Abstract
Purpose of Review
In December 2019, there was an outbreak of viral disease in Wuhan, China which raised the concern across the whole world. The viral disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or novel coronavirus or COVID-19 (CoV-19) is known as a pandemic. After SARS-CoV and Middle East respiratory syndrome (MERS)–related CoV, COVID-19 is the third most pathogenic virus, hazardous to humans which have raised worries concerning the capacity of current security measures and the human services framework to deal with such danger.
Recent Findings
According to WHO, the mortality rate of COVID-19 exceeded that of SARS and MERS in view of which COVID-19 was declared as public health emergency of international concern. Coronaviruses are positive-sense RNA viruses with single stranded RNA and non-segmented envelopes. Recently, genome sequencing confirmed that COVID-19 is similar to SARS-CoV and bat coronavirus, but the major source of this pandemic outbreak, its transmission, and mechanisms related to its pathogenicity to humans are not yet known.
Summary
In order to prevent the further pandemic and loss to humanity, scientists are studying the development of therapeutic drugs, vaccines, and strategies to cure the infections. In this review, we present a brief introduction to emerging and re-emerging pathogens, i.e., coronavirus in humans and animals, its taxonomic classification, genome organization, its replication, pathogenicity, impact on socioeconomic growth, and drugs associated with COVID-19.
Keywords: Coronavirus, RNA genomes, SARS-CoV, MERS-CoV
Introduction
Coronaviruses have been studied for more than 50 years and have infected many species of animals, which include birds and humans, and its first reported strain was isolated from the prototype murine coronavirus strain JHM [1]. The increasing population, frequent mixing of animals, deforestation, and urbanization have increased the population of other viruses too along with coronavirus. Coronaviruses belong to the largest group of viruses which are positive-sense RNA viruses, have spike-like projections on the surface, and contain a large unusual genome which has a unique self-replication phenomenon with high rate of mutation and recombination. Because of these special characteristics, this virus needs to cross the species barrier and find new hosts to survive and replicate. In 2005, Susan and Sonia [2] reported an avian infectious bronchitis virus (IBV), bovine coronavirus (BCoV), and porcine transmissible gastroenteritis virus (TGEV) as the examples of animal viruses which are of great importance in veterinary research methodology. In 2015, reported severe acute respiratory syndrome coronavirus (SARS-CoV), porcine epidemic diarrhea virus (PEDV), and Middle East respiratory syndrome coronavirus (MERS-CoV) are the best examples of coronaviruses which caused epidemic diseases in animals with huge economic loss [3]. From the last two decades, these viruses caused lethal respiratory infections and serious problems in different classes of mammals and birds which include humans, dogs, chickens, pigs, and cows [3, 4].
Taxonomic Classification and Structure
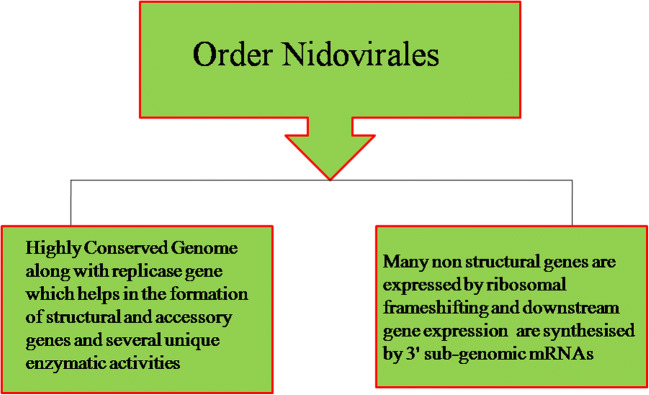
In 1968, Tyrrel et al. coined the name coronavirus, which was further derived by cryo-electron tomography microscopy techniques, has a crown-like structure, belongs to the order Nidovirales and family Coronavirinae which are non-segmented enveloped positive-sense RNA, and contains a large genome of 30 kb in size for RNA viruses [4]. The Nidovirales order includes families Coronaviridae (SARS, PEDV), Arteriviridae (swine and equine pathogens), and Roniviridae (invertebrate viruses) [5, 6•]. The other common characteristics of Nidoviriales are shown in Fig. 1. The major differences in the family of Nidovirus are in its number, sizes, and type of structural proteins which leads to morphological and structural changes of the virus structure. Coronavirus has always been in controversy as it belongs to SARS-CoV. However, in 2004, Goebel et al. [7] and Gorbalenya et al. [8] listed it in Group II which is tabulated in Table 1. In feral pigeon, graylag goose, and mallard [9], coronavirus sequences had been detected using reverse transcription and phylogenetic analysis of replicase enzyme and nucleocapsid sequences whose cellular receptors are not determined yet [10]. Coronavirus is rounded, enveloped, and non-segmented with approximately 80 to 125 nm in size. It consists of positive-sense RNA of genome size of ∼ 30 kb and has four structural proteins which are encoded within the 3′ end of the viral genome [11–14]. The nucleocapsid protein (N) is helically symmetrical which forms a helical capsid inside the viral membrane and contains three viral proteins. There is club shape–like spike (S) projections of ~ 150 kDa type I glycoproteins, forming peplomers from the surface of virion, which gave them crown-like structure which is depicted by electron microscopy. The virus also contains membrane (M) proteins which are extended three times to the outer surface, a small membrane protein (E), and short N-terminal ectodomain along with cytoplasmic tail and a highly hydrophobic protein [15–19, 20•]. The detailed classification of coronavirus group with its host, viruses, diseases, and cellular responses of CoV is given in Table 1 and the detailed view of structural genes of coronavirus is summarized in Table 2. Figure 2 depicts the structure of respiratory syndrome causing human coronavirus.
Fig. 1.
Characteristics of Nidoviriales family of coronavirus
Table 1.
Classification of coronavirus group with its host, viruses, diseases, and cellular responses
| Group | Host | Virus | Diseases | Cellular responses |
|---|---|---|---|---|
| I (Animal Pathogens) | Human | 229E and NL-63 | Respiratory infections | Human APN and ACE2 |
| Pig | TGEV, PRCoV | Respiratory and enteric infection | Porcine APN | |
| Cat | Canine coronavirus, FeCoV, FIPV | Respiratory, enteric, and neurologic infection, and hepatitis | Canine and feline APN | |
| II (Veterinary Pathogens) | Human | OC43, HKU1, and SARS-CoV | Respiratory infection, possibly enteric infection | Neu5,9Ac2-containing moiety |
| Mouse | MHV | Enteric and neurologic infection and hepatitis | Murine CEACAM1 | |
| Rat | Sialodacryoadenitis coronavirus | Neurologic infection | Not determined | |
| Pig | Hemagglutinating encephalomyocarditis | Respiratory, enteric, and neurologic infection, and hepatitis | Neu5,9Ac2-containing moiety | |
| Cow | BCoV | Enteric infection | Neu5,9Ac2-containing moiety | |
| III (Avian Pathogens) | Turkey | Turkey coronavirus | Respiratory and enteric infection | Not determined |
| Chicken | IBV | Respiratory infection, hepatitis | Not determined |
Table 2.
Detailed view of structural genes of coronavirus
| Structural genes | Composition | Functions | Reference |
|---|---|---|---|
| Nucleocaspid (N) | Two separate domains, i.e., N-terminal and C-terminal. Highly phosphorylated, TRSs and genomic packaging signals are two specific RNA substrates, consists of nsp3 (component of M protein and replicase complex) | Domains required to bind RNA, phosphorylation leads to a structural change which increases the affinity of viral RNA versus non-viral RNA and this viral genome binds with N protein forming beads like conformation. Genomic packaging signal bind specifically to the second or C-terminal RNA binding domain. Protein interaction helps in the formation of replicase-transcriptase complex (RTC) and packaging of encapsulated genome into the viral particles | [21–28] |
| Membrane (M) | Most abundant, small in size (~ 25–30 kDa), 3 transmembrane domain structural protein, contains small N- and large C-terminal glycosylated ectodomain which ranges from 6 to 8 nm inside the virus | Helps to give shape to virion and exists in dimer from different conformations, so that membrane curvatures and nucleocapsid can bind very well. Most of the M protein does not contain signal sequences although they are translationally inserted in the endoplasmic reticulum membrane | [29, 30] |
| Envelope (E) | Present in small amount (∼ 8–12 kDa), transmembrane protein in the virion. Highly divergent but consists of a common architect. Also consists of a N-ectodomain and C-endodomain terminal with ion channel activity | Membrane topology of E protein is not completely resolved. Helps in assembly and release of the virus and ion channel in SARS-CoV; this protein is required for pathogenesis but not for viral replication | [31–33] |
| Spike (S) | Heavily N-linked glycosylated protein of ~ 150 kDa which uses N-terminal sequence for the functioning to the ER, consists of S glyco-trimeric class I fusion protein in which S1 helps in the formation of the large receptor-binding domain and S2 in stalk of the spike molecule | Helps in the formation of homotrimers encoded by S protein spike like structure on the outer surface of the virion, Helps in attachment to the host receptor | [15–20] |
| Hemagglutinin-esterase (HE) | Subset of β-coronaviruses, acts as a hemagglutinin which binds sialic acids on the surface of glycoproteins and also contains acetyl-esterase activity | Enhances murine hepatitis virus (MHV) neurovirulence and the S protein-mediated cell entry through which virus spread through mucosa. It is also used against tissue culture whose function is still unknown | [29, 30] |
Fig. 2.
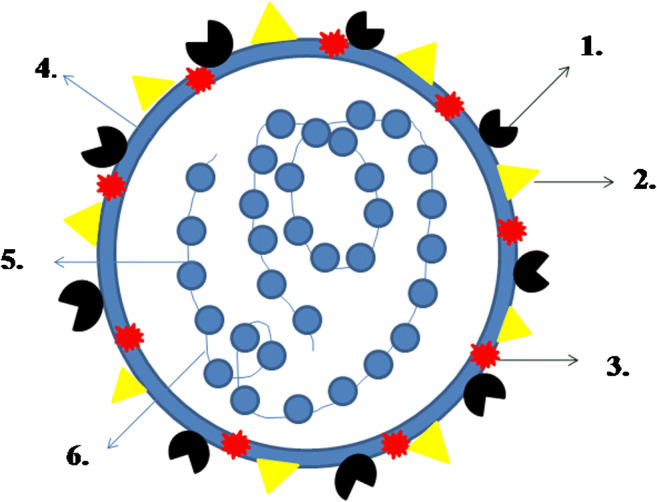
Structure of coronavirus: (1) spike (S), (2) membrane protein (M), (3) envelope protein (E), (4) lipid bilayer, (5) nucleocaspid (N), (6) RNA
Genome Organization and Life Cycle
The spike protein in coronavirus plays an important role in the entry of virus for its attachment with its receptor and also determines the tissue tropism of the virus. This entry is not dependent on pH and its cell-to-cell spread, so it is believed that it occurs directly through the plasma membrane although some viruses may utilize endosomal route but it is not through endosomal route for coronaviruses [34, 35]. In 1995, Holmes and Compton identified carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM 1) as first CoV receptor binding domain at the N-terminal which was utilized by murine coronavirus (MHV) whereas SARS-CoV has receptors at the C-terminal of S1 fusion protein [36, 37]. As the virus enters into the cell, the 5′ end of the RNA genome which contains the open reading frames 1a and 1b (ORF 1a and 1b) is translated into poly protein 1a (pp la) (frameshift mechanism which occurs at a very high speed) and pp1ab ORF 1a codes for one to two papain-like proteases and picoronavirus (group of related RNA viruses which infect vertebrates including mammals and birds) which further helps in the formation of mature replicase proteins through pp1a and pp1ab. ADP-ribose 1-phosphatase activity is also encoded in the X domain of ORF1 and a RNA-dependent RNA polymerase (RdRp) and a helicase are encoded in the X domain of 3′ to 5′ exonuclease ORF 1a [38–40]. Also, cyclic phosphodiesterase putative enzymatic is encoded downstream in ORF 2a. Along with this, multiple other enzymes like poly (U)-specific endoribonuclease (XendoU) and (putative) S-adenosylmethionine-dependent ribose 2_-O-methyltranferase play an important role in metabolism of coronavirus RNA and with the host cell process [39–41]. The replication of genome leads to the full-length negative-strand RNA synthesis (present in low concentration) which serves as template (mRNA 75–78 nt) for transcription along with leader and lagging strand at 5′ and 3′ ends of mRNA. The synthesis of positive and negative strands which involves a unique discontinuous transcription mechanism is still not completely understood. The replicase enzyme is translated from the 5′-end of the genomic RNA, and ORFs are mediated by an internal ribosomal entry site. The endoplasmic reticulum forms a compartment with M and E proteins of Golgi bodies which are near the intracellular membrane, and is an actual site of budding. During assembly, after budding process the intracellular and plasma membrane having spike protein interacts with transmembrane of M protein and N protein which forms helical structures with genomic RNA [42]. The nucleocapsid interacts with M protein and budding into vesicles takes place from where the virus is transported to cell surface, and it leaves the cell at this stage [43].
Pathogenicity and Transmission
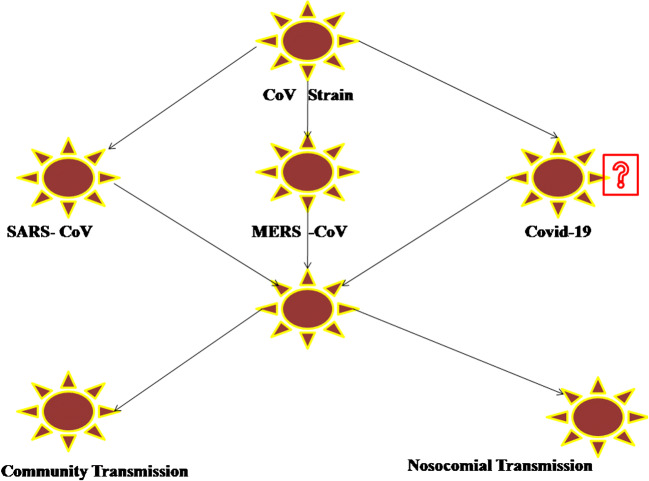
Today, it is well known that SARS-CoV-2 induces clinical spectrum from asymptomatic forms to severe respiratory failures requiring mechanical ventilation and treatment in intensive care unit to fatal cases of sepsis and multi-organ dysfunction syndromes [44]. The pathogenic mechanisms underlying the most common serious manifestation, i.e., virus-produced pneumonia characterized mainly by fever, cough, sore throat, fatigue, headache, and shortness of breath, are very complex [44, 45]. Infection with SARS-CoV-2 can induce an extensive immune response in the host organism leading in some cases to massive tissue damage [44]. One of the most important players in this reaction is interleukin-6 (IL-6), as this cytokine initiates a series of inflammatory events [44]. Besides this, elevated levels of other inflammatory cytokines including interleukin-2, interleukin-7, interleukin-10, (IL-2, IL-7, IL-10), and tumor necrosis factor-α (TNF-α) have also been reported in the laboratory analyses of patient’s plasma being related to disease severity [45, 46••]. Viral pneumonia appears typically bilaterally, involving mostly the lower lobes, and is more severe in elderly patients older than 70 years of age and those suffering from various comorbidities [44]. However, most children and younger adults with SARS-CoV-2 present mild to moderate flu-like symptoms, having a good prognosis [45]. In addition to the aforementioned clinical manifestations, some patients have complained also about gastrointestinal problems, such as vomiting and diarrhea [45]. The illustrations of reservoirs and transmission CoV strain are shown in Fig. 3.
Fig. 3.
Illustrations of reservoirs and TransmissionCoV Strain
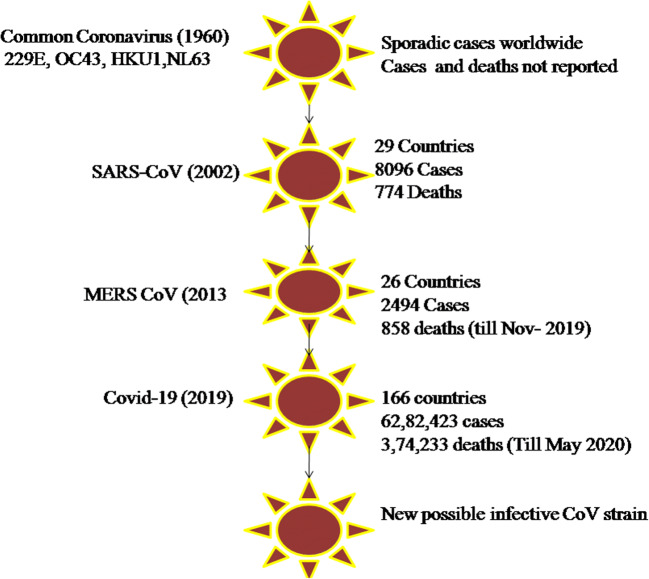
The virus can enter the body through the mucous membranes, particularly nasal and larynx mucosa, passes the respiratory tract, reaches to the lungs, and replicates rapidly [44, 47]. Although there are several possible transmission ways, respiratory droplets spread by coughing or sneezing is the principal route for virus distribution, whereas it can occur also by means of asymptomatic persons [45, 47, 48, 49•]. Getting the infection usually necessitates a direct close contact (within 2 m) with a virus-positive person for a prolonged period of time [44]. In addition, it is possible that SARS-CoV-2 can spread also via fecal–oral transmission, as the virus has been detected in the stool and urine of patients [47, 48]. Incubation period for COVID-19 has been reported by WHO to be between 2 and 10 days [44]. After entering the body, the virus can potentially attack the target tissues expressing angiotensin-converting enzyme 2 (ACE2), including the lungs, heart, kidneys, and gastrointestinal tract [44, 46••, 48]. Despite the first recovery, the virus typically induces a second attack, associated with the aggravation of patient’s condition about 7 to 14 days after onset. The median time from the onset of disease to the most serious clinical outcome, death, has been demonstrated to be 14 days [44]. The systematic comparison and outbreak of common coronavirus in terms of cases and deaths in 1960 to COVID-19 is shown in Fig. 4.
Fig. 4.
Systematic comparison and outbreak of common coronavirus (1960) with COVID (2019)
Detection Techniques
The symptomatic presentation of COVID-19 patients is non-specific and does not warrant precise diagnosis of patients. This is mainly attributed to the clinical symptoms which may be a manifestation of respiratory infections. This is supported by findings from the study which mentioned that 89% of patients presented with fever later when hospitalized in comparison with 44% of patients who presented with fever upon entering hospital [50]. In view of the limitations of symptomatic diagnosis, CT scans and nucleic acid testing are currently being used widely for COVID-19 screening and diagnosis. The strength of molecular techniques for precise diagnosis is attributed to their capability of identifying target specific pathogens.
Nucleic Acid Test (RT-PCR)
The primary method for diagnosis of COVID-19 is nucleic acid testing technique which is based on RT-PCR. In this method, according to laboratory testing for COVID-19, the suspected human samples are taken from upper respiratory tract (nasopharyngeal swab, nasal aspirate, or pharyngeal swab) or the lower respiratory tract (sputum, tracheal aspirate) for RNA extraction followed by reverse transcription and cDNA amplification of a specific region [51]. The primers for RT-PCR are designed against the conserved sequence of SARS-CoV-2 viral genome with RdRP in the ORF1ab, envelop protein gene (E) with high analytical sensitivity with a detection limit of as low as 3.6 and 3.9 copies/reaction, respectively, in both primers and nucleocapsid protein gene (N) with detection limit as 8.3 copies/reaction with poor analytical sensitivity [52]. The RT-PCR method is a two-step method in which the single-step RT-PCR is quick, currently being used by the USA for screening COVID-19 patients with cycling conditions and recommended by CDC, Atlanta, GA, 2020, and also provides the positive control nCoVPC sequence for reference and adequate to address the increased need for quick and timely COVID-19 detection. On the other hand, the two-step assay is more sensitive than one-step assay; however, its main limitation is the requirement of standardization of additional experimental parameters and also it is more time consuming [53]. A number of different SARS-CoV-2 RT-PCR detection kits have been marketed from different companies and research groups and are summarized in Table 3.
Table 3.
Number of SARS-Cov-2 RT-PCR detection kits from companies and research groups
| Detection platform | Sample type | Number of samples | Technology | Detection technology | Reference |
|---|---|---|---|---|---|
| Nucleic acid testing | |||||
| RT-PCR | Throat swabs | 1014 | RT-PCR | Reverse-transcription amplification and fluorescent signal detection | [54] |
| RT-PCR | Oropharyngeal/nasopharyngeal swabs | 176 | RT-PCR | Reverse-transcription amplification and fluorescent signal detection | [55] |
| RT-PCR | Oro-nasopharyngeal swabs or endotracheal aspirate | 32 | RT-PCR | Reverse-transcription amplification and fluorescent signal detection | [56] |
| RT-PCR | Placental and fetal membrane samples | 11 | RT-PCR | Reverse-transcription amplification and fluorescent signal detection | [57] |
| RT-PCR | Nasopharyngeal, throat swab, sputum, saliva | 59 | RT-PCR | Reverse-transcription amplification and fluorescent signal detection | [58] |
| RCA | Serum | 7 | Rolling circle amplification | Circular primer repeated amplification | [59] |
| RPA | Fecal and nasal swabs | 30 | RPA | DNA blinded with forward and reverse primers and amplified | [60] |
| NASBA | Nasal swabs | 138 | REAL TIME-NASBA | RNA target—transcription amplification | [61] |
| RT-LAMP | Nasopharyngeal aspirates | 59 | LAMP | Reverse transcriptase isothermal cDNA amplification | [62] |
| RT-LAMP | Throat swabs | 16 | LAMP | Reverse transcriptase isothermal cDNA amplification | [63] |
| RT-LAMP | Throat swabs | 56 | LAMP | Reverse transcriptase isothermal cDNA amplification | [64] |
| LAMP | Throat swabs | 53 | LAMP | Isothermal DNA amplification | [65] |
| iLACO | Not specified | 248 | LAMP | Isothermal LAMP | [66] |
| CRISPR | Nasopharyngeal swabs | 384 | RT-RPA | SHERLOCK fluorescence multiplexed signal detection | [67] |
| CRISPR | Serum | 110 | RPA | CRISPR/Ca9-mediated lateral flow nucleic assay (CASLFA)-PCR | [68] |
| Magnetic bead | Stool | 17 | Magnetic | Magnetic bead isolation for PCR detection | [69] |
| Quantum dot barcode | Serum | 72 | Barcode | RPA detection of viral DNA captured using multiplexed quantum beads | [70] |
| Paramagnetic bead | Serum | 12 | Magnetic biosensor | Protein targets—magnetic separation | [71] |
| Smartphone dongle | Blood | 96 | ELISA | ELISA operated through microfluidic-based cassette | [72] |
| Protein testing | |||||
| Rapid antigen test | Serum | 117 | Lateral flow | Colorimetric signal produced by gold-coated particles if SARS-CoV-2 positive on paper | [73] |
| ELISA | Serum | 30 | ELISA | Colored product from enzymatic reaction | [74] |
| Biobarcode assay | Serum | 18 | DNA-assisted immunoassay | Gold nanoparticle conjugated DNA is amplified followed by signal detection | [75] |
| SIMOA | Serum | 30 | Digital ELISA | Digital readout of colored product from enzymatic reaction | [76] |
The other method of nucleic acid testing is isothermal amplification test because of its high specificity [77] and is conducted at a single temperature, i.e., reverse transcription-loop-mediated isothermal amplification (RT-LAMP) [66, 78–80]. The major limitation of RT-LAMP is optimization of specific reaction condition and primer combination to be used in view of which other isothermal techniques for amplification are being looked into [81]. In addition, SHERLOCK, a Cas13a ribonuclease based RNA sensing detection strategy, is also being used with SARS-CoV-2 detection protocol [67, 79] This is further being explored for other Cas13a-based detection tests for SARS-CoV-2 detection [82].
CT Scan
In view of short supply of COVID-19 detection kits and false-negative rate of RT-PCR, CT scans are also temporarily being used for COVID-19 clinical diagnosis. The non-invasive chest CT scans are cross-sectional images captured based on measurement of X-rays at different angles across the patient’s chest and analyzed by radiologists for abnormal presentation [83, 84]. Imaging studies using CT scans in COVID-19 patients presented with diverse features in scans which mainly varied depending on the time of onset of symptoms and stage of infection [85, 86]. It has been reported that patients presenting with early stage of COVID-19 were diagnosed with normal findings in CT scan in 56% of the COVID-19 patients [85] and had lung involvement after 10 days of infection or onset of symptoms [86]. The most commonly observed hallmark of COVID-19 manifestation included consolidation of lungs and peripheral and bilateral ground-glass opacity [85, 86]. It was reported that the ground-glass opacity was more evident 0–4 days after onset of symptoms. As the infection progressed, it led to irregular paved stone pattern followed by lung consolidation (solid or fluid in compressible lung tissue) [85, 86]. Interestingly, CT scans have also been the method of choice in view of false-negative rate of RT-PCR and 86–98% sensitivity [54, 87, 88]. A major limitation of using CT scans extensively for COVID-19 diagnosis is its low specificity which overlaps with viral presentations like pneumonia [54].
Protein Testing
According to WHO (2020), the primary priority is to improve patient screening with integration of nucleic acid testing and serological testing based on protein quantification. The main advantage of protein testing is time-point-based screening from diagnosis to recovery which is not possible with nucleic acid testing; it is cost-effective and can be implemented in rural areas without instrumental infrastructure facilities [89]. For protein testing, the levels of viral protein antibodies are used for detection which provides a larger window for detection in comparison with viral load with fluctuates from initial weeks to later [90]. However, a major challenge with development of serological tests was to address the issue of SARS-CoV-2 antibody cross-reactivity against other known strains of coronavirus. High cross-reactivity frequency was observed when 15 COVID-19 plasma samples were tested against S protein of SARS-CoV and SARS-CoV-2 [91]. Different serological tests are being developed for viral specific testing [79, 92–94]. Zhang et al. used the nucleocapsid protein SARS-CoV-2 Rp3 and used ELISA for detection of immunoglobulin M (IgM) and immunoglobulin G (IgG) in serum of COVID-19 patients [79]. Xian et al. also detected SARS-CoV-2 IgM and IgG antibody levels [94]. In an interesting study, COVID-19-infected patients were reported to have high D-dimer and C-reactive protein (CRP) levels and low levels of blood platelets, leukocytes, and lymphocytes [88].
Challenges with Nucleic Acid Testing and CT Scan
COVID-19 is presently being screened using CT scans and diagnosed with RT-PCR worldwide. However, both techniques have shortfalls. A major limitation of CT scan is the requirement of technical expertise to perform and analyze scans; it is expensive and cannot precisely diagnose COVID-19. With RT-PCR, the major challenge at this time is the development and availability of COVID-19 detection kits which is falling short in view of the exponential increase in patient infection rate. Second, lack of appropriate infrastructure and RT-PCR instruments in medical hospitals in sub-urban and rural areas are to comprehend high sample throughput. Next, as RT-PCR is based on detection of SARS-CoV-2 RNA, there have been cases reported where an asymptomatic patient recovered from SARS-CoV-2 infections and hence RT-PCR was not able to detect previous infection or cases where patients were in incubation state when samples were taken for screening and developed symptoms later on. Hence, using these two techniques simultaneously may help to achieve a more accurate diagnosis of COVID-19 patients. Besides these techniques, other rapid detection technologies are also being investigated to address these limitations of SARS-CoV-2 detection.
Drugs and Treatment Therapy for COVID-19
As COVID-19 pandemic has entered a dangerous new phase and wreaking havoc, the world is trying to discover approaches to slow the spread of the novel coronavirus and to discover viable medications. It will prompt the decrease of burden on the healthcare system of the nation by restricting the number of individuals who are seriously sick by COVID-19 and will diminish the span of infection carriage so as to confine the transmission in the network [95••, 96]. So, there is an earnest requirement for therapeutics focusing on SARS-CoV-2. Shockingly, there are no FDA-approved drugs for COVID-19 yet; however, they have made a unique crisis program for potential treatments, the coronavirus treatment acceleration program (CTAP). It utilizes each accessible strategy to move new medications to patients as fast as could be expected under the circumstances while simultaneously seeing if they are useful or harmful (FDA 2020). At present, treatment provided to the affected people is mostly symptomatic and the critically ill individuals are provided with organ support [97]. Utilization of old antiviral medications will be an intriguing technique on account of information on security profile, reactions, and phonology, and medication communications are notable [98]. Here, we investigate a portion of the significant medications that may help in the fight against COVID-19.
A recent paper revealed an inhibitory impact of remdesivir (a new antiviral medication) and chloroquine (an old antimalarial medicate) on the development of SARS-CoV-2 in vitro [99••]. Thus, following the in vitro outcomes, 20 clinical examinations were propelled in a few Chinese emergency clinics [100]. Results demonstrated the predominance of chloroquine (500 mg chloroquine two times per day for 10 days) contrasted with treatment of the control group as far as decrease of pneumonia, length of indications, and deferral of viral freedom, all without extreme reactions [101]. Similarly, hydroxychloroquine (an analogue of chloroquine) has been shown to have an inhibitory effect on SARS-CoV activity in vitro [102]. A few other pre-clinical in vitro examinations propose that both chloroquine and hydroxychloroquine have activity against SARSCoV-2 [95, 102, 103], despite the fact that one in vitro investigation proposes that hydroxychloroquine might be stronger than chloroquine and displayed a higher in vitro antiviral impact as compared with chloroquine [102, 104]. Further, hydroxychloroquine clinical welfare profile is better than that of chloroquine (during long haul use) and permits higher daily dose and has fewer apprehensions about drug–drug interactions [105]. Both medications are accounted for to hinder significant viral replicating enzymes like viral DNA and RNA polymerase and processes, such as viral protein glycosylation, virus assembly, new infection molecule transport, and infection discharge. A few reports proposed that these medications may lead to inhibition of angiotensin-converting enzyme 2 (ACE2) cell receptor, acidification at the surface of the cell membrane impeding fusion of the virus, and immunomodulation of cytokine release [106, 107].
A nucleoside (adenosine) analogue remdesivir is a broad-spectrum antiviral agent. It was produced by Gilead Sciences in 2017 as a treatment for Ebola infection. In vitro studies demonstrated that remdesivir can repress coronaviruses, for example, SARS-CoV and MERS-CoV replication [108•]. Information propose remdesivir represses movement of SARS-CoV, MERS-CoV, and bat CoV strains that can replicate in human epithelial cells and acts as an intermediate channel by means of human CoV receptors. Remdesivir has indicated prophylactic and remedial adequacy against 2002 SARS-CoV in a mouse model [108, 109]. It had been recommended that remdesivir may be a possibility for the treatment of patients with COVID-19 [110]. In pre-clinical trials, remdesivir has demonstrated noteworthy activity against coronavirus and a high genetic barrier to resistance [108•]. In vitro studies have also shown that remdesivir exerts intense antiviral action against a clinical isolate of SARS-CoV-2, given the broad-spectrum anti-COV action of remdesivir that were shown in pre-clinical investigations. A randomized, controlled, twofold visually impaired clinical preliminary study is planned to assess the safety and efficacy of remdesivir in hospitalized patients with mild or moderate COVID-19 respiratory disease [111]. In a case report, remdesivir treatment was begun intravenously on day 7 of a patient with COVID-19. It was observed that 68% of patients demonstrated clinical improvement when treated with remdesivir [112]. Remdesivir acts as an inhibitor of RNA-dependent RNA polymerases. It is reported to compete with adenosine triphosphate for incorporation into nascent viral RNA chains. When fused into the viral RNA at position i, it ends RNA amalgamation at position i + 3. Since RDV-TP does not cause quick chain end (i.e., three extra nucleotides are joined after RDV-TP), the drug appears to evade proof-reading by viral exoribonuclease (a catalyst thought to extract nucleotide simple inhibitors) [99, 108, 113–115].
A randomized, controlled, open-label trial on hospitalized patients with affirmed SARS-CoV-2 disease was directed to check the adequacy of two HIV Protease Inhibitor medicines, i.e., lopinavir and ritonavir, toward SARS-CoV-2 infection [116]. Based on prior in vitro and animal model examinations, both these medications have indicated action against coronaviruses (SARS-CoVand MERS-CoV) [54, 104]. Clinical improvement was seen in patients treated with lopinavir and ritonavir; however, no distinction was noted in the duration of viral shedding after treatment [116]. Both these medications are reported to bind to Mpro, a key enzyme for coronavirus replication and help in suppression of coronavirus action [117]. Another broad-range antiviral medicine, favipiravir, with known in vitro action against RNA viruses is clinically under scrutiny for treatment against SARS-CoV-2 disease (Peking University 2020) [118]. Favipiravir is a RNA-dependent RNA polymerase inhibitor that represses viral RNA synthesis [118].
In a clinical trial, azithromycin was also used in combination with hydroxychloroquine to prevent bacterial contamination in patients suffering from COVID-19 [119••]. Initial results show the potential advantage of azithromycin as an adjunct therapy. Previously, azithromycin has also been utilized as an adjunct therapy in patients with MERS-CoV in combination with antiviral treatment [120]. Azithromycin may stop bacterial infection; furthermore, macrolides have immunomodulatory properties to be used as adjunct therapy [119, 121]. Macrolides have demonstrated immunomodulatory properties in pneumonic inflammatory disorders, which may down control provocative reactions and lessen the over-the-top cytokine production related with respiratory viral contaminations. Azithromycin may help in lessening chemotaxis of neutrophils (PMNs) to the lungs by hindering cytokines (i.e., IL-8) [121, 122]. Another glycopeptide antibiotic, teicoplanin, a routinely utilized anti-microbial to treat bacterial contamination, was seen as dynamic in vitro against SARS-CoV and has joined the rundown of particles that could be utilized as restorative agent against COVID-19 [123]. This antibiotic, as of now utilized in the treatment of Gram-positive bacterial disease, particularly in Staphylococcal infections, has just demonstrated adequacy against different infections, for example, Ebola, flu virus, flavivirus, hepatitis C virus, HIV virus, and on coronavirus (MERS-CoV and SARS-CoV) [98].
Patients with COVID-19 have demonstrated expanded plasma convergences of inflammatory cytokines, for example, TNF-α and IL-2, 7, and 10, particularly in ICU patients, which suggested that a cytokine storm happened [124••]. In light of these discoveries, an interleukin-6 (IL-6) receptor-inhibiting monoclonal antibody named tocilizumab may prove to be successful for COVID-19. In this way, patients analyzed as critical COVID-19 were given tocilizumab treatment. Patients received standard treatment as per treatment protocol for COVID-19 in combination with tocilizumab. The results with tocilizumab treatment were promising. The temperature of the considerable number of patients came back to normal rapidly. The respiratory capacity and every other manifestation improved amazingly. Among these 21 patients, 20 patients have been recouped and released within 14 days post-tocilizumab treatment [125]. Additional information with respect to clinical viability for COVID-19 is being assessed [99, 114]. Tocilizumab hinders IL-6-interceded motioning by competitively binding to both solvent and film bound IL-6 receptors (sIL-6R and mIL-6R) [126]. Thus, another interleukin-6 (IL-6) receptor-inhibiting monoclonal antibodies, namely, sarilumab and siltuximab, are recommended for COVID-19, as it has been accounted for to help in charge of cytokine discharge disorder which is a segment of extreme cases in COVID-19 patients [93, 127]. An investigation of 21 patients with COVID-19 prompted pneumonia/ARDS was being examined who received treatment with siltuximab [128].
Recombinant human interleukin-1 (IL-1) receptor antagonist anakinra and januskinase (JAK) inhibitor baricitinib answered to work in cytokine discharge condition are being assessed for its adequacy against COVID-19 (Swedish Orphan Biovitrum 2020) [93]. Anakinra also acts to the local interleukin-1 receptor adversary (IL-1Ra) by seriously hindering the authoritative IL-1, explicitly IL-1alpha and IL-1beta, to the interleukin-1 sort 1 receptor (IL-1R1). IL-1 is a genius incendiary cytokine that intervenes different fiery and immunological reactions, including actuation of IL-6 (Swedish Orphan Biovitrum 2018), while Janus kinases are intracellular chemicals that transmit signals emerging from cytokine or development factor receptor communications on the cellular membrane to impact cell procedures of invulnerable cell capacity and hematopoiesis (Olumiant 2019). Another monoclonal antibody, namely, leronlimab, answered to improve insusceptible reaction while relieving cytokine storm is additionally assessed for potential treatment of COVID-19 (CytoDyn. Public statement 2020) [129].
Clinical trials are being directed to assess the utilization of COVID-19 convalescent plasma to treat patients with extreme or immediately life-threatening COVID-19 diseases. COVID-19 convalescent plasma is not expected for prevention of the disease. Clinical patients are treated with plasma gathered from people who have recuperated from COVID-19 that may contain antibodies to SARS-CoV-2 for clinical preliminaries (FDA 2020). Rousing outcomes were achieved in patients who were administered convalescent plasma. After plasma imbuement, body temperature normalized inside 3 days in four of five patients. Sequential organ failure assessment score diminished and PAO2/FIO2 increased inside 12 days [130••].
The FDA continues to explore the utilization of NSAIDs in patients with COVID-19 (FDA 2020). Worry for potential worsening of COVID-19 symptoms has been proposed, yet corroborative clinical information is missing as of now [131]. The job of healthful enhancements for the treatment or prevention of COVID-19 is obscure. A few enhancements are under scrutiny in combination with other treatment modalities (e.g., zinc, nutrient C, nutrient D) for both treatment and prophylaxis (Progena Biome 2020).
As there is no approved drug for COVID-19 disease and it will require some investment for potential immunization improvement, along these lines, it is the ideal opportunity for all the residents to hold hands together to battle against coronavirus by rehearsing self-cleanliness and social distancing.
Global Impact of Season on Coronavirus
COVID-19, caused by SARS-CoV-2, subsequently spreads to many other regions in the world through global travel [124, 132]. Because of geographical proximity and significant travel connections, epidemiological modeling of the epicenter predicted that regions in Europe, America, and Southeast Asia would follow Wuhan and China in the epidemic [133, 134]. Temperature and humidity are known factors in SARS-CoV, MERS-CoV, and influenza survival, new outbreaks, and the increase in the risk of infection [21, 135, 136, 137•, 138]. Besides potentially prolonging half-life and viability of the virus, other potential mechanisms associated with cold temperature and low humidity include stabilization of the droplet and enhanced propagation in nasal mucosa, as has been demonstrated with other respiratory viruses [139, 140]. Although most studies have focused on relative humidity that can be affected by temperature, few focused on specific humidity to assess the effect of humidity as variable. The researchers have found that low specific humidity is a key factor in laboratory transmission of influenza as well as the onset of seasonal influenza in the USA [139]. It was reported that high temperature and relative humidity reduce the transmission of COVID-19 with 1% significance levels of evidence that high temperature and high humidity reduce the transmission of influenza [141–145]. This suggested that the arrival of summer and rainy season in the northern hemisphere can effectively reduce the transmission of the COVID-19, and it is unlikely that the COVID-19 pandemic diminishes by summer since the central US, northwest China, and countries in the southern hemisphere (e.g. Australia and South Africa) still have a high coronavirus transmission. Therefore, other measures such as social distancing are still important for blocking the COVID-19 transmission.
Role of Mass Media in Prevention and Management of COVID-19
The COVID-19 pandemic has created a worldwide challenging emergency that has deeply affected the regular life of every individual and their existence. The COVID-19 pandemic is to be known as a more serious public health risk than influenza pandemic of the early twentieth century that killed over 50 million people worldwide. The number of infected patients and lethalities of COVID-19 augmenting exponentially and its ultimate global impacts are still mysterious. The countries facing novel corona pandemic worldwide have implemented various forms of social distancing to reduce the virus spread until specific medicines and vaccines will be available in the market [146, 147]. Most of the countries face unique challenges to combat the COVID-19 pandemic. More specifically, developing countries have poor healthcare resources, limited state capacity, a large population below the poverty line, and monetary issues that are posing a challenge to save life against this pandemic. Therefore, countrywide lockdown by keeping in mind the physical danger, and social and physical distancing policy has forced huge stress on the population below the poverty line. Meanwhile, the media came in front in qualitative and quantitative means and play a remarkable role in providing the latest updates to people by spreading awareness among them. The effect of social distancing are apparent in India through data and “individual-based simulation model” comparative account with China, Italy, America, etc. [148, 149]. The data which have been combined with data on the early and swift commencement of COVID-19 associated to demographic pattern, rate of infection and dispersal, frequency of hospitalization with critical care facility, and mortality might be highly influential in the policy arena. The improvement suggested by Ferguson et al. (2020) in moderate form of social distancing includes a 7-day isolation of anyone suspected with coronavirus symptoms and a 14-day entirely voluntary household quarantine to spectacularly reduce social contact is fabulously helpful in lowering COVID-19 significantly. All these measures that began in India since late February 2020 may flatten the curve of infection but unable to stop exclusively. This suppression scenario makes projections for strict and repeated imposition of social distancing throughout the year until a vaccine will be developed for drastic reduction in cases and rebound epidemic. In every step of this pandemic, the media always came with the latest updates and safety measures, multiple stories, progressive headlines, and politicians’ addresses to nation across the past few months. In any case, this consistent blast of new data, new cases, and new counsel has been trying to increase awareness. It not just makes the story hard to stay aware of from a columnist’s viewpoint, it makes it prone to misunderstanding for anybody attempting to follow the story. A news piece you read one day could be totally obsolete by the following morning, and this has implied that there have been numerous inquiries from the general society encompassing the flare-up and the infection. Furthermore, as supplementary data have risen over the previous weeks, specialists and general well-being authorities have reconsidered their conclusions, exhortation, and proposals in accordance with this, and it has been concluded that these updates have made it difficult to construct trust.
Effect of COVID-19 on the Economy and Supply Chain
The influence of novel coronavirus disease along with mortality and morbidity has become apparent since the outbreak in highly interconnected global capita. The worldwide act of lockdown was associated with interruptions to production amidst the slowing down of global economy. Consecutively, the global supply chain has also been discomfited which leads to great socioeconomic hike. The discoveries uncover that the expanding number of lockdown days, fiscal arrangement choices, and travel limitations seriously influenced the degree of financial growth and the end, opening, most minimal, and most noteworthy stock cost of significant securities exchange records. Conversely, the forced limitation on inward development and higher monetary approach spending positively affected the degree of financial growth despite the fact that the expanding number of affirmed coronavirus cases did not significantly affect the degree of financial growth [150]. Companies worldwide, irrespective of size, have started experiencing reductions in production and export because bounded transport in restricted countries further slowed down global economic exchange activities. The economic stock markets have also been approachable to changes and global stock indices through various direct and indirect economic costs since novel corona outbreak, disease burden, and associated illness. The time, income, and direct financial loss of bearer on medical care and associated services are extra in quantitative asset estimate of the economic loss because of COVID-19. The conformist comes up with underestimation of the true economic costs of infectious pandemic diseases for which there is no vaccine until today so far (e.g., COVID-19, HIV/AIDS, pandemic influenza) [151, 152••, 153–156]. The exercises of the earlier reported pandemics might be helpful in the assessment of valuable information for reducing and combating current pandemic outbreak implications. The horror and psychological effects of this unknown deadly virus are quite more like biological terrorism threats which cause a high level of stress, with longer-term consequences [157]. Social distancing, travel restrictions, and self-isolation not only decrease economic sectors across the world but also caused loss to many educational institutions which have locked down also. On the contrary, the demand for medical supplies and food resources increases due to panic-buying and stockpiling of medical supplements and food products, which is also a big challenge [158].
Conclusion
In 2002, SARS CoV-1 epidemic created havoc in Guangdong region of China, but in December 2019, SARS-Cov-2, novel coronavirus had emerged in Wuhan, China, which caused an epidemic in the whole world. This virus transmitted so quickly that it affected more than hundreds of thousands of people over the world. Although the source, origin, and transmission mechanism of this virus are not yet clear, there are preventive measures like social distancing, washing of hands, and sanitization of hands in public places accepted worldwide. Scientists and researchers are rapidly studying on this topic and helping people to overcome this pandemic. Also, the doctors, border security officers, army men, police officers, nurses, and municipal workers are trying their level best to save the world from this disease. Until now, there is no vaccine for coronavirus, but scientists are hopefully finding and managing an effective vaccine for this virus. In the meantime, various antiviral drugs mainly in combination with azithromycin are used for short-term benefits. So, in the end, it is a lesson to be learned on the basis of this viral disaster in terms of global and public health for any future pandemic like this.
Footnotes
This article is part of the Topical Collection on Clinical Pharmacology
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Bailey OT, Pappenheimer AM, Cheever FS, Daniels JB. A murine virus (Jhm) causing disseminated encephalomyelitis with extensive destruction of myelin: ii. Pathology J Exp Med. 1949;90(3):195–212. doi: 10.1084/jem.90.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau SK, Chan JF. Coronaviruses: emerging and re-emerging pathogens in humans and animals. Virol J. 2015;12:209. doi: 10.1186/s12985-015-0432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses. Springer. 2015:1–23. [DOI] [PMC free article] [PubMed]
- 5.Cowley JA, Dimmock CM, Spann KM, Walker PJ. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J Gen Virol. 2000;81(Pt 6):1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148(11):2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goebel SJ, Taylor J, Masters PS. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J Virol. 2004;78(14):7846–7851. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorbalenya AE, Snijder EJ, Spaan WJ. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol. 2004;78(15):7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonassen CM, Kofstad T, Larsen IL, Lovland A, Handeland K, Follestad A, et al. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos) J Gen Virol. 2005;86(Pt 6):1597–1607. doi: 10.1099/vir.0.80927-0. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh D, Mawditt K, Welchman Dde B, Britton P, Gough RE. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 2002;31(1):81–93. doi: 10.1080/03079450120106651. [DOI] [PubMed] [Google Scholar]
- 11.Lomniczi B. Biological properties of avian coronavirus RNA. J Gen Virol. 1977;36(3):531–533. doi: 10.1099/0022-1317-36-3-531. [DOI] [PubMed] [Google Scholar]
- 12.Lee HJ, Shieh CK, Gorbalenya AE, Koonin EV, La Monica N, Tuler J, et al. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180(2):567–582. doi: 10.1016/0042-6822(91)90071-i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, Milligan RA, Yeager M, Buchmeier MJ. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80(16):7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barcena M, Oostergetel GT, Bartelink W, Faas FG, Verkleij A, Rottier PJ, et al. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc Natl Acad Sci U S A. 2009;106(2):582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins AR, Knobler RL, Powell H, Buchmeier MJ. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell–cell fusion. Virology. 1982;119(2):358–371. doi: 10.1016/0042-6822(82)90095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luytjes W, Sturman LS, Bredenbeek PJ, Charite J, van der Zeijst BA, Horzinek MC, et al. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987;161(2):479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham S, Kienzle TE, Lapps W, Brian DA. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990;176(1):296–301. doi: 10.1016/0042-6822(90)90257-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol. 1990;64(11):5367–5375. doi: 10.1128/JVI.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beniac DR, Andonov A, Grudeski E, Booth TF. Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol. 2006;13(8):751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, Lee SJ, Hsiao HH, Wu WJ, Chang WL, Lin CH, Huang TH. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13(1):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sturman LS, Holmes KV, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33(1):449–462. doi: 10.1128/JVI.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurst KR, Koetzner CA, Masters PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. 2013;87(16):9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurst KR, Koetzner CA, Masters PS. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J Virol. 2009;83(14):7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo L, Masters PS. Functional analysis of the murine coronavirus genomic RNA packaging signal. J Virol. 2013;87(9):5182–5192. doi: 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molenkamp R, Spaan WJ. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239(1):78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stohlman SA, Baric RS, Nelson GN, Soe LH, Welter LM, Deans RJ. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62(11):4288–4295. doi: 10.1128/JVI.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stohlman SA, Lai MM. Phosphoproteins of murine hepatitis viruses. J Virol. 1979;32(2):672–675. doi: 10.1128/JVI.32.2.672-675.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nal B, Chan C, Kien F, Siu L, Tse J, Chu K, Kam J, Staropoli I, Crescenzo-Chaigne B, Escriou N, van der Werf S, Yuen KY, Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S. M and E J Gen Virol. 2005;86(Pt 5):1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 30.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhn P, Buchmeier MJ. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godet M, L'Haridon R, Vautherot JF, Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188(2):666–675. doi: 10.1016/0042-6822(92)90521-p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieto-Torres JL, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Castaño-Rodriguez C, Fernandez-Delgado R, Torres J, Aguilella VM, Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeDiego ML, Alvarez E, Almazan F, Rejas MT, Lamirande E, Roberts A, et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81(4):1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizzen L, Hilton A, Cheley S, Anderson R. Attenuation of murine coronavirus infection by ammonium chloride. Virology. 1985;142(2):378–388. doi: 10.1016/0042-6822(85)90345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kooi C, Cervin M, Anderson R. Differentiation of acid-pH-dependent and -nondependent entry pathways for mouse hepatitis virus. Virology. 1991;180(1):108–119. doi: 10.1016/0042-6822(91)90014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubo H, Yamada YK, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol. 1994;68(9):5403–5410. doi: 10.1128/JVI.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng PK, Wong DA, Tong LK, Ip SM, Lo AC, Lau CS, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363(9422):1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorbalenya AE. Big nidovirus genome. When count and order of domains matter. Adv Exp Med Biol. 2001;494:1–17. doi: 10.1007/978-1-4615-1325-4_1. [DOI] [PubMed] [Google Scholar]
- 39.Ziebuhr J, Thiel V, Gorbalenya AE. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J Biol Chem. 2001;276(35):33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331(5):991–1004. doi: 10.1016/s0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci U S A. 2004;101(34):12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Haan CA, Smeets M, Vernooij F, Vennema H, Rottier PJ. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J Virol. 1999;73(9):7441–7452. doi: 10.1128/JVI.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo L, Masters PS. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J Virol. 2002;76(10):4987–4999. doi: 10.1128/jvi.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Gennaro F, Pizzol D, Marotta C, Antunes M, Racalbuto V, Veronese N, et al. Coronavirus diseases (COVID-19) current status and future perspectives: a narrative review. Int J Environ Res Public Health. 2020;17(8). 10.3390/ijerph17082690. [DOI] [PMC free article] [PubMed]
- 45.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil Med Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020;55(5):105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin Y, Yang H, Ji W, Wu W, Chen S, Zhang W, et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12(4). 10.3390/v12040372. [DOI] [PMC free article] [PubMed]
- 49.Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int. 2020;139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freeman WM, Walker SJ, Vrana KE. Quantitative RT-PCR: pitfalls and potential. Biotechniques. 1999;26(1):112–122. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 52.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3). 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed]
- 53.Wong ML, Medrano JF. One-step versus two-step real-time PCR. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 54.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases [published online ahead of print February 26, 2020]. Radiology. 10. [DOI] [PMC free article] [PubMed]
- 55.Norz D, Fischer N, Schultze A, Kluge S, Mayer-Runge U, Aepfelbacher M, et al. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J Clin Virol. 2020;128:104390. doi: 10.1016/j.jcv.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He JL, Luo L, Luo ZD, Lyu JX, Ng MY, Shen XP, Wen Z. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan. China Respir Med. 2020;168:105980. doi: 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penfield CA, Brubaker SG, J. L Detection of severe acute respiratory syndrome coronavirus 2 in placental and fetal membrane samples. American Journal of Obstetrics &Gynecology MFM. 2020;8:100133. doi: 10.1016/j.ajogmf.2020.100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yip CC, Ho CC, Chan JF, To KK, Chan HS, Wong SC, et al. Development of a novel, genome subtraction-derived, SARS-CoV-2-specific COVID-19-nsp2 real-time RT-PCR assay and its evaluation using clinical specimens. Int J Mol Sci. 2020;21(7). 10.3390/ijms21072574. [DOI] [PMC free article] [PubMed]
- 59.Martel N, Gomes SA, Chemin I, Trepo C, Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA) J Virol Methods. 2013;193(2):653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 60.Amer HM, Abd El Wahed A, Shalaby MA, Almajhdi FN, Hufert FT, Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J Virol Methods. 2013;193(2):337–340. doi: 10.1016/j.jviromet.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, Evans R, Doull I. The role of respiratory viruses in cystic fibrosis. J Cyst Fibros. 2008;7(4):320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shirato K, Nishimura H, Saijo M, Okamoto M, Noda M, Tashiro M, Taguchi F. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2007;139(1):78–84. doi: 10.1016/j.jviromet.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang WE, Lim B, Hsu CC, Xiong D, Wu W, Yu Y, Jia H, Wang Y, Zeng Y, Ji M, Chang H, Zhang X, Wang H, Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J Mol Sci. 2020;21(8). 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed]
- 65.Imai M, Ninomiya A, Minekawa H, Notomi T, Ishizaki T, Van Tu P, et al. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J Virol Methods. 2007;141(2):173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Yu L, Wu S, Hao X, Dong X, Mao L, Pelechano V, Chen WH, Yin X. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Xiong E, Tian T, Cheng M, Lin W, Wang H, Zhang G, Sun J, Zhou X. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14(2):2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 69.Nilsson H-O, Aleljung P, Nilsson I, Tyszkiewicz T, Wadström T. Immunomagnetic bead enrichment and PCR for detection of helicobacter pylori in human stools. J Microbiol Methods. 1996;27(1):73–79. doi: 10.1016/0167-7012(96)00930-X. [DOI] [Google Scholar]
- 70.Kim J, Biondi MJ, Feld JJ, Chan WC. Clinical validation of quantum dot barcode diagnostic technology. ACS Nano. 2016;10(4):4742–4753. doi: 10.1021/acsnano.6b01254. [DOI] [PubMed] [Google Scholar]
- 71.Aytur T, Foley J, Anwar M, Boser B, Harris E, Beatty PR. A novel magnetic bead bioassay platform using a microchip-based sensor for infectious disease diagnosis. J Immunol Methods. 2006;314(1–2):21–29. doi: 10.1016/j.jim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 72.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci Transl Med. 2015;7(273):273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 73.Bosch I, de Puig H, Hiley M, Carre-Camps M, Perdomo-Celis F, Narvaez CF, et al. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci Transl Med. 2017;9(409). 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed]
- 74.Rowe T, Abernathy RA, Hu-Primmer J, Thompson WW, Lu X, Lim W, Fukuda K, Cox NJ, Katz JM. Detection of antibody to avian influenza a (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol. 1999;37(4):937–943. doi: 10.1128/JCM.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, Mirkin CA. Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy. Proc Natl Acad Sci U S A. 2009;106(44):18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12(14):2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. MedRxiv. 2020.
- 80.Lamb L, Bartolone S, Ward E, Chancellor M. Rapid detection of novel coronavirus (COVID-19) by reverse transcription-loop-mediated isothermal amplification. medRxiv. 2020. 10.1101/2020.02.19.20025155.
- 81.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289(1):150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 82.Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, et al. Development and evaluation of a CRISPR-based diagnostic for 2019-novel coronavirus. medRxiv. 2020.
- 83.Whiting P, Singatullina N, Rosser J. Computed tomography of the chest: I. Basic principles. Bja Education. 2015;15(6):299–304. doi: 10.1093/bjaceaccp/mku063. [DOI] [Google Scholar]
- 84.Lee EYP, Ng MY, Khong PL. COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis. 2020;20(4):384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, Li S, Shan H, Jacobi A, Chung M. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295(3):200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;200432:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mahmoudi T, de la Guardia M, Baradaran B. Lateral flow assays towards point-of-care cancer detection: a review of current progress and future trends. TrAC Trends Anal Chem. 2020;125:115842. doi: 10.1016/j.trac.2020.115842. [DOI] [Google Scholar]
- 90.To KK. Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lv H, Wu NC, Tsang OT, Yuan M, Perera R, Leung WS et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. bioRxiv. 2020. 10.1101/2020.03.15.993097.
- 92.Cai X, Chen J, Hu J, Long Q, Deng H, Fan K, et al. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of corona virus disease 2019 (COVID-19). medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 93.Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, Wang J, Qin Y, Zhang X, Yan X, Zeng X, Zhang S. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the perspectives of clinical immunologists from China. Clin Immunol. 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiang J, Yan M, Li H, Liu T, Lin C, Huang S, et al. Evaluation of enzyme-linked immunoassay and colloidal gold-immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). MedRxiv. 2020.
- 95.Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020;55(4):105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vellingiri B, Jayaramayya K, Iyer M, Narayanasamy A, Govindasamy V, Giridharan B, Ganesan S, Venugopal A, Venkatesan D, Ganesan H, Rajagopalan K, Rahman PKSM, Cho SG, Kumar NS, Subramaniam MD. COVID-19: a promising cure for the global panic. Sci Total Environ. 2020;725:138277. doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zumla A, Hui DS, Azhar EI, Memish ZA, Maeurer M. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet. 2020;395(10224):e35–ee6. doi: 10.1016/S0140-6736(20)30305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Colson P, Raoult D. Fighting viruses with antibiotics: an overlooked path. Int J Antimicrob Agents. 2016;48(4):349–352. doi: 10.1016/j.ijantimicag.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhi ZJHHHXZ. Expert consensus on chloroquine phosphate for the treatment of novel coronavirus pneumonia. 2020;43(3):185–8. 10.3760/cma.j.issn.1001-0939.2020.03.009. [DOI] [PubMed]
- 101.Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 102.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed]
- 103.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 104.Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus—a possible reference for coronavirus disease-19 treatment option. J Med Virol. 2020;92(6):556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marmor MF, Kellner U, Lai TY, Melles RB, Mieler WF. American Academy of O. Recommendations on screening for chloroquine and hydroxychloroquine retinopathy (2016 revision) Ophthalmology. 2016;123(6):1386–1394. doi: 10.1016/j.ophtha.2016.01.058. [DOI] [PubMed] [Google Scholar]
- 106.Ben-Zvi I, Kivity S, Langevitz P, Shoenfeld Y. Hydroxychloroquine: from malaria to autoimmunity. Clin Rev Allergy Immunol. 2012;42(2):145–153. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6(2):67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.• Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2). 10.1128/mBio.00221-18The paper highlights the mechanistic insight of remdesivir against COVID 19. [DOI] [PMC free article] [PubMed]
- 109.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396). 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed]
- 110.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cao B. A trial of remdesivir in adults with mild and moderate COVID-19. Capital Medical University. 2020.
- 112.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox LA, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.WAng G. Favipiravir combined with tocilizumab in the treatment of corona virus disease 2019. 2020.
- 115.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, Braun MR, Flint M, McMullan LK, Chen SS, Fearns R, Swaminathan S, Mayers DL, Spiropoulou CF, Lee WA, Nichol ST, Cihlar T, Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Li H, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, Wang J, Liu X, Wang S, Wu X, Ge Q, He J, Zhan H, Qiu F, Guo L, Huang C, Jaki T, Hayden FG, Horby PW, Zhang D, Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu X, Wang XJ. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genomics. 2020;47(2):119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shiraki K, Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.•• Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020:105949. 10.1016/j.ijantimicag.2020.105949The study explains clinical utility of Hydroxychloroquine and azithromycin in COVID 19 treatment. [DOI] [PMC free article] [PubMed] [Retracted]
- 120.Arabi YM, Deeb AM, Al-Hameed F, Mandourah Y, Almekhlafi GA, Sindi AA, et al. Macrolides in critically ill patients with Middle East respiratory syndrome. Int J Infect Dis. 2019;81:184–190. doi: 10.1016/j.ijid.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zarogoulidis P, Papanas N, Kioumis I, Chatzaki E, Maltezos E, Zarogoulidis K. Macrolides: from in vitro anti-inflammatory and immunomodulatory properties to clinical practice in respiratory diseases. Eur J Clin Pharmacol. 2012;68(5):479–503. doi: 10.1007/s00228-011-1161-x. [DOI] [PubMed] [Google Scholar]
- 122.Beigelman A, Mikols CL, Gunsten SP, Cannon CL, Brody SL, Walter MJ. Azithromycin attenuates airway inflammation in a mouse model of viral bronchiolitis. Respir Res. 2010;11:90. doi: 10.1186/1465-9921-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baron SA, Devaux C, Colson P, Raoult D, Rolain JM. Teicoplanin: an alternative drug for the treatment of COVID-19? Int J Antimicrob Agents. 2020;55(4):105944. doi: 10.1016/j.ijantimicag.2020.105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu X, Han M, Li T, Sun W, Wang D, Fu B, Zhou Y, Zheng X, Yang Y, Li X, Zhang X, Pan A, Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Genentech . Actemra (tocilizumab) injection package insert. 2013. [Google Scholar]
- 127.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gritti G, Raimondi F, Ripamonti D, Riva I, Landi F, Alborghetti L, et al. Use of siltuximab in patients with COVID-19 pneumonia requiring ventilatory support. MedRxiv. 2020.
- 129.CytoDyn. Treatment with CytoDyn’s leronlimab indicates significant trend toward immunological restoration in severely ill COVID-19 patients 2020.
- 130.•• Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA, 2020. 10.1001/jama.2020.4783The study reveals the role of convalescent plasma therapy in COVID-19 treatment. [DOI] [PMC free article] [PubMed]
- 131.Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, Pham DH, Deif H, LaMere EA, Chang M, Kain KC, Farcas GA, Ferguson P, Latchford M, Levy G, Dennis JW, Lai EKY, Fish EN. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA. 2003;290(24):3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- 132.Grais RF, Ellis JH, Glass GE. Assessing the impact of airline travel on the geographic spread of pandemic influenza. Eur J Epidemiol. 2003;18(11):1065–1072. doi: 10.1023/a:1026140019146. [DOI] [PubMed] [Google Scholar]
- 133.CSSE. Coronavirus COVID-19 global cases by Johns Hopkins University. 2020.
- 134.Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MUG, Khan K. Potential for global spread of a novel coronavirus from China. J Travel Med. 2020;27(2). 10.1093/jtm/taaa011. [DOI] [PMC free article] [PubMed]
- 135.Tan J, Mu L, Huang J, Yu S, Chen B, Yin J. An initial investigation of the association between the SARS outbreak and weather: with the view of the environmental temperature and its variation. J Epidemiol Community Health. 2005;59(3):186–192. doi: 10.1136/jech.2004.020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Doremalen N, Bushmaker T, Munster VJ. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. 2013;18(38). 10.2807/1560-7917.es2013.18.38.20590. [DOI] [PubMed]
- 137.Otter JA, Donskey C, Yezli S, Douthwaite S, Goldenberg SD, Weber DJ. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. 2016;92(3):235–250. doi: 10.1016/j.jhin.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chan KH, Peiris JS, Lam SY, Poon LL, Yuen KY, Seto WH. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv Virol. 2011;2011:734690–734697. doi: 10.1155/2011/734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lowen AC, Steel J. Roles of humidity and temperature in shaping influenza seasonality. J Virol. 2014;88(14):7692–7695. doi: 10.1128/JVI.03544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dan S, Sharma D, Mandal M, Sharma D. Incidence of COVID-19 and its correlation between temperature and population density. Int J Sci Res Biol Sci. 2020;7(2).
- 141.Lipsitch M, Viboud C. Influenza seasonality: lifting the fog. Proc Natl Acad Sci U S A. 2009;106(10):3645–3646. doi: 10.1073/pnas.0900933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Steel J, Palese P, Lowen AC. Transmission of a 2009 pandemic influenza virus shows a sensitivity to temperature and humidity similar to that of an H3N2 seasonal strain. J Virol. 2011;85(3):1400–1402. doi: 10.1128/JVI.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Park JE, Son WS, Ryu Y, Choi SB, Kwon O, Ahn I. Effects of temperature, humidity, and diurnal temperature range on influenza incidence in a temperate region. Influenza Other Respir Viruses. 2020;14(1):11–18. doi: 10.1111/irv.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lowen AC, Steel J, Mubareka S, Palese P. High temperature (30 degrees C) blocks aerosol but not contact transmission of influenza virus. J Virol. 2008;82(11):5650–5652. doi: 10.1128/JVI.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shaman J, Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci U S A. 2009;106(9):3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hatzius J, Philips A, Mericle D, Struyven D. US Daily: a sudden stop for the US economy. Economic Research in Goldman Sachs. 2020.
- 147.Hilsenrath J, Armour S. As economic toll mounts, nation ponders trade-offs. Cost of confronting pandemic is millions of jobs, trillions in wealth lost to save potentially millions of lives. 2020. [Google Scholar]
- 148.Thunström L, Newbold SC, Finnoff D, Ashworth M, Shogren JF. The benefits and costs of using social distancing to flatten the curve for COVID-19. Journal of Benefit-Cost Analysis. 2020:1–27.
- 149.Ferguson N, Laydon D, Nedjati Gilani G, Imai N, Ainslie K, Baguelin M, et al. Report 9: impact of non-pharmaceutical interventions (NPIs) to reduce COVID19 mortality and healthcare demand. 2020. [Google Scholar]
- 150.Ozili PK, Arun T. Spillover of COVID-19: impact on the global economy. Available at SSRN 3562570. 2020.
- 151.Bell C, Devarajan S, Gersbach H. Thinking about the long-run economic costs of AIDS. The macroeconomics of HIV/AIDS. 2004;96:128–129. [Google Scholar]
- 152.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10(12 Suppl):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Siu A, Wong YR. Economic impact of SARS: the case of Hong Kong. Asian Economic Papers. 2004;3(1):62–83. doi: 10.1162/1535351041747996. [DOI] [Google Scholar]
- 154.McKibbin WJ, Sidorenko A. Global macroeconomic consequences of pandemic influenza. Australia: Lowy Institute for International Policy Sydney; 2006. [Google Scholar]
- 155.Barro RJ. Convergence and modernisation. Econ J. 2015;125(585):911–942. doi: 10.1111/ecoj.12247. [DOI] [Google Scholar]
- 156.Chou J, Kuo N-F, Peng S-L. Potential impacts of the SARS outbreak on Taiwan’s economy. Asian Economic Papers. 2004;3(1):84–99. doi: 10.1162/1535351041747969. [DOI] [Google Scholar]
- 157.Hyams KC, Murphy FM, Wessely S. Responding to chemical, biological, or nuclear terrorism: the indirect and long-term health effects may present the greatest challenge. J Health Polit Policy Law. 2002;27(2):273–291. doi: 10.1215/03616878-27-2-273. [DOI] [PubMed] [Google Scholar]
- 158.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int J Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]