Abstract
Human adenovirus (HAdV) can cause severe disease in certain at-risk populations such as newborns, young children, the elderly and individuals with a compromised immune system. Unfortunately, no FDA-approved antiviraldrug is currently available for the treatment of HAdV infections. Within the nucleus of infected cells, the HAdV genome associates with histones and forms a chromatin-like structure during early infection, and viral gene expression appears to be regulated by cellular epigenetic processes. Thus, one potential therapeutic strategy to combat HAdV disease may be to target the cellular proteins involved in modifying the viral nucleoprotein structure and facilitating HAdV gene expression and replication. We have screened a panel of small molecules that modulate the activity of epigenetic regulatory proteins for compounds affecting HAdV gene expression. Several of the compounds, specifically chaetocin, gemcitabine and lestaurtinib, reduced HAdV recovery by 100- to 1000-fold, while showing limited effects on cell health, suggesting that these compounds may indeed be promising as anti-HAdV therapeutics.
Keywords: Adenovirus, Small-molecule screen, Antiviral compounds, Epigenetic regulators
1. Introduction
Human adenovirus (HAdV) has a remarkable capacity to spread with patients contracting disease from as few as 5 virus particles (Musher, 2003). To place this in perspective, sputum from infected individuals typically contain 106 and 107 virus particles per milliliter. As a result, HAdV can spread rapidly through confined populations such as schools, daycares, long-term care facilities and military training venues. HAdV mainly causes minor illnesses in most infected individuals, but can lead to significant disease, and even death, in certain groups, including neonates, young children, the elderly, and immunocompromised individuals (Bhatti and Dhamoon, 2017; Binder et al., 2017; Kajon and Ison, 2016; Kojaoghlanian et al., 2003; Lion, 2014; Ying et al., 2014). Due to significant loss of recruit-hours to HAdV disease, the US military has recently re-established a mandatory anti-HAdV vaccination program for all new recruits (Hoke and Snyder, 2013). During the 2 years after re-introduction of the anti-HAdV vaccine for military personnel, there was more than a 100-fold decline in HAdV disease burden (from 5.8 to 0.02 cases per 1000 weeks attended by recruits), eliminating approximately 1 death, 1100–2700 hospitalizations, and 13,000 febrile HAdV cases each year (Radin et al., 2014). This vaccine is not available to civilians and no specific therapeutic exists to combat HAdV-induced disease. Antivirals typically used to treat severe HAdV infections include several drugs approved for other viruses, such as cidofovir, ganciclovir and ribavirin (Keyes et al., 2016; Lenaerts et al., 2008; Ronchi et al., 2014). However, these drugs often have low efficacy against HAdV and can be associated with serious side effects (Caruso Brown et al., 2015; Keyes et al., 2016; Ronchi et al., 2014). Thus, new and more effective drugs that more directly target HAdV or HAdV-induced disease are needed.
Several research groups have performed small molecule screens to identify novel anti-HAdV compounds (Duffy et al., 2013; Grosso et al., 2017; Hartline et al., 2018; Marrugal-Lorenzo et al., 2018; Sanchez-Cespedes et al., 2016; Wang et al., 2018). To facilitate and simplify high-throughput screening for anti-HAdV compounds, we recently developed a wildtype-like HAdV type 5 (HAdV-5) reporter construct that contained the red fluorescent protein (RFP) gene under regulation of the HAdV major late promoter (MLP), such that the level of RFP expression correlated with viral late gene expression and DNA replication (Saha and Parks, 2019). Thus, quantification of fluorescence intensity in cells infected with this virus could be used to easily identify compounds that affect virus infection, gene expression and/or replication. Using this platform, a screen of ~1200 FDA-approved drugs was conducted and a number of compounds that reduced HAdV replication were found, including several cardiotonic steroids, corticosteroids and chemotherapeutic agents (Saha et al., 2019). These novel anti-HAdV compounds, in addition to those identified in successful screens previously conducted by other research groups, could be developed as antiviral agents to treat HAdV-induced disease in patients.
HAdV is a nuclear DNA virus, encoding early and late genes which are expressed before and after the onset of viral DNA replication, respectively (Berk, 2007). As with all viruses, HAdV is exquisitely adept at manipulating host cellular proteins and pathways to effectively achieve viral gene expression, genome replication, and progeny virion production. In the virion, the HAdV genome associates with three highly basic proteins (VII, V and mu), which form a condensed nucleoprotein structure that is refractory to viral gene expression (Korn and Horwitz, 1986; Matsumoto et al., 1993). Thus, this structure must be remodeled to allow gene expression and DNA replication to proceed efficiently in the newly-infected cell nucleus. We and others have shown that shortly after reaching the nucleus, HAdV DNA associates with histones and assembles into a chromatin-like structure similar to that of the host genome (Giberson et al., 2012, 2018; Komatsu and Nagata, 2012; Lynch et al., 2019; Ross et al., 2011; Wong et al., 2013). Virus-encoded genes are expressed from this template, and expression can be regulated by cellular epigenetic modifiers (Hsu et al., 2018; Komatsu et al., 2011; Soriano et al., 2019). Although only a subset of cellular proteins involved in regulating the HAdV genome has been identified to date, these observations raise the possibility of targeting such proteins in order to inhibit viral gene expression and replication. Indeed, a similar approach has been effective against several other viruses (Arbuckle et al., 2017; Hill et al., 2014; Liang et al., 2009, 2013a, 2013b). For HAdV, we previously showed that treatment of infected cells with histone deacetylase (HDAC) inhibitors reduced viral gene expression, DNA replication and virus recovery (Saha and Parks, 2019). The pan-HDAC inhibitor vorinostat (or SAHA) had efficacy against types 4, 5 and 7. Similarly, inhibition of methyltransferases EZH2 and EZH1 (EZH2/1) and histone H3K9 demethylase LSD1 was shown to attenuate HAdV gene expression (Arbuckle et al., 2017; Liang et al., 2013a). Thus, epigenetic regulators are valid therapeutic targets to combat HAdV infection.
In this study, we performed a screen of 150 small molecules that modulate the activity of epigenetic regulatory proteins, including methyltransferases, demethylases, histone acetyltransferases and deacetylases, and acetylated lysine readers. We identified 19 compounds that reduced gene expression from our reporter HAdV by at least 50% and, consistent with our previous reports, most of these positive hits were pan-HDAC inhibitors. Here, we have conducted follow-up studies on three compounds with non-HDAC targets: chaetocin, gemcitabine and lestaurtinib. All three compounds significantly reduced HAdV gene expression from several HAdV types, suggesting potential broad applicability as anti-HAdV agents.
2. Materials and methods
2.1. Cell and virus culture
The drug screen and follow-up studies were performed in human lung adenocarcinoma A549 cells (CCL-185, ATCC). Cells were cultured in Minimum Essential Medium (MEM, Sigma-Aldrich) containing 10% (v/v) Fetal Bovine Serum (FBS, Sigma Aldrich), 2 mM GlutaMAX (Invitrogen) and 1x antibiotic-antimycotic (Invitrogen).
Ad-late/RFP was used to perform the initial screen, and the construction of this virus was described previously (Saha and Parks, 2019). Ad-late/RFP is based on HAdV-5, and contains an intact E1 region, but has the E3 region replaced by the monomeric RFP coding sequence with an upstream splice acceptor site, which places RFP expression under the control of the viral MLP (Del Papa et al., 2019; Saha and Parks, 2019). The virus was propagated in 293 cells and purified by cesium chloride buoyant density centrifugation using standard procedures (Ross and Parks, 2009). Viral titer (in plaque forming units or PFU) was determined by plaque assay on 293 cells as reported previously (Saha and Parks, 2017). HAdV-4 (VR-4, ATCC) and HAdV-7 (VR-7, ATCC) stocks were propagated and titered on A549 cells. HAdV-5 was obtained from Dr. John Bell (Ottawa Hospital Research Institute), and was grown and titered on 293 cells.
2.2. Infection and drug treatment
For all infections other than the compound screen (described below), medium was removed from confluent monolayers of A549 cells before adding HAdV-5 diluted in phosphate-buffered saline (PBS, Sigma-Aldrich). The multiplicity of infection (MOI) was calculated as PFU per cell, and an MOI of 10 was used for all experiments. After incubation for 1 h at 37 °C, medium containing vehicle/drug was added. The infected cells with drug/vehicle were incubated in a humidified CO2 incubator at 37 °C until protein/DNA extraction at the indicated time points.
2.3. Chemical library and screen
The Epigenetics Screening Library (cat# 11076, batch 0468238–2) was obtained from Cayman Chemical. Each compound in the library was provided as a 10 mM stock in dimethyl sulfoxide (DMSO). For validation and follow-up experiments, aliquots from the original library were used: chaetocin (item number 13156, ≥95% purity), gemcitabine (item number 11690, ≥98% purity), lestaurtinib (item number 12094, ≥98% purity). All compounds were dissolved in DMSO to prepare stock solutions and diluted in cell media for treatments.
The protocol for the small molecule screen has been described in detail previously (Saha et al., 2019). A549 cells were seeded in 96-well plates at a density of 20,000 cells/well. The next day, compounds from the library were diluted in PBS containing 10% DMSO, and immediately added to the cells to achieve a final concentration of 1 μM or 0.25 μM. After 2 h of treatment, the cells were infected at an MOI of 10 with Ad-late/RFP prepared in MEM. The infected, drug-treated cells were incubated at 37 °C until 24 h post infection (hpi). The media was aspirated, the cell monolayer was washed once with PBS, and fixed with 4% w/v paraformaldehyde (pH 7) for 15 min. The fixed cells were stored in PBS at 4 °C overnight. All drug treatments were performed in triplicate (e.g. three 96-well plates containing the same compound treatments, and one well per treatment per plate). Prior to RFP quantification, the cell nuclei were stained by a 15–20 min incubation with 10 μg/ml Hoechst 33342 (Life Technologies) diluted in PBS, followed by replacement with fresh PBS. RFP quantification was performed using the Cellomics High Content Screening (HCS) Platform and the Cellomics navigator version of the HCS Studio software (Thermo Fisher Scientific). Images were obtained using the 10x objective, the 386excitation (ex) nm filter for Hoechst, and the 549ex nm filter for RFP. Roughly 1,000 cells (identified by Hoechst-stained intact nuclei) per well were analyzed for RFP quantification. The RFP intensity in each drug-treated well was calculated as a percentage of the intensity in vehicle-treated wells. The mean data from the three replicate plates was used for analysis.
2.4. Immunoblot analysis
Cells were lysed in SDS–PAGE protein loading buffer (62.5 mM Tris of pH 6.8, 10% glycerol, 2% SDS, 0.1% bromophenol blue, 5% 2-mercaptoethanol), boiled for 10–20 min, and separated by electrophoresis on a 15% SDS–polyacrylamide gel. The separated proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) using the Trans-Blot SD Semi-Dry Transfer System (Bio-Rad). The membrane was blocked with 5% milk in tris-buffered saline containing 0.2% Tween 20 (Thermo Fisher Scientific), and probed with antibodies diluted in the 5% milk solution. The following primary antibodies were used: HAdV-5 E1A (1/5000; MA5-13643, Invitrogen), HAdV-5 capsid protein antibody (1/10,000; ab6982, Abcam), vinculin (1/10,000; ab129002, Abcam), and actin (1/10,000; A1978, Sigma Aldrich). The antibody raised against HAdV-5 virion proteins detects all of the major HAdV-5 structural proteins, and also cross-reacts with some of the HAdV-4 and HAdV-7 capsid proteins. Following exposure to antibody, the membranes were washed three times in TBST and incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (HRP). Blots were developed using the Immobilon Classico Western HRP Substrate (Millipore).
2.5. Quantitative real-time PCR (qPCR)
A549 cells were infected and treated with drug as described above. Medium was removed from the infected cells at the indicated time points and the cells were incubated overnight in SDS-proteinase K buffer (10 mM Tris-HCl of pH 7.4, 10 mM EDTA, 1% w/v SDS, 1 mg/ml proteinase K) at 37 °C. DNA was extracted with phenol-chloroform, precipitated with ethanol and NaCl, and the resulting DNA pellet was dissolved in 1x Tris-EDTA (TE) buffer. qPCR was conducted as described previously (Ross et al., 2011) using 200 ng of genomic DNA per reaction, and the following primers directed to the hexon region of the genome: 5′-CTT ACC CCC AAC GAG TTT GA and 5′-GGA GTA CAT GCG GTC CTT GT. Viral genome copy numbers were calculated from the Ct values using a standard curve obtained using serial dilutions of pRP3089, a bacterial HAdV genomic plasmid containing the hexon gene.
2.6. Plaque assay for virus yield
A549 cells in 35 mm dishes were infected with HAdV-5 (MOI 10) for 1 h. The virus inoculum was removed and the cells were washed with PBS to remove any unattached virions prior to replacement of fresh medium containing vehicle or compounds. After 24 h of infection, the cells were collected by scraping into the medium, sucrose (diluted in 10 mM Tris) was added to a final concentration of 4% w/v, and the samples were flash-frozen in ethanol and dry ice. The resulting crude stock of virus was serially diluted and used to infect monolayers of A549 cells. After 1 h of infection, the cells were overlaid with medium containing agarose (50% v/v of a 1% w/v agarose solution, 43% clear 2x MEM, 5% FBS, 1% GlutaMAX and 1% antibiotic-antimycotic) and placed in a 37 °C incubator. Plaques were counted 8–10 days later.
2.7. MTS metabolic activity assays
A549 cells in 96-well plates were infected with HAdV-5, treated with the indicated dilutions of compounds and 24 h later, the metabolic activity was determined using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega). The assay was conducted according to the manufacturer's instructions. Briefly, cells were incubated for 1 h at 37 °C with 20 μl of the MTS substrate, and absorbance readings were obtained at 490 nm using the SpectraMax 190 plate spectrophotometer (Molecular Devices).
3. Results
3.1. Screening the Cayman Chemicals Epigenetics Screening Library for compounds exhibiting anti-HAdV activity
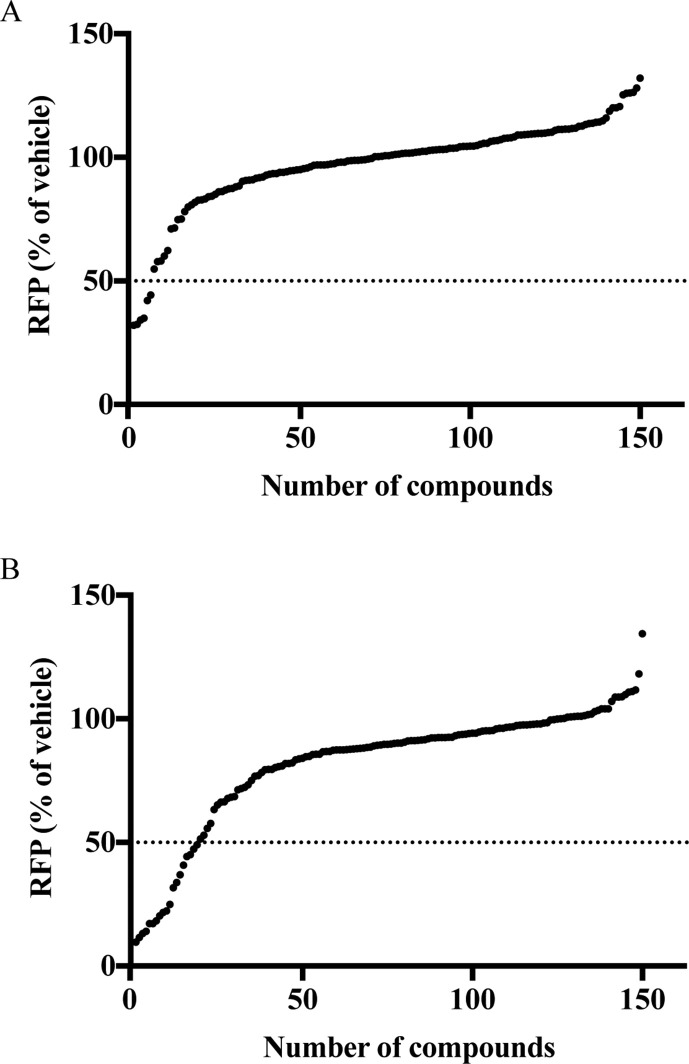
Recently, we generated a reporter HAdV construct (designated Ad-late/RFP) and established a protocol to perform small-molecule screens for compounds affecting HAdV gene expression and replication (Saha et al., 2019). We used this approach again to screen the Cayman Chemicals Epigenetics Screening Library. A549 cells were infected with Ad-late/RFP, from which, RFP expression serves as an effective surrogate marker to monitor and quantify drug-induced changes in viral gene expression and replication. The infected cells were treated with the various compounds at two different concentrations (1 μM or 0.25 μM), and RFP levels were determined 24 h later by high-content imaging. As shown in Fig. 1 A, treatment of cells with 0.25 μM drug allowed the identification of 6 compounds that reduced RFP expression by at least 50%. Some of these compounds yielded the greatest effects on RFP expression at the 1 μM screening concentration, in addition to the identification of 13 other compounds that reduced RFP expression by 50% or more (Fig. 1B). A summary of the positive hits from the screen is provided in Table 1 .
Fig. 1.
Screen of the Cayman Epigenetics Screening Library. A549 cells in 96-well plates were pretreated with vehicle or compounds at a concentration of 0.25 μM (panel A) or 1 μM (panel B) for 2 h prior to infection with Ad-late/RFP (MOI 10). Cells were fixed at 24 hpi and RFP intensities were measured using the Cellomics ArrayScan VTI High Content Screening Platform. Changes in RFP levels were calculated as a percentage of vehicle treatment. Each data point represents the mean RFP percentage from three independent assays for one compound – error bars have been omitted for clarity. The RFP values for all 150 compounds were plotted in order of increasing magnitude (i.e. the compound numbers in the x-axis are arbitrary). While most compounds did not alter RFP expression from Ad-late/RFP, at both screen concentrations, several compounds were found to either significantly reduce or moderately enhance RFP levels.
Table 1.
Positive hits from the Cayman Epigenetics Screening Library. These compounds reduced RFP by at least 50% at 24 hpi - the percentage values indicate the RFP levels in cells treated with 1 μM drug relative to vehicle-treated cells (100% RFP) in the screen.
| Compound | % RFPa | Mode of action |
|---|---|---|
| 1) Chaetocinb | 10 | Methyltransferase inhibitor |
| 2) Apicidinb | 12 | HDAC inhibitor |
| 3) HC Toxinb | 13 | HDAC inhibitor |
| 4) Panobinostatb | 14 | HDAC inhibitor |
| 5) Givinostat | 17 | HDAC inhibitor |
| 6) Oxamflatin | 17 | HDAC inhibitor |
| 7) Gemcitabineb | 18 | DNA synthesis inhibitor |
| 8) Trifluoroacetate salt | 29 | HDAC inhibitor |
| 9) Pracinostat | 20 | HDAC inhibitor |
| 10) TSA | 22 | HDAC inhibitor |
| 11) Lestaurtinib | 22 | Tyrosine kinase inhibitor |
| 12) Decitabine | 25 | DNMT-1 inhibitor |
| 12) LAQ824b | 32 | HDAC inhibitor |
| 14) CAY10603 | 34 | HDAC inhibitor |
| 15) CUDC-101 | 37 | Multi-targeted inhibitor of HDACs, EGFR, HER2 |
| 16) M344 | 41 | HDAC inhibitor |
| 17) 4-iodo-SAHA | 45 | HDAC inhibitor |
| 18) CBHA | 47 | HDAC inhibitor |
| 19) Scriptaid | 49 | HDAC inhibitor |
Values represent the mean of three experimental replicates. The CV for all compounds was <35%, with the exception of CUDC-101 (61%) and LAQ824 (111%).
These compounds were also identified as positive hits when the screen was conducted with all compounds at 0.25 μM. The RFP percentage for chaetocin, apicidin, HC toxin, panobinostat, gemcitabine and LAQ824 were 34%, 42%, 35%, 32%, 44% and 32%, respectively.
Of the 19 compounds found to reduce RFP expression from our HAdV-based reporter construct at 1 μM, 15 are HDAC inhibitors (Table 1). This result is unsurprising since we and others previously showed that HDAC inhibitors are potent inhibitors of HAdV gene expression and replication (Hoti et al., 2006; Saha and Parks, 2019). Chaetocin, which reduced RFP expression to 10% of vehicle-treated cells, inhibits histone methyltransferases (HMTs), primarily the H3 Lys9 HMTs SUV39H1 and G9a. Gemcitabine (18% RFP expression relative to vehicle) is a nucleoside analog that induces termination of chain elongation when incorporated into DNA, but can also inhibit DNA methyltransferase protein DNMT1 (Gailhouste et al., 2018; Gray et al., 2012). Lestaurtinib (22% RFP expression relative to vehicle) inhibits JAK2 and PRK1, which can phosphorylate H3 Tyr41 and H3 Thr 11, respectively. Finally, decitabine (25% RFP expression relative to vehicle) inhibits DNA methyltransferases. We performed validation and follow up studies on chaetocin, gemcitabine and lestaurtinib.
3.2. Effect of chaetocin, gemcitabine and lestaurtinib on cellular metabolic activity
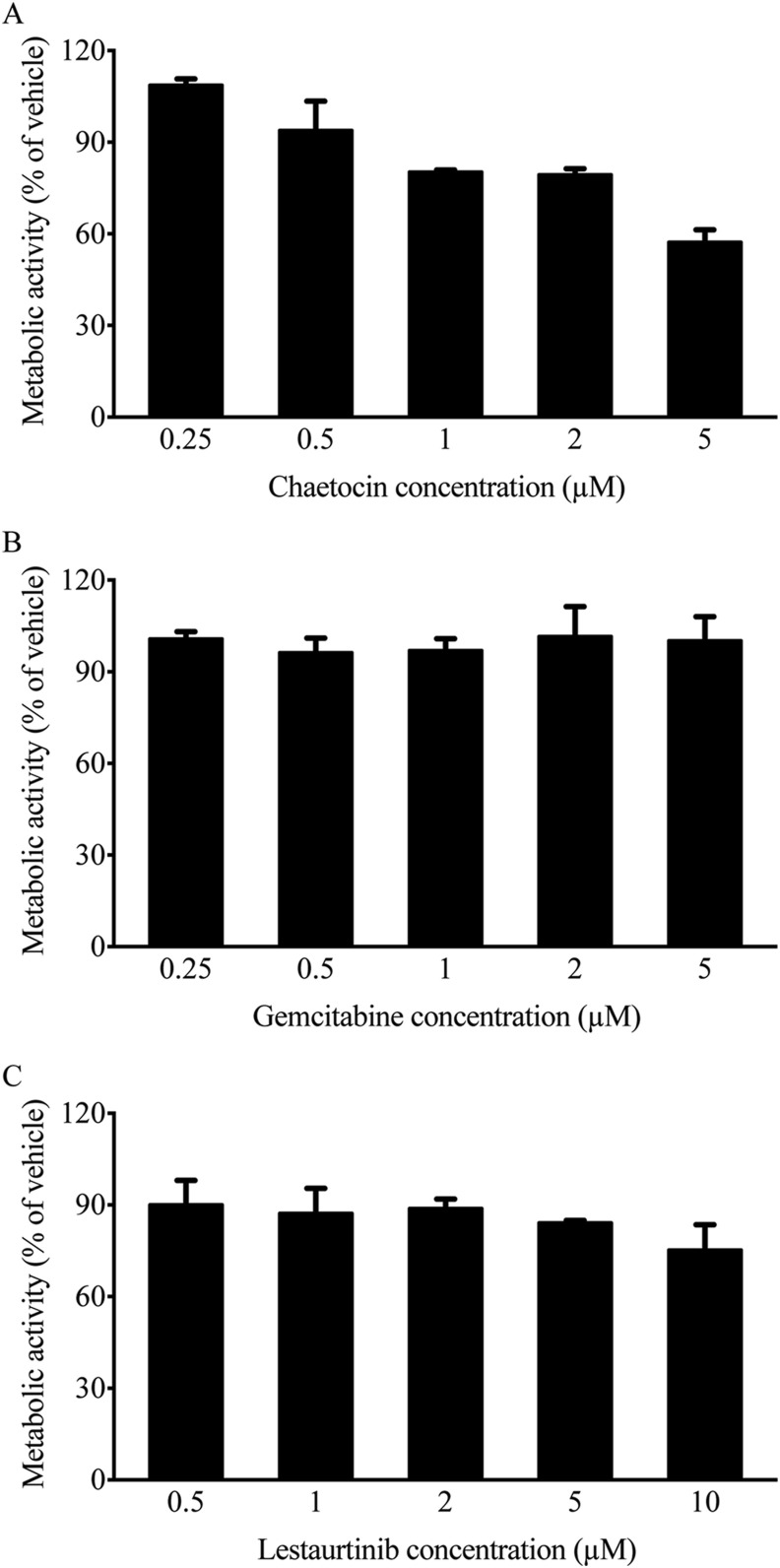
The various drugs identified in our screen could be directly affecting virus function/biology, or could be acting indirectly through effects on cell health. We therefore examined how these drugs alter cellular metabolic activity. A549 cells were infected with HAdV-5 at an MOI of 10, treated with vehicle (DMSO) or each compound at the indicated concentrations for 24 h, and cellular metabolic activity was assessed. As shown in Fig. 2 , chaetocin, lestaurtinib and gemcitabine all had minimum impact on cell health over the range of concentrations used in our assay, with a reduction in metabolic activity of no more than 20% within the range of 0.25–2 μM. At 5 μM, chaetocin induced a 40% reduction in metabolic activity in treated cells, while gemcitabine had no noticeable impact, and lestaurtinib led to only a 25% reduction at 10 μM.
Fig. 2.
Dose-dependent effects of selected positive hits on cellular metabolic activity. A549 cells were infected with HAdV-5 (MOI 10), and treated with vehicle (DMSO) or each compound at the indicated concentrations for 24 h. Cellular metabolic activity was determined by MTS assay, and calculated as a percentage of vehicle treatment. Error bars represent standard deviation (SD) of experimental replicates (n = 2).
3.3. Effect of chaetocin, gemcitabine and lestaurtinib on HAdV gene expression
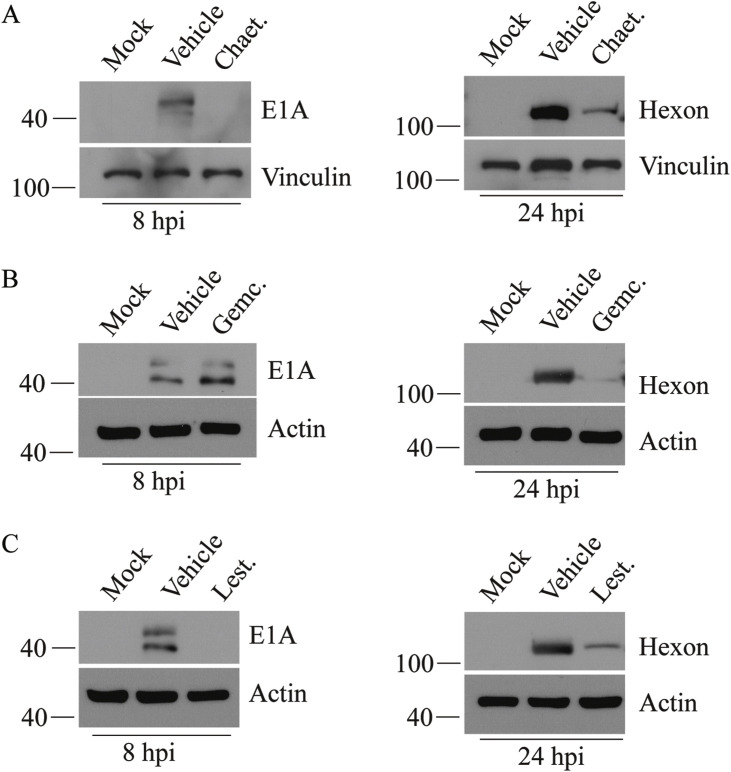
We first performed a dose response assay to determine the effect of drug concentration on HAdV late gene expression, as gauged by RFP expression from our reporter construct. Based on this preliminary analysis (data not shown), drug concentrations used in all subsequent studies are: chaetocin at 1 μM, gemcitabine at 0.5 μM and lestaurtinib at 5 μM. At these concentrations, cellular metabolic activity was >80% relative to vehicle treatment (Fig. 2). To determine whether these compounds interfered with the early or late stage of the virus lifecycle, we examined the quantity of viral proteins E1A (a representative early protein) and hexon (a representative late protein) in infected and treated cells. A549 cells were infected with HAdV-5 at an MOI of 10 for 1 h, at which time, complete medium or medium containing vehicle or drug was placed on the cells. Crude protein samples were collected at 8 hpi to assess E1A protein levels by immunoblot, and at 24 hpi for examination of the late protein hexon. As shown in Fig. 3 , treatment of cells with both chaetocin and lestaurtinib reduced E1A protein levels, whereas gemcitabine had no effect on E1A expression. All three of the drugs significantly reduced the levels of the late hexon protein in treated cells at the 24 h timepoint. These results indicate that unlike chaetocin and lestaurtinib, gemcitabine exerts its effects in the later stages of HAdV lifecycle, during or after viral DNA replication.
Fig. 3.
Chaetocin, gemcitabine and lestaurtinib reduce expression of viral early and late proteins. A549 cells were infected at an MOI of 10 of HAdV-5 for 1 h, and medium containing vehicle or compound was added. Compound concentrations used were chaetocin at 1 μM, gemcitabine at 0.5 μM and lestaurtinib at 5 μM. Cell lysates were collected at the indicated times and subjected to immunoblot analysis for detection of early (E1A) and late (hexon) proteins.
3.4. Effect of chaetocin, gemcitabine and lestaurtinib on HAdV genome replication in treated cells
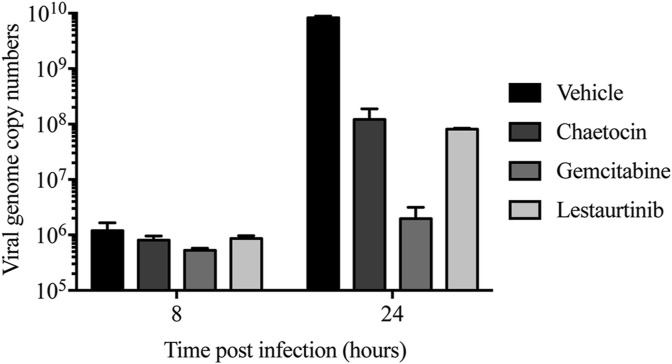
We next evaluated the effect of chaetocin, gemcitabine and lestaurtinib on viral genome copy numbers within treated cells. Infection and drug treatment of A549 cells were performed as described above. At 8 and 24 hpi, the cells were treated with SDS-proteinase K, and total cellular DNA was isolated using standard methods. Quantitative PCR analysis with primers specific to the viral hexon region of the genome showed that in untreated cells, there was a 4-log increase in viral genome copy number between 8 and 24 hpi. Treatment of cells with gemcitabine had a significant negative effect on the quantity of viral DNA produced within the cell, with a relatively minor 3-fold increase between 8 and 24 hpi (Fig. 4 ). Chaetocin and lestaurtinib led to a 2-log reduction in genome copy number within the treated cells at 24 hpi, relative to infected, vehicle-treated cells.
Fig. 4.
Chaetocin, gemcitabine and lestaurtinib reduce virus genome copy numbers within treated cells. A549 cells were infected and treated as described in Fig. 3. At the indicated times, the infected cells were treated with SDS-proteinase K, and the DNA was isolated by phenol/chloroform extraction and ethanol precipitation. The resulting genomic DNA was subjected to qPCR using primers specific to hexon. HAdV genome copy numbers per 200 ng of total gDNA are reported. Error bars represent SD of experimental replicates (n = 2).
3.5. Treatment of cells with chaetocin, gemcitabine and lestaurtinib significantly reduce HAdV yield
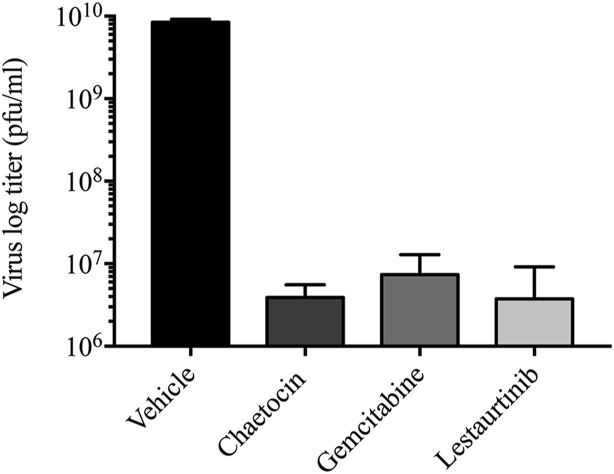
As chaetocin, gemcitabine and lestaurtinib all decreased genome copy numbers (Fig. 4) and late gene expression (Fig. 3) of HAdV-5 at 24 hpi, we predicted that these compounds would also affect yield of virus from the infected cells. A549 cells were infected at an MOI of 10 with HAdV-5 for 1 h, the cells were washed with PBS to remove residual, unattached virus, and complete medium containing appropriate drug or vehicle was added. At 24 hpi, the infected cells were scraped into the medium, freeze/thawed to release virus, and titer of the virus was determined on A549 cells. Treatment with all three drugs resulted in more than a 3-log reduction in virus recovery (Fig. 5 ). Indeed, the quantity of recovered virus from drug treated cells likely represents the residual amount of virus typically present in cells from the infecting inoculum (Sargent et al., 2004), suggesting minimal progeny virions were recovered from the drug-treated cells. Taken together, our results clearly show that treatment of HAdV-5-infected cells with chaetocin, gemcitabine and lestaurtinib reduces viral gene expression, genome replication and virus recovery.
Fig. 5.
Chaetocin, gemcitabine and lestaurtinib reduce virus yield from infected cells. A549 cells were infected at an MOI of 10 of HAdV for 1 h, and washed with PBS to remove unbound virus before adding medium containing vehicle or compound (chaetocin at 1 μM, gemcitabine at 0.5 μM and lestaurtinib at 5 μM). At 24 hpi, the infected cells were collected into the medium and the titer of virus in the cell lysates were determined by plaque assay. Error bars represent SD of experimental replicates (n = 2).
3.6. Chaetocin, gemcitabine and lestaurtinib inhibit clinically-relevant HAdV types
Our studies show that all three drugs we validated reduced recovery of HAdV-5. However, HAdV-5 is not the most prevalent serotype associated with human disease, and accounted for only 3.7% of all HAdV infection cases reported through the United States National Adenovirus Type Reporting System in the years spanning 2003–2016 (Binder et al., 2017). HAdV-4 and HAdV-7 are typically associated with more severe disease in human patients, and account for 12.4 and 8.5%, respectively, of all reported cases during the same time period (Binder et al., 2017). Indeed, these two types of HAdV were such a burden on the US Army, with 80% of new recruits becoming infected and 20% requiring hospitalization, that an anti-HAdV vaccination program for HAdV-4 and HAdV-7 was recently re-established (Clemmons et al., 2017; Hoke and Snyder, 2013; Radin et al., 2014). We thus tested chaetocin, gemcitabine and lestaurtinib against these two HAdV types. A549 cells were infected with HAdV-4 or HAdV-7 and treated with drug as previously described, crude protein samples were collected at 24 hpi, and examined by immunoblot for expression of a late protein, penton. Since late proteins are not expressed at appreciable levels until after the onset of viral DNA replication (Akusjarvi and Persson, 1981; Larsson et al., 1992; Thomas and Mathews, 1980), evaluating the levels of penton acts as a surrogate marker for overall effects on viral fitness. As shown in Fig. 6 , all three compounds significantly reduced the quantity of penton protein in infected cells, with a similar trend for both HAdV types, indicating that each compound exhibited anti-HAdV activity. Both chaetocin and lestaurtinib effectively abrogated penton expression from both HAdV at the 24 h timepoint, while gemcitabine reduced penton protein levels to ~5% of that observed in vehicle-treated cells at 24 hpi. Taken together, these data indicate that chaetocin, gemcitabine and lestaurtinib act as effective anti-HAdV compounds, and are effective against multiple different types.
Fig. 6.
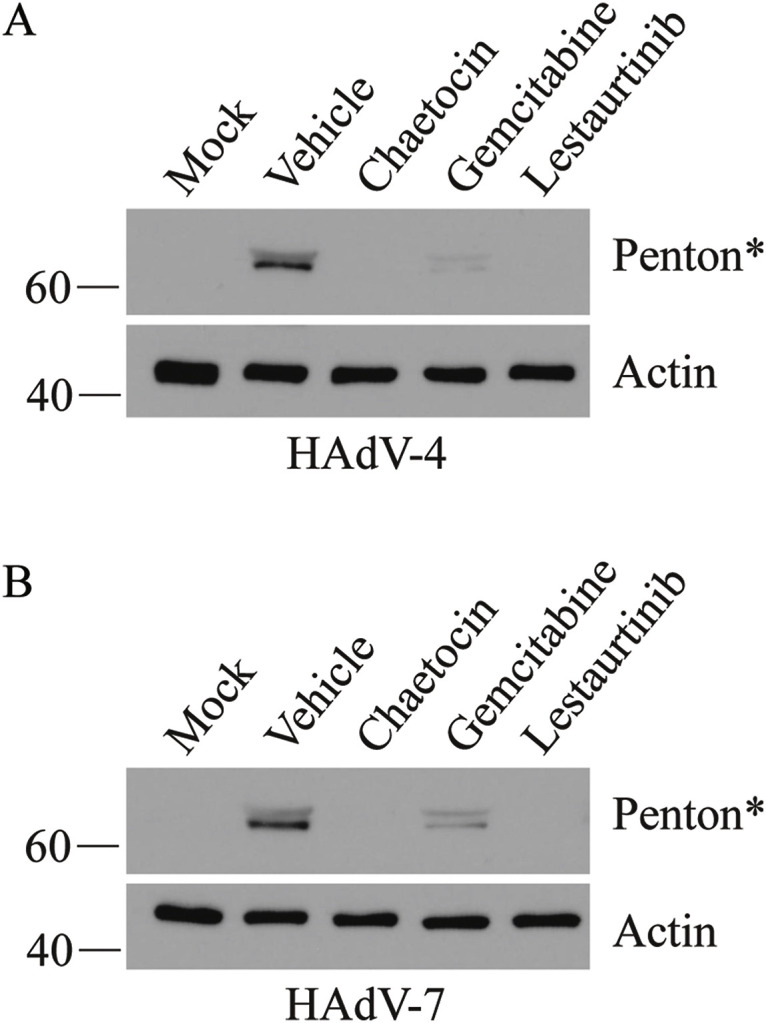
Chaetocin, gemcitabine and lestaurtinib reduce late gene expression from HAdV-4 and HAdV-7. A549 cells were infected at an MOI of 10 of HAdV-4 or HAdV-7 for 1 h, and overlaid with medium containing vehicle or compound (chaetocin at 1 μM, gemcitabine at 0.5 μM and lestaurtinib at 5 μM). Cell lysates were collected at 24 hpi for analysis of late protein expression by immunoblot with an antibody raised to HAdV-5 capsid proteins. *The protein detected using the anti-HAdV-5 antibody, which cross-reacts the HAdV-4 and HAdV-7 capsid proteins, is most likely penton based on its size of ~60 kDa.
4. Discussion
Although HAdV is generally considered a relatively innocuous virus, infections can be associated with high morbidity and mortality in some cases. HAdV can lead to severe disease in seemingly healthy individuals, with no obvious susceptibility factors that impact disease manifestation (Bhatti and Dhamoon, 2017; Binder et al., 2017; Kojaoghlanian et al., 2003; Lion, 2014). A recent outbreak that gained much attention in the popular press involved HAdV-7 at the Wanaque Center for Nursing and Rehabilitation in New Jersey, infecting ~35 people, including 23 children, and resulting in 11 deaths within the pediatric population (Crenshaw et al., 2019). There are no approved anti-HAdV therapeutics. Many approved antivirals for other viruses have been used to combat HAdV infections in patients, such as cidofovir, ganciclovir and ribavirin (Keyes et al., 2016; Lenaerts et al., 2008; Ronchi et al., 2014), however, with variable efficacy.
During early infection, within the cell nucleus, HAdV DNA forms a nucleosome-bound structure that is similar to cellular chromatin (Giberson et al., 2012, 2018; Komatsu and Nagata, 2012; Ross et al., 2011; Wong et al., 2013). This nucleoprotein structure serves as the template for early gene expression and initiation of DNA replication. Previously, we showed that expression of genes from HAdV-based vectors (Ross et al., 2009) or wildtype virus (Saha and Parks, 2019) can be influenced by drugs targeting epigenetic regulatory proteins that were not previously known to play a role in HAdV infection. Similarly, drugs that target other epigenetic regulators could prevent transition of the viral DNA to a transcriptionally active template, which may consequently prevent the virus from establishing a productive infection within a newly-infected cell. We thus performed a screen to identify compounds that affect HAdV function, using the Cayman Chemical Epigenetic Screening Library, which is comprised of compounds with described abilities to modulate activities of cellular epigenetic regulatory proteins. Of the ~150 drugs present in the library, 19 were identified to have anti-HAdV-5 activity. Among those, 15 were HDAC inhibitors. The identification of HDAC inhibitors as anti-HAdV compounds is consistent with data from our previous studies showing that such compounds were effective in reducing HAdV gene expression, DNA replication and virus yield (Saha and Parks, 2019). This effect was primarily due to the loss of class I HDAC activity, mainly HDAC2. Interestingly, we showed that E1A physically interacts with HDAC2, suggesting that E1A may naturally function to modulate the activity or gene targeting of HDAC2. HAdV E1A protein has been shown to interact with and redirect another cellular epigenetic regulatory protein, p300/CBP histone acetyltransferase, and epigenetically reprogram cellular chromatin, thereby conferring a global change in cellular gene expression (Ferrari et al., 2008; Horwitz et al., 2008). E1A was also shown to bind the hBre1/RNF20 complex to prevent monoubiquitination of histone 2B (H2B) at lysine 120, an event necessary for activation of interferon stimulated genes (Fonseca et al., 2012). Thus, E1A, the first protein expressed during viral infection, may serve as a generalized factor that interacts with key epigenetic regulatory proteins to achieve global changes in gene expression to mold the cellular microenvironment for optimal virus replication.
The 4 remaining compounds identified in our screen that reduced HAdV replication were chaetocin (inhibits histone methyltransferases), gemcitabine (a nucleoside analog and DNA methyltransferases 1 inhibitor), lestaurtinib (inhibits JAK2 and PRK1 kinases which can phosphorylate H3), and decitabine (inhibits DNA methyltransferases). We performed follow-up experiments on 3 of these compounds, and all showed efficacy in reducing viral gene expression (Fig. 3), DNA replication (Fig. 4) and virus yield (Fig. 5), while having minimal impact on cell health (Fig. 2). These compounds were also effective against multiple HAdV types (Fig. 6).
Gemcitabine is a nucleoside analog approved for treatment of various cancers, including breast, ovarian, non-small cell lung, pancreatic, and bladder cancers. This drug inhibits cell growth and induces death through multifaceted functions. Within the cell, gemcitabine (a prodrug) is converted to a triphosphate form that, when incorporated into DNA, prevents DNA chain elongation (Hertel et al., 1990). Gemcitabine is also a potent inhibitor of ribonucleotide reductase, which results in the depletion of deoxynucleoside triphosphate pools within the cell (Heinemann et al., 1990). Lastly, gemcitabine interferes with epigenetic regulation by inhibiting and destabilizing the DNA methyltransferase protein DNMT1 (Gailhouste et al., 2018; Gray et al., 2012). Gemcitabine has been explored in combination therapies with HAdV-based vectors in a number of studies for treatment of cancer (Nattress and Hallden, 2018), but not as an HAdV-directed antiviral therapeutic. However, it does show broad spectrum antiviral activity against many RNA viruses, such as Middle East respiratory syndrome (MERS) coronavirus, severe acute respiratory syndrome (SARS) coronavirus, Zika virus, hepatitis C virus (HCV), poliovirus, influenza A virus, human immunodeficiency virus (HIV), and enteroviruses (Shin et al., 2018), as well as the dsDNA-containing Herpes simplex virus type I (HSV1) (Denisova et al., 2012). Other nucleoside analogs have shown efficacy against HAdV, including cidofovir, ganciclovir and ribavirin (Keyes et al., 2016; Lenaerts et al., 2008; Ronchi et al., 2014). Thus, we have now extended this class of potential anti-HAdV therapeutic to include gemcitabine. Since this drug did not impact early protein expression, but drastically reduced HAdV genome copy numbers (>1000-fold, Fig. 4), its antiviral activity is likely due to the inhibition of viral DNA replication. However, inhibition of DNMT1 could be a contributing factor as well.
At the concentration of chaetocin used in our study, the drug would primarily inhibit H3 Lys9 HMTs SUV39H1 and G9a, but other members of the family can be inhibited at concentrations above 90 μM (Greiner et al., 2005). Although the library contains several other compounds that target H3 Lys9 HMTs, none of them were detected as positive hits (i.e. led to >50% reduction in RFP from Ad-late/RFP) in our screen. However, all of these other compounds appear to be highly specific for G9a and the related G9a-like protein (GLP) HMTs (Liu et al., 2009, 2010, 2011, 2013), suggesting that the effects of chaetocin may be due to impact on SUV39H1 specifically, or due to other non-epigenetic effects of the drug. For example, chaetocin can act as a competitive and selective substrate for thioredoxin reductase-1, leading to oxidative stress-induced cell death (Isham et al., 2007). As with many compounds that target epigenetic regulators, chaetocin has been investigated for its chemotherapeutic properties and has shown some efficacy in in vitro and in vivo models of cancer (Isham et al., 2007; Spannhoff et al., 2009).
Lestaurtinib inhibits JAK2 and PRK1 which, from an epigenetic standpoint, phosphorylate H3 Tyr41 and H3 Thr11, respectively (Hexner et al., 2008; Kohler et al., 2012). Phosphorylation of H3 Tyr41 by JAK2 prevent binding of heterochromatin protein 1alpha (HP1alpha) to this region of H3, thereby preventing HP1alpha-mediated repression (Dawson et al., 2009). Phosphorylation of H3 Thr11 by PRK1 is required for activation of androgen receptor-dependent transcription (Metzger et al., 2008). PRK1 also phosphorylates HDAC-5, -7 and -9, which inhibits their nuclear import (Harrison et al., 2010). However, both JAK2 and PRK1 have many other non-histone natural cellular substrates (Perner et al., 2019; Smythe and Ayscough, 2003). JAK2 is more commonly known for its role in the JAK/STAT pathway, a pleiotropic signaling cascade involved in the cellular response to many cytokines and growth factors, and in controlling cell proliferation, differentiation, cell migration and apoptosis (Rawlings et al., 2004). PRK1 plays a role in controlling the pyrin inflammasome, an innate immune sensor that detects bacterial toxin-induced Rho guanosine triphosphatase (Rho GTPase)-inactivation (Schnappauf et al., 2019), and in regulating intermediate filaments of the actin cytoskeleton (Matsuzawa et al., 1997). Thus, like gemcitabine and chaetocin, lestaurtinib could exert its effect on HAdV replication through either epigenetic or non-epigenetic mechanisms as well.
The proteins that are targeted by chaetocin, lestaurtinib and gemcitabine mostly confer epigenetic modifications that typically reduce gene expression. If these repressive modifications were occurring on the viral genome, inhibition of these epigenetic regulatory proteins would be predicted to enhance virus gene expression, which was not observed in our study. Thus, the post-translational modifications made by these proteins that naturally allow efficient virus replication likely occurs on host cell chromatin. As mentioned earlier, HAdV E1A can engage cellular epigenetic regulators to induce global changes in gene expression within the host cell, (Ferrari et al., 2008; Fonseca et al., 2012; Horwitz et al., 2008), but it is not the only HAdV protein that can mediate such changes. The HAdV E4-ORF3 protein has been shown to induce SUV39H1/2-mediated H3K9 trimethylation and heterochromatin formation at a subset of cellular genes, including p53, leading to transcriptional repression (Soria et al., 2010). In this case, treatment with chaetocin may lead to enhanced levels of p53 and other key proteins, which adversely affect HAdV replication. Modulation of cellular epigenetic regulators to promote virus replication is not unique to HAdV. Inhibition of the methyltransferases EZH2 and EZH1 (EZH2/1), which deposit the repressive histone H3K27 tri-methylation mark, unexpectedly reduced HSV1 replication due to the induction of an enhanced antiviral state within the cell (Arbuckle et al., 2017). Thus, HSV1 likely naturally engages EZH2/1 to suppress this antiviral state. The antiviral state induced through inhibition of EZH2/1 was effective against several other viruses as well (e.g. Zika virus, human cytomegalovirus and HAdV (Arbuckle et al., 2017)), suggesting that engaging epigenetic regulators to rewrite host gene expression may be a common mechanism for many viruses. Again, it should be noted that the antiviral activities of chaetocin, lestaurtinib and gemcitabine on HAdV could be a consequence of their effects on other cellular processes/pathways unrelated to epigenetic regulation. Further investigation is required to understand the mechanisms underlying the anti-HAdV activity of all 3 compounds. Gemcitabine and lestaurtinib have been investigated in clinical studies for the treatment of various cancers, and thus, existing clinical data may facilitate the testing of the drug as an antiviral in clinical trials.
Our work has identified several compounds that inhibit cellular epigenetic regulatory proteins that led to reduced HAdV gene expression, DNA replication and yield. Some of these compounds have been investigated or are approved for use in humans for other health conditions, suggesting that they could rapidly progress to testing in humans for efficacy against HAdV. Additional preclinical studies are warranted to determine the mechanism of action and efficacy of these compounds in animal models of HAdV-induced disease.
CRediT authorship contribution statement
Bratati Saha: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Visualization, Writing - original draft, Writing - review & editing. Robin J. Parks: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing - original draft, Writing - review & editing.
Declarations of competing interest
All authors disclose no conflicts of interest.
Acknowledgements
We thank Dr. William Stanford (Ottawa Hospital Research Institute) for providing access to Cellomics ArrayScan platform and the HCS Studio software. We also thank Ms. Kathy Poulin for excellent technical assistance and Ryan Clarkin for valuable discussions. Funding was provided by grants to Robin J. Parks from the Canadian Institutes of Health Research (MOP-136898, MOP-142316) and the Natural Sciences and Engineering Research Council of Canada (RGPIN-2014-04810, RGPIN-2019-04786). Bratati Saha was supported by the Ontario Graduate Scholarship (OGS) and a Queen Elizabeth II Graduate Scholarship in Science and Technology from the Government of Ontario, Canada over the duration of this study.
References
- Akusjarvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981;292:420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Arbuckle J.H., Gardina P.J., Gordon D.N., Hickman H.D., Yewdell J.W., Pierson T.C., Myers T.G., Kristie T.M. Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. mBio. 2017;8 doi: 10.1128/mBio.01141-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A.J. In: Fields Virology. 5 ed. Knipe D.M., Howley P.M., editors. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. Adenoviridae: the viruses and their replication; pp. 2355–2394. [Google Scholar]
- Bhatti Z., Dhamoon A. Fatal adenovirus infection in an immunocompetent host. Am. J. Emerg. Med. 2017;35:1034.e1031–1034.e1032. doi: 10.1016/j.ajem.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Binder A.M., Biggs H.M., Haynes A.K., Chommanard C., Lu X., Erdman D.D., Watson J.T., Gerber S.I. Human adenovirus surveillance - United States, 2003-2016. MMWR. Morbidity and mortality weekly report. 2017;66:1039–1042. doi: 10.15585/mmwr.mm6639a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso Brown A.E., Cohen M.N., Tong S., Braverman R.S., Rooney J.F., Giller R., Levin M.J. Pharmacokinetics and safety of intravenous cidofovir for life-threatening viral infections in pediatric hematopoietic stem cell transplant recipients. Antimicrob. Agents Chemother. 2015;59:3718–3725. doi: 10.1128/AAC.04348-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmons N.S., McCormic Z.D., Gaydos J.C., Hawksworth A.W., Jordan N.N. Acute respiratory disease in US Army trainees 3 Years after reintroduction of adenovirus vaccine (1) Emerg. Infect. Dis. 2017;23:95–98. doi: 10.3201/eid2301.161297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crenshaw B.J., Jones L.B., Bell C.R., Kumar S., Matthews Q.L. Perspective on adenoviruses: epidemiology, pathogenicity, and gene therapy. Biomedicines. 2019;7 doi: 10.3390/biomedicines7030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., Bannister A.J., Gottgens B., Foster S.D., Bartke T., Green A.R., Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Papa J., Petryk J., Bell J.C., Parks R.J. An oncolytic adenovirus vector expressing p14 FAST protein induces widespread syncytium formation and reduces tumor growth rate in vivo. Mol Ther Oncolytics. 2019;14:107–120. doi: 10.1016/j.omto.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisova O.V., Kakkola L., Feng L., Stenman J., Nagaraj A., Lampe J., Yadav B., Aittokallio T., Kaukinen P., Ahola T., Kuivanen S., Vapalahti O., Kantele A., Tynell J., Julkunen I., Kallio-Kokko H., Paavilainen H., Hukkanen V., Elliott R.M., De Brabander J.K., Saelens X., Kainov D.E. Obatoclax, saliphenylhalamide, and gemcitabine inhibit influenza a virus infection. J. Biol. Chem. 2012;287:35324–35332. doi: 10.1074/jbc.M112.392142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M.R., Parker A.L., Kalkman E.R., White K., Kovalskyy D., Kelly S.M., Baker A.H. Identification of novel small molecule inhibitors of adenovirus gene transfer using a high throughput screening approach. J. Contr. Release : official journal of the Controlled Release Society. 2013;170:132–140. doi: 10.1016/j.jconrel.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Ferrari R., Pellegrini M., Horwitz G.A., Xie W., Berk A.J., Kurdistani S.K. Epigenetic reprogramming by adenovirus e1a. Science (New York, N.Y.) 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca G.J., Thillainadesan G., Yousef A.F., Ablack J.N., Mossman K.L., Torchia J., Mymryk J.S. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe. 2012;11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Gailhouste L., Liew L.C., Hatada I., Nakagama H., Ochiya T. Epigenetic reprogramming using 5-azacytidine promotes an anti-cancer response in pancreatic adenocarcinoma cells. Cell Death Dis. 2018;9:468. doi: 10.1038/s41419-018-0487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giberson A.N., Davidson A.R., Parks R.J. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res. 2012;40:2369–2376. doi: 10.1093/nar/gkr1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giberson A.N., Saha B., Campbell K., Christou C., Poulin K.L., Parks R.J. Human adenoviral DNA association with nucleosomes containing histone variant H3.3 during the early phase of infection is not dependent on viral transcription or replication. Biochem. Cell. Biol. 2018;96:797–807. doi: 10.1139/bcb-2018-0117. [DOI] [PubMed] [Google Scholar]
- Gray S.G., Baird A.M., O'Kelly F., Nikolaidis G., Almgren M., Meunier A., Dockry E., Hollywood D., Ekstrom T.J., Perry A.S., O'Byrne K.J. Gemcitabine reactivates epigenetically silenced genes and functions as a DNA methyltransferase inhibitor. Int. J. Mol. Med. 2012;30:1505–1511. doi: 10.3892/ijmm.2012.1138. [DOI] [PubMed] [Google Scholar]
- Greiner D., Bonaldi T., Eskeland R., Roemer E., Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat. Chem. Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Grosso F., Stoilov P., Lingwood C., Brown M., Cochrane A. Suppression of adenovirus replication by cardiotonic steroids. J. Virol. 2017;91 doi: 10.1128/JVI.01623-16. e01623-01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B.C., Huynh K., Lundgaard G.L., Helmke S.M., Perryman M.B., McKinsey T.A. Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Lett. 2010;584:1103–1110. doi: 10.1016/j.febslet.2010.02.057. [DOI] [PubMed] [Google Scholar]
- Hartline C.B., Keith K.A., Eagar J., Harden E.A., Bowlin T.L., Prichard M.N. A standardized approach to the evaluation of antivirals against DNA viruses: orthopox-, adeno-, and herpesviruses. Antivir. Res. 2018;159:104–112. doi: 10.1016/j.antiviral.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Heinemann V., Xu Y.Z., Chubb S., Sen A., Hertel L.W., Grindey G.B., Plunkett W. Inhibition of ribonucleotide reduction in CCRF-CEM cells by 2',2'-difluorodeoxycytidine. Mol. Pharmacol. 1990;38:567–572. [PubMed] [Google Scholar]
- Hertel L.W., Boder G.B., Kroin J.S., Rinzel S.M., Poore G.A., Todd G.C., Grindey G.B. Evaluation of the antitumor activity of gemcitabine (2',2'-difluoro-2'-deoxycytidine) Canc. Res. 1990;50:4417–4422. [PubMed] [Google Scholar]
- Hexner E.O., Serdikoff C., Jan M., Swider C.R., Robinson C., Yang S., Angeles T., Emerson S.G., Carroll M., Ruggeri B., Dobrzanski P. Lestaurtinib (CEP701) is a JAK2 inhibitor that suppresses JAK2/STAT5 signaling and the proliferation of primary erythroid cells from patients with myeloproliferative disorders. Blood. 2008;111:5663–5671. doi: 10.1182/blood-2007-04-083402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.M., Quenelle D.C., Cardin R.D., Vogel J.L., Clement C., Bravo F.J., Foster T.P., Bosch-Marce M., Raja P., Lee J.S., Bernstein D.I., Krause P.R., Knipe D.M., Kristie T.M. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci. Transl. Med. 2014;6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke C.H., Jr., Snyder C.E., Jr. History of the restoration of adenovirus type 4 and type 7 vaccine, live oral (Adenovirus Vaccine) in the context of the Department of Defense acquisition system. Vaccine. 2013;31:1623–1632. doi: 10.1016/j.vaccine.2012.12.029. [DOI] [PubMed] [Google Scholar]
- Horwitz G.A., Zhang K., McBrian M.A., Grunstein M., Kurdistani S.K., Berk A.J. Adenovirus small e1a alters global patterns of histone modification. Science (New York, N.Y.) 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoti N., Chowdhury W., Hsieh J.T., Sachs M.D., Lupold S.E., Rodriguez R. Valproic acid, a histone deacetylase inhibitor, is an antagonist for oncolytic adenoviral gene therapy. Mol. Ther. 2006;14:768–778. doi: 10.1016/j.ymthe.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Hsu E., Pennella M.A., Zemke N.R., Eng C., Berk A.J. Adenovirus E1A activation domain regulates H3 acetylation affecting varied steps in transcription at different viral promoters. J. Virol. 2018;92 doi: 10.1128/JVI.00805-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isham C.R., Tibodeau J.D., Jin W., Xu R., Timm M.M., Bible K.C. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood. 2007;109:2579–2588. doi: 10.1182/blood-2006-07-027326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajon A.E., Ison M.G. Severe infections with human adenovirus 7d in 2 adults in family, Illinois, USA, 2014. Emerging Infectious Disease journal. 2016;22:730. doi: 10.3201/eid2204.151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes A., Mathias M., Boulad F., Lee Y.J., Marchetti M.A., Scaradavou A., Spitzer B., Papanicolaou G.A., Wieczorek I., Busam K.J. Cutaneous involvement of disseminated adenovirus infection in an allogeneic stem cell transplant recipient. Br. J. Dermatol. 2016;174:885–888. doi: 10.1111/bjd.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler J., Erlenkamp G., Eberlin A., Rumpf T., Slynko I., Metzger E., Schule R., Sippl W., Jung M. Lestaurtinib inhibits histone phosphorylation and androgen-dependent gene expression in prostate cancer cells. PloS One. 2012;7 doi: 10.1371/journal.pone.0034973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojaoghlanian T., Flomenberg P., Horwitz M.S. The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 2003;13:155–171. doi: 10.1002/rmv.386. [DOI] [PubMed] [Google Scholar]
- Komatsu T., Haruki H., Nagata K. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res. 2011;39:889–901. doi: 10.1093/nar/gkq783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T., Nagata K. Replication-uncoupled histone deposition during adenovirus DNA replication. J. Virol. 2012;86:6701–6711. doi: 10.1128/JVI.00380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn R., Horwitz M.S. Adenovirus DNA synthesis in vitro is inhibited by the virus-coded major core protein. Virology. 1986;150:342–351. doi: 10.1016/0042-6822(86)90299-0. [DOI] [PubMed] [Google Scholar]
- Larsson S., Svensson C., Akusjarvi G. Control of adenovirus major late gene expression at multiple levels. J. Mol. Biol. 1992;225:287–298. doi: 10.1016/0022-2836(92)90922-7. [DOI] [PubMed] [Google Scholar]
- Lenaerts L., De Clercq E., Naesens L. Clinical features and treatment of adenovirus infections. Rev. Med. Virol. 2008;18:357–374. doi: 10.1002/rmv.589. [DOI] [PubMed] [Google Scholar]
- Liang Y., Quenelle D., Vogel J.L., Mascaro C., Ortega A., Kristie T.M. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio. 2013;4 doi: 10.1128/mBio.00558-12. e00558-00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Vogel J.L., Arbuckle J.H., Rai G., Jadhav A., Simeonov A., Maloney D.J., Kristie T.M. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci. Transl. Med. 2013;5:167ra165. doi: 10.1126/scitranslmed.3005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Vogel J.L., Narayanan A., Peng H., Kristie T.M. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 2014;27:441–462. doi: 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Barsyte-Lovejoy D., Allali-Hassani A., He Y., Herold J.M., Chen X., Yates C.M., Frye S.V., Brown P.J., Huang J., Vedadi M., Arrowsmith C.H., Jin J. Optimization of cellular activity of G9a inhibitors 7-aminoalkoxy-quinazolines. J. Med. Chem. 2011;54:6139–6150. doi: 10.1021/jm200903z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Barsyte-Lovejoy D., Li F., Xiong Y., Korboukh V., Huang X.P., Allali-Hassani A., Janzen W.P., Roth B.L., Frye S.V., Arrowsmith C.H., Brown P.J., Vedadi M., Jin J. Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J. Med. Chem. 2013;56:8931–8942. doi: 10.1021/jm401480r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Chen X., Allali-Hassani A., Quinn A.M., Wasney G.A., Dong A., Barsyte D., Kozieradzki I., Senisterra G., Chau I., Siarheyeva A., Kireev D.B., Jadhav A., Herold J.M., Frye S.V., Arrowsmith C.H., Brown P.J., Simeonov A., Vedadi M., Jin J. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J. Med. Chem. 2009;52:7950–7953. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Chen X., Allali-Hassani A., Quinn A.M., Wigle T.J., Wasney G.A., Dong A., Senisterra G., Chau I., Siarheyeva A., Norris J.L., Kireev D.B., Jadhav A., Herold J.M., Janzen W.P., Arrowsmith C.H., Frye S.V., Brown P.J., Simeonov A., Vedadi M., Jin J. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J. Med. Chem. 2010;53:5844–5857. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K.L., Gooding L.R., Garnett-Benson C., Ornelles D.A., Avgousti D.C. Epigenetics and the dynamics of chromatin during adenovirus infections. FEBS Lett. 2019;593:3551–3570. doi: 10.1002/1873-3468.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrugal-Lorenzo J.A., Serna-Gallego A., Gonzalez-Gonzalez L., Bunuales M., Poutou J., Pachon J., Gonzalez-Aparicio M., Hernandez-Alcoceba R., Sanchez-Cespedes J. Inhibition of adenovirus infection by mifepristone. Antivir. Res. 2018;159:77–83. doi: 10.1016/j.antiviral.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Nagata K., Ui M., Hanaoka F. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 1993;268:10582–10587. [PubMed] [Google Scholar]
- Matsuzawa K., Kosako H., Inagaki N., Shibata H., Mukai H., Ono Y., Amano M., Kaibuchi K., Matsuura Y., Azuma I., Inagaki M. Domain-specific phosphorylation of vimentin and glial fibrillary acidic protein by PKN. Biochem. Biophys. Res. Commun. 1997;234:621–625. doi: 10.1006/bbrc.1997.6669. [DOI] [PubMed] [Google Scholar]
- Metzger E., Yin N., Wissmann M., Kunowska N., Fischer K., Friedrichs N., Patnaik D., Higgins J.M., Potier N., Scheidtmann K.H., Buettner R., Schule R. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat. Cell Biol. 2008;10:53–60. doi: 10.1038/ncb1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musher D.M. How contagious are common respiratory tract infections? N. Engl. J. Med. 2003;348:1256–1266. doi: 10.1056/NEJMra021771. [DOI] [PubMed] [Google Scholar]
- Nattress C.B., Hallden G. Advances in oncolytic adenovirus therapy for pancreatic cancer. Canc. Lett. 2018;434:56–69. doi: 10.1016/j.canlet.2018.07.006. [DOI] [PubMed] [Google Scholar]
- Perner F., Perner C., Ernst T., Heidel F.H. Roles of JAK2 in aging, inflammation, Hematopoiesis and malignant transformation. Cells. 2019:8. doi: 10.3390/cells8080854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin J.M., Hawksworth A.W., Blair P.J., Faix D.J., Raman R., Russell K.L., Gray G.C. Dramatic decline of respiratory illness among US military recruits after the renewed use of adenovirus vaccines. Clin. Infect. Dis. 2014;59:962–968. doi: 10.1093/cid/ciu507. [DOI] [PubMed] [Google Scholar]
- Rawlings J.S., Rosler K.M., Harrison D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- Ronchi A., Doern C., Brock E., Pugni L., Sanchez P.J. Neonatal adenoviral infection: a seventeen year experience and review of the literature. J. Pediatr. 2014;164:529–535. doi: 10.1016/j.jpeds.2013.11.009. e521-524. [DOI] [PubMed] [Google Scholar]
- Ross P.J., Kennedy M.A., Christou C., Risco Quiroz M., Poulin K.L., Parks R.J. Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J. Virol. 2011;85:3950–3958. doi: 10.1128/JVI.01787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P.J., Kennedy M.A., Parks R.J. Host cell detection of noncoding stuffer DNA contained in helper-dependent adenovirus vectors leads to epigenetic repression of transgene expression. J. Virol. 2009;83:8409–8417. doi: 10.1128/JVI.00796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P.J., Parks R.J. Construction and characterization of adenovirus vectors. Cold Spring Harbor protocols 2009. 2009 May;4(5):1–12. doi: 10.1101/pdb.prot5011. pdb.prot5011. [DOI] [PubMed] [Google Scholar]
- Saha B., Parks R.J. Human adenovirus type 5 vectors deleted of early region 1 (E1) undergo limited expression of early replicative E2 proteins and DNA replication in non-permissive cells. PloS One. 2017;12 doi: 10.1371/journal.pone.0181012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B., Parks R.J. Histone deacetylase inhibitor suberoylanilide Hydroxamic acid suppresses human adenovirus gene expression and replication. J. Virol. 2019;93 doi: 10.1128/JVI.00088-19. e00088-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B., Varette O., Stanford W.L., Diallo J.S., Parks R.J. Development of a novel screening platform for the identification of small molecule inhibitors of human adenovirus. Virology. 2019;538:24–34. doi: 10.1016/j.virol.2019.09.005. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes J., Martinez-Aguado P., Vega-Holm M., Serna-Gallego A., Candela J.I., Marrugal-Lorenzo J.A., Pachon J., Iglesias-Guerra F., Vega-Perez J.M. New 4-Acyl-1-phenylaminocarbonyl-2-phenylpiperazine derivatives as potential inhibitors of adenovirus infection. Synthesis, Biological evaluation, and structure-activity relationships. J. Med. Chem. 2016;59:5432–5448. doi: 10.1021/acs.jmedchem.6b00300. [DOI] [PubMed] [Google Scholar]
- Sargent K.L., Meulenbroek R.A., Parks R.J. Activation of adenoviral gene expression by protein IX is not required for efficient virus replication. J. Virol. 2004;78:5032–5037. doi: 10.1128/JVI.78.10.5032-5037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappauf O., Chae J.J., Kastner D.L., Aksentijevich I. The pyrin inflammasome in health and disease. Front. Immunol. 2019;10:1745. doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.J., Kim C., Cho S. Gemcitabine and nucleos(t)ide synthesis inhibitors are broad-spectrum antiviral drugs that activate innate immunity. Viruses. 2018:10. doi: 10.3390/v10040211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe E., Ayscough K.R. The Ark1/Prk1 family of protein kinases. Regulators of endocytosis and the actin skeleton. EMBO Rep. 2003;4:246–251. doi: 10.1038/sj.embor.embor776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria C., Estermann F.E., Espantman K.C., O'Shea C.C. Heterochromatin silencing of p53 target genes by a small viral protein. Nature. 2010;466:1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano A.M., Crisostomo L., Mendez M., Graves D., Frost J.R., Olanubi O., Whyte P.F., Hearing P., Pelka P. Adenovirus 5 E1A interacts with E4orf3 to regulate viral chromatin organization. J. Virol. 2019;93 doi: 10.1128/JVI.00157-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannhoff A., Sippl W., Jung M. Cancer treatment of the future: inhibitors of histone methyltransferases. Int. J. Biochem. Cell Biol. 2009;41:4–11. doi: 10.1016/j.biocel.2008.07.024. [DOI] [PubMed] [Google Scholar]
- Thomas G.P., Mathews M.B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980;22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang Q., Zhou Z., Liu M., Chen Y., Li J., Xu L., Guo J., Li Q., Yang J., Wang S. Retinoic acid receptor beta, a potential therapeutic target in the inhibition of adenovirus replication. Antivir. Res. 2018;152:84–93. doi: 10.1016/j.antiviral.2018.01.014. [DOI] [PubMed] [Google Scholar]
- Wong C.M., McFall E.R., Burns J.K., Parks R.J. The role of chromatin in adenoviral vector function. Viruses. 2013;5:1500–1515. doi: 10.3390/v5061500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B., Tollefson A.E., Spencer J.F., Balakrishnan L., Dewhurst S., Capella C., Buller R.M., Toth K., Wold W.S. Ganciclovir inhibits human adenovirus replication and pathogenicity in permissive immunosuppressed Syrian hamsters. Antimicrob. Agents Chemother. 2014;58:7171–7181. doi: 10.1128/AAC.03860-14. [DOI] [PMC free article] [PubMed] [Google Scholar]