integrated in the shoot via the transcription factor TCP20 and cytokinin (CK) signaling pathways. Local NH4+ stimulates LR emergence in an AMT1;3-dependent manner. OsNAR2.1 and ZmPIN9 modulate polar auxin transport that promotes LR initiation under local high NO3– in rice and maize, respectively. The dashed outline indicates hypothetical functions or signaling steps that require further experimental validation. tZ, trans-zeatin; IPT, isopentenyl transferase.
Localized nitrate promotes lateral root elongation and density
When nitrate is locally supplied to an otherwise poorly N-supplemented root system, plants preferentially allocate root proliferation to NO3–-rich soil patches by stimulating lateral root elongation and/or branching; the extent of this response may vary across plant species (Drew et al., 1975; Zhang and Forde; 1998; Remans et al., 2006; Guan et al., 2014; Yan et al., 2014; Yu et al., 2015a). There is plenty of experimental evidence that NO3–per se acts as a potent signaling molecule regulating lateral root proliferation in NO3–-rich patches (Zhang and Forde, 1998; Zhang et al., 1999; Ruffel et al., 2011). In Arabidopsis, detailed analyses in split-root growth systems revealed that adaptive responses of lateral roots to local NO3– involves both local and systemic signals (Zhang and Forde, 1998; Zhang et al., 1999; Remans et al., 2006; Ruffel et al., 2011; Guan et al., 2014; Mounier et al., 2014). Localized NO3– modulates lateral root elongation by regulating meristematic activity in the lateral root tip via the nitrate transceptor NRT1.1/NPF6.3 (Zhang et al., 1999; Remans et al., 2006; Krouk et al., 2010; Mounier et al., 2014). In the absence of NO3–, NRT1.1/NPF6.3 acts as an NO3–-controlled auxin transporter that decreases auxin levels in the lateral root apex to suppress meristematic activity and lateral root elongation. This process acts upstream of the MADS-box transcription factor ANR1, which elicits a signaling cascade controlling lateral root elongation into zones of high NO3– (Remans et al., 2006; Bouguyon et al., 2015, 2016). Information on local NO3– availability is translocated to the shoot via long-distance mobile signals that include cytokinin (CK) and small peptides. In particular, trans-zeatin has been suggested to relay a long-distance systemic N demand signal that redirects lateral root growth from low-NO3– to high-NO3– zones (Ruffel et al., 2011; Poitout et al., 2018). In this case, local NO3– provision stimulates isopentenyl transferase (IPT)-dependent trans-zeatin accumulation in the root and ABCG14-mediated root to shoot translocation (Ko et al., 2014). The perception of trans-zeatin in the shoot then leads to transcriptional reprogramming and a shoot to root signal that induces nitrate transporters and root proliferation into the NO3– patch. Remarkably, in the shoot, transcriptome genes involved in glutamate and glutamine biosynthesis were found to be significantly enriched, prompting a model in which shoot-derived amino acids may transmit CK-dependent N demand signals down to the roots (Kiba et al., 2011; Poitout et al., 2018). In a parallel signaling pathway, local N deprivation triggers synthesis of C-terminally encoded peptides (CEPs) in the root, which travel to the shoot via the xylem and are perceived by two leucine-rich repeat (LRR)-receptor kinases, CEP Receptor 1 and 2 (CEPR1/2; Tabata et al., 2014). This triggers expression of two glutaredoxin-like small polypeptides, namely CEP Downstream1 and 2 (CEPD1/2), which are translocated to the roots and promote compensatory NO3– uptake through up-regulation of the high-affinity NO3– transporter gene NRT2.1 in the NO3–-replete tissue (Ohkubo et al., 2017). Although suggested, it remains elusive whether the CEP signaling pathway also regulates lateral root elongation in response to local NO3–. In addition, it has been shown that the transcription factor TCP20 functions as a systemic signaling regulator that directs lateral root foraging under heterogeneous NO3– supply (Guan et al., 2014). Thereby, TCP20 might act as an integrator of N and CK signaling, because this transcription factor can bind to promoters of type-A response regulators such as ARR5/7 that are up-regulated by NO3– in shoots (Ruffel et al., 2011). Early genetic studies using abscisic acid (ABA) biosynthesis and signaling mutants suggested that ABA also participates in the lateral root elongation response to local NO3– (Signora et al., 2001). However, it still remains open how ABA regulates lateral root responses to localized nitrate. More recently, it has been shown that the mutant abi2-2, defective in the ABA co-receptor ABI2, phenocopies the attenuated lateral root response of the NRT1.1/NPF6.3 mutant allele chl1-5 defective in nitrate perception. ABI2 modulates lateral root elongation possibly through interaction with CIPK23 and CBL1, because abi2 mutant analysis and co-expression in oocytes showed that ABI2 largely decreased the phosphorylation state of CIPK23 and CBL1, which in turn stimulated NRT1.1/NPF6.3-dependent transport, and sensing and signaling of NO3– (Leran et al., 2015). The CIPK/CBL proteins are major decoders of intracellular calcium (Ca2+) signatures (Dodd et al., 2010). In this context, Riveras et al. (2015) proposed Ca2+ as a secondary messenger in the primary NO3– response in Arabidopsis roots, raising the possibility that Ca2+ signaling exerts an early function in localized NO3–-induced lateral root elongation.
Despite the evolutionary distance between Arabidopsis and graminaceous plant species, lateral root growth in cereal crops is also strongly responsive to localized NO3– (Wang et al., 2002; Yu et al., 2015a). In rice, local NO3– supply promotes lateral root elongation and density in seminal roots, whereas in adult maize plants the extent of morphological plasticity of lateral roots depends on the root type and initiation time of shoot-borne roots (Wang et al., 2002; Yan et al., 2014; Yu et al., 2015a). For instance, although local nitrate stimulates lateral root elongation primarily in embryonic primary and seminal roots, it can also increase the length and density of laterals on shoot-borne roots (Yu et al., 2015a). In rice, evidence has been provided that ANR1-like MADS-box genes also regulate lateral root elongation in response to localized NO3– (Yan et al., 2014). The rice genome harbours five ANR1-like MADS-box transcription factor genes (OsMADS23, 25, 27, 57, and 61), four of which (OsMADS23, 25, 27, and 57) are expressed in roots and are targets of the monocot-specific miRNA miRNA444a (Puig et al., 2013; Yan et al., 2014). Overexpressing miR444a in rice lowers expression of the target MADS-box genes and impairs their stimulatory effect on NO3–-dependent lateral root elongation (Yan et al., 2014). This finding suggests that one or more of these ANR1-like genes play a similar role to that of Arabidopsis ANR1 and that further ANR1-like genes may play evolutionarily conserved roles in NO3–-regulated lateral root elongation across dicots and monocots. Notably, here care should be taken as in rice miRNA444a has additional targets, whose activities have not yet been formally excluded as participating in lateral root adaptation to localized NO3–. More recently, the rice NO3– transceptor NRT1;1B/OsNPF6.5, which is orthologous to AtNRT1;1/NFP6.3, has been discovered to control the natural genetic variation of N use efficiency between indica and japonica rice (Hu et al., 2015). However, its role in NO3–-regulated lateral root growth and its potential link to ANR1-like genes are still unclear. In addition, another nitrate transporter in rice, OsNAR2.1, appears to modulate the responsiveness of lateral roots to localized nitrate (Huang et al., 2015). In maize, increased growth rate and appearance of lateral roots under local NO3– supply correlated positively with auxin levels in those root segments that were supplied with nitrate (Sattelmacher et al., 1993). In accordance with distinct NO3– responses of lateral roots in different root types, recent cell type-specific RNA sequencing approaches revealed root type-specific transcriptomes and a unique transcriptomic landscape in pericycle cells of brace roots (Yu et al., 2015b, 2016). Detailed analysis showed that the stimulatory effect of local NO3– on lateral root initiation in shoot-borne roots depends on ZmPIN9-mediated auxin efflux and subsequent cell cycle activation facilitated by auxin/SCFSKP2B-mediated repression of Kip-related proteins (KRPs) (Yu et al., 2015b).
Local ammonium stimulates lateral root branching
NH4+ is a preferential N source for most crop plants (Gu et al., 2013), and stimulation of lateral branching by local NH4+ has been observed for decades (Drew et al., 1975; Hodge, 2004). Although enhanced lateral root proliferation under banded (i.e. localized) NH4+ supply is part of agricultural practice and contributes to enhanced fertilizer use efficiency (Ma et al., 2013), it is not understood which mechanism underlies this adaptive response. A significant advance was made when Lima et al. (2010) discovered that local NH4+ and NO3– act synergistically on lateral root proliferation and that in Arabidopsis NH4+-stimulated lateral root branching involves the NH4+ transporter AMT1;3. It was observed that NH4+-induced lateral root branching was significantly suppressed in the amt1;3 mutant but almost absent in a quadruple NH4+ transporter mutant (qko; amt1;1 amt1;2 amt1;3 amt2;1). Interestingly, reconstituted expression of either AMT1;3 or AMT1;1 in qko showed that only AMT1;3 could restore lateral root proliferation, although both transporters share similar cell type-specific localization and NH4+ transport properties (Lima et al., 2010). At that time, it remained open whether AMT1;3 acts as an NH4+ transceptor or whether the sensing event occurs downstream of AMT1;3, since AMT1;3 is consistently expressed in rhizodermal cells and rapidly feeds NH4+ into the symplast (Lima et al., 2010; Duan et al., 2018).
Responses of root system architecture to homogeneous N signals
Root growth responses to homogenous NO3– supply underlie dose-dependent regulation. Moderate NO3– supply stimulates the growth of primary and lateral roots, whereas excess nitrate suppresses them (Zhang et al., 1999; Vidal et al., 2010; Liu et al., 2017; Fig. 2). In addition to NO3–, evidence has been provided that NH4+ and the amino acid l-glutamate (l-Glu) inhibit root elongation (Walch-Liu et al., 2006; Liu et al., 2013; Fig. 2).
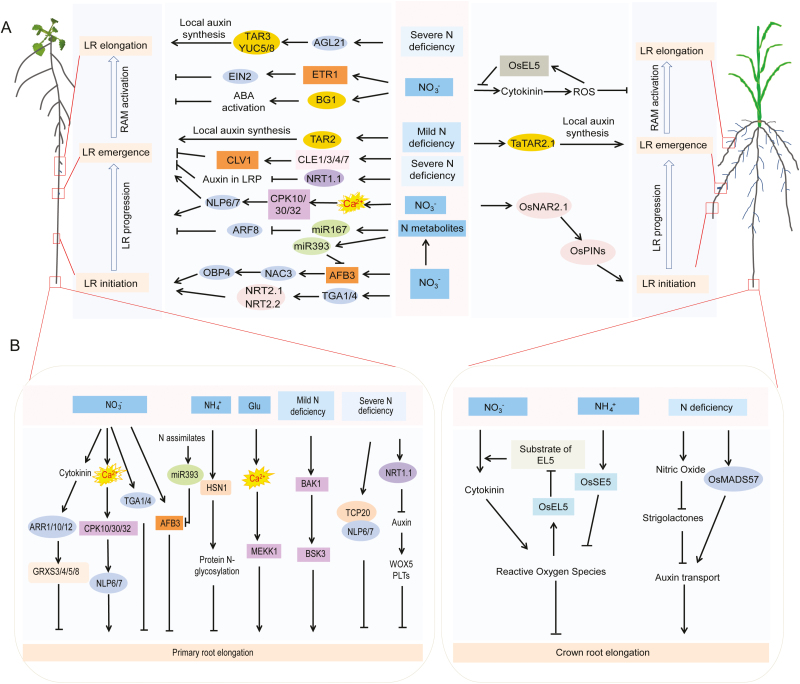
Fig. 2.
Signaling pathways shaping lateral root or primary root development in response to N deficiency or N supply in Arabidopsis and graminaceous species. (A) Signaling pathways involved in N-dependent lateral root (LR) formation in Arabidopsis and graminaceous species. Nitrate modulates LR growth in almost all developmental phases. Nitrate controls LR initiation through transcription factors TGA1/4 and the miR393/AFB3–NAC3–OBP4 signaling cascade. It also employs the miR167/ARF8 module and Ca2+–CPK10/30/32–NLP6/7 signaling to modulate LR progression/emergence. After the emergence, nitrate controls LR elongation through ETR1–EIN2-dependent ethylene and BG1-dependent ABA signalling pathways. In rice, OsNAR2.1 modulates polar auxin transport to control LR initiation, and OsEL5 interacts with NO3–-dependent cytokinin signaling to regulate LR elongation. Regarding N deficiency, severe N deficiency prevents LR emergence through the CLE/CLV1 peptide signaling module and NRT1.1-modulated auxin removal from LR primodia (LRP). It also positively regulates the MADS-box transcription factor AGL21 to regulate LR elongation, possibly through regulating local auxin biosynthesis. Mild N deficiency stimulates TAR2-dependent local auxin biosynthesis in the vasculature and pericycle to promote LR emergence, which is a rather conserved signaling cascade also discovered in wheat. (B) Signaling pathways shaping primary root development in Arabidopsis and graminaceous species under varying N availabilities. In addition to their roles in LR growth, transcription factors TGA1/4, miR393/AFB3, and Ca2+–CPK10/30/32–NLP6/7 signaling in Arabidopsis and OsEL5 in rice modulate primary root growth under nitrate supply. Nitrate also suppresses primary root elongation through glutaredoxins (GRXS3/4/5/8) acting downstream of cytokinin signaling. The inhibitory effect of ammonium on primary root elongation depends on HSN1-mediated protein N-glycosylation in Arabidopsis and production of ROS. In rice, OsSE5 can counteract the inhibition of ammonium by ROS detoxification. Glutamate (l-Glu) inhibits primary root elongation via the signaling kinase MEKK1. Severe N deficiency regulates root meristem size and distal stem cell differentiation, which involves TCP20–NLP6/7 and NRT1.1–auxin–WOX5/PLTs signaling pathways. In Arabidopsis, mild N deficiency enhances brassinosteroid signaling mediated by BAK1 and BSK3 to stimulate primary root elongation. In parallel, low N promotes crown root elongation in rice, which depends on polar auxin transport tuned by the intricate interaction of nitric oxide, strigolactones, and OsMADS57. ROS, reactive oxygen species; RAM, root apical meristem.
The amount of homogeneously distributed nitrate modulates lateral root formation
Depending on the available concentration and growth context, NO3– either stimulates or represses lateral root formation. These responses are orchestrated by spatial and temporal changes of gene expression in roots perceiving NO3– signals (Gifford et al., 2008; Walker et al., 2017). Ca2+ signaling plays a central role in the stimulatory effect of NO3– on lateral root formation, since intracellular NO3– permeated by NRT1.1/NPF6.3 induces cytosolic Ca2+ waves that can be decoded by the subgroup III Ca2+-sensor protein kinases CPK10/30/32. These CPKs phosphorylate NIN-LIKE PROTEIN 6/7 (NLP6/7) to retain their nuclear localization, which promotes progression and emergence of lateral root primordia (Liu et al., 2017). Excess NO3– (≥10 mM) modulates phytohormone homeostasis and/or signaling to down-regulate most, if not all, developmental processes of lateral root formation (Zhang et al., 2007; Krouk, 2016). In Arabidopsis, elevated NO3– modulates lateral root formation via the auxin signaling modules miR167/ARF8 and miR393/AFB3 (Gifford et al., 2008; Vidal et al., 2010). Cell type-specific transcriptome profiling revealed that downstream of NO3–, the N metabolites glutamine/glutamate repress the expression of miR167 in pericycle cells, allowing accumulation of ARF8 transcripts in the pericycle to control the key developmental checkpoint between lateral root initiation and emergence (Gifford et al., 2008). Unlike ARF8 that is not regulated by NO3–, NO3–per se strongly induces expression of the auxin receptor gene AFB3, whose transcript levels are feedback repressed by downstream N assimilates via miR393 that targets the AFB3 transcript for degradation (Vidal et al., 2010). Mutation of AFB3 or overexpression of miR393 markedly abrogates the stimulatory effect of NO3– on lateral root initiation (Vidal et al., 2010). This pathway has been further extended by the finding that the transcription factor gene NAC4 and its target OBP4 act downstream of AFB3 to specifically promote lateral root initiation and emergence (Vidal et al., 2013). Using a systems biology approach, the two bZIP transcription factor genes TGA1 and TGA4 have been identified. TGA1/4 act downstream of the nitrate transceptor NRT1.1/NFP6.3 and bind to promoters of the two high-affinity NO3– transporter genes NRT2.1/NRT2.2 to regulate lateral root initiation (Alvarez et al., 2014).
After emergence from the parental root, the meristem of lateral roots is activated to commence elongation. Excess supply of NO3– (≥10 mM) interferes with this developmental process by inducing a systemic signal to repress the elongation of lateral roots (Zhang et al., 1999). Analysis of ABA biosynthesis and signaling mutants suggested the involvement of ABI4- and ABI5-dependent ABA signaling as well as of an ABA-independent pathway mediating this systemic repression (Signora et al., 2001). Recently, NO3– provision (30 mM) has been shown to stimulate the release of ABA from a conjugated form (ABA-glucose) by the induction of a β-glucosidase (BG1), thereby eliciting downstream an ABA response that represses lateral root growth (Ondzighi-Assoume et al., 2016). Exposure to high NO3– (10 mM) rapidly induces ethylene production and represses lateral root growth. Lateral root growth in etr1-3 and ein2-1 mutants, both impaired in ethylene signaling, is insensitive to high NO3–, suggesting a regulatory role for ethylene in the systemic repression of lateral root growth by high NO3– (Tian et al., 2009). Compared with these extensive studies in Arabidopsis, it is poorly understood how excess NO3– regulates lateral root branching in crop plants, except for the characterization of OsEL5, a ubiquitin ligase that interacts with NO3–-dependent cytokinin signaling to repress lateral root formation (Mochizuki et al., 2014).
Effects of homogeneously supplied nitrate on primary root elongation
Apart from lateral roots, NO3– also regulates primary root elongation. The auxin signaling module miRNA393/AFB3 and the transcription factors TGA1/4 have been shown to control the inhibitory effect of NO3– on primary root elongation, while the signaling cascade NO3––Ca2+–CPKs–NLP6/7 promotes primary root growth (Vidal et al., 2010; Alvarez et al., 2014; Liu et al., 2017). Thereby, a link has been established between glutaredoxins and NO3–. NO3– up-regulates expression of a group of glutaredoxin genes (AtGRXS1/3/4/5/6/8/11) in shoots, while RNA silencing of AtGRXS3/4/5/8 reverses the inhibitory effect of elevated NO3– on primary root growth (Patterson et al., 2016). NO3–-dependent up-regulation of AtGRXS3/4/5/8 appears to be mediated by CKs, because this response becomes almost completely lost in an arr1,10,12 mutant that is defective in CK signaling (Patterson et al., 2016). These results collectively suggest that NO3– provision stimulates biosynthesis of CKs in roots and their translocation to the shoot, where CKs activate ARR1/10/12-mediated expression of GRX genes, which may be transported rootwards to repress primary root elongation. Likewise, in primary, seminal, and crown roots of maize, high NO3– (≥10 mM) inhibits cell elongation, which coincides with decreased auxin levels in apical root segments (Tian et al., 2008). Physiological approaches further suggest auxin, CKs, and nitric oxide (NO) to trigger NO3–-inhibited root elongation (Tian et al., 2005, 2008; Zhao et al., 2007); however, the underlying molecular mechanisms remain to be discovered.
Elevated ammonium inhibits root elongation
Whenever NH4+ is supplied to plants as the sole N source, it inhibits the elongation of primary and lateral roots. Evidence from supplying NH4+ to different root zones suggested that NH4+ is sensed in the root apex (Li et al., 2010). At the cellular level, NH4+ considerably represses cell proliferation and expansion (Liu et al., 2013). In an earlier study, auxin has been suggested to be involved in NH4+-mediated inhibition of root elongation, because mutants impaired either in auxin transport (aux1) or in signaling (axr1 and axr2) are more resistant to NH4+ (Cao et al., 1993). However, a more recent study showed that in the presence of NH4+, primary root elongation of aux1 is still as sensitive as in the wild type (Liu et al., 2013), leaving it open whether auxin participates in NH4+-dependent inhibition of root elongation. One component in the molecular mechanism underlying NH4+-inhibited primary root elongation has been unmasked by the isolation of the hsn1-1 mutant that is hypersensitive to NH4+ (Qin et al., 2008). HSN1, allelic to VTC1, encodes a GDP-mannose pyrophosphorylase (GMPase). The lack of HSN1 and subsequent N-glycosylation of proteins has been associated with root hypersensitivity to NH4+ (Qin et al., 2008; Barth et al., 2010). NH4+ triggers the production of reactive oxygen species (ROS) that act as signaling molecules in primary root elongation (Patterson et al., 2010; Tsukagoshi et al., 2010; Bloch et al., 2011). In rice, the heme–heme oxygenase OsSE5, a novel antioxidant regulatory enzyme, is strongly induced by elevated NH4+, while inhibition of root elongation by NH4+ is markedly mitigated by up-regulation of OsSE5 activity using a chemical heme oxygenase inducer or by overexpression of OsSE5 in Arabidopsis or rice (Xie et al., 2015). It has been reported that ROS levels do not differ between wild-type and vtc1 mutant plants, suggesting that GDP-mannose-dependent protein N-glycosylation and ROS signaling act independently in the regulation of primary root elongation in the presence of NH4+. In addition, a mutant defective in Indeterminate Domain 10 (IDD10) was also shown to be hypersensitive to NH4+, while the detailed mechanism is unknown (Xuan et al., 2013).
l-Glutamate as a signal suppressor of primary root growth
Although NO3– and NH4+ are the predominant N forms in most soils, organic N forms can also account for a major fraction of the available N pool, for instance in bog soils, and interfere with root growth processes. Previous work has shown that even very low concentrations (≤0.05 mM) of external l-Glu can remarkably inhibit primary root elongation and stimulate root branching, consequently resulting in a shorter but more branched root system (Walch-Liu et al., 2006). The sensing of l-Glu appears to take place in the root tip, because the inhibitory effect of l-Glu supplied locally to primary root tips was as strong as that supplied to the whole root system. In this case, NRT1;1/NPF6.3-dependent NO3– signaling could antagonistically suppress l-Glu-mediated inhibition of primary root growth (Walch-Liu and Forde, 2008). With respect to auxin, l-Glu has been shown to reduce expression of the auxin-responsive reporter DR5–GFP (green fluorescent protein) in the primary root apex, while in auxin mutants (aux1-7, axr1-3, and axr1-12), the sensitivity of primary root growth to l-Glu is altered (Walch-Liu et al., 2006). However, the exact role of auxin in l-Glu-mediated root architectural changes remains to be demonstrated. Regarding the mode of action, l-Glu-elicited signal transduction involves the mitogen-activated protein (MAP) signaling kinase MEKK1 (Forde et al., 2013). Very recently, quantitative trait locus (QTL) analysis using recombinant inbred lines derived from reciprocal crosses between the Arabidopsis accessions C24 and Col-0 mapped a major locus, named GluS1, controlling root sensitivity to l-Glu (Walch-Liu et al., 2017). Future studies isolating the causal gene are required to extend our understanding on the mechanistic action by which l-Glu inhibits primary root growth.
Dual effects of N deficiency on root system architecture
A systematic comparison of root architectural changes under diverse nutrient deficiencies revealed a dose dependency of root architectural traits on external N supply and on the plant N nutritional status (Gruber et al., 2013; Fig. 2). Thereby, mild and severe N deficiency can be distinguished by shoot fresh weight and shoot N concentration responses to external N, which both decreased between 550 µM and 275 µM N supply, indicative of mild N deficiency, while at ≤100 µM external N (i.e. severe N deficiency), shoot N concentrations dropped below the critical deficiency level (Gruber et al., 2013). Accordingly, root growth was stimulated in a concentration range of 200–550 µM N, whereas at <100 μM external N, plants adopt a ‘survival strategy’ by inhibiting the elongation of both primary and lateral roots as well as the emergence of new lateral roots (Gruber et al., 2013; Giehl and von Wirén, 2014). This morphological response restricts root growth to nutritionally unfavorable environments to economize on the cost for root development in favor of plant survival. In contrast, at external N levels of 200–550 µM that induce mild deficiency, plants expand their root system by increasing the emergence of lateral roots (Ma et al., 2014; Shao et al., 2017) and in particular the length of primary and lateral roots (Chun et al., 2005; Gruber et al., 2013; Sun et al., 2014). This foraging response enhances the capacity of the root system to explore deeper soil horizons where N may be more abundant.
Severe N deficiency restricts root branching and elongation
Under persistent N deficiency, early lateral root development is modulated by a regulatory pathway that involves CLAVATA3/ESR-related (CLE) signaling peptides and their receptor protein, the LRR receptor-like kinase CLAVATA1 (CLV1) (Araya et al., 2014a, b, 2016). While expression of several CLE homologs, including CLE3, is up-regulated by low N (≤0.1 mM) in pericycle cells, the receptor protein CLV1 localizes to phloem companion cells (Araya et al., 2014a). Whether this indicates CLE peptide cycling via the shoot or radial short-distance transport within the root remains open. Focusing on lateral root primordia, N deprivation favors the accumulation of NRT1.1/NFP6.3, which facilitates the shootward movement of auxin, thereby lowering auxin accumulation in lateral root primordia to inhibit their emergence (Krouk et al., 2010; Bouguyon et al., 2015, 2016). Low nitrate further modulates stem cell dynamics in the root meristem with profound impacts on root system architecture. TCP20, a systemic master regulator in nitrate signaling, physically interacts with NLP6/7 to regulate the expression of the cell cycle gene CYCB1;1 and cell division in the root apical meristem (Guan et al., 2017). In addition to cell division, low nitrate (0.05 mM) is also found to promote differentiation of distal stem cells (Wang et al., 2019). At low nitrate, when auxin accumulation in the root apex is significantly suppressed, expression levels of the stem cell identity genes WOX5 and PLT1/2 become down-regulated. This response involves NRT1.1/NFP6.3, because roots of chl1-12 plants, defective in NRT1.1/NFP6.3 function, are less sensitive to low N in terms of distal stem cell differentiation and show higher expression levels of WOX5 and PLT genes. These observations led to a working model, in which low N restricts auxin accumulation in the root tips via NRT1.1/NFP6.3-dependent signaling and subsequently represses expression of WOX5 and PLT genes to promote distal stem cell differentiation. Considering that in the context of low nitrate, NRT1.1/NFP6.3 prevents auxin accumulation in the lateral root primordium via its auxin transport activity (Krouk et al., 2010), distal stem cell differentiation is more likely to be due to the activity of NRT1.1/NFP6.3 in auxin transport rather than in nitrate sensing. Further analyses on distal stem cell differentiation in chl1-9 or T101A/D substitution lines that can decouple nitrate sensing from auxin transport will refine the mechanism by which NRT1.1/NFP6.3 modulates stem cell differentiation (Bouguyon et al., 2015). N deprivation also induces expression of the MADS-box transcription factor gene AGL21 (Yu et al., 2014). Mutation in AGL21 leads to impaired lateral elongation especially under severe N limitation, probably through modulated local auxin biosynthesis.
Mild N deficiency stimulates root elongation and branching
The stimulation of root growth by mild N deficiency is of particular interest as it increases the soil volume that can be explored for nutrient acquisition. The positive effect of mild N deficiency on lateral root formation requires the auxin biosynthesis gene TAR2. The corresponding protein catalyzes the first step in the main auxin biosynthesis route by converting tryptophan to indole-3-pyruvic acid (Zhao, 2012; Ma et al., 2014). Mild N deficiency induces expression of TAR2 in the pericycle and vasculature of mature root zones, whilst tar2 mutants show reduced lateral root numbers especially under mild N deficiency (Ma et al., 2014). So far, it is less likely that TAR2-mediated auxin biosynthesis is also necessary for lateral root elongation, because TAR2-dependent auxin production cannot explain the phenotype of lateral root elongation. Recently, Brumos et al. (2018) showed that precise spatial expression of auxin biosynthesis genes is critical for root development and root responses to ethylene. It is very likely that the inability of TAR2-dependent auxin biosynthesis to modulate lateral root elongation is due to its spatially restricted expression in the root maturation zone (Ma et al., 2014; Ursache et al., 2014). More recently, a role for brassinosteroid (BR) signaling has been discovered to regulate root foraging under mild N deficiency. By employing genome-wide association mapping, Jia et al. (2019a) identified Brassinosteroid Signaling Kinase 3 (BSK3) as being associated with a QTL of primary root length and promoting cell elongation in response to mild N deficiency. Interestingly, allelic variation caused by a single amino acid substitution from proline to leucine enhances plant BR sensitivity and signaling, which increases the root foraging response. Whereas N deficiency has no impact on the transcriptional regulation of BSK3, transcript levels of the BR co-receptor gene BAK1 increase under N deficiency, suggesting systemic N deficiency signals entering BR signaling in roots via BAK1 (Jia et al., 2019a). Like Arabidopsis, crop plants also develop longer roots under N deficiency, albeit with considerable genotypic variation (Chun et al., 2005; Sun et al., 2014; Melino et al., 2015; Shao et al., 2017). Although BR signaling components have been well characterized in cereals, for example in rice, maize, and barley, their roles in modulating root foraging under N deficiency await discovery (Dockter et al., 2014; Kir et al., 2015; Tong and Chu, 2018). In rice, a combined role for strigolactones and auxin has been implicated in low N-induced seminal and adventitious root elongation (Sun et al., 2014). It has been reported that low N enhances strigolactone biosynthesis and signaling, involving the genes D10, D27, and D3, which in turn reduces OsPIN1b-mediated rootward polar auxin transport and attenuates seminal root growth (Sun et al., 2014, 2018). Upstream of strigolactone signaling, low nitrate stimulates NO production. NO targets the strigolactone signaling repressor D53 for degradation, thereby allowing elongation of seminal and adventitious roots (Sun et al., 2016). Most recently, OsMADS57 has been shown to modulate seminal and adventitious root elongation under low nitrate by modulating PIN-directed auxin accumulation in the root tips (Huang et al., 2019). So far, it has been shown that OsMADS57 modulates tillering in rice through D14-dependent strigolactone signaling (Guo et al., 2013). The role of OsMADS57 in strigolactone-modulated polar auxin transport and in root elongation, however, awaits further investigation.
An inconsistency across plant species has been observed regarding the regulation of lateral root density by low N. On the one hand, it has been reported that low N increases lateral root emergence in Arabidopsis and wheat (Ma et al., 2013; Shao et al., 2017). On the other hand, low N decreased lateral root density in rice and maize (Sun et al., 2014; Gao et al., 2015). As mentioned above, low N stimulates vasculature-expressed TAR2 to promote lateral root emergence in Arabidopsis (Ma et al., 2014). A conserved function of its orthologous gene TaTAR2.1 has been reported in wheat, in which transgenic lines with reduced or enhanced expression grow fewer or more lateral roots, respectively, under low N (Shao et al., 2017). In rice, low nitrate elevates strigolactone levels and decreases lateral root density via D3, an F-box protein mediating strigolactone perception (Sun et al., 2014). Furthermore, knockdown of OsNAR2.1, encoding a partner protein of the high-affinity nitrate transporters OsNRT2.1/2.2/2.3a, inhibits lateral root formation in response to nitrate, probably through decreased PIN-mediated rootward transport of auxin (Huang et al., 2015).
Signaling mechanisms underlying N-dependent root hair formation and root gravitropic responses
Root hairs greatly expand the absorptive surface area of roots and express a number of nutrient transporters, which facilitates the uptake of water and minerals. The formation and elongation of root hairs are highly responsive to environmental N signals, indicating that root hair growth can be seen as a read-out for N-sensing events. Mechanisms shaping N-dependent root hair plasticity are just beginning to emerge. In a recent study, high NO3– availability has been shown to increase root hair density mainly by suppressing the longitudinal elongation of trichoblasts (Canales et al., 2017). The mechanism of action involves the NO3– transceptor NRT1;1/NPF6.3 and the signal transducers TGA1/4, which directly regulate expression of the root hair-specifying gene CPC to increase root hair density. In the case of NH4+, the tonoplast-localized Ca2+-associated protein kinase CAP1 has been proven essential for NH4+-regulated root hair growth (Bai et al., 2014). In this case, CAP1 acts as modulator of cytoplasmic NH4+ homeostasis that establishes tip-focused cytoplasmic Ca2+ and pH gradients as prerequisite for root hair growth. Regardless of N forms, root hair length is continuously increasing with decreasing N levels in the growth substrate (Vatter et al., 2015). Auxin and ethylene have been shown to play key roles in root hair formation and to be responsible for root developmental changes elicited by nutrient signals (Tian et al., 2009; Bhosale et al., 2018). To what extent these phytohormones also modulate root hair elongation under N-deficient conditions remains to be discovered. Although a number of regulators of root hair development have been isolated in crops, including rice and maize (Giri et al., 2018; Hochholdinger et al., 2018), it remains completely unknown which mechanisms mediate root hair responses to varying N availabilities in cereal crops.
Root growth angle is critical for the spatial distribution of the root system in soils and thus for nutrient and water uptake. For instance, it has been shown that the steeper root growth angle conferred by DRO1 in rice plants increases tolerance to drought (Uga et al., 2013). Regarding responses of the root growth angle to N availability, it has been observed that moderate levels of NH4+ increase gravitropism while excess NH4+ causes agravitropism (Zou et al., 2012). Notably, this phenomenon is independent of the inhibitory effect of NH4+ on root elongation (Liu et al., 2013). NH4+-induced agravitropism is associated with repression of the auxin efflux carrier PIN2 and subsequent failure of asymmetric auxin distribution in the root elongation zone (Zou et al., 2012; Liu et al., 2013). In addition, Altered Response to Gravity 1 (ARG1) has been shown to sustain PIN3-mediated lateral auxin gradients across the lateral root cap and AUX1-facilitated basipetal auxin movement to antagonize the role of PIN2, thereby protecting root gravitropism under excess NH4+ (Zou et al., 2013). In response to low N, the root growth angle becomes steeper, probably for improved N foraging in deeper soil layers (Trachsel et al., 2013). However, the underlying mechanisms are still unknown.
N-dependent root architectural changes dependent on other nutrient signals
Although extensive studies have been carried out to elucidate how roots respond to N signals, phenotypic changes in root traits depend not only on the supplied N but also on the level of supply of other nutrients (Fig. 3). Indeed, this is an expected scenario for plant roots growing in natural heterogeneous soils, where roots are exposed to multiple facets of nutrient gradients at the same time. Recent exciting studies reveal intricate interactions between NO3– and phosphate (Pi) signaling in shaping plant physiological and developmental processes (Medici et al., 2015, 2019; Hu et al., 2019). The reduction in primary root elongation under Pi deficiency depends on NO3– signalling. In this case, the GRAS family transcription factor gene HRS1 together with its paralog HHO1 have been reported to be transcriptionally induced by NO3– but post-translationally regulated by Pi starvation before coordinating an NO3–-dependent primary root response to Pi deficiency (Medici et al., 2015). Evidence also points to crosstalk between N and potassium (K) signaling. For example, primary root elongation is strongly repressed under low K, which in turn depends on NH4+ (Xu et al., 2006). Another angle of N–K interaction appears with regard to the modulation of lateral root branching. It shows that low NO3– can largely suppress second-order lateral root formation induced by K starvation (Kellermeier et al., 2014). Analysis of root systems in mutants defective in genes with known roles in K and NO3– transport and/or signaling suggests crucial roles for the NO3– transceptor NTR1.1/NPF6.3 and the K channel AKT1 in this root response (Kellermeier et al., 2014). In this case, CIPK23 appears to be a connective node integrating NO3– and K signaling through phosphorylation of NTR1.1 and AKT1 to regulate lateral root formation (Tsay et al., 2011; Kellermeier et al., 2014).
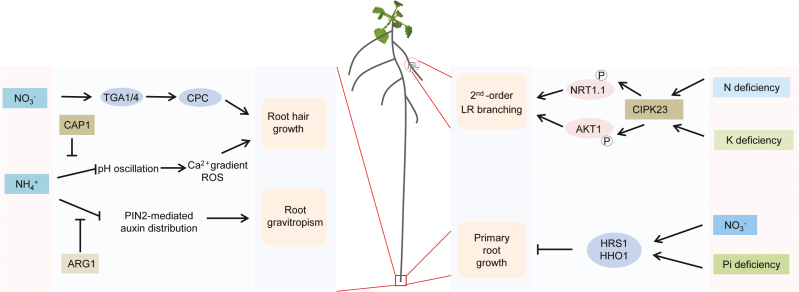
Fig. 3.
Nitrogen-coordinated root hair growth, gravitropism, and root architectural changes by interaction with other nutrient signals in Arabidopsis. Nitrate activates a signaling cascade involving TGA1/4 and CPC to increase root hair density. Ammonium induces intracellular pH imbalance, which disturbs cytosolic Ca2+ gradients and activates reactive oxygen species (ROS), leading to swelling of root hairs. This process can be alleviated by a tonoplast-localized receptor kinase CAP1, which maintains intracellular ammonium homeostasis by vacuolar compartmentation. In addition, ammonium inhibits PIN2-mediated asymmetric auxin distribution and prevents root gravitropism, which can be counteracted by ARG1. CIPK23 is an integrator of low N–K signals into the regulation of second-order lateral root (LR) branching. This process requires the nitrate transceptor NRT1.1 and potassium channel AKT1; both are phosphorylation targets of CIPK23. Nitrate also coordinates the primary root response to phosphate deficiency by regulating the activity of HRS1/HHO1.
Conclusion and perspectives
Over the past couple of years, several root responses to N have been investigated across plant species and found to be highly conserved. Significant progress has been made in elucidating the molecular and cellular basis of how roots sense environmental N signals, in particular by the discovery of phytohormones and peptides conveying local and systemic N signals in the model plant Arabidopsis (Tabata et al., 2014; Ohkubo et al., 2017; Poitout et al., 2018; Jia et al., 2019a). In crops, however, much work remains to be done as compared with the knowledge gained in Arabidopsis, as most studies in graminaceous species do not go far beyond the stage of physiological description. Particularly successful examples, such as modulating the expression of Arabidopsis-orthologous ANR1-like genes in rice or TAR2 in wheat (Yan et al., 2014; Shao et al., 2017), indicate conserved roles for these genes in N-driven root architectural changes, underscoring the possibility to translate findings from Arabidopsis to crop species. Thus, ongoing efforts in characterizing orthologous genes in cereals will further advance our understanding of N sensing and downstream root responses in crops. With regard to recent studies showing that root responses to N are integrated with responses to other environmental stimuli (Kellermeier et al., 2014; Medici et al., 2015, 2019), future challenges will lie in the identification of regulators intersecting these responses. In particular, genomic studies in combination with systems biology approaches have great potential to uncover the most relevant players in the corresponding regulatory networks. There is a large intraspecific variation in root system architectural traits in Arabidopsis as well as in crop species (Rosas et al., 2013; Maccaferri et al., 2016; Jia et al., 2019b). Obviously, with the development of high-throughput, non-destructive root phenotyping platforms and advanced imaging tools (Kenobi et al., 2017), genome-wide association studies will become an even more powerful tool for decoding the genetic complexity of RSA responses to N signals and the identification of useful natural alleles. Carried further, identified favorable alleles can be used for root tissue- or cell type-specific expression and allele-specific modification in crops, taking advantage of CRISPR/Cas9 technology for the benefit of improved N use efficiency.
Acknowledgements
The research of the authors related to this review is supported by a fellowship from China Scholarship Council to ZJ (no. 201406350062) and by the Deutsche Forschungsgemeinschaft (DFG) with a grant (WI1728/13-2) to NvW.
References
- Alvarez JM, Riveras E, Vidal EA, et al. 2014. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal 80, 1–13. [DOI] [PubMed] [Google Scholar]
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H. 2014a CLE–CLAVATA1 peptide–receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proceedings of the National Academy of Sciences, USA 111, 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T, von Wirén N, Takahashi H. 2014b CLE peptides regulate lateral root development in response to nitrogen nutritional status of plants. Plant Signaling & Behavior 9, e29302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T, von Wirén N, Takahashi H. 2016. CLE peptide signaling and nitrogen interactions in plant root development. Plant Molecular Biology 91, 607–615. [DOI] [PubMed] [Google Scholar]
- Bai L, Ma X, Zhang G, Song S, Zhou Y, Gao L, Miao Y, Song CP. 2014. A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. The Plant cell 26, 1497–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, Gouzd ZA, Steele HP, Imperio RM. 2010. A mutation in GDP-mannose pyrophosphorylase causes conditional hypersensitivity to ammonium, resulting in Arabidopsis root growth inhibition, altered ammonium metabolism, and hormone homeostasis. Journal of Experimental Botany 61, 379–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale R, Giri J, Pandey BK, et al. 2018. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nature Communications 9, 1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D, Monshausen G, Singer M, Gilroy S, Yalovsky S. 2011. Nitrogen source interacts with ROP signalling in root hair tip-growth. Plant, Cell & Environment 34, 76–88. [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, et al. 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1, 15015. [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Perrine-Walker F, Pervent M, et al. 2016. Nitrate controls root development through posttranscriptional regulation of the NRT1.1/NPF6.3 transporter/sensor. Plant Physiology 172, 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN. 2018. Local auxin biosynthesis is a key regulator of plant development. Developmental Cell 47, 306–318.e5. [DOI] [PubMed] [Google Scholar]
- Canales J, Contreras-López O, Álvarez JM, Gutiérrez RA. 2017. Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. The Plant Journal 92, 305–316. [DOI] [PubMed] [Google Scholar]
- Cao Y, Glass AD, Crawford NM. 1993. Ammonium inhibition of Arabidopsis root growth can be reversed by potassium and by auxin resistance mutations aux1, axr1, and axr2. Plant Physiology 102, 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun L, Mi GH, Li JS, et al. 2005. Genetic analysis of maize root characteristics in response to low nitrogen stress. Plant and Soil 276, 369–382. [Google Scholar]
- Dockter C, Gruszka D, Braumann I, et al. 2014. Induced variations in brassinosteroid genes define barley height and sturdiness, and expand the green revolution genetic toolkit. Plant Physiology 166, 1912–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. 2010. The language of calcium signaling. Annual Review of Plant Biology 61, 593–620. [DOI] [PubMed] [Google Scholar]
- Duan F, Giehl RFH, Geldner N, Salt DE, von Wirén N. 2018. Root zone-specific localization of AMTs determines ammonium transport pathways and nitrogen allocation to shoots. PLoS Biology 16, e2006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC. 1975. Comparison of effects of a localized supply of phosphate, nitrate, ammonium and potassium on growth of seminal root system, and shoot, in barley. New Phytologist 75, 479–490. [Google Scholar]
- Forde BG. 2014. Nitrogen signalling pathways shaping root system architecture: an update. Current Opinion in Plant Biology 21, 30–36. [DOI] [PubMed] [Google Scholar]
- Forde BG, Cutler SR, Zaman N, Krysan PJ. 2013. Glutamate signalling via a MEKK1 kinase-dependent pathway induces changes in Arabidopsis root architecture. The Plant Journal 75, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeteg U, Ahmad I, Jämtgård S, Aguetoni-Cambui C, Inselsbacher E, Svennerstam H, Schmidt S, Näsholm T. 2017. Amino acid transporter mutants of Arabidopsis provides evidence that a non-mycorrhizal plant acquires organic nitrogen from agricultural soil. Plant, Cell & Environment 40, 413–423. [DOI] [PubMed] [Google Scholar]
- Gao K, Chen F, Yuan L, Zhang F, Mi G. 2015. A comprehensive analysis of root morphological changes and nitrogen allocation in maize in response to low nitrogen stress. Plant, Cell & Environment 38, 740–750. [DOI] [PubMed] [Google Scholar]
- Giehl RF, Gruber BD, von Wirén N. 2014. It’s time to make changes: modulation of root system architecture by nutrient signals. Journal of Experimental Botany 65, 769–778. [DOI] [PubMed] [Google Scholar]
- Giehl RF, von Wirén N. 2014. Root nutrient foraging. Plant Physiology 166, 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. 2008. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences, USA 105, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri J, Bhosale R, Huang G, et al. 2018. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nature Communications 9, 1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N. 2013. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiology 163, 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R, Duan F, An X, Zhang F, von Wirén N, Yuan L. 2013. Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays L.). Plant & Cell Physiology 54, 1515–1524. [DOI] [PubMed] [Google Scholar]
- Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM. 2017. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proceedings of the National Academy of Sciences, USA 114, 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Wang R, Nacry P, Breton G, Kay SA, Pruneda-Paz JL, Davani A, Crawford NM. 2014. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proceedings of the National Academy of Sciences, USA 111, 15267–15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K. 2013. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nature Communications 4, 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuermann D, Gentsch N, Boy J, Schweneker D, Feuerstein U, Groß J, Bauer B, Guggenberger G, von Wirén N. 2019. Interspecific competition among catch crops modifies vertical root biomass distribution and nitrate scavenging in soils. Scientific Reports 9, 11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Yu P, Marcon C. 2018. Genetic control of root system development in maize. Trends in Plant Science 23, 79–88. [DOI] [PubMed] [Google Scholar]
- Hodge A. 2004. The plastic plant: root responses to heterogeneous supplies of nutrients. New Phytologist 162, 9–24. [Google Scholar]
- Hodge A, Robinson D, Griffiths BS, Fitter AH. 1999. Why plants bother: root proliferation results in increased nitrogen capture from an organic patch when two grasses compete. Plant Cell & Environment 22, 811–820. [Google Scholar]
- Hu B, Jiang Z, Wang W, et al. 2019. Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nature Plants 5, 401–413. [DOI] [PubMed] [Google Scholar]
- Hu B, Wang W, Ou S, et al. 2015. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nature Genetics 47, 834–838. [DOI] [PubMed] [Google Scholar]
- Huang S, Chen S, Liang Z, Zhang C, Yan M, Chen J, Xu G, Fan X, Zhang Y. 2015. Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Scientific Reports 5, 18192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Liang Z, Chen S, Sun H, Fan X, Wang C, Xu G, Zhang Y. 2019. A transcription factor, OsMADS57, regulates long-distance nitrate transport and root elongation. Plant Physiology 180, 882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Giehl RFH, Meyer RC, Altmann T, von Wirén N. 2019a Natural variation of BSK3 tunes brassinosteroid signaling to regulate root foraging under low nitrogen. Nature Communications 10, 2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Liu Y, Gruber BD, Neumann K, Kilian B, Graner A, von Wirén N. 2019b Genetic dissection of root system architectural traits in spring barley. Frontiers in Plant Science 10, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A. 2014. Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. The Plant Cell 26, 1480–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenobi K, Atkinson JA, Wells DM, Gaju O, De Silva JG, Foulkes MJ, Dryden IL, Wood ATA, Bennett MJ. 2017. Linear discriminant analysis reveals differences in root architecture in wheat seedlings related to nitrogen uptake efficiency. Journal of Experimental Botany 68, 4969–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H. 2011. Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. Journal of Experimental Botany 62, 1399–1409. [DOI] [PubMed] [Google Scholar]
- Kielland K, Mcfarland JW, Ruess RW, Olson K. 2007. Rapid cycling of organic nitrogen in taiga forest ecosystems. Ecosystems 10, 360–368. [Google Scholar]
- Kir G, Ye H, Nelissen H, Neelakandan AK, Kusnandar AS, Luo A, Inzé D, Sylvester AW, Yin Y, Becraft PW. 2015. RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiology 169, 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D, Kang J, Kiba T, et al. 2014. Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proceedings of the National Academy of Sciences, USA 111, 7150–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Bohner A, Gassert B, Yuan L, von Wirén N. 2007. AtDUR3 represents the major transporter for high-affinity urea transport across the plasma membrane of nitrogen-deficient Arabidopsis roots. The Plant Journal 52, 30–40. [DOI] [PubMed] [Google Scholar]
- Komarova NY, Thor K, Gubler A, Meier S, Dietrich D, Weichert A, Suter Grotemeyer M, Tegeder M, Rentsch D. 2008. AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiology 148, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G. 2016. Hormones and nitrate: a two-way connection. Plant Molecular Biology 91, 599–606. [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell 18, 927–937. [DOI] [PubMed] [Google Scholar]
- Lark RM, Milne AE, Addiscott TM, et al. 2004. Scale- and location-dependent correlation of nitrous oxide emissions with soil properties: an analysis using wavelets. European Journal of Soil Science 55, 611–627. [Google Scholar]
- Léran S, Edel KH, Pervent M, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Tillard P, Gojon A, Kudla J, Lacombe B. 2015. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Science Signaling 8, ra43. [DOI] [PubMed] [Google Scholar]
- Li Q, Li BH, Kronzucker HJ, Shi WM. 2010. Root growth inhibition by NH4+ in Arabidopsis is mediated by the root tip and is linked to NH4+ efflux and GMPase activity. Plant, Cell & Environment 33, 1529–1542. [DOI] [PubMed] [Google Scholar]
- Lima JE, Kojima S, Takahashi H, von Wirén N. 2010. Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. The Plant Cell 22, 3621–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Niu Y, Konishi M, et al. 2017. Discovery of nitrate–CPK–NLP signalling in central nutrient-growth networks. Nature 545, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lai N, Gao K, Chen F, Yuan L, Mi G. 2013. Ammonium inhibits primary root growth by reducing the length of meristem and elongation zone and decreasing elemental expansion rate in the root apex in Arabidopsis thaliana. PLoS One 8, e61031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, von Wirén N. 2017. Ammonium as a signal for physiological and morphological responses in plants. Journal of Experimental Botany 68, 2581–2592. [DOI] [PubMed] [Google Scholar]
- Ma QH, Zhang FS, Rengel Z, Shen JB. 2013. Localized application of NH4+-N plus P at the seedling and later growth stages enhances nutrient uptake and maize yield by inducing lateral root proliferation. Plant and Soil 372, 65–80. [Google Scholar]
- Ma W, Li J, Qu B, He X, Zhao X, Li B, Fu X, Tong Y. 2014. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. The Plant Journal 78, 70–79. [DOI] [PubMed] [Google Scholar]
- Maccaferri M, El-Feki W, Nazemi G, Salvi S, Canè MA, Colalongo MC, Stefanelli S, Tuberosa R. 2016. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. Journal of Experimental Botany 67, 1161–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A, Marshall-Colon A, Ronzier E, Szponarski W, Wang R, Gojon A, Crawford NM, Ruffel S, Coruzzi GM, Krouk G. 2015. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nature Communications 6, 6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A, Szponarski W, Dangeville P, et al. 2019. Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. The Plant Cell 31, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino VJ, Melino AD, Fiene G, Enju A, Cai J, Buchner P, Heuer S. 2015. Genetic diversity for root plasticity and nitrogen uptake in wheat seedlings. Functional Plant Biology 42, 942–956. [DOI] [PubMed] [Google Scholar]
- Mochizuki S, Jikumaru Y, Nakamura H, Koiwai H, Sasaki K, Kamiya Y, Ichikawa H, Minami E, Nishizawa Y. 2014. Ubiquitin ligase EL5 maintains the viability of root meristems by influencing cytokinin-mediated nitrogen effects in rice. Journal of Experimental Botany 65, 2307–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, et al. 2017. Shaping 3D root system architecture. Current Biology 27, R919–R930. [DOI] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P. 2014. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant, Cell & Environment 37, 162–174. [DOI] [PubMed] [Google Scholar]
- Ondzighi-Assoume CA, Chakraborty S, Harris JM. 2016. Environmental nitrate stimulates abscisic acid accumulation in Arabidopsis root tips by releasing it from inactive stores. The Plant Cell 28, 729–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa-Ohnishi M, Matsubayashi Y. 2017. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nature Plants 3, 17029. [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. 2007. Hidden branches: developments in root system architecture. Annual Review of Plant Biology 58, 93–113. [DOI] [PubMed] [Google Scholar]
- Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA. 2010. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell & Environment 33, 1486–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K, Walters LA, Cooper AM, Olvera JG, Rosas MA, Rasmusson AG, Escobar MA. 2016. Nitrate-regulated glutaredoxins control Arabidopsis primary root growth. Plant Physiology 170, 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitout A, Crabos A, Petřík I, Novák O, Krouk G, Lacombe B, Ruffel S. 2018. Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-zeatin in shoots. The Plant Cell 30, 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig J, Meynard D, Khong GN, Pauluzzi G, Guiderdoni E, Gantet P. 2013. Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expression Patterns 13, 160–170. [DOI] [PubMed] [Google Scholar]
- Qin C, Qian W, Wang W, Wu Y, Yu C, Jiang X, Wang D, Wu P. 2008. GDP-mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 105, 18308–18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. 2006. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proceedings of the National Academy of Sciences, USA 103, 19206–19211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA. 2015. The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiology 169, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Hodge A, Griffiths BS, Fitter AH. 1999. Plant root proliferation in nitrogen-rich patches confers competitive advantage. Proceedings of the Royal Society B: Biological Sciences 266, 431–435. [Google Scholar]
- Rogers ED, Benfey PN. 2015. Regulation of plant root system architecture: implications for crop advancement. Current Opinion in Biotechnology 32, 93–98. [DOI] [PubMed] [Google Scholar]
- Rosas U, Cibrian-Jaramillo A, Ristova D, et al. 2013. Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proceedings of the National Academy of Sciences, USA 110, 15133–15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein DE. 2009. Soil amino-acid availability across a temperate-forest fertility gradient. Biogeochemistry 92, 201–215. [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. 2011. Nitrogen economics of root foraging: transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences, USA 108, 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengwilai P, Tian X, Lynch JP. 2014. Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiology 166, 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattelmacher B, Gerendas J, Thoms K, et al. 1993. Interaction between root-growth and mineral-nutrition. Environmental and Experimental Botany 33, 63–73. [Google Scholar]
- Shao A, Ma W, Zhao X, Hu M, He X, Teng W, Li H, Tong Y. 2017. The auxin biosynthetic TRYPTOPHAN AMINOTRANSFERASE RELATED TaTAR2.1-3A increases grain yield of wheat. Plant Physiology 174, 2274–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H. 2001. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. The Plant Journal 28, 655–662. [DOI] [PubMed] [Google Scholar]
- Sun H, Bi Y, Tao J, et al. 2016. Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant, Cell & Environment 39, 1473–1484. [DOI] [PubMed] [Google Scholar]
- Sun H, Tao J, Bi Y, et al. 2018. OsPIN1b is involved in rice seminal root elongation by regulating root apical meristem activity in response to low nitrogen and phosphate. Scientific Reports 8, 13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G. 2014. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. Journal of Experimental Botany 65, 6735–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. 2014. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. [DOI] [PubMed] [Google Scholar]
- Tian Q, Chen F, Liu J, Zhang F, Mi G. 2008. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. Journal of Plant Physiology 165, 942–951. [DOI] [PubMed] [Google Scholar]
- Tian QY, Chen FJ, Zhang FS, Mi GH. 2005. Possible involvement of cytokinin in nitrate-mediated root growth in maize. Plant and Soil 277, 185–196. [Google Scholar]
- Tian QY, Sun P, Zhang WH. 2009. Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytologist 184, 918–931. [DOI] [PubMed] [Google Scholar]
- Tong H, Chu C. 2018. Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends in Plant Science 23, 1016–1028. [DOI] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. 2013. Maize root growth angles become steeper under low N conditions. Field Crops Research 140, 18–31. [Google Scholar]
- Tsay YF, Ho CH, Chen HY, Lin SH. 2011. Integration of nitrogen and potassium signaling. Annual Review of Plant Biology 62, 207–226. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. 2010. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. [DOI] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, et al. 2013. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Ursache R, Miyashima S, Chen Q, Vatén A, Nakajima K, Carlsbecker A, Zhao Y, Helariutta Y, Dettmer J. 2014. Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 141, 1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatter T, Neuhäuser B, Stetter M, Ludewig U. 2015. Regulation of length and density of Arabidopsis root hairs by ammonium and nitrate. Journal of Plant Research 128, 839–848. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. 2010. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras–López O, Gutiérrez RA. 2013. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proceedings of the National Academy of Sciences, USA 110, 12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG. 2008. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises l-glutamate-induced changes in root architecture. The Plant Journal 54, 820–828. [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Liu LH, Remans T, Tester M, Forde BG. 2006. Evidence thatl-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant & Cell Physiology 47, 1045–1057. [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Meyer RC, Altmann T, Forde BG. 2017. QTL analysis of the developmental response to l-glutamate in Arabidopsis roots and its genotype-by-environment interactions. Journal of Experimental Botany 68, 2919–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L, Boddington C, Jenkins D, et al. 2017. Root architecture shaping by the environment is orchestrated by dynamic gene expression in space and time. Plant Cell 29, 2393–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu P, Xia M, Wu Z, Chen Q, Liu F. 2002. Identification of genes enriched in rice roots of the local nitrate treatment and their expression patterns in split-root treatment. Gene 297, 93–102. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gong Z, Friml J, Zhang J. 2019. Nitrate modulates the differentiation of root distal stem cells. Plant Physiology 180, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Mao Y, Xu S, Zhou H, Duan X, Cui W, Zhang J, Xu G. 2015. Heme–heme oxygenase 1 system is involved in ammonium tolerance by regulating antioxidant defence in Oryza sativa. Plant, Cell & Environment 38, 129–143. [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. 2006. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. [DOI] [PubMed] [Google Scholar]
- Xuan YH, Priatama RA, Huang J, et al. 2013. Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytologist 197, 791–804. [DOI] [PubMed] [Google Scholar]
- Yan Y, Wang H, Hamera S, Chen X, Fang R. 2014. miR444a has multiple functions in the rice nitrate-signaling pathway. The Plant Journal 78, 44–55. [DOI] [PubMed] [Google Scholar]
- Yu LH, Miao ZQ, Qi GF, Wu J, Cai XT, Mao JL, Xiang CB. 2014. MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Molecular Plant 7, 1653–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Baldauf JA, Lithio A, Marcon C, Nettleton D, Li C, Hochholdinger F. 2016. Root type-specific reprogramming of maize pericycle transcriptomes by local high nitrate results in disparate lateral root branching patterns. Plant Physiology 170, 1783–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Eggert K, von Wirén N, Li C, Hochholdinger F. 2015b Cell type-specific gene expression analyses by RNA sequencing reveal local high nitrate-triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle regulation. Plant Physiology 169, 690–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Hochholdinger F, Li C. 2015a Root-type-specific plasticity in response to localized high nitrate supply in maize (Zea mays). Annals of Botany 116, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences, USA 96, 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Rong HL, Pilbeam D. 2007. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. Journal of Experimental Botany 58, 2329–2338. [DOI] [PubMed] [Google Scholar]
- Zhao DY, Tian QY, Li LH, Zhang WH. 2007. Nitric oxide is involved in nitrate-induced inhibition of root elongation in Zea mays. Annals of Botany 100, 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. 2012. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Molecular Plant 5, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou N, Li B, Chen H, Su Y, Kronzucker HJ, Xiong L, Baluška F, Shi W. 2013. GSA-1/ARG1 protects root gravitropism in Arabidopsis under ammonium stress. New Phytologist 200, 97–111. [DOI] [PubMed] [Google Scholar]
- Zou N, Li B, Dong G, Kronzucker HJ, Shi W. 2012. Ammonium–induced loss of root gravitropism is related to auxin distribution and TRH1 function, and is uncoupled from the inhibition of root elongation in Arabidopsis. Journal of Experimental Botany 63, 3777–3788. [DOI] [PubMed] [Google Scholar]