Abstract
Nucleic acids play a central role in all domains of life, either as genetic blueprints or as regulators of various biochemical pathways. The chemical makeup of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA), generally represented by a sequence of four monomers, also provides precise instructions for folding and higher-order assembly of these biopolymers that, in turn, dictate biological functions. The sequence-based specific 3D structures of nucleic acids led to the development of the directed evolution of oligonucleotides, SELEX (systematic evolution of ligands by exponential enrichment), against a chosen target molecule. Among the variety of functions, selected oligonucleotides named aptamers also allow targeting of cell-specific receptors with antibody-like precision and can deliver functional RNAs without a transfection agent. The advancements in the field of customizable nucleic acid nanoparticles (NANPs) opened avenues for the design of nanoassemblies utilizing aptamers for triggering or blocking cell signaling pathways or using aptamer–receptor combinations to activate therapeutic functionalities. A recent selection of fluorescent aptamers enables real-time tracking of NANP formation and interactions. The aptamers are anticipated to contribute to the future development of technologies, enabling an efficient assembly of functional NANPs in mammalian cells or in vivo. These research topics are of top importance for the field of therapeutic nucleic acid nanotechnology with the promises to scale up mass production of NANPs suitable for biomedical applications, to control the intracellular organization of biological materials to enhance the efficiency of biochemical pathways, and to enhance the therapeutic potential of NANP-based therapeutics while minimizing undesired side effects and toxicities.
Keywords: SELEX, aptamers, NANPs, therapeutic nucleic acids, immunotherapy, exosomes, RNA nanotechnology, nucleic acid delivery
Graphical Abstract
In the last three decades, the continuous discovery of different classes of noncoding RNAs (ncRNAs) involved in many crucial cellular pathways and regulations tremendously boosted and expanded the field of RNA biology.1 In contrast to coding parts of mRNA, ncRNAs’ folding and overall resulting 3D structures determine their functions. Our increasing understanding of the roles that ncRNAs play in biology not only engenders intriguing questions about the extent of ncRNA importance for cellular life but also encourages the exploration of various therapeutic strategies where RNAs can be targets for therapies and also serve as therapeutics themselves.2
Through the simple interactions between two nucleic acid strands (RNA–RNA, DNA–RNA, DNA–DNA) that are dictated by the canonical Watson–Crick base-pairing rules, it is plausible to target aberrant RNA transcripts (e.g., mRNA) with high specificity. Subsequently, the targeted strand, in the form of duplex RNA–RNA or hybrid DNA–RNA, becomes diverted or completely removed from the original biological pathway. This approach, based on the complementarity between therapeutic and disease-specific nucleic acids, is promising for the treatment of a variety of diseases.3 Additionally, the interactions between therapeutic nucleic acids (TNAs) and disease-relevant proteins or metabolites are also of importance, whereas combining further two or more different TNAs in a single formulation represents a promising combinatorial strategy for synergistic therapeutics.4–6
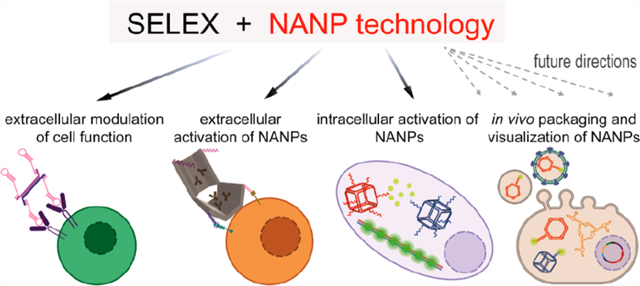
Aptamers are becoming a key component of TNA technology, facilitating cell-specific delivery or high-affinity recognition of target molecules. Aptamers are single-stranded oligonucleotides that can be considered nucleic acid analogues of monoclonal antibodies. The coupling of aptamers with multiple TNAs via assembly of programmable nucleic acid nanoparticles (NANPs) provides a modular platform to simultaneously target different biological pathways for enhanced therapeutic effects. In this review, we will discuss the selection of aptamers, their mechanisms of actions, therapeutic potential, and use as experimental tools to promote the field of therapeutic nucleic acid nanotechnology (Figure 1).
Figure 1.
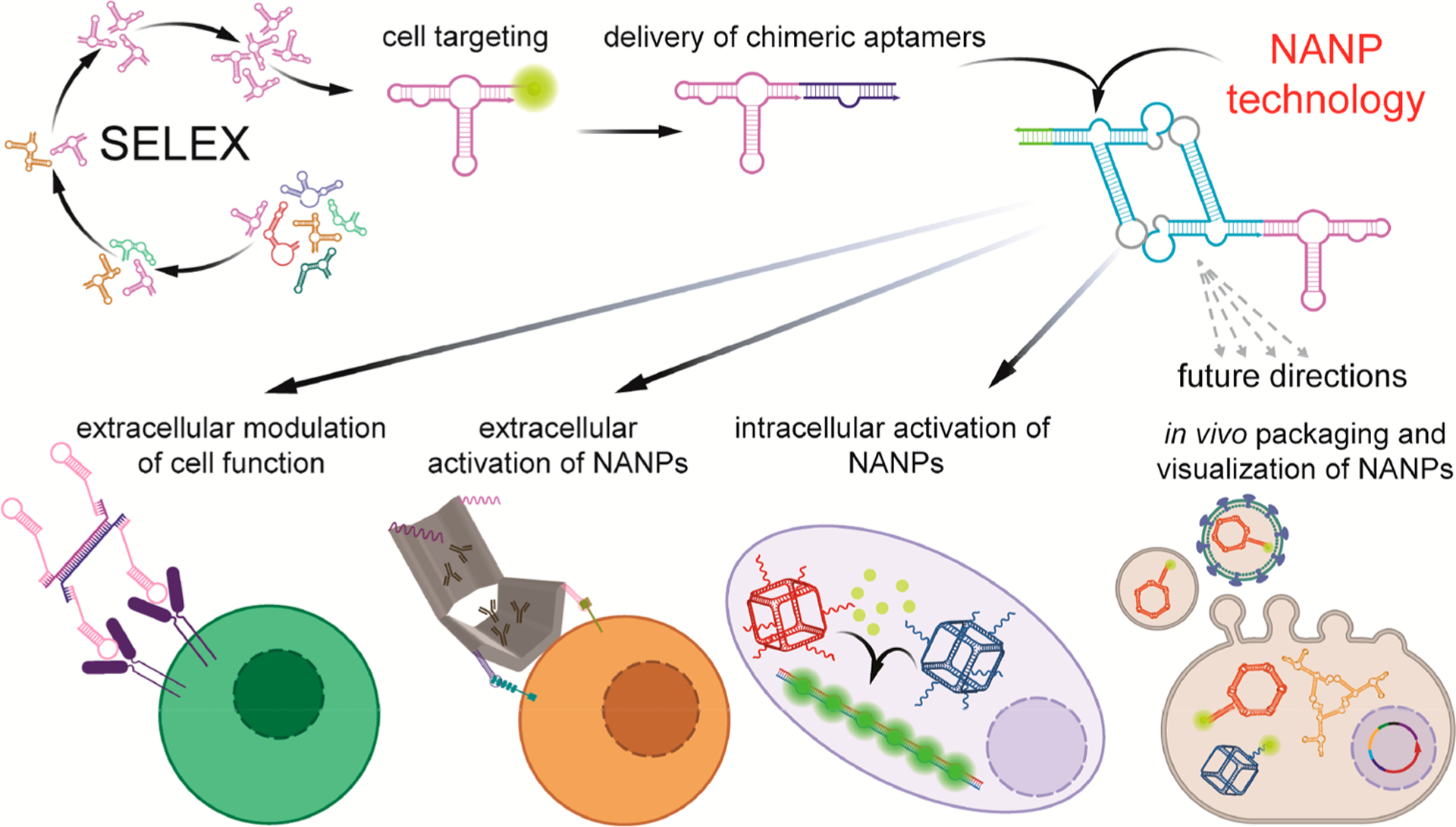
Schematic description of growing structural and functional complexity of aptamer involvements into nucleic acid nanotechnology. Fluorescently labeled aptamers that are specific to cell receptors can be used for cell detection. Their interactions with receptors often result in modulation of the receptor signaling. Later development led to the design of aptamer chimeras, where aptamers deliver the functional RNA or DNA moieties to target cells. Inclusion of aptamers to NANPs enhances the combinatorial applications of aptamers in changing cellular pathways and allowing for NANPs to logically respond to the presence of key triggers. In addition, light-up aptamers are potentially suitable reporters of NANP assembly or real-time monitoring of mutual interactions of NANPs in vivo. One of the future applications of aptamers in NANP technology could be a transport of NANPs to cell vesicles or viral vectors that would be mediated by aptamers targeted to vesicle- or virus-specific proteins.
Generation of Aptamers via SELEX (Systematic Evolution of Ligands by Exponential Enrichment). All single-stranded RNAs adopt more or less complex tertiary structures which interact with other cellular components and most importantly with proteins. Those interactions are either essential for RNA maturation or RNA itself assumes a crucial part of active RNA–protein complexes. Nucleic acids interact with proteins in varying degrees through physical forces, among which are electrostatic and hydrophobic interactions and hydrogen bonding. However, as not all proteins evolved to naturally interact with RNAs, a technique for the selection of specific RNA sequences that can adopt a particular tertiary structure which dictates its high binding affinity to a protein of interest was of great demand.
The in vitro selection process termed SELEX has been available since 1990, when two laboratories independently developed the same strategy, which is a bona fide directed evolution of oligonucleotides that leads to their recognition by a molecule of interest.7,8 During SELEX, a library of ~1012–1015 short (<100 nt) single-stranded randomized sequences of nucleic acids is subjected to iterative cycles of in vitro incubation with the target molecule, which ultimately leads to the isolation of just a few sequences termed “aptamers” that exhibit the highest affinity for the molecular target (Figure 2).7,8 Depending on the desired mechanism of action for the aptamers, a variety of SELEX methods have been developed.9 The spectrum of reported aptamer targets spans from small molecules, through proteins and viruses, up to individual bacterial or eukaryotic cells. Although it is important to understand that when work refers to aptamers selected against viruses and cells as targets, the aptamers still selectively bind to undetermined molecular complexes or biomolecules within the context of a cell or viral surface. By recent analysis of 1003 in vitro experiments, Dunn et al. found that the majority of aptamers are selected against proteins (584 aptamers), then small molecules (234 aptamers), and cells (141 aptamers). Only 22 aptamers were generated against viruses and 22 aptamers against other nucleic acids.10 Broad recognition of aptamers translates to broad therapeutic applications. The most widely used SELEX approaches are protein-based and cell-based, with some variations in experimental settings.9
Figure 2.
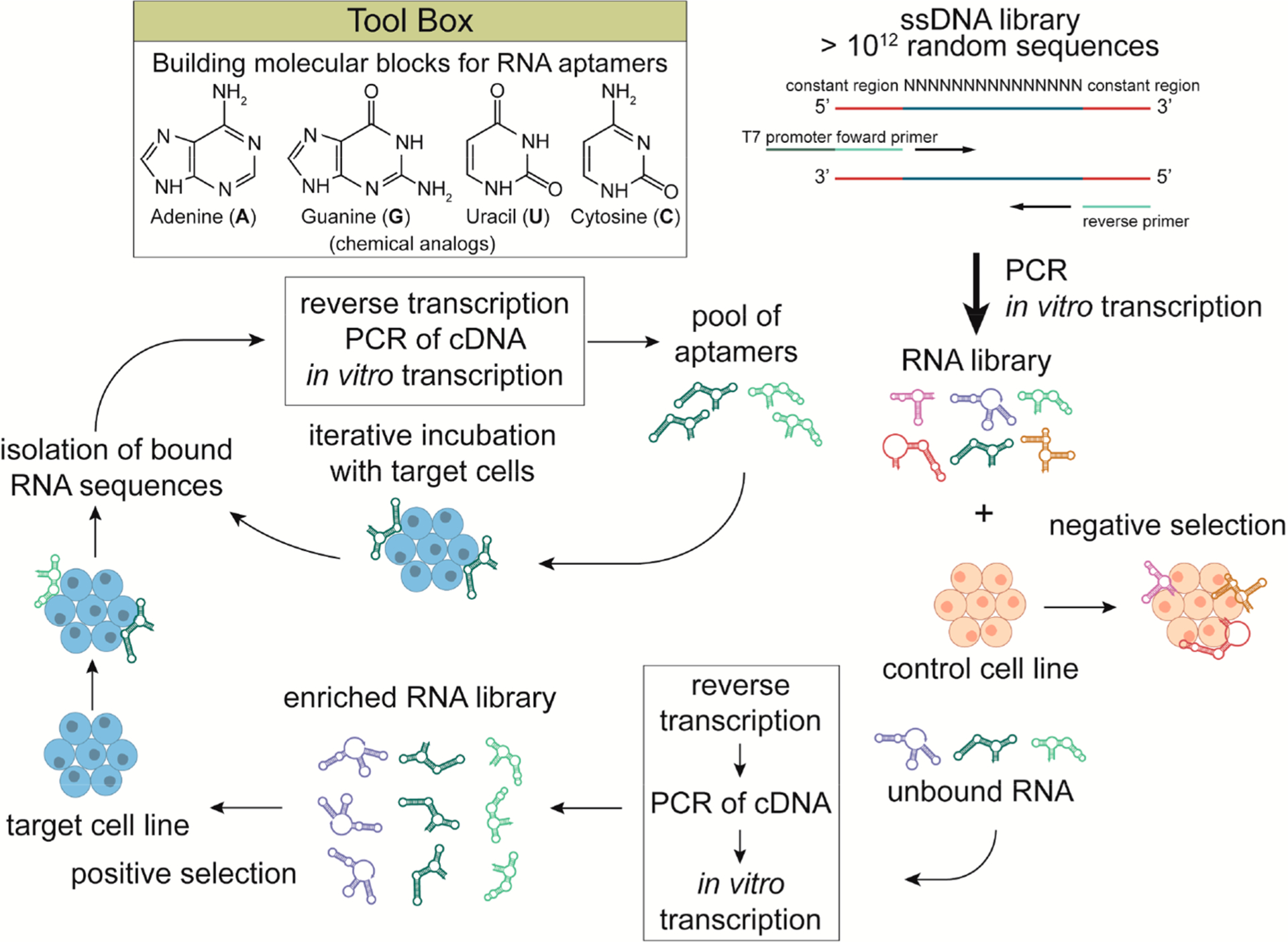
Schematic illustration of whole-cell SELEX. A library of commercially available ssDNAs is amplified by PCR and subsequently in vitro transcribed to an RNA library. This is possible due to the constant 5′ and 3′ sequences that are the same for each ssDNA and contain complementary sites for PCR as well as a T7 promoter for transcription. The variable body of aptamers that is unique for each strand is located between common 5′ and 3′ sequences required for PCR amplification. In the first step, the RNA library is incubated with the control cell population that does not express target receptors. In the next step, the unbound sequences are recovered and reverse transcribed to cDNA that is amplified by PCR. The subsequent in vitro transcribed RNA library is enriched with sequences with low or no affinity to the control cell line. The library is then incubated with target cells, unbound RNA is washed out, and bound strands are isolated and again reamplified. In vitro transcribed RNA in this step is used for the next round of selection. With each cycle, specific aptamers should prevail in the reamplified population. The final sequence(s) with the highest affinity is identified by sequencing analysis. RNA structures can be visualized using the NUPACK Web Application.20–27
Protein-based SELEX consists of an initial subtraction step aiming to remove nucleic acid sequences with significant affinity to the separation platform or background used during selection (e.g., magnetic particles, nitrocellulose filter, or chip-based supports). Subsequently, the recovered unbound oligonucleotides are incubated with the target molecule, against which SELEX is performed. After the unbound oligonucleotides are washed out, the bound population is recovered and amplified for the next selection round.9
Recently, another variation of SELEX (based on multiplexed poly target selection) against a panel of primate lentiviral reverse transcriptases (RTs) utilizing libraries pre-enriched against a single RT resulted in the identification of a broad spectrum of aptamers. Through this process, researchers identified additional, rare, and known motifs able to suppress, to varying degrees, the replication of viral constructs expressing phylogenetically diverse RTs. Therefore, this modified protein-based SELEX approach is advantageous for selecting cross-reactive aptamers.11
In cell SELEX, the target for aptamer selection is a specific cell subpopulation (Figure 2). Again, the first step is the negative selection achieved by incubation of the nucleic acid library with cells that do not express the target antigen (known or putative). Next, the pool of recovered unbound sequences is incubated with cells expressing the cell surface antigen of interest. The recovered target-bound sequences are isolated, reamplified, and used for the next round of incubation with cells.12 Cell SELEX has the primary advantage of selecting aptamers against receptors in the natural biological environment of the cell membrane. Additionally, a modified version of the described technology, termed cell internalization SELEX, allows the selection of aptamers that bind cell surface antigens and are rapidly internalized into the cellular interior under physiological conditions.13 The cell internalization SELEX method is ideal for selecting the aptamers that can deliver therapeutic cargo with intracellular functions into the target cells.
Despite the several unquestionable advantages of using living cells in SELEX, manipulation with cells can change their physiology and ultimately lead to cell death, which is associated with unspecific uptake that would lead to enrichment with false specific sequences.14 Similarly, modifications of the oligonucleotide backbone may affect the binding of oligonucleotides.15
Though the principle of SELEX is simple, the process is complex with specific technological requirements for individual steps as well as opportunities to advance.16 Since the SELEX discovery, many SELEX protocols were or are being developed with the aim to increase the specificity and affinity while maintaining minimum costs and time. Future studies will show if one or many approaches from different protocols such as in vivo SELEX, cross-over, toggle, chimerix SELEX, etc. will become the preferred choice of aptamer selection, though the intended application of an aptamer may determine the most suitable approach for its selection.17 Furthermore, there is a need to establish minimal sets of controls for the selection and functional description of aptamers.
Aptamers versus Antibodies.
Aptamers can be considered as nucleic acid analogues of monoclonal antibodies, although aptamer targets do not have to be immunogenic. These properties make aptamers very promising candidates for targeted therapy. In fact, the first aptamer-based therapy, pegaptanib (Macugen), was approved by the U.S. Food and Drug Administration (FDA) in 2004 for the treatment of age-related macular degeneration.18 Despite its initial success, Macugen was outcompeted by monoclonal antibody fragments, ranibizumab (Lucentis) and bevacizumab (Avastin), crediting a higher effectivity and lower cost, respectively.19 Although antibodies are more developed and understood, synthetic aptamers have several advantages in comparison to antibodies such as time and control over production together with the flexibility of designs, which is probably one of the most important benefits of aptamer technology. Whereas antibody design and manufacturing can take months and requires a biological system with intrinsic stochasticity, selection of aptamers is carried out within a few weeks under precisely controlled experimental conditions and can be potentially automated. The further production of selected aptamers can be entirely synthetic, which minimizes batch-to-batch variability and can be scaled up. Another important advantage of aptamers over antibodies is their great shelf life due to the heat stability and ability of renaturation. With these factors, transportation and storage of aptamers does not require cold-chain. Furthermore, from a biomedical point of view, aptamers are much less immunogenic than antibodies.
However, one primary concern for the clinical use of RNA aptamers is their sensitivity to nuclease degradation and, therefore, short half-life in vivo. The phosphodiester backbone and the 5′ and 3′ termini are extremely sensitive to serum ribonucleases and exonucleases, respectively. This sensitivity would effectively prevent any systemic application of therapeutic aptamers and allow just their local and/or temporary use; however, substitution of natural ribonucleo-tides with chemically modified analogues during or after SELEX increases RNA aptamers’ chemical stability and prolongs their bioavailability.28 Additionally, locked nucleic acids (LNAs), modifications of phosphodiester bonds, and polyethylene glycol (PEG) can be incorporated to stabilize aptamer structures and prolong circulation times.28,29 Collectively, this greatly expands the therapeutic potential of RNA aptamers.
In comparison to antibodies, the relatively lower affinity of aptamers to their targets is caused by a limited number of natural building blocks where four nucleotides with similar physicochemical properties create less diverse chemical motifs than 20 amino acids with a broader range of physicochemical properties, thus providing a higher heterogeneity of interactions. The advancements in synthetic biology provide us with toolkits in molecular biology reactions to expand the genetic alphabet with unnatural forms of DNA such as artificially expanded genetic information systems (AEGISs) for its use in aptamer selection. The artificial bases base pair by hydrogen bonding or via shape complementarity.30–32 The application of unnatural bases in NANP technology can potentially boost the field of nucleic acid nanotechnology and lead to NANP structures with antibody-like avidity to antigens, while preserving nucleic acid advantages.
Organization of Aptamers into Complex Structures.
Aptamers as a stand-alone molecule can inhibit (antagonize) or activate (function as agonists) target receptors in diverse conditions. Linking together aptamers that bind to disease-specific receptors and TNAs designed to silence the gene of interest led to the development of so-called aptamer chimera technology.33 Generally, the term aptamer chimera refers to an aptamer fused with another functional oligonucleotide. Since their first description in 2006, chimeric aptamers have been shown as a vital combination for receptor-specific delivery of functionally diverse nucleic-acid-based molecules (e.g., siRNAs, anti-miRs, decoys, DNAzymes) (Figure 3A).34 Besides the assistance in TNA intracellular localization, aptamers can also accommodate the delivery of chemotherapeutics and bioactive peptides.35,36 Aptamers may be connected to functional TNAs in several ways: (i) complementary single-stranded extension,37 (ii) sticky bridge,4 (iii) carbon linker,4 (iv) disulfide linker,36 or (v) by incorporation of aptamers directly within the functional structure38 (Figure 3B).
Figure 3.
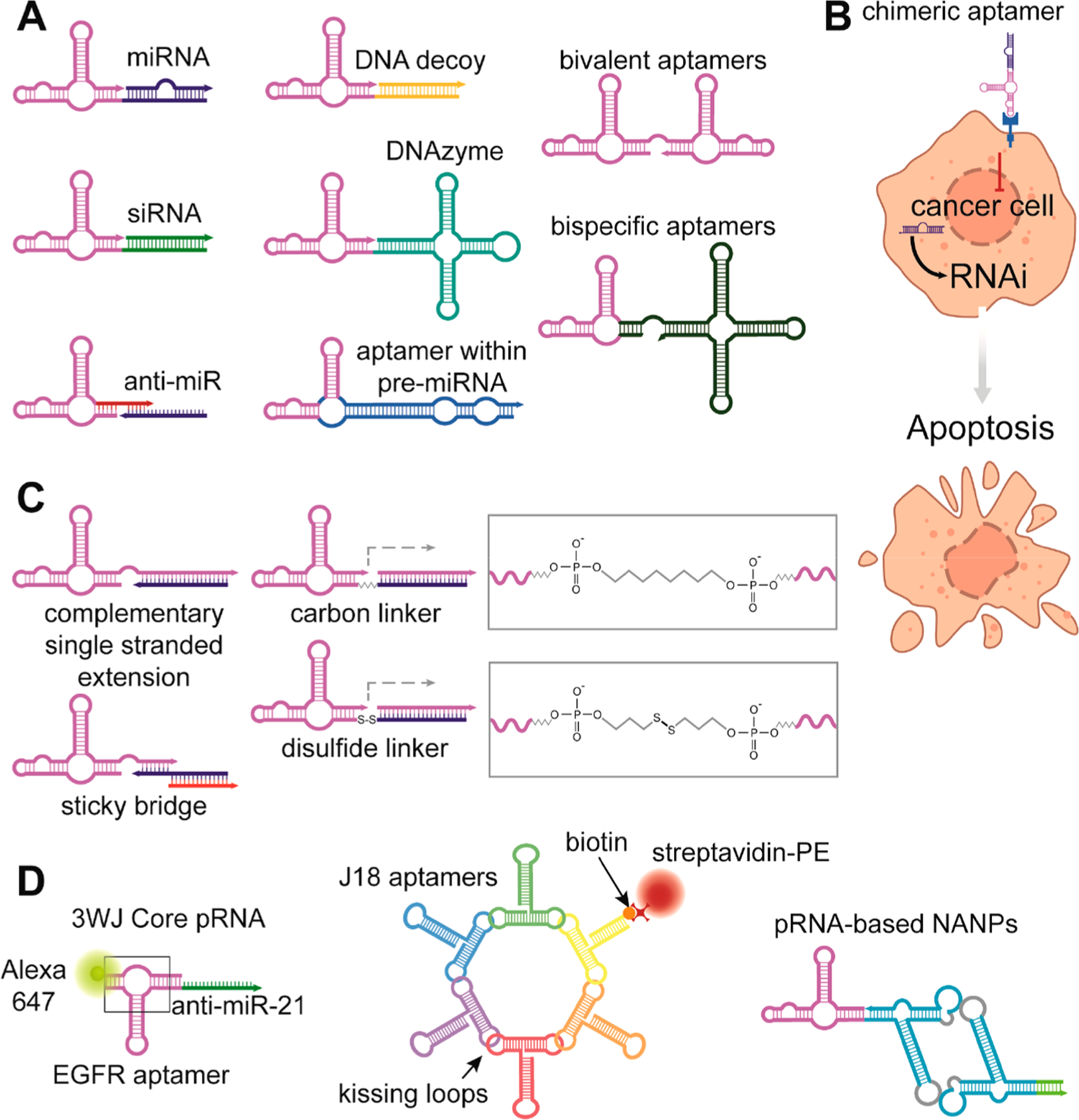
Schematic depiction of various chimeric aptamers. Multiple nucleic-acid-based functionalities can be linked to cell-specific aptamers with many different approaches that are beyond the scope of the article. (A) Post-transcriptional silencing of gene expression is achieved by delivery of miRNA or siRNA. Transcription of aptamers within pre-miRNA from gene constructs offers prolonged production of chimeric RNA. Downregulation of some genes by endogenous miRNAs during tumorigenesis can be reverted by the delivery of anti-miRs that block binding of miRNAs to target mRNAs. Delivery of a DNAzyme that cleaves specific mRNA is another way to repress gene expression. Conjugation of two identical aptamers is used for oligomerization of receptors in comparison with two different aptamers that may attach to cell surface proteins from intercellular space or the bloodstream as well as interconnect two different cells. (B) Schematic illustration of cell targeting by chimeric aptamer with a synergistic effect. An aptamer blocks signaling, while the therapeutic payload silences genes crucial for cell survival. Thus, simultaneously both functional parts promote apoptosis. (C) Most utilized ways of chimeric TNA conjugations. (D) From left to right: 3WJ-EGFR aptamer/anti-miR-21 nanoparticles harboring three functional modules: EGFR RNA aptamer for targeted delivery, anti-miR-21 LNA for therapy, and Alexa-647 dye for imaging. The RNA nanoring carrying five J18 aptamers for cell targeting, connected to the RNA ring. One biotinylated oligonucleotide provides fluorescent readout after coupling to a streptavidin–phycoerythrin conjugate. The 3WJ pRNA motif can be used to multiply assemble scaffolds.
The next logical step in the development of complex TNAs was the design of NANPs to accommodate multiple functionalities within the same complex. NANPs are rationally designed nanoscaffolds assembled through canonical and noncanonical base pairings.39–61 The multifunctional versatility of NANPs can be easily expanded by various functional DNA and RNA moieties such as aptamers, miRNAs, siRNAs ribozymes, and decoy oligonucleotides as well as other chemical components such as fluorophores, chemotherapeutics, toxins, and peptides, all introduced in a precisely controlled fashion (Figure 3C). Furthermore, aptamers embedded in rationally designed nanoscaffolds can enhance the interaction between the cells.62 Additionally, in vivo assembled NANPs promise to scale up mass production of the nanoparticles for biomedical applications and to control the intracellular organization of biological materials with the aim to enhance the efficiency of biochemical pathways.63–65
Mechanisms of Action for Stand-Alone Therapeutic Aptamers.
Antagonistic aptamers appear to have a dominant role in therapeutic research. Currently, all aptamers in clinical trials belong to this category.66 The benefits of repressing the function of disease-associated receptors have been shown in many human diseases such as cancer,67 ocular and bleeding disorders,68,69 cardiovascular disease,70,71 autoimmunity,72,73 degenerative neurological disease, and infectious disease.74 An aptamer binding a pathological cell receptor can block the interaction of the receptor with its ligand, resulting in disruption of downstream signaling. This antagonistic function has a direct therapeutic effect by itself. Aptamers can function as antagonists by targeting either the receptor or the ligand.75,76
Manipulation of interplay between the tumor and immune cells has potential to stimulate the immune defense against carcinogenesis. The immune system is regulated by an intricate network of molecular interactions that maintain self-tolerance and balance the immune response to keep damage in unintended targets at minimal levels while encountering infectious agents. Erroneous gene expression and regulation are inherent to cancer cells. The negligent control mechanisms in malignant cells thus provide a vast spectrum of antigens that can be recognizable by immune cells. However, cancer cells can effectively intervene against the attack of immune cells by inhibiting co-stimulatory pathways and stimulating restriction signaling. The therapeutic blockade of these interactions or immune checkpoints can be game-changing treatments in cancer therapy (Figure 3A).77 The importance of this strategy gained momentum in 2018 by awarding a shared Nobel prize to James P. Allison and Tasuku Honjo for their work focused on “…stimulating the inherent ability of our immune system to attack tumor cells… where they established an entirely different principle for cancer therapy”.78
One of the first studies that demonstrated aptamers manipulating the immune cells in vitro and in vivo targeted murine CTLA-4 (Figure 4A,B). The binding of the aptamer to T cells in vitro enhanced T cell proliferation and inhibited tumor growth in vivo. The study also showed that the bioactivity of this aptamer can be increased by the generation of a tetravalent form. Enhanced bioactivity in vivo was likely due to the higher avidity of the aptamer complex to CTLA-4. This improvement led to the decrease of the dose requirement.79 Additionally, the immunosuppressive tumor microenvironment is established by activating co-inhibitory receptors on infiltrating effector T cells by the cell surface expression of co-inhibitory ligands (i.e., PD-L1) within the tumor microenvironment. One of the essential immune inhibitory pathways involves the PD-1–PD-L1 interaction (Figure 4A,B).80 Recently, a DNA aptamer MP7 specific to PD-1 blocked interaction with PD-L1 in primary T cells. To increase the MP7 half-life in vivo, the 5′ end of the aptamer was linked to polyethylene glycol (PEG). PEGylated MP7 restored, in a murine model of colon carcinoma, antitumor immune responses and suppressed the tumor growth with a potency comparable to that of an antagonistic anti-PD-1 antibody.75
Figure 4.
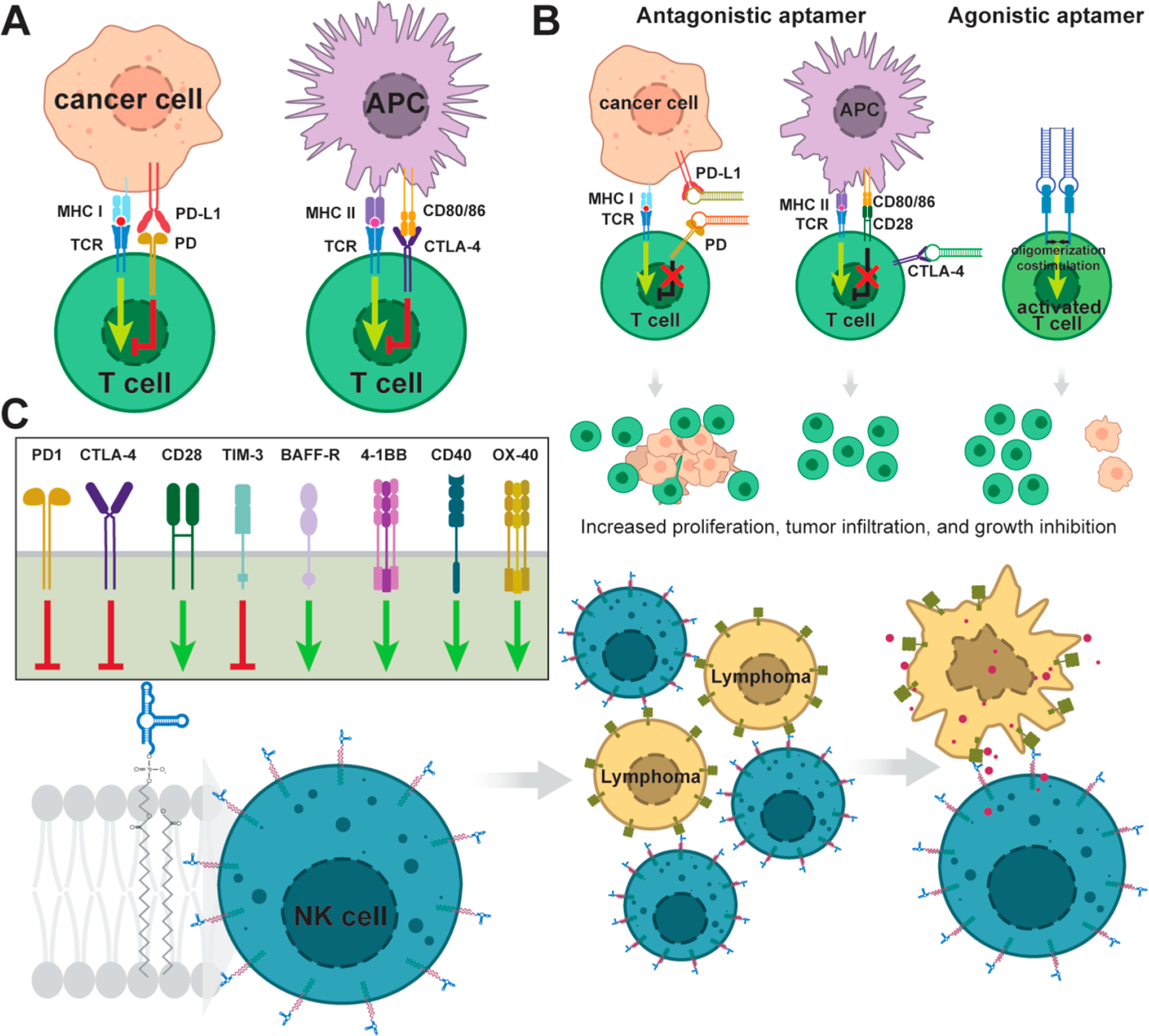
Illustration of aptamer involvement in cancer immunotherapy. (A) Two most crucial (most known) immune checkpoints. (B) Antagonistic aptamers, selected against either receptor or ligand, inhibit their interaction. Therefore, the downstream pathway is not triggered. Agonistic aptamers dimerize receptor dimers that result in switching on respective signaling. (C) Selected immune system receptors that have already been targeted by aptamers. (D) Whereas standard approaches use aptamers as targeting agents to deliver therapeutic cargo to cells, aptamer-engineered NK cells are directed by cell-membrane-embedded aptamers to lymphoma cells.
Second, aptamers can function as agonists to dimerize receptors, resulting in stimulation of downstream signaling (Figure 4B). Stimulation of receptors by agonist aptamers shows promising results especially in the field of cancer immunotherapy and antiviral treatment. The lack of or weak immune response to tumor cells is one of the hallmarks of anticancer therapy that results from the absence of co-stimulatory ligands able to activate cell surface receptors by oligomerization. In an effort to cross-link relevant receptors, many agonistic aptamers were designed in the form of bivalent molecules. Efficient initiation of signaling cascades by cross-linking requires the rational design of the linker that connects monomeric aptamers with respect to the distance between target domains, orientation, and rotational freedom of aptamers.
This was observed when the bivalent but not the monovalent form of the 4–1BB RNA aptamer co-stimulated CD8+ (cytotoxic) T cell activation by binding the 4–1BB major co-stimulatory receptor in vitro and resulted in tumor rejection in a murine model. The correct orientation and pairing of the two 4–1BB aptamers were achieved through complementary 3′ side 21 nt long extensions, creating a double-stranded linker. The distance was assessed based on the approximate length between the two variable domains of the 4–1BB-specific antibody.81 A DNA scaffold composed from two 20 bp long oligonucleotides connected with an 18 carbon polyethylene spacer, which provides flexibility to the assembled complex, represents another option to connect two OX40 RNA aptamers through 3′ side extensions base pairing to the DNA scaffold.82
Interestingly, dual functionality was shown in 2′-fluoropyrimidine-modified RNA aptamers selected to target CD28, a protein required for co-stimulation in T cell activation. From two selected aptamers in monomeric forms, one (CD28Apt2) had an antagonistic effect, preventing the co-stimulatory signal, whereas the second aptamer (CD28Apt7) upon binding was neutral. Surprisingly, after assembly of aptamers into the bivalent form, both functioned as an agonist triggering co-stimulatory signal. Dimeric aptamers were engineered by two methods using a 21 bp double-stranded linker or by fusing both aptamer strands into one linear sequence and transcribing the dimer as a single RNA strand. The single molecule CD28Apt7 dimer had the strongest co-stimulatory effect, even higher than that of a CD28-specific antibody.83
The SELEX process can also result in several high-affinity aptamers, and each aptamer can trigger a different outcome in cell signaling. This situation allows for evolving different therapeutic concepts. For example, an agonist aptamer for dimeric CD40, needed for activation of antigen-presenting cells, was shown to improve the recovery from bone marrow aplasia, which can occur in oncological patients upon chemotherapy or radiotherapy treatment. The recovery is not compromised even when agonist CD40 aptamer is used as a chimera with SMG-1-shRNA in B-cell lymphoma where shRNA activates RNA interference (RNAi) to specifically silence nonsense-mediated mRNA decay. Inhibition results in enhanced tumor antigenicity and higher tumor infiltration by lymphocytes. Similar overall mice survival was achieved by application of stand-alone CD40 antagonistic aptamer.84
To date, several aptamers have been selected against receptors involved in immune signaling (Figure 4C).85 Aptamers as nucleic-acid-based ligands of various membrane proteins involved in immune cross-talk can stimulate a patient’s immune system against malignant cells. Interestingly, by embedding aptamers into the membranes of immune cells, they can serve as nucleic acid receptors and target them to cancer cells. This strategy is relatively straightforward and can represent an alternative to chimeric antigen receptor therapy that relies on genetically engineered T or NK cells (Figure 4E).86–88
In addition to the cancer therapeutics discussed above, aptamer agonists can be applied to the activation of cellular defenses to viral infections as many viruses have developed different molecular strategies to evade or suppress the immune system.89 The retinoic acid inducible gene I (RIG-I) is a pattern recognition cytoplasmic receptor that detects nonself RNAs. RIG-I receptors recognize RNAs of evolutionarily distinct viruses and mediate immune responses against infections through IFNα/β production.90 Although located in the cytosol, RIG-I receptors may present a target for RIG-I agonist aptamers (Cl9) to block viral infection especially in nonimmune cells with restricted expression patterns of Toll-like receptors (TLR).91 Targeting additional receptors including but not limited to TLR 3, TLR 7/8, TLR9, MDA5, STING, and cGAS has the potential to identify therapeutic aptamers for treatment of pathogen infections. Previous investigations also indicate adjuvants targeting nucleic acid sensors activate humoral and cellular immune responses.92 As such, aptamers targeting nucleic acid sensors would be strong adjuvant candidates.
The potential application of an agonist RIG-I aptamer in cancer therapy is suggested by observations of immune response to tumor cells through RIG-I activation.93–95 The absence of RIG-I in human tumor cells increases their resistance to irradiation and chemotherapeutics. Radiation induces translocation of small nuclear RNAs U1 and U2 to the cytoplasm, where they form a complex with RIG-I and trigger downstream signaling, leading to activation of interferon genes. Transcription of IFN genes can sensitize cancer cells and mobilize immune cells to the tumor microenvironment.93 Despite the effectivity of cancer immunotherapies, some tumors do not react to this treatment; however, they can be reverted by induction of RIG-I that can activate tumor cell death.94 Furthermore, in hepatocellular carcinoma, upregulated RIG-I expression blocked the proliferation and spread of cancer cells.95
Although bivalent aptamers have the ability to dimerize targeted receptors (Figure 2), conjugation of two different aptamers into bispecific constructs is a strategy to direct cell–cell interactions, such as to cross-link tumor cells with immune cells and to activate the cognate receptors.62,83 In a similar strategy, a bispecific aptamer targeting the abundant tumor stromal protein, VEGF, on one side and 4–1BB cellular receptor on the other side enhanced therapeutic indexes in several unrelated tumors in a murine model.96
Generally, selecting aptamers against infectious agents has better potential to replace antibody-based applications including pathogen diagnosis, vaccine potency testing, and research. The selection of aptamer-discriminating pathogens or toxins may be less prone to isolating false positive sequences than cell SELEX. Currently, several bacterial, viral, and parasitic pathogens were subjected for aptamer selection to detect or inhibit infection.73,97,98 An extensive aptamer library for identification of specific pathogens as well as a means to discriminate pathogens expressing or lacking key virulence factors could greatly improve the current diagnostic process and therapy. Given the physicochemical and economic advantages of aptamers over antibodies, the development of treatment and diagnosis against infectious organisms based on aptamers may prove as viable in less developed countries.
Chimeric Aptamers To Deliver TNAs.
The conjugation of aptamers and other functional oligonucleotides into one chimeric construct is a way to specifically deliver TNAs without a need for a transfection reagent. The majority if not all of chimeric constructs are designed to affect post-transcriptional regulation upon binding to the specific receptor and successful trafficking to the cytoplasm.
Knocking down the expression of certain genes is an intensely researched approach in the context of nucleic-acid-based therapies. The very first FDA approval of RNAi therapy (ONPATTRO) as a treatment option against polyneuropathy in patients with hereditary transthyretin-mediated amyloidosis makes all relevant research of timely importance.99 Depending on the type of linked interfering RNAs, it becomes possible to silence either just one specific gene (by siRNA) or repress an expression of multiple genes simultaneously (by miRNA). The first proof-of-concept for in vivo (in a xenograft model) targeted delivery of therapeutic miRNA relied on antagonistic aptamer GL21.T targeting Axl, a receptor tyrosine kinase (an oncogene overexpressed in several human cancers) and tumor suppressor let-7g miRNA. Synergistic chimeras downregulated let-7g target genes in tumor cells, leading to apoptosis, decreased cell proliferation, and a reduction in tumor size. The versatility of this approach was confirmed by using alternative linking strategies and a combination of diverse aptamers and miRNAs.37
Given the importance of RNAi in control of gene expression, it becomes evident that, during malignant transformation, certain miRNAs participate in aberrant post-transcriptional regulation. Increases in the expression of the target gene repressed by the aberrant miRNA can be restored by outcompeting mRNA with synthetic miRNA antagonistic oligonucleotides (anti-miR), which mimic fragments of target mRNAs and base pair with corresponding miRNAs. A double-stranded miRNA–anti-miR complex, therefore, does not bind target mRNAs.100,101 The first study reporting aptamer–anti-miR chimeras delivered a phosphorothiolated anti-miR LNA targeting the miR-21 linked to the nucleolin-binding DNA aptamer (AS1411) at the 3′ end with three thymidine nucleotides.102
Inhibition of one miRNA type (miR-222) by anti-miR can be mediated by aptamers selected to target different receptors (Axl or PDGFRβ). Interestingly, no increase in antagonizing potential was observed when two identical anti-miR-222s connected in tandem were co-delivered, but the tandem of two different anti-miRs (anti-miR-222 and anti-miR-10b) reduced respective miRNAs and increased target protein levels.103 The additive inhibitory effect on tumors can be achieved by combined co-treatment with two independent aptamer chimeras targeting two independent tumor-specific receptors (Axl and PDGFRβ). While one chimera silences target mRNAs (GL21.T-miR 137), the other upregulates expression by inhibiting miRNA (Gint4.T-anti-miR-10b).104
Additionally, aptamers in conjunction with TNAs can be used to enhance antiviral responses. Every phase of the viral life cycle depends on the host cell. Aptamer-mediated blocking of either a cellular receptor or a viral glycoprotein is a viable strategy to prevent the infection of cells.105,106 Delivery of aptamer–siRNA chimeras via cellular receptor or viral envelope proteins on the cell surface may enhance antiviral effects through downregulation of viral and/or virus-required host genes. Together with aptamers targeting proteins and enzymes crucial for virus replication, we can combat virus infection throughout the whole viral replication cycle.107 Furthermore, the simultaneous silencing of both the virus and cellular transcripts by a cocktail of several aptamer chimeras has the potential to prevent the formation of viral escape mutants. The feasibility of this idea has been tested in the experimental model of humanized mice by targeting the expression of HIV-1 viral proteins tat and rev and host CD4 receptor that is required for HIV-1 entry as well as transportin 3 (TNPO3) necessary for viral integration.4 In a follow-up study, an RNA aptamer (G-3) targeting the human CCR5 protein alone decreased R5 HIV-1 virus replication regardless of the application prior or postinfection. Enhanced suppression of HIV-1 replication was also observed in human primary PBMCs treated with G-3-TNPO3 siRNA chimera.5
Recently, a strategy for the stable expression of an aptamer targeting HIV integrase and reverse transcriptase was introduced. The long-term inhibition of HIV replication in a cell culture system relies on the cellular transcription of aptamers incorporated into the terminal loop of an shRNA targeting the tat–rev region.38 Regarding cellular expression of aptamers, the flanking sequences of the transcript are important for proper folding. However, on one side, these sequences can abrogate functional conformations of an aptamer, but on the other side, proper design and experimental validation of aptamer adjacent sequences can stabilize transcripts and increase the resistance to degradation.108,109
How to prevent viral infections is a primary concern. If the integrity of the human body is not compromised by wounds, bites, etc., the most probable routes for infection are accessible tissues, that is, upper respiratory tract, lower airways, or reproductive organs. Application of a formulation of aerosol containing therapeutic aptamers or a gel with aptamers or aptamer–siRNA chimeras might protect against viral infections. In that context, interesting results were observed in a humanized murine model where the application of hydroxyethyl cellulose gel formulated with CD4+ (T helper cells)-specific aptamers linked to siRNAs silenced either the host CCR5 gene or the viral gag–vif sequences in order to prevent HIV infection up to 4 days before intravaginal exposure to the virus. In the following 10 weeks postinfection, no p24 antigen or viral RNA was detected in animal plasma, and the number of CD4+ cells remained unchanged.6
In addition to aptamer delivery of oligonucleotides affecting ncRNAs involved in RNAi, there are minor but inspirational nucleic-acid-based constructs including DNAzymes, splice-switching oligonucleotides (SSOs), and decoy oligonucleotides that allow destruction or alternative splicing of target mRNA or inhibit transcription factors, respectively. DNAzymes are artificial single-stranded DNAs selected to catalyze specific chemical reactions such as RNA ligation, DNA phosphorylation, or cleavage of RNA including specific mRNAs.110 The first attempt at employing aptamer-mediated (AS1411-surviving mRNA specific DNAzyme) delivery of DNAzyme resulted in inconclusive results.111 Rather than in targeted therapy, aptamer–DNAzyme chimeras can potentially find some applications in biosensing.112
Splice-switching oligonucleotides are single-stranded oligonucleotides that target nuclear pre-mRNA molecules to alter mRNA splicing, resulting in alternative protein isoforms.113 A prerequisite of successful therapy is the conjugation of SSOs to an aptamer whose target protein (e.g., nucleolin) is translocated to the nucleus. It seems that even a small percentage of repaired pre-mRNA becomes sufficient to generate phenotypic effects.114
Another rapidly developing class of TNAs are decoy oligonucleotides that are double-stranded DNAs with a specific or consensus transcription factor binding sequence. Decoy oligonucleotides bind to a corresponding transcription factor in order to inhibit binding to its specific target genome sequences present in promoters and enhancers, thus attenuating transcription of corresponding genes.115 Delivery of a decoy mimicking the NF-κB consensus promoter sequence by an RNA aptamer specific to human transferrin receptor in pancreatic carcinoma MIA PaCa-2 cells reduced translocation of NF-κB from the cytoplasm to the nucleus. A synergistic effect was achieved by elongation of the aptamer with a doxorubicin-binding short GC-rich double-stranded DNA tail that hybridized to an anti-tail sequence conjugated through a disulfide linker to an NF-κB self-complementary decoy oligonucleotide.36 Most of the stand-alone aptamers can function just by binding to their receptors on the cell surface. However, for chimeric aptamers carrying TNA, rapid internalization and release to the cytoplasm are crucial processes. Although many aptamers that efficiently delivered TNA into the cytoplasm were described, the process of their passage through the membrane without help from external carriers remains elusive and is represented with a complex set of limitations.116 Therefore, studies requiring high-resolution microscopy together with functional tests are necessary to elucidate chimeric aptamer trafficking.
Aptamers as Modular Components of Nucleic Acid Nanoparticles.
At the end of the 1990s and the beginning of this millennium, several pioneering studies introduced the notion of the programmable design of RNA nanoparticles assembled using long-range RNA tetraloop-receptor interacting motifs117–121 or structural RNA motifs derived from the DNA-packaging motor of bacteriophage phi29122 that can serve as a building block to engineer NANPs via a bottom-up assembly123,124 and implement therapeutic aptamers embedded into their structures. In the latter case, the independent folding of several domains is the main hallmark of packaging RNA (pRNA) monomers for their further controlled assembly.41 The 5′ and 3′ ends of pRNA located in the double-stranded helical domain can be replaced with aptamers or other TNAs without affecting the pRNA structure. This spatial arrangement has been proven, in multiple in vitro or in vivo studies, as an effective vector of diverse functional RNAs. In one of the first experiments, the pRNA sequence was fused with a hammerhead ribozyme cleaving hepatitis B virus polyA mRNA or U7 snRNA-targeting ribozyme in vitro or in cell culture.125
The ability of dimerization mediated by interlocking domains leads to the assembly of NANPs with gene- and cell-targeting properties. CD4-specific aptamer or folate on one subunit delivered to targeted cells can introduce an interlocked subunit carrying siRNA against the survivin or luciferase genes. Ex vivo treatment of nasopharyngeal epidermal carcinoma cells with dimers containing folate and siRNA–survivin suppressed tumor formation in athymic nude mice after the axillary injection of targeted cells.126 Engineering the central domain of bacteriophage phi29 pRNA forms a stable X-motif that can display up to four functional RNA oligonucleotides such as a malachite green aptamer, folate displaying sequence, luciferase siRNA, and survivin siRNA.127 Similarly, dimeric pRNA with the FB4 aptamer directed against the mouse transferrin receptor delivered siRNA targeting intercellular adhesion molecule-1 (ICAM-1) mRNA. Uptake of chimeric dimers in murine brain-derived endothelial cells stimulated by TNF-α or under oxygen-glucose deprivation/reoxygenation, an in vitro inflammatory cell model, blocked the increase of ICAM-1 expression and inhibited the adhesion of monocytes. The treatment has the potential to inhibit inflammation in ischemic strokes and other neuroinflammatory diseases.128
Generalized use of 3WJ motifs of the phi29 DNA packaging motor allows different options for bottom-up self-assembly of simple or more complex multifunctional RNA NANPs carrying aptamers in their structures.129 For example, the pRNA-3WJ core was adapted to display EGFR aptamers for specific targeting and delivery of anti-miR-21 upon systemic injection in orthotopic TNBC tumor-bearing mice (Figure 2C). The knockdown of miR-21 upregulated PTEN and PDCD4, leading to the inhibition of tumor growth. In vivo biodistribution of RNA nanoparticles was tumor specific with negligible or no accumulation in healthy organs and tissues.130 In a similar study, Endo28 aptamers incorporated to the core of pRNA-3WJs targeted annexin A2, which is expressed in the vasculature of ovarian tumors. The doxorubicin intercalated into the pRNA-3WJ scaffold with high loading efficiency and then selectively entered annexin A2 positive cells through receptor-mediated endocytosis. The polyvalency of the pRNA-3WJ scaffold allowed for an imaging probe Alexa 647 for tracking. Doxorubicin associated with pRNA scaffolds is slowly released, which is advantageous upon systematic application.131
Computer-assisted rational design tremendously expands the possibilities for generating versatile NANPs, thus creating an alternative approach to pRNA-based designs.61,132–134 The compact multifunctional RNA NANPs designed in silico,135 extensively tested in vitro,136–138 and then demonstrated in animal models60 have the capacity to simultaneously carry on a combination of several aptamers (Figure 5B) and/or RNAi inducers designed to simultaneously silence up to six different genes required, for example, in HIV-1 replication.60 Recently, the simple modification of NANP structures allowed for doubling the number of functionalities per the same construct.139 The exceptional diversity of functional NANPs is achieved by base pairing the TNAs directly to the NANP strands, and as the study has also shown, the use of biotinylated sequences allows the direct coupling of NANPs to streptavidin-conjugated proteins. In that work, the J18 RNA aptamer with affinity to EGFR provided specificity to target A431 cells,140 but the intracellular trafficking of the particles remains to be elucidated (Figure 3C).60 The flow cytometry data, however, suggest that the increase in number of aptamers over three per single NANP had no dramatic increase in specific cell binding. This means that the remaining positions can be used for displaying other functional moieties. Additionally, some recent data suggest that aptamer-mediated delivery of naked, large, functional RNA NANPs leads to the internalization of payloads in a nondisrupted state for more than 2 h.141 Perseverance of NANPs in correct folding gives an opportunity to couple NANPs with fusogenic peptides or proteins that can facilitate the endosome escape of NANPs. Similarly, as for chimeric aptamers, the release of NANPs from endosomes currently represents one of the main obstacles in the application of naked NANPs in vivo. It would be interesting if we could engineer nucleic acid structures that would respond to acidic environment analogically as certain viral fusogenic proteins.142 Whereas RNA structures dynamically responding to changes in pH have been already described,143 such effort would probably require the application of unnatural bases. Ultimately, such features could lead to development of multifunctional NANPs synthesized purely from nucleic acids (Figure 5).
Figure 5.
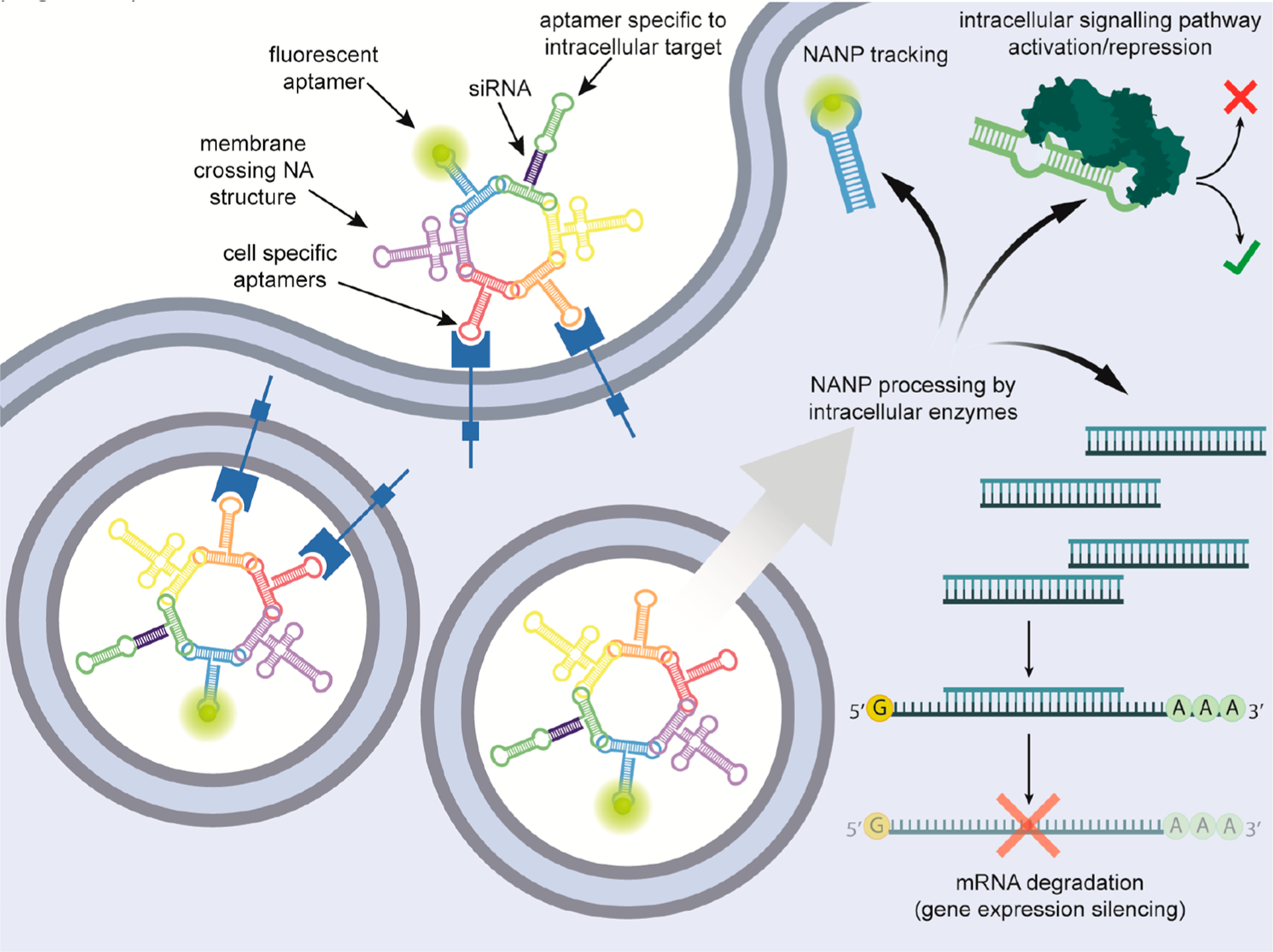
Vision of uptake of multifunctional NANPs composed purely from nucleic acids (NA). Cell-specific aptamers facilitate targeted delivery of NANPs. Fluorescent aptamers would provide tracking of NANPs from the cell surface, through the endosome compartment, to cytoplasm. Membrane crossing NAs, currently hypothetical in structure, promote escape of NANPs to the cytoplasm. Subsequently, intracellular enzyme machineries would “liberate” functional parts—siRNAs and aptamers against intracellular targets—resulting in a synergistic effect.
Use of Aptamers in Nanorobot Construction.
Besides NANP technology, significant progress has been demonstrated in the experimental application of targeted DNA origamis. DNA origami is a groundbreaking technology used to create customizable DNA assemblies by folding multi-kilobase ssDNA via interactions with computationally designed short “staple” ssDNAs.144
The RNA NANPs upon specific cell targeting mostly rely on cellular uptake and endosomal escape to exert their function. Although this is true also for many reported NANPs delivered by aptamers, DNA origami-based structures promise a cell-specific delivery and logical release of therapeutic cargo without entering the diseased cell. The following works are illustrative of the potential of aptamers for the creation of nanorobots combining diagnostics with therapeutics that are based on the aptamer logic-gated checks minimalizing off-target effects. The logical trigger is activated only in the presence of cellular receptors that are recognized by a pair of NANP-associated aptamers. The first DNA origami-based logic-gated nanorobot in the form of a hexagonal barrel consisting of two halves linked together by single-stranded scaffold hinges was locked by two DNA aptamers (Figure 4D).145,146 The aptamers base pair to partially complementary strands attached to the opposite half-barrel domains. In the inactive state, therapeutic cargo is hidden inside the barrel and the functional payload is exposed and activated only when both aptamers simultaneously recognize target proteins (protein keys), which will cause dissociation of aptamer lock duplexes. The process operates equivalently to a logic AND gate. The specificity of the aptamer-encoded logic gating strategy has been proven by six different robots displaying pairwise combinations of aptamer locks from a set of three aptamer sequences: 41t, TE17, and sgc8c. Several experiments tested the functionality and robustness of nanorobots. For example, the fluorescent signal was emitted from fluorescently labeled Fab′ fragments only after an aptamer pair recognized two protein keys on the cell surface. Nanorobots with specific aptamer pairs were able to distinguish matching antigens on target NKL cells in a mixture of 106 receptor-negative cells. Furthermore, nanorobots loaded with a combination of an antibody to human CD33 and antibody to human CDw328 Fab′ fragments induced growth arrest in target leukemic cells. The result showed an induced growth arrest in NKL cells in a dose-dependent fashion. Finally, logic-gated nanorobots induced T cell activation in response to human CD3e Fab′ and flagellin Fab′ antibodies.
Recently, another study tested the feasibility of logic-gated nanorobots targeting various tumors in vivo. In the open state DNA, the origami nanorobot adopts sheet conformation, whereas the closed particle has a cylindrical shape. The tube structure encapsulates four blood coagulation factor thrombin molecules, effectively hiding them from circulating platelets and plasma fibrinogen. The closed inactive state is ensured by several fastener two-strand sequences containing duplex fibers and modified DNA aptamers (AS1411) targeting the solid tumor-specific molecular marker nucleolin. Four AS1411 aptamers were located also at both ends of the tube. The robustness and functionality were tested in a mouse model of breast and lung cancer or melanoma as well as in Bama miniature pigs. As hypothesized, the nanorobots accumulated at the tumor where binding of aptamer fasteners to nucleolin unlocked the nanotubes. Thus, exposed thrombin triggered the formation of intravascular blood clots specifically at the tumor site. Clogged vessels resulted finally in tumor infarction.147
Currently, there is no known nanorobot similar to the described nanorobot made from RNA. This is partly due to the nonexistence of large-scale productions of RNA via synthetic or biological processes which we use for DNA.148
Light-up Aptamers as Trackers of NANP Assembly.
Efficient assembly of functional RNA NANPs in mammalian and bacterial cells is a top priority in the rapidly evolving field of RNA nanotechnology. Establishing a model for RNA NANPs’ assembly in vivo can enhance the therapeutic potential of RNA-based therapeutics while minimizing their undesired side effects and toxicities.39 To date, several natural fluorescent proteins were discovered and even more were derived from them, but no natural or rationally engineered RNA molecule with intrinsic fluorescence is known. Therefore, tracking of RNA or monitoring intracellular RNA NANPs’ assembly is challenging with limited tools available.
One of the most promising strategies would be to employ RNA aptamers that emit a fluorescent signal upon binding a fluorophore.149 The biggest advantage of light-up RNA aptamers is their ability to serve as genetically encoded tags. RNA aptamer–fluorophore pairs should feature five essential properties: (i) it should be as bright as possible to be sensitive up to one molecule resolution. Therefore, the dye should have an elevated absorption coefficient (ε), and its quantum yield should be close to 1. (ii) Fluorogen in its free form should display the lowest possible quantum yield to minimize background fluorescence and avoid any nonspecific activation by any biological molecules. (iii) The aptamer–fluorogen interaction should be highly specific, so the assay will not interfere with the other compounds. (iv) Moreover, the interaction should occur with a high affinity in order to use a low concentration of the dye to keep the fluorescence background low.150 (v) Finally, the RNA aptamer plus fluorophore should be photostable to allow for prolonged data acquisition.
Various types of fluorogens are used for attachment to the RNA aptamers: environment-sensitive fluorogens, molecular rotor fluorogens, and quenched fluorogens. Environmental dyes include Hoechst 33258, which binds directly to DNA in the minor groove at AT-rich regions. However, the nonspecific DNA binding capacity of these dyes can lead to significant unwanted background fluorescence when used in the cell-based assay. This can be suppressed by creating chemical derivatives from initial dyes no longer capable of nonspecific DNA binding, but preserving their fluorogenic capacity and becoming fluorescent only upon specific interaction with DNA and RNA aptamers.151,152 Fluorogenicity of molecular rotors is the result of intramolecular movements allowing for non-radiative relaxation of the dye upon excitation. They are poorly fluorescent in their unbound form in a fluid environment. However, fluorescence can be restored by restricting intramolecular movements either by strongly increasing medium viscosity or upon specific interaction with a nucleic acid. Examples of such molecular rotors are malachite green, patent blue,153,154 thiazole orange,155,156 and others. Quenched fluorogens are dyes obtained by appending a quenching group to a fluorescent organic dye. The fluorescence of the dye is quenched by a photoinduced electron transfer,157 Förster resonance energy transfer,158 or contact-mediated quenching.159 In these approaches, fluorescence of the free dye is quenched and emitted only upon binding to an aptamer.
Recently, another generation of RNA aptamers mimicking the green fluorescent protein has been generated and their potential tested in vitro and in vivo. Spinach, the first member of GFP-like aptamers, was selected to bind and activate the GFP-derived chromophore (Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-1,2-dimethyl-1H-imidazol-5(4H)-one (DFHBI). The brightness of the Spinach aptamer reaches approximately 50% of GFP, and its transcription in fusion with 5S rRNA was successfully visualized in HEK293T cells.160 In a follow-up study in Escherichia coli, authors developed RNA sensors for small metabolites such as adenosine, ADP, SAM, guanine, or guanosine 5′-triphosphate (GTP). The detection system consisted of small-molecule-binding aptamers linked to Spinach. Binding of small metabolites induced binding of Spinach to DFHBI, leading to fluorescence.161 Using a similar setup allows for detection of small proteins.162 Later, the authors rationally designed, by systematic mutagenesis, a Spinach2 aptamer that has enhanced thermostability and brightness compared to that of Spinach.163 Structural studies elucidated the molecular basis of Spinach fluorescence and found structure-driven design and tuning of fluorescent RNAs exemplified by an optimized miniaturized version of Spinach, called Baby Spinach.164
Even with extensive optimization, it is challenging to select aptamers intended for in vivo applications using in vitro SELEX. The cellular interior presents several obstacles for efficient function of in vitro selected aptamers, including but not limited to RNA degradation and incorrect folding. To comply with intracellular conditions, Jaffrey’s group developed a modified SELEX protocol generating RNA–fluorophore complexes adapted to the cellular environment. The protocol begins with standard SELEX using DFHBI bound to a sepharose matrix for RNA aptamer selection. Reverse transcribed aptamers were then cloned into a plasmid fused with tRNALys. After induction of transcription in transformed E. coli cells and incubation with a fluorophore, cells expressing the aptamer with the highest fluorescence can be sorted with flow cytometry. The selected aptamer (a 49 nt long Broccoli) was less dependent on magnesium ions and exhibited higher thermostability than Spinach or Spinach2. In addition, Broccoli green fluorescence is brighter upon binding DFHBI or its improved version (Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-1-(2,2,2-trifluoroethyl)-1H-imidazol-5(4H)-one) (DFHBI-1T).165,166 So-called directed viral evolution is another application that is based on selection in a cellular environment, thereby resulting in enhanced brightness of the Spinach2 aptamer. Improved Spinach2 was subsequently used for virus tracking.167
Proper folding and stability of synthetic RNA expressed in cells is ensured by a fusion of RNA with the natural scaffold. Characterization of Broccoli stability in the context of various scaffolds showed that tRNA scaffold, and not Broccoli, triggers RNA cleavage both in bacterial and mammalian cells. In E. coli cells, using additional scaffolds—the Vibrio proteolyticus 5S rRNA scaffold (V5) and the F29 RNA three-way junction motif (F29) —showed reduced cleavage of both scaffolds in comparison to the tRNA scaffold. Interestingly, in mammalian HEK293T cells, V5-Broccoli was expressed as the full-length transcript, but two bands were observed for F29-Broccoli. The deeper analysis found that the F29 scaffold contains a sequence similar to the transcription terminator. Despite the presence of intact V5-Broccoli transcript in-gel analysis, the fluorescent signal in mammalian cells was outcompeted by F29-Broccoli. Re-engineering of F29 resulted in an F30 scaffold that, together with Broccoli, is transcribed as a single strand adopting a conformation with robust fluorescence. Furthermore, both F30 arms can incorporate at least two additional Broccoli aptamers, which increases the fluorescence of the construct.168
One of the main drawbacks of aptamers binding DFHBI is their rapid photobleaching, which makes quantitative analysis impossible. Therefore, for monitoring of Pol III transcription dynamics, another RNA aptamer, Corn, with markedly improved photostability was selected. Corn selectively binds 3,5-difluoro-4-hydroxybenzylideneimidazolinone-2-oxime (DFHO), a chromophore found in the red fluorescent protein, and induces its fluorescence in the yellow spectrum.169 Even though Corn folding is not dependent on magnesium ions, it is dependent on potassium ions. In a solution, this aptamer unexpectedly functions as a homodimer with the quasi-symmetric structure of the ligand-binding site. That means that even though the two units exhibit an identical primary structure, there are certain minor differences in conformations. In this case, the three adenosines that are at the DFHO-binding site adopt different conformations in each subunit. Interestingly, there is no intermolecular Watson–Crick base pairing at the interface of dimers.170
Since the selection of Spinach, the trend leads to the reduction of the sequence length from 98 nt in Spinach, 95 nt Spinach2, and 49 nt for Broccoli to 36 nt for the Corn monomer or 25 nt for Mango. The decrease in size can potentially make shorter aptamers more favorable for fusion with transcripts of interest or incorporation into RNA nanoparticles as the smaller structure has a lower probability to interfere with the rest of the tagged molecule.
The conundrum of real-time visualization of RNA NANP co-transcriptional assembly in vivo may be resolved by incorporation of fluorogenic split aptamers. The strategy of using the split aptamer system is based on the physical separation of the aptamer strand into two separate parts. When both nonfunctional halves reassociate together, the functional aptamer is formed, which is detected by a fluorescent signal.171 Currently, experimental work has not utilized fluorescent split aptamers to monitor the assembly of RNA NANPs in vivo. However, co-transcriptional assembly of RNA NANPs was successfully confirmed in vitro using the split aptamer approach.172
The ncRNAs predominantly exist as single-stranded molecules, but in comparison with coding RNA (e.g., mRNA or viral RNA+ genome) or DNA, where carrying information encoded by their primary sequence is their functionality, ncRNAs’ functionality originates from their 3D shape. How RNA folds and what processes influence it remains at the forefront of RNA biology. Therefore, a model system such as split Spinach or split Broccoli represents an important tool for monitoring the dynamics of RNA reassembly in vitro.173–175
Besides studying the conditions for the reassociation of RNA, a fluorescent aptamer can be used to detect nucleic acid sequences.176 As a first intracellular E. coli “split Broccoli” RNA system working as a stand-alone AND logic gate and as a device fused to an RNA toehold switch upstream of a fluorescent protein, split Broccoli fluorescence was observed ahead of the red mCherry translation.177 The self-assembling hexameric nanorings represent a widely studied model of RNA NANP.137,138 Fully assembled nanorings consist of six dumbbell-like units connected by RNA kissing loop–loop interactions.137 Formation of the functional light-up split aptamer confirmed the assembly of nanorings, whereas the maximum fluorescence occurred only when a fully assembled ring was formed. As split strands of aptamers were linked only to two monomers that adopt, in the hexamer, an opposite position, the reassembly of Spinach has the ability to detect/ confirm interactions of units that are not directly base-paired to an aptamer (Figure 6A).178 To prove that light-up aptamers can track the co-transcriptional assembly of RNA nanoparticles (cubes), which is a situation resembling the endogenous transcription, Afonin et al. embedded a split MG-binding aptamer into two strands (Figure 6B).172
Figure 6.
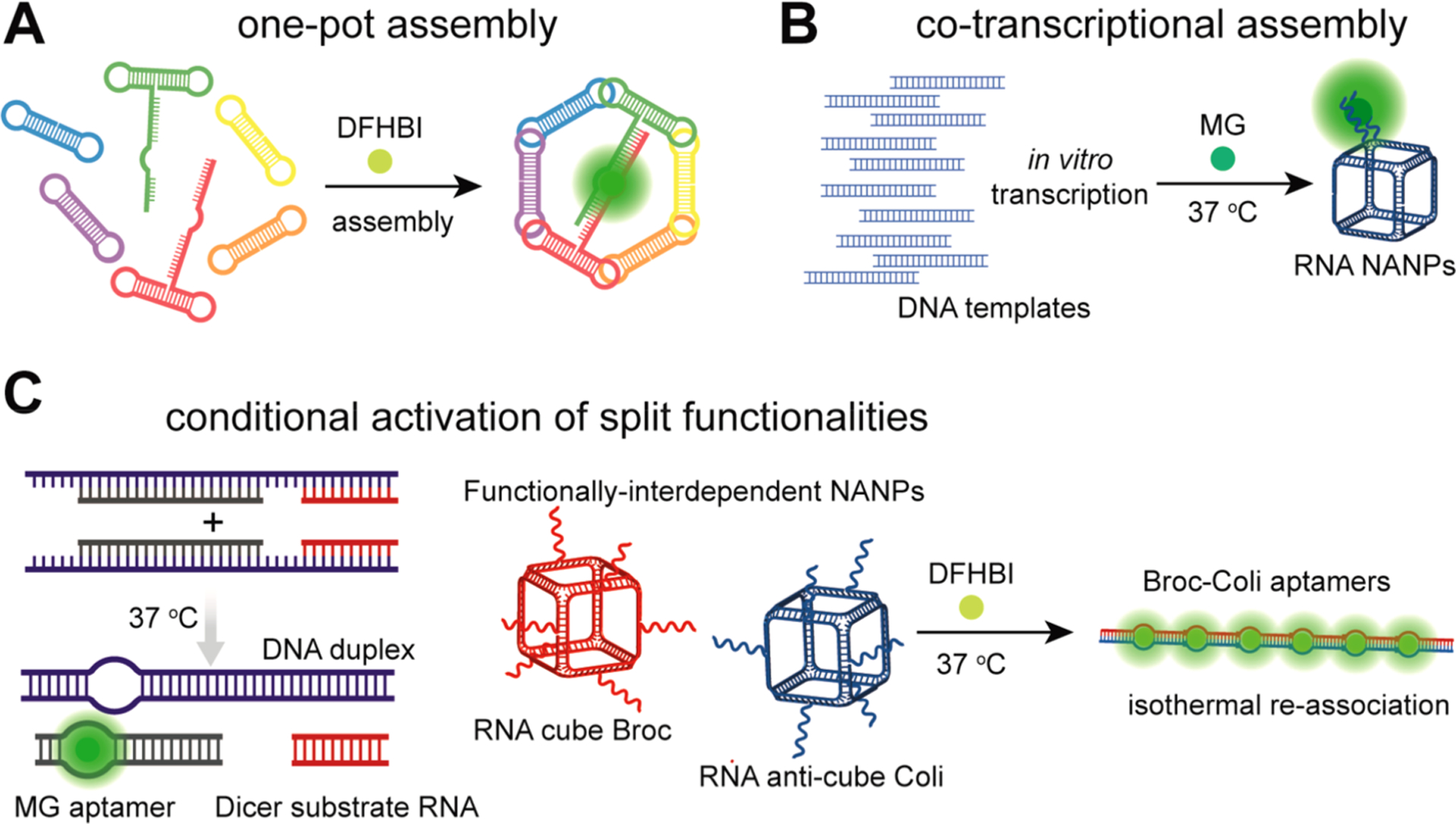
Fluorescent aptamers as a tool in NANP assembly. (A) Signal from individually reassembled nanoparticles’ parts forming complete NANPs correlates with full ring assembly. (B) Assembly of 3D nanostructures is possible during the parallel transcription of all templates.(C) Embedding split light-up aptamers into conditionally pairwise reshaping NANPs enables the monitoring of their interactions in real time.
Light-up Aptamers as Trackers for Intracellular Activation of NANPs.
Cell-specific delivery of fully functional nucleic acid nanoparticles is ideal, but the need for a more controllable system is of immediate concern. In our laboratory, we are developing a system that is conditionally activated where functional RNA/DNA parts are divided and carried by two particles. Inspired by split protein systems, our research group and collaborators developed a technique based on a pair of RNA–DNA hybrids with embedded split functionalities. Separated hybrids are inactive, and to become functional, hybrids require reassociation of cognate double strands. Versatility of the technique was confirmed in mammalian cells and in vivo with the triggering of various functionalities and silencing of several genes (HIV-1 genes, eGFP, or glutathione S-transferaseP1), fluorescence resonance energy transfer (FRET), and aptamer binding the malachite green (Figure 6C).179
Physical interactions of sense and antisense parts leads to the interaction of respective complementary counterparts by toehold interactions and restores functionality.180 One of the ways to monitor this process is the splitting of the Broccoli aptamer sequence into two separate nonfunctional strands (called Broc and Coli). When functionally interdependent shape-switching NANPs interact together, the formation of Broc and Coli reassemble into an active fluorescent aptamer (Figure 6C).56 Modularity of multifunctional RNA NANPs can be used by itself for creation of logic circuits in eukaryotic or prokaryotic cells.181 Combination of light-up RNA aptamers of various colors into a synthetic RNA network can provide a relatively simple read-out for RNA logic gates.182
FRET is one of the tools for monitoring reassembly of various NANPs.53,56 Traditionally, oligonucleotides had to be conjugated with dyes before cell transfection, and no genetically encoded system was known. Recently, RNA aptamers termed apta-FRET constructs have been generated with endogenous FRET. In this single-stranded RNA origami183 construct, Spinach and Mango aptamers are positioned in close proximity to induce high FRET signals between the two fluorophores. Due to the versatility of nanoparticles to respond to conformational changes, two different nanomechanical constructs were designed. In the first nanoparticle, the reversible change in FRET resulted after incubation of the apta-FRET construct with a complementary invading RNA oligonucleotide strand. In the second case, a FRET-based conformational sensor responding to the S-adenosylmethionine (SAM) was integrated into the nanoparticle. The unbound apta-FRET construct exhibits low FRET, and upon binding to SAM, the FRET signal is increased. The ability of the apta-FRET system to induce FRET during the co-transcriptional assembly was observed in E. coli cells.184 Collectively, this establishes a genetically encodable RNA origami system capable of reporting conformational changes in vivo. Therefore, this system has potential to expand our knowledge of basic RNA biology in vivo and for broader applications regarding nanomedicine.
Aptamer-Assisted Packaging, Visualization, and Delivery of NANPs Using Natural Pathways.
Natural vehicles for the delivery of functional RNAs can open a perspective field of diverse aptamer technologies. Upon intracellular assembly, genetically encoded TNAs or NANPs can be trafficked into the natural vesicles or engineered virus-like particles. We envisage that NANP loading will be based on and specifically mediated by aptamers binding to proteins characteristic for vesicles. In this section, we discuss described observations and experimental efforts loading the vesicles with RNA.
Transportation of extracellular RNAs (exRNAs) by extracellular vesicles (EVs) appears to be very abundant in all kingdoms of life. However, the relevance and importance of intercellular delivery of EVs enriched with small ncRNAs remains to be elucidated. Even more puzzling is the inter- and trans-kingdom exchange of EVs containing exRNAs that may have global evolutional and ecological influence.185 One major advantage of aptamer-targeted exosomes is their natural fusogenicity that promotes the efficient delivery of cargo to cells via endocytosis followed by the escape of the cargo from the endosomal compartment to the cytosol. Once in the cytosol, small ncRNAs could then perform a given function. Therefore, employment of EVs in biotechnology and therapy may revolutionize respective fields of biology.
Extracellular vesicles are a heterogeneous group of small phospholipid membrane-enclosed vesicles that are released into the extracellular environment by various cell types. In humans, EVs are secreted from healthy, malignant, or virus-infected cells and are involved in many physiological as well pathophysiological processes, such as cancer progression, the immune response, cell proliferation, cell migration, angio-genesis, and extracellular matrix remodeling. Based on the size and biogenesis, EVs can be further organized into apoptotic bodies, microvesicles, and exosomes that are the subject of a growing area of research. The protein, mRNA, and miRNA composition of exosomes is dependent on the exosome cell origin. However, it seems that some proteins and RNAs are specifically enriched in exosomes.186,187 Transmission of exosome cargo between the cells may occur via endocytosis or by direct fusion. Our current understanding proposes that exosomes constitute a sophisticated system of cell-to-cell communication.
Unraveling the processes governing EV biogenesis and cell tropism can lead to the development of vehicles for targeted delivery of genetically encoded RNA NANPs. Knowledge of the types, selection, and incorporation of RNA into exosomes would therefore be instrumental for engineering exosomes with genetically encoded RNA NANPs. Two approaches exist for incorporation of RNAs with therapeutically relevant properties into exosomes. Either vesicles isolated from cell lines are loaded with siRNAs connected to a lipid compound such as cholesterol (Figure 7A) or endogenous siRNAs are selectively taken up by exosomes from the transgenic construct (Figure 7B).
Figure 7.
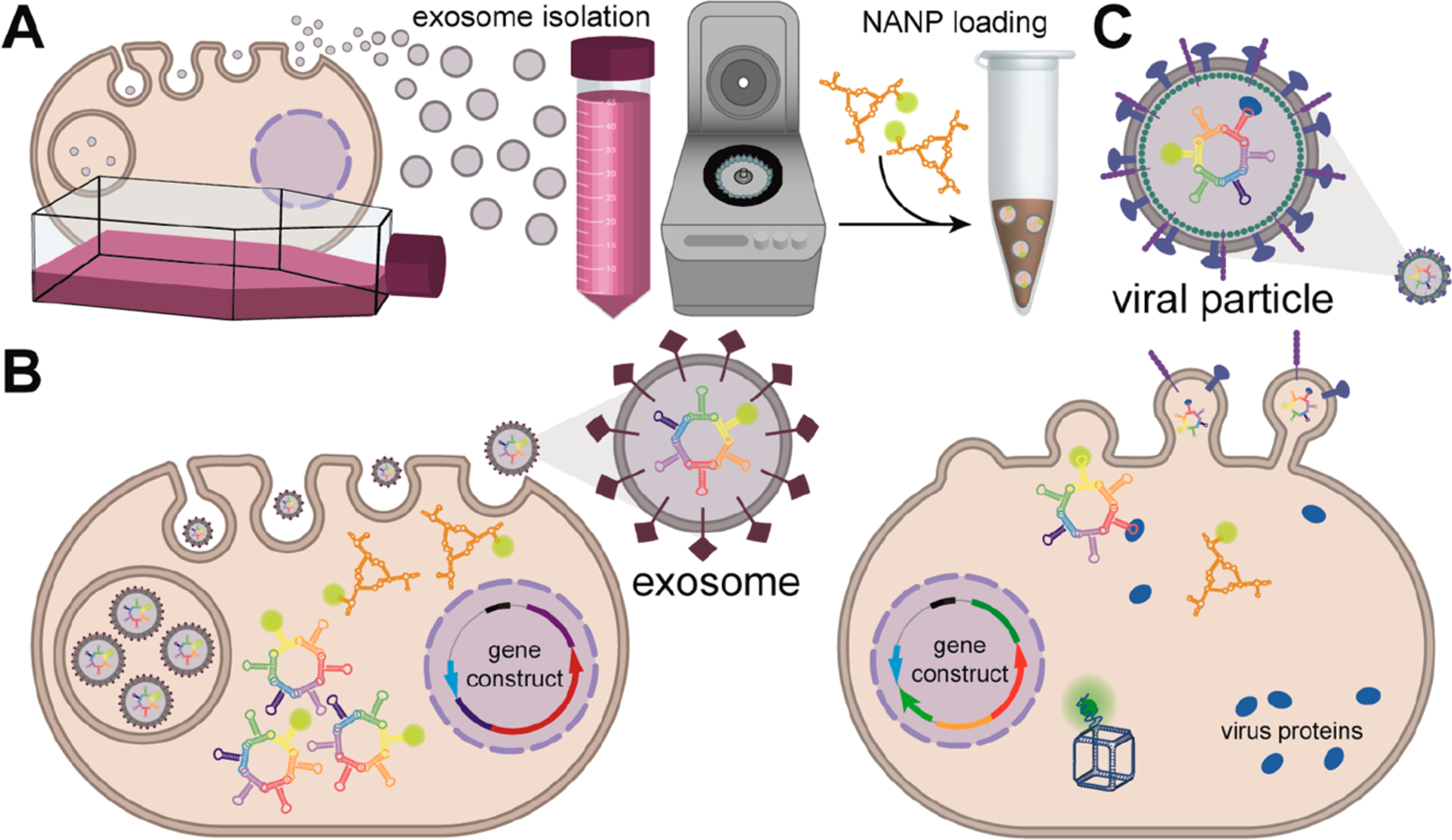
Schematic description proposing NANP loading into exosomes or lentiviruses. (A) In vitro loading of NANPs to isolated exosomes.(B) Genetically encoded NANPs are loaded to exosomes. (C) NANPs are loaded into viral particles via the aptamers binding to virus proteins.
One concern regarding the former is the method of cholesterol conjugation to siRNA which may affect the loading of siRNA into vesicles and potentially interfere with the gene-silencing ability of siRNA. Therefore, the optimization of siRNA chemical modification patterns is necessary for efficient gene downregulation and reproducible and scalable exosome-mediated delivery. Recently, research has investigated the effect of backbone, 5′-phosphate, and chemical modifications of the ribose and linker on hydrophobically modified asymmetric siRNAs. The combination of 5′-(E)-vinylphosph-onate with 2′-F and 2′-O-methyl ribose modifications surpassed siRNAs with partially modified bases. Furthermore, of the two common linkers of cholesterol to siRNA—triethyl glycol (TEG) and 2-aminobutyl-1–3-propanediol (C7)—the TEG linker provided better loading with a saturation number of ~3000 siRNA copies per exosome.188
Interestingly, the position of cholesterol on RNA NANPs has an impact on the loading of arrow-shaped pRNA-3WJ nanostructures into the exosomes. Placing cholesterol on the arrowhead resulted in partial loading of RNA into the vesicles. In contrast, cholesterol on the arrowtail of the RNA resulted in loading onto the surface of exosomes isolated from HEK293T cells. The authors used the later system for linking arrowheads with aptamers and folate for targeting exosomes loaded with siRNA against survivin in three cancer models. Exosomes displaying aptamers for EGFR or PSMA inhibited orthotopic breast cancer models or prostate cancer xenografts, respectively. Similarly, folate-displaying exosomes suppressed colorectal cancer xenografts.189
However, the mechanisms underlying small RNA enrichment in exosomes are not well-defined. Therefore, it is necessary to understand mechanistically how genetically encoded RNAs are directed and enriched in exosomes in order to develop effective exosome-based therapeutics. Current observations suggest the existence of an active process for loading RNAs even with the ability to discriminate among mRNA, miRNA, and other small RNAs. It seems that the sequence motif of miRNAs or “zipcode”-like sequence of mRNAs may facilitate their selective loading into vesicles.190,191 In addition to this, an alternative model proposes that sorting specific miRNAs into exosomes is necessary for maintaining balance with a cell. In this model, miRNAs in excess compared to their cellular target mRNAs are passively transported to exosomes for disposal.192 Despite these and other reports, there is no general concept of RNA loading into exosomes. In fact, more processes may be involved.
As discussed in previous sections, aptamers can function as a delivery agent. Therefore, an aptamer could potentially target delivery of therapeutic RNA to exosomes.189,193,194 However, strategies using aptamers for active uptake to exosomes are undeveloped. In a recent effort to identify exosome-tropic exogenous RNA sequence aptamers by SELEX, an RNA library was transfected into B16-BL6 murine melanoma cells. After subsequent isolation of exosomes, isolated RNAs were subjected to the next round of SELEX. Of the 56 exosome-associated clones that were found, 29 shared a similar sequence.195 However, it is currently not known if these common sequences are sufficient to drive sorting and enrichment of RNAs into the vesicles.
Alternative to SELEX-developed aptamers or natural sequences, specific protein-binding RNA motifs may be fused to sequences intended for exosome uploading. An engineered system for active RNA loading into vesicles might help to elucidate fundamental questions of RNA sorting to exosomes as well as lead to the development of therapeutic RNA delivery by exosomes. To meet these criteria, a recent platform named targeted and modular EV loading (TAMEL) was established. The TAMEL system is based on the recombinant EV-loading protein fused with its luminal terminus to an RNA-binding domain, the MS2 bacteriophage coat protein. Therefore, RNAs of interest displaying the simple stem-loop binding motif for MS2 should be selectively incorporated into the EV interior.196
The analysis provided by the TAMEL system revealed that the length of RNA is one of the parameters for its efficient upload. Another important variable that impacts active loading of mRNA is the choice of EV-loading protein. The highest enrichment of MS2-binding mRNA was observed in vesicles produced by cells expressing the viral VSV-G protein. Interestingly, despite the differences in the amount of incorporated RNA due to loading, no difference in the numbers of produced vesicles was seen.196 The analogical system was engineered by fusion of a lysosomal-associated membrane, Lamp2a protein, with the TAT peptide/HIV-1 transactivation response (TAR) RNA interacting peptide, and the modified TAR RNA loop was inserted into the gene for pre-miR-199a.197 Even if both systems are functional, the effect on target cells upon delivery is minimal in comparison with results obtained by application of exosomes with exogenously loaded siRNAs. One explanation would be that there are almost ~106 more RNA copies per vesicle when exosomes are loaded with RNA by transfection. Alternative explanations include RNA’s inability to escape the endosomal compartment, resulting in degradation within the lysosomes or degradation of released RNA cargo by cytoplasmic nucleases.
Viruses offer an additional option for delivery of therapeutic RNA NANPs. The viral life cycle requires effective packaging of its genome into a virion within host cells and virion transfer to other host cells. After three decades of research, virus-mediated gene therapy is slowly translating into a promising therapeutic option.198 If temporary or persistent gene expression is not compatible with the expected outcome, virus-mediated delivery of ready-to-work molecules is an option. Protein transduction is the delivery of proteins without a genome by pseudotyped lentivirus-like nanoparticles. Delivered proteins execute their function immediately and therefore host-mediated gene expression is not required.199,200 It is tempting to expect that direct delivery of fully assembled multifunctional RNA nanoparticles could carry out instantaneous action. Again, the incorporation of RNA NANPs would require the identification of sequences that are preferentially packed into the virion. Such sequences could be embedded into the RNA nanoparticle and drive hitchhiking of therapeutic RNAs to the budding virus particle. Extensive research focused on the biology of retroviruses revealed that several diverse RNAs (mostly ncRNAs) are encapsidated during virion assembly, and this RNA may reach up to 30% of total RNA mass in the virus particle. It seems that, for some host RNAs, the process of packaging is nonrandom, but the common mechanism has yet to be fully elucidated.201 Aptamers selected against internal viral proteins are effectively encapsidated upon expression in virus-producing cells as has been shown for aptamers targeting HIV reverse transcriptase.202,203 The method of expressing an RNA library inside the virus-producing cells followed by the iterative process of isolation, reverse transcription, and again transfection of other virus-producing cells can lead to the selection of aptamers with high affinities to structural or functional components of the virus interior.204 This method may provide a mechanism for enrichment of therapeutic RNAs into virions and increase the potential of viral-mediated gene therapy (Figure 5C). A platform for recruitment of RNA to virus particles via binding of a 19 nt long MS2 RNA motif to MS2 proteins is another option. Fusion of the MS2 protein sequence to retroviral nucleocapsid proteins compared to the fusion with one of its zinc finger domains drafted around six mRNAs with MS2 motifs per virus particle. Delivery of engineered mRNAs was confirmed in vitro and in vivo where the signal was still detectable after 5 h.205 In a similar study, several retroviral proteins were tested for fusion with the MS2 coat protein. From viral protein R, negative regulatory factor, nucleocapsid protein (NC), and matrix protein, the transduction of cells with particles containing the fusion of MS2 with NC led to the highest number of cells with expected phenotypes. The number and proper insertion of MS2-binding RNA motifs into the 3′ UTR of mRNA has also had a profound impact on the production of functional viral particles. More than one MS2 motif decreases the stability of mRNA. To improve mRNA endurance, authors inserted two 3′ UTR copies of human β-hemoglobin between the stop codon and MS2 aptamer.206
Because biogenesis and biochemistry of EVs resemble retroviruses, combining our knowledge of exosomes and viral replication may yield RNA motifs that drive efficient packaging into these therapeutic delivery agents. Moreover, a very blurred line exists between EVs and defective retroviruses, leading to the hypothesis that EVs and viruses may be related entities.207 Viruses manipulate exosome composition and production to help their spread into uninfected tissue, where exosomes carry on similar functions as they have in cancerogenesis by preparing target tissue for receiving metastases—arranging the conditions for virus infection. Therefore, exploitation of viral factors for the production of exosomes specifically delivering RNA cargo would be another strategy in the development of nanotherapeutics.208,209 Although, as mentioned earlier, aptamers have potential in virus targeting and inhibition, the opposite idea to retarget the virus by aptamers was suggested for vectors of lentiviral origin, and aptamers were used to improve transduction of glioma cells by an adenovirus.210,211 The concept is to develop a general approach to alter the targeting specificity of vectors using aptamers. This approach has two main components: a vector containing a blinded envelope protein and an interchangeable cell-specific aptamer attached to an envelope protein.
Transfecting or endogenously loading NANPs into EVs or viruses would offer an appealing mode of delivery, but as our recent research shows, the situation is more complicated. Whereas naked NANPs are immunoquiescent, upon formulation with transfection reagents, they may become immunogenic. Immunogenicity of NANPs depends on, but is not limited to, shape, connectivity, and composition. In comparison to traditional TNAs, NANPs may activate different pathways of TLR signaling and cytokine responses.51–54,212 How encapsidation of NANPs by natural membranes would compare with delivery by transfection agents is unknown, but we might expect a similar response. In this case, the delivery of immunostimulatory NANPs within exosomes or viruses may represent promising approaches in immunotherapy and should definitely be investigated further.
The idea of intracellular assembly of less or more complex genetically encoded NANPs is very appealing, but transcription of aptamers and other functional or scaffold NANP strands is challenging. Transcription from several cassettes would be required to be rapid and efficient before individual vectors will be separated due to cell division. On the other side, expression from one vector would require optimization of transcription to obtain all transcripts in an equimolar ratio. After transcription, it is important to maintain proper folding and stability. This will require implementing in silico designed or naturally derived flanking sequences to individual NANP strands. However, the presence of such flanking sequences may interfere with the ability to assemble into NANPs. Recently, an innovative approach for expression of aptamers was presented. The Tornado (Twister-optimized RNA for durable overexpression) expression system creates stable RNA aptamers by quick circularization that is mediated by flanking ribozymes. Free termini are subsequently ligated by cellular RNA ligase RtcB.213 The resulting structure resembles a dumbbell. The loop sequences therefore can be used for kissing loop interactions, creating NANPs (Figure 8).
Figure 8.
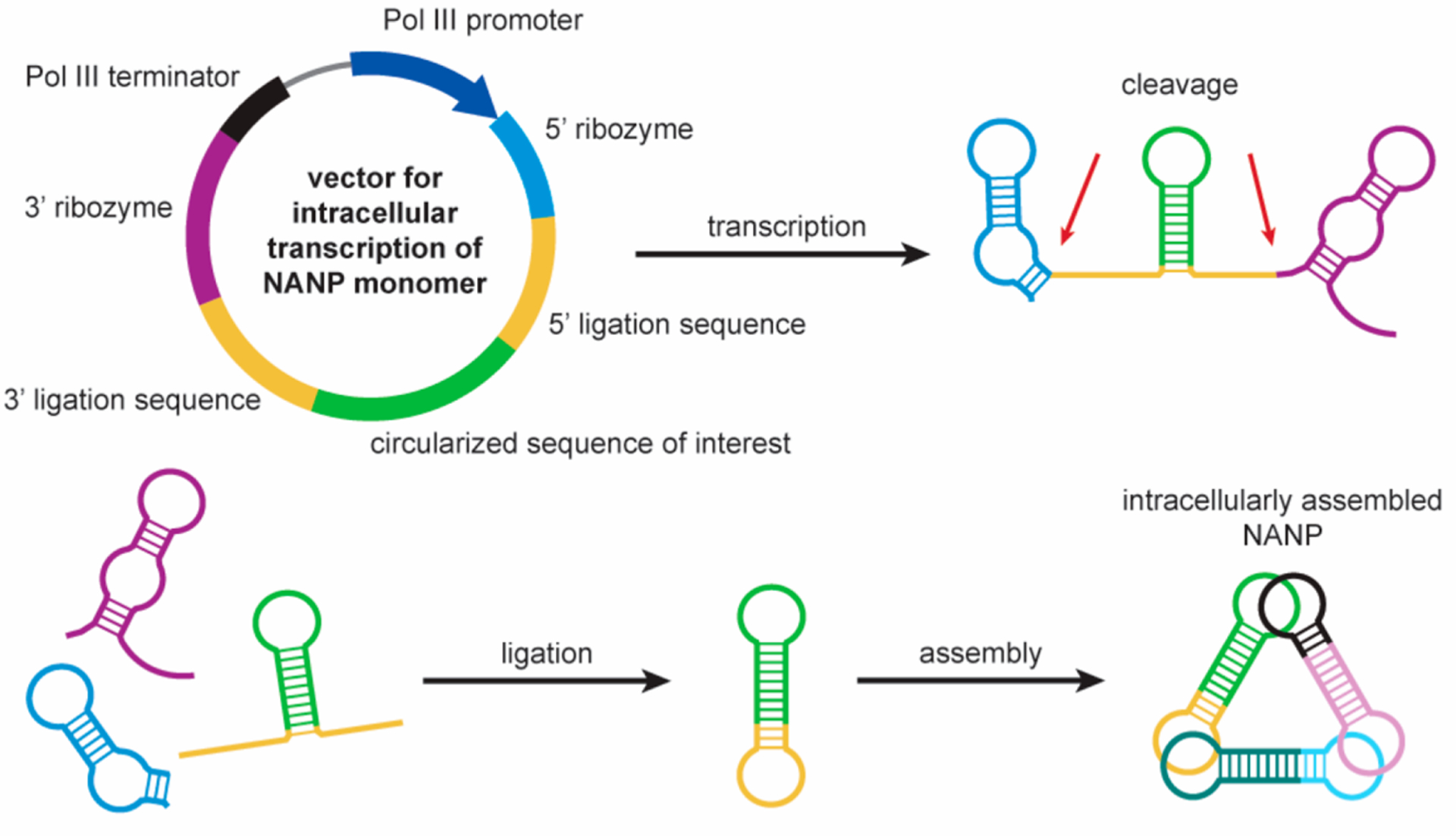
Intracellular assembly of NANPs can be designed on the basis of the Tornado system.213 NANP building blocks transcribed from one or several vectors are flanked by ribozymes that, after self-cleavage, create termini—a 5′ hydroxyl and a 2′,3′-cyclic phosphate at the 3′ end that are recognized by endogenous RtcB ligase. The resulting circularized sequence is chemically stable and, in addition to the Broccoli aptamer, can also code for another functional aptamer (e.g., NF-κB). This system is inspiring for the creation of intracellular structures that can assemble into NANPs.
Stable and efficient in vivo transcription of NANPs has potential to revolutionize the field of NANP technology with the promise of scaled up production either for NANP isolation or formulation within biologically derived vesicles.
ACKNOWLEDGMENTS
Research reported in this publication was partially supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM120487 (to K.A.A). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. M.P.’s work on the subject was partially supported by the MediPark, Košice - Phase II ITMS2014+: 313011D103 supported by the Operational Programme Research and Development Programme, funded by the ERDF; and NEXO II (Network of Excellence in Oncology) under project ITMS 26220120039.
VOCABULARY
- SELEX
systematic evolution of ligands by exponential enrichment, a technique that allows selecting oligonucleotides, called aptamers, to bind to specific targets with the highest affinity
- NANPs
nucleic acid nanoparticles composed of rationally designed short oligonucleotides that self-assemble into various programmable geometric shapes
- TNAs
therapeutic nucleic acids
- immune checkpoints
immune system pathways that regulate the extent of immune responses by either stimulating or inhibiting immune progress
- EV
extracellular vesicles are lipid-bilayer-enclosed particles with complex cargos that are naturally released by most cells to act as mediators of intercellular communication
- exosomes
extracellular vesicles ranging from 30 to 100 nm that contain nucleic acids (mostly RNA) and proteins which are released into surrounding body fluids to facilitate cell–cell communication
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Cech TR; Steitz JA The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [DOI] [PubMed] [Google Scholar]
- (2).Lieberman J Tapping the RNA World for Therapeutics. Nat. Struct. Mol. Biol 2018, 25, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chakraborty C; Sharma AR; Sharma G; Doss CGP; Lee S-S Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther.–Nucleic Acids 2017, 8, 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhou J; Neff CP; Swiderski P; Li H; Smith DD; Aboellail T; Remling-Mulder L; Akkina R; Rossi JJ Functional In Vivo Delivery of Multiplexed Anti-HIV-1 siRNAs via a Chemically Synthesized Aptamer with a Sticky Bridge. Mol. Ther 2013, 21, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhou J; Satheesan S; Li H; Weinberg MS; Morris KV; Burnett J; Rossi J Cell-Specific RNA Aptamer Against Human CCR5 Specifically Targets HIV-1 Susceptible and Inhibits HIV-1 Infectivity. Chem. Biol 2015, 22, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wheeler LA; Vrbanac V; Trifonova R; Brehm MA; Gilboa-Geffen A; Tanno S; Greiner DL; Luster AD; Tager AM; Lieberman J Durable Knockdown and Protection from HIV Transmission in Humanized Mice Treated with Gel-Formulated CD4 Aptamer-siRNA Chimeras. Mol. Ther 2013, 21, 1378–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ellington AD; Szostak JW In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- (8).Tuerk C; Gold L Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- (9).Darmostuk M; Rimpelova S; Gbelcova H; Ruml T Current Approaches in SELEX: An Update to Aptamer Selection Technology. Biotechnol. Adv 2015, 33, 1141–1161. [DOI] [PubMed] [Google Scholar]
- (10).Dunn MR; Jimenez RM; Chaput JC Analysis of Aptamer Discovery and Technology. Nat. Rev. Chem 2017, 1, 0076. [Google Scholar]
- (11).Alam KK; Chang JL; Lange MJ; Nguyen PDM; Sawyer AW; Burke DH Poly-Target Selection Identifies Broad-Spectrum RNA Aptamers. Mol. Ther.–Nucleic Acids 2018, 13, 605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Catuogno S; Esposito CL Aptamer Cell-Based Selection: Overview and Advances. Biomedicines 2017, 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Thiel WH; Thiel KW; Flenker KS; Bair T; Dupuy AJ; McNamara JO; Miller FJ; Giangrande PH Cell-Internalization SELEX: Method for Identifying Cell-Internalizing RNA Aptamers for Delivering siRNAs to Target Cells. Methods Mol. Biol 2015, 1218, 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Avci-Adali M; Metzger M; Perle N; Ziemer G; Wendel HP Pitfalls of Cell-Systematic Evolution of Ligands by Exponential Enrichment (SELEX): Existing Dead Cells During In Vitro Selection Anticipate the Enrichment of Specific Aptamers. Oligonucleotides 2010, 20, 317–323. [DOI] [PubMed] [Google Scholar]
- (15).Zhao Q; Matson S; Herrera CJ; Fisher E; Yu H; Krieg AM Comparison of Cellular Binding and Uptake of Antisense Phosphodiester, Phosphorothioate, and Mixed Phosphorothioate and Methylphosphonate Oligonucleotides. Antisense Res. Dev 1993, 3, 53–66. [DOI] [PubMed] [Google Scholar]
- (16).Wang T; Chen C; Larcher LM; Barrero RA; Veedu RN Three Decades of Nucleic Acid Aptamer Technologies: Lessons Learned, Progress and Opportunities on Aptamer Development. Biotechnol. Adv 2019, 37, 28–50. [DOI] [PubMed] [Google Scholar]
- (17).Yan J; Xiong H; Cai S; Wen N; He Q; Liu Y; Peng D; Liu Z Advances in Aptamer Screening Technologies. Talanta 2019, 200, 124–144. [DOI] [PubMed] [Google Scholar]
- (18).Gilboa E; McNamara JN; Pastor F Use of Oligonucleotide Aptamer Ligands to Modulate the Function of Immune Receptors. Clin. Cancer Res 2013, 19, 1054–1062. [DOI] [PubMed] [Google Scholar]
- (19).Stein CA; Castanotto D FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther 2017, 25, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zadeh JN; Steenberg CD; Bois JS; Wolfe BR; Pierce MB; Khan AR; Dirks RM; Pierce NA NUPACK: Analysis and Design of Nucleic Acid Systems. J. Comput. Chem 2011, 32, 170–173. [DOI] [PubMed] [Google Scholar]
- (21).Dirks RM; Bois JS; Schaeffer JM; Winfree E; Pierce NA Thermodynamic Analysis of Interacting Nucleic Acid Strands. SIAM Rev 2007, 49, 65–88. [Google Scholar]
- (22).Dirks RM; Pierce NA An Algorithm for Computing Nucleic Acid Base-Pairing Probabilities Including Pseudoknots. J. Comput. Chem 2004, 25, 1295–1304. [DOI] [PubMed] [Google Scholar]
- (23).Dirks RM; Pierce NA A Partition Function Algorithm for Nucleic Acid Secondary Structure Including Pseudoknots. J. Comput. Chem 2003, 24, 1664–1677. [DOI] [PubMed] [Google Scholar]
- (24).Wolfe BR; Porubsky NJ; Zadeh JN; Dirks RM; Pierce NA Constrained Multistate Sequence Design for Nucleic Acid Reaction Pathway Engineering. J. Am. Chem. Soc 2017, 139, 3134–3144. [DOI] [PubMed] [Google Scholar]
- (25).Wolfe BR; Pierce NA Sequence Design for a Test Tube of Interacting Nucleic Acid Strands. ACS Synth. Biol 2015, 4, 1086–1100. [DOI] [PubMed] [Google Scholar]
- (26).Zadeh JN; Wolfe BR; Pierce NA Nucleic Acid Sequence Design via Efficient Ensemble Defect Optimization. J. Comput. Chem 2011, 32, 439–452. [DOI] [PubMed] [Google Scholar]
- (27).Dirks RM; Lin M; Winfree E; Pierce NA Paradigms for Computational Nucleic Acid Design. Nucleic Acids Res. 2004, 32, 1392–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Keefe AD; Cload ST SELEX with Modified Nucleotides. Curr. Opin. Chem. Biol 2008, 12, 448–456. [DOI] [PubMed] [Google Scholar]
- (29).Kovacevic KD; Gilbert JC; Jilma B Pharmacokinetics, Pharmacodynamics and Safety of Aptamers. Adv. Drug Delivery Rev 2018, 134, 36–50. [DOI] [PubMed] [Google Scholar]
- (30).Sefah K; Meng L; Lopez-Colon D; Jimenez E; Liu C; Tan W DNA Aptamers as Molecular Probes for Colorectal Cancer Study. PLoS One 2010, 5, No. e14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kimoto M; Matsunaga KI; Hirao I Evolving Aptamers with Unnatural Base Pairs. Curr. Protoc. Chem. Biol 2017, 9, 315–339. [DOI] [PubMed] [Google Scholar]
- (32).Hamashima K; Kimoto M; Hirao I Creation of Unnatural Base Pairs for Genetic Alphabet Expansion Toward Synthetic Xenobiology. Curr. Opin. Chem. Biol 2018, 46, 108–114. [DOI] [PubMed] [Google Scholar]
- (33).McNamara JO; Andrechek ER; Wang Y; Viles KD; Rempel RE; Gilboa E; Sullenger BA; Giangrande PH Cell Type-Specific Delivery of siRNAs with Aptamer-siRNA Chimeras. Nat. Biotechnol 2006, 24, 1005–1015. [DOI] [PubMed] [Google Scholar]
- (34).Panigaj M; Reiser J Aptamer Guided Delivery of Nucleic Acid-Based Nanoparticles. DNA and RNA Nanotechnology 2016, 2,42. [Google Scholar]
- (35).Romanelli A; Affinito A; Avitabile C; Catuogno S; Ceriotti P; Iaboni M; Modica J; Condorelli G; Catalucci D An Anti-PDGFRβ Aptamer for Selective Delivery of Small Therapeutic Peptide to Cardiac Cells. PLoS One 2018, 13, No. e0193392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Porciani D; Tedeschi L; Marchetti L; Citti L; Piazza V; Beltram F; Signore G Aptamer-Mediated Codelivery of Doxorubicin and NF-κB Decoy Enhances Chemosensitivity of Pancreatic Tumor Cells. Mol. Ther.–Nucleic Acids 2015, 4, No. e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Esposito CL; Cerchia L; Catuogno S; De Vita G; Dassie JP; Santamaria G; Swiderski P; Condorelli G; Giangrande PH; de Franciscis V Multifunctional Aptamer-miRNA Conjugates for Targeted Cancer Therapy. Mol. Ther 2014, 22, 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Pang KM; Castanotto D; Li H; Scherer L; Rossi JJ Incorporation of Aptamers in the Terminal Loop of shRNAs Yields an Effective and Novel Combinatorial Targeting Strategy. Nucleic Acids Res. 2018, 46, No. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jasinski D; Haque F; Binzel DW; Guo P Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shu Y; Pi F; Sharma A; Rajabi M; Haque F; Shu D; Leggas M; Evers BM; Guo P Stable RNA Nanoparticles as Potential New Generation Drugs for Cancer Therapy. Adv. Drug Delivery Rev 2014, 66, 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Shu Y; Haque F; Shu D; Li W; Zhu Z; Kotb M; Lyubchenko Y; Guo P Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting without Accumulation in Normal Organs. RNA 2013, 19, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Shukla GC; Haque F; Tor Y; Wilhelmsson LM; Toulmé J-J; Isambert H; Guo P; Rossi JJ; Tenenbaum SA; Shapiro BA A Boost for the Emerging Field of RNA Nanotechnology. ACS Nano 2011, 5, 3405–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Guo P The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Dibrov SM; McLean J; Parsons J; Hermann T Self-Assembling RNA Square. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 6405–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Osada E; Suzuki Y; Hidaka K; Ohno H; Sugiyama H; Endo M; Saito H Engineering RNA-Protein Complexes with Different Shapes for Imaging and Therapeutic Applications. ACS Nano 2014, 8, 8130–8140. [DOI] [PubMed] [Google Scholar]
- (46).Ohno H; Kobayashi T; Kabata R; Endo K; Iwasa T; Yoshimura SH; Takeyasu K; Inoue T; Saito H Synthetic RNA-Protein Complex Shaped Like an Equilateral Triangle. Nat. Nanotechnol 2011, 6, 116–120. [DOI] [PubMed] [Google Scholar]
- (47).Shiohara T; Saito H; Inoue T A Designed RNA Selection: Establishment of a Stable Complex between a Target and Selectant RNA via Two Coordinated Interactions. Nucleic Acids Res. 2009, 37, No. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Saito H; Inoue T Synthetic Biology with RNA Motifs. Int. J. Biochem. Cell Biol 2009, 41, 398–404. [DOI] [PubMed] [Google Scholar]
- (49).Roh YH; Lee JB; Shopsowitz KE; Dreaden EC; Morton SW; Poon Z; Hong J; Yamin I; Bonner DK; Hammond PT Layer-by-Layer Assembled Antisense DNA Microsponge Particles for Efficient Delivery of Cancer Therapeutics. ACS Nano 2014, 8, 9767–9780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lee JB; Hong J; Bonner DK; Poon Z; Hammond PT Self-Assembled RNA Interference Microsponges for Efficient siRNA Delivery. Nat. Mater 2012, 11, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Hong E; Halman JR; Shah A; Cedrone E; Truong N; Afonin KA; Dobrovolskaia MA Toll-Like Receptor–Mediated Recognition of Nucleic Acid Nanoparticles (NANPs) in Human Primary Blood Cells. Molecules 2019, 24, 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Rackley L; Stewart JM; Salotti J; Krokhotin A; Shah A; Halman JR; Juneja R; Smollett J; Lee L; Roark K; Viard M; Tarannum M; Vivero-Escoto J; Johnson PF; Dobrovolskaia MA; Dokholyan NV; Franco E; Afonin KA RNA Fibers as Optimized Nanoscaffolds for siRNA Coordination and Reduced Immunological Recognition. Adv. Funct. Mater 2018, 28, 1805959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia MA; Afonin KA RNA-DNA Fibers and Polygons with Controlled Immunorecognition Activate RNAi, FRET and Transcriptional Regulation of NF-κB in Human Cells. Nucleic Acids Res. 2019, 47, 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hong E; Halman JR; Shah AB; Khisamutdinov EF; Dobrovolskaia MA; Afonin KA Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Lett. 2018, 18, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Johnson MB; Halman JR; Satterwhite E; Zakharov AV; Bui MN; Benkato K; Goldsworthy V; Kim T; Hong E; Dobrovolskaia MA; Khisamutdinov EF; Marriott I; Afonin KA Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small 2017, 13, 1701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN; Lu JS; Miller J; Khisamutdinov EF; Shapiro BA; Dobrovolskaia MA; Afonin KA Functionally-Interdependent Shape-Switching Nanoparticles with Controllable Properties. Nucleic Acids Res. 2017, 45, 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Bui MN; Brittany Johnson M; Viard M; Satterwhite E; Martins AN; Li Z; Marriott I; Afonin KA; Khisamutdinov EF Versatile RNA Tetra-U Helix Linking Motif as a Toolkit for Nucleic Acid Nanotechnology. Nanomedicine 2017, 13, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Stewart JM; Viard M; Subramanian HK; Roark BK; Afonin KA; Franco E Programmable RNA Microstructures for Coordinated Delivery of siRNAs. Nanoscale 2016, 8, 17542–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Parlea L; Puri A; Kasprzak W; Bindewald E; Zakrevsky P; Satterwhite E; Joseph K; Afonin KA; Shapiro BA Cellular Delivery of RNA Nanoparticles. ACS Comb. Sci 2016, 18, 527–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA Multifunctional RNA Nanoparticles. Nano Lett. 2014, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Afonin KA; Kasprzak WK; Bindewald E; Kireeva M; Viard M; Kashlev M; Shapiro BA In Silico Design and Enzymatic Synthesis of Functional RNA Nanoparticles. Acc. Chem. Res 2014, 47, 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Liu X; Yan H; Liu Y; Chang Y Targeted Cell-Cell Interactions by DNA Nanoscaffold-Templated Multivalent Bispecific Aptamers. Small 2011, 7, 1673–1682. [DOI] [PubMed] [Google Scholar]
- (63).Delebecque CJ; Lindner AB; Silver PA; Aldaye FA Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science 2011, 333, 470–474. [DOI] [PubMed] [Google Scholar]
- (64).Sachdeva G; Garg A; Godding D; Way JC; Silver PA In Vivo Co-Localization of Enzymes on RNA Scaffolds Increases Metabolic Production in a Geometrically Dependent Manner. Nucleic Acids Res. 2014, 42, 9493–9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Li M; Zheng M; Wu S; Tian C; Liu D; Weizmann Y; Jiang W; Wang G; Mao C In Vivo Production of RNA Nanostructures via Programmed Folding of Single-Stranded RNAs. Nat. Commun 2018, 9, 2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Zhou J; Rossi J Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discovery 2017, 16, 181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Hori S.-i.; Herrera A; Rossi JJ; Zhou J Current Advances in Aptamers for Cancer Diagnosis and Therapy. Cancers 2018, 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Waters EK; Genga RM; Schwartz MC; Nelson JA; Schaub RG; Olson KA; Kurz JC; McGinness KE Aptamer ARC19499 Mediates a Procoagulant Hemostatic Effect by Inhibiting Tissue Factor Pathway Inhibitor. Blood 2011, 117, 5514–5522. [DOI] [PubMed] [Google Scholar]
- (69).Drolet DW; Green LS; Gold L; Janjic N Fit for the Eye: Aptamers in Ocular Disorders. Nucleic Acid Ther. 2016, 26, 127–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Haberland A; Wallukat G; Dahmen C; Kage A; Schimke I Aptamer Neutralization of Beta1-Adrenoceptor Autoantibodies Isolated from Patients with Cardiomyopathies. Circ. Res 2011, 109, 986–992. [DOI] [PubMed] [Google Scholar]
- (71).Li J; Yousefi K; Ding W; Singh J; Shehadeh LA Osteopontin RNA Aptamer Can Prevent and Reverse Pressure Overload-Induced Heart Failure. Cardiovasc. Res 2017, 113, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Li W; Lan X Aptamer Oligonucleotides: Novel Potential Therapeutic Agents in Autoimmune Disease. Nucleic Acid Ther. 2015, 25, 173–179. [DOI] [PubMed] [Google Scholar]
- (73).Davydova A; Vorobjeva M; Pyshnyi D; Altman S; Vlassov V; Venyaminova A Aptamers Against Pathogenic Microorganisms. Crit. Rev. Microbiol 2016, 42, 847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Bouvier-Muller A; Duconge F Nucleic Acid Aptamers for Neurodegenerative Diseases. Biochimie 2018, 145, 73–83. [DOI] [PubMed] [Google Scholar]
- (75).Prodeus A; Abdul-Wahid A; Fischer NW; Huang EHB; Cydzik M; Gariépy J Targeting the PD-1/PD-L1 Immune Evasion Axis with DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers. Mol. Ther.–Nucleic Acids 2015, 4, No. e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Lai W-Y; Huang B-T; Wang J-W; Lin P-Y; Yang P-C A Novel PD-L1-Targeting Antagonistic DNA Aptamer with Antitumor Effects. Mol. Ther.–Nucleic Acids 2016, 5, No. e397. [DOI] [PubMed] [Google Scholar]
- (77).Pardoll DM The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Press release: The Nobel Prize in Physiology or Medicine 2018, https://www.nobelprize.org/prizes/medicine/2018/press-release/ (accessed Oct 19, 2019).
- (79).Santulli-Marotto S; Nair SK; Rusconi C; Sullenger B; Gilboa E Multivalent RNA Aptamers That Inhibit CTLA-4 and Enhance Tumor Immunity. Cancer Res. 2003, 63, 7483–7489. [PubMed] [Google Scholar]
- (80).Reiss KA; Forde PM; Brahmer JR Harnessing the Power of the Immune System via Blockade of PD-1 and PD-L1: A Promising New Anticancer Strategy. Immunotherapy 2014, 6, 459–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).McNamara JO; Kolonias D; Pastor F; Mittler RS; Chen L; Giangrande PH; Sullenger B; Gilboa E Multivalent 4–1BB Binding Aptamers Costimulate Cd8+ T Cells and Inhibit Tumor Growth in Mice. J. Clin. Invest 2008, 118, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Dollins CM; Nair S; Boczkowski D; Lee J; Layzer JM; Gilboa E; Sullenger BA Assembling OX40 Aptamers on a Molecular Scaffold to Create a Receptor-Activating Aptamer. Chem. Biol 2008, 15, 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Pastor F; Kolonias D; McNamara JO; Gilboa E Targeting 4–1BB Costimulation to Disseminated Tumor Lesions with Bi-Specific Oligonucleotide Aptamers. Mol. Ther 2011, 19, 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Soldevilla MM; Villanueva H; Bendandi M; Inoges S; López-Díaz de Cerio A; Pastor F 2-Fluoro-RNA Oligonucleotide CD40 Targeted Aptamers for the Control of B Lymphoma and Bone-Marrow Aplasia. Biomaterials 2015, 67, 274–285. [DOI] [PubMed] [Google Scholar]
- (85).Soldevilla MM; Villanueva H; Pastor F Aptamers: A Feasible Technology in Cancer Immunotherapy. J. Immunol. Res 2016, 2016, 1083738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Yang S; Wen J; Li H; Xu L; Liu Y; Zhao N; Zeng Z; Qi J; Jiang W; Han W; Zu Y Aptamer-Engineered Natural Killer Cells for Cell-Specific Adaptive Immunotherapy. Small 2019, 15, 1900903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Oelsner S; Waldmann A; Billmeier A; Röder J; Lindner A; Ullrich E; Marschalek R; Dotti G; Jung G; Große-Hovest L; Oberoi P; Bader P; Wels WS Genetically Engineered Car NK Cells Display Selective Cytotoxicity against FLT3-Positive B-ALL and Inhibit In Vivo Leukemia Growth. Int. J. Cancer 2019, 145, 1935–1945. [DOI] [PubMed] [Google Scholar]
- (88).Miliotou AN; Papadopoulou LC Car T-Cell Therapy: A New Era in Cancer Immunotherapy. Curr. Pharm. Biotechnol 2018, 19, 5–18. [DOI] [PubMed] [Google Scholar]
- (89).Meyer B; Ly H Inhibition of Innate Immune Responses Is Key to Pathogenesis by Arenaviruses. J. Virol 2016, 90, 3810–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Kell AM; Gale M RIG-I in RNA Virus Recognition. Virology 2015, 479–480, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Hwang S-Y; Sun H-Y; Lee K-H; Oh B-H; Cha YJ; Kim BH; Yoo J-Y 5′-Triphosphate-RNA-Independent Activation of RIG-I via RNA Aptamer with Enhanced Antiviral Activity. Nucleic Acids Res. 2012, 40, 2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Duthie MS; Windish HP; Fox CB; Reed SG Use of Defined TLR Ligands as Adjuvants within Human Vaccines. Immunol. Rev 2011, 239, 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Ranoa DR; Parekh AD; Pitroda SP; Huang X; Darga T; Wong AC; Huang L; Andrade J; Staley JP; Satoh T; Akira S; Weichselbaum RR; Khodarev NN Cancer Therapies Activate RIG-I-Like Receptor Pathway Through Endogenous Non-Coding RNAs. Oncotarget 2016, 7, 26496–26515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Elion DL; Jacobson ME; Hicks DJ; Rahman B; Sanchez V; Gonzales-Ericsson PI; Fedorova O; Pyle AM; Wilson JT; Cook RS Therapeutically Active RIG-I Agonist Induces Immunogenic Tumor Cell Killing in Breast Cancers. Cancer Res. 2018, 78, 6183–6195. [DOI] [PubMed] [Google Scholar]
- (95).Liu Z; Dou C; Jia Y; Li Q; Zheng X; Yao Y; Liu Q; Song T RIG-I Suppresses the Migration and Invasion of Hepatocellular Carcinoma Cells by Regulating MMP9. Int. J. Oncol 2015, 46, 1710–1720. [DOI] [PubMed] [Google Scholar]
- (96).Schrand B; Berezhnoy A; Brenneman R; Williams A; Levay A; Kong L-Y; Rao G; Zhou S; Heimberger AB; Gilboa E Targeting 4–1BB Costimulation to the Tumor Stroma with Bispecific Aptamer Conjugates Enhances the Therapeutic Index of Tumor Immunotherapy. Cancer Immunol. Res 2014, 2, 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Teng J; Yuan F; Ye Y; Zheng L; Yao L; Xue F; Chen W; Li B Aptamer-Based Technologies in Foodborne Pathogen Detection. Front. Microbiol 2016, 7, 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Hong KL; Sooter LJ Single-Stranded DNA Aptamers Against Pathogens and Toxins: Identification and Biosensing Applications. BioMed Res. Int 2015, 2015, 419318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Adams D; Gonzalez-Duarte A; O’Riordan WD; Yang CC; Ueda M; Kristen AV; Tournev I; Schmidt HH; Coelho T; Berk JL; Lin KP; Vita G; Attarian S; Planté-Bordeneuve V; Mezei MM; Campistol JM; Buades J; Brannagan TH; Kim BJ; Oh J; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med 2018, 379, 11–21. [DOI] [PubMed] [Google Scholar]
- (100).Adams BD; Kasinski AL; Slack FJ Aberrant Regulation and Function of microRNAs in Cancer. Curr. Biol 2014, 24, R762–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Stenvang J; Petri A; Lindow M; Obad S; Kauppinen S Inhibition of microRNA Function by Antimir Oligonucleotides. Silence 2012, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Pofahl M; Wengel J; Mayer G Multifunctional Nucleic Acids for Tumor Cell Treatment. Nucleic Acid Ther. 2014, 24, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Catuogno S; Rienzo A; Di Vito A; Esposito CL; de Franciscis V Selective Delivery of Therapeutic Single Strand Antimirs by Aptamer-Based Conjugates. J. Controlled Release 2015, 210, 147–159. [DOI] [PubMed] [Google Scholar]
- (104).Esposito CL; Nuzzo S; Kumar SA; Rienzo A; Lawrence CL; Pallini R; Shaw L; Alder JE; Ricci-Vitiani L; Catuogno S; de Franciscis V A Combined microRNA-Based Targeted Therapeutic Approach to Eradicate Glioblastoma Stem-Like Cells. J. Controlled Release 2016, 238, 43–57. [DOI] [PubMed] [Google Scholar]
- (105).Khati M; Schüman M; Ibrahim J; Sattentau Q; Gordon S; James W Neutralization of Infectivity of Diverse R5 Clinical Isolates of Human Immunodeficiency Virus Type 1 by gp120-binding 2’F-RNA Aptamers. J. Virol 2003, 77, 12692–12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Dey AK; Khati M; Tang M; Wyatt R; Lea SM; James W An Aptamer That Neutralizes R5 Strains of Human Immunodeficiency Virus Type 1 Blocks gp120-CCR5 Interaction. J. Virol 2005, 79, 13806–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Wandtke T; Woźniak J; Kopinński P Aptamers in Diagnostics and Treatment of Viral Infections. Viruses 2015, 7, 751–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Martell RE; Nevins JR; Sullenger BA Optimizing Aptamer Activity for Gene Therapy Applications Using Expression Cassette SELEX. Mol. Ther 2002, 6, 30–34. [DOI] [PubMed] [Google Scholar]
- (109).Lange MJ; Sharma TK; Whatley AS; Landon LA; Tempesta MA; Johnson MC; Burke DH Robust Suppression of HIV Replication by Intracellularly Expressed Reverse Transcriptase Aptamers Is Independent of Ribozyme Processing. Mol. Ther 2012, 20, 2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Baum DA; Silverman SK Deoxyribozymes: Useful DNA Catalysts In Vitro and In Vivo. Cell. Mol. Life Sci 2008, 65, 2156–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Subramanian N; Kanwar JR; Akilandeswari B; Kanwar RK; Khetan V; Krishnakumar S Chimeric Nucleolin Aptamer with Survivin DNAzyme for Cancer Cell Targeted Delivery. Chem. Commun 2015, 51, 6940–6943. [DOI] [PubMed] [Google Scholar]
- (112).Knudsen SM; Ellington AD Aptazymes: Allosteric Ribozymes and Deoxyribozymes as Biosensors In The Aptamer Handbook: Functional Oligonucleotides and Their Applications; Klussmann S, Ed.; Wiley-VCH: Weinheim, Germany, 2006; pp 290–310. [Google Scholar]
- (113).Havens MA; Hastings ML Splice-Switching Antisense Oligonucleotides as Therapeutic Drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Kotula JW; Pratico ED; Ming X; Nakagawa O; Juliano RL; Sullenger BA Aptamer-Mediated Delivery of Splice-Switching Oligonucleotides to the Nuclei of Cancer Cells. Nucleic Acid Ther. 2012, 22, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Hecker M; Wagner AH Transcription Factor Decoy Technology: A Therapeutic Update. Biochem. Pharmacol 2017, 144, 29–34. [DOI] [PubMed] [Google Scholar]
- (116).Tawiah KD; Porciani D; Burke DH Toward the Selection of Cell Targeting Aptamers with Extended Biological Functionalities to Facilitate Endosomal Escape of Cargoes. Biomedicines 2017, 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Westhof E; Masquida B; Jaeger L RNA Tectonics: Towards RNA Design. Folding Des. 1996, 1, R78–88. [DOI] [PubMed] [Google Scholar]
- (118).Jaeger L; Leontis NB Tecto-RNA: One-Dimensional Self-Assembly Through Tertiary Interactions. Angew. Chem., Int. Ed 2000, 39, 2521–2524. [DOI] [PubMed] [Google Scholar]
- (119).Jaeger L; Westhof E; Leontis NB TectoRNA: Modular Assembly Units for the Construction of RNA Nano-Objects. Nucleic Acids Res. 2001, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Chworos A; Severcan I; Koyfman AY; Weinkam P; Oroudjev E; Hansma HG; Jaeger L Building Programmable Jigsaw Puzzles with RNA. Science 2004, 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- (121).Jaeger L; Chworos A The Architectonics of Programmable RNA and DNA Nanostructures. Curr. Opin. Struct. Biol 2006, 16, 531–543. [DOI] [PubMed] [Google Scholar]
- (122).Guo P; Zhang C; Chen C; Garver K; Trottier M Inter-RNA Interaction of Phage Phi29 pRNA to Form a Hexameric Complex for Viral DNA Transportation. Mol. Cell 1998, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- (123).Shu D; Huang LP; Hoeprich S; Guo P Construction of Phi29 DNA-Packaging RNA Monomers, Dimers, and Trimers with Variable Sizes and Shapes as Potential Parts for Nanodevices. J. Nanosci. Nanotechnol 2003, 3, 295–302. [DOI] [PubMed] [Google Scholar]
- (124).Shu D; Moll W-D; Deng Z; Mao C; Guo P Bottom-Up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology. Nano Lett. 2004, 4, 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Hoeprich S; Zhou Q; Guo S; Shu D; Qi G; Wang Y; Guo P Bacterial Virus Phi29 pRNA as a Hammerhead Ribozyme Escort to Destroy Hepatitis B Virus. Gene Ther. 2003, 10, 1258–1267. [DOI] [PubMed] [Google Scholar]
- (126).Guo S; Tschammer N; Mohammed S; Guo P Specific Delivery of Therapeutic RNAs to Cancer Cells via the Dimerization Mechanism of Phi29 Motor pRNA. Hum. Gene Ther 2005, 16, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Haque F; Shu D; Shu Y; Shlyakhtenko LS; Rychahou PG; Evers BM; Guo P Ultrastable Synergistic Tetravalent RNA Nanoparticles for Targeting to Cancers. Nano Today 2012, 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Hu J; Xiao F; Hao X; Bai S; Hao J Inhibition of Monocyte Adhesion to Brain-Derived Endothelial Cells by Dual Functional RNA Chimeras. Mol. Ther.–Nucleic Acids 2014, 3, No. e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Guo S; Piao X; Li H; Guo P Methods for Construction and Characterization of Simple or Special Multifunctional RNA Nanoparticles Based on the 3WJ of Phi29 DNA Packaging Motor. Methods (Amsterdam, Neth.) 2018, 143, 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Shu D; Li H; Shu Y; Xiong G; Carson WE; Haque F; Xu R; Guo P Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano 2015, 9, 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Pi F; Zhang H; Li H; Thiviyanathan V; Gorenstein DG; Sood AK; Guo P RNA Nanoparticles Harboring Annexin A2 Aptamer Can Target Ovarian Cancer for Tumor-Specific Doxorubicin Delivery. Nanomedicine 2017, 13, 1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Dao BN; Viard M; Martins AN; Kasprzak WK; Shapiro BA; Afonin KA Triggering RNAi with Multifunctional RNA Nanoparticles and Their Delivery. DNA and RNA Nanotechnology 2015, 1, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Grabow WW; Jaeger L RNA Self-Assembly and RNA Nanotechnology. Acc. Chem. Res 2014, 47, 1871–1880. [DOI] [PubMed] [Google Scholar]
- (134).Geary C; Chworos A; Verzemnieks E; Voss NR; Jaeger L Composing RNA Nanostructures from a Syntax of RNA Structural Modules. Nano Lett. 2017, 17, 7095–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Yingling YG; Shapiro BA Computational Design of an RNA Hexagonal Nanoring and an RNA Nanotube. Nano Lett. 2007, 7, 2328–2334. [DOI] [PubMed] [Google Scholar]
- (136).Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L Design and Self-Assembly of siRNA-Functionalized RNA Nanoparticles for Use in Automated Nanomedicine. Nat. Protoc 2011, 6, 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Grabow WW; Zakrevsky P; Afonin KA; Chworos A; Shapiro BA; Jaeger L Self-Assembling RNA Nanorings Based on RNAI/II Inverse Kissing Complexes. Nano Lett. 2011, 11, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Afonin KA; Kireeva M; Grabow WW; Kashlev M; Jaeger L; Shapiro BA Co-Transcriptional Assembly of Chemically Modified RNA Nanoparticles Functionalized with siRNAs. Nano Lett. 2012, 12, 5192–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Sajja S; Chandler M; Fedorov D; Kasprzak WK; Lushnikov A; Viard M; Shah A; Dang D; Dahl J; Worku B; Dobrovolskaia MA; Krasnoslobodtsev A; Shapiro BA; Afonin KA Dynamic Behavior of RNA Nanoparticles Analyzed by AFM on a Mica/Air Interface. Langmuir 2018, 34, 15099–15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Li N; Ebright JN; Stovall GM; Chen X; Nguyen HH; Singh A; Syrett A; Ellington AD Technical and Biological Issues Relevant to Cell Typing with Aptamers. J. Proteome Res 2009, 8, 2438–2448. [DOI] [PubMed] [Google Scholar]
- (141).Porciani D; Cardwell LN; Tawiah KD; Alam KK; Lange MJ; Daniels MA; Burke DH Modular Cell-Internalizing Aptamer Nanostructure Enables Targeted Delivery of Large Functional RNAs in Cancer Cell Lines. Nat. Commun 2018, 9, 2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Staring J; Raaben M; Brummelkamp TR Viral Escape from Endosomes and Host Detection at a Glance. J. Cell Sci 2018, 131, jcs216259. [DOI] [PubMed] [Google Scholar]
- (143).Modi S; S MG; Goswami D; Gupta GD; Mayor S; Krishnan YA DNA Nanomachine That Maps Spatial and Temporal pH Changes inside Living Cells. Nat. Nanotechnol 2009, 4, 325–330. [DOI] [PubMed] [Google Scholar]
- (144).Rothemund PWK Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. [DOI] [PubMed] [Google Scholar]
- (145).Andersen ES; Dong M; Nielsen MM; Jahn K; Subramani R; Mamdouh W; Golas MM; Sander B; Stark H; Oliveira CL; Pedersen JS; Birkedal V; Besenbacher F; Gothelf KV; Kjems J Self-Assembly of a Nanoscale DNA Box with a Controllable Lid. Nature 2009, 459, 73–76. [DOI] [PubMed] [Google Scholar]
- (146).Douglas SM; Bachelet I; Church GM A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [DOI] [PubMed] [Google Scholar]
- (147).Li S; Jiang Q; Liu S; Zhang Y; Tian Y; Song C; Wang J; Zou Y; Anderson GJ; Han J-Y; Chang Y; Liu Y; Zhang C; Chen L; Zhou G; Nie G; Yan H; Ding B; Zhao Y A DNA Nanorobot Functions as a Cancer Therapeutic in Response to a Molecular Trigger In Vivo. Nat. Biotechnol 2018, 36, 258–264. [DOI] [PubMed] [Google Scholar]
- (148).Praetorius F; Kick B; Behler KL; Honemann MN; Weuster-Botz D; Dietz H Biotechnological Mass Production of DNA Origami. Nature 2017, 552, 84–87. [DOI] [PubMed] [Google Scholar]
- (149).Bouhedda F; Autour A; Ryckelynck M Light-Up RNA Aptamers and Their Cognate Fluorogens: From Their Development to Their Applications. Int. J. Mol. Sci 2018, 19, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Dolgosheina EV; Unrau PJ Fluorophore-Binding RNA Aptamers and Their Applications. Wiley Interdiscip. Rev.: RNA 2016, 7, 843–851. [DOI] [PubMed] [Google Scholar]
- (151).Sando S; Narita A; Aoyama Y Light-Up Hoechst–DNA Aptamer Pair: Generation of an Aptamer-Selective Fluorophore from a Conventional DNA-Staining Dye. ChemBioChem 2007, 8, 1795–1803. [DOI] [PubMed] [Google Scholar]
- (152).Sando S; Narita A; Hayami M; Aoyama Y Transcription Monitoring Using Fused RNA with a Dye-Binding Light-Up Aptamer as a Tag: A Blue Fluorescent RNA. Chem. Commun 2008, 3858. [DOI] [PubMed] [Google Scholar]
- (153).Grate D; Wilson C Laser-Mediated, Site-Specific Inactivation of RNA Transcripts. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 6131–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Babendure JR; Adams SR; Tsien RY Aptamers Switch on Fluorescence of Triphenylmethane Dyes. J. Am. Chem. Soc 2003, 125, 14716–14717. [DOI] [PubMed] [Google Scholar]
- (155).Lee LG; Chen C-H; Chiu LA Thiazole Orange: A New Dye for Reticulocyte Analysis. Cytometry 1986, 7, 508–517. [DOI] [PubMed] [Google Scholar]
- (156).Nygren J; Svanvik N; Kubista M The Interactions Between the Fluorescent Dye Thiazole Orange and DNA. Biopolymers 1998, 46, 39–51. [DOI] [PubMed] [Google Scholar]
- (157).Sparano BA; Koide K A Strategy for the Development of Small-Molecule-Based Sensors That Strongly Fluoresce When Bound to a Specific RNA. J. Am. Chem. Soc 2005, 127, 14954–14955. [DOI] [PubMed] [Google Scholar]
- (158).Murata A; Sato S.-i.; Kawazoe Y; Uesugi M Small-Molecule Fluorescent Probes for Specific RNA Targets. Chem. Commun. (Cambridge, U. K.) 2011, 47, 4712. [DOI] [PubMed] [Google Scholar]
- (159).Arora A; Sunbul M; Jäschke A Dual-Colour Imaging of RNAs Using Quencher- and Fluorophore-Binding Aptamers. Nucleic Acids Res. 2015, 43, gkv718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Paige JS; Wu KY; Jaffrey SR RNA Mimics of Green Fluorescent Protein. Science 2011, 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Paige JS; Nguyen-Duc T; Song W; Jaffrey SR Fluorescence Imaging of Cellular Metabolites with RNA. Science 2012, 335, 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Song W; Strack RL; Jaffrey SR Imaging Bacterial Protein Expression Using Genetically Encoded Sensors Composed of RNA. Nat. Methods 2013, 10, 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Strack RL; Disney MD; Jaffrey SR A Superfolding Spinach2 Reveals the Dynamic Nature of Trinucleotide Repeat-Containing RNA. Nat. Methods 2013, 10, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Warner KD; Chen MC; Song W; Strack RL; Thorn A; Jaffrey SR; Ferré-D’Amaré AR Structural Basis for Activity of Highly Efficient RNA Mimics of Green Fluorescent Protein. Nat. Struct. Mol. Biol 2014, 21, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Filonov GS; Moon JD; Svensen N; Jaffrey SR Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J. Am. Chem. Soc 2014, 136, 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Song W; Strack RL; Svensen N; Jaffrey SR Plug-and-Play Fluorophores Extend the Spectral Properties of Spinach. J. Am. Chem. Soc 2014, 136, 1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Nilaratanakul V; Hauer DA; Griffin DE Development and Characterization of Sindbis Virus with Encoded Fluorescent RNA Aptamer Spinach2 for Imaging of Replication and Immune-Mediated Changes in Intracellular Viral RNA. J. Gen. Virol 2017, 98, 992–1003. [DOI] [PubMed] [Google Scholar]
- (168).Filonov GS; Kam CW; Song W; Jaffrey SR In-Gel Imaging of RNA Processing Using Broccoli Reveals Optimal Aptamer Expression Strategies. Chem. Biol 2015, 22, 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Song W; Filonov GS; Kim H; Hirsch M; Li X; Moon JD; Jaffrey SR Imaging RNA Polymerase III Transcription Using a Photostable RNA–Fluorophore Complex. Nat. Chem. Biol 2017, 13, 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Warner KD; Sjekloća L; Song W; Filonov GS; Jaffrey SR; Ferré-D’Amaré AR A Homodimer Interface without Base Pairs in an RNA Mimic of Red Fluorescent Protein. Nat. Chem. Biol 2017, 13, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Kolpashchikov DM Binary Malachite Green Aptamer for Fluorescent Detection of Nucleic Acids. J. Am. Chem. Soc 2005, 127, 12442–12443. [DOI] [PubMed] [Google Scholar]
- (172).Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L In Vitro Assembly of Cubic RNA-Based Scaffolds Designed In Silico. Nat. Nanotechnol 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Rogers TA; Andrews GE; Jaeger L; Grabow WW Fluorescent Monitoring of RNA Assembly and Processing Using the Split-Spinach Aptamer. ACS Synth. Biol 2015, 4, 162–166. [DOI] [PubMed] [Google Scholar]
- (174).Chandler M; Lyalina T; Halman J; Rackley L; Lee L; Dang D; Ke W; Sajja S; Woods S; Acharya S; Baumgarten E; Christopher J; Elshalia E; Hrebien G; Kublank K; Saleh S; Stallings B; Tafere M; Striplin C; Afonin KA Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology. Molecules 2018, 23, 3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Sajja S; Chandler M; Striplin CD; Afonin KA Activation of Split RNA Aptamers: Experiments Demonstrating the Enzymatic Synthesis of Short RNAs and Their Assembly as Observed by Fluorescent Response. J. Chem. Educ 2018, 95, 1861–1866. [Google Scholar]
- (176).Kikuchi N; Kolpashchikov DM A Universal Split Spinach Aptamer (USSA) for Nucleic Acid Analysis and DNA Computation. Chem. Commun 2017, 53, 4977–4980. [DOI] [PubMed] [Google Scholar]
- (177).Alam KK; Tawiah KD; Lichte MF; Porciani D; Burke DH A Fluorescent Split Aptamer for Visualizing RNA-RNA Assembly In Vivo. ACS Synth. Biol 2017, 6, 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).O’Hara J; Marashi D; Morton S; Jaeger L; Grabow W Optimization of the Split-Spinach Aptamer for Monitoring Nanoparticle Assembly Involving Multiple Contiguous RNAs. Nanomaterials 2019, 9, 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Afonin KA; Viard M; Martins AN; Lockett SJ; Maciag AE; Freed EO; Heldman E; Jaeger L; Blumenthal R; Shapiro BA Activation of Different Split Functionalities on Re-Association of RNA-DNA Hybrids. Nat. Nanotechnol 2013, 8, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Afonin KA; Viard M; Tedbury P; Bindewald E; Parlea L; Howington M; Valdman M; Johns-Boehme A; Brainerd C; Freed EO; Shapiro BA The Use of Minimal RNA Toeholds to Trigger the Activation of Multiple Functionalities. Nano Lett. 2016, 16, 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (181).Benenson Y RNA-Based Computation in Live Cells. Curr. Opin. Biotechnol 2009, 20, 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Goldsworthy V; LaForce G; Abels S; Khisamutdinov E Fluorogenic RNA Aptamers: A Nano-Platform for Fabrication of Simple and Combinatorial Logic Gates. Nanomaterials 2018, 8, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Geary C; Rothemund PW; Andersen ES RNA Nanostructures. A Single-Stranded Architecture for Cotranscriptional Folding of RNA Nanostructures. Science 2014, 345, 799–804. [DOI] [PubMed] [Google Scholar]
- (184).Jepsen MDE; Sparvath SM; Nielsen TB; Langvad AH; Grossi G; Gothelf KV; Andersen ES Development of a Genetically Encodable FRET System Using Fluorescent RNA Aptamers. Nat. Commun 2018, 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Tsatsaronis JA; Franch-Arroyo S; Resch U; Charpentier E Extracellular Vesicle RNA: A Universal Mediator of Microbial Communication? Trends Microbiol. 2018, 26, 401–410. [DOI] [PubMed] [Google Scholar]
- (186).Xiao D; Ohlendorf J; Chen Y; Taylor DD; Rai SN; Waigel S; Zacharias W; Hao H; McMasters KM Identifying mRNA, microRNA and Protein Profiles of Melanoma Exosomes. PLoS One 2012, 7, No. e46874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Ohshima K; Inoue K; Fujiwara A; Hatakeyama K; Kanto K; Watanabe Y; Muramatsu K; Fukuda Y; Ogura S.-i.; Yamaguchi K; Mochizuki T Let-7 microRNA Family Is Selectively Secreted into the Extracellular Environment via Exosomes in a Metastatic Gastric Cancer Cell Line. PLoS One 2010, 5, No. e13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Haraszti RA; Miller R; Didiot M-C; Biscans A; Alterman JF; Hassler MR; Roux L; Echeverria D; Sapp E; DiFiglia M; Aronin N; Khvorova A Optimized Cholesterol-siRNA Chemistry Improves Productive Loading onto Extracellular Vesicles. Mol. Ther 2018, 26, 1973–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (189).Pi F; Binzel DW; Lee TJ; Li Z; Sun M; Rychahou P; Li H; Haque F; Wang S; Croce CM; Guo B; Evers BM; Guo P Nanoparticle Orientation to Control RNA Loading and Ligand Display on Extracellular Vesicles for Cancer Regression. Nat. Nanotechnol 2018, 13, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (190).Villarroya-Beltri C; Gutiérrez-Vázquez C; Sánchez-Cabo F; Pérez-Hernández D; Vázquez J; Martin-Cofreces N; Martinez-Herrera DJ; Pascual-Montano A; Mittelbrunn M; Sánchez-Madrid F Sumoylated hnRNPA2B1 Controls the Sorting of miRNAs into Exosomes through Binding to Specific Motifs. Nat. Commun 2013, 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Bolukbasi MF; Mizrak A; Ozdener GB; Madlener S; Ströbel T; Erkan EP; Fan J-B; Breakefield XO; Saydam O miR-1289 and ″Zipcode″-Like Sequence Enrich mRNAs in Microvesicles. Mol. Ther.–Nucleic Acids 2012, 1, No. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Squadrito ML; Baer C; Burdet F; Maderna C; Gilfillan GD; Lyle R; Ibberson M; De Palma M Endogenous RNAs Modulate microRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell Rep. 2014, 8, 1432–1446. [DOI] [PubMed] [Google Scholar]
- (193).Wang Y; Chen X; Tian B; Liu J; Yang L; Zeng L; Chen T; Hong A; Wang X Nucleolin-Targeted Extracellular Vesicles as a Versatile Platform for Biologics Delivery to Breast Cancer. Theranostics 2017, 7, 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Zou J; Shi M; Liu X; Jin C; Xing X; Qiu L; Tan W Aptamer-Functionalized Exosomes: Elucidating the Cellular Uptake Mechanism and the Potential for Cancer-Targeted Chemotherapy. Anal. Chem 2019, 91, 2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Yamashita T; Shinotsuka H; Takahashi Y; Kato K; Nishikawa M; Takakura Y SELEX-Based Screening of Exosome-Tropic RNA. Biol. Pharm. Bull 2017, 40, 2140–2145. [DOI] [PubMed] [Google Scholar]
- (196).Hung ME; Leonard JN A Platform for Actively Loading Cargo RNA to Elucidate Limiting Steps in EV-Mediated Delivery. J. Extracell. Vesicles 2016, 5, 31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (197).Sutaria DS; Jiang J; Elgamal OA; Pomeroy SM; Badawi M; Zhu X; Pavlovicz R; Azevedo-Pouly ACP; Chalmers J; Li C; Phelps MA; Schmittgen TD Low Active Loading of Cargo into Engineered Extracellular Vesicles Results in Inefficient miRNA Mimic Delivery. J. Extracell. Vesicles 2017, 6, 1333882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Lundstrom K Viral Vectors in Gene Therapy. Diseases 2018, 6, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (199).Link N; Aubel C; Kelm JM; Marty RR; Greber D; Djonov V; Bourhis J; Weber W; Fussenegger M Therapeutic Protein Transduction of Mammalian Cells and Mice by Nucleic Acid-Free Lentiviral Nanoparticles. Nucleic Acids Res. 2006, 34, No. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Aoki T; Miyauchi K; Urano E; Ichikawa R; Komano J Protein Transduction by Pseudotyped Lentivirus-Like Nanoparticles. Gene Ther. 2011, 18, 936–941. [DOI] [PubMed] [Google Scholar]
- (201).Telesnitsky A; Wolin SL The Host RNAs in Retroviral Particles. Viruses 2016, 8, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Joshi P; Prasad VR Potent Inhibition of Human Immunodeficiency Virus Type 1 Replication by Template Analog Reverse Transcriptase Inhibitors Derived by SELEX (Systematic Evolution of Ligands by Exponential Enrichment). J. Virol 2002, 76, 6545–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Lange MJ; Nguyen PDM; Callaway MK; Johnson MC; Burke DH RNA-Protein Interactions Govern Antiviral Specificity and Encapsidation of Broad Spectrum Anti-HIV Reverse Transcriptase Aptamers. Nucleic Acids Res. 2017, 45, 6087–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Hill KJ; Swain AD; Lange MJ; Burke DH Viral Hitchhiking to Increase Therapeutic Potential: Autonomously Packaged Elements (APEs). Mol. Ther 2015, 23, S56. [Google Scholar]
- (205).Prel A; Caval V; Gayon R; Ravassard P; Duthoit C; Payen E; Maouche-Chretien L; Creneguy A; Nguyen TH; Martin N; Piver E; Sevrain R; Lamouroux L; Leboulch P; Deschaseaux F; Bouillé P; Sensébé L; Pagès JC Highly Efficient In Vitro and In Vivo Delivery of Functional RNAs Using New Versatile MS2-Chimeric Retrovirus-Like Particles. Mol. Ther.–Methods Clin. Dev 2015, 2, 15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Lu B; Javidi-Parsijani P; Makani V; Mehraein-Ghomi F; Sarhan WM; Sun D; Yoo KW; Atala ZP; Lyu P; Atala A Delivering SaCas9 mRNA by Lentivirus-Like Bionanoparticles for Transient Expression and Efficient Genome Editing. Nucleic Acids Res. 2019, 47, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Nolte-’t Hoen E; Cremer T; Gallo RC; Margolis LB Extracellular Vesicles and Viruses: Are They Close Relatives? Proc. Natl. Acad. Sci. U. S. A 2016, 113, 9155–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Koppers-Lalic D; Hogenboom MM; Middeldorp JM; Pegtel DM Virus-Modified Exosomes for Targeted RNA Delivery; a New Approach in Nanomedicine. Adv. Drug Delivery Rev 2013, 65, 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Raab-Traub N; Dittmer DP Viral Effects on the Content and Function of Extracellular Vesicles. Nat. Rev. Microbiol 2017, 15, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Chen H; Zheng X; Di B; Wang D; Zhang Y; Xia H; Mao Q Aptamer Modification Improves the Adenoviral Transduction of Malignant Glioma Cells. J. Biotechnol 2013, 168, 362–366. [PubMed] [Google Scholar]
- (211).Panigaj M; Marino M; Reiser J Cell-Specific Targeting of Lentiviral Vectors Mediated by RNA Aptamers. Mol. Ther 2014, 22, S21–S21. [Google Scholar]
- (212).Chandler M; Afonin KA Smart-Responsive Nucleic Acid Nanoparticles (NANPs) with the Potential to Modulate Immune Behavior. Nanomaterials 2019, 9, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Litke JL; Jaffrey SR Highly Efficient Expression of Circular RNA Aptamers in Cells Using Autocatalytic Transcripts. Nat. Biotechnol 2019, 37, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]


