Abstract
Objective:
To investigate the function of a novel primate-specific long non-coding RNA (lncRNA), named FLANC, based on its genomic location, and to characterize its potential as a biomarker and therapeutic target.
Design:
FLANC expression was analyzed in 349 tumors from four cohorts and correlated to clinical data. In a series of multiple in vitro and in vivo models and molecular analyses, we characterized the fundamental biological roles of this lncRNA. We further explored the therapeutic potential of targeting FLANC in a mouse model of CRC metastases.
Results:
FLANC, a primate-specific lncRNA feebly expressed in normal colon cells, was significantly up-regulated in cancer cells compared with normal colon samples in two independent cohorts. High levels of FLANC were associated with poor survival in two additional independent CRC patient cohorts. Both in vitro and in vivo experiments demonstrated that the modulation of FLANC expression influenced cellular growth, apoptosis, migration, angiogenesis and metastases formation ability of CRC cells. In vivo pharmacological targeting of FLANC by administration of DOPC-nanoparticles loaded with a specific siRNA, induced significant decrease in metastases, without evident tissue toxicity or pro-inflammatory effects. Mechanistically, FLANC up-regulated and prolonged the half-life of phosphorylated STAT3, inducing the overexpression of VEGFA, a key regulator of angiogenesis.
Conclusions:
Based on our findings, we discovered FLANC as a novel primate specific lncRNA that is highly up-regulated in CRC cells and regulates metastases formation. Targeting primate-specific transcripts such as FLANC may represent a novel and low toxic therapeutic strategy for the treatment of CRC patients.
Keywords: Colorectal cancer, metastases, non-coding RNA, RNA therapeutics, primate-specific, pyknons
INTRODUCTION
Cancer mortality represents a huge medical issue 1, in spite of major advances in the characterization of the genomic alterations in most cancers types 2 and the development of a plethora of biomarkers 3 and targeted therapies 4. Colorectal cancer (CRC) is the third most common malignancy in developed countries 5 and a substantial number of patients initially present or sequentially develop distant metastases, leading to a poor 5-year cancer-specific survival rate of about 10–20% 6. Treatment options for metastatic CRC are limited, and include combination cytotoxic chemotherapy with 5-Fluorouracil, irinotecan and/or oxaliplatin, as well as the addition of anti-angiogenic (anti-vascular endothelial growth factor (VEGF) or its receptor) or epidermal growth factor receptor (EGFR)-directed drugs 7.
Within the last decade, non-coding (nc)RNA alterations have been identified to be involved in the progression from adenoma-to-carcinoma in colorectal carcinogenesis 8, 9. Several ncRNAs, which include long non-coding RNAs (lncRNAs) and short microRNAs (miRNAs), have recently been functionally characterized 10, 11, 12, 13, 14. Because of their cancer specific expression pattern, lncRNAs carry tremendous potential for novel targeted treatment approaches 15,16. Although the number of lncRNAs, including the primate/human-specific ones, 17, 18, 19, 20, 21, 22 probably will surpass the number of coding genes 23, 24, at present time only few of the former are proved to be involved in cancers 25, as the main focus of the translational research nowadays is on genes (coding or non-coding) highly conserved in evolution 8, 13, 26. Recently, we reported that multiple genomic non-coding loci harboring human/primate specific motifs 27 are differentially transcribed between CRC and normal colon and between ZAP-70 positive and ZAP-70 negative CLL, and the expression levels correlated with patients’ overall survival 25. We further dissected the function of one of these transcripts in CRC, the lncRNA N-BLR, and we observed that it plays a key role in metastases. Herein, we functionally characterized a novel primate-specific lncRNA from one of these regions and we demonstrated the therapeutic potential of targeting this transcript, through RNA-based strategies, without evident tissue toxicity or pro-inflammatory effects in vivo.
METHODS
Patient samples
Four different patient cohorts were studied with the staging performed by Dukes classification: cohorts A 28 and B 25 for the evaluation of FLANC expression in CRC tissue compared with matched normal colon, and two larger CRC cohorts, C (n = 170) and D (n=126), for testing the prognostic value of FLANC (online supplementary table 1 to 3). The patients’ clinicopathological data were collected from medical records at the same institutions. All cases were reviewed based on pathology reports and histological slides. Written informed consent was obtained from each patient for these four cohorts, and the studies were approved by the institutional review boards of all the involved institutions.
In vivo treatment of colorectal cancer metastases
Male athymic nude mice were purchased from Taconic Farms (Hudson, NY). All animals were 6–8 weeks old at the time of injection. Metastatic liver models of colorectal cancer were developed as described previously 29. For all animal experiments, HCT116 cells (labelled with a stable expression luciferase gene) were used. For all therapeutic experiments, the dose of FLANC small interfering RNA (siRNA) was 200 μg/kg, as described previously 30. The oligos were incorporated into neutral 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) nanoliposomes. These were administered via intravenous injection twice weekly beginning two weeks after tumor cell injection and continued for 4 weeks. All mouse studies were approved and supervised by the MD Anderson Cancer Center (MDACC) Institutional Animal Care and Use Committee.
Additional methods are presented in the supplementary methods section.
RESULTS
A novel primate-specific long non-coding RNA resides within the first intron of CELSR1
Following our initial report about enrichment of primates specific motifs in ncRNAs involved in CRC 25, we identified a novel transcript located at chromosome 22q13.31 in the first intron of the cadherin EGF LAG seven-pass G-type receptor 1 (CELSR1) gene (online supplementary figure 1A). The protein encoded by this gene is a member of the flamingo subfamily of the cadherin superfamily 31 and has been described to be associated with the Wnt signaling pathway in gastrointestinal malignancies and leukemia 32, 33. According to its location, we named this novel transcript flamingo non-coding RNA (FLANC). We studied FLANC following the workflow presented in figure 1A. By strand-specific qRT-PCR we proved that FLANC is transcribed in the antisense direction compared to CELSR1 (online supplementary figure 1B), being therefore an independent transcript from the CELSR1 messenger RNA or primary RNA.
Figure 1. FLANC is over expressed in colorectal cancer and correlates with patients’ overall survival.

(A) Schematic work flow of the identification and comprehensive functional characterization of FLANC in colorectal carcinogenesis. (B-C) Expression levels of FLANC in normal colon mucosa, adenocarcinoma of the colon, metastases of colorectal cancer, benign colon polyp and inflammatory bowel disease and specifically detected by in situ hybridization on a tissue microarray. Overall, in adenocarcinoma (p<0.01) and metastatic tissue (p<0.001), FLANC is significantly up-regulated in comparison to normal, benign polyps and inflamed colon tissue. The corresponding tissue slides indicate a nuclear and cytoplasmic up-regulation and location of FLANC in epithelial cancer cells. (D) Using two independent cohorts, cancer tissue contained significantly higher expression levels of FLANC than corresponding normal colon tissue. (E) In addition, two large cohorts of colorectal cancer patients were analyzed where high expression levels of FLANC were associated with poor survival in both independent cohorts (log-rank test). Data are presented as means ± s.d. (**p < 0.01; ***p < 0.001)
By using Rapid Amplification of cDNA Ends (RACE) on RNA isolated from the HCT116 cell line, we cloned a mono-exonic 873 nucleotides (nts) transcript from the CELSR1 intron 1 (online supplementary figure 2A). Using in silico analysis, we were only able to identify the transcript in primates, with 98% homology with Pan troglodytes and 87% homology with Gorilla genomes. Further analysis of the sequence and conservation of this genomic region suggested that the evolution of this gene started within the class of mammalians and further evolved by the introduction of two Alu elements in primates (online supplementary figure 2B).
We next analyzed the protein coding potential of FLANC using multiple methods. First, an in vitro transcription-translation assay suggested lack of protein coding potential (online supplementary figure 3A). We further used mass spectrometry for the identification of large proteins or micro-peptides (mPEP less than 100 aminoacids long), and could not identify any of the peptides predicted by an open reading frame (ORF) analysis of this transcript (online supplementary figure 3B). All these data support the lack of protein-coding potential of FLANC.
FLANC expression in colorectal cancer correlates with patients’ overall survival
We first measured the expression of FLANC in a panel of eight CRC cell lines (online supplementary table 4). In five of them FLANC expression was significantly higher than in five pooled normal colon tissues (p<0.05, online supplementary figure 1C). Additionally, we measured the level of FLANC in seven different organs and peripheral blood mononuclear cells (PBMCs) from healthy controls. FLANC was not detectable in healthy brain and liver tissues, was expressed at very low levels in normal colon, ovary, spleen and PBMCs tissues, and was expressed at low levels in lung and testicular tissue (online supplementary figure 1D). Hence, we concluded that FLANC is a CRC specific transcript.
To evaluate the levels of FLANC and its tissue location in CRC, we performed in situ hybridization (ISH) on a commercially available tissue microarray (TMA). Significantly higher levels of FLANC were observed in cancerous (primary adenocarcinoma and metastatic tumors) compared with normal colon tissues (metastatic CRC versus normal colon, p<0.001, primary adenocarcinoma versus normal colon, p<0.01). In contrast, there were no significant differences between colitis and benign/polyp lesions versus normal tissue, suggesting that the up-regulation of FLANC probably occurs in epithelial malignant cells, and not in stroma cells, premalignant or in inflammatory lesions (figures 1B and 1C and online supplementary figure 4). This observation is supported by the fact that FLANC has very low expression in healthy tissues and PBMCs (online supplementary figure 1D), but further clarification by performing microdissection studies on CRC specimens is needed. Epithelial CRC cells of adenocarcinoma and metastatic tissue showed an enriched localization of FLANC in both cytoplasm and nucleus when compared with non-malignant cells (figure 1C and online supplementary figure 4–5). Biochemical separation of nuclear and cytoplasmic RNA fractions confirms the ISH findings of expression in both cell compartments (online supplementary figure 5A and B). In line with the results from ISH analysis, FLANC was significantly up-regulated, measured by qRT-PCR, in CRC tissue compared to paired normal colon tissue using two independent cohorts (p<0.01, A and B, see Methods). Of note, most of the normal tissues show no expression or very low expression of FLANC (figure 1D).
To test the value of FLANC as a prognostic biomarker, we performed clinical correlation in two additional independent CRC cohorts (C and D, see Methods and an overview about the two cohorts in online supplementary table 1): high expression levels of FLANC were associated with high pathological tumor stage in both cohorts and with vessel invasion in cohort C (in cohort D, information about vessel invasion was not available) (online supplementary table 2 and 3). In the screening cohort C (n=170) high expression of FLANC was associated with poor overall survival using a Kaplan Meier curve (p=0.013, log-rank test, figure 1E left). Using univariate and multivariate Cox proportional models, FLANC expression represented an independent prognostic factor even after adjustment for other well-known prognostic factors including tumor differentiation and stage (table 1). For independent confirmation, we measured the FLANC expression in a validation cohort D (n = 126) and confirmed the association of poor survival with high FLANC expression (figure 1E right, p<0.032), which prevailed again in multivariate analyses (table 1).
Table 1.
Univariate and multivariate analyses for predictors of overall survival in cohorts C and D.
| Cohort C (n=170) | Cohort D (n=126) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Univariate | Multivariate | Univariate | Multivariate | ||||||||
| HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | |
| Gender (Female vs. Male) | 0.94 | 0.53–1.66 | 0.828 | 0.97 | 0.53–1.78 | 0.933 | 1.22 | 0.63–2.35 | 0.549 | 1.02 | 0.50–2.09 | 0.941 |
| Age (≧68 (median) vs. <68) | 0.59 | 0.33–1.04 | 0.071 | 1.77 | 0.97–3.23 | 0.059 | 0.78 | 0.41–1.48 | 0.450 | 1.20 | 0.59–2.44 | 0.600 |
| Location (Colon vs Rectum) | 0.87 | 0.49–1.55 | 0.648 | 0.95 | 0.53–1.72 | 0.888 | 1.35 | 0.69–2.64 | 0.370 | 1.44 | 0.69–2.96 | 0.323 |
| Histological type (Differentiated vs. Undifferentiated) |
3.88 | 1.98–7.62 | <0.001 | 4.27 | 2.09–8.73 | <0.001 | 1.85 | 0.89–3.83 | 0.097 | 2.77 | 1.27–6.03 | 0.010 |
| Stage classification (stage I-III vs stage IV) | 6.76 | 3.83–11.93 | <0.001 | 7.65 | 4.26–13.7 | <0.001 | 7.47 | 3.9–14.30 | <0.001 | 9.97 | 4.82–20.6 | <0.001 |
| FLANC expression (low versus high) | 2.00 | 1.14–3.50 | 0.015 | 2.08 | 1.17–3.70 | 0.012 | 2.37 | 1.05–5.39 | 0.038 | 2.38 | 1.02–5.55 | 0.044 |
HR: hazard ratio
CI: confidence interval
in bold - p<0.05.
Knock-down of FLANC inhibits cell proliferation, migration, and induces apoptosis in colorectal cancer cells
In order to identify the biological roles of FLANC in colorectal carcinogenesis, we established two independent siRNAs targeting FLANC (plus strand, see online supplementary figure 1A). A ~40–50% knock-down efficiency was obtained at 96 hours, whereas the host CELSR1 gene expression (located on the minus strand) was not significantly affected by this treatment (HCT116 cell line, online supplementary figure 6A to 6D). Afterwards, we evaluated the biological effect of transient FLANC knock-down in a panel of three CRC cell lines expressing high levels of FLANC: the microsatellite instability (MSI) positive HCT116 and DLD-1 cells and the microsatellite stable (MSS) SW480 cells (online supplementary table 4 and online supplementary figure 1C). Reduction of FLANC expression induced a significant decrease in cellular growth in all tested CRC cell lines after 120 hours; this was verified by CCK8 assay (figure 2A). To confirm the ability of FLANC to regulate cell growth, we used a clonogenic assay as a second independent method. We measured a significant reduction in the number of colonies by FLANC knock-down after 10 to 14 days (about 40% for HCT116 (p<0.01), and 50–70% reduction of colonies in SW480 (p<0.001) and DLD-1 cells (p<0.05), figures 2B and 2C). In contrast, in HT-29 cells, which contained almost undetectable levels of endogenous FLANC and we regarded it as a negative control, FLANC knock-down did not affect cell growth ability (online supplementary figure 7A and 7B). Next, we proved that the anchorage-independent growth ability of CRC cells substantially decreased when FLANC levels were reduced by siRNA (p<0.001 for all three cell lines, figure 2D). Moreover, the number and size of tumor spheres were significantly reduced in HCT116 and DLD-1 cells (p<0.001, supplementary figures 7C and 7D). Because SW480 cells did not form tumor spheres under the selected conditions we could not generate data for this cell line. In addition, knock-down of FLANC decreased migration of CRC cells (reduction between 40% to 50% for both, the HCT116 and SW480 cells, p<0.05, respectively, online supplementary figure 7E and 7F).
Figure 2. FLANC is an oncogene in colorectal cancer cell lines.
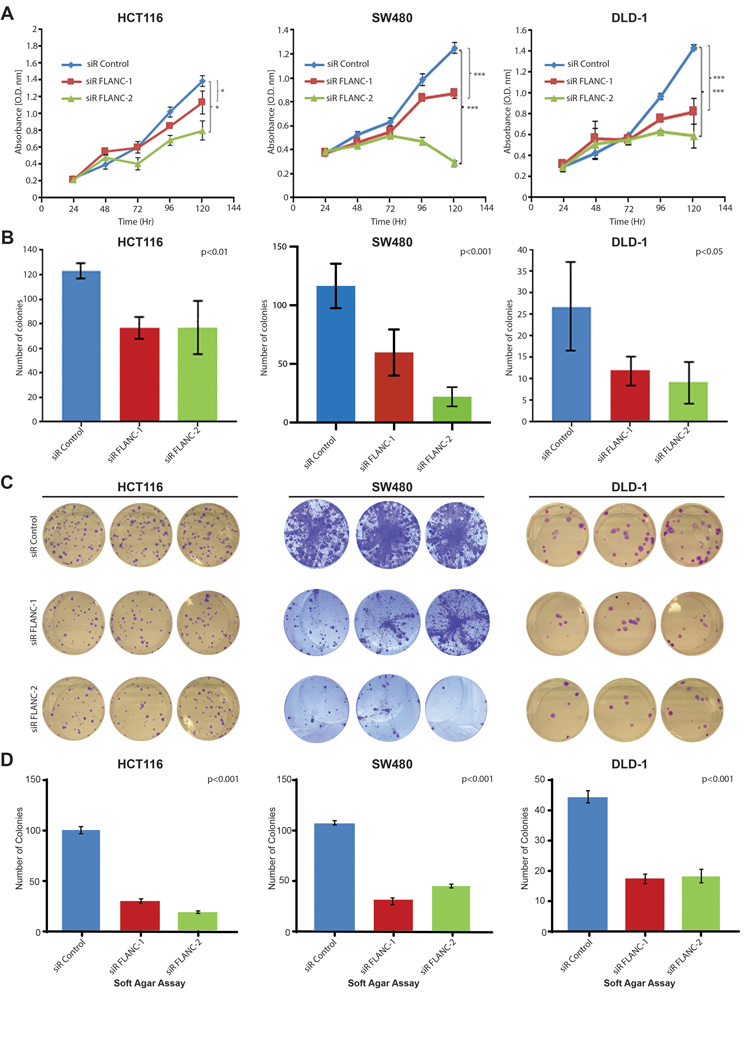
(A) Cellular growth assays (CCK8) demonstrate that knock-down of FLANC leads to decreased proliferation in three independent colorectal cancer cell lines, which (B-C) is confirmed by lower numbers of colonies in an independent clonogenic growth assay. ANOVA test was used to test for differences through all groups. (D) Knock-down of FLANC decreased the ability of these three colorectal cancer cell lines to grow under anchorage-independent conditions in soft agar. Data are presented as means ± s.d. (*p < 0.05; ***p < 0.001).
We further explored whether reduced levels of FLANC can induce apoptosis. We performed a multi-parametric apoptosis assay which evaluated the different steps in the apoptotic cascade (i.e. mitochondrial depolarization, caspase 3/7 activation, phosphatidylserine exposure on the external leaflet of the plasma membrane and chromatin condensation) after siRNA-mediated knock-down. Reduction of FLANC levels induced an increase of apoptosis in HCT116 cells, as measured by the increase of the apoptotic markers evaluated (figure 3A and online supplementary figure 8). In order to confirm these findings with a second alternative assay, we established a second multi-parametric apoptosis assay using fluorescence-dye based single cell high resolution microscopy (online supplementary figure 9). Using this approach, we could confirm the activation of all steps of apoptosis induced by siRNA-mediated knock-down of FLANC, chi-square=341.4, p<0.0001 (figure 3B and online supplementary figures 10). Finally, we performed a third alternative assay (caspase activity) and we found that reduced levels of FLANC induced activation of caspase 3 and caspase 7 in HCT116, SW480 and DLD-1 cells (figures 3C to 3E). Our findings clearly indicated that knock-down of FLANC has a pro-apoptotic effect in CRC cells.
Figure 3. FLANC has an anti-apoptotic effect in CRC cells.

(A) A multi-parametric apoptosis marker FACS assay shows that knock-down of FLANC increased all indicators of apoptosis, (B) which is confirmed by single cell high-resolution multi-parametric apoptosis assay. Chi-square test was used to calculate differences between groups (C) Caspase 3/7 assays confirmed the pro-apoptotic phenotype after knock-down of FLANC in all three colorectal cancer cell lines. Data are presented as means ± s.d. (Student’s t-test: **p < 0.01; ***p < 0.001; ****p<0.0001).
FLANC induces growth and metastatic spread of colorectal cancer in vivo
To evaluate the role of FLANC in vivo, we generated HCT116 clones which had either a stable knock-down of FLANC by two independent short hairpin RNAs (shRNA FLANC-1 and shRNA FLANC-2) or stable overexpression of FLANC transcript. The stable expression of shRNA targeting FLANC reduced FLANC levels by about 60 to 80% compared with the empty vector control (online supplementary figure 11A). We performed cell growth, clonogenic growth, and caspase 3/7 assays to confirm the results we observed after transiently knock-down of FLANC by siRNAs. As shown in online supplementary figure 11B to 11D, cell growth rate and number of colonies decreased significantly in shRNA FLANC-1 and shRNA FLANC-2 clones, whereas apoptosis increased significantly. Conversely, HCT116 cells stably overexpressing FLANC (online supplementary figure 12A) had significantly increased proliferation rates (confirmed by two independent methods), increased ability to form colonies, and decreased apoptosis (online supplementary figure 12B to 12E). To confirm these data in a second cell line, we generated FLANC overexpressing HT-29 cells. Here, we measured a significant increase in cellular growth (online supplementary figure 12F and 12G).
For in vivo confirmation of the observed phenotype, we subcutaneously injected 1×106 HCT116 stably expressing cells with: shRNA FLANC-1, shRNA control, FLANC overexpressing, and empty vector control, to the flank of male nude mice and measured the tumor volume for 7 weeks. The shRNA-mediated knock-down of FLANC led to a significant decrease in tumor volume in comparison with shRNA control (median/25th-75th percentiles: 295/282–341mm3 and 458/369–825mm3, p<0.01, figure 4A). Conversely, FLANC overexpression generated larger tumors compared to empty vector control (median/25th-75th percentiles: 766/497–1001mm3, 631/523–794mm3, respectively, p=ns, online supplementary figure 13A).
Figure 4. In vivo xenograft colon cancer model for the role of FLANC in tumor growth.
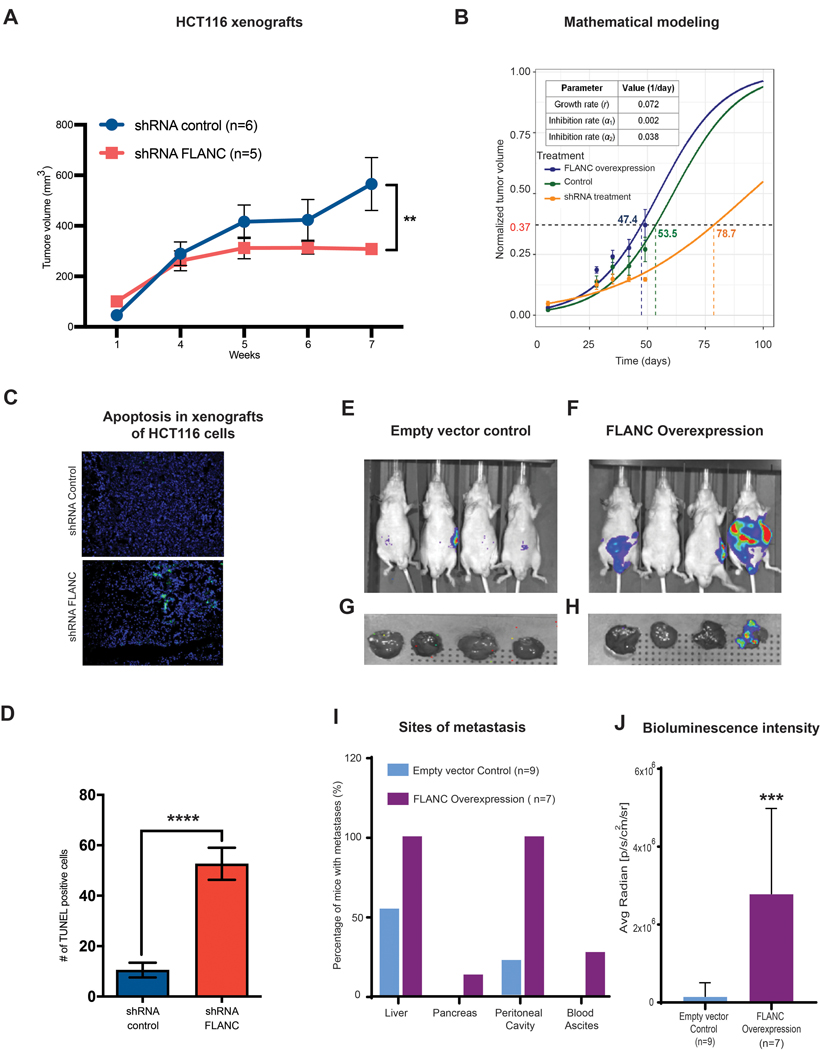
(A) Mice injected with HCT116-shRNA-FLANC cells showed significantly smaller tumors than the shRNA control (**p<0.01). (B) Prediction of tumor volume in different experimental conditions using a mathematical model. (See supplementary materials for modeling details). Tumor growth rate was first estimated (r = 0.072 day−1), and then drug inhibition rates for the shRNA-mediated empty control (α1 = 0.002 day−1) and the shRNA-mediated knock-down of FLANC groups (α2 = 0.038 day−1) were estimated. With these estimated parameters, the model successfully predicts experimentally measured tumor volume change over time. It can be observed that the model predicts the outcome with acceptable accuracy in all three conditions: FLANC overexpression (blue), shRNA empty control (green), and shRNA treatment (orange). (C-D) The TUNEL assay (marks apoptotic bodies) suggests that knock down by shRNA-FLANC is able to induce apoptosis, compared to shRNA control. Colorectal cancer metastases formation was assessed by applying intra-splenic injection of FLANC-expression manipulated HCT116 cells (E-J). Representative chemiluminescent images from whole body nude mice of empty control (E), FLANC overexpression (F), and from removed livers of the empty control (G) and FLANC overexpression cells (H) after approximately seven weeks of intra-splenic injection with empty vector and FLANC overexpressing HCT116 clones are shown. (I) Overall numbers of mice positive by macroscopically exploration of different sites (liver, pancreas, intra-cavity of peritoneum and formation of bloody ascites) (J) The higher number of metastasis correlated with significantly higher intensity of bioluminescence signal in the livers of the group of FLANC overexpressing mice compared with those expressing empty vector control. Data are presented as means ± s.d. (Student’s t-test: **p < 0.01; ***p < 0.001; ****p<0.0001).
Next, we modeled the tumor response to drug treatment based on our prior work on mathematical modeling of cancer 34, 35, 36. We first fit the model (where the inhibition rate (α) = 0) to the data for the FLANC overexpression group in order to determine the tumor growth rate (r). We then fit the model to drug treatment data to obtain tumor inhibition rate α for the empty vector control group and the shRNA FLANC group. The model successfully predicted treatment outcome (change of tumor volume over time) and reproduced experimentally obtained values with acceptable accuracy in all three conditions: no treatment, empty vector control, and shRNA FLANC (figure 4B). According to this model, to reach a tumor volume of 775 mm3 at the end time point (corresponding to normalized value of 0.37), it takes 47.4 days for the no treatment group, 53.5 days for the empty vector control group, and 78.7 days for the shRNA FLANC treatment group (note these are model-predicted values).
Microscopic examination showed that tumors with FLANC knock-down induced higher number of apoptotic cells compared with shRNA control (p<0.0001) (TUNEL assay, figures 4C and 4D), but no difference could be observed between FLANC overexpression and empty vector control (p=ns) (online supplementary figures 13B and 13C).
Furthermore, we tested whether FLANC expression influences metastases formation in vivo. We performed intra-splenic injection of 1×106 HCT116 cells (empty vector control and FLANC overexpressing cells) and monitored metastases formation by luciferase intensity imaging and pathological exploration (figures 4E to 4H). After 6 weeks, we observed a higher number of macro-metastases detected by anatomical exploration of the metastatic sites in mice injected with HCT116 cells overexpressing FLANC compared with HCT116 cells expressing empty vector control (figure 4I). The higher number of metastasis correlated with significantly higher intensity of bioluminescence signal in the livers of the group of FLANC overexpressing mice compared with those expressing empty vector control (figure 4J). Overall, our data support that moderately increased levels of FLANC (HCT116 shRNA control versus shRNA FLANC) regulate tumor growth and highly increased levels (HCT116 FLANC overexpression versus FLANC empty vector control), regulate metastatic spread in vivo and the effects can be modelled for improving therapeutic efficacy.
FLANC expression modulation has genome-wide effects on transcripts and protein expression
The mechanisms of action of ncRNAs are numerous and still poorly understood 8, 13 Therefore, we performed gene expression analyses (GEA) from HCT116 clones with either downregulation (shRNA) or up-regulation (overexpression) FLANC (GEO accession#: GSE127785 and GSE127786) (figure 5A to 5D) and combined the results with reverse phase protein array (RPPA) data (online supplementary figure 14A to 14D). On GEA, we identified 25 cancer-related pathways with negatively correlation between the two types of clones; the most significantly dysregulated signaling pathways includes the WNT signaling pathway and transcriptional regulation by TP53 (Figure 5B to D), both well-known drivers of colon tumorigenesis 37, 38. We investigated the protein expression changes by RPPA 39 (supplementary figure 14A). We detected 34 proteins to be upregulated and 43 downregulated in HCT116 FLANC overexpressing cells when compared with empty vector control (supplementary table 5). The FLANC upregulated proteins were grouped into pathways and the top 20 stimulated pathways are showed in online supplementary figure 14B. Correspondingly, pathway analysis was also performed for the proteins inhibited by FLANC and the top 20 inhibited pathways are represented in online supplementary figure 14C. In order to visualize the proteins regulated by FLANC, we characterized the molecular network controlled by FLANC. According to the highest p-values, the top 5 inhibited and top 5 activated pathways, with the corresponding proteins, are shown in online supplementary figure 14D. When combining the RPPA data with GEA data (online supplementary figure 15A), we identified 11 signaling pathways in common between the various types of profiling performed, with the most significantly abnormal being “Integrated cancer pathways”, “DNA damage response”, ‘Retinoblastoma in Cancer” and “Transcriptional regulation by TP53” (online supplementary Figure 15B). All these data proved a widespread effect of FLANC deregulation on both gene expression and protein regulation levels.
Figure 5. Genome-wide gene expression variation after FLANC levels modulation.
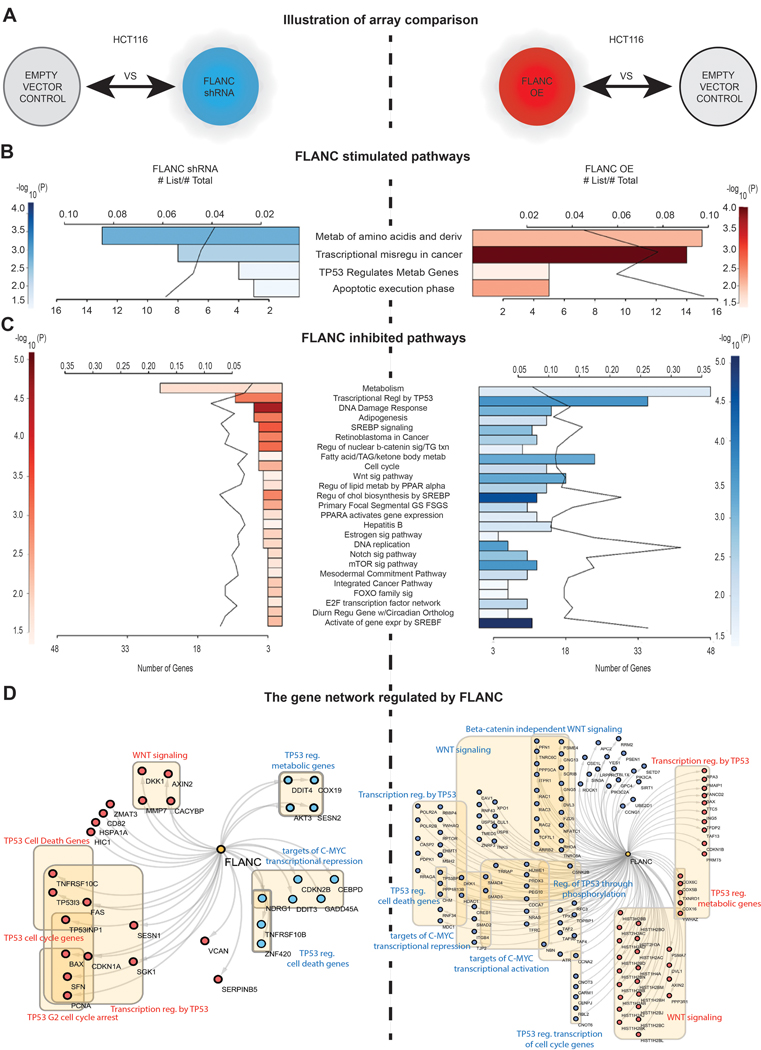
(A) Illustrative representation of the HCT116 clones used for the array comparison. (B) FLANC stimulated signaling pathways negatively correlated in FLANC knock-down (inhibited - blue) and overexpression clones (activated - red) of HCT116 cells. (C) FLANC inhibited signaling pathways negatively correlated in FLANC knock-down (activated - red) and overexpression clones (inhibited - blue) of HCT116 cells. (D) The genes regulated by FLANC are key components of the TP53, WNT and C-MYC pathway, which negatively correlate in FLANC knock-down versus overexpression clones (red = activation, blue = inhibition).
siRNA therapeutics targeting FLANC leads to decreased proliferation and metastasis in vivo
To study FLANC as a potential therapeutic target, we evaluated whether siRNA-mediated targeting of FLANC might result in effective antitumor activity. We determined the effects of systemic delivery of FLANC siRNA #2 on metastasis formation using FLANC siRNA incorporated into DOPC nanoliposomes. This vehicle is currently used for small RNAs delivery in clinical trials of cancer patients (see ClinicalTrials.gov Identifier: NCT01591356). For this experiment, we injected HCT116 cells into the spleen of mice (n=10 each group) and monitored liver metastases formation by in vivo imaging. After two weeks, mice were randomly assigned to the following groups: untreated, treated with scrambled siRNA control, and treated with FLANC siRNA, and we started administering the treatments twice a week (200 μg of siRNA/kg per injection). After 6 weeks, the body weight of mice treated with FLANC siRNA was significantly higher when compared with mice treated with the scramble siRNA (median/(25th–75th percentiles) body weight: 25.3/(24.41–27.76) g (control siRNA) versus 28.92/(25.75–29.86) g (FLANC siRNA, p=0.045), respectively. Regarding tumor growth-related parameters, we observed a significant decrease in bioluminescence signals in the whole mice (figure 6A), as well as in the isolated livers (figure 6B), in the FLANC siRNA group compared with siRNA scramble control (figures 6C). Overall, 5 out of 10 mice in the untreated group, 5 out of 10 mice in the siRNA scramble control group, and 4 out of 8 mice in the siRNA FLANC group showed macroscopically visible liver metastases. The macroscopically countable liver metastases were 26 in the untreated mice, 21 in the siRNA control group, and 6 in the siRNA FLANC group (p<0.05, ANOVA test), respectively (figures 6D and 6E). The successful knock-down of FLANC was confirmed in vivo by performing ISH for FLANC (online supplementary figure 16). We next examined the effects of the treatment on proliferation (Ki67 staining) and apoptosis (TUNEL assay) in vivo. Administration of siRNA FLANC resulted in reduced tumor cell proliferation (figure 6F and 6G). Significant apoptosis induction was measured after the treatment with siRNA FLANC (p<0.05) compared with the corresponding siRNA scramble control (figure 6F and 6H). In summary, the treatment with siRNA FLANC reduced the number of macro-metastases in the liver, tumor burden in the liver, and the tumor cell proliferation and apoptosis.
Figure 6. Therapeutically In vivo intra-splenic colon cancer model.
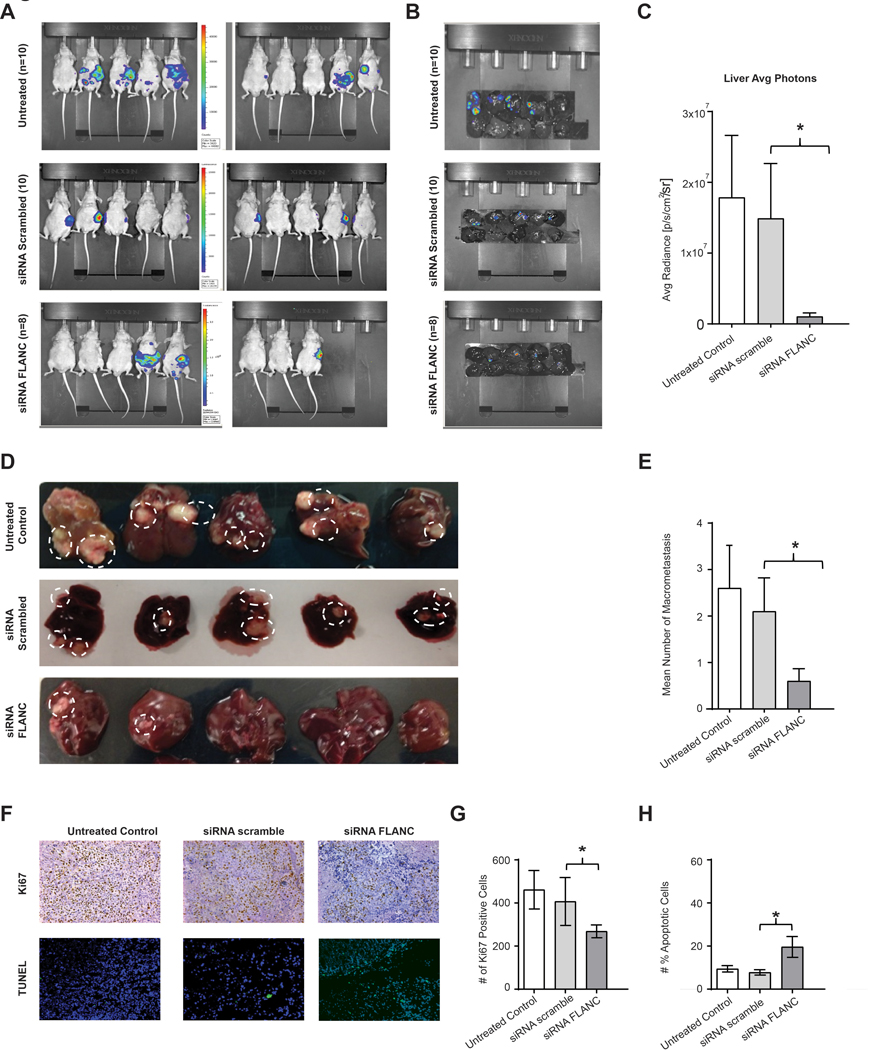
(A) Representative chemiluminescent images from nude mice of untreated, siRNA scramble and siRNA-FLANC groups, and (B) liver metastases after approximately seven weeks of intra-splenic injection with HCT116 cells are shown. (C) Weekly imaging was performed using the Xenogen IVIS spectrum system within 12 min following injection of D-Luciferin (150 mg/mL). Living image 4.1 software was used to determine the regions of interest (ROI), and average photon radiance (p/s/cm2/sr) was measured for each mouse. (C-E) Representative images from liver macro-metastasis and quantification of macro-metastasis. (F) Immunohistochemistry for Ki-67 (proliferation marker) and the TUNEL assay (marks apoptotic bodies) suggest that knock-down FLANC is able to reduce cell proliferation, and to induce apoptosis. (G-H) Quantification analysis of ki-67 positive cells and number of positive apoptotic cell. Data were log-transformed before analysis. n=10. Data are presented as means ± s.d. (Student’s t-test: *p < 0.05).
Absence of acute tissue toxicity or inflammatory response after anti-FLANC therapy
Additionally, we studied the effect of therapy on physiological parameters and evaluated the possible acute toxicity of the treatment (e.g.: impaired organ function or inflammatory cytokine responses). Male C57BL/6J mice were either untreated or treated with single i.v. injections of either siRNA scramble control or FLANC siRNA and serum was collected after 72 hours. As shown in online supplementary figure 17A, there were no differences in parameter measuring liver (Aspartate aminotransferase, AST) and kidney function (Blood Urea Nitrogen, BUN) or high cell turnover parameters (Lactate Dehydrogenase, LDH). In addition, we did not detect significant differences in levels of 12 pro-inflammatory cytokines (LIX, MIP-2, KC, IL-10, IL-6, IL-2, IL-1β, IL-1β, M-CSF, TNFα, GM-CSF, G-CSF), using a Luminex assay (online supplementary figure 17B). Tissue samples were also obtained for post-mortem histopathology studies (brain, spleen, liver and kidney). H&E staining of the various tissues were analyzed by Histopathology Core’s veterinary pathologist and no inflammatory changes were observed in the tissues studied (online supplementary figure 18). In summary, the administration of FLANC siRNA resulted in an effective decrease of metastatic disease without any signs for acute toxicity or inflammatory response.
FLANC promotes angiogenesis in vitro and in vivo
We also checked the effect of FLANC inhibition on angiogenesis by analyzing the microvessel density (CD31 expression) in fresh frozen specimens from the therapeutic in vivo mouse study. Administration of FLANC siRNA induced a significant reduction of endothelial cells compared to siRNA scramble or untreated control (p<0.01) (figure 7A). In vitro, we observed that siRNA mediated knock-down of FLANC in EC-RF24 cells determined a reduction in endothelial tube formation (p<0.05 for siRNA FLANC-1 and p<0.01 for siRNA FLANC-2 (figure 7B). These data corroborated with the GEA data, made us focus on VEGFA, which is one of the top down-regulated molecules in the shRNA FLANC vs. shRNA control cell lines and a key regulator of angiogenesis 40. Of note, the RPPA antibody panel did not include anti-VEGFA.
Figure 7. FLANC induces angiogenesis in vitro and in vivo.
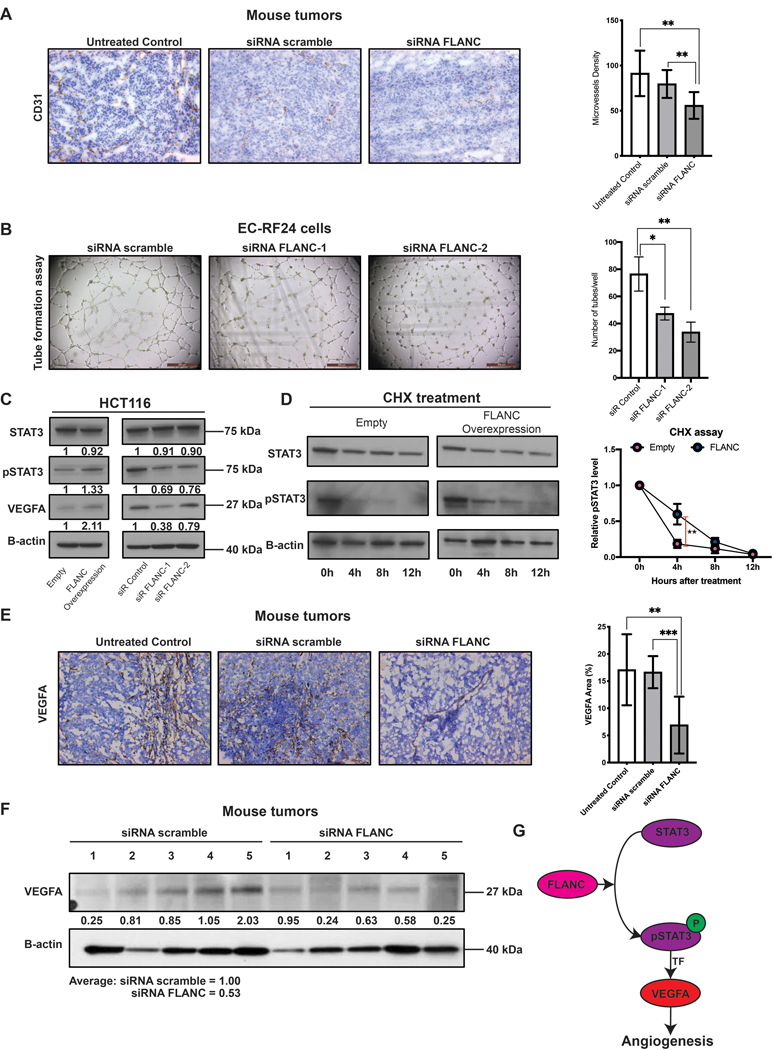
(A) Immunohistochemistry for CD31 (endothelial cell marker) suggested that knock-down of FLANC is able to reduce angiogenesis in vivo. (B) Endothelial cell tube formation assay showing that siRNA-FLANC-1 and siRNA FLANC-2 decreased the number of tubes compared to siRNA scramble in vitro. (C) Immunoblotting analysis showed that FLANC overexpression compared to empty vector control increases the protein level of phosphorylated STAT3 Tyr 705 (pSTAT3) and VEGFA, but not of total STAT3 (left panel). The revers was observed after siRNA mediated knock-down of FLANC, both siRNA FLANC-1 and FLANC-2 decrease the level of pSTAT3 and VEGFA compared to siRNA control (right panel). (D) Cycloheximide chase assay suggested that the half-life of pSTAT3, but not of total STAT3 in HCT116 cells with FLANC overexpression was prolonged compared to empty control vector (left panel). ImageJ quantification analysis showed that 4 hours after adding cycloheximide to the culture media the protein expression of pSTAT3 was higher in FLANC overexpression clones (right panel). (E) Immunohistochemistry expression of VEGFA in mouse tumors proved that siRNA FLANC treatment reduces the protein expression of VEGFA compared to siRNA scramble and untreated control. (F) Immunoblotting analysis showing that VEGFA protein level in mouse tumors is on average higher in the siRNA scramble group (average intensity = 1) compared with the siRNA FLANC group (average intensity = 0.53). (G) Schematic representation of the pathway regulated by FLANC. FLANC increases the expression level of pSTAT3 by prolonging its half-life and pSTAT3 is an established transcription factor for VEGFA, which in turn induces neovascularization of the tumor. Data are presented as means ± s.d. (Student’s t-test: *p < 0.05; **p < 0.01; ***p < 0.001).
Indeed, we confirmed by western-blot that VEGFA protein expression is positively correlated with FLANC modulation (figure 7C). Because STAT3 is an established transcription factor for VEGFA 41 we hypothesized that FLANC regulates VEGFA via STAT3. Western-blot data showed that FLANC overexpression induced a higher level of the phosphorylated STAT3 (pSTAT3 at Tyr 705), the active form that induces VEGFA transcription 42, 43, but not of total STAT3. FLANC knock-down confirmed the reverse effect (figure 7C). It is known that lncRNAs are capable to alter the half-life of proteins 44. Therefore, we checked the half-life of STAT3 and pSTAT3 in empty vector control and FLANC overexpression clones. We observed that high levels of FLANC prolonged the half-life of pSTAT3, but not total STAT3 (figure 7D).
In vivo IHC data showed that siRNA FLANC treatment induces a down-regulation of VEGFA in tumor tissue compared to siRNA scramble (p<0.001) and untreated control (p<0.01) (figure 7E). These results were confirmed by western-blot of in vivo tumors where despite samples heterogeneity, a tendency of down-regulation of VEGFA by siRNA FLANC treatment was observed (average density of scramble siRNA = 1 and of siRNA FLANC = 0.53) (figure 7F). In summary, FLANC induces angiogenesis via STAT3/VEGFA axis (figure 7G).
DISCUSSION
Our study addresses an important novel therapeutic concept that could be exploited to kill malignant cells in vivo with less toxicities: targeting a primate-specific lncRNA, expressed at very low levels in normal cells, but at significantly higher levels in tumor cells. We identified and functionally characterized FLANC, a novel primate-specific lncRNA that is located in the first intron, and expressed in the antisense direction of the CELSR1 protein coding gene. FLANC expression levels do not influence considerably the host gene expression levels, as we saw no significant difference in CELSR1 expression after FLANC knock-down. This suggests that FLANC is not an antisense exonic transcript acting in cis to regulate the sense host gene transcript. This novel approach has the potential to become a mainstream of cancer gene targeting. Recent studies identified about 11,000 primate specific lncRNAs by analyzing the expression patterns in tetrapods 45 and found that about 20% of the human long intergenic ncRNAs are not expressed beyond closely related primates such as chimpanzee or rhesus monkey46. Several of such hominid-specific genes have a coordinated expression under the effect of extracellular factors involved in cancers, such as estrogens47.
In order to identify therapeutically-actionable primate-specific transcripts, we used a category of very short (16 nucleotides) DNA motifs with species-specific sequences named “pyknons” 27. We proved earlier that pyknons can serve as “proxies” for transcripts that comprise them and can be functionally consequential 25. The majority of the more than 209,000 pyknons reported for the human genome 27, 48 are specific to humans/primates. This suggests the possibility that a vast spectrum of primate-specific lncRNAs exist, which are important for our understanding of human diseases and still remain uncharacterized. The absence of sequence conservation suggests that pyknon-containing transcripts can potentially reveal human-specific biology that the use of mouse models cannot uncover.
FLANC has a useful characteristic for therapeutic RNA targeting: it is expressed at very low levels in normal colonic cells, and therefore the cell death induced by RNA-targeting agents will be restrained mainly to the cancer cells with low toxicity. To test the therapeutic potential of FLANC, we used an intra-splenic injection approach commonly used as a CRC liver metastases model 49. As ncRNAs are involved in all steps of immune system and inflammatory cell fate 50, and a previous first-in-human miRNA restoration trial exhibited toxicity due to proinflammatory side effects 51, we further evaluated cellular or immunological changes for the proposed treatment. Though we did not find any toxicities, further dose-response studies need to be carried out to determine the optimal pharmacologic therapeutic window to optimize the efficacy. Because previously we reported that pyknons can also differentiate ZAP-70 positive from ZAP-70 negative CLL 25, we do not exclude the hypothesis that FLANC or other primate-specific transcripts could be overexpressed also in other cancer types and could represent ideal therapeutic targets.
In summary, in this study we described a novel treatment approach in CRC and beyond, based on the identification of a novel primate-specific transcript FLANC, which is involved in colorectal carcinogenesis. Mechanistically, we observed that FLANC regulates the STAT3/VEGFA axis being involved in angiogenesis. FLANC function cannot be studied by genetically engineered mouse models of colorectal carcinogenesis 49, as FLANC is primate-specific and thus not conserved in rodents. Because of the wide effect of FLANC on transcription and translation we consider that also other pathways involved in cancer are regulated by FLANC and need to be characterized. Due to the potential large number of primate/human specific transcripts that define the “humanity” fingerprint of cancers, this approach could represent a future mainstream of therapeutics.
Supplementary Material
Significance of this study.
What is already known about this subject?
Primate specific long non-coding RNAs represents a newly discovered class of transcripts in need for functional characterization in human carcinogenesis.
RNA-based targeting strategies have entered clinical trials in humans in different diseases.
What are the new findings?
Using data about genomic patterns of DNA motifs in cancer-associated regions, we were able to identify a novel primate-specific long non-coding RNA, that we termed FLANC.
FLANC is up-regulated in CRC tissue and poorly expressed in normal colon cells, and possesses prognostic potential in CRC.
FLANC influences cellular growth, migration, anchorage-independent growth, angiogenesis and metastases-formation in vivo.
Mechanistically, FLANC induces angiogenesis via STAT3/VEGFA pathway.
Targeting FLANC by siRNA carrying nanoparticles reduces angiogenesis and represents a potential novel cancer therapeutic approach with higher specificity for malignant cells.
How might it impact on clinical practice in the foreseeable future?
The primate-specific RNA FLANC represents a novel CRC driver, a prognostic factor and a therapeutic target.
Acknowledgments
Funding.
Dr. Calin is the Felix L. Endowed Professor in Basic Science. Work in Dr. Calin’s laboratory is supported by National Institutes of Health (NIH/NCATS) grant UH3TR00943-01 through the NIH Common Fund, Office of Strategic Coordination (OSC), the NCI grants 1R01 CA182905-01 and 1R01CA222007-01A1, an NIGMS 1R01GM122775-01 grant, a U54 grant #CA096297/CA096300 - UPR/MDACC Partnership for Excellence in Cancer Research 2016 Pilot Project, a Team DOD (CA160445P1) grant, a Ladies Leukemia League grant, a Chronic Lymphocytic Leukemia Moonshot Flagship project, a Sister Institution Network Fund (SINF) 2017 grant, and the Estate of C. G. Johnson Jr., Dr. Guoliang Huang was supported by China scholarship Council, Dr. Cristian Rodriguez-Aguayo was supported by the NIH through the Ovarian SPORE Career Enhancement Program, the NCI grants FP00000019. Martin Pichler was supported by an Erwin Schroedinger Scholarship of the Austrian Science Funds (No. J3389-B23). Zhihui Wang and Vittorio Cristini acknowledge support from the National Science Foundation Grant DMS-1716737, the NIH Grants 1U01CA196403, 1U01CA213759, 1R01CA226537, 1R01CA222007, and U54CA210181. Dr. Lopez-Berestein is the John Q. Gaines Professor of Cancer Research. Dr. Goel’s work was supported by the grants CA72851, CA181572, CA184792 and CA187956 from the National Cancer Institute, National Institute of Health. Isidore Rigoutsos is the Richard Hevner Professor in Computational Medicine at Thomas Jefferson University. Dr. Rigoutsos’ work was partially supported by a William M. Keck Foundation grant and by Institutional Funds. The Functional Proteomics RPPA Core facility and the Flow Cytometry and Cellular Imaging Core Facility (FCCICF) are supported by NCI Cancer Center Support Grant P30CA16672.
Abbreviations list:
- CRC
colorectal cancer
- VEGF
vascular endothelial growth factor
- EGFR
epidermal growth factor receptor
- ncRNAs
non-coding RNAs
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- CLL
chronic lymphatic leukemia
- FLANC
flamingo non-coding RNA
- siRNA
small interfering RNA
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphatidylcholine
- MDACC
MD Anderson Cancer Center
- CELSR1
cadherin EGF LAG seven-pass G-type receptor 1
- RACE
Rapid Amplification of cDNA Ends
- nts
nucleotides
- mPEP
micro-peptides
- ORF
open reading frame
- ISH
in situ hybridization
- TMA
tissue microarray
- MSI
microsatellite instability
- MSS
microsatellite stable
- shRNA
short hairpin RNA
- α
inhibition rate
- r
growth rate
- GEA
gene expression analysis
- OE
overexpression
- RPPA
reverse phase protein array
- AST
aspartate aminotransferase
- BUN
blood urea nitrogen
- LDH
lactate dehydrogenase
- EST
expressed sequence tag
Footnotes
Competing financial interests.
The authors declare no competing financial interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol 2017;14:235–246. [DOI] [PubMed] [Google Scholar]
- 3.Anfossi S, Babayan A, Pantel K, et al. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol 2018. [DOI] [PubMed] [Google Scholar]
- 4.Dang CV, Reddy EP, Shokat KM, et al. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer 2017;17:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177–193. [DOI] [PubMed] [Google Scholar]
- 6.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers 2015;1:15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puccini A, Lenz HJ. Colorectal cancer in 2017: Practice-changing updates in the adjuvant and metastatic setting. Nat Rev Clin Oncol 2018;15:77–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015;34:5003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 10.Carotenuto P, Fassan M, Pandolfo R, et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut 2017;66:1268–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almeida MI, Nicoloso MS, Zeng L, et al. Strand-specific miR-28–5p and miR-28–3p have distinct effects in colorectal cancer cells. Gastroenterology 2012;142:886–896 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling H, Pickard K, Ivan C, et al. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut 2016;65:977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferdin J, Nishida N, Wu X, et al. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ 2013;20:1675–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragomir MP, Knutsen E, Calin GA. SnapShot: Unconventional miRNA Functions. Cell 2018;174:1038–1038 e1. [DOI] [PubMed] [Google Scholar]
- 15.Adams BD, Parsons C, Walker L, et al. Targeting noncoding RNAs in disease. J Clin Invest 2017;127:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah MY, Ferrajoli A, Sood AK, et al. microRNA Therapeutics in Cancer - An Emerging Concept. EBioMedicine 2016;12:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Awan HM, Shah A, Rashid F, et al. Primate-specific Long Non-coding RNAs and MicroRNAs. Genomics Proteomics Bioinformatics 2017;15:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durruthy-Durruthy J, Sebastiano V, Wossidlo M, et al. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat Genet 2016;48:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szell M, Danis J, Bata-Csorgo Z, et al. PRINS, a primate-specific long non-coding RNA, plays a role in the keratinocyte stress response and psoriasis pathogenesis. Pflugers Arch 2016;468:935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Li Y, Lin B, et al. HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev Cell 2017;43:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Cheng M, Niu Y, et al. Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int J Biol Sci 2017;13:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Q, Yu B, Anderson C, et al. LncEGFL7OS regulates human angiogenesis by interacting with MAX at the EGFL7/miR-126 locus. Elife 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pertea M, Salzberg SL. Between a chicken and a grape: estimating the number of human genes. Genome Biol 2010;11:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abascal F, Juan D, Jungreis I, et al. Loose ends: almost one in five human genes still have unresolved coding status. Nucleic Acids Res 2018;46:12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigoutsos I, Lee SK, Nam SY, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol 2017;18:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calin GA, Trapasso F, Shimizu M, et al. Familial cancer associated with a polymorphism in ARLTS1. N Engl J Med 2005;352:1667–76. [DOI] [PubMed] [Google Scholar]
- 27.Rigoutsos I, Huynh T, Miranda K, et al. Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc Natl Acad Sci U S A 2006;103:6605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spaventi R, Pecur L, Pavelic K, et al. Human tumour bank in Croatia: a possible model for a small bank as part of the future European tumour bank network. Eur J Cancer 1994;30A:419. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Ye X, Fan F, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 2013;23:171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura M, Jung EJ, Shah MY, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov 2013;3:1302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatin F, Taddei A, Weston A, et al. Planar cell polarity protein Celsr1 regulates endothelial adherens junctions and directed cell rearrangements during valve morphogenesis. Dev Cell 2013;26:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh M, Katoh M. Comparative integromics on non-canonical WNT or planar cell polarity signaling molecules: transcriptional mechanism of PTK7 in colorectal cancer and that of SEMA6A in undifferentiated ES cells. Int J Mol Med 2007;20:405–9. [PubMed] [Google Scholar]
- 33.Kaucka M, Plevova K, Pavlova S, et al. The planar cell polarity pathway drives pathogenesis of chronic lymphocytic leukemia by the regulation of B-lymphocyte migration. Cancer Res 2013;73:1491–501. [DOI] [PubMed] [Google Scholar]
- 34.Pascal J, Bearer EL, Wang Z, et al. Mechanistic patient-specific predictive correlation of tumor drug response with microenvironment and perfusion measurements. Proc Natl Acad Sci U S A 2013;110:14266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Kerketta R, Chuang YL, et al. Theory and Experimental Validation of a Spatio-temporal Model of Chemotherapy Transport to Enhance Tumor Cell Kill. PLoS Comput Biol 2016;12:e1004969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brocato TA, Brown-Glaberman U, Wang Z, et al. Predicting breast cancer response to neoadjuvant chemotherapy based on tumor vascular features in needle biopsies. JCI Insight 2019;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen P, Pichler M, Chen M, et al. To Wnt or Lose: The Missing Non-Coding Linc in Colorectal Cancer. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015;149:1177–1190 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y, Ling S, Hegde AM, et al. Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Semin Oncol 2016;43:476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apte RS, Chen DS, Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019;176:1248–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 2002;21:2000–8. [DOI] [PubMed] [Google Scholar]
- 42.Chen SH, Murphy DA, Lassoued W, et al. Activated STAT3 is a mediator and biomarker of VEGF endothelial activation. Cancer Biol Ther 2008;7:1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 2018;37:4094–4109. [DOI] [PubMed] [Google Scholar]
- 44.Shah MY, Ferracin M, Pileczki V, et al. Cancer-associated rs6983267 SNP and its accompanying long noncoding RNA CCAT2 induce myeloid malignancies via unique SNP-specific RNA mutations. Genome Res 2018;28:432–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014;505:635–40. [DOI] [PubMed] [Google Scholar]
- 46.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 2014;24:616–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin CY, Kleinbrink EL, Dachet F, et al. Primate-specific oestrogen-responsive long non-coding RNAs regulate proliferation and viability of human breast cancer cells. Open Biol 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsirigos A, Rigoutsos I. Human and mouse introns are linked to the same processes and functions through each genome’s most frequent non-conserved motifs. Nucleic Acids Res 2008;36:3484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romano G, Chagani S, Kwong LN. The path to metastatic mouse models of colorectal cancer. Oncogene 2018;37:2481–2489. [DOI] [PubMed] [Google Scholar]
- 50.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol 2016;16:279–94. [DOI] [PubMed] [Google Scholar]
- 51.van Zandwijk N, Pavlakis N, Kao SC, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol 2017;18:1386–1396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


