Abstract
Calculation of radiation dosimetry in targeted nuclear medicine therapies is traditionally resource-intensive, requiring multiple posttherapy SPECT acquisitions. An alternative approach is to take advantage of existing pharmacokinetic data from these smaller cohorts to enable dose computation from a single posttreatment scan in a manner that may be applied to a much broader patient population. Methods: In this work, a technical description of simplified dose estimation is presented and applied to the assessment of 177Lu-prostate-specific membrane antigen (PSMA)-617 therapy for metastatic prostate cancer. By normalizing existing time–activity curves to a single measurement time, it is possible to calculate a mean and range of time-integrated activity values that relate to absorbed radiation dose. To assist with accurate pharmacokinetic modeling of the training cohort, a method for contour-guided image registration was developed. Results: Tissue-specific dose conversion factors for common posttreatment imaging times are reported along with a characterization of added uncertainty in comparison to a traditional serial imaging protocol. Single-time-point dose factors for tumor were determined to be 11.0, 12.1, 13.6, and 15.2 Gy per MBq/mL at image times of 24, 48, 72, and 96 h, respectively. For normal tissues, parotid gland factors were 6.7, 9.4, 13.3, and 19.3 Gy per MBq/mL at those times, and kidneys were 7.1, 10.3, 15.0, and 22.0 Gy per MBq/mL. Tumor dose estimates were most accurate using delayed scanning at times beyond 72 h. Dose to healthy tissues is best characterized by scanning patients in the first 2 d of treatment because of the larger degree of tracer clearance in this early phase. Conclusion: This work demonstrates a means for efficient dose estimation in 177Lu-PSMA-617 therapy. By providing methods to simplify and potentially automate radiation dosimetry, we hope to accelerate the understanding of radiobiology and development of dose–response models in this unique therapeutic context.
Keywords: 177Lu-PSMA-617, radiation dosimetry, radiobiology, image processing
See an invited perspective on this article on page 1028.
The accurate determination of absorbed radiation dose in normal tissues and tumor is key to the optimal development of all forms of radiotherapy. In external-beam radiotherapy, where the dose in tissues and tumor can be reliably controlled, dose–response relationships have been well established (1) and shown to determine the likelihood of response and toxicity. Radiation dosimetry is also key to the optimal development of directed radionuclide therapy but carries different logistical challenges that limit its application in routine practice. As a result, many response relationships in targeted therapies are not clearly established.
A novel form of therapy for prostate cancer uses radiolabeled small molecules targeting prostate-specific membrane antigen (PSMA). This is overexpressed in most prostate cancers (2,3) and limited in nonprostatic tissues (4,5), making it a highly attractive target for prostate cancer. Nevertheless, clinically, high uptake is observed in the salivary glands, duodenum, and kidneys. Of these, radiation sialadenitis with failure of adequate saliva production poses a significant issue with respect to the quality of life of patients receiving this therapy (6).
Treatment with 177Lu-PSMA-617, which is labeled with a β-emitting radionuclide, has demonstrated high activity and low acute toxicity in men with advanced cancer in both retrospective (7–12) and prospective (13) clinical trials. Detailed voxel-based radiation dosimetry was performed in the latter cohort with SPECT/CT imaging at 3 time points: 4, 24, and 96 h (14). This showed that whole-body tumor-absorbed dose was a predictor of biochemical response and that absorbed dose in normal tissues varied widely among patients. High uptake and retention of radiation occurred within tumor, whereas the normal tissues receiving the highest radiation dose included the salivary and lacrimal glands and the kidneys, findings concordant with other reported dosimetry data (7,14–20).
To date, most radiation dosimetry using radiolabeled PSMA has incorporated the MIRD schema (21–23). This involves computation of time–activity curves that are integrated to quantify disintegrations and converted into absorbed dose using a factor that approximates radiation transport for a given isotope and physical material (24,25). With β-emitting isotopes, the absorbed dose is closely proportional to the integral number of disintegrations in a region of uptake (26,27). Typical application of MIRD requires sequential scans and manual drawing of multiple regions of interest, a process that can be overly time- and resource-intensive. Techniques to simplify dose estimation from a single measurement have been described by Madsen et al. (28) and Hänscheid et al. (29). These works offer similar approaches to infer time-integrated activity based on a single measurement by using a generic effective half-time or a mixture of exponential and biexponential kinetic parameters.
The aim of this work is to expand on previous methods for single-time point dosimetry with application to tumors and at-risk organs followed administration of 177Lu-PSMA-617. The developed methodology determines local time-integrated activity for any posttreatment image time and preserves the kinetic parameters of all training cases to permit assessment according to population mean with an associated error analysis. In its development, the work also illustrates an efficient method to measure uptake on serial imaging by focusing on each region individually to improve spatial accuracy.
MATERIALS AND METHODS
177Lu-PSMA Therapy and Image Acquisition
Thirty patients were enrolled and underwent therapy with 177Lu-PSMA-617 for metastatic castration-resistant prostate cancer (30). The trial was registered with the Australian New Zealand Clinical Trials Registry (ANZCTR12615000912583) and reviewed by the local institutional review board. All patients gave written informed consent before entry. After the first cycle of therapy, patients were imaged with serial posttreatment quantitative SPECT at 4, 24, and 96 h to characterize radiotracer pharmacokinetics in tumor and normal tissues of interest. Twenty-nine patients were available for analysis, with complete multiphase image series. Each imaging system (Siemens Symbia T6 or Intevo 16) was calibrated for quantitative imaging with 177Lu according to a previously described method (31), yielding volumetric images in pseudo-PET format: voxel intensity in units of Bq/mL.
Image Alignment and Biodistribution Analysis
The pharmacokinetics for each organ was characterized with a triexponential model to account for an initial phase of uptake and for rapid and long-term clearance phases (27). The model of activity concentration, A(t), is given in Equation 1, with the amplitude of the initial uptake phase (−A1) equivalent to the negative sum of the 2 clearance phases (A2 + A3), yielding curves that pass through zero at t = 0. The selection of model and posttreatment imaging time points considered both the physical half-life of 177Lu (6.7 d) and the variety of target and at-risk tissues relevant to PSMA-directed therapy.
| Eq. 1 |
On the basis of a previous evaluation (32), regions that exhibited reliable image-based measurements for characterizing tissue kinetics included the tumor, parotid glands, submandibular glands, liver, kidney, and spleen. The lacrimal glands were outside the field of view at some time points and subject to significant partial-volume effects, rendering them unreliable for developing a population model. Red bone marrow was omitted from the analysis because most patients displayed osseous disease infiltration that did not permit consistent measurement of reproducible marrow regions.
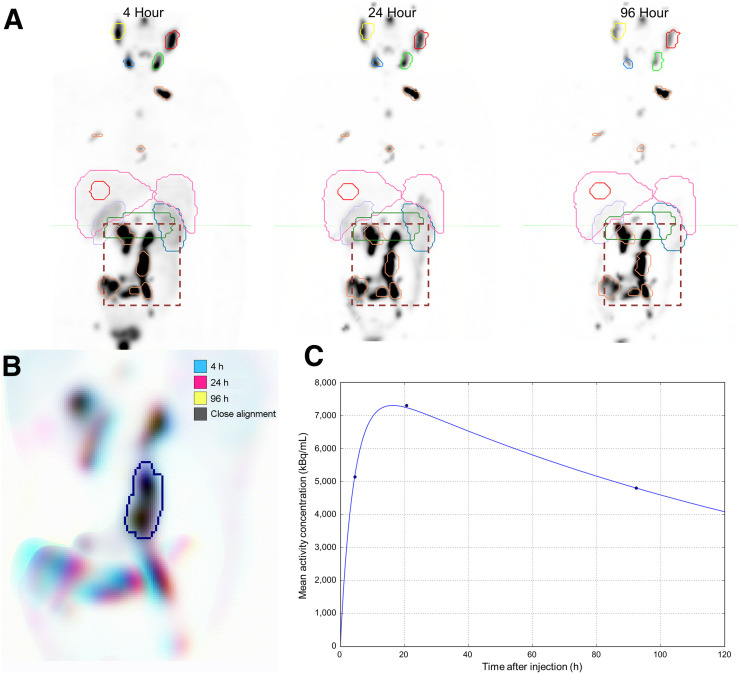
To evaluate serial measurements, 3-dimensional volumes of interest were defined on the 4-h image volume. Subsequent images were aligned for each contour independently. The 2-stage process first used a rigid CT-to-CT registration of the full image volume to achieve a gross initial alignment (33). Each volume of interest was then selected as a masked subregion, and a fine-level registration was performed using only the voxels in the structure plus an expanded range of 5–8 cm to provide boundary information. Contours with focal uptake on functional imaging, such as tumor and parotid glands, were guided by intensity on SPECT, whereas regions with clear structural margins and relatively uniform uptake, such as kidney and liver, were fused on the basis of CT Hounsfield values. After alignment, each contour was reviewed for accuracy at each of the 3 time points by colored overlay of a maximum-intensity-projection image as shown in Figure 1. The computer codes to automate sequential contour-guided image registration and generation of maximum-intensity-projection images have been made available online for general use (34).
FIGURE 1.
(A) Initial coarse alignment of full image volume for serial 4-, 24-, and 96-h quantitative SPECT images. (B) Output of focused image registration based on contour-defined mask (dashed region of serial maximum-intensity projections in A). Three-color maximum-intensity projection is used to verify alignment for serial quantitative SPECT images, with 3-dimensional contour region highlighted. Black indicates good alignment whereas color-fringing shows areas with spatial offset. (C) Three-phase exponential curve is generated and parameters are used for subsequent population analysis.
Single-Time-Point Pharmacokinetics Normalization
The curve-fitting parameters from each subject or organ pair with visually acceptable alignment were tabulated to generate a population pharmacokinetic model for each tissue type. Those with unsatisfactory agreement were excluded from further analysis. In total, 226 tumor regions were included, and 45, 43, 57, 27, and 28 regions were evaluated for parotid gland, submandibular gland, kidney, liver, and spleen, respectively. To normalize for a single posttherapy image measurement, each time–activity curve in the population model was multiplied by a single scaling factor, which modified the relative height, or amplitude, of the curve while preserving the kinetic parameters k1–3 and the relative contribution of each uptake and clearance phase. Using this method, the curves may be normalized to any postinjection time point, including intermediate values that lie between the times used for initial modeling.
| Eq. 2-1 |
| Eq. 2-2 |
| Eq. 2-3 |
| Eq. 2-4 |
| Eq. 2-5 |
where Ap,1–3 and kp,1–3 are the best-fit parameters for an organ of interest for patient p. The predicted function based on a single reference patient is given as Act(t). The scaling factor Sp(tN) modifies the height of the curve to match the measured activity at normalization time tN for a single measurement time as shown in Figure 2. For each patient reference curve, a unique scaling factor can be calculated according to Equation 2-2. The integral time-integrated activity, Ã, is then taken for all of the normalized curves as shown in Equation 2-4. A detailed derivation is provided in the supplemental material (available at http://jnm.snmjournals.org). This integral factor will determine time-integrated activity (concentration) Ã from a measurement at an arbitrary time point, tN, a value that equals or is equivalent to Ã/A(tN). As a population model, the mean and average error for each patient and time point relative to the original triple-time-point measured value is reported. To avoid subtle bias, this model is recalculated omitting each patient’s own pharmacokinetic data and given as the mean absolute deviation. Calculating the population mean values for every time point by averaging over all time-integrated activity factors for tN will reproduce the functions in Figure 3. For images reconstructed and calibrated in units of activity concentration (Bq/mL), multiplying by the time-integrated activity factor (hours) yields a measure of disintegrations per unit volume (Bq × h/mL). Absorbed dose (Gy) may be calculated by multiplying by a radiation transport S factor to a representative tissue medium. For tissue with a nominal density of 1.0 g/mL with 177Lu, the factor is approximated at 8.67E−5 Gy/(kBq × h/mL) by the OLINDA sphere model, which exhibits little dependence on physical size (Supplemental Fig. 1) (23).
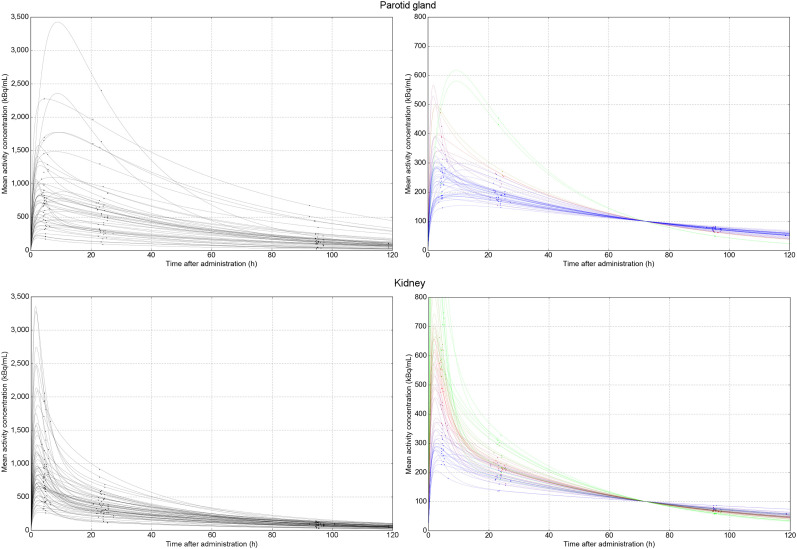
FIGURE 2.
Population time–activity curves for parotid gland (top left) and kidney (bottom left) for 177Lu-PSMA-617 based on serial posttreatment imaging. To calculate time-integrated activity based on single imaging time point, all curves are normalized by amplitude to single measurement value (in this example, 100 kBq/mL at 72 h after administration [top and bottom right]).
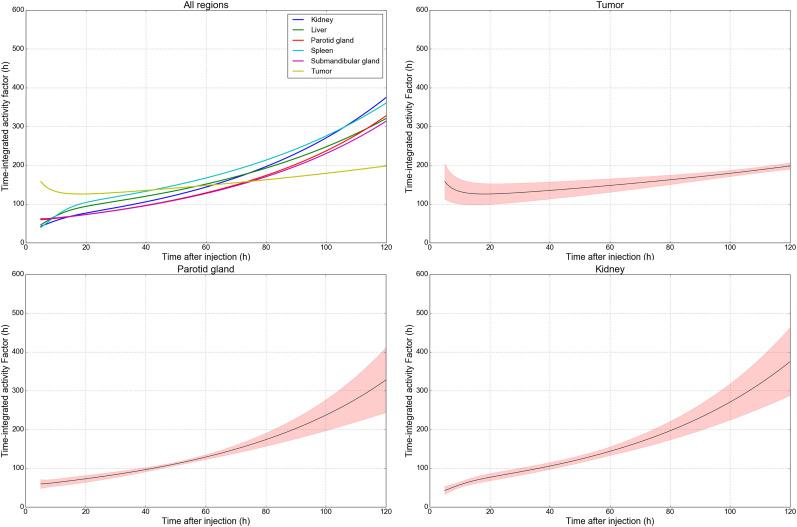
FIGURE 3.
Single-time-point factors to translate local tissue activity concentration (Bq/mL) into disintegrations per unit volume (Bq × h/mL) for dosimetry of 177Lu-PSMA-617. Dose factors tumor, parotid glands, and kidneys are shown, and all 7 considered tissues are illustrated in Supplemental Figure 2. Individual plots include overlay of mean absolute error based on range of modeled time-integrated activity values when normalized to specified measurement time. Estimates of tumor dosimetry are most accurate using delayed imaging, whereas at-risk tissues benefit from imaging in first 48 h after administration.
RESULTS
The described image measurement method provided reliable contour matching across serial time points for most images in both tumor and healthy organs. For each patient, between 8 and 12 individual tumor contours were delineated. Of the initial 260 regions, 226 (87%) displayed visually clear 3-dimensional alignment across all 3 posttreatment imaging time points and were included in the pharmacokinetics analysis (34). The results for all included organs are reported in Table 1.
TABLE 1.
Number of Patients and Contours Included for Each Tissue Type in Population Pharmacokinetic Model
| Organ | Patients | Total regions in analysis | Available contours |
| Tumor | 29 | 226 | 86.9% |
| Parotid gland | 24 | 45 | 77.6% |
| Submandibular gland | 23 | 43 | 74.1% |
| Kidney | 29 | 57 | 98.3% |
| Liver | 27 | 27 | 93.1% |
| Spleen | 28 | 28 | 96.6% |
From initial cohort, only regions with satisfactory 3-dimensional spatial alignment at all imaging time points and normal-tissue regions free from tumor infiltration were considered reliable for analysis.
For tumor and normal organs, measurements of activity concentration on a single image were correlated with time-integrated activity as estimated by serial imaging. Organ- and time-specific conversion factors are presented in Table 2 to multiply activity concentration into an estimate of decays per unit volume (Bq × h/mL). With the addition of a radiation transport S factor, it is possible to directly estimate absorbed radiation dose (Gy) from quantitative SPECT images at a given time after administration. Time-integrated activity conversion factors for tumor with images acquired at 24, 48, and 96 h after injection equate to 127, 140, and 176 h, respectively. For parotid glands, factors at those time points are 77, 108, and 222 h, indicative of the relative difference in pharmacologic clearance between tissue types.
TABLE 2.
Time-Integrated Activity Factor (h) to Convert Single Measurement (Bq/mL) into Estimated Time-Integrated Activity Concentration (Bq × h/mL, disintegrations per unit volume)
| Image time (h) | Tumor | Parotid gland | Submandibular gland | Kidney | Liver | Spleen |
| 12 | 129 ± 22% | 65 ± 13% | 66 ± 14% | 62 ± 12% | 76 ± 24% | 80 ± 25% |
| 18 | 126 ± 21% | 71 ± 11% | 71 ± 13% | 74 ± 12% | 91 ± 22% | 100 ± 24% |
| 24 | 127 ± 19% | 77 ± 10% | 77 ± 12% | 82 ± 11% | 99 ± 19% | 110 ± 21% |
| 36 | 133 ± 17% | 91 ± 7% | 91 ± 9% | 99 ± 9% | 115 ± 15% | 127 ± 16% |
| 48 | 140 ± 14% | 108 ± 4% | 107 ± 6% | 119 ± 8% | 131 ± 11% | 145 ± 12% |
| 60 | 148 ± 12% | 129 ± 5% | 127 ± 5% | 144 ± 8% | 151 ± 9% | 167 ± 10% |
| 72 | 156 ± 9% | 154 ± 8% | 151 ± 6% | 173 ± 10% | 175 ± 8% | 193 ± 11% |
| 96 | 176 ± 5% | 222 ± 17% | 217 ± 14% | 254 ± 16% | 235 ± 12% | 262 ± 19% |
| 120 | 198 ± 4% | 328 ± 28% | 314 ± 23% | 375 ± 22% | 321 ± 20% | 361 ± 28% |
For normalized image time and tissue-of-interest, mean error of residence time as percentage based on modeled cohort is reported.
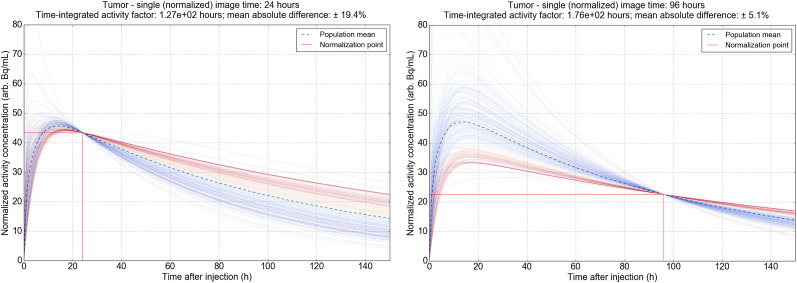
The selection of imaging time is related to the precision of dose estimate and varies by region. In tumor, the predicted single-time-point dose is more reliable with delayed acquisitions at 2 d or beyond. At 48 h, the mean absolute difference between the simplified single-measurement model and serial imaging is 14.3% of the predicted time-integrated activity value. At 72 and 96 h, the error as a percentage decreases to 9.3 and 5.1, respectively. This decrease is indicative of the prolonged retention of 177Lu-PSMA in tumor, where the proportion of disintegrations for the 6.7-d half-life isotope is largely influenced by the slow washout phase. The range of normalized time–activity curves for tumor are illustrated in Figure 4, showing the variety of temporal shapes across the population, as well as the representative mean and integral shaded area as a blend of all modeled curves.
FIGURE 4.
Population pharmacokinetic models for tumor based on serial imaging of 226 individual lesions. Each chart is normalized to mean of all curves at 24 h (left) and 96 h (right). Individual time–activity curves are shaded according to agreement with mean population integral time-integrated activity estimate.
By contrast, healthy tissues with primary uptake followed by relatively efficient washout were poorly characterized for dosimetry on delayed-phase imaging, as retention at 96 h was not representative of early activity concentrations. The minimum uncertainty using the single-time-point model for parotid glands occurs at approximately 48 h after administration (3.9% of mean), increasing to over 20% beyond 96 h. Figure 3 illustrates dose factors for each tissue, along with the relative uncertainty in applying a single-time-point model compared with serial imaging as a function of imaging time. A tabulated list of common imaging time points is given in Table 2, and values for all intermediate time values are reported in Supplemental Tables 1–3. To illustrate, for a lesion with a measured mean activity concentration of 1,500 kBq/mL at 96 h after injection, the time-integrated activity factor is determined to be 176 h ± 5%, and using a nominal soft-tissue S factor of 8.7E−5 Gy per (kBq × h/mL), the radiation dose is estimated at 22.8 ± 1.2 Gy.
DISCUSSION
Using data from patients treated with 177Lu-PSMA-617 in a prospective trial with serial imaging (30), we describe 2 novel methodologies for the application of radiation dosimetry. The first provides an automated means to perform contour-guided alignment of functional images using DICOM radiotherapy structure sets. When linked to a pharmacokinetic interpolation algorithm, this methodology provides a reliable means of characterizing tracer clearance. Even for small structures such as the parotid and submandibular glands, for which activity measurements are unreliable with displacements of several millimeters, the automated workflow achieved satisfactory coregistration across all 3 time points at a rate of 75%.
The second reports the ability of a single-time-point measurement to assess dosimetry in normal tissues and tumor. This method takes advantage of previously recorded pharmacokinetic data from a representative cohort in a manner that is similar to the works of Madsen and Hänscheid (28,29,35). Although previous works determined generic pharmacokinetic parameters for each tissue, in this method all input curves are preserved and normalized independently, facilitating analysis of mean and associated uncertainties at any time point. The model may be updated with data derived from images acquired at different postadministration timelines and may, in principle, incorporate data from any continuous pharmacokinetic function. The average deviation from single- and multiple-time-point measurements indicates that the most reliable range of image times to predict radiation dose varies with the organ or tissue of interest. This finding is in accordance with a previous work that found normal-tissue clearance to be largely completed within the first 48 h after administration (36), whereas tumor retention is more closely reflected by the physical half-life of 177Lu.
If coupled to image segmentation algorithms, such as those emerging through developments in convolutional neural network image recognition, there is a straightforward road map to fully automate dose reporting based on routine imaging follow-up (37–39). The long-term aim of these investigations is to provide individualized therapy based on a patient’s known physiology using a validated rationale to administer a personalized maximum tolerated dose and so optimize the therapeutic ratio of treatment (40). We have demonstrated in previous work that biochemical response is related to whole-body tumor dose, but we have not clearly reported a relationship between dose and acute normal-tissue toxicity; however, this may be due to the lack of acute toxicity that has been observed in the clinical application of 177Lu-PSMA-617 to date (13). Additionally, the pharmacokinetic models may be decoupled from the physical half-life of 177Lu and applied to other therapeutic tracers by decay-correcting and using an appropriate radiation transport dose factor. In this manner, the same methodology may be applied to single-time-point dose factors to generate factors for, for example, 90Y-PSMA-617 therapy under the assumption that the tracer has a comparable pharmacologic biodistribution and clearance kinetics (41).
In the long term, it is hoped that this methodology can be expanded with additional multiple-time-point data either locally or from other centers to improve the overall accuracy of the model and potentially identify predictors of which patients will be either slow-clearing or fast-clearing, which may become apparent across a broader population of sampled data. It would also be worthwhile to investigate whether organ time–activity curves are interdependent within a patient. That is, in this evaluation it is assumed that organ clearance is not affected by the kinetic models in other tissues.
CONCLUSION
This work presents a means of estimating radiation dosimetry for 177Lu-PSMA therapy based on readily available posttreatment imaging for a single time point. A novel method of predicting time-integrated activity using a population of normalized pharmacokinetic curves is illustrated—one that may incorporate data from a variety of imaging timelines and interpolate dose coefficients for any intermediate times. The aim is to improve dose–response modeling by enabling simplified radiation dose estimates from a broad cohort of patients receiving molecular radiotherapy, as opposed to only those in trials involving comprehensive follow-up imaging.
DISCLOSURE
Price Jackson is supported by the Victorian Cancer Agency. Michael Hofman is supported by a Clinical Fellowship Award from the Peter MacCallum Foundation and receives research support from Endocyte (a Novartis company). Rodney Hicks is the recipient of a National Health and Medical Research Council Practitioner Fellowship (APP1108050). 177Lu was supplied by the Australian Nuclear Science and Technology Organization (ANSTO) and PSMA-617 by Endocyte and Advanced Biochemical Compounds (ABX). Rodney Hicks holds stock in Telix Pharmaceuticals on behalf of the Peter MacCallum Cancer Centre, with all dividends donated to his department. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Is it possible to obtain reliable radiation dose estimates to tumor and at-risk organs based on a single follow-up quantitative SPECT/CT scan in the treatment of prostate cancer with 177Lu-PSMA-617?
PERTINENT FINDINGS: Using a pilot cohort of 29 patients with serial posttreatment imaging, a novel method of incorporating existing pharmacokinetic models was developed to predict dosimetry from a single posttherapy image. The method presents conversion factors to estimate the absorbed dose from a measured activity concentration, along with a characterization of added uncertainty by using the simplified model. Findings show that the ideal imaging time to yield accurate dose estimates across tissue types is in the window of 2–3 d after administration of 177Lu-PSMA-617.
IMPLICATIONS FOR PATIENT CARE: The technique permits radiation dose calculation in routine follow-up of PSMA therapy, as would be standard at many clinical centers. Absorbed dose may be considered a more robust metric to inform ongoing management and optimize prescription of subsequent treatment cycles than are traditional measures of uptake such as SUV. Additionally, by collecting dosimetry from a broader cohort of patients, this technique should accelerate the understanding of radiobiology and potential for treatment personalization in targeted radionuclide therapies.
Supplementary Material
REFERENCES
- 1.Marks LB, Yorke ED, Jackson A, et al. Use of normal tissue complication probability models in the clinic. Int J Radiat Oncol Biol Phys. 2010;76(suppl):S10–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith-Jones PM, Vallabhajosula S, Navarro V, Bastidas D, Goldsmith SJ, Bander NH. Radiolabeled monoclonal antibodies specific to the extracellular domain of prostate-specific membrane antigen: preclinical studies in nude mice bearing LNCaP human prostate tumor. J Nucl Med. 2003;44:610–617. [PubMed] [Google Scholar]
- 3.Ghosh A, Heston WD. Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. J Cell Biochem. 2004;91:528–539. [DOI] [PubMed] [Google Scholar]
- 4.Troyer JK, Beckett ML, Wright GL., Jr Detection and characterization of the prostate-specific membrane antigen (PSMA) in tissue extracts and body fluids. Int J Cancer. 1995;62:552–558. [DOI] [PubMed] [Google Scholar]
- 5.Sokoloff RL, Norton KC, Gasior CL, Marker KM, Grauer LS. A dual-monoclonal sandwich assay for prostate-specific membrane antigen: levels in tissues, seminal fluid and urine. Prostate. 2000;43:150–157. [DOI] [PubMed] [Google Scholar]
- 6.Taïeb D, Foletti J-M, Bardiès M, Rocchi P, Hicks RJ, Haberkorn U. PSMA-targeted radionuclide therapy and salivary gland toxicity: why does it matter? J Nucl Med. 2018;59:747–748. [DOI] [PubMed] [Google Scholar]
- 7.Zechmann CM, Afshar-Oromieh A, Armor T, et al. Radiation dosimetry and first therapy results with a 124I/131I-labeled small molecule (MIP-1095) targeting PSMA for prostate cancer therapy. Eur J Nucl Med Mol Imaging. 2014;41:1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmadzadehfar H, Rahbar K, Kurpig S, et al. Early side effects and first results of radioligand therapy with 177Lu-DKFZ-617 PSMA of castrate-resistant metastatic prostate cancer: a two-centre study. EJNMMI Res. 2015;5:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahbar K, Ahmadzadehfar H, Kratochwil C, et al. German multicenter study investigating 177Lu-PSMA-617 radioligand therapy in advanced prostate cancer patients. J Nucl Med. 2017;58:85–90. [DOI] [PubMed] [Google Scholar]
- 10.Kratochwil C, Giesel FL, Stefanova M, et al. PSMA-targeted radionuclide therapy of metastatic castration-resistant prostate cancer with 177Lu-labeled PSMA-617. J Nucl Med. 2016;57:1170–1176. [DOI] [PubMed] [Google Scholar]
- 11.Heck MM, Retz M, D’Alessandria C, et al. Systemic radioligand therapy with 177Lu labeled prostate specific membrane antigen ligand for imaging and therapy in patients with metastatic castration resistant prostate cancer. J Urol. 2016;196:382–391. [DOI] [PubMed] [Google Scholar]
- 12.Yadav MP, Ballal S, Tripathi M, et al. 177Lu-DKFZ-PSMA-617 therapy in metastatic castration resistant prostate cancer: safety, efficacy, and quality of life assessment. Eur J Nucl Med Mol Imaging. 2017;44:81–91. [DOI] [PubMed] [Google Scholar]
- 13.Hofman MS, Violet J, Hicks RJ, et al. [177Lu]-PSMA-617 radionuclide treatment in patients with metastatic castration-resistant prostate cancer (LuPSMA trial): a single-centre, single-arm, phase 2 study. Lancet Oncol. 2018;19:825–833. [DOI] [PubMed] [Google Scholar]
- 14.Violet J, Jackson P, Ferdinandus J, et al. Dosimetry of 177Lu-PSMA-617 in metastatic castration-resistant prostate cancer: correlations between pretherapeutic imaging and whole-body tumor dosimetry with treatment outcomes. J Nucl Med. 2019;60:517–523. [DOI] [PubMed] [Google Scholar]
- 15.Hohberg M, Eschner W, Schmidt M, et al. Lacrimal glands may represent organs at risk for radionuclide therapy of prostate cancer with [Lu]DKFZ-PSMA-617. Mol Imaging Biol. 2016;18:437–445. [DOI] [PubMed] [Google Scholar]
- 16.Delker A, Fendler WP, Kratochwil C, et al. Dosimetry for 177Lu-DKFZ-PSMA-617: a new radiopharmaceutical for the treatment of metastatic prostate cancer. Eur J Nucl Med Mol Imaging. 2016;43:42–51. [DOI] [PubMed] [Google Scholar]
- 17.Kabasakal L, AbuQbeitah M, Aygun A, et al. Pre-therapeutic dosimetry of normal organs and tissues of 177Lu-PSMA-617 prostate-specific membrane antigen (PSMA) inhibitor in patients with castration-resistant prostate cancer. Eur J Nucl Med Mol Imaging. 2015;42:1976–1983. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto S, Thieme A, Allmann J, et al. Radiation dosimetry for 177Lu-PSMA-I&T in metastatic castration-resistant prostate cancer: absorbed dose in normal organs and tumor lesions. J Nucl Med. 2017;58:445–450. [DOI] [PubMed] [Google Scholar]
- 19.Yadav MP, Ballal S, Tripathi M, et al. Post-therapeutic dosimetry of 177Lu-DKFZ-PSMA-617 in the treatment of patients with metastatic castration-resistant prostate cancer. Nucl Med Commun. 2017;38:91–98. [DOI] [PubMed] [Google Scholar]
- 20.Fendler WP, Kratochwil C, Ahmadzadehfar H, et al. 177Lu-PSMA-617 therapy, dosimetry and follow-up in patients with metastatic castration-resistant prostate cancer [in German]. Nucl Med (Stuttg). 2016;55:123–128. [PubMed] [Google Scholar]
- 21.Hindorf C, Glatting G, Chiesa C, Linden O, Flux G. EANM Dosimetry Committee guidelines for bone marrow and whole-body dosimetry. Eur J Nucl Med Mol Imaging. 2010;37:1238–1250. [DOI] [PubMed] [Google Scholar]
- 22.Siegel JA, Thomas SR, Stubbs JB, et al. MIRD pamphlet no. 16: techniques for quantitative radiopharmaceutical biodistribution data acquisition and analysis for use in human radiation dose estimates. J Nucl Med. 1999;40(suppl):37S–61S. [PubMed] [Google Scholar]
- 23.Stabin MG, Sparks RB, Crowe E. OLINDA/EXM: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med. 2005;46:1023–1027. [PubMed] [Google Scholar]
- 24.Bolch WE, Bouchet LG, Robertson JS, et al. MIRD pamphlet no. 17: the dosimetry of nonuniform activity distributions—radionuclide S values at the voxel level. J Nucl Med. 1999;40(suppl):11S–36S. [PubMed] [Google Scholar]
- 25.Watson EE, Stabin MG, Siegel JA. MIRD formulation. Med Phys. 1993;20:511–514. [DOI] [PubMed] [Google Scholar]
- 26.Bolch WE, Eckerman KF, Sgouros G, Thomas SR. MIRD pamphlet no. 21: a generalized schema for radiopharmaceutical dosimetry—standardization of nomenclature. J Nucl Med. 2009;50:477–484. [DOI] [PubMed] [Google Scholar]
- 27.Jackson PA, Beauregard JM, Hofman MS, Kron T, Hogg A, Hicks RJ. An automated voxelized dosimetry tool for radionuclide therapy based on serial quantitative SPECT/CT imaging. Med Phys. 2013;40:112503. [DOI] [PubMed] [Google Scholar]
- 28.Madsen MT, Menda Y, O’Dorisio TM, O’Dorisio MS. Single time point dose estimate for exponential clearance. Med Phys. 2018;45:2318–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänscheid H, Lapa C, Buck AK, Lassmann M, Werner RA. Dose mapping after endoradiotherapy with 177Lu-DOTATATE/DOTATOC by a single measurement after 4 days. J Nucl Med. 2018;59:75–81. [DOI] [PubMed] [Google Scholar]
- 30.Hofman MS, Violet J, Hicks RJ, et al. [177Lu]-PSMA-617 radionuclide treatment in patients with metastatic castration-resistant prostate cancer (LuPSMA trial): a single-centre, single-arm, phase 2 study. Lancet Oncol. 2018;19:825–833. [DOI] [PubMed] [Google Scholar]
- 31.Beauregard J-M, Hofman MS, Pereira JM, Eu P, Hicks RJ. Quantitative 177Lu SPECT (QSPECT) imaging using a commercially available SPECT/CT system. Cancer Imaging. 2011;11:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Violet J, Jackson P, Ferdinandus J, et al. Dosimetry of 177Lu-PSMA-617 in metastatic castration-resistant prostate cancer: correlations between pretherapeutic imaging and whole-body tumor dosimetry with treatment outcomes. J Nucl Med. 2019;60:517–523. [DOI] [PubMed] [Google Scholar]
- 33.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. [DOI] [PubMed] [Google Scholar]
- 34.Contour-guided registration. GitHub website. https://github.com/jacksonmedphysics/contour-guided-registration. Accessed March 10, 2020.
- 35.Hänscheid H, Lapa C, Buck AK, Lassmann M, Werner RA. Absorbed dose estimates from a single measurement one to three days after the administration of 177Lu-DOTATATE/-TOC. Nuklearmedizin. 2017;56:219–224. [DOI] [PubMed] [Google Scholar]
- 36.Kratochwil C, Giesel FL, Stefanova M, et al. PSMA-targeted radionuclide therapy of metastatic castration-resistant prostate cancer with 177Lu-labeled PSMA-617. J Nucl Med. 2016;57:1170–1176. [DOI] [PubMed] [Google Scholar]
- 37.Belal SL, Sadik M, Kaboteh R, et al. Deep learning for segmentation of 49 selected bones in CT scans: first step in automated PET/CT-based 3D quantification of skeletal metastases. Eur J Radiol. 2019;113:89–95. [DOI] [PubMed] [Google Scholar]
- 38.Jackson P, Hardcastle N, Dawe N, Kron T, Hofman MS, Hicks RJ. Deep learning renal segmentation for fully automated radiation dose estimation in unsealed source therapy. Front Oncol. 2018;8:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sjostrand K, Anand A, Richter J, et al. Automated detection and quantification of prostatic PSMA uptake in SPECT/CT using a deep learning algorithm for segmentation of pelvic anatomy [abstract]. J Nucl Med. 2018;59(suppl 1):30. [Google Scholar]
- 40.Kabasakal L, Türkay Toklu NY, Demirci E, et al. Lu-177-PSMA-617 prostate-specific membrane antigen inhibitor therapy in patients with castration-resistant prostate cancer: stability, bio-distribution and dosimetry. Mol Imaging Radionucl Ther. 2017;26:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rathke H, Flechsig P, Mier W, et al. Dosimetry estimate and initial clinical experience with 90Y-PSMA-617. J Nucl Med. 2019;60:806–811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.