Abstract
Objectives
This manuscript reviews the current state of veterinary medical electronic health records and the ability to aggregate and analyze large datasets from multiple organizations and clinics. We also review analytical techniques as well as research efforts into veterinary informatics with a focus on applications relevant to human and animal medicine. Our goal is to provide references and context for these resources so that researchers can identify resources of interest and translational opportunities to advance the field.
Methods and Results
This review covers various methods of veterinary informatics including natural language processing and machine learning techniques in brief and various ongoing and future projects. After detailing techniques and sources of data, we describe some of the challenges and opportunities within veterinary informatics as well as providing reviews of common One Health techniques and specific applications that affect both humans and animals.
Discussion
Current limitations in the field of veterinary informatics include limited sources of training data for developing machine learning and artificial intelligence algorithms, siloed data between academic institutions, corporate institutions, and many small private practices, and inconsistent data formats that make many integration problems difficult. Despite those limitations, there have been significant advancements in the field in the last few years and continued development of a few, key, large data resources that are available for interested clinicians and researchers. These real-world use cases and applications show current and significant future potential as veterinary informatics grows in importance. Veterinary informatics can forge new possibilities within veterinary medicine and between veterinary medicine, human medicine, and One Health initiatives.
Keywords: informatics, one health, translational science, veterinary medicine, medicine
INTRODUCTION
Biomedical informatics has become well-known within the human medical community for its breadth and depth of integration between medicine, computer science, statistics, and biology. This cross-disciplinary approach has led to discoveries in diseases, trends in success and failures for diagnosis and therapies, and evidence-based decision-making. It is now a regular part of physician’s training. While there has been significant progress integrating informatics with human medicine both in practice and education, veterinary medicine lags in both areas. However, informatics in veterinary medicine offers significant possibilities for translational research.
Use of biomedical informatics in veterinary medicine, what we term veterinary informatics, combines the knowledge and techniques currently utilized within the human medical field of biomedical informatics with veterinary domain knowledge. There has been some examination of veterinary informatics within the veterinary sphere,1–4 and the benefits of this combination are starting to emerge with a recent increase in the number of publications.5–8 Even though advances in veterinary informatics offer significant possibilities for translational research, this space remains far behind many other areas of informatics.2–4,9–13
One key factor limiting the advancement and integration of veterinary informatics into everyday clinical practice is a lack of external drivers to enforce structure on veterinary data. Unlike in human medical practice, adoption of insurance coverage for pets is still rare (<1% of pets are covered),14 leading most owners to pay for care out of pocket. This removes one of the primary incentives for coding in the medical profession, insurance companies. As a result, veterinary records have billing codes, but they are not standardized between hospitals unless they are in a shared practice group. Depending on whether you use the total veterinary positions available or those specifically identified as a practicing veterinarian, approximately 65–95% of veterinary data is contained within private organizations based on estimates of veterinarians in private practice,15 which represents a data access challenge for non-affiliated groups.
Additional limitations exist in the lack of formal, organized support for robust informatics training and methods development from national veterinary organizations, which limits the ability to set basic standards for data organization that would allow for its reuse. The concept of FAIR (Findable, Accessible, Interoperable, and Reusable) data16 has not become an integral part of management practice for veterinary data. This will change only when clinicians see improvements in patient care facilitated by access to large, organized veterinary clinical databases with standardized data formats and ontologies. Increasing the access of veterinary resources to cutting edge informatics research and highlighting use cases that cross the artificial divide between human medicine and veterinary medicine will bring additional opportunities and incentives for making the data reusable and accessible.
Thus, the applications mentioned below are focused on those that represent the opportunity to improve care for the patients we serve. We highlight a set of diverse use cases, including biosurveillance for zoonotic disease, antibiotic resistance, and disease outbreak prediction, as well as environmental health effects and insights into the biology of rare diseases. Key resources for these applications as well as methods are reviewed for analysis of electronic health records, genetics, medical imaging, and clinical laboratory data. We draw parallels with other medical disciplines that have approached similar problems through informatics and demonstrate opportunities and possible future directions that could be explored through veterinary informatics for the benefit of both humans and animals.
Scope of veterinary small animal practice
While specific data points are sparse, a recent survey by GfK (the Society for Consumer Research) found that 57% of consumers own pets of which dogs are the most popular followed by cats. Some of the highest ownership rates were in Argentina, Mexico, and Brazil (82%, 81%, and 76%, respectively). The United States ranks fifth in dog ownership and third in cat ownership.17
Specifics by country are difficult to acquire, but as of 2016 in the United States, there were approximately 89.7 million owned dogs and 94.2 million owned cats with an average of 3 veterinary visits for dogs and 2.4 veterinary visits for cats per a year. This is estimated to have generated about 269 million and 226 million visits per year, respectively.18–20 Comparatively, in human medicine, according to CDC statistics, there are approximately 864 million hospital outpatient visits per year in the United States.21 Even though there is approximately 50% fewer visits for veterinary patients, the average human patient visits about 5.4 medical professionals per year, and the estimated number of veterinarians seen per year is approximately 2.6 for dogs and 1.6 for cats.22,23 This is 25–50% of the number of human physicians interactions per patient per year. This could allow for increased intra- and inter-operator agreement in veterinary medicine for a given patient.24–26
Standardization of veterinary medical records have been isolated to individual private organizations that require specific techniques for extraction, but there have been few attempts to integrate structured terminology in veterinary clinical practice.27 In human medicine, many medical records are encoded either during practice or as a post-processing technique once the record is complete.28–31 This is often due to common drivers such as effective insurance billing and documented medical information retrieval, which veterinary medicine currently lacks. The number of animals covered by pet insurance is increasing, but <1% of animals are currently covered limiting the impact it has on encoding in veterinary practice.14
For those looking to integrate standard veterinary terminologies, one such example is the Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) Veterinary Extension maintained by the Virginia Tech Veterinary Terminology Services Laboratory (United States). Any company in a country that contributes to SNOMED can utilize it, after registration; however, outside of those countries, it might require a commercial license. There are also proprietary terminologies available, such as VeNOM that is based in the United Kingdom. However, the use of these terminologies is challenging and rarely integrated into a regular workflow. One reason is the need for most workflows to be centered on billing as this is a key concern for private practitioners, who represent approximately 91% of the clinically practicing veterinarians in the United States. Even in systems where it is possible to utilize standard terms, it is challenging to have clinicians use them consistently as veterinarians have similar time pressures to their human counterparts.32–34
We divide this review into 3 components: a description of available data sources, their characteristics, and research into their development and expansion, some examples of current analytical strategies applied to the veterinary data sources with parallels to human medicine, and finally, some highlights of current applications of veterinary informatics to One Health and translations science.
DATA TYPES AND SOURCES
Electronic medical records
As previously discussed, there are approximately 495 million clinical records for dogs and cats generated by about 80 000 veterinarians each year in the United States. The number of records for large animals, food animals, exotics, and zoological animals is challenging to quantify. For most dogs and cats, their records, in general, are stored as free text as most systems are mainly designed for efficient charging and processing of transactions throughout a patient visit. These systems, called Practice Information Management Systems (PIMS), can organize lab results as separate retrievable analytes, but there is little standardization in naming. It is important to compare the features of a PIMS to human electronic medical record systems as they have different features (Table 1).
Table 1.
Comparison of common features between EMRs and PIMS
| Activity | EMR (human medicine) | PIMS |
|---|---|---|
| Billing and finance | ± | X |
| Reminders | X | X |
| Tracking procedures | X | ± |
| Free text medical notes | X | X |
| Structured medical notes | X | ± |
| Decision support systems | X | – |
| Integration with LIMS | X | X |
| Integration with PACS | X | ± |
| Integration with RIS | X | ± |
| Computer physician order entry | X | – |
| Adverse event detection | X | – |
Note: Human EMR features from following citations.35–38 Comparison of PIMS capability is from experience combined with availability of features.39–42
–: means no current ability or requires 3rd party tool; ±: partial compatibility or function; EMR: electronic medical record; LIMS: Laboratory Information Management System; PACS: Picture Archiving and Communication System; PIMS: Practice Information Management System; RIS: Radiological Information System; X: native capability.
Like human medicine, many medical and operational drives increased PIMS adoption within veterinary medicine. As of 2014, a limited survey reported that 80% of veterinary medicine respondents utilized a PIMS though to a varying degree (63% combined electronic with paper, while 17% used only electronic records). Common uses were: billing, estimate generation, legal document retention, lab ordering and receiving (when integrated), and client account operations.43 Some of these systems have capabilities to add diagnoses to the medical record, but the reported adoption rate is difficult to validate.
Deidentification requirements are also very different. In human medicine, significant data points need to be censored for retrospective studies30,44,45; however, in veterinary medicine, there is little regulation governing sharing of records for both practice and research, with the exception of a few states.46 In these states, some hospitals have adopted a release form like those used in human medicine. This introduces additional challenges to acquiring data from clinical practices.
When looking to acquire data, on a pure numerical basis, more pets have records in smaller practices (defined as <10 veterinarians) than in larger practices, even though recent years has seen many small practices consolidated by a few larger veterinary practice groups.47 While clinical data pertaining to veterinary patients is not covered under current HIPAA regulations, owner data is considered sensitive and can sometimes be included in free text fields within medical documentation, making anonymization of veterinary medical records difficult.48
Electronic medical records are currently either being aggregated into accessible databases or through services that allow analytics. Though out of scope for this article, we have provided a list of resources and descriptions within the Supplementary Information to aid in their use for clinical research.
Imaging
Most imaging studies employ the Digital Imaging and Communications in Medicine (DICOM) imaging standard for image requests and communication; however, the way the images are stored can vary from vendor to vendor. Most radiographs generated by the practice of veterinary medicine are stored either on-premise or within a private cloud system. Commercial cloud-based systems are usually more rigorous in the implementation of the DICOM standard and can have similar features and capabilities to the Picture Archiving and Communication System implemented within the human space. The on-premise systems tend to be quite varied in implementation ranging from an index folder structure to more sophisticated storage techniques.
In veterinary medicine, the use of magnetic resonance imaging, computed tomography, fluoroscopy, and ultrasound is expanding both in available sites and studies performed per year. This type of data represents new challenges and new opportunities for study. The main source of these imaging modalities is within the specialty veterinary medicine arena, which is an increasingly popular source of care. These specialty centers are also where advanced research and training occurs offering tremendous opportunity for future collaboration.49–53
Laboratory
Diagnostic laboratory testing is an increasingly crowded arena within veterinary medicine. The two largest private veterinary diagnostic lab services are Antech Diagnostics and IDEXX Laboratories, but there are other smaller vendors. Diagnostic laboratories have various integration capabilities that change frequently. Until recently, only larger providers were “integrated” into the PIMS, meaning that requests could be submitted, and the results could be received digitally. Many reference laboratories that are integrated transmit their results via either an online portal or an interface that uses a proprietary data structure or digital documents such as a PDF. Neither the reference laboratories nor receiving PIMS utilize a standardized nomenclature for tagging data (SNOMED-CT, LOINC, etc.) during transmission, resulting in challenges during extraction and analysis. Some of them are required to annotate records for disease surveillance (eg, state diagnostic laboratories).54–58 This large, rich cohort of information represents a significant resource for future work.
One area of diagnostics, the in-house clinic, is frequently overlooked. The retention rate of patients within a veterinary hospital means that sampling variability should be significantly less. Even within a single hospital, the opportunity to correlate diagnostics with textual notes and bill codes (that can be analogous for both disease state and procedure codes) offers advantages that can be challenging to replicate within human medicine.
Genetics
The National Human Genome Research Institute recognized the value of the canine genome (sequenced in 2004) in the study of inherited disorders in man.59–61 Subsequently, an increasing number of Mendelian traits and traits associated with more complex diseases have been identified.62–66 One source for genetic information in dogs, as well as many other animals, is the Online Mendelian Inheritance in Animals.67 As of this publication, there were over 800 Mendelian traits with likely causal variants listed for many different species including dogs, cattle, cats, sheep, horse, chickens, and pigs.
Private companies have created genetic panels for varying purposes,68–70 and canine cancer genetics have been particularly well-researched in terms of translational potential for humans.71 While these companies could be a potential source of information, to date, there have been few resources developed for public use. Some companies have genotyped more than 800,000 dogs, and tens of thousands of tests have been reported by the newer canine sequencing companies. Most tests are sold directly to consumers and are used primarily for breed identity, although a few companies offer disease testing. The underlying technologies and tested markers vary, but the technology is generally based on Illumina microarrays that are proprietary.
Inherited disease screening tests are also now more freely available to help breeders and owners determine if their animals are carriers for disease(s). The University of Pennsylvania maintains a database of available genetic tests that include commercial as well as academic laboratories.72 However, available phenotypic data to pair with genetic data is sparse and is generally based on customer-provided surveys on specific traits and diseases. This is an area of active interest for some genetics companies as many of the inherited diseases do not show complete dominance even in the presence of homogeneity.
In addition, there are genetics testing companies and academic laboratories offering genetic testing for horses and livestock, reporting on genetic markers for physical traits, desirable genetic characteristics, and potential disease-linked traits. These may consist of simple parentage analysis, single-gene testing, single-nucleotide polymorphism assays, or microarrays. Livestock genetic testing is too extensive to cover here, but there has been a recent review on this topic.73
Animal genetic testing has impacts beyond companion and livestock. The National Institutes of Health recently funded the Arizona Cancer Evolution Center to integrate data, including genomics, from a broad array of species to understand the evolutionarily conserved basis for cancer.74 This group is integrating human clinical data with animal clinical and pathology data from zoological collections to examine a multitude of traits such as treatment protocols, pharmaceutical and procedure side-effects, and patient outcomes that are not captured by pathology reports. They hope to achieve a better understanding of the natural history of shared diseases across species and to allow us to better interpret genomic information being produced from “non-traditional” disease models.
Sensor data
The use of sensors has recently expanded from research applications to track animal migration patterns and behavior and for disease surveillance into the commercial and medical markets. Data collection from wearables is expanding rapidly, and market share for pet wearables is estimated to be greater than $3 billion by 2025.75 Wearable sensors can provide a rich source of potential information including physio markers, such as heart rate, respiratory rate, and temperature, as well as gyroscopic measurements reflecting activity levels, sleep metrics, and estimates of calories burned. In some cases, these wearable devices can sample the animal’s interstitial glucose. The non-invasive and continuous method of data collection is relatively inexpensive, longitudinal, and removes the influence of stress induced during veterinary visits. As more research is done, this information can be utilized for more informed health decisions by the owner and veterinarian.
Sensor data is commonly used for assessment of response to specific therapies,76 and, in conjunction with robust machine learning algorithms, it has the powerful potential to allow early identification of at-risk veterinary and human patients, especially when used in shared naturally occurring diseases. Larger companies are storing pet information for additional behavioral and health studies. Data from wearables, in a manner similar to telematics or driver monitoring devices, may be used by insurance companies as an incentive for rates adjustment with certain known preventative behaviors such as activity levels.77
Databases and other resources
There are generally 3 types of databases and resources available within veterinary medicine: Medical Record Aggregation, Laboratory (including Genomic) Information, and Research or Mission Based data sources. For Medical Record Aggregation, one of the primary examples would be the Veterinary Medical Database (www.vmdb.org). They contain annotated medical records from 26 American university veterinary hospitals amounting to over 7 million patient records. Research and mission-based data sources represent the largest set of possible resources and include resources like ESCRA (www.escra.org), an exotic species tumor database, as well as VetCOT (www.vetcot.org), a trauma data collection initiative by the American College of Veterinary Emergency and Critical Care. They are collecting information about trauma cases and their outcomes from veterinary trauma centers nationwide and compiling it into a single resource with more than 27 000 trauma case records to date. There are many other databases and resources available, and we have expanded upon their details and included other resources within the Supplementary Information.
SOME KEY METHODS FOR VETERINARY INFORMATICS
Expert-based rule text extraction and recording
Initial research used expert-based rules for inclusion and exclusion of veterinary text for biosurveillance.78,79 In addition to rules-based identification, experts built data dictionaries with key terms for diseases of interest.80 This approach was often successful for the disease being studied, but the approach was challenging to scale to thousands of additional disease diagnoses, such as those in the SNOMED-CT veterinary extension. Expert-based rules require a significant number of knowledge bases, which often reflect local biases, similarly seen in rule-based systems used in human medicine.81,82 This also creates challenges for scalability and reliability when extending them into other areas.83–85 There have been rule-based machine learning methods that combine both expert and machine learning-based rules for text extraction, but there has been little application within the veterinary space.86,87
Natural language processing
To gain access to the large amount of clinical data within free text inside medical records (human and veterinary), researchers and companies have focused on utilizing natural language processing (NLP) to extract the information.88,89 NLP techniques have been generally developed in the human medical domain, using various patterns and algorithms like n-grams and long- and short-term memory networks to increase performance.90–92
There are many examples that use NLP tools within the human domain such as improving pneumonia screening in the emergency department, assisting in adenoma detection, and simplifying hospital processes by identifying billing codes from clinical notes.93,94 Within veterinary medicine, only a handful of publications and proceedings describe using NLP. Use cases include identifying the reasons race horses were retired,95 extracting medication information from veterinary message boards,96 and emerging efforts at biosurveillance across large veterinary clinical repositories.97 More recent efforts have demonstrated success in leveraging academic, coded clinical datasets to apply diagnosis labels to private practice data.98
Machine learning and artificial intelligence
More and more pipelines utilizing machine learning and artificial intelligence are being applied to human clinical records and healthcare data, with a variable use of training data or supervision depending on the datasets used to develop these systems.85,99–107 We have visualized one common workflow in biomedical informatics for using common veterinary clinical data types for a variety of applications in Figure 1. One problem with utilizing these approaches with veterinary healthcare datasets is the lack of labeled, open-source, or shareable training data. Most studies focus on single-system or hospital analysis, establishing a gold-standard within a single institution.27,108,109 Newer, “noisy” labeling techniques may be able to help provide training data for machine learning algorithms for specific clinical tasks, and this is an area of possible significant impact.110 Examining the extent to which the few labeled data that exist can be used to build algorithms to help with prediction task from other domains (private practice, specialty practice) will be important to help advance the field without the massive number of medical coders who work in human healthcare. Research into optimized feature selection and feature engineering for machine learning techniques is ongoing within human healthcare but lags significantly for veterinary applications.101,111–114
Figure 1.
Roadmap of veterinary data inputs, preprocessing, analysis, and applications. NLP, natural language processing.
Use of machine learning in radiology has been ongoing for many years.115–124 These applications range from generic clinical decision support to specific mammography-based problems. Recently, there is also an effort to apply image processing and analysis within the veterinary sphere.108,125,126 As we see in Figure 2, you can apply segmentation or edge detection to outline areas of interest in a canine thoracic radiograph, but there is much more work to be done. While nascent, many of the same problems in veterinary medicine represent opportunities for application and development of newer algorithms.
Figure 2.
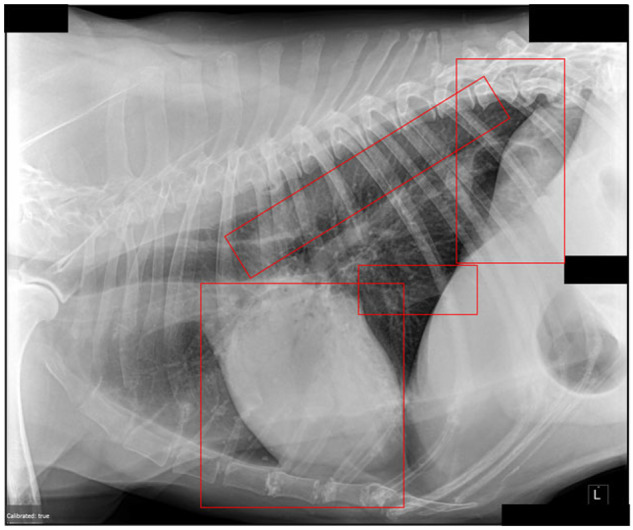
An example of anatomical structures highlighted by machine learning.
Since there is a substantial body of veterinary literature that is not indexed in PubMed, researchers have challenges finding examples of these types of applications. An application of machine learning and NLP can aid in bringing the breadth of the published veterinary knowledge to the forefront from the large, unstructured corpus of clinical texts.
APPLICATIONS OF VETERINARY INFORMATICS
Biosurveillance
A classic example of informatics application is through a zoonotic disease surveillance and reporting system.80 Studies have shown the importance of having these systems interconnected and that there are many barriers to successful implementation.127,128 Research has shown that although almost 90% of veterinarians report encountering a zoonotic disease, 70% believe they do not have access to a surveillance system, and <50% have reported a zoonotic disease to state or federal agencies. With the inability to share much of the data electronically, integrating into human platforms may overcome some of these barriers.129 Such efforts would potentially lead to earlier detection of zoonotic or environmental diseases and improved diagnosis and treatment of human disease as these diseases often occur in animals prior to human disease detection.130–132 Most approaches in biosurveillance either create a pipeline to encode medical text using NLP techniques mentioned above or directly utilized known diagnoses and combine them with a machine learning model for prediction.133–136
An example of these techniques, the CDC has been tracking Chronic Wasting Disease (CWD), a prion disease in free-ranging deer, elk, and moose for years. The CDC combines data from various sources and tracks the outbreak of the disease across the United States by county.137 There is concern of increased risk of transmission of CWD as it spreads among various countries, with one author recommending a One Health approach to monitoring, culling, and tracking to decrease this outbreak.138
Zoonosis for food animal
As stated by Doherr and Audige139 in their 2001 paper, “The health and safety of the animal and human generations depends on our continuous ability to detect, monitor and control newly emerging or re-emerging livestock diseases and zoonoses rapidly.” Need for monitoring has increased in recent years with major areas of food animal surveillance including rare and exotic disease outbreaks (e.g., foot and mouth disease) and spread of antimicrobial resistance, a significant public health concern.130,132,140–142
Best practices likely include utilizing both passive and active data collection for early identification of important rare and newly emerging diseases. The sources of information are many from general passive surveillance to laboratory samples to targeted surveillance and sentinel networks.139 Collecting and integrating the data in a cost-effective and maximally protective manner presents many challenges and requires an understanding of behavioral considerations of all players involved in the livestock supply chain.143 Software and techniques for using data for livestock production and livestock disease surveillance have shown proof of concept in several papers.143–146 These papers utilize various approaches for analysis within a population or institution, but all wrestle with deidentification of point locations so that livestock companies and locations are willing to share information.147
Antimicrobial resistance in food animal populations
The spread of antimicrobial resistance presents an emerging crisis across both human and veterinary medicine.148,149 In particular, there has been significant research into the presence of antimicrobial resistance in food and how it impacts human health.150–155 This area of study has a significant opportunity for more collaborative and expansive work using data from veterinary and human arenas particularly from a global perspective.156–160
Applications to clinical health
In addition to enhanced and coordinated monitoring of zoonotic, biosecurity, and production-based disease outbreaks, improvements in veterinary informatics can provide mechanisms for enhanced clinical disease monitoring.78,109 Current mechanisms rely solely on the individual, busy practitioner to recognize disease trends. Using machine learning techniques to allow early and accurate identification of increased incidence of clinical disease can lead to improved outcomes through recognition of spatial relationships and by informing targeted research. As discussed below, inclusion of location data in the monitoring can help identify possible environmental associations to animal disease that may precede similar human outcomes.
Extended outbreak detection
Cross-species diseases are increasing as is the need to both detect them and prevent transmission. These diseases (e.g., Lyme disease, Leptospirosis) are not necessarily transmitted between 2 species but can be indicative of the environment that they share.130,161–163 Work is relatively nascent, but sharing environmental data provides multiple opportunities to inform on animal and human health.142
Research into methicillin-resistant Staphylococcus aureus (MRSA) in relation to pet ownership provides an example of this work. Specifically, researchers evaluated pets as possible reservoirs of MRSA within the patient population.164–166 Other diseases have also been examined (eg, Toxoplasmosis, Rocky Mountain Spotted Fever, and Hantavirus),141,167,168 but many require additional research efforts to allow for extended detection and treatment in animals and humans. Recently, there has been an increased focus on carbapenem resistance and its transmission both between humans and humans and animals.169–177 There is much more research that needs to be done in this area as this represents a challenge in the practice of both human and veterinary medicine.
Disease outcome prediction
Disease outcome prediction is of increasing interest as reflected by publication volume alone: more than 20 000 publications since 2018. In human medicine, disease outcome prediction research not only has to handle the difficulties of the disease but also the challenge of data access and specificity.101,113,178,179 Many of the techniques can be utilized within veterinary medicine to train the machine learning algorithms prior to utilization within the human healthcare sphere. As previously established, the accessibility and specificity is less of an issue within veterinary medicine and can lead to, in some cases, transfer learning where veterinary medicine directly informs on the knowledge base and algorithms used in human medicine.11,180,181
Environmental effects on pets and people
Companion animals share homes and food with humans, which in addition to providing comfort and companionship, makes them excellent sentinels for environmental health hazards and bioterrorism.132,182 In addition, data show that greater than 60% of emerging infectious diseases in people start as zoonoses.183 Pets are exposed to environmental contaminants, infectious agents, and pollutants to which they may be more susceptible than humans. Several studies have shown the efficacy of using animal data as environmental indicators outside of the well-known zoonotic risks.162,168 This is particularly true within the aquatic environment where animals can be keys to understanding the current environmental status.184,185
Rare diseases
Rare diseases, defined as affecting 1 in 10 (or fewer) Americans, are being identified at increasing rates and present diagnostic and treatment challenges. A recent study discusses combining existing rare disease knowledge resources and data within the electronic medical record to assist in identifying rare disease phenotypes.186 In addition, identification of naturally occurring disease models in companion animals that are representative of rare human diseases can result in translation therapies that improve both animal and human lives. For example, osteosarcoma (OSA) is a type of bone cancer that while the most common bone tumor in humans, is very rare with <1000 new cases diagnosed in the United States each year and half of those diagnosed are under the age of 25.70 Even with diagnosis, they generally require surgery and chemotherapy for disease control, with the continued risk of metastasis. In dogs, however, OSA is the most common primary bone tumor with an incidence 27 times that of humans.70 There is ongoing research to bridge the gap between OSA in dogs and humans, and this case provides an example of how veterinary informatics, particularly machine learning and statistical analysis techniques, to identify cases of rare diseases to serve as models of human disease has the potential to greatly improve human and veterinary health.
CONCLUSIONS
There are many possibilities for use of the available veterinary data types through both the creation of new algorithms and application of current algorithms within veterinary informatics; we have only been able to touch on a small fraction of initiatives and possibilities within this space. Expansion of veterinary informatics has significant potential benefits for both human and veterinary medicine, and the potential impact will only increase as more work is done in this arena.
FUNDING
This work was supported by the Association for Veterinary Informatics and the CTSA One Health Alliance directly.
AUTHOR CONTRIBUTIONS
JLL—initiated the paper and was primary for conceptualization, primarily responsible for writing significant components of the document and editing the text. He was a primary writer for electronic medical records, machine learning, biosurveillance, and contributed to the introduction and the conclusions. He is also responsible for review, primary correspondence, formatting, creating, and organization of the references. AZ—involved in initial paper conceptualization. Contributed to multiple sections of the document: primary writer on natural language processing, figure development, and contributed significantly to the introduction and conclusions. WS—contributed to the database detail discovery, write up of the data types and sources as well as their use in current veterinary informatics-based projects. EAG—responsible for text for the zoonotic and infectious diseases as they translate from animals to humans. She wrote and edited much of the applications to human medicine that has been interspersed throughout the document. TLW—involved in initial paper conceptualization; primary writer for the environmental impact, sensor data, rare diseases, clinical health application, and contributed significantly to the zoonotic and biosurveillance sections. All authors contributed to the editing of the manuscript and added pertinent references to the paper.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to acknowledge the Association for Veterinary Informatics, the COHA Pilot Grant Program, and the COHA Advancing One Health Datasets Workshop for inspiration and support of this article. We also want to acknowledge the Department of Biomedical Data Science at Stanford University for its support of this article. We additionally wish to thank Amanda E. Chase, MA, for assisting in editing.
CONFLICT OF INTEREST STATEMENT
AZ reports salary from Fauna Bio Inc., outside the submitted work. TLW reports grants from COHA during the conduct of the study. No other authors report a conflict of interest.
REFERENCES
- 1. Fricke S. Veterinary Informatics: State-of-the-Art and the Role of Librarians. Budapest, Hungary: International Conference of Animal Health Information Specialists; 2018. [Google Scholar]
- 2. Johnson LM, Ames TR, Jacko JA, Watson LA.. The informatics imperative in veterinary medicine: collaboration across disciplines. J Vet Med Educ 2011; 38 (1): 5–9. [DOI] [PubMed] [Google Scholar]
- 3. Santamaria SL, Zimmerman KL.. Uses of informatics to solve real world problems in veterinary medicine. J Vet Med Educ 2011; 38 (2): 103–9. [DOI] [PubMed] [Google Scholar]
- 4. Smith RD, Williams M.. Applications of informatics in veterinary medicine. Bull Med Libr Assoc 2000; 88 (1): 49–55. [PMC free article] [PubMed] [Google Scholar]
- 5. Raina MacIntyre C, Engells TE, Scotch M, et al. Converging and emerging threats to health security. Environ Syst Decis 2018; 38 (2): 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burkom H, Estberg L, Akkina J, Elbert Y, Zepeda C, Baszler T.. Equine syndromic surveillance in Colorado using veterinary laboratory testing order data. PLoS One 2019; 14 (3): e0211335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beard R, Wentz E, Scotch M.. A systematic review of spatial decision support systems in public health informatics supporting the identification of high risk areas for zoonotic disease outbreaks. Int J Health Geogr 2018; 17 (1): 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singleton DA, Noble PJM, Sánchez-Vizcaíno F, et al. Pharmaceutical prescription in canine acute diarrhoea: a longitudinal electronic health record analysis of first opinion veterinary practices. Front Vet Sci 2019; 6: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anholt RM, Berezowski J, Maclean K, Russell ML, Jamal I, Stephen C.. The application of medical informatics to the veterinary management programs at companion animal practices in Alberta, Canada: a case study. Prev Vet Med 2014; 113 (2): 165–74. [DOI] [PubMed] [Google Scholar]
- 10. Bellamy JE. Veterinary informatics—why are we dragging our feet? Can Vet J 1999; 40 (12): 861–3. [PMC free article] [PubMed] [Google Scholar]
- 11. Bilic P, Kules J, Galan A, et al. Proteomics in veterinary medicine and animal science: neglected scientific opportunities with immediate impact. Proteomics 2018; 18 (14): e1800047. [DOI] [PubMed] [Google Scholar]
- 12. Smith-Akin KA, Bearden CF, Pittenger ST, Bernstam EV.. Toward a veterinary informatics research agenda: an analysis of the PubMed-indexed literature. Int J Med Inform 2007; 76 (4): 306–12. [DOI] [PubMed] [Google Scholar]
- 13. Talbot RB. Veterinary medical informatics. J Am Vet Med Assoc 1991; 199 (1): 52–7. [PubMed] [Google Scholar]
- 14.North American Pet Health Insurance Association. State of the Industry 2019. In: Watson WT, ed. State of the Industry. Boise, ID: North American Pet Health Insurance Association; 2019: 8.
- 15.American Veterinary Medical Association. Market Research Statistics: U.S. Veterinarians 2018. Secondary Market Research Statistics: U.S. Veterinarians 2018. 2018. https://www.avma.org/resources-tools/reports-statistics/market-research-statistics-us-veterinarians-2018. Accessed December 23, 2019.
- 16. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016; 3 (1): 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Industry P. INFOGRAPHIC: Most of World Owns Pets; Dogs are Tops. Secondary INFOGRAPHIC: Most of World Owns Pets; Dogs are Tops. 2016. https://www.petfoodindustry.com/articles/5845-infographic-most-of-world-owns-pets-dogs-are-tops? utm_source=KnowledgeMarketing&utm_medium=Watt%20Products&utm_campaign=Pet%20Weekly%20Roundup%20All%20Others%20List&eid=237316200&bid=1430132. Accessed May 13, 2019.
- 18. Burns K. Vital Statistics. Secondary Vital Statistics 2/1/2013. 2013. https://www.avma.org/News/JAVMANews/Pages/130201a.aspx. Accessed August 9, 2019.
- 19.American Veterinary Medical Association. AVMA pet ownership and demographics sourcebook executive summary. In: AVMA, ed. AVMA Pet Ownership and Demographics Sourcebook. Schaumburg, IL: AVMA; 2018: 5.
- 20.American Pet Products Association. 2015-2016 National Pet Owners Survey. Greenwich, UK: American Pet Products Manufacturers Association; 2016. http://www.americanpetproducts.org/pubs_survey.asp. Accessed July 19, 2019.
- 21. Rui P, Okeyode T. National Ambulatory Medical Care Survey: 2016 National Summary Tables. Secondary National Ambulatory Medical Care Survey: 2016 National Summary Tables. 2016. https://www.cdc.gov/nchs/data/ahcd/namcs_summary/2016_namcs_web_tables.pdf. Accessed August 12, 2019.
- 22. O’Hara B, Caswell K. Health Status, Health Insurance, and Medical Services Utilization: 2010. Household Economic Studies. Washington, DC: United States Census Bureau; 2013: 15.
- 23.American Animal Hosptial Association. 2015 AAHA state of the industry fact sheet. In: AAHA, ed. AAHA State of the Industry. Lakewood, CO: American Animal Hospital Association; 2015: 4.
- 24. Patel J, Mowery D, Krishnan A, Thyvalikakath T.. Assessing information congruence of documented cardiovascular disease between electronic dental and medical records. AMIA Annu Symp Proc 2018; 2018: 1442–50. [PMC free article] [PubMed] [Google Scholar]
- 25. South BR, Mowery D, Suo Y, et al. Evaluating the effects of machine pre-annotation and an interactive annotation interface on manual de-identification of clinical text. J Biomed Inform 2014; 50: 162–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Popovic ZB, Thomas JD.. Assessing observer variability: a user’s guide. Cardiovasc Diagn Ther 2017; 7 (3): 317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones-Diette JS, Brennan ML, Cobb M, Doit H, Dean RS.. A method for extracting electronic patient record data from practice management software systems used in veterinary practice. BMC Vet Res 2016; 12 (1): 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terry AL, Chevendra V, Thind A, Stewart M, Marshall JN, Cejic S.. Using your electronic medical record for research: a primer for avoiding pitfalls. Fam Pract 2010; 27 (1): 121–6. [DOI] [PubMed] [Google Scholar]
- 29. Sarkies MN, Bowles KA, Skinner EH, et al. Data collection methods in health services research: hospital length of stay and discharge destination. Appl Clin Inform 2015; 6 (1): 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Danciu I, Cowan JD, Basford M, et al. Secondary use of clinical data: the Vanderbilt approach. J Biomed Inform 2014; 52: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stausberg J, Koch D, Ingenerf J, Betzler M.. Comparing paper-based with electronic patient records: lessons learned during a study on diagnosis and procedure codes. J Am Med Inform Assoc 2003; 10 (5): 470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khullar D, Wolfson D, Casalino LP.. Professionalism, performance, and the future of physician incentives. JAMA 2018; 320 (23): 2419–20. [DOI] [PubMed] [Google Scholar]
- 33. Meier R, Muheim L, Senn O, Rosemann T, Chmiel C.. The impact of financial incentives to improve quality indicators in patients with diabetes in Swiss primary care: a protocol for a cluster randomised controlled trial. BMJ Open 2018; 8 (6): e023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pandya A, Asch DA, Volpp KG, et al. Cost-effectiveness of financial incentives for patients and physicians to manage low-density lipoprotein cholesterol levels. JAMA Netw Open 2018; 1 (5): e182008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.III RJJ. A comprehensive review of an electronic health record system soon to assume market ascendancy: EPIC®. J Healthcare Commun 2016; 1 (4): 9. [Google Scholar]
- 36. Bodagh N, Archbold RA, Weerackody R, et al. Feasibility of real-time capture of routine clinical data in the electronic health record: a hospital-based, observational service-evaluation study. BMJ Open 2018; 8 (3): e019790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Al Alawi S, Al Dhaheri A, Al Baloushi D, Al Dhaheri M, Prinsloo EA.. Physician user satisfaction with an electronic medical records system in primary healthcare centres in Al Ain: a qualitative study. BMJ Open 2014; 4 (11): e005569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pogue JM, Potoski BA, Postelnick M, et al. Bringing the “power” to Cerner’s PowerChart for antimicrobial stewardship. Clin Infect Dis 2014; 59 (3): 416–24. [DOI] [PubMed] [Google Scholar]
- 39.Solutions IV. ImproMed Veterinary Practice Management Software. Secondary ImproMed Veterinary Practice Management Software. 2019. https://www.impromed.com/impromed-veterinary-software. Accessed July 18, 2019.
- 40.Software ezyVet. ezyVet Features. Secondary ezyVet Features. 2019. https://www.ezyvet.com/features/. Accessed July 18, 2019.
- 41.IDEXX Laboratories I. Cornerstone Software—Features, Diagnostic Integration, Apps and Services. Secondary Cornerstone Software—Features, Diagnostic Integration, Apps and Services. 2019. https://www.idexx.com/en/veterinary/software-services/cornerstone-software/. Accessed July 12, 2019.
- 42.Covetrus. AVImark Software Features. Secondary AVImark Software Features. 2019. https://avimark.net/avimark-software/product-features/. Accessed August 5, 2019.
- 43. Krone LM, Brown CM, Lindenmayer JM.. Survey of electronic veterinary medical record adoption and use by independent small animal veterinary medical practices in Massachusetts. J Am Vet Med Assoc 2014; 245 (3): 324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fernandes AC, Cloete D, Broadbent MT, et al. Development and evaluation of a de-identification procedure for a case register sourced from mental health electronic records. BMC Med Inform Decis Mak 2013; 13 (1): 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kayaalp M, Browne A, Dodd Z, Sagan P, McDonald C.. Clinical Text De-Identification Research Bethesda, MD: The Lister Hill National Center for Biomedical Communications; 2013: 71.
- 46.American Veterinary Medical Association. Confidentiality of Veterinary Patient Records. Secondary Confidentiality of Veterinary Patient Records. 2019. https://www.avma.org/advocacy/state-local-issues/confidentiality-veterinary-patient-records. Accessed December 10, 2019.
- 47.American Veterinary Medical Association. Confidentiality of Veterinary Patient Records. Secondary Confidentiality of Veterinary Patient Records. 2019. https://www.avma.org/Advocacy/StateAndLocal/Pages/sr-confidentiality-patient-records.aspx. Accessed December 10, 2019.
- 48. Iyengar A, Kundu A, Pallis G.. Healthcare informatics and privacy. IEEE Internet Comput 2018; 22 (2): 29–31. [Google Scholar]
- 49. Bentley RT. Magnetic resonance imaging diagnosis of brain tumors in dogs. Vet J 2015; 205 (2): 204–16. [DOI] [PubMed] [Google Scholar]
- 50. Cook CR, Cook JL.. Diagnostic imaging of canine elbow dysplasia: a review. Vet Surg 2009; 38 (2): 144–53. [DOI] [PubMed] [Google Scholar]
- 51. LeBlanc AK, Daniel GB.. Advanced imaging for veterinary cancer patients. Vet Clin North Am Small Anim Pract 2007; 37 (6): 1059–77. [DOI] [PubMed] [Google Scholar]
- 52. Marino DJ, Loughin CA.. Diagnostic imaging of the canine stifle: a review. Vet Surg 2010; 39 (3): 284–95. [DOI] [PubMed] [Google Scholar]
- 53. Mattoon JS, Bryan JN.. The future of imaging in veterinary oncology: learning from human medicine. Vet J 2013; 197 (3): 541–52. [DOI] [PubMed] [Google Scholar]
- 54. Jones PH, Dawson S, Gaskell RM, et al. Surveillance of diarrhoea in small animal practice through the Small Animal Veterinary Surveillance Network (SAVSNET). Vet J 2014; 201 (3): 412–8. [DOI] [PubMed] [Google Scholar]
- 55. Radford AD, Noble PJ, Coyne KP, et al. Antibacterial prescribing patterns in small animal veterinary practice identified via SAVSNET: the small animal veterinary surveillance network. Vet Rec 2011; 169 (12): 310. [DOI] [PubMed] [Google Scholar]
- 56. Liu Y, Watson SC, Gettings JR, et al. A Bayesian spatio-temporal model for forecasting Anaplasma species seroprevalence in domestic dogs within the contiguous United States. PLoS One 2017; 12 (7): e0182028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Self SCW, McMahan CS, Brown DA, Lund RB, Gettings JR, Yabsley MJ.. A large-scale spatio-temporal binomial regression model for estimating seroprevalence trends. Environmetrics 2018; 29 (8): e2538. [Google Scholar]
- 58. Watson SC, Liu Y, Lund RB, et al. A Bayesian spatio-temporal model for forecasting the prevalence of antibodies to Borrelia burgdorferi, causative agent of Lyme disease, in domestic dogs within the contiguous United States. PLoS One 2017; 12 (5): e0174428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ostrander EA, Wayne RK.. The canine genome. Genome Res 2005; 15 (12): 1706–16. [DOI] [PubMed] [Google Scholar]
- 60. Spencer G. Researchers publish dog genome sequence: analysis sheds light on human disease; differences among canine breeds. NIH News; 2005: 1.
- 61. Wayne RK, Ostrander EA.. Out of the dog house: the emergence of the canine genome. Heredity (Edinb) 2004; 92 (4): 273–4. [DOI] [PubMed] [Google Scholar]
- 62. Wilbe M, Kozyrev SV, Farias FHG, et al. Multiple changes of gene expression and function reveal genomic and phenotypic complexity in SLE-like disease. PLoS Genet 2015; 11 (6): e1005248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bianchi M, Dahlgren S, Massey J, et al. A multi-breed genome-wide association analysis for canine hypothyroidism identifies a shared major risk locus on CFA12. PLoS One 2015; 10 (8): e0134720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Truvé K, Dickinson P, Xiong A, et al. Utilizing the dog genome in the search for novel candidate genes involved in glioma development—genome wide association mapping followed by targeted massive parallel sequencing identifies a strongly associated locus. PLoS Genet 2016; 12 (5): e1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dodman NH, Ginns EI, Shuster L, et al. Genomic risk for severe canine compulsive disorder, a dog model of human OCD. Int J Appl Res Vet Med 2016; 14 (1): 18. [Google Scholar]
- 66. Plassais J, Kim J, Davis BW, et al. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat Commun 2019; 10 (1): 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Science SSoV. Online Mendelian Inheritance in Animals, OMIA. World Wide Web, 2019.
- 68. Mellersh C. DNA testing and domestic dogs. Mamm Genome 2012; 23 (1–2): 109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Linde Forsberg C, Persson G.. A survey of dystocia in the Boxer breed. Acta Vet Scand 2007; 49 (1): 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS.. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand 2017; 59 (1): 71–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schiffman JD, Breen M.. Comparative oncology: what dogs and other species can teach us about humans with cancer. Philos Trans R Soc Lond B Biol Sci 2015; 370 (1673): 13. doi:10.1098/rstb.2014.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Slutsky J, Raj K, Yuhnke S, et al. A web resource on DNA tests for canine and feline hereditary diseases. Vet J 2013; 197 (2): 182–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ghosh M, Sharma N, Singh AK, Gera M, Pulicherla KK, Jeong DK.. Transformation of animal genomics by next-generation sequencing technologies: a decade of challenges and their impact on genetic architecture. Crit Rev Biotechnol 2018; 38 (8): 1157–75. [DOI] [PubMed] [Google Scholar]
- 74. Nunney L, Maley CC, Breen M, Hochberg ME, Schiffman JD.. Peto’s paradox and the promise of comparative oncology. Philos Trans R Soc Lond B Biol Sci 2015; 370 (1673): 8. doi: 10.1098/rstb.2014.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Research GV. Pet wearable market size, share & trends analysis report by technology (RFID, GPS, Sensors), by application (identification & tracking, medical diagnosis & treatment), and segment forecasts, 2019–2025. In: Laura Wood SPM, ed. Pet Wearable Market Size, Share & Trends Analysis Report. San Francisco, CA: Grand View Research; 2019: 120.
- 76. Hssayeni MD, Burack MA, Jimenez-Shahed J, Ghoraani B.. Assessment of response to medication in individuals with Parkinson’s disease. Med Eng Phys 2019; 67: 33–43. [DOI] [PubMed] [Google Scholar]
- 77. Dinh-Le C, Chuang R, Chokshi S, Mann D.. Wearable health technology and electronic health record integration: scoping review and future directions. JMIR Mhealth Uhealth 2019; 7 (9): e12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anholt RM, Berezowski J, Jamal I, Ribble C, Stephen C.. Mining free-text medical records for companion animal enteric syndrome surveillance. Prev Vet Med 2014; 113 (4): 417–22. [DOI] [PubMed] [Google Scholar]
- 79. Anholt RM, Berezowski J, Ribble CS, Russell ML, Stephen C.. Using informatics and the electronic medical record to describe antimicrobial use in the clinical management of diarrhea cases at 12 companion animal practices. PLoS One 2014; 9 (7): e103190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kass PH, Weng HY, Gaona MA, et al. Syndromic surveillance in companion animals utilizing electronic medical records data: development and proof of concept. PeerJ 2016; 4: e1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Norman GR, Monteiro SD, Sherbino J, Ilgen JS, Schmidt HG, Mamede S.. The causes of errors in clinical reasoning: cognitive biases, knowledge deficits, and dual process thinking. Acad Med 2017; 92 (1): 23–30. [DOI] [PubMed] [Google Scholar]
- 82. Jacob VS, Gaultney LD, Salvendy G.. Strategies and biases in human decision-making and their implications for expert systems. Behav Inform Technol 1986; 5 (2): 119–40. [Google Scholar]
- 83. Wencheng Sun ZC, Yangyang L, Liu F, Fang S, Wang G.. Data processing and text mining technologies on electronic medical records: a review. J Healthcare Eng 2018; 2018: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hadi W, Al-Radaideh QA, Alhawari S.. Integrating associative rule-based classification with Naïve Bayes for text classification. Appl Soft Comput 2018; 69: 344–56. [Google Scholar]
- 85. Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH.. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci 2018; 1 (1): 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Koopman B, Zuccon G, Nguyen A, Bergheim A, Grayson N.. Extracting cancer mortality statistics from death certificates: A hybrid machine learning and rule-based approach for common and rare cancers. Artif Intell Med 2018; 89: 1–9. [DOI] [PubMed] [Google Scholar]
- 87. Awaysheh A, Wilcke J, Elvinger F, Rees L, Fan W, Zimmerman KL.. Review of medical decision support and machine-learning methods. Vet Pathol 2019; 56 (4): 512–25. [DOI] [PubMed] [Google Scholar]
- 88. Velupillai S, Mowery D, South BR, Kvist M, Dalianis H.. Recent advances in clinical natural language processing in support of semantic analysis. Yearb Med Inform 2015; 10 (1): 183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Khachidze M, Tsintsadze M, Archuadze M.. Natural language processing based instrument for classification of free text medical records. Biomed Res Int 2016; 2016: 8313454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sun S, Luo C, Chen J.. A review of natural language processing techniques for opinion mining systems. Inform Fusion 2017; 36: 10–25. [Google Scholar]
- 91. Jurafsky D, Martin JH.. Speech and Language Processing: An Introduction to Natural Language Processing, Computational Linguistics, and Speech Recognition. 2nd ed.Upper Saddle River, NJ: Pearson Prentice Hall; 2009. [Google Scholar]
- 92. Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 93. Demner-Fushman D, Elhadad N.. Aspiring to unintended consequences of natural language processing: a review of recent developments in clinical and consumer-generated text processing. Yearb Med Inform 2016; (1): 224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pivovarov R, Perotte AJ, Grave E, Angiolillo J, Wiggins CH, Elhadad N.. Learning probabilistic phenotypes from heterogeneous EHR data. J Biomed Inform 2015; 58: 156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lam K, Parkin T, Riggs C, Morgan K.. Use of free text clinical records in identifying syndromes and analysing health data. Vet Rec 2007; 161 (16): 547–51. [DOI] [PubMed] [Google Scholar]
- 96.Extracting information about medication use from veterinary discussions. In: proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Denver, Colorado: Association for Computational Linguistics; 2015.
- 97. McGreevy P, Thomson P, Dhand NK, et al. VetCompass Australia: a national big data collection system for veterinary science. Animals (Basel) 2017; 7 (10): 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhang Y, Nie A, Zehnder A, Page RL, Zou J.. VetTag: improving automated veterinary diagnosis coding via large-scale language modeling. NPJ Digit Med 2019; 2 (1): 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digital Med 2018; 1 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gopalakrishnan V, Lustgarten JL, Visweswaran S, Cooper GF.. Bayesian rule learning for biomedical data mining. Bioinformatics 2010; 26 (5): 668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Beam AL, Kohane IS.. Big data and machine learning in health care. JAMA 2018; 319 (13): 1317–18. [DOI] [PubMed] [Google Scholar]
- 102. Kavuluru R, Rios A, Lu Y.. An empirical evaluation of supervised learning approaches in assigning diagnosis codes to electronic medical records. Artif Intell Med 2015; 65 (2): 155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Subotin M, Davis AR.. A method for modeling co-occurrence propensity of clinical codes with application to ICD-10-PCS auto-coding. J Am Med Inform Assoc 2016; 23 (5): 866–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lustgarten JL. Machine Learning with Supervised Discretization Aids ‘Omic’-Data Analysis 2006.
- 105. Lustgarten JL, Gopalakrishanan V, Malehorn D, Bigbee WL.. Assigning Putative Protein Identifications to Selected Lung Cancer Biomarkers from Surface-Enhanced Laser Desorption/Ionization Time-of-Flight Mass Spectrometry of Blood Serum Advancing Practice, Instruction, and Innovation Through Informatics (APIII 2008) Conference. Washington, DC: Archives of Pathology & Laboratory Medicine; 2008: 1148–65. [Google Scholar]
- 106. Lustgarten JL. A Bayesian Rule Generation Framework for ‘Omic’ Biomedical Data Analysis 2009.
- 107. Lustgarten JL, Balasubramanian JB, Visweswaran S, Gopalakrishnan V.. Learning parsimonious classification rules from gene expression data using Bayesian networks with local structure. Data (Basel) 2017; 2 (1): 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Banzato T, Bernardini M, Cherubini GB, Zotti A.. A methodological approach for deep learning to distinguish between meningiomas and gliomas on canine MR-images. BMC Vet Res 2018; 14 (1): 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Dorea FC, Muckle CA, Kelton D, et al. Exploratory analysis of methods for automated classification of laboratory test orders into syndromic groups in veterinary medicine. PLoS One 2013; 8 (3): e57334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ratner A, Bach SH, Ehrenberg H, Fries J, Wu S, Re C.. Snorkel: rapid training data creation with weak supervision. Proceedings VLDB Endowment 2017; 11 (3): 269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Dai Y, Wang G.. A deep inference learning framework for healthcare. Pattern Recognit Lett 2018; doi: 10.1016/j.patrec.2018.02.009. [Google Scholar]
- 112.Muniasamy A, Tabassam S, Hussain MA, Sultana H, Muniasamy V, Bhatnagar R. Deep Learning for Predictive Analytics in Healthcare. In: A H, A A, T G, R B, M FT, eds. The International Conference on Advanced Machine Learning Technologies and Applications (AMLTA2019). Cairo, Egypt: Springer International Publishing; 2019: 32–42. [Google Scholar]
- 113. Ngiam KY, Khor IW.. Big data and machine learning algorithms for health-care delivery. Lancet Oncol 2019; 20 (5): e262–73. [DOI] [PubMed] [Google Scholar]
- 114. Abdelaziz A, Elhoseny M, Salama AS, Riad AM.. A machine learning model for improving healthcare services on cloud computing environment. Measurement 2018; 119: 117–28. [Google Scholar]
- 115. Antropova N, Huynh B, Li H, Giger ML.. Breast lesion classification based on dynamic contrast-enhanced magnetic resonance images sequences with long short-term memory networks. J Med Imaging (Bellingham) 2019; 6 (1): 011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rodriguez P, Cucurull G, Gonalez J, et al. Deep pain: exploiting long short-term memory networks for facial expression classification. IEEE Transactions on Cybernetics; 2017;PP(99): 11; doi: 10.1109/TCYB.2017.2662199. [DOI] [PubMed] [Google Scholar]
- 117. Akata Z, Perronnin F, Harchaoui Z.. Good practice in large-scale learning for image classification. IEEE Trans Pattern Anal Mach Intell. 2014; 36 (3): 507–20. [DOI] [PubMed] [Google Scholar]
- 118. Kutlu H, Avci E.. A novel method for classifying liver and brain tumors using convolutional neural networks, discrete wavelet transform and long short-term memory networks. Sensors (Basel) 2019; 19 (9): 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yoon Y, Hwang T, Lee H.. Prediction of radiographic abnormalities by the use of bag-of-features and convolutional neural networks. Vet J 2018; 237: 43–48. [DOI] [PubMed] [Google Scholar]
- 120. Boone JM, Sigillito VG, Shaber GS.. Neural networks in radiology: an introduction and evaluation in a signal detection task. Med Phys 1990; 17 (2): 234–41. [DOI] [PubMed] [Google Scholar]
- 121. Wu Y, Giger ML, Doi K, Vyborny CJ, Schmidt RA, Metz CE.. Artificial neural networks in mammography: application to decision making in the diagnosis of breast cancer. Radiology 1993; 187 (1): 81–7. [DOI] [PubMed] [Google Scholar]
- 122. Forsstrom JJ, Dalton KJ.. Artificial neural networks for decision support in clinical medicine. Ann Med 1995; 27 (5): 509–17. [DOI] [PubMed] [Google Scholar]
- 123. Nakamura K, Yoshida H, Engelmann R, et al. Computerized analysis of the likelihood of malignancy in solitary pulmonary nodules with use of artificial neural networks. Radiology 2000; 214 (3): 823–30. [DOI] [PubMed] [Google Scholar]
- 124. Chen CM, Chou YH, Han KC, et al. Breast lesions on sonograms: computer-aided diagnosis with nearly setting-independent features and artificial neural networks. Radiology 2003; 226 (2): 504–14. [DOI] [PubMed] [Google Scholar]
- 125. Song RB, Vite CH, Bradley CW.. Postmortem evaluation of 435 cases of intracranial neoplasia in dogs and relationship of neoplasm with breed, age, and body weight. J Vet Intern Med 2013; 27 (5): 1143–52. [DOI] [PubMed] [Google Scholar]
- 126. Banzato T, Bonsembiante F, Aresu L.. Use of transfer learning to detect diffuse degenerative hepatic diseases from ultrasound images in dogs: a methodological study. Vet J 2018; 233: 35–40. [DOI] [PubMed] [Google Scholar]
- 127. Scotch M, Mattocks K, Rabinowitz P, Brandt C.. A qualitative study of state-level zoonotic disease surveillance in new England. Zoonoses Public Health 2011; 58 (2): 131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Scotch M, Rabinowitz P, Brandt C.. State-level zoonotic disease surveillance in the United States. Zoonoses Public Health 2011; 58 (8): 523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Meidenbauer KL. Animal surveillance: use of animal health data to improve global disease surveillance. Online J Public Health Inform 2017; 9 (1): e147. [Google Scholar]
- 130. Bowser NH, Anderson NE.. Dogs (Canis familiaris) as sentinels for human infectious disease and application to Canadian populations: a systematic review. Vet Sci 2018; 5 (4): 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zinsstag J, Schelling E, Roth F, Bonfoh B, de Savigny D, Tanner M.. Human benefits of animal interventions for zoonosis control. Emerg Infect Dis 2007; 13 (4): 527–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Neo JPS, Tan BH.. The use of animals as a surveillance tool for monitoring environmental health hazards, human health hazards and bioterrorism. Vet Microbiol 2017; 203: 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Pakhomov SV, Buntrock JD, Chute CG.. Automating the assignment of diagnosis codes to patient encounters using example-based and machine learning techniques. J Am Med Inform Assoc 2006; 13 (5): 516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Dhakal S, Burrer SL, Winston CA, Dey A, Ajani U, Groseclose SL.. Coding of electronic laboratory reports for biosurveillance, selected United States hospitals, 2011. Online J Public Health Inform 2015; 7 (2): e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lee DH, Lau FY, Quan H.. A method for encoding clinical datasets with SNOMED CT. BMC Med Inform Decis Mak 2010; 10 (1): 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lucero CA, Oda G, Cox K, et al. Enhanced health event detection and influenza surveillance using a joint Veterans Affairs and Department of Defense biosurveillance application. BMC Med Inform Decis Mak 2011; 11 (1): 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Centers for Disease Control and Prevention. Chronic Wasting Disease. Secondary Chronic Wasting Disease 11-15-2019. 2019. https://www.cdc.gov/prions/cwd/occurrence.html. Accessed December 23, 2019.
- 138. Osterholm MT, Anderson CJ, Zabel MD, Scheftel JM, Moore KA, Appleby BS.. Chronic wasting disease in cervids: implications for prion transmission to humans and other animal species. MBio 2019; 10 (4): e01091-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Doherr MG, Audige L.. Monitoring and surveillance for rare health-related events: a review from the veterinary perspective. Philos Trans R Soc Lond B Biol Sci 2001; 356 (1411): 1097–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Barrett D. The potential for big data in animal disease surveillance in Ireland. Front Vet Sci 2017; 4: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. McQuiston JH, Guerra MA, Watts MR, et al. Evidence of exposure to spotted fever group rickettsiae among Arizona dogs outside a previously documented outbreak area. Zoonoses Public Health 2011; 58 (2): 85–92. [DOI] [PubMed] [Google Scholar]
- 142. Merrill MM, Boughton RK, Lord CC, et al. Wild pigs as sentinels for hard ticks: a case study from south-central Florida. Int J Parasitol Parasites Wildl 2018; 7 (2): 161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rich KM, Denwood MJ, Stott AW, Mellor DJ, Reid SW, Gunn GJ.. Systems approaches to animal disease surveillance and resource allocation: methodological frameworks for behavioral analysis. PLoS One 2013; 8 (11): e82019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Van Metre DC, Barkey DQ, Salman MD, Morley PS.. Development of a syndromic surveillance system for detection of disease among livestock entering an auction market. J Am Vet Med Assoc 2009; 234 (5): 658–64. [DOI] [PubMed] [Google Scholar]
- 145. VanderWaal KL, Picasso C, Enns EA, et al. Network analysis of cattle movements in Uruguay: Quantifying heterogeneity for risk-based disease surveillance and control. Preventive Veterinary Medicine 2016; 123: 12–22. [DOI] [PubMed] [Google Scholar]
- 146. Yazdanbakhsh O, Zhou Y, Dick S.. An intelligent system for livestock disease surveillance. Information Sciences 2017; 378: 26–47. [Google Scholar]
- 147. Martin MK, Helm J, Patyk KA.. An approach for de-identification of point locations of livestock premises for further use in disease spread modeling. Prev Vet Med 2015; 120 (2): 131–40. [DOI] [PubMed] [Google Scholar]
- 148. Verraes C, Van Boxstael S, Van Meervenne E, et al. Antimicrobial resistance in the food chain: a review. Int J Environ Res Public Health 2013; 10 (7): 2643–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.World Health Organization. Antimicrobial Resistance in the Food Chain. Secondary Antimicrobial Resistance in the Food Chain. 2017. https://www.who.int/foodsafety/areas_work/antimicrobial-resistance/amrfoodchain/en/. Accessed May 20, 2019.
- 150. Marshall BM, Levy SB.. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 2011; 24 (4): 718–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Landers TF, Cohen B, Wittum TE, Larson EL.. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep 2012; 127 (1): 4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. McDermott PF, Zhao S, Wagner DD, Simjee S, Walker RD, White DG.. The food safety perspective of antibiotic resistance. Anim Biotechnol 2002; 13 (1): 71–84. [DOI] [PubMed] [Google Scholar]
- 153. Mathew AG, Cissell R, Liamthong S.. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis 2007; 4 (2): 115–33. [DOI] [PubMed] [Google Scholar]
- 154. Tellez G, Laukova A, Latorre JD, Hernandez-Velasco X, Hargis BM, Callaway T.. Food-producing animals and their health in relation to human health. Microb Ecol Health Dis 2015; 26: 25876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Parkunan T, Ashutosh M, Sukumar B, et al. Antibiotic resistance: a cross-sectional study on knowledge, attitude, and practices among veterinarians of Haryana state in India. Vet World 2019; 12 (2): 258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Van Boeckel TP, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA 2015; 112 (18): 5649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Thakur S, Gray GC.. The mandate for a global “One Health” approach to antimicrobial resistance surveillance. Am J Trop Med Hyg 2019; 100 (2): 227–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Gilbert M, Nicolas G, Cinardi G, et al. Global distribution data for cattle, buffaloes, horses, sheep, goats, pigs, chickens and ducks in 2010. Sci Data 2018; 5 (1): 180227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist 2018; 11: 1645–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Aenishaenslin C, Häsler B, Ravel A, Parmley J, Stärk K, Buckeridge D.. Evidence needed for antimicrobial resistance surveillance systems. Bull World Health Organ 2019; 97 (4): 283–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Peggy L, Schmidt DM.. Companion animals as sentinels for public health. Vet Clin North Am Small Anim Pract 2009; 39 (2): 241–500. [DOI] [PubMed] [Google Scholar]
- 162. Movalli P, Krone O, Osborn D, Pain D.. Monitoring contaminants, emerging infectious diseases and environmental change with raptors, and links to human health. Bird Study 2018; 65 (sup1): S96–109. [Google Scholar]
- 163. Ellwanger JH, Kaminski V. D L, Chies JAB.. Emerging infectious disease prevention: where should we invest our resources and efforts? J Infect Public Health 2019; 12 (3): 313–16. [DOI] [PubMed] [Google Scholar]
- 164. Bramble M, Morris D, Tolomeo P, Lautenbach E.. Potential role of pet animals in household transmission of methicillin-resistant Staphylococcus aureus: a narrative review. Vector Borne Zoonotic Dis 2011; 11 (6): 617–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Iverson SA, Brazil AM, Ferguson JM, et al. Anatomical patterns of colonization of pets with staphylococcal species in homes of people with methicillin-resistant Staphylococcus aureus (MRSA) skin or soft tissue infection (SSTI). Vet Microbiol 2015; 176 (1–2): 202–8. [DOI] [PubMed] [Google Scholar]
- 166. Morris DO, Lautenbach E, Zaoutis T, Leckerman K, Edelstein PH, Rankin SC.. Potential for pet animals to harbour methicillin-resistant Staphylococcus aureus when residing with human MRSA patients. Zoonoses Public Health 2012; 59 (4): 286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Nicholas B, Ravel A, Leighton P, et al. Foxes (Vulpes vulpes) as sentinels for parasitic zoonoses, Toxoplasma gondii and Trichinella nativa, in the northeastern Canadian Arctic. Int J Parasitol Parasites Wildl 2018; 7 (3): 391–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Tian H, Stenseth NC.. The ecological dynamics of hantavirus diseases: From environmental variability to disease prevention largely based on data from China. PLoS Negl Trop Dis 2019; 13 (2): e0006901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Sullivan T, Ichikawa O, Dudley J, Li L, Aberg J.. The rapid prediction of carbapenem resistance in patients with Klebsiella pneumoniae bacteremia using electronic medical record data. Open Forum Infect Dis 2018; 5 (5): ofy091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Pedersen T, Sekyere JO, Govinden U, et al. Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and pandrug-resistant clinical Enterobacteriaceae in Durban. South Africa 2018; 62 (5): e02178–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Katchanov J, Asar L, Klupp E-M, et al. Carbapenem-resistant Gram-negative pathogens in a German university medical center: prevalence, clinical implications and the role of novel β-lactam/β-lactamase inhibitor combinations. PLoS One 2018; 13 (4): e0195757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Garg A, Garg J, Kumar S, Bhattacharya A, Agarwal S, Upadhyay GC.. Molecular epidemiology & therapeutic options of carbapenem-resistant Gram-negative bacteria. Indian J Med Res 2019; 149 (2): 285–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Smith A, Wayne AS, Fellman CL, Rosenbaum MH.. Usage patterns of carbapenem antimicrobials in dogs and cats at a veterinary tertiary care hospital. J Vet Intern Med 2019; 33 (4): 1677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Fernández J, Guerra B, Rodicio MR.. Resistance to carbapenems in non-typhoidal Salmonella enterica serovars from humans, animals and food. Vet Sci 2018; 5 (2): E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Sellera FP, Fernandes MR, Ruiz R, et al. Identification of KPC-2-producing Escherichia coli in a companion animal: a new challenge for veterinary clinicians. J Antimicrob Chemother 2018; 73 (8): 2259–61. [DOI] [PubMed] [Google Scholar]
- 176. Grönthal T, Österblad M, Eklund M, et al. Sharing more than friendship—transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill 2018; 23 (27): 1700497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Gentilini F, Turba ME, Pasquali F, et al. Hospitalized pets as a source of carbapenem-resistance. Front Microbiol 2018; 9: 2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Mertz L. Machine learning takes on health care: Leonard D’Avolio’s Cyft employs big data to benefit patients and providers. IEEE Pulse 2018; 9 (1): 10–11. [DOI] [PubMed] [Google Scholar]
- 179. Bzdok D, Meyer-Lindenberg A.. Machine learning for precision psychiatry: opportunities and challenges. Biol Psychiatry Cogn Neurosci Neuroimaging 2018; 3 (3): 223–30. [DOI] [PubMed] [Google Scholar]
- 180. Thums M, Fernández-Gracia J, Sequeira AMM, Eguíluz VM, Duarte CM, Meekan MG.. How big data fast tracked human mobility research and the lessons for animal movement ecology. Front Mar Sci 2018; 5: 21. [Google Scholar]
- 181. Plekhanova E, Nuzhdin SV, Utkin LV, Samsonova MG.. Prediction of deleterious mutations in coding regions of mammals with transfer learning. Evol Appl 2019; 12 (1): 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Reif JS. Animal sentinels for environmental and public health. Public Health Rep 2011; 126 (Suppl 1): 50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Mackenzie JS, Jeggo M.. Reservoirs and vectors of emerging viruses. Curr Opin Virol 2013; 3 (2): 170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Sievers M, Hale R, Parris KM, Swearer SE.. Impacts of human-induced environmental change in wetlands on aquatic animals. Biol Rev 2018; 93 (1): 529–54. [DOI] [PubMed] [Google Scholar]
- 185. Blanco-Rayón E, Guilhermino L, Marigómez I, Izagirre U.. Collection and transport of sentinel mussels in biomarker-based coastal pollution monitoring: Current flaws and reliable practices. Ecological Indicators 2019; 103: 722–34. [Google Scholar]
- 186. Shen F, Zhao Y, Wang L, et al. Rare disease knowledge enrichment through a data-driven approach. BMC Med Inform Decis Mak 2019; 19 (1): 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



