Abstract
APOE4 is a major risk factor for sporadic Alzheimer’s disease; however, it is unclear how it exerts its pathological effects. Others and we have previously shown that autophagy is impaired in APOE4 compared to APOE3 astrocytes, and demonstrated differences in the expression of mitochondrial dynamics proteins in brains of APOE3 and APOE4 transgenic mice. Here, we investigated the effect of APOE4 expression on several aspects of mitochondrial function and network dynamics, including fusion, fission, and mitophagy, specifically in astrocytes. We found that APOE3 and APOE4 astrocytes differ in their mitochondrial dynamics, suggesting that the mitochondria of APOE4 astrocytes exhibit reduced fission and mitophagy. APOE4 astrocytes also show impaired mitochondrial function. Importantly, the autophagy inducer rapamycin enhanced mitophagy and improved mitochondrial functioning in APOE4 astrocytes. Collectively, the results demonstrate that APOE4 expression is associated with altered mitochondrial dynamics, which might lead to impaired mitochondrial function in astrocytes. This, in turn, may contribute to the pathological effects of APOE4 in Alzheimer’s disease.
Subject terms: Biochemistry, Neurological disorders
Introduction
Alzheimer’s disease (AD) is characterized by brain Aβ plaques and neurofibrillary tangles, neuronal loss and cognitive decline. The ε4 allele of APOE gene, a major AD risk factor, is associated with increased Aβ deposition and neuronal degeneration1. Nonetheless, the mechanisms mediating APOE4 pathological effects remain unknown1. Mitochondrial dysfunction plays a fundamental role in AD pathogenesis2 and represents an early event2. Mitochondrial dynamics include fusion, fission, and degradation events, linked to mitochondrial function3,4. Fusion and fission regulate mitochondrial network size/shape depending on cellular demands. Damaged mitochondria are degraded by selective autophagy, termed mitophagy, following fission5. Mitochondrial dynamics were shown to be abnormal in AD brains6.
Several proteins are required for mitochondrial fusion, including the mitofusins (Mfn1/Mfn2)3,4. This process is triggered by increased mitochondrial membrane potential (MMP) and involves the oligomerization of mitofusins on the outer mitochondrial membrane (OMM)4. Mitochondrial fusion is enhanced under stress and high energetic demands, allowing increased bioenergetic efficiency7. During mitochondrial fission, cytosolic Drp1 is recruited to mitochondrial receptors (e.g., Fis1)3,4, allowing mitochondrial partitioning, turnover and reduction of network size under low energetic demand4,7. Mitophagy proceeds mitochondrial fission and is triggered by decreased MMP5. It allows the removal of damaged/excessive mitochondria via the lysosome. Several pathways mediate mitophagy, including the PINK1/parkin pathway5,8. The mitochondrial kinase PINK1 is stabilized on the OMM leading to the recruitment and activation of parkin (E3 ubiquitin ligase), which ubiquitinates several mitochondrial proteins leading to mitophagy5,8. Several studies demonstrated a link between APOE4 and mitochondrial dysfunction, mainly in neurons9–11; yet the effect of APOE4 on mitochondrial dynamics was less studied. Also, although astrocytes play a fundamental role in AD pathology12, little is known about APOE4 effect on their mitochondria.
Previously, others and we have found that APOE4 is associated with impaired autophagy in astrocytes13,14, which is linked to reduced clearance of Aβ plaques and protein aggregates13,14. Also, our recent findings revealed differences in the expression of mitochondrial dynamics proteins in APOE4 transgenic mouse brains15. Here, we further investigated the effect of APOE4 expression on various mechanistic aspects of mitochondrial dynamics, including mitophagy, and mitochondrial function specifically in astrocytes. Our results show that APOE4 expression is associated with altered mitochondrial dynamics, namely, reduced mitochondrial fission and parkin-mediated mitophagy, under basal and mitochondrial stress conditions. Furthermore, mitochondrial function is compromised in the APOE4 astrocytes. Notably, induction of mitophagy by rapamycin restored mitochondrial function. Taken together, the results suggest that impaired mitochondrial dynamics in APOE4 astrocytes is involved in mitochondrial dysfunction and contribute to AD pathology.
Materials and methods
The antibodies, buffers, and reagents used are described in the supplementary methods.
Cell lines and primary astrocytes
The human APOE3 and APOE4 targeted-replacement astrocyte cell lines were previously described16. The study was conducted according to the NIH Guidelines for Use and Care of Laboratory Animals, following the approval by Animal Care Committee of TAU (#L-04-16-002). Mouse primary astrocytes were prepared and grown as previously described13.
Brain sections preparation and IHC
APOE3/APOE4 targeted-replacement mice were perfused with PBS followed by 4% paraformaldehyde. Brains were removed, post-fixed for overnight at 4 °C in 4% paraformaldehyde followed by 20% sucrose in PBS for 48 h at 4 °C. Coronal serial cryostat sections were cut (20 µm) and stained as described previously17. Sections were imaged using fluorescence microscopy (Olympus motorized inverted research microscope Model IX81; ×20 magnifications).
Lysate preparation, immunoblot and immunoprecipitation
Preparation of cell lysates and Western Blot analysis was performed as previously described13. For the immunoprecipitation, agarose beads conjugated to mouse anti-ubiquitin antibodies (Santa Cruz Biotechnology) were used. For no IgG control samples, protein G PLUS-agarose beads (Santa Cruz Biotechnology) were used.
Immunoreactive bands were detected using enhanced chemiluminescence reagent (Immobilon Crescendo substrate, Millipore). Bands were visualized by Amersham Imager 600 within the linear range, employing the Automatic/Semi-autmatic exposure options, which allows optimal exposure times below saturation to enable accurate quantification (according to manufacturer instructions). Target protein band intensities were quantified using the ImageJ software and normalized to loading controls.
Mitochondrial fractionation
Mitochondrial fractionation of the cells was performed using the ProteoExtract® Cytosol/Mitochondria Fractionation Kit (EMD Millipore), according to manufacturer protocol.
Analysis of mitochondrial network morphology
APOE3/APOE4 astrocytes cell line was incubated with 100 nM MitoTracker™ Deep Red FM (MTDR, Invitrogen) for 30 min at 37 °C or infected with Mito-GFP as described below, nuclei were stained with 1 µg/ml Hoechst 33342 and fixed in ice-cold methanol for 15 min (MTDR) or in 4% paraformaldehyde for 30 min at RT (Mito-GFP). Mitochondrial network morphology of individual cells was visualized by Leica TCS SP8 confocal microscope (×63 magnifications) and analyzed using the ImageJ macro tool MiNa18.
Retroviral infection and generation of APOE3/APOE4 cell lines stably expressing Mito-GFP
HEK-293T cells were transfected using the calcium-phosphate method with the following retroviral vectors: pCMV-VSV-G, pCMV-Gag-Pol and pQCXIP containing the EGFP-TA-MAO sequence (Mito-GFP; EGFP-tagged OMM localization signal peptide), which was a gift from Prof. Reuven Stein, Tel Aviv University. Medium containing viral particles was collected and used to infect the APOE3/APOE4 astrocytes cell line. Astrocytes stably expressing Mito-GFP were selected using 2 µg/ml puromycin.
Transient transfection with the LC3-EGFP-mRFP vector
Transient transfection of APOE3/APOE4 astrocytes cell line with the tandem LC3-EGFP-mRFP (Addgene ptfLC3) was performed using Lipofectamine® 2000 reagent (Invitrogen). Nuclei were stained; the cells were fixed and microscopically analyzed as described above.
Electron microscopy
Preparation of cellular TEM samples from APOE3/APOE4 astrocytes cell line is described in the supplementary. The samples were analyzed using G-12 Spirit FEI electron microscope.
MTT, ATP levels, and cell number measurement
MTT and CellTiter-Glo (Promega) assays were used in APOE3/APOE4 cell line to assess mitochondrial metabolism and ATP levels, respectively, and the results were normalized to cell number using the methylene blue assay according to the manufacturer instructions and as previously described19,20.
Measurement of MMP by flow cytometry
The APOE3/APOE4 cell line was incubated with either 100 nM MitoTracker™ Red CMXRox (MTR, Invitrogen) or 50 nM MitoTracker™ Deep Red FM (MTDR, Invitrogen) to assess MMP or mitochondrial mass, respectively, by flow cytometry, as described21–25. The cells were then trypsinized, washed and resuspended in PBS. MTDR/MTR fluorescence was collected using Stratedigm S1000EXi Flow Cytometer (mCherry and APC filters, respectively), and analyzed using Cyflogic software.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from APOE3/APOE4 astrocytes cell line, reverse-transcribed into cDNA and used for RT–PCR as described previously19. The primers used are described in the supplementary.
Medium pH measurement
The medium was collected from APOE3/APOE4 cell line, placed in sealed tubes, and pH was measured using Sartorius PB-11 Basic Benchtop pH Meter. The number of cells in the cultures was simultaneously measured.
Statistical analysis
All experiments were performed at least 3 times. Experimental differences were tested for statistical significance using one-sided Student’s t test. p-value of < 0.05 was considered as significant.
Results
Altered mitochondrial dynamics proteins expression and morphology in APOE4 astrocytes
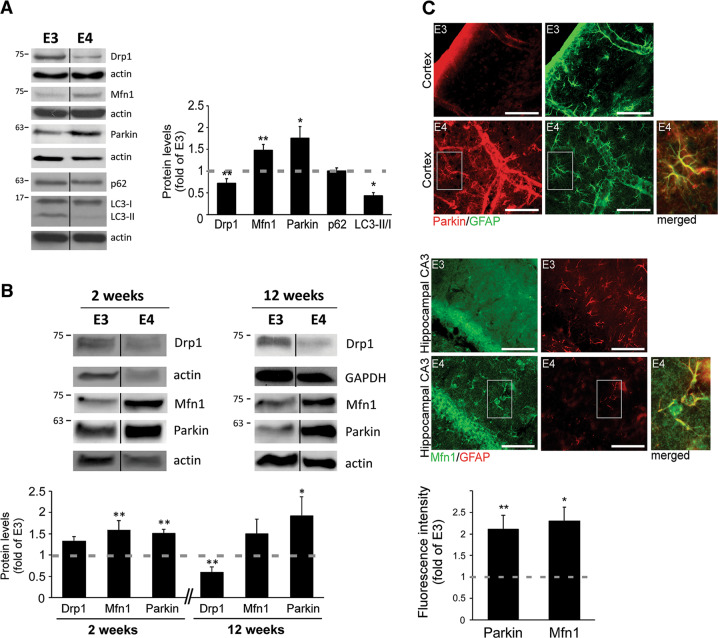
To investigate the link between APOE4 and mitochondrial dynamics, we first examined the expression of fission, fusion and mitophagy proteins in astrocytes expressing human APOE4 or APOE3 (Fig. 1a). As shown, APOE4 astrocytes exhibit lower Drp1 levels (fission), higher Mfn1 levels (fusion) and increased parkin levels (targets mitochondria for mitophagy, followed by proteasomal/lysosomal degradation26–29). In addition, we measured the levels of autophagic markers, SQSTM1/p62 and LC330. APOE4 had no effect on p62 levels; however, it reduced LC3-II/LC3-I ratio, indicating decreased autophagy30. Importantly, similar pattern of these mitochondrial dynamics proteins was detected in primary astrocytes generated from APOE4 mice (differences in Drp1 levels were detected only in 12 weeks cultures) and brain sections from these mice demonstrated increased levels of parkin and Mfn1, specifically in astrocytes (Fig. 1b, c). These results imply that APOE4 astrocytes exhibit altered mitochondrial dynamics.
Fig. 1. Levels of mitochondrial dynamics proteins in APOE4 astrocytes.
a APOE3/APOE4 astrocytes were subjected to Immunoblot, using the indicated antibodies. Left panel, representative results; right panel, densitometric analysis of APOE4 cells as fold of APOE3 (dashed line; n ≥ 3) b Primary astrocytes generated from APOE3/APOE4 targeted-replacement mice were cultured for the indicated time period and subjected to Immunoblot using the indicated antibodies. Upper panel, representative results. Lower panel, quantification of the results as described in (a). c Brain sections of APOE3/APOE4 mice (5 months old; n = 3 mice/group) were co-stained using anti-GFAP (astrocytic marker) with anti-parkin or anti-Mfn1 antibodies. Right panel, co-expression of parkin (red) and GFAP (green) in the cortex area; Mfn1 (green) and GFAP (red) in the hippocampus CA3 area. (scale bars, 50 μm). Magnified inset: merged GFAP with parkin/Mfn1 in APOE4 astrocytes. Left panel, fluorescence intensity was quantified using ImageJ only in GFAP-positive cells, (fold of APOE3, dashed line). a–c Means ± SE; *p < 0.05 and **p < 0.01.
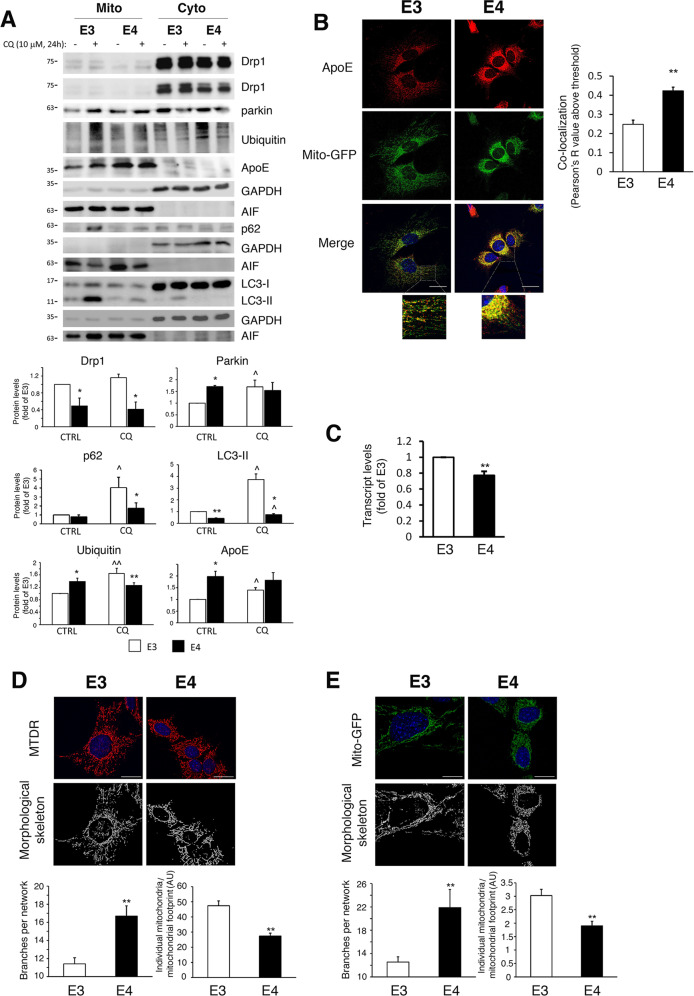
To further explore mitochondrial dynamics, we examined the mitochondrial fraction of the APOE3/APOE4 astrocytes. As shown (Fig. 2a), APOE3 astrocytes exhibited higher levels of mitochondrial Drp1 and LC3-II, indicating less mitochondrial fission and association of LC3 with mitochondria in APOE4 astrocytes. Moreover, increased levels of parkin and ubiquitin were detected in APOE4 astrocytes mitochondria, indicating either increased recruitment of parkin and ubiquitination of mitochondria, or accumulation due to reduced mitochondrial degradation. Therefore, we measured the mitochondrial recruitment of mitophagy/autophagy proteins by examining the mitochondrial fraction following chloroquine treatment24,30. As shown in Fig. 2a, increased accumulation (differences between CQ treated and untreated cells) of parkin, p62, LC3-II and ubiquitin was observed in the mitochondrial fraction of the APOE3 astrocytes following treatment, suggesting their degradation is reduced in APOE4 astrocytes. Interestingly, although the levels of LC3-II, p62 and ubiquitin in the mitochondrial fraction of CQ-treated APOE3 cells were higher compared with APOE4 cells, the levels of parkin did not differ (Fig. 2a), suggesting that the recruitment of parkin to the mitochondria of the APOE4 astrocytes is intact. Also, fractionation and immunostaining revealed that mitochondrial apoE4 levels are higher compared to apoE3 (Fig. 2a, b), which is not due to increased APOE4 mRNA synthesis (Fig. 2c). Chloroquine induced accumulation only of mitochondrial apoE3 (Fig. 2a), suggesting increased accumulation of mitochondrial apoE4.
Fig. 2. Mitochondrial fraction and mitochondrial morphology in APOE4 astrocytes.
a APOE3/APOE4 astrocytes were treated with 10 μM chloroquine (CQ) for 24 h. Mitochondrial and cytosolic fractions were subjected to Immunoblot using the indicated antibodies. AIF and GAPDH were used as markers for cytosol and mitochondria, respectively. Upper panel, representative results; lower panel, densitometric analysis of proteins in the mitochondrial fraction (normalized to AIF; fold of APOE3; n ≥ 3). b APOE3/APOE4 astrocytes expressing Mito-GFP were stained with anti-apoE antibodies (scale bars 25 μm). Left panel, representative results. Right panel, co-localization analysis of Mito-GFP (green) and apoE (red) using the Pearson’s R value above threshold as calculated by the ImageJ Coloc 2 plugin after thresholding for apoE; n ≥ 50 cells; means ± SE; **p < 0.01. c APOE3/APOE4 astrocytes were subjected to real-time PCR for APOE expression (fold of APOE3; n ≥ 3); means ± SE; **p < 0.01. d, e APOE3/APOE4 astrocytes were incubated with MitoTracker Deep Red (MTDR, d) or infected with Mito-GFP (e). Upper panels, representative images and morphological skeleton analysis as generated by the MiNa tool (Scale bars, 25 μm); Lower panels, quantification of branches per network (mean network size) and individual mitochondria relative to the mitochondrial network footprint (n ≥ 50 cells). a–e Means ± SE; *p < 0.05 and **p < 0.01, APOE3-expressing compared with APOE4-expressing cells; ^p < 0.05, CQ-treated compared to untreated cells.
To further explore whether APOE4 is associated with decreased mitochondrial fission and degradation, the mitochondrial network morphology was visualized using MitoTracker Deep Red staining or Mito-GFP expression. As demonstrated (Fig. 2d, e), the mitochondrial network of the APOE4 astrocytes is indeed more hyperfused (exhibits more branching and less individual mitochondria).
APOE4-expressing astrocytes exhibit impaired mitophagy
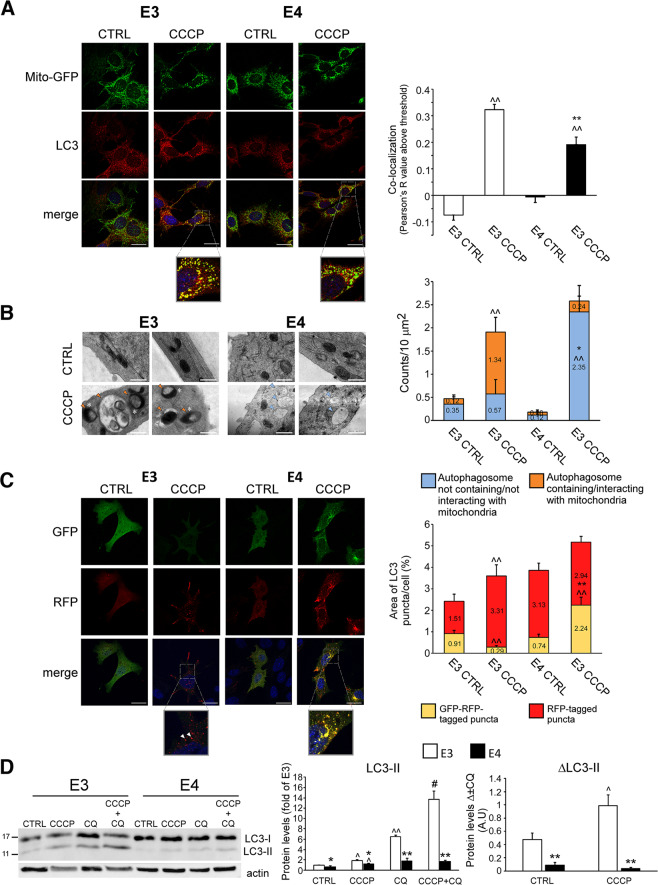
The reduced fission/mitophagy observed in the APOE4 astrocytes may result from abnormal metabolic state, or may represent mitophagy impairment. Thus, we next examined the effect of mitochondrial damage induced by mitochondrial uncoupler CCCP8 on mitochondrial dynamics. CCCP markedly increased LC3 puncta (red), as well as their co-localization with mitochondria (green), suggestive of mitophagy induction (Fig. 3a), which was higher in APOE3 astrocytes. Likewise, EM analysis revealed that in the APOE3 astrocytes, CCCP increased the number of autophagosomes containing mitochondria and the formation of spheroid mitochondria, which represents mitochondria interacting with autophagosomes/lysosomes31,32 (Fig. 3b). In contrast, in APOE4 astrocytes the treatment induced significant formation of autophagosomes that do not contain mitochondria (Fig. 3b). Furthermore, using the tandem LC3-GFP-RFP reporter30, we found that following CCCP treatment, ~90% of autophagosomes in the APOE3 astrocytes were fused with lysosomes, compared to ~55% in the APOE4 astrocytes (Fig. 3c). Accordingly, APOE3 astrocytes treated with CCCP combined with chloroquine showed increased accumulation of LC3-II compared to APOE4 astrocytes, indicating higher autophagic flux30 (Fig. 3d). Thus, damaged mitochondria recognition by autophagosomes and their delivery to lysosomes is hampered in APOE4 astrocytes. Consistently, in APOE3/APOE4 astrocytes stably expressing Mito-GFP, treatment with CCCP resulted in decreased number of branches per network, which was more profound in the APOE3 astrocytes (Fig. S1a).
Fig. 3. CCCP-induced mitophagy in APOE3/APOE4 astrocytes.
a APOE3/APOE4 astrocytes expressing Mito-GFP were treated with 15 μM CCCP for 1 h and stained with anti-LC3 antibodies (scale bars 25 μm). Left panel, representative results. Right panel, co-localization analysis of Mito-GFP (green) and LC3 puncta (red) using the Pearson’s R value above threshold as calculated by the ImageJ Coloc 2 plugin after thresholding for LC3 puncta; n ≥ 50 cells/treatment; means ± SE; **p < 0.01, APOE3 compared with APOE4; ^^p < 0.01, CCCP-treated compared to untreated cells. b APOE3/APOE4 astrocytes were treated with 25 μM CCCP for 4 h and analyzed by electron microscopy. Left panel, representative micrographs; right panel, the number of autophagosomes containing/interacting (orange arrows) or not (blue arrows) with mitochondria was measured per 10 μm2 fields (scale bars, 500 nm); n ≥ 10 fields/treatment; means ± SE; *p < 0.05 and **p < 0.01, APOE3 compared with APOE4; ^^p < 0.01, CCCP-treated compared to untreated cells.; white asterisks, spheroid/ring-shaped mitochondria (presumably mitochondria interacting with autophagosome/lysosome) c APOE3/APOE4 astrocytes expressing LC3-EGFP-mRFP were treated with 10 μM CCCP for 4 h. Left panel, representative results (scale bars 25 μm); right panel, ImageJ analysis of %area of LC3-EGFP-mRFP (arrows) or LC3-mRFP (arrow heads) positive puncta; n ≥ 50 cells/treatment; means ± SE; *p < 0.05 and **p < 0.01, APOE3- compared with APOE4; ^^p < 0.01, CCCP-treated compared to untreated cells. d APOE3/APOE4 astrocytes were treated with 10 μM chloroquine (CQ) for 48 h, with or without 7.5 μM CCCP for the last 24 h. Levels of LC3 were determined by Immunoblot. Left panel, representative results; right panel, densitometric analysis of the results is presented as fold of APOE3 (left graph) and as the difference between measured values of LC3 levels with or without CQ (right graph, Δ ± CQ); n = 3; means ± SE; *p < 0.05 and **, APOE3 compared with APOE4; ^p < 0.05 and ^^p < 0.01, treated compared with untreated cells, ##p < 0.01, combined CCCP and CQ treatment compared to each treatment alone.
We next studied the effect of CCCP on fission/mitophagy at the protein level. As shown, CCCP treatment induced a dose-dependent decrease of parkin and Mfn1 levels (indicating their degradation) and increased LC3-II/LC3-I ratio (autophagosomes formation), consistent with mitophagy induction (Fig. S1b); yet, the effect was more pronounced in the APOE3 cells. In addition, 12 µM CCCP treatment reduced p62 levels only in the APOE3 astrocytes, indicating increased p62 degradation in these cells (Fig. S1b). Examination of the mitochondrial fractions revealed that CCCP treatment increased mitochondrial Drp1 levels only in the APOE3 astrocytes, and elevated mitochondrial LC3-II, which was more prominent in the APOE3 astrocytes (Fig. S1c). This suggests that following treatment, less fission and association of LC3-II with damaged mitochondria occurs in APOE4 astrocytes. Taken together, the results further demonstrate dysfunctional fission and mitophagy in the APOE4 astrocytes following mitochondrial damage.
APOE4-expressing astrocytes exhibit altered synthesis, ubiquitination and proteasomal/lysosomal degradation of mitochondrial dynamics proteins
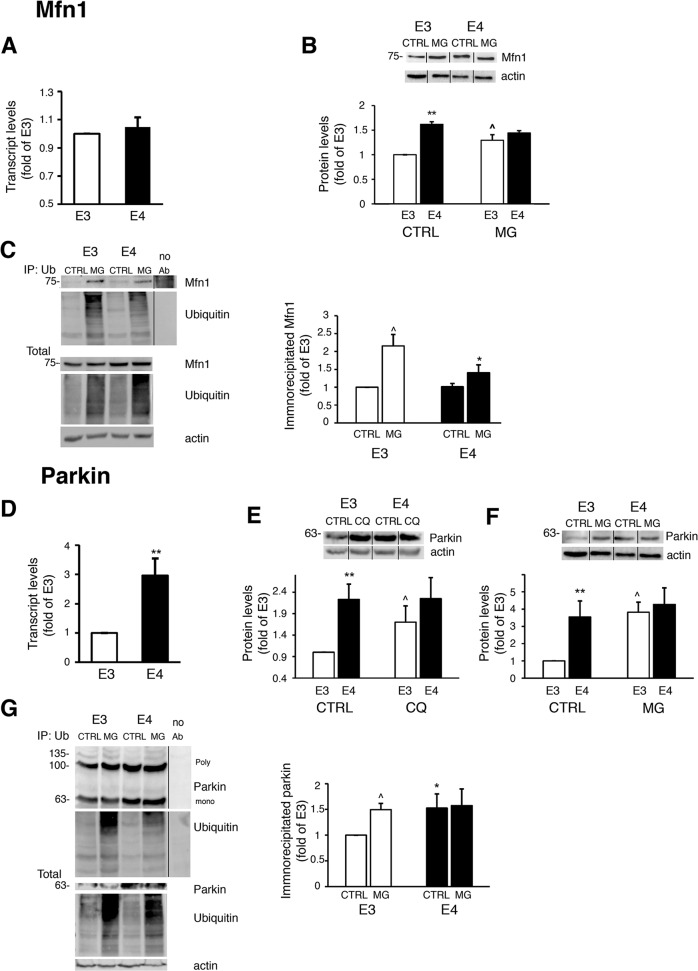
We next characterized the levels of synthesis, ubiquitination and turnover of mitochondrial dynamics proteins. According to qRT-PCR (Fig. 4a), the observed elevation of Mfn1 protein in APOE4 astrocytes (Fig. 1) is not due to increased MFN1 transcription. However, blockage of proteasomal degradation (MG-132), led to accumulation of Mfn1, only in the APOE3 astrocytes (Fig. 4b). Therefore, we next measured the levels of Mfn1 ubiquitination (Fig. 4c). Following MG-132 treatment the total amount and accumulation of ubiquitinated Mfn1 was higher in APOE3 compared to APOE4 astrocytes. During mitophagy, Mfn1 undergoes ubiquitination by parkin followed by proteasomal degradation, which suppresses mitochondrial fusion and allows the removal of mitochondria from the network8. Thus, it appears that the basal differences of Mfn1 levels observed in APOE4 result from reduced Mfn1 ubiquitination and proteasomal degradation, possibly contributing to their hyperfused mitochondrial network (Fig. 2d, e).
Fig. 4. Synthesis/degradation of Mfn1 (a-c) and Parkin (d-g) in APOE3/APOE4 astrocytes.
a, d APOE3/APOE4 astrocytes were subjected to real-time PCR of MFN1 (a) or PARK2/PRKN (parkin) (d) expression (fold of APOE3; n ≥ 3). b Cells were treated with 1 μM MG-132 (MG) for the indicated times and the levels of Mfn1 were determined by Immunoblot. Upper panel, representative results; lower panel, densitometric analysis of Mfn1 levels is presented as fold of APOE3 (left graph) (n ≥ 3). c, g Cells were treated with 1 μM MG-132 for 8 h and cell lysates were subjected to immunoprecipitation with anti-ubiquitin antibodies. Left panel, representative results. Right panel, densitometric analysis of immunoprecipitated ubiquitinated Mfn1 (c) or ubiquitinated parkin (g) (fold of APOE3; n ≥ 3). e, f Cells were treated with 10 μM chloroquine (CQ; e) or 1 μM MG-132 (MG; f) for 24 h or 8 h, respectively, and the parkin levels were determined by Immunoblot. Upper panel, representative results; lower panel, densitometric analysis of parkin levels (fold of APOE3, n ≥ 3). a–g Means ± SE; *p < 0.05 and **p < 0.01, APOE3 compared to APOE4; ^p < 0.05 and ^^p < 0.01, CQ/MG-132 treated compared to untreated cells.
The reduced levels of Drp1 in APOE4 astrocytes (Fig. 1) could result from decreased DNM1L (Drp1) mRNA synthesis (Fig. S2a), while degradation does not appear to contribute to this difference, since MG-132/chloroquine had no effect of Drp1 protein levels (Fig. S2b, c).
Interestingly, according to qRT-PCR, the increased parkin levels in APOE4 astrocytes are at least partially due to increased PARK2/PARKN mRNA synthesis (Fig. 4d). Additionally, treatment with chloroquine/MG-132 led to parkin accumulation only in the APOE3 astrocytes (Fig. 4e, f), indicating its reduced mitophagic and proteasomal degradation, respectively, in the APOE4 astrocytes. Proteasomal degradation of parkin was demonstrated during mitophagy due to autoubiquitination, halting its activity28,29,33,34. Immunoprecipitation assay revealed that the levels of ubiquitinated parkin (Fig. 4g) are higher in APOE4 astrocytes, and that treatment with MG-132 induces accumulation of ubiquitinated parkin only in APOE3 astrocytes (Fig. 4g). Therefore, the results indicate that parkin undergoes ubiquitination in APOE4 astrocytes, yet it is not degraded by the proteasome. It is possible that the high levels of ubiquitinated parkin in the APOE4 astrocytes interfere with its ability to ubiquitinate other proteins (e.g., Mfn1), resulting in their reduced labeling for proteasomal/mitophagic degradation. Indeed, the levels of two other mitochondrial parkin substrates, namely Tom40 and Tom2035, exhibited higher levels in APOE4 astrocytes (Fig. S2d). The elevated levels of Tom40 were not due to mRNA synthesis (Fig. S2e), and following MG-132 treatment, Tom40 accumulated only in APOE3 astrocytes (Fig. S2f), suggesting that reduced ubiquitination of Tom40 underlies its increased levels in APOE4 astrocytes. Collectively, the results indicate that parkin activity is altered in the APOE4 astrocytes.
APOE4 astrocytes exhibit impaired mitochondrial function
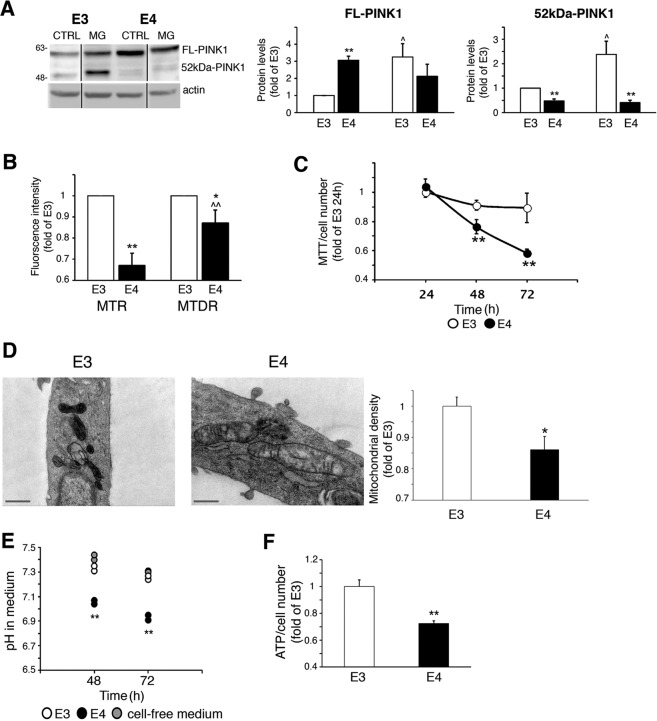
Since mitochondrial dynamics are related to mitochondrial quality control, we next investigated several aspects of mitochondrial function. We first focused on PINK1, which accumulates on the OMM following MMP reduction, while it undergoes cleavage in healthy mitochondria due to proteasomal degradation5,8. As shown, APOE4 astrocytes express higher levels of full-length PINK1 (Fig. 5a), which does not result from increased mRNA synthesis (Fig. S3a), and lower levels of cleaved PINK1 compared to APOE3 astrocytes, which were increased following MG-132 treatment only in APOE3 astrocytes (Fig. 5a). Collectively, the results indicate that PINK1 cleavage is reduced in APOE4 astrocytes, indicating reduced MMP. Likewise, the fluorescence of the MMP-sensitive MitoTracker Red22,23 was significantly lower in the APOE4 astrocytes (Fig. 5b). This probably does not reflect reduced mitochondrial content in the APOE4 astrocytes, since their fluorescence of MitoTracker Deep Red, which is less sensitive to MMP21,24,25, was significantly greater compared to MitoTracker Red (Fig. 5b); and the levels of several abundant mitochondrial proteins and transcripts were similar in the APOE3/APOE4 astrocytes (Fig. S3b, c).
Fig. 5. Mitochondrial function in APOE3/APOE4.
a Cells were treated with 1 μM MG-132 (MG) for 8 h and PINK1 levels were determined by Immunoblot. Left panel, representative results; Right panels, densitometric analysis of PINK1 levels (fold of APOE3; n ≥ 3). FL-PINK1, full-length PINK1; 52kDa-PINK1, cleaved PINK1; *p < 0.05 and **p < 0.01, APOE3 compared to APOE4; ^p < 0.05 and ^^p < 0.01, treated compared to untreated cells. b Cells were stained with MitoTracker Red (MTR) or MitoTracker Deep Red (MTDR), and fluorescence was measured (fold of APOE3; n ≥ 3); *p < 0.05 and **p < 0.01, APOE3-expressing compared to APOE4 cells; ^p < 0.05, MTDR compared with MTR. c, g Cells were subjected to MTT assay normalized to cell number at the indicated time points (c) (n = 6/treatment). d Cells were processed and analyzed by electron microscopy. Left panels, representative results; right panel, analysis of mitochondrial density was measured using ImageJ (fold of APOE3; n ≥ 20 mitochondria; scale bars, 500 nm); means ± SE; **p < 0.01. e, f Cells were cultured for 48 h, after which, media pH (e) or ATP levels (f) were measured (n ≥ 3); means ± SE; **p < 0.01, APOE3-expressing compared to APOE4-expressing cells.
Similarly, the APOE4 astrocytes exhibit reduced mitochondrial metabolism as measured by MTT20,36 (Fig. 5c), and by less dense cristae, which is another indicator of impaired mitochondrial activity37,38 (Fig. 5d). Interestingly, the APOE4 astrocytes also exhibit lower medium pH, suggestive of increased compensatory glycolysis39 (Fig. 5e) and reduced ATP levels (Fig. 5f).
Rapamycin induces mitophagy and restores mitochondrial function in APOE4-expressing astrocytes
We next examined whether non-toxic mitophagy induction could counteract mitochondrial dysfunction in APOE4 astrocytes. For this aim we have used rapamycin, which promotes autophagosome formation30, and was shown to induce mitophagy by upregulating the parkin/PINK1 pathway25,40–42.
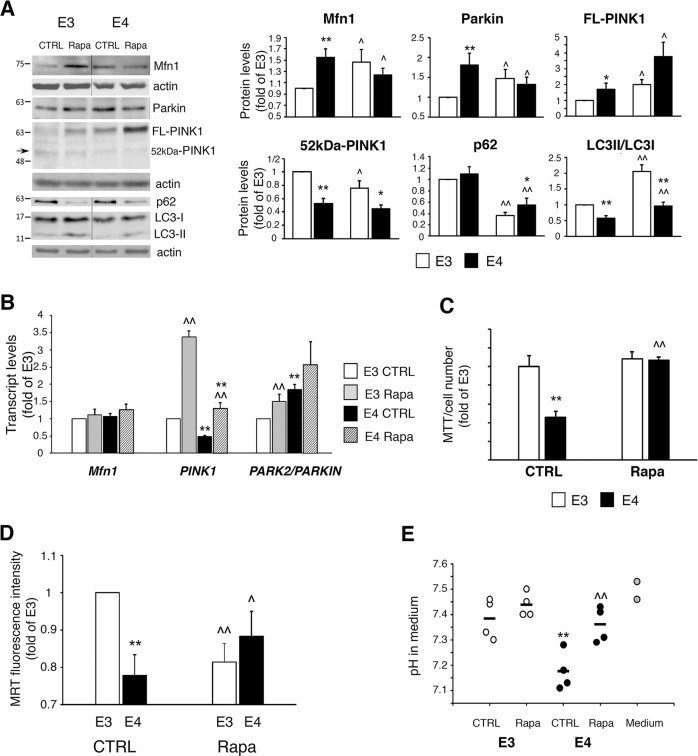
Rapamycin treatment increased LC3-II/LC3-I ratio and decreased p62 levels, in both the APOE3/APOE4 astrocytes (Fig. 6a), indicating enhanced autophagosome formation and cargo degradation. Furthermore, in the APOE4 astrocytes, rapamycin decreased Mfn1 and parkin levels, which is not the result of reduced gene expression (Fig. 6a, b), and is indicative for mitophagy induction (Fig. S1b). Accordingly, rapamycin decreased APOE4 astrocytes’ mitochondrial content, as demonstrated by MitoTracker Deep Red (Fig. S3d). Rapamycin had no effect on cleaved PINK1 levels in APOE4 astrocytes, indicating no reduction in MMP, yet it elevated FL-PINK1 levels (Fig. 6a), which probably results from enhanced PINK1 mRNA synthesis (Fig. 6b). Notably, in the APOE4 astrocytes, rapamycin restored mitochondrial metabolism (MTT), augmented MMP (MitoTracker Red), and corrected the altered medium pH without affecting cell number (Fig. 6c–e). Taken together, these results demonstrate that rapamycin restores mitochondrial function in the APOE4 astrocytes.
Fig. 6. Effect of rapamycin on APOE3/APOE4 astrocytes.
a APOE3/APOE4 astrocytes were treated with 150 nM rapamycin (Rapa) for 48 h and subjected to Immunoblot. Left panels, representative results. Right panels, densitometric analysis of protein levels (fold of APOE3; n ≥ 3). b Cells were treated with 150 nM rapamycin for 24 h and subjected to real-time PCR (fold of APOE3; n ≥ 3). c Cells were treated with 150 nM rapamycin for the last 24 h (72 h culture) and subjected to MTT assay normalized to cell number (fold of APOE3; n = 6/treatment). d Cells were treated with 150 nM rapamycin for 72 h, followed by staining with MitoTracker Red (MTR). Fluorescence intensity of MTR was measured using flow cytometry (fold of APOE3; n ≥ 3/treatment). e Cells were treated with 150 nM rapamycin for 72 h, after which, media pH was measured (n = 3/treatment). a–e Means ± SE; *p < 0.05 and **p < 0.01, APOE3 compared with APOE4; ^p < 0.05 and ^^p < 0.01, treated compared with untreated cells.
Discussion
Changes in mitochondrial dynamics and mitochondrial dysfunction are recognized as hallmarks of AD, and although mitochondrial deterioration in astrocytes occurs early in the disease development, the mechanism is vague12. Among the risk factors for AD is the APOE4 allele1; however, its effects on mitochondrial dynamics and dysfunction are unclear. Previously, it was shown that APOE4 is associated with impaired autophagy in astrocytes13,14, as well as with changes in mitochondrial dynamics proteins expression in transgenic mice brains15. This prompted us to further explore the mechanisms involved in the effect of APOE4 on mitochondrial dynamics and function specifically in astrocytes.
Using primary astrocytes and GFAP-stained brain sections generated from APOE3/APOE4 targeted-replacement mice, we demonstrated differences in mitochondrial dynamics proteins expression similar to those observed in APOE3 and APOE4-expressing immortalized astrocytes. We further demonstrated that APOE4 expression in astrocytes is associated with altered synthesis, recruitment, ubiquitination and degradation of mitochondrial dynamics proteins. Drp1 protein and mRNA levels and its mitochondrial recruitment, are decreased in APOE4 astrocytes, indicating reduced mitochondrial fission. Mitochondrial fusion is mediated by mitofusins, which are regulated by transcription and protein modification, including ubiquitination3. During mitophagy, parkin ubiqitinates Mfn1, leading to its proteasomal degradation and fusion inhibition8,43. We found that Mfn1 ubiquitination/degradation is reduced in APOE4 astrocytes, leading to increased Mfn1 levels and fusion, possibly interfering with mitophagy.
Parkin labels mitochondria for mitophagy by ubiquitinating specific mitochondrial proteins; parkin is then degraded by the proteasome/lysosome due to autoubiquitination26–29. APOE4 astrocytes exhibit increased levels of parkin, possibly due to reduced degradation of ubiquitinated mitochondrial parkin. This inefficient turnover of parkin might impair its activity, leading to deficient labeling of mitochondria for mitophagy. In fact, abnormal parkin ubiquitination was shown to prevent its proteasomal degradation and impede parkin-mediated mitophagy44. Consistently, the ubiquitination/proteasomal degradation of Mfn1 and Tom408,35, as well as of total mitochondrial ubiquitin, are reduced in the APOE4 astrocytes, further indicating impaired parkin activity. Moreover, the interaction of the autophagic proteins LC3-II and p62 with the mitochondria, and the lysosomal degradation of mitochondrial LC3-II and p62 are reduced in APOE4 astrocytes. Therefore, the results indicate that mitochondrial dynamics are altered in APOE4 astrocytes, and specifically suggest reduced parkin-mediated mitophagy.
To investigate whether these findings represent mitophagy impairment, we examined mitochondrial dynamics following induction of mitochondrial damage (CCCP), which normally activates mitophagy8. Mitophagy induction following damage was impaired in APOE4 compared to APOE3 astrocytes. Consistently, other studies have demonstrated impaired mitophagy and parkin accumulation in AD neurons45,46. Interestingly, we also detected less autophagosomes-lysosomes fusion in APOE4 astrocytes following CCCP treatment, which is consistent with other lysosomal dysfunctions associated with APOE447. The mechanism by which APOE4 affects mitochondrial dynamics is yet to be investigated; it may involve a detrimental interaction of apoE4 itself with the mitochondria, as was shown in neurons10,11 and is consistent with our findings. Alternatively, apoE4 may affect mitochondrial dynamics indirectly, by interfering with the autophagy/lysosomal machineries, which affects upstream processes (e.g., mitochondrial fission and labeling) as was demonstrated in other neurodegenerative disorders47,48.
The hyperfused mitochondrial network morphology observed in APOE4 astrocytes may reflect reduced mitophagy and fission, and correlates with findings from AD fibroblasts6. Yet, we did not detect a greater mitochondrial mass in APOE4 astrocytes, suggesting that their mitochondrial network is in a static hyperfused state49; i.e., less fission/degradation occurs together with reduced mitochondrial biogenesis. Therefore, less quality control and mitochondrial turnover is possible, leading to increased population of dysfunctional mitochondria50. Accordingly, we found impaired mitochondrial activity in APOE4 astrocytes. Possibly, this mitochondrial dysfunction reduces energy production, which, in turn, represses Drp1 expression and mitochondrial fission to maintain high mitochondrial volume51,52.
Assuming that impaired removal of damaged mitochondria underlies APOE4 astrocytes mitochondrial dysfunction, it is plausible that mitophagy induction, without causing mitochondrial damage, will improve mitochondrial function. Indeed, rapamycin treatment, which was shown to stimulate mitophagy40,41,53, partially improved APOE4 astrocytes’ mitochondrial function. Hence, non-toxic mitophagy might induce efficient removal of dysfunctional mitochondria, therefore improving mitochondrial activity, which in turn, improves AD-related function in APOE4 astrocytes.
Taken together, our results show that APOE4 astrocytes exhibit altered mitochondrial dynamics, specifically impaired mitophagy. In addition, these cells display a mitochondrial dysfunction that might be linked to the impaired mitophagy. Thus, the results suggest that altered mitochondrial dynamics in astrocytes plays a role in the pathological effects of APOE4.
Supplementary information
Acknowledgements
This work was supported by The Prajs-Drimmer Institute for the Development of Anti-Degenerative Drugs; SAIA Fund for HIV and Parkinson’s Diseases Research, Tel Aviv University. We would like to thank Prof Holtzman D.M. from the Washington University for kindly providing us the APOE3/APOE4 astrocytes.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Verkhratsky
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Eran Schmukler, Shira Solomon
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-02776-4).
References
- 1.Michaelson, D. M. APOE epsilon4: the most prevalent yet understudied risk factor for Alzheimer’s disease. Alzheimer’s Dement.10, 861–868 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow, R. H. Mitochondria and mitochondrial cascades in Alzheimer’s disease. J. Alzheimer’s Dis.: JAD62, 1403–1416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisner, V., Picard, M. & Hajnoczky, G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol.20, 755–765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilokani, L., Nagashima, S., Paupe, V. & Prudent, J. Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem.62, 341–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickles, S., Vigie, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol.28, R170–R185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu, X., Perry, G., Smith, M. A. & Wang, X. Abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Alzheimer’s Dis.33, S253–262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westermann, B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta1817, 1833–1838 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Truban, D., Hou, X., Caulfield, T. R., Fiesel, F. C. & Springer, W. PINK1, parkin, and mitochondrial quality control: what can we learn about Parkinson’s disease pathobiology? J. Parkinson’s Dis.7, 13–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dose, J., Huebbe, P., Nebel, A. & Rimbach, G. APOE genotype and stress response—a mini review. Lipids Health Dis.15, 121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura, T., Watanabe, A., Fujino, T., Hosono, T. & Michikawa, M. Apolipoprotein E4 (1-272) fragment is associated with mitochondrial proteins and affects mitochondrial function in neuronal cells. Mol. Neurodegeneration4, 35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H. K. et al. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J. Biol. Chem.286, 5215–5221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampinen, R., Belaya, I., Boccuni, I., Malm, T., Kanninen, K. M. in Astrocyte – Physiology and Pathology (ed. D’Amato MTGaLC), INTECH 139–162 (2018). 10.5772/intechopen.71825.
- 13.Simonovitch, S. et al. Impaired autophagy in APOE4 astrocytes. J. Alzheimers Dis.51, 915–927 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Parcon, P. A., et al. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimer’s Demen.10.1016/j.jalz.2017.07.754 (2017). [DOI] [PMC free article] [PubMed]
- 15.Simonovitch, S., Schmukler, E., Masliah, E., Pinkas-Kramarski, R. & Michaelson, D. M. The effects of APOE4 on mitochondrial dynamics and proteins in vivo. J. Alzheimers Dis.70, 861–875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morikawa, M. et al. Production and characterization of astrocyte-derived human apolipoprotein E isoforms from immortalized astrocytes and their interactions with amyloid-beta. Neurobiol. Dis.19, 66–76 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Wolfson, E., Solomon, S., Schmukler, E. & Goldshmit, Y. Pinkas-Kramarski R. Nucleolin and ErbB2 inhibition reduces tumorigenicity of ErbB2-positive breast cancer. Cell Death Dis.9, 47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valente, A. J., Maddalena, L. A., Robb, E. L., Moradi, F. & Stuart, J. A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochemica119, 315–326 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Schmukler, E., Wolfson, E., Elazar, Z., Kloog, Y. & Pinkas-Kramarski, R. Continuous treatment with FTS confers resistance to apoptosis and affects autophagy. PLoS ONE12, e0171351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai, Y. et al. Mitochondrial biogenesis and metabolic hyperactivation limits the application of MTT assay in the estimation of radiation induced growth inhibition. Sci. Rep.8, 1531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteban-Martinez, L., Villarejo-Zori, B. & Boya, P. Cytofluorometric assessment of mitophagic flux in mammalian cells and tissues. Methods Enzymol.588, 209–217 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Shibata, T. et al. T-2307 causes collapse of mitochondrial membrane potential in yeast. Antimicrobial Agents Chemother.56, 5892–5897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Mata, M. et al. Pharmacological chaperones and coenzyme Q10 treatment improves mutant beta-glucocerebrosidase activity and mitochondrial function in neuronopathic forms of gaucher disease. Sci. Rep.5, 10903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauro-Lizcano, M. et al. New method to assess mitophagy flux by flow cytometry. Autophagy11, 833–843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebrahimi-Fakhari, D. et al. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep.17, 2162 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Vives-Bauza, C. et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA107, 378–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol.183, 795–803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seirafi, M., Kozlov, G. & Gehring, K. Parkin structure and function. FEBS J.282, 2076–2088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakovic, A. et al. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J. Biol. Chem.288, 2223–2237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy12, 1–222 (2016). [DOI] [PMC free article] [PubMed]
- 31.Cook, K. L. et al. Mitochondria directly donate their membrane to form autophagosomes during a novel mechanism of parkin-associated mitophagy. Cell Biosci.4, 16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding, W. X. et al. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J. Biol. Chem.287, 42379–42388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaugule, V. K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J.30, 2853–2867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrd, R. A. & Weissman, A. M. Compact Parkin only: insights into the structure of an autoinhibited ubiquitin ligase. EMBO J.32, 2087–2089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshii, S. R., Kishi, C., Ishihara, N. & Mizushima, N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem.286, 19630–19640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguer, C. et al. Galactose enhances oxidative metabolism and reveals mitochondrial dysfunction in human primary muscle cells. PLoS ONE6, e28536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woods, D. C. Mitochondrial heterogeneity: evaluating mitochondrial subpopulation dynamics in stem cells. Stem Cells Int.2017, 7068567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen, J. et al. Plasticity in mitochondrial cristae density allows metabolic capacity modulation in human skeletal muscle. J. Physiol.595, 2839–2847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Celotto, A. M., Chiu, W. K., Van Voorhies, W. & Palladino, M. J. Modes of metabolic compensation during mitochondrial disease using the Drosophila model of ATP6 dysfunction. PLoS ONE6, e25823 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartolome, A. et al. MTORC1 regulates both general autophagy and mitophagy induction after oxidative phosphorylation uncoupling. Mol. Cellular Biol.37, e00441 (2017). [DOI] [PMC free article] [PubMed]
- 41.Li, Q. et al. Rapamycin attenuates mitochondrial dysfunction via activation of mitophagy in experimental ischemic stroke. Biochem. Biophys. Res. Commun.444, 182–188 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Wang, C. et al. The post-therapeutic effect of rapamycin in mild traumatic brain-injured rats ensuing in the upregulation of autophagy and mitophagy. Cell Biol. Int.41, 1039–1047 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Glauser, L., Sonnay, S., Stafa, K. & Moore, D. J. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem.118, 636–645 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Durcan, T. M. et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J.33, 2473–2491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerr, J. S. et al. Mitophagy and Alzheimer’s disease: cellular and molecular mechanisms. Trends Neurosci.40, 151–166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye, X., Sun, X., Starovoytov, V. & Cai, Q. Parkin-mediated mitophagy in mutant hAPP neurons and Alzheimer’s disease patient brains. Hum. Mol. Genet.24, 2938–2951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmukler, E., Michaelson, D. M. & Pinkas-Kramarski, R. The interplay between apolipoprotein E4 and the autophagic-endocytic-lysosomal axis. Mol. Neurobiol.55, 6863–6880 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Seranova, E. et al. Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem.61, 733–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoitzing, H., Johnston, I. G. & Jones, N. S. What is the function of mitochondrial networks? A theoretical assessment of hypotheses and proposal for future research. BioEssays: N. Rev. Mol., Cell. Dev. Biol.37, 687–700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andres, A. M., Stotland, A., Queliconi, B. B. & Gottlieb, R. A. A time to reap, a time to sow: mitophagy and biogenesis in cardiac pathophysiology. J. Mol. Cell. Cardiol.78, 62–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomes, L. C., Di Benedetto, G. & Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol.13, 589–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambold, A. S., Kostelecky, B., Elia, N. & Lippincott-Schwartz, J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl Acad. Sci. USA108, 10190–10195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebrahimi-Fakhari, D. et al. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep.17, 1053–1070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.