Reprogramming the genetic regulation of nitrogen fixation and assimilation in a root-associated diazotroph to produce novel strains can restore ammonium production in the presence of exogenous nitrogen inputs.
Keywords: Ammonium production, biological nitrogen fixation, corn rhizosphere, diazotroph, Kosakonia sacchari, nitrogenase expression, regulatory nodes
Abstract
Plants depend upon beneficial interactions between roots and root-associated microorganisms for growth promotion, disease suppression, and nutrient availability. This includes the ability of free-living diazotrophic bacteria to supply nitrogen, an ecological role that has been long underappreciated in modern agriculture for efficient crop production systems. Long-term ecological studies in legume–rhizobia interactions have shown that elevated nitrogen inputs can lead to the evolution of less cooperative nitrogen-fixing mutualists. Here we describe how reprogramming the genetic regulation of nitrogen fixation and assimilation in a novel root-associated diazotroph can restore ammonium production in the presence of exogenous nitrogen inputs. We isolated a strain of the plant-associated proteobacterium Kosakonia sacchari from corn roots, characterized its nitrogen regulatory network, and targeted key nodes for gene editing to optimize nitrogen fixation in corn. While the wild-type strain exhibits repression of nitrogen fixation in conditions replete with bioavailable nitrogen, such as fertilized greenhouse and field experiments, remodeled strains show elevated levels in the rhizosphere of corn in the greenhouse and field even in the presence of exogenous nitrogen. Such strains could be used in commercial applications to supply fixed nitrogen to cereal crops.
Introduction
One of the most significant agricultural inputs is nitrogen fertilizer, especially in the production of cereal crops. In the last 40 years, grain yield in corn (Zea mays) in the USA has increased linearly at a relative gain of 1.2% per year (Cassman and Liska, 2007). This increase in genetic gain is supported by technological advances in hybrid breeding, biotechnology, irrigation, conservation tillage, and fertilization practices. One of the earliest innovations contributing to increased cereal crop yields was the availability of synthetic nitrogen fertilizer via the Haber–Bosch process, which converts relatively non-reactive nitrogen gas to biologically available ammonia using high temperature and pressure (Erisman et al., 2008). Today, commercial fertilizer is responsible for 40–60% of the world’s food production, and global demand for nitrogen fertilizer remains strong (Roberts, 2009). While inorganic fertilizers have become fundamental to ensuring global food supplies through the 21st century, future increases in sustainable food production must be accomplished in an environmentally safe manner through ecological intensification (Roberts, 2009).
Increases in cereal crop yields in the second half of the 20th century necessitated a massive increase in the production and application of synthetic nitrogen fertilizer, of which about half is lost to the environment as aquatic or atmospheric pollutants (Lassaletta et al., 2014; Zhang et al., 2015). Since the 1960s, human use of synthetic nitrogen fertilizers has increased 9-fold, with further substantial increases of ~40–60% expected over the next 40 years (Sutton et al., 2013)). The manufacture of synthetic nitrogen fertilizer consumes ~2% of the world’s energy production (Glendining et al., 2009), and its intensified use has disrupted the world’s biogeochemical nitrogen cycle, causing major environmental, health, and economic problems (Rockstrom et al., 2009; Kanter, 2018). The European Nitrogen Assessment, a comprehensive assessment of the massive anthropogenic increase in reactive nitrogen in the environment, has identified five key societal threats: water quality, air quality, greenhouse gas balance, ecosystems, and biodiversity and soil quality (Sutton et al., 2011).
Crop-associated microbiota represent an attractive alternative to deliver nitrogen to cereal crops in an efficient and environmentally friendly manner. Microbes within the rhizosphere, endosphere, and phyllosphere of plants interact with crops through numerous mechanisms including nitrogen fixation, nutrient solubilization, metabolism of plant growth hormones, secretion of biocontrol metabolites, and stimulation of plant systemic defenses (Compant et al., 2019). In the past two decades, tools for probing crop–microbe interactions have become more sophisticated (Cardinale, 2014; Haichar et al., 2016; Kaul et al., 2016), and the diversity of microbial species that can be domesticated and commercially formulated has expanded. Numerous efforts are underway to develop novel strains that confer useful agronomic properties and improve the efficiency and reliability of microbial-based products for large-scale farming operations (Kaminsky et al., 2019). Realizing the broad potential of crop-associated microbes would be accelerated by leveraging crop–microbe interaction data to rationally and rapidly improve microbial genotypes and phenotypes. We have recently described a guided microbial remodeling platform by which microbes with plant-beneficial traits can be isolated, optimized, and developed for use as inoculants in agriculture (Bloch et al., 2019b).
A small group of bacteria and archaea, known as diazotrophs, are capable of reducing dinitrogen to ammonium using nitrogenase enzymes (de Bruijn, 2015). This process of biological nitrogen fixation (BNF) is the major contributor to the nitrogen economy of the biosphere, accounting for 30–50% of the total nitrogen in crop fields (Ormeno-Orrillo et al., 2013). While legumes (Fabaceae) form specialized symbiotic associations with diazotrophic rhizobia in root nodules, non-legume crops associate with diazotrophic bacteria which reside in the rhizosphere at the surface or interior of the root (de Bruijn, 2015). A root-associated, nitrogen-fixing bacterium could theoretically provide significant nitrogen to a cereal crop given adequate abundance and nitrogen-fixing activity in the rhizosphere, reducing producers’ dependence on synthetic nitrogen. Several recent reviews discuss strategies and challenges associated with the use of diazotrophic bacteria for agricultural benefit in non-legumes (Mus et al., 2016; Batista and Dixon, 2019; Bloch et al., 2019a; Pankievicz et al., 2019). However, due to the high energetic cost of BNF, requiring 16 ATPs per molecule of N2 reduced, the abundance of synthetic nitrogen selects against and represses BNF in modern agricultural settings, preventing significant contributions of nitrogen by natural soil bacteria (Chapman et al., 1949; Stewart, 1969; Dixon and Kahn, 2004; Weese et al., 2015; Smercina et al., 2019). Additionally, nitrogen-fixing bacteria efficiently assimilate fixed nitrogen into microbial biomass, preventing it from being released to the rhizosphere (Batista and Dixon, 2019). While several plant-associated diazotrophs with altered ammonium assimilation pathways have been shown to support the growth of algae (Ortiz-Marquez et al., 2014; Barney et al., 2015) and plants (Pankievicz et al., 2015; Ambrosio et al., 2017) in lab and greenhouse settings, few have been applied in the field (Bageshwar et al., 2017), probably due to a lack of fitness in the rhizosphere (Colnaghi et al., 1997). Recently, synthetic biology has been applied to refactor the regulation of genes encoding the nitrogenase enzyme complex (known as the nif cluster), enabling more control over nitrogenase expression in a range of species (Temme et al., 2012; Wang et al., 2013; Smanski et al., 2014). In one instance, the transgenic nitrogen-fixing endophyte Pseudomonas protogens Pf-5 X940 led to improved nitrogen and biomass accumulation in inoculated corn and wheat (Triticum aestivum) plants in greenhouse conditions without nitrogen fertilizer (Fox et al., 2016). Despite these advances, reports of diazotrophs that can overcome repression by exogenous fertilizer and express nitrogenase genes in fertilized field conditions are lacking.
As a step toward overcoming this challenge, we used our guided microbial remodeling platform (Bloch et al., 2019b) to optimize the nitrogen fixation and ammonium assimilation regulatory networks in a corn root-associated bacterium. A strain of Kosakonia sacchari was isolated from corn roots and rewired to express nitrogenase and fix and release nitrogen in the presence of exogenous sources of nitrogen. The resulting strains are capable of expressing nitrogenase genes in the rhizosphere of corn in fertilized conditions, both in the greenhouse and in the field. Additionally, they are non-transgenic, potentially leading to accelerated regulatory approvals for commercialization. This work constitutes a critical proof of concept that rewiring the regulatory networks of microbes has the potential to up-regulate plant-beneficial phenotypes in the root microbiome.
Materials and methods
Media
Minimal medium contains (per liter) 25 g of Na2HPO4, 0.1 g of CaCl2·2H2O, 3 g of KH2PO4, 0.25 g of MgSO4·7H2O, 1 g of NaCl, 2.9 mg of FeCl3, 0.25 mg of Na2MoO4·2H2O, and 20 g of sucrose. Growth medium is defined as minimal medium supplemented with glutamine to a final concentration of 10 mM. Super Optimal Broth (SOB) contains (per liter) 20 g of casein hydrolysate, 5 g of yeast extract, 0.5 g of NaCl, 2.4 g of MgSO4, and 0.186 g of KCl (RPI, P/N S25040-1000).
Isolation of Kosakonia sacchari PBC6.1
Corn seedlings were grown from seed (DKC 66-40, DeKalb, IL, USA) for 2 weeks in a greenhouse environment controlled from 22 °C (night) to 26 °C (day) and exposed to 16 h light cycles in agricultural soil collected from San Joaquin County, CA, USA. Roots were harvested and washed with sterile deionized water to remove bulk soil. Root tissues were homogenized, and the samples were centrifuged for 1 min at 13 000 rpm to separate tissue from root-associated bacteria. Supernatants were diluted and plated on NfB medium supplemented with 1.5% agar (Baldani et al., 2014). Plates were incubated at 30 °C for 5–7 d. Colonies that emerged were tested for the presence of the nifH gene by colony PCR with primers Ueda19f and Ueda407r (Gaby and Buckley, 2012). Genomic DNA from strains with a positive nifH colony PCR was isolated (QIAamp DNA Mini Kit, Cat No. 51306, QIAGEN, Germany) and sequenced (Illumina MiSeq v3, SeqMatic, Fremont, CA, USA). Reads were assembled and annotated using a5 (https://academic.oup.com/bioinformatics/article/31/4/587/2748163) and Prokka (https://academic.oup.com/bioinformatics/article/30/14/2068/2390517), respectively, and the isolates containing nitrogen fixation gene clusters were utilized in downstream research.
Fluorescence microscopy
The K. sacchari PBC6.1 strain was selected for further study and gene editing. For fluorescence microscopy, corn seedlings were cultivated on unsterilized 1% Murashige and Skoog agar. PBC6.1 was transformed with plasmid PB114-RFP, a plasmid containing the pSC101 origin of replication, chloramphenicol resistance cassette, and a gene encoding red fluorescent protein (RFP) under the control of a strong constitutive promoter. Transformed cells were cultured overnight in SOB with 50 µg ml–1 chloramphenicol at 30 °C with shaking, and 1 ml of cell suspension was inoculated directly on the seed at the time of seeding. Seven days after germination, roots were removed from agar, gently washed, and cut into sections using a sterile razor blade. Root sections were mounted on microscope slides and imaged on a 6D Widefield Nike TI inverted fluorescence microscope located at the Nikon Imaging Center at the University of California, San Francisco.
Initial field trial with isolated diazotrophs
Of the first 49 strains isolated and genome sequenced, six were selected for a field study of corn root colonization, which was carried out near San Luis Obispo, CA, USA in the summer of 2015. Corn seed was coated with a culture suspension of each isolated diazotroph, resulting in ~107 viable bacterial cells per seed. Treated and untreated control (UTC) seeds of two maize genotypes (BR56M30 and P0876R) were planted on 24 June 2015. Root samples from three replicate plants per treatment per genotype were collected at 12 weeks after planting. Root samples were processed and assayed for colonization of the inoculant diazotrophs as described below.
Extraction of the root-associated microbiome
Roots were shaken gently to remove loose particles, and root systems were separated and soaked in an RNA stabilization solution (Thermo Fisher P/N AM7021) for 30 min. The roots were then briefly rinsed with sterile deionized water. Samples were homogenized using bead beating with 1/2 inch stainless steel ball bearings in a tissue lyser (TissueLyser II, Qiagen P/N 85300) in 2 ml of lysis buffer (Qiagen P/N 79216). Genomic DNA (gDNA) extraction was performed with the ZR-96 Quick-gDNA kit (Zymo Research P/N D3010), and RNA extraction was performed using the RNeasy kit (Qiagen P/N 74104).
Root colonization assay
Genomic DNA extracted from root samples was used to quantify root colonization using quantitative PCR (PCR) with primers designed by Primer Blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to amplify unique regions of the wild-type (WT) genome. For the initial field colonization experiment, qPCR was carried out using the SYBR GreenER qPCR SuperMix Universal (Thermo Fisher P/N 11762100) kit, using only a forward and reverse amplification primer; for all other assays, the Kapa Probe Force kit (Kapa Biosystems P/N KK4301) was used with amplification primers and a TaqMan probe containing a FAM dye label at the 5' end, an internal ZEN quencher, and a minor groove binder and fluorescent quencher at the 3' end (Integrated DNA Technologies). Primer and probe sequences used for qPCR are listed in Supplementary Table S1 at JXB online. qPCR efficiency was measured using a standard curve generated from a known quantity of gDNA from the target genome. Data were normalized to genome copies per g FW using the tissue weight and extraction volume. For each experiment, the colonization numbers were compared with UTC seedlings.
Acetylene reduction assay (ARA)
A modified version of the ARA (Temme et al., 2012) was used to measure nitrogenase activity in pure culture conditions. Strains were propagated from a single colony in 3 ml of SOB for 24 h (30 °C, 200 rpm) and then subcultured 1:25 into growth medium and grown aerobically for 24 h (30 °C, 200 rpm). A 1 ml aliquot of the growth medium culture was then added to 4 ml of minimal medium supplemented with 0–10 mM glutamine in airtight Hungate tubes and grown anaerobically for 4 h (30 °C, 200 rpm). Headspace (10%) was removed then replaced by an equal volume of acetylene by injection, and incubation continued for 1 h. Subsequently, 2 ml of headspace was removed via a gas-tight syringe for quantification of ethylene production using an Agilent 6850 gas chromatograph equipped with a flame ionization detector (FID).
Ammonium excretion assay
Excretion of fixed nitrogen in the form of ammonium was measured using batch fermentation in anaerobic bioreactors. Strains were propagated from a single colony in 1 ml per well of SOB in a 96-well DeepWell plate. The plate was incubated for 24 h (30 °C, 200 rpm) and then diluted 1:25 into a fresh plate containing 1 ml per well of growth medium. Cells were incubated for 24 h (30 °C, 200 rpm) and then diluted 1:10 into a fresh plate containing minimal medium. The plate was transferred to an anaerobic chamber (Coy) with a gas mixture of >98.5% nitrogen, 1.2–1.5% hydrogen, and <30 ppm oxygen, and incubated at 1350 rpm, at room temperature for 66–70 h. Initial culture biomass was compared with end biomass by measuring the optical density (OD) at 590 nm. Cells were then separated by centrifugation, and supernatant from the reactor broth was assayed for free ammonium using the Megazyme Ammonia Assay kit (P/N K-AMIAR) normalized to biomass at each time point.
Gene editing
The genome modifications described in Fig. 2 were generated using genome editing methods described in a recently published patent application on our guided microbial remodeling platform (Bloch et al., 2019b). Genome-edited strains were cured of all plasmids used to carry out genome editing by repeated subculturing followed by sequence verification of the desired edits. Primers used to verify the edits are listed in Supplementary Table S2.
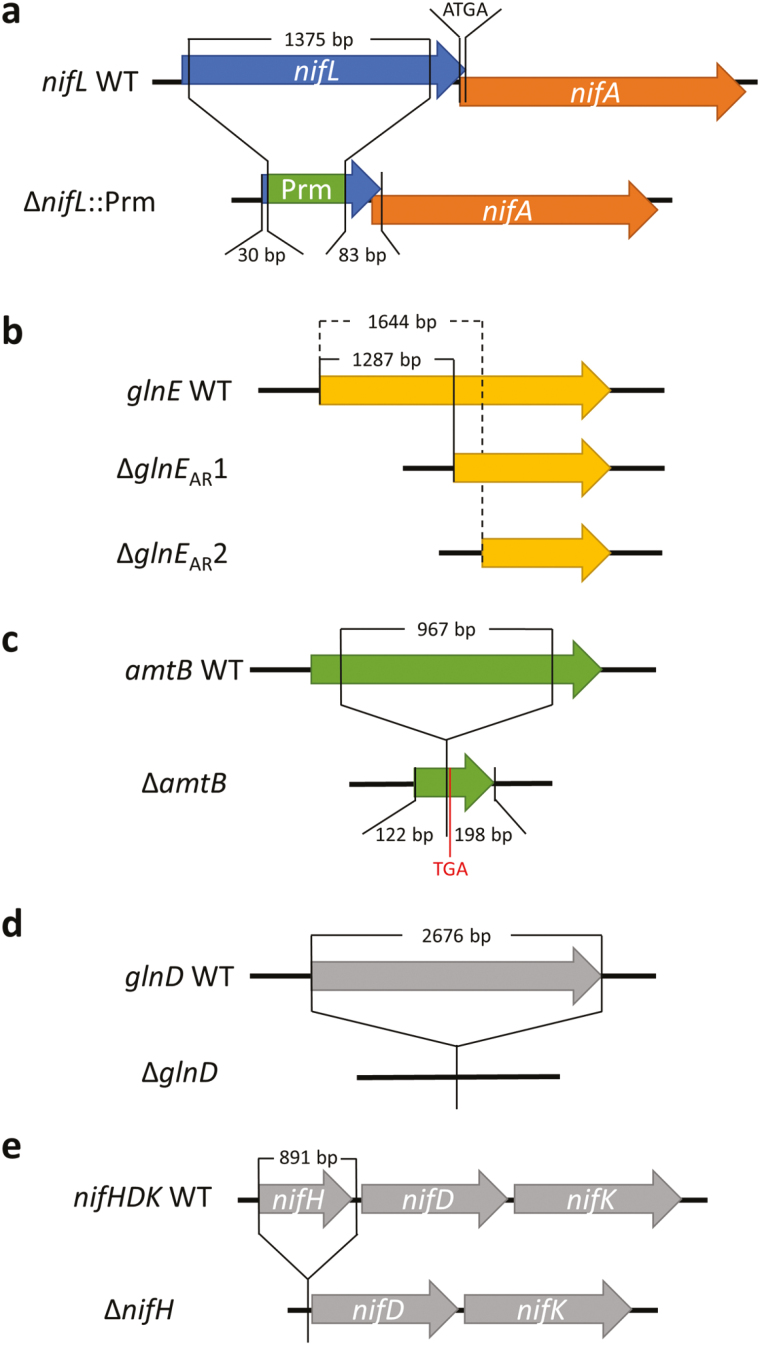
Fig. 2.
Mutated sequences of the key genes of the nitrogen fixation and assimilation regulatory network of PBC6.1. (a) Representation of the ΔnifL::Prm mutations. The majority of the nifL gene was disrupted, leaving some nucleotides at the 5' and 3' ends. The start codon of nifA, which overlaps with the stop codon of nifL, was left intact. Promoter (Prm) sequences from two different loci in the PBC6.1 genome were inserted into the disrupted portion of the nifL gene to drive nifA transcription. (b) Representation of the ΔglnEAR mutations. The 5' region of the glnE gene was deleted, to remove the N-terminal adenylyl-removing (AR) domain of the resulting protein. A start codon was re-inserted at the 5' end of the truncated sequences to allow protein translation. (c) Representation of the ΔamtB mutation. The majority of the amtB gene was deleted, leaving some nucleotides at the 5' and 3' end. The deletion resulted in a frameshift, causing a stop codon just after the point of deletion. (d) Representation of the ΔglnD mutation. The entire coding region of the glnD gene was deleted, starting at the A of the ATG start codon and ending immediately after the gene’s stop codon. No flanking regions were deleted. (e) Representation of the nifHDK operon and the ΔnifH mutation. The entire nifH coding region was deleted starting at the A of the ATG start codon and ending 9 bp downstream of the stop codon.
Greenhouse assays to measure nifH transcription in PBC6.29, PBC6.99, PBC6.38, and PBC6.94
A planting medium with minimal background nitrogen was prepared using either pure sand (Fig. 5A–C) or a mixture of vermiculite and washed sand (Fig. 5D, E). The sand mixture was autoclaved for 1 h at 122 °C and ~600 g was measured out into a D40 Deepot (Stuewe and Sons) before planting corn seeds (DKC 66-40) at a depth of ~1 cm. For the 2 week assays, each seed was inoculated with either sterile phosphate-buffered saline (PBS; UTC controls) or an equal volume of microbial suspension using cells diluted to a set OD. For the 4 week assay, seedlings were inoculated with a suspension of cells drenched directly over the emerging coleoptile at 5 d after planting. Inoculum was prepared from 5 ml of overnight cultures in SOB, which was spun down and resuspended twice in 5 ml of PBS to remove residual SOB before final dilution to an OD of 1.0. The plants were maintained under standard growth room conditions using fluorescent lamps and a 16 h daylength with a 26 °C day temperature and 22 °C night temperature. Plants were fertilized twice per week with a modified Hoagland’s fertilizer solution containing 2 mM KNO3. Additional watering during the assay was performed using either deionized water (Fig. 5A–C) or the modified Hoagland’s solution with no KNO3 The fertilizer solution contained (per liter) 3 mmol of CaCl2, 0.5 mmol of KH2PO4, 2 mmol of MgSO4, 17.9 µmol of FeSO4, 2.86 mg of H3BO3, 1.81 mg of MnCl2·4H2O, 0.22 mg of ZnSO4·7H2O, 51 µg of CuSO4·5H2O, 0.12 mg of Na2MoO4·2H2O, and 0.14 nmol of NiCl2. All pots were watered with sterile deionized H2O as needed to maintain consistent soil moisture. At 2 or 4 weeks after planting, plants were harvested, and nucleic acids were extracted from root tissue. Colonization of each strain was measured as described in the ‘Extraction of the root-associated microbiome’ and ‘Root colonization assay’ sections above. For microbial RNA quantification, roots were chopped into ~1 cm sections, and 5 g of root tissues was placed in conical tubes containing 10 ml of RNAlater solution (Invitrogen). Samples were vortexed for 10 min at 4 °C, and the solution containing the microbial cells was separated from root tissue using a 100 µm strainer. Microbial cells were then pelleted by centrifugation and resuspended in PBS. After digestion with lysozyme and proteinase K, microbial RNA was extracted using the RNeasy Kit (Qiagen) according to the standard protocol. The extracted RNA was used for NanoString analysis of nifA, nifH, and rpoB microbial transcript on an nCounter Sprint (Core Diagnostics, Hayward, CA, USA).
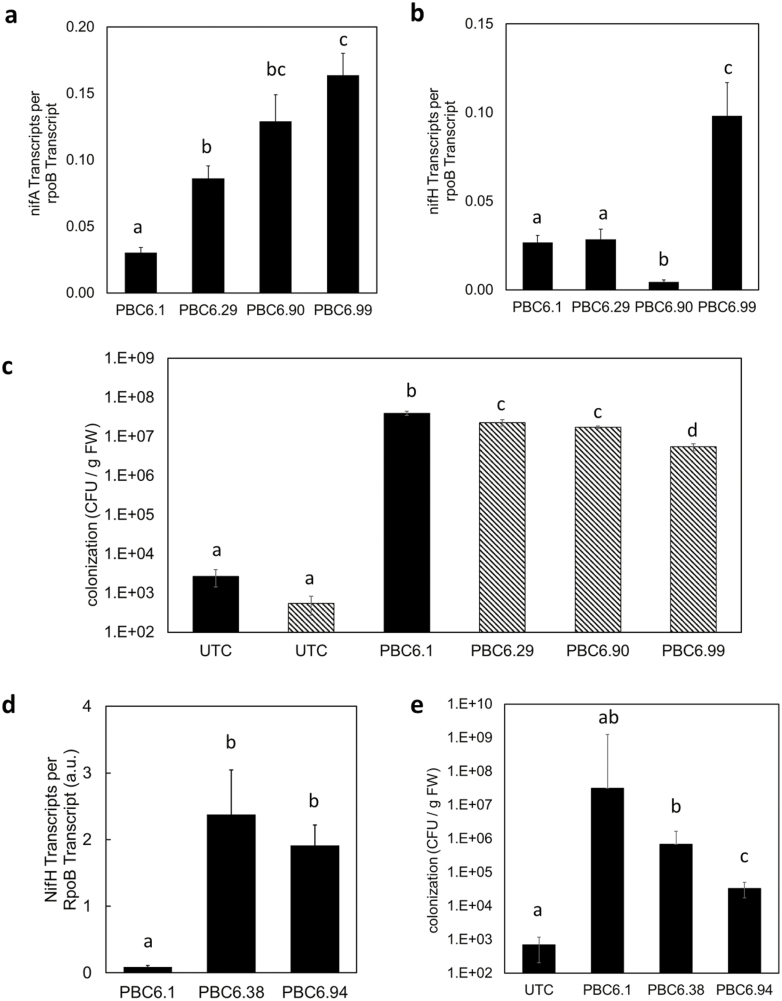
Fig. 5.
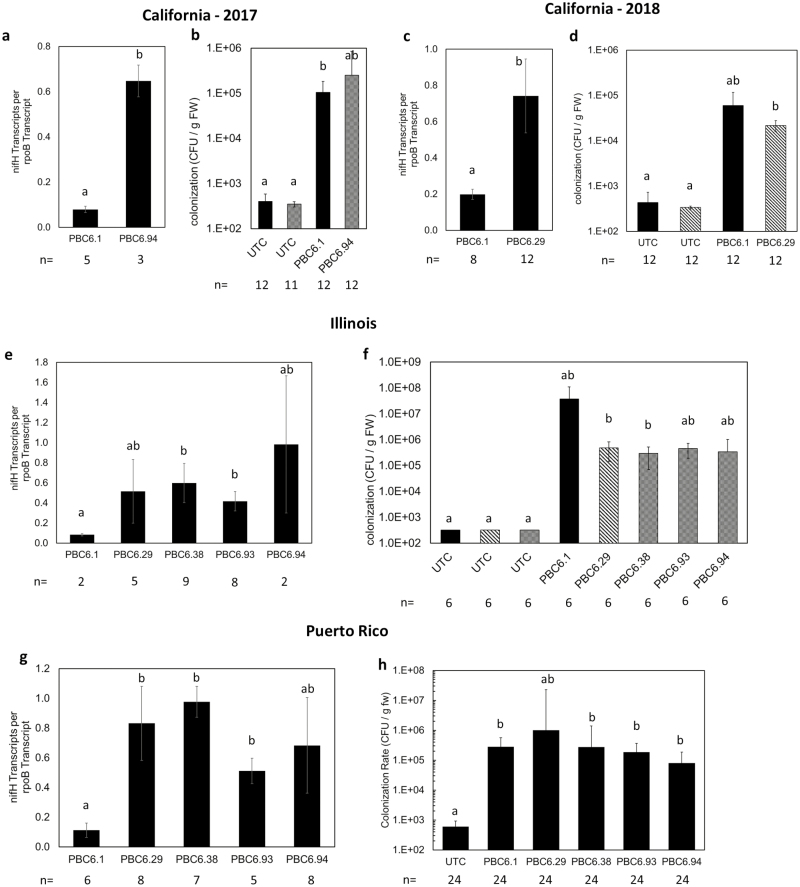
Greenhouse experiments demonstrate rhizosphere nifH transcription in fertilized corn. (A–C) Corn was planted in a controlled-environment growth chamber and inoculated with PBC6.1, PBC6.29, or PBC6.99. Upon the emergence of the second leaf collar, plants were harvested, and nucleic acids were extracted from the rhizosphere to quantify nifA (A) and nifH (B) transcript levels and colonization (C) of each strain. Data are presented as the mean of five root samples pooled from five plants each. (D and E) In a second experiment, 4 weeks after inoculation with PBC6.1 derivative strains, the corn root-associated microbiome was extracted. (D) In planta transcription of nifH was measured. The remodeled strains showed higher nifH transcription than PBC6.1 in the corn root environment. Error bars show the SE of the mean of at least three biological replicates. (E) Colonization is reported as the geometric means of at least eight biological replicates. In (C) and (E), black bars represent samples analyzed using primers targeting the nifH–por2 intergenic region, and hatched bars represent samples analyzed using primers targeting the ∆nifL::Prm5 genotype. Error bars show the SE of the mean. Within each panel, letters indicate groups for which P was <0.05 according to a two-tailed, two-sample unequal variance t-test.
Field trials to measure colonization and nifH transcription
California
Two small-scale field trials were carried out in the late summer season in southern California. For the 2017 field trial, plots were prepared by fertilization to a level of 123 kg N ha–1, and treated and UTC seeds were planted in individual plots. Root samples from 12 replicate plants were collected at 13 d after planting for colonization analysis; at the same time, root samples were collected from 108 replicate plants to measure nifH transcript. Colonization samples were chilled to 4 °C and processed within 3 d of collection as described above. From each root sample, a 0.25 g tissue sample was collected from inner nodal and seminal roots, gDNA was prepared using the standard protocol described above, and the resulting sample was subjected to a colonization qPCR assay as described above. Root samples to be processed for RNA extraction were immediately packed in RNAlater at the field and frozen on arrival at the laboratory. Tissue samples of 1 g each were then homogenized, RNA was prepared using our standard protocol described above, and samples from three replicate plants were pooled to increase signal for downstream analysis, generating a total of 36 transcript samples per strain. The resulting RNA samples were analyzed via Nanostring for nifH and housekeeping genes. The 2018 field trial was carried out as above, except that the root samples were collected 6 weeks after planting, RNA was extracted from root samples immediately after sampling without RNAlater treatment or freezing, and RNA samples were not pooled but analyzed as individual plant samples. These trials were designed as small-scale trials to measure colonization and nifH transcript only; therefore, yield data were not collected.
Puerto Rico
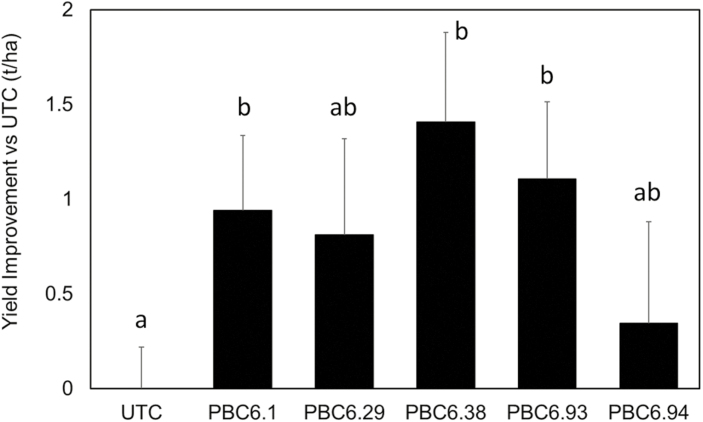
Field trials were carried out during the winter season of Puerto Rico, USA. Treated and UTC seeds were planted in four sets of four replicate plots, each set fertilized with a different level of nitrogen (0, 67.25, 134.5, or 201.75 kg N ha–1 urea application 14 d after planting) for a total of 16 plots per microbial treatment. Root samples from 24 replicate plants were collected at 30 d after planting. Colonization samples were chilled to 4 °C and processed within 3 d of collection as described above. From each root sample, a 0.25 g tissue sample was collected from inner nodal and seminal roots and gDNA was prepared using the standard protocol described above. All root samples were subjected to a colonization qPCR assay, and a subset of these samples were subjected to microbiome profiling as described above. Root samples to be processed for RNA extraction were immediately packed in RNAlater at the field and frozen on arrival at the laboratory. Tissue samples of 1 g each were then homogenized, and RNA was prepared using our standard protocol described above. The resulting RNA samples were analyzed via Nanostring for nifH and housekeeping genes. All plots were measured for yield at harvest 118 d after planting. Though the nitrogen fertilizer level is expected to be correlated with yield, no such correlation was observed (R2=0.00336, P=0.4319). However, yield was significantly correlated with plot row and range (R2=0.147, P=6.4×10–8, ANOVA), suggesting that nitrogen mobilization occurred during the growing season. We therefore averaged colonization, transcript, and yield data across all 16 plots for each treatment.
Illinois
Field trials were carried out during the standard corn-growing season in southern Illinois. Treated seeds and UTC seeds were planted in six replicate unfertilized plots. Root samples from one plant per plot were collected at 21 d after planting for colonization analysis; at the same time, root samples were collected from two replicate plants per plot to measure nifH transcript. Colonization samples were chilled to 4 °C and processed within 3 d of collection as described above. From each root sample, a 0.25 g tissue sample was collected from inner nodal and seminal roots, gDNA was prepared using the standard protocol described above, and the resulting sample was subjected to a colonization qPCR assay as described above. Root samples to be processed for RNA extraction were immediately packed in RNAlater at the field and frozen on arrival at the laboratory. Tissue samples of 1 g each were then homogenized, RNA was prepared using our standard protocol described above, and the resulting samples were analyzed via Nanostring for nifH and housekeeping genes.
Re-isolation of edited strains from field corn root samples
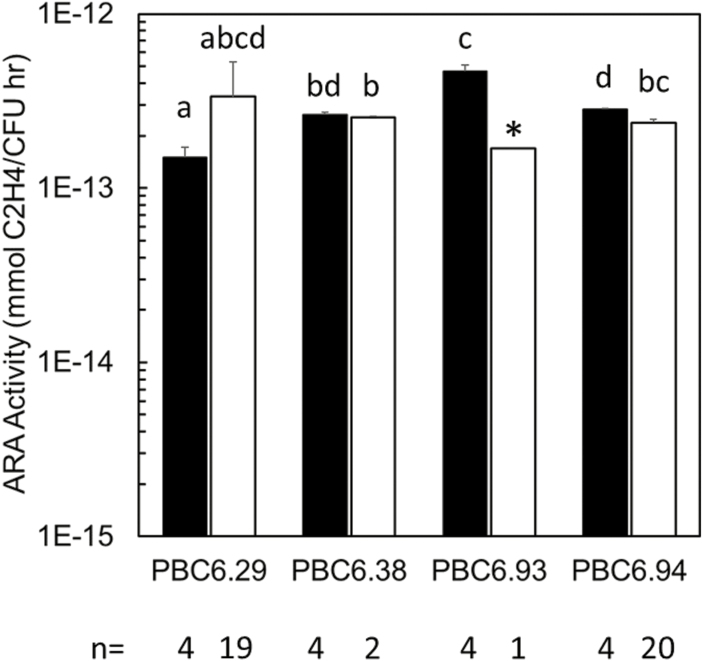
Roots from field samples taken 41 d after plating were homogenized as described above and dilution-plated to single colonies, and colonies were selected for screening based on morphotypic similarity to the parent inoculum. Isolates were screened using PCR primers specific to the mutations, and those with the correct band sizes were further verified by Sanger sequencing of the 16S rRNA region. Between two and 20 clones of each re-isolated strain were then purified and assessed for ARA activity relative to the original inoculant strain.
Results and discussion
We hypothesized that we could improve nitrogen contributions by BNF through identification and isolation of diazotrophs that closely associate with key crops, followed by gene editing to disrupt regulatory networks linking nitrogen sensing, fixation, and assimilation. To this end, corn plants were grown in agricultural soils, roots were collected, and microbial populations were extracted from the rhizosphere and endosphere. Isolates capable of growth on nitrogen-free media were purified, their genomes were sequenced, and the presence of the nif cluster was confirmed. Because the success of a microbial inoculant product depends on the ability of the strain to establish and persist in field conditions, selected strains were inoculated onto corn seeds which were grown in a field trial to identify strains capable of highly colonizing the root system. Genomic DNA from root samples taken 12 weeks after planting was extracted and subjected to a qPCR assay to quantify the colonization of inoculant strains. One of the most robust colonizers in the field study was a bacterium we classified through 16S rRNA and whole-genome sequencing as Kosakonia sacchari (strain PBC6.1), which was detected in 60% of inoculated plants and was not detected in any UTC plants. When detected, PBC6.1 colonized to ~2×105 CFU g–1 of root FW. Kosakonia sacchari is known to be associated with sugarcane (Saccharum officinarum) (Chen et al., 2014; Gu et al., 2014) and sweet potato (Ipomoea batatas) (Shinjo et al., 2016), while other members of the genus Kosakonia (formerly part of Enterobacter) are beneficially associated with cereal crops including wheat (Kämpfer et al., 2005), sugarcane (Taule et al., 2016; Beracochea et al., 2019; Taule et al., 2019), rice (Oryza sativa) (Li et al., 2016; Li et al., 2017), and corn (Kämpfer et al., 2016). Using fluorescence microscopy of roots inoculated with a strain of PBC6.1 expressing RFP, we observed colonization of the corn root surface along the perimeters of cells (Supplementary Fig. S1).
Genome sequencing revealed that PBC6.1 has a genome of at least 5.4 Mbp, and a nif gene cluster and a nitrogen metabolic regulatory network similar to those described in the literature (Fig. 1). The functions and regulation of the nif genes have been well studied, and nitrogen metabolism pathways in PBC6.1 are similar to those of the model organism for non-symbiotic nitrogen fixation, Klebsiella oxytoca M5A1 (Batista and Dixon, 2019). The nifLA operon directly regulates the rest of the nif cluster through transcriptional activation by NifA and nitrogen- and oxygen-dependent repression of NifA by NifL (Dixon and Kahn, 2004). Glutamine synthetase (GS) is responsible for rapid assimilation of newly fixed nitrogen in nitrogen-limiting conditions (van Heeswijk et al., 2013). GS is reversibly regulated by the two-domain adenylyltransferase (ATase) enzyme GlnE through the adenylylation and deadenylylation of GS to attenuate and restore activity, respectively (Jiang and Ninfa, 2009). The nifLA operon and GS (encoded by the glnA gene) are regulated at the transcriptional level by the PII protein regulatory cascade, which originates with the glutamine-sensing protein GlnD (van Heeswijk et al., 2013; Batista and Dixon, 2019). In nitrogen starvation conditions, GlnD covalently modifies PII proteins, which leads to the up-regulation of nitrogen fixation and assimilation pathways. In the presence of exogenous nitrogen, GlnD removes the covalent modification from the PII proteins, leading to the repression of nitrogen fixation and assimilation genes (Batista and Dixon, 2019).
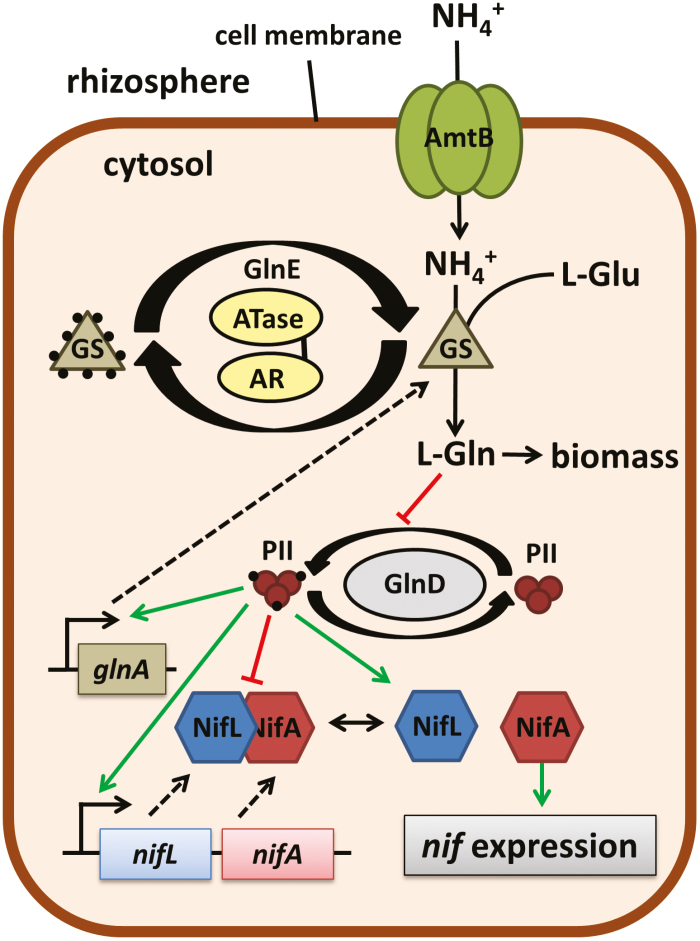
Fig. 1.
Kosakonia sacchari PBC6.1 contains well-characterized nitrogen regulatory pathways. The regulatory network controlling nitrogen fixation and assimilation in PBC6.1 is shown, including the key nodes via AmtB, glutamine synthetase (GS; encoded by the glnA gene), GlnE depicted as the two-domain ATase-AR enzyme, GlnD, and PII proteins depicted as small trimeric proteins, NifL and NifA.
To assess the sensitivity of PBC6.1 to exogenous nitrogen and to predict activity in a fertilized field, nitrogenase activity in pure culture was measured with the classical ARA (Temme et al., 2012) in the presence and absence of fixed nitrogen. PBC6.1 exhibited a high level of nitrogenase activity in nitrogen-free media, and exogenous fixed nitrogen in the form of 5 mM glutamine or ammonium repressed acetylene reduction (strain PBC6.1, Fig. 3). To evaluate how well PBC6.1 might transfer nitrogen to the roots of a host plant, we measured the release of newly fixed nitrogen into the media in an ammonium excretion assay. Strains were cultured in anaerobic conditions in nitrogen-free media, and the concentration of ammonium in the media was quantified after 3 d. Minimal ammonium from PBC6.1 could be detected (Fig. 4), suggesting that transfer of fixed nitrogen from the strain to a host crop might be limited by both nif repression by fertilizer and the microbe’s rapid assimilation of newly fixed nitrogen.
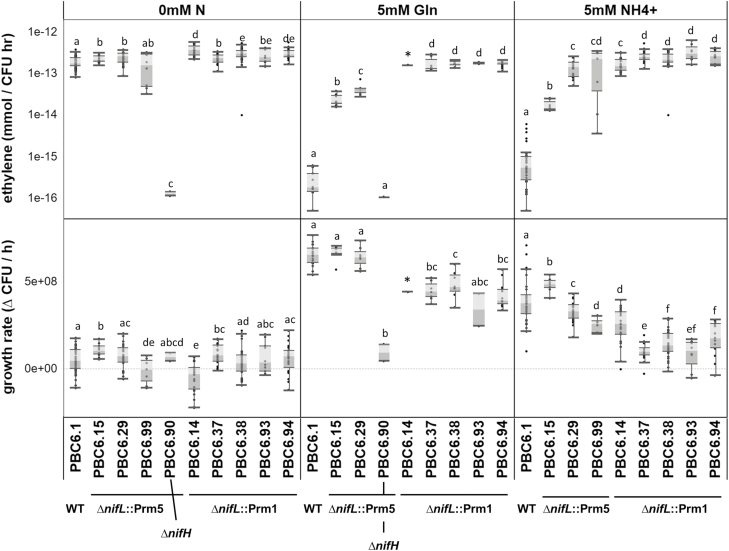
Fig. 3.
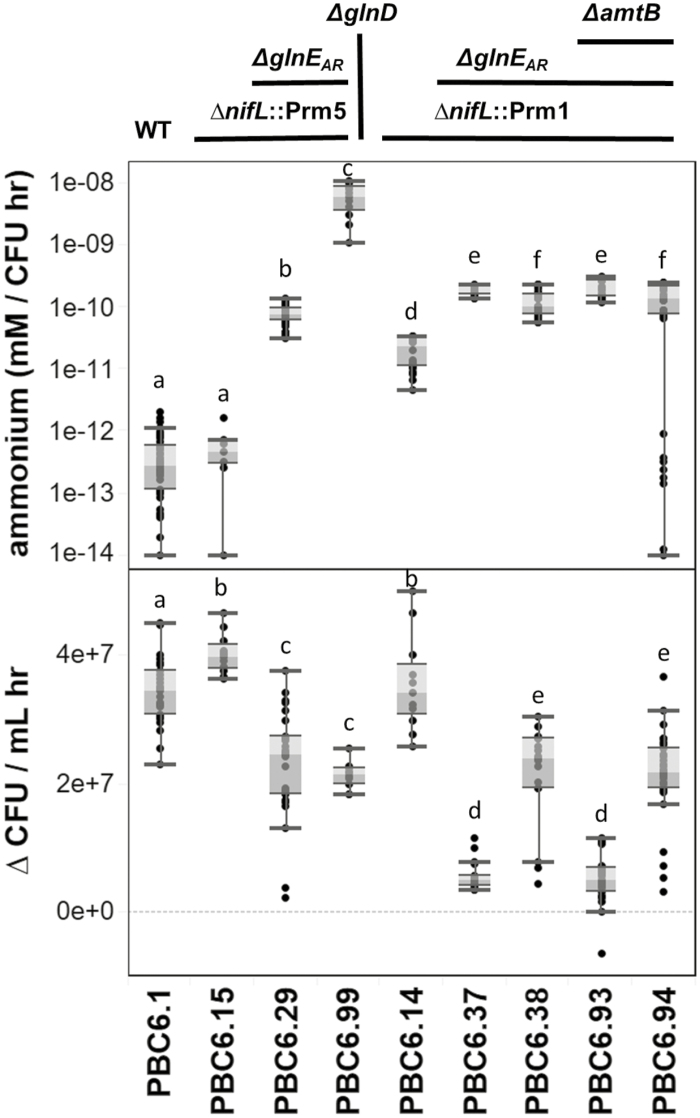
Edited strains of PBC6.1 fix nitrogen independently of nitrogen status. Nitrogenase activity under varying concentrations and forms of exogenous nitrogen was measured with the acetylene reduction assay. The wild-type strain exhibits repression of nitrogenase activity as glutamine concentrations increase, while remodeled strains show nitrogenase activity in the presence of 5 mM glutamine (Gln) or NH4+. Each dot represents a single biological replicate; shaded boxes and whiskers represent the quartiles and range of the data, respectively; dots outside of whiskers are statistical outliers. Within each panel, letters indicate groups for which P was <0.05 as determined by a two-tailed, two-sample unequal variance t-test. An asterisk indicates where only one sample was analyzed. PBC6.90 was not tested in 5 mM NH4+; PBC6.99 was not tested in 5 mM glutamine.
Fig. 4.
Edited strains of PBC6.1 excrete fixed nitrogen into their environment. PBC6.1 and derived strains were grown in minimal nitrogen-free media in anaerobic conditions, and the resulting supernatant was cleared of cells and assayed for the presence of ammonium. While the WT strain cultured had little to no ammonium present, several gene-edited strains exhibited excretion of ammonium into the culture medium. Each dot represents a single biological replicate; shaded boxes and whiskers represent the quartiles and range of the data, respectively; dots outside of whiskers are statistical outliers. Data from multiple replicated experiments were combined. Within each panel, letters indicate groups for which P was <0.05 according to a two-tailed, two-sample unequal variance t-test.
Because of its robust colonization of corn roots, its genome containing the nif cluster, and its demonstrated nitrogenase activity in ARA, PBC6.1 was selected for further study and optimization for nitrogen fixation. We hypothesized that we could remodel the regulatory networks of PBC6.1 through intragenomic gene editing to enable nitrogen fixation and excretion of ammonium in high exogenous concentrations of fixed nitrogen. We targeted several critical nodes within the nitrogen fixation and assimilation gene regulatory network to increase nitrogen fixation and ammonium excretion in PBC6.1 (Fig. 2). First, disruption of nifL can abolish inhibition of NifA and improve nif expression in the presence of both oxygen and exogenous fixed nitrogen (Brewin et al., 1999; Ortiz-Marquez et al., 2014). Furthermore, expressing nifA under the control of a nitrogen-independent promoter could decouple nitrogenase biosynthesis from the PII protein regulatory cascade (Fig. 2a) (Bageshwar et al., 2017). Secondly, truncation of the GlnE protein to delete its adenylyl-removing (AR) domain would lead to constitutively adenylylated GS, limiting ammonium assimilation by the microbe and leading to ammonium build up and release (Fig. 2b) (Clancy et al., 2007). Thirdly, abolishing expression of AmtB, the transporter responsible for uptake of ammonium, could lead to greater extracellular ammonium by preventing reuptake of excreted ammonium (Fig. 2c) (Zhang et al., 2012; Barney et al., 2015). Finally, deletion of the GlnD protein would lead to a constant nitrogen sufficiency signal by eliminating the ability of the cells to covalently modify PII proteins (Fig. 2d). In combination with a nifA gene whose transcription is decoupled from the PII regulatory cascade, this genotype should lead to a cell with constitutively up-regulated nitrogen fixation and down-regulated nitrogen assimilation, causing a net excess of ammonium and thereby ammonium excretion. To generate rationally designed microbial phenotypes without the use of transgenes, we employed two approaches: creating markerless and scarless deletions of genomic sequences encoding protein domains or whole genes, and rewiring regulatory networks by intragenomic promoter (Prm) rearrangement (Fig. 2). Through an iterative gene editing process, we generated several non-transgenic variant strains of PBC6.1 (Table 1), including a strain with the nifH gene deleted to serve as a negative control for nitrogenase expression (Fig. 2e).
Table 1.
List of isolated and remodeled K. sacchari strains used in this work
| Strain | Genotype |
|---|---|
| PBC6.1 | WT |
| PBC6.15 | ΔnifL::Prm5 |
| PBC6.29 | ΔnifL::Prm5 ΔglnEAR1 |
| PBC6.99 | ΔnifL::Prm5 ΔglnEAR1 ΔglnD |
| PBC6.90 | ΔnifL::Prm5 ΔglnEAR1 ΔnifH |
| PBC6.14 | ΔnifL::Prm1 |
| PBC6.37 | ΔnifL::Prm1 ΔglnE AR2 |
| PBC6.38 | ΔnifL::Prm1 ΔglnEAR1 |
| PBC6.93 | ΔnifL::Prm1 ΔglnE AR2 ΔamtB |
| PBC6.94 | ΔnifL::Prm1 ΔglnE AR1 ΔamtB |
Prm, promoter sequence derived from the PBC6.1 genome; ΔglnEAR1 and ΔglnEAR2, different truncated versions of the glnE gene removing the adenylyl-removing domain sequence.
Several in vitro assays were performed to characterize the edited strains. While PBC6.1 exhibited repression of nitrogenase activity at high exogenous glutamine concentrations, the edited strains reduced acetylene robustly at 5 mM glutamine and ammonium (Fig. 3). Strains containing the ∆nifL::Prm5 mutation show partial restoration of nitrogenase activity in the presence of 5 mM N, while strains containing the ∆nifL::Prm1 mutation show full restoration of nitrogenase activity in the presence of 5 mM N, reflecting the relative strength of the promoters inserted upstream of nifA. Additionally, some strains with the ∆nifL::Prm mutations show a decrease in growth rate in the presence of nitrogen (Fig. 3), suggesting that high levels of nitrogen fixation in nitrogen-replete conditions may carry a fitness cost. PBC6.90, the ∆nifH strain, exhibits no nitrogen fixation in any condition tested, as expected. We next measured whether edited strains excreted ammonium into the culture medium under nitrogen-fixing conditions (Fig. 4). While the ∆nifL::Prm5 mutation alone (PBC6.15) was insufficient to confer an ammonium excretion phenotype, the ∆nifL::Prm1 mutation alone (PBC6.14) led to significant excretion of ammonium. This suggests that an increase in nitrogenase expression and activity may be sufficient to generate an excess of ammonium ions within the cell which are not assimilated by GS, leading to passive excretion across the cell membrane, as has previously been observed in Azotobacter vinelandii (Bali et al., 1992; Brewin et al., 1999; Ortiz-Marquez et al., 2014; Barney et al., 2015). The ΔglnEAR mutations led to an increase in ammonium excretion when stacked with the ∆nifL::Prm mutations, supporting our hypothesis that down-regulation of GS activity would lead to ammonium excretion. Furthermore, the ∆glnD mutation led to an additional increase in ammonium excretion, probably by causing a decrease in glnA expression. Notably, the ammonium excretion phenotype conferred by the ΔglnEAR and ∆glnD mutations came with a corresponding decrease in growth rates, similar to what was observed in the acetylene reduction assay (Fig. 4). The ∆amtB mutations had no apparent effect on ammonium excretion or growth rate when stacked with the ∆nifL::Prm and ΔglnEAR mutations. These results suggest that the edited strains may be able to fix nitrogen and transfer it to the crop in fertilized field conditions.
To determine whether the edited microbes were able to express nitrogenase in the rhizosphere of fertilized plants, we inoculated corn plants in greenhouse assays with PBC6.1 and a subset of edited strains to measure colonization of the corn rhizosphere and nifH transcription therein. In a first experiment, corn plants fertilized with 2 mM nitrate were harvested after the emergence of the second leaf collar, and the root metagenome and metatrascriptome were isolated. A probe-based method was used to measure the presence of PBC6.1 nifA and nifH transcripts relative to the housekeeping gene rpoB. The transcription of nifA was significantly up-regulated compared with the WT in all strains tested, all of which contained the ∆nifL::Prm5 mutation. Interestingly, PBC6.99 showed a significantly increased level of nifA (Fig. 5A) and nifH (Fig. 5B) transcript compared with its direct parent PBC6.29, suggesting that the ∆glnD mutation has an impact on nifA transcription. This is unexpected in the context of the ∆nifL::Prm5 mutation, which was hypothesized to confer a consistent level of nifA transcript in a variety of backgrounds. This suggests that the deletion of glnD has activated an unknown pathway which regulates nifA transcription, at least in the context of the inserted promoter. PBC6.29 showed no significant increase in nifH transcript in the corn rhizosphere, despite the significant increase in nifA transcription—this may be due to a basal level of nifH transcription in the WT strain in the relatively low nitrogen conditions of the assay. As expected, nifH transcript was not detected in the nifH knockout strain (PBC6.90). Overall colonization was significantly higher than observed in the initial field study, probably reflecting both the earlier growth stage of the plant and the more controlled conditions of the greenhouse assay (Fig. 5C). All remodeled strains exhibited a decrease in colonization compared with the WT, with the greatest decrease observed in PBC6.99, suggesting that the high expression of nifA and nifH in that strain led to a fitness cost. However, the decrease in colonization of the nifH knockout strain (PBC6.90) suggests that this fitness cost is not entirely due to nitrogenase expression and may be a function of the nitrogen assimilation edits present in the strain.
In another study, we measured microbial colonization of root tissues and expression of nifH in strains containing the ∆nifL::Prm1 (PBC6.38 and PBC6.94) mutation 4 weeks after inoculation of the corn rhizosphere. Measurement of microbial transcription within root tissues showed that nifH expression was significantly increased in both edited strains compared with the WT (Fig. 5D), suggesting that the ∆nifL::Prm1 mutation (in the absence of the ∆glnD mutation) confers a stronger nitrogenase up-regulation phenotype than the ∆nifL::Prm5 mutation in the rhizosphere. Both edited strains showed a decrease in colonization compared with the WT, with PBC6.94 exhibiting significantly reduced abundance (Fig. 5E), again suggesting a fitness cost to high levels of nitrogenase expression and/or disruption of nitrogen assimilation.
In the past, translating microbial inoculants from the greenhouse to field conditions has posed significant challenges and, in some cases, has thwarted successful implementation of beneficial microbes in large-scale agriculture. Therefore, to measure the presence and nitrogenase expression of PBC6.1 and its derivatives in a variety of field conditions, we conducted replicated plot field trials using seeds coated with selected edited strains at three locations with a wide range of climates, soil types, and soil nitrogen levels. The Illinois trials represent typical growing conditions in a major corn-producing region of the USA, while California and Puerto Rico trials represent arid and tropical corn-producing regions, respectively. Because these trials were designed to study the persistence and nitrogenase expression of the bacteria in the rhizosphere, yield data were not collected, except for the Puerto Rico trial (Fig. 7). Root samples were collected between 2 and 5 weeks after planting for nucleic acid extraction to verify the presence of the strain and expression of the nifH gene (Fig. 6). PBC6.1 and edited strains colonize consistently across most of the locations tested, at levels of ~105 CFU g–1 of root FW (Fig. 6b, d, h). One exception was the Illinois location, at which the WT PBC6.1 strain colonized ~108 CFU g–1 of root FW, though this was not statistically significantly higher than the mutant strains (Fig. 6f). While this shows robust colonization by the strain in the field, these levels are significantly lower than values observed in the greenhouse (Fig. 5), possibly due to the combination of harsher abiotic conditions or the presence of a robust competing microbiome in the field. We observed consistently low normalized transcript levels of nifH in the WT parent strain PBC6.1 in all conditions tested, which ranged from unfertilized with residual nitrates measured at 16 kg nitrate N ha–1 to a field fertilized to a level of 123 kg N ha–1. All edited strains show 5- to 10-fold increased transcription of nifH compared with the WT, though transcript results varied between strains and locations. PBC6.38 and PBC6.93 showed significantly increased nifH transcript at both locations where they were tested (Fig. 6e, g). While PBC6.29 did not show a significant increase in nifH transcript over PBC6.1 in the Illinois trial (Fig. 6e), it did show a significantly increased nifH transcript level in the 2018 California trial (Fig. 6d) and in the Puerto Rico trial (Fig. 6g), which is in contrast to the greenhouse results (Fig. 6b). The results correlate with differences in soil nitrogen levels—the greenhouse study was fertilized at low levels, and the Illinois trial was unfertilized, while the California and Puerto Rico trials were fertilized at levels consistent with farmer practice in the region. Strains with the ∆nifL::Prm1 mutation showed significant increases in nifH transcript compared with PBC6.1 at all locations, except PBC6.94, which showed high variability in the Illinois and Puerto Rico trials (Fig. 6a, e, g). In the Puerto Rico trial, yield data were collected. Because yield correlated with plot location rather than fertilizer treatment, we assumed significant nitrogen mobilization across the field and therefore averaged the yield data across all plots and nitrogen treatments (see the Materials and methods). The UTC plots produced a yield of 6.3 t ha–1, and PBC6.1 and some of the edited strains showed significant increases in yield compared with the UTC (Fig. 7). While these data are highly variable and only represent a single field season, they suggest that these strains may have potential to improve the growth of corn. Additionally, clones of edited strains were re-isolated from the roots of several plants in the Puerto Rico trial and found to be genetically consistent and to exhibit similar in vitro nitrogenase activity levels to the original inoculant strains (Fig. 8). These data confirm a true association between PBC6.1 derivatives and corn plants and that the strains retained their nitrogen-fixing capability in the field. Taken together, these data show that PBC6.1 and its derivatives colonize corn plants and survive during the growing season in diverse environments, and that nitrogenase expression is de-repressed in edited strains in a broad range of soil nitrogen levels.
Fig. 7.
Corn inoculated with PBC6.1 and its derivatives exhibited an increase in grain yield above UTC in the Puerto Rico field trial. Error bars represent the SE of the mean across 16 plots. Letters indicate groups for which P was <0.05 according to a two-tailed, two-sample unequal variance t-test.
Fig. 6.
Remodeled strains colonize corn roots and express nitrogenase in diverse locations, soil types, and nitrogen levels. Three replicated plot field trials were conducted by planting corn seed coated with PBC6.1 and derived strains. Root samples were collected, the root-associated microbiome was extracted, and nifH transcription (a, c, e, g) and colonization (b, d, f, h) were quantified. The California 2017 trial (a, b) was fertilized to a level of 123 kg N ha–1 prior to planting, and root samples were collected 13 d after planting; the California 2018 trial (c, d) was planted at the same location and fertilizer level as the previous year, and root samples were collected 6 weeks after planting. In Illinois (e, f), an unfertilized trial was planted in a field found to have 16 kg ha–1 residual nitrates, and root samples were collected 21 d after planting. In Puerto Rico (g, h), a fertilized trial was planted in a field with pre-plant nitrate N levels measured at 89 kg ha–1, and root samples were collected 5 weeks after planting; because of heavy rains causing apparent leaching and pooling of fertilizer in fields, the samples were averaged across all fertilizer treatments. At all locations, remodeled strains show increased normalized nifH transcript levels and similar colonization when compared with PBC6.1. In (b, d, f, and h), black bars represent samples analyzed using primers targeting the nifH–por2 intergenic region; shaded and hatched bars represent samples analyzed using primers targeting the ∆nifL::Prm1 and ∆nifL::Prm5 genotypes, respectively. Colonization values (b, d, and f) are reported as geometric means, and error bars on all panels represent the SE of the mean. Within each panel, n indicates the number of replicate plants; letters indicate groups for which P was <0.05 according to a two-tailed, two-sample unequal variance t-test.
Fig. 8.
Nitrogenase activity is observed in clones re-isolated from field-grown root samples. White bars represent the input strains used to inoculate the field trial; black bars represent clones reisolated from root tissues. Re-isolated clones show activity similar to the original inoculant strains. Error bars show the SE of the mean of at least two clones, except for PBC6.93, for which only one clone was re-isolated and analyzed. Letters indicate groups for which P was <0.05 as determined by a two-tailed, two-sample unequal variance t-test. Asterisk indicates the data where only one sample was analyzed.
Conclusions
Long-term ecological studies in legume–rhizobia interactions have shown that elevated nitrogen inputs can lead to the evolution of less cooperative nitrogen-fixing mutualists (Weese et al., 2015). In this study, we have shown that a non-transgenic microbe can express nitrogenase in close association with corn roots in the presence of nitrogen fertilizer and can thrive in typical field conditions. Through gene editing, we generated strains in which nitrogenase biosynthesis is decoupled from the regulatory networks that sense and respond to cellular nitrogen status. These strains fix and excrete significant quantities of nitrogen into their environment at various levels of exogenous nitrogen (Figs 3, 4). By utilizing plant-by-plant measurements of colonization and transcription, we were able to study plant association and nitrogenase expression in these microbes on a molecular level (Figs 5, 6). We observed robust colonization and expression of nitrogenase in diverse field environments and soil types, and that functional clones can be re-isolated from field-grown roots 12 weeks after planting (Fig. 8). The persistence of gene-edited, ammonium-excreting strains in a field trial is a promising finding because translation to field conditions has been a challenge for the development of microbial products for production-scale agriculture, especially for ammonium-excreting diazotrophs in cereal cropping systems. Designing bacteria that fix nitrogen in the presence of exogenously fertilizer is a first step toward developing strains that can replace synthetic fertilizers in cereal crop production.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Fluorescence micrograph of an RFP-expressing PBC6.1 strain colonizing the surface of corn roots.
Table S1. Primers used in this study for genotypic detection.
Table S2. Primers used in this study for colonization assays.
Acknowledgements
We acknowledge the efforts of Pivot Bio managerial and support staff who contributed to the completion of lab work, as well as Pivot Bio scientists who provided feedback and scientific input. This work was funded by Pivot Bio, Inc.
Conflict of interest
The authors declare a competing financial interest: Pivot Bio has filed patent applications and is developing products based on the microbial strains in this study.
References
- Ambrosio R, Ortiz-Marquez JCF, Curatti L. 2017. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metabolic Engineering 40, 59–68. [DOI] [PubMed] [Google Scholar]
- Bageshwar UK, Srivastava M, Pardha-Saradhi P, et al. 2017. An environment friendly engineered Azotobacter can replace substantial amount of urea fertilizer and yet sustain same wheat yield. Applied and Environmental Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldani JI, Reis VM, Videira SS, Boddey LH, Baldani VLD. 2014. The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant and Soil 384, 413–431. [Google Scholar]
- Bali A, Blanco G, Hill S, Kennedy C. 1992. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Applied and Environmental Microbiology 58, 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barney BM, Eberhart LJ, Ohlert JM, Knutson CM, Plunkett MH. 2015. Gene deletions resulting in increased nitrogen release by Azotobacter vinelandii: application of a novel nitrogen biosensor. Applied and Environmental Microbiology 81, 4316–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista MB, Dixon R. 2019. Manipulating nitrogen regulation in diazotrophic bacteria for agronomic benefit. Biochemical Society Transactions 47, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beracochea M, Taule C, Battistoni F. 2019. Draft genome sequence of Kosakonia radicincitans UYSO10, an endophytic plant growth-promoting bacterium of sugarcane (Saccharum officinarum). Microbiology Resource Announcements 8, doi: 10.1128/MRA.01000-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch SE, Ryu MH, Ozaydin B, Broglie R. 2019a Harnessing atmospheric nitrogen for cereal crop production. Current Opinion in Biotechnology 62, 181–188. [DOI] [PubMed] [Google Scholar]
- Bloch SE, Temme K, Tamsir A, Higgins D, Davis-Richardson A, Clark R, Gottlieb S. 2019b Guided microbial remodeling, a platform for the rational improvement of microbial species for agricultureInternational patent publication number WO/2020/006246, published 1 February 2020, assigned to PIVOT BIO, INC. https://patentscope.wipo.int/beta/en/detail.jsf?docId=WO2020006246. [Google Scholar]
- Brewin B, Woodley P, Drummond M. 1999. The basis of ammonium release in nifL mutants of Azotobacter vinelandii. Journal of Bacteriology 181, 7356–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale M. 2014. Scanning a microhabitat: plant–microbe interactions revealed by confocal laser microscopy. Frontiers in Microbiology 5, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassman KG, Liska AJ. 2007. Food and fuel for all: realistic or foolish? Biofuels, Bioproducts and Biorefining 1, 18–23. [Google Scholar]
- Chapman HD, Liebig GF, Rayner DS. 1949. A lysimeter investigation of nitrogen gains and losses under various systems of covercropping and fertilization, and a discussion of error sources. Hilgaridia 19, 57–128. [Google Scholar]
- Chen M, Zhu B, Lin L, Yang L, Li Y, An Q. 2014. Complete genome sequence of Kosakonia sacchari type strain SP1(T.). Standards in Genomic Sciences 9, 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy P, Xu Y, van Heeswijk WC, Vasudevan SG, Ollis DL. 2007. The domains carrying the opposing activities in adenylyltransferase are separated by a central regulatory domain. FEBS Journal 274, 2865–2877. [DOI] [PubMed] [Google Scholar]
- Colnaghi R, Green A, He L, Rudnick P, Kennedy C. 1997. Strategies for increased ammonium production in free-living or plant associated nitrogen fixing bacteria. Plant and Soil 194, 145–154. [Google Scholar]
- Compant S, Samad A, Faist H, Sessitsch A. 2019. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. Journal of Advanced Research 19, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn FJ. 2015. Biological nitrogen fixation. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nature Reviews. Microbiology 2, 621–631. [DOI] [PubMed] [Google Scholar]
- Erisman JW, Sutton MA, Galloway J, Klimont Z,Winiwarter W. 2008. How a century of ammonia synthesis changed the world. Nature Geoscience 1, 636–639. [Google Scholar]
- Fox AR, Soto G, Valverde C, et al. 2016. Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen-fixing bacterium Pseudomonas protegens Pf-5 X940. Environmental Microbiology 18, 3522–3534. [DOI] [PubMed] [Google Scholar]
- Gaby JC, Buckley DH. 2012. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7, e42149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendining MJ, Dailey AG, Williams AG, van Evert FK, Goulding KWT, Whitmore AP. 2009. Is it possible to increase the sustainability of arable and ruminant agriculture by reducing inputs? Agricultural Systems 99, 117–125. [Google Scholar]
- Gu CT, Li CY, Yang LJ, Huo GC. 2014. Enterobacter xiangfangensis sp. nov., isolated from Chinese traditional sourdough, and reclassification of Enterobacter sacchari Zhu et al. 2013 as Kosakonia sacchari comb. nov. International Journal of Systematic and Evolutionary Microbiology 64, 2650–2656. [DOI] [PubMed] [Google Scholar]
- Haichar FEZ, Heulin T, Guyonnet JP, Achouak W. 2016. Stable isotope probing of carbon flow in the plant holobiont. Current Opinion in Biotechnology 41, 9–13. [DOI] [PubMed] [Google Scholar]
- Jiang P, Ninfa AJ. 2009. Reconstitution of Escherichia coli glutamine synthetase adenylyltransferase from N-terminal and C-terminal fragments of the enzyme. Biochemistry 48, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky LM, Trexler RV, Malik RJ, Hockett KL, Bell TH. 2019. The inherent conflicts in developing soil microbial inoculants. Trends in Biotechnology 37, 140–151. [DOI] [PubMed] [Google Scholar]
- Kämpfer P, McInroy JA, Doijad S, Chakraborty T, Glaeser SP. 2016. Kosakonia pseudosacchari sp. nov., an endophyte of Zea mays. Systematic and Applied Microbiology 39, 1–7. [DOI] [PubMed] [Google Scholar]
- Kämpfer P, Ruppel S, Remus R. 2005. Enterobacter radicincitans sp. nov., a plant growth promoting species of the family Enterobacteriaceae. Systematic and Applied Microbiology 28, 213–221. [DOI] [PubMed] [Google Scholar]
- Kanter DR. 2018. Nitrogen pollution: a key building block for addressing climate change. Climatic Change 147, 11–21. [Google Scholar]
- Kaul S, Sharma T, K Dhar M. 2016. ‘Omics’ tools for better understanding the plant–endophyte interactions. Frontiers in Plant Science 7, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaletta L, Billen G, Grizzett B, Anglade J, Garnier J. 2014. 50 year trends in nitrogen use efficiency of world cropping systems: the relationship between yield and nitrogen input to cropland. Environmental Research Letters 9, 105011. [Google Scholar]
- Li XX, Liu Q, Liu XM, Shi HW, Chen SF. 2016. Using synthetic biology to increase nitrogenase activity. Microbial Cell Factories 15, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li S, Chen M, Peng G, Tan Z, An Q. 2017. Complete genome sequence of Kosakonia oryzae type strain Ola 51T. Standards in Genomic Sciences 12, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Crook MB, Garcia K, et al. 2016. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Applied and Environmental Microbiology 82, 3698–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormeno-Orrillo E, Hungria M, Martínez-Romero E. 2013. Dinitrogen-fixing prokaryotes. In: Rosenberg E, de Long EF, Lory S, Stackebrandt E., Thompson F., eds. The prokaryote. Berlin Heidelberg: Springer-Verlag, 427–451. [Google Scholar]
- Ortiz-Marquez JC, Do Nascimento M, Curatti L. 2014. Metabolic engineering of ammonium release for nitrogen-fixing multispecies microbial cell-factories. Metabolic Engineering 23, 154–164. [DOI] [PubMed] [Google Scholar]
- Pankievicz VC, do Amaral FP, Santos KF, et al. 2015. Robust biological nitrogen fixation in a model grass–bacterial association. The Plant Journal 81, 907–919. [DOI] [PubMed] [Google Scholar]
- Pankievicz VCS, Irving TB, Maia LGS, Ané JM. 2019. Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biology 17, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL. 2009. The role of fertilizer in growing the world’s food. Better Crops 93, 12–15. [Google Scholar]
- Rockström J, Steffen W, Noone K, et al. 2009. A safe operating space for humanity. Nature 461, 472–475. [DOI] [PubMed] [Google Scholar]
- Shinjo R, Uesaka K, Ihara K, Loshakova K, Mizuno Y, Yano K, Tanaka A. 2016. Complete genome sequence of Kosakonia sacchari strain BO-1, an endophytic diazotroph isolated from a sweet potato. Genome Announcements 4, doi: 10.1128/genomeA.00868-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Bhatia S, Zhao D, et al. 2014. Functional optimization of gene clusters by combinatorial design and assembly. Nature biotechnology 32, 1241–1249. [DOI] [PubMed] [Google Scholar]
- Smercina DN, Evans SE, Friesen ML, Tiemann LK. 2019. To fix or not to fix: controls on free-living nitrogen fixation in the rhizosphere. Applied and Environmental Microbiology 85, doi: 10.1128/AEM.02546-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart WD. 1969. Biological and ecological aspects of nitrogen fixation by free-living micro-organisms. Proceedings of the Royal Society B: Biological sciences 172, 367–388. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Bleeker A, Howard CM, et al. 2013. Our nutrient world: the challenge to produce more food and energy with less pollution. Edinburgh, UK: Centre for Ecology and Hydrology (CEH), on behalf of the Global Partnership on Nutrient Management (GPNM) and the International Nitrogen Initiative (INI). [Google Scholar]
- Sutton MA, Howard CA, Erisman JW, Billen G, Bleeker A, Grennfelt P, van Grinsven P and Grizzetti B, eds. 2011. The European Nitrogen Assessment. Cambridge: Cambridge University Press. [Google Scholar]
- Taule C, Castillo A, Villar S, Olivares F, Battistoni F. 2016. Endophytic colonization of sugarcane (Saccharum officinarum) by the novel diazotrophs Shinella sp. UYSO24 and Enterobacter sp. UYSO10. Plant and Soil 403, 403–418. [Google Scholar]
- Taule C, Luizzi H, Beracochea M, Mareque C, Platero R, Battistoni F. 2019. The Mo- and Fe-nitrogenases of the endophyte Kosakonia sp. UYSO10 are necessary for growth promotion of sugarcane. Annals of Microbiology 69, 741–750. [Google Scholar]
- Temme K, Zhao D, Voigt CA. 2012. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proceedings of the National Academy of Sciences, USA 109, 7085–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeswijk WC, Westerhoff HV, Boogerd FC. 2013. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiology and Molecular Biology Reviews 77, 628–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang JG, Chen L, Wang JL, Cheng Q, Dixon R, Wang YP. 2013. Using synthetic biology to distinguish and overcome regulatory and functional barriers related to nitrogen fixation. PLoS One 8, e68677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weese DJ, Heath KD, Dentinger BT, Lau JA. 2015. Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution 69, 631–642. [DOI] [PubMed] [Google Scholar]
- Zhang T, Yan Y, He S, et al. 2012. Involvement of the ammonium transporter AmtB in nitrogenase regulation and ammonium excretion in Pseudomonas stutzeri A1501. Research in Microbiology 163, 332–339. [DOI] [PubMed] [Google Scholar]
- Zhang X, Davidson EA, Mauzerall DL, Searchinger TD, Dumas P, Shen Y. 2015. Managing nitrogen for sustainable development. Nature 528, 51–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.