Abstract
Purpose
The aim of this study was to study the roles and potential mechanism of LINC00520 in the progression of lung cancer.
Methods
The expression of LINC00520 and miR-3175 in lung cancer tissues and cells was detected by qRT-PCR. The relationship between LINC00520 level and disease stage was also calculated. Kaplan–Meier survival curve was drawn to observe the survival difference between high and low expression patients. Lipofectamine 2000 was used to transfect siLINC00520, miR-3175 inhibitor and their controls in lung cancer cells. CCK8 and colony formation assay were processed for cell proliferation. Transwell assay was undertaken for migration and invasion of lung cancer cells. MiRDB predicts the combination of LINC00520 and miR-3175. Luciferase and RNA pulldown assay were applied to verify the binding site. Correlation analysis of miR-3175 and LINC00520 expression in lung cancer tissues was shown.
Results
LINC00520 was highly expressed in lung cancer tissues and cells. Patients at III+IV stage were always with higher LINC00520 level than patients at I+II stage. Patients with high expression of lncRNA LINC00520 have short survival time (hazard ratio=1.7). Knockdown of LINC00520 inhibited proliferation, invasion and migration of lung cancer cells. LINC00520 targeted and negatively regulated miR-3175 (r=−0.528; P<0.001). MiR-3175 inhibitor rescued the effect of si-LINC00520 on lung cancer progression.
Conclusion
LncRNA LINC00520 could predict poor prognosis and promote progression of lung cancer by inhibiting miR-3175 expression.
Keywords: lncRNA LINC00520, lung cancer, prognosis, miR-3175
Introduction
The incidence and mortality of lung cancer in China rank first among malignant tumors, and the 5-year survival rate is only 19.7%.1 According to different degrees of differentiation and morphological characteristics, there are two main types of lung cancer, including non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC).2 At present, lung cancer examination methods mainly include bronchoscopy, chest tomography, CT and MRI, sputum cytology, lung puncture, superficial lymph node aspiration, radionuclide inspection and positron emission tomography.3 Notably, more than 50% of lung cancer patients may relapse within 5 years.4 Even for primary tumors with no lymph node metastasis and tumors smaller than 1 cm, nearly 8% of patients die from this disease within 5 years after complete resection.5 The reason is that the disease cannot be detected in the early stages in a timely and early manner.6 To solve the many problems of lung cancer prevention and treatment, the key is to further promote and improve lung cancer screening and early diagnosis and treatment strategies.7
The occurrence of lung cancer is a long-term, multi-stage process. Similar to most tumor characteristics, the occurrence and development of lung cancer are affected by many factors, such as the regulation of apoptosis-related gene expression, post-translational protein modification, molecular interactions, signal pathways and feedback, DNA damage repair and removal of damaged cells, activation of oncogenes and silencing of tumor suppressor genes.8,9 In the process of exploring the pathogenesis of lung cancer, the close relationship between non-coding RNAs including micro-RNAs and long-chain non-coding RNAs (lncRNA) and tumors has been identified as a new research direction.10,11 The latest research results also suggest that a variety of miRNAs and lncRNAs play an important role in the occurrence and development of lung cancer. Hundreds of lncRNAs were associated with lung cancer through gene expression profiling and large-scale RNA sequencing studies on lung cancer tissues and their adjacent control tissues.12,13 LncRNA has important regulatory roles in key cellular processes of lung cancer, such as proliferation, invasion, and survival.14 For instance, lncRNA-SNHG7 could increase expression of FAIM2, and then accelerates apoptosis and inhibits proliferation.15 In addition, abnormal expression of lncRNA is closely related to lung cancer metastasis, progression of pathological stages, and poor prognosis.16 As reported in previous study, lncRNA NNT-AS1 was referred to be related with stage of lung tumor and metastasis, which also could improve proliferation and invasion by regulating the expression of miR-129-5p in vitro.17 However, more effective and convenient lncRNAs need to be explored for lung cancer diagnosis.
Recent years, LINC00520 was confirmed to play a pivotal role in various cancers.18 For example, the expression of LINC00520 was up-regulated in breast cancer, which plays a proto-oncogene role in breast cancer and promotes tumor invasion and metastasis.18 Previous researchers also used the lncRNA expression profile chip to study the expression profile of lncRNA in laryngeal squamous cell carcinoma tissues, and found that the expression of LINC00520 was significantly up-regulated in laryngeal cancer tissues.19 However, the molecular mechanism of LINC00520 has not been researched so far.
Materials and Methods
Patients and Tissues
A total of 52 lung cancer patients were included in this study. All patients had not received chemotherapy or molecular target therapy before surgery for diagnosis. In all patients, 26 patients were at stage I+II, and the other patients were at stage III+IV. Besides, 21 patients were with aggressive tumor, while 31 patients were with non-aggressive tumor. Based on the media expression of lncRNA LINC00520, all patients could be divided into high expressed group (n=27) and low expressed group (n=25). This study was approved by the ethics committee of Qilu Hospital of Shandong University. All experiments were performed in accordance with the Declaration of Helsinki. All patients signed informed consent. During the operation, lung cancer tissues and paired adjacent tissues were obtained and quickly frozen and stored in −80°C refrigerator.
Cell Culture and Transfection
The cell lines used in the study were four lung cancer cell lines (H460, H1975, H1299 and A549) and a human normal lung epithelial cell line (BEAS-2B), which were obtained from American type culture collection (ATCC, USA). The cells were cultured in a DMEM medium containing 10% FBS in a 37°C, 5% CO2 incubator. Lung cancer cells in log phase growth were seeded into 6-well plates before transfection. When the degree of cell fusion was 80% to 90%, the cells were transfected. LncRNA LINC00520 siRNA, miR-3175 mimic, miR-3175 inhibitor and their controls were designed and synthesized by Shanghai Ruisai Biotechnology Co., Ltd. lncRNA LINC00520 siRNA and LipofectamineTM 2000 (Invitrogen, Carlsbad, USA) were diluted in serum-free medium, then mixed and incubated at room temperature for 5 minutes. The transfection mixed reagent was added to the culture plate for transfection. After 6–8 h of culture, the transfection culture solution was aspirated, and fresh DMEM medium containing serum was added to continue the culture. MiR-3175 mimic or miR-3175 inhibitor was also transfected by above steps.
QRT-PCR Assay
Lung cancer tissue and adjacent tissue specimens were taken and ground on ice, then 1 mL of TRIzol reagent (Invitrogen, USA) was added. Similarly, the total RNA of each group of cells was extracted with TRIzol reagent on ice, and the purity of the RNA was detected with Ultra trace nucleic acid protein analyzer (Nanodrop 2000, Thermo Fisher Scientific, Waltham, MA, USA). Total RNA was reversely transcribed into cDNA using an OneStep RT-PCR kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer’s protocol. SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Dalian, China) was used to perform qPCR. The primers used in the experiments were designed by Primer 3. The primers were shown as follows: lncRNA LINC00520 (forward) 5ʹ-GTGCTAGGAGGACCCATACG-3ʹ, (reverse) 5ʹ-GCCACCAATGTGCTGTCAAG-3ʹ; miR-3175 (forward) 5ʹ-GATACTCACGGGGAGAGAACGCAG-3ʹ, (reverse) 5ʹ-GTGCAGGGTCCGAGGT-3ʹ.
CCK8 Assay for Proliferation
Logically staged H460 cells were transfected with specific siLINC00520 and/or miR-3175 or controls. After 8 h of incubation, 100 μL of medium containing 2000 cells was added to a 96-well plate. After 24, 48, 72 h and 96 h of incubation, 10 μL of CCK8 reagent (Dojindo, Japan) was added to each well, and the incubation was continued for 2 h. The absorbance (D) at 450 nm was measured with a microplate reader.
RNA Pulldown
MiR-3175 sequence was labeled with Biotin and performed RNA pulldown using Pierce Magnetic RNA Pull-Down Kit (Thermo Fisher, USA). qRT-PCR assay was used to detect LINC00520 enrichment in the RNA fraction.
Colony Formation Assay
After transfection, a single cell suspension was prepared by digestion with 0.25% trypsin. Subsequently, the cell suspension was repeatedly blown with a pipette tip until more than 95% of the cells became single, and the cell counting plate was used to count living cells after trypan blue staining. 100 cells were seeded in a six-well plate, 2 mL of complete medium was added, and the cells were shaken evenly. Three parallel samples were set, and the liquid was changed every other day, and cultured in a 37°C, 5% CO2 incubator for 7 days. Finally, the number of clone formation was calculated.
Transwell Assay for Migration and Invasion
The brief experimental steps of invasion were as follows. Total 20 μL of 1 mg/mL matrigel was added to the upper chamber, while 600 μL of DMEM medium containing 20% serum was added to the lower chamber of a 24-well plate. Then, 100 μL of a serum-free medium was added to the upper chamber. After 48 h of incubation, the cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, and treated with 0.1% crystal violet for 0.5 h, photographed under an inverted microscope, and counted. For migration experiments, Matrigel was not used.
Luciferase Assay
The biological information predicts that there is a miR-3175 binding site on lncRNA LINC00520. A fragment with both binding sites was amplified by RT-PCR and inserted into the PGL-3-REPORT luciferase vector to construct a LINC00520 wild plasmid. At the same time, the cells were transfected with lncRNA LINC00520 wild plasmid, lncRNA LINC00520 mutant plasmid, and miR-3175 separately or at the same time. According to the instructions of the Dual Luciferase Reporter Kit, luciferase activity was measured in the dark.
Statistical Analysis
Data were processed using SPSS 16.0 software, measurement data were expressed as mean ± standard deviation. Un-paired t-test was used to compare the differences between the two groups of data. The association between LINC00520 expression and overall survival was analyzed by K-M method and compared by Log-rank test. ANOVA test was used for analysis of over two groups. Pearson correlation analysis evaluates miR-3175 and LINC00520 expression correlation in lung cancer tissues. The difference was statistically significant with P <0.05.
Results
LINC00520 Expression and Prognosis of Lung Cancer
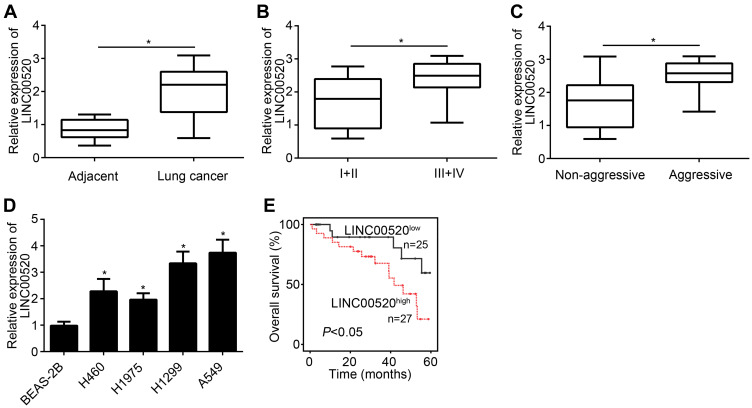
QRT-PCR was processed to detect the expression of LINC00520 in lung cancer tissues and cell lines. Relative expression of LINC00520 was significantly higher in lung cancer tissue than adjacent normal tissue (Figure 1A). Besides, the LINC00520 expression was upregulated as the increase of tumor stage and aggressive (Figure 1B and C). The LINC00520 level was higher in all lung cancer cell lines (H460, H1975, H1299 and A549) than that in BEAS-2B cell line (Figure 1D). Obviously, LINC00520 was highest expressed in H1299 and A549 cell lines. Thereby, the two cell lines were chosen for the following experiments. Furthermore, patients with high expressed LINC00520 were always with poor overall survival (Figure 1E). According to the above results, we inferred that high LINC00520 expression predicted poor prognosis of lung cancer.
Figure 1.
LINC00520 expression and prognosis of lung cancer. (A) The expression of LINC00520 in lung cancer tissue and adjacent normal tissue. (B) The expression of LINC00520 in lung cancer tissues with different stages (I+II, N=26; III+IV, N=26). (C) The expression of LINC00520 in metastatic or non-metastatic lung cancer tissues (Aggressive, N = 21; Non aggressive, N = 31. (D) LINC00520 expression in tumor cell lines. (E) Survival curve (Low expression, N = 25; high expression, N = 27. *p < 0.05.
Knockdown of LINC00520 Inhibited Progression of Lung Cancer Cells
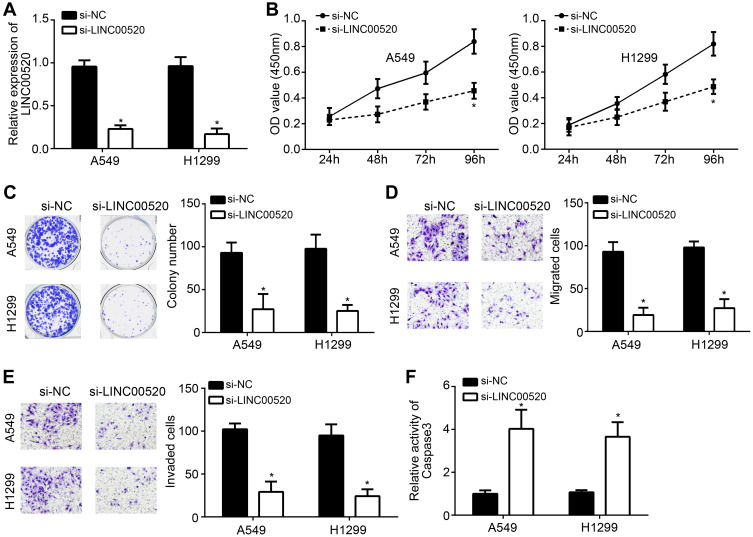
After siLINC00520 transfection, the expression of LINC00520 was significantly lower, which indicated the transfection was successful (Figure 2A). CCK8 assay and colony formation assay were processed to detect the proliferation of lung cancer cells after transfection. As the results shown in Figure 2B and C, the proliferation was significantly suppressed. Simultaneously, migration and invasion processes were also inhibited (Figure 2D and E). Importantly, relative activity of Caspase 3 was significantly increased, which indicating that si-LINC00520 was significantly accelerate apoptotic process (Figure 2F).
Figure 2.
Knockdown of LINC00520 inhibited progression of lung cancer cells. (A) Knockdown of LINC00520. (B) CCK8 assay for proliferation after siLINC00520 transfection. (C) Colony formation experiment. (D) Transwell assay for migration. (E) Transwell assay for invasion. (F) Caspase-3 activity detection for apoptosis. *p < 0.05.
LINC00520 Negatively Regulated the Expression of MiR-3175
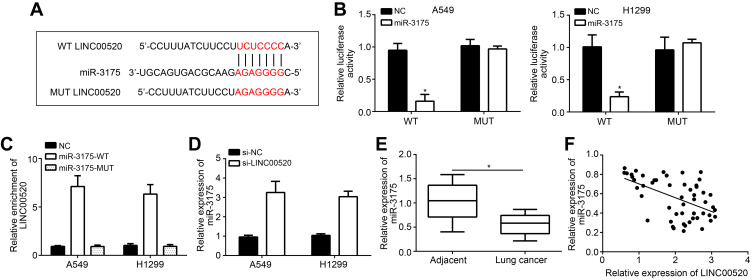
MiRDB database was applied to predict the binding between LINC00520 and miR-3175, and the binding site is shown in Figure 3A. Luciferase assay and RNA pulldown assay were processed to confirm the binding. The relative luciferase activity was significantly decreased by miR-3175 transfection in LINC00520 WT cells, while that of LINC00520 MUT cells were not influenced (Figure 3B). Moreover, LINC00520 was enriched by miR-3175 (Figure 3C). Si-LINC00520 could also up-regulate the expression of miR-3175 in both A549 and H1299 cells (Figure 3D). Above results confirmed that miR-3175 was the target of LINC00520. In addition, miR-3175 was lower expressed in lung cancer tissues than normal (Figure 3E). There was negative correlation between LINC00520 and miR-3175 expression (r=−0.528; P<0.001; Figure 3F).
Figure 3.
LINC00520 negatively regulated the expression of miR-3175. (A) MiRDB database was applied to predict the combination of LINC00520 and miR-3175. (B) Luciferase experiment for verifying the binding of LINC00520 and miR-3175. (C) RNA pulldown analysis of LINC00520/miR-3175 binding. (D) MiR-3175 expression detection in lung cancer cells. (E) MiR-3175 level in lung cancer tissues. (F) Correlation between miR-3175 and LINC00520 expression in lung cancer tissues. *p < 0.05.
MiR-3175 Inhibitor Rescued the Effect of Si-LINC00520
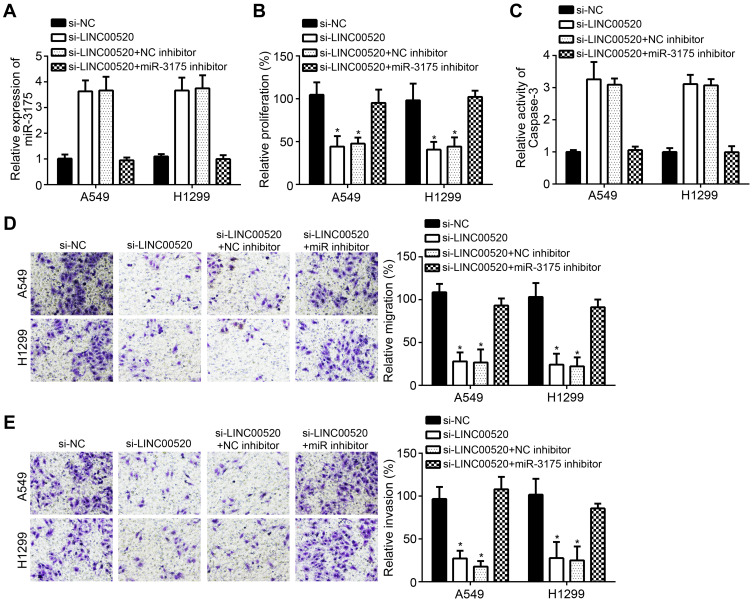
All cell lines were grouped into siLINC00520, siNC, siLINC00520+NC inhibitor and si-LINC00520+miR-3175 inhibitor groups for rescue assay. Si-LINC00520 could increase the expression of miR-3175, while miR-3175 inhibitor could decrease miR-3175 expression (Figure 4A). Moreover, siLINC00520 could inhibit proliferation, migration and invasion. In addition, siLINC00520 increased the relative activity of caspase 3, which indicating that siLINC00520 accelerated apoptotic process. Interestingly, miR-3175 inhibitor rescued the effect of siLINC00520 in the progress of lung cancer cell proliferation, migration, invasion and apoptosis (Figure 4B–E).
Figure 4.
MiR-3175 inhibitor rescued the effect of si-LINC00520. All cell lines were grouped into siLINC00520, siNC, siLINC00520+NC inhibitor and si-LINC00520+miR-3175 inhibitor groups for rescue assay. (A) QRT-PCR assay for miR-3175 expression. (B) CCK8 assay. (C) Caspase 3 activity detection for apoptosis. (D) Transwell assay for migration. (E) Transwell assay for invasion. *p < 0.05.
Discussion
Genetic testing has a good guiding function in the screening and targeted treatment of lung cancer patients. The role of lncRNA LINC00520 in the progression of various cancers has been confirmed, but its function in lung cancer has not been researched so far. In this study, lncRNA LINC00520 was highly expressed in lung cancer tissues and cells. Patients at III+IV stage were always with higher lncRNA LINC00520 level than patients at I+II stage. Besides, patients with high expression of lncRNA LINC00520 have short survival time. Knockdown of LINC00520 inhibited proliferation, invasion and migration of lung cancer cells. MiR-3175 inhibitor rescued the effect of si-LINC00520 on lung cancer progression.
LncRNA LINC00520, as a new type of lncRNA, is approximately 112 kb from the kinesin receptor KTN1 and approximately 321 kb from the E3 ubiquitin ligase family member 2-PELI2.18 In previous studies, the role of lncRNA LINC00520 has been referred. In breast cancer, the oncogenes Src, PIK3CA, and STAT3 could regulate the high expression of LINC00520 and thus affect the progression of breast cancer.18 Chen et al referred that activation of PI3K/AKT pathway affected migration and invasion of lung cancer cell.20 Inhibition of PI3K/AKT signaling pathway could suppress Gefitinib resistance during lung cancer treatment.21 Thereby, LINC00520 may play an important role in development of lung cancer.
Furthermore, miRDB database and experiments results in this study referred that lncRNA LINC00520 targeted and negatively regulated miR-3175. So far, few studies reported the role of miR-3175 in cancer. In 2015, Liang et al suggested that down-regulated HOXB1 could accelerate the progression of glioma by regulating HOXB1 level.22 Besides, miR-3175 could also participate in PI3K/AKT signaling pathway and modulate development of glioma cells.23 Furthermore, carboplatin was verified to suppress invasion of lung cancer cells by increasing Smad7 level.24 Additionally, miR-3175 inhibitor rescued the effect of si-LINC00520 on lung cancer progression in this study. According to the above evidence, we concluded that lncRNA LINC00520 promote progression of lung cancer by inhibiting miR-3175 expression. We also showed that LINC00520 inhibited the level of miR-3175. However, how LINC00520 regulates miR-3175 expression remains investigation in the future.
However, there are some limitations in this study. Firstly, the detailed molecular mechanism was only researched in vitro. In vivo research will be the focus of our future study. Secondly, fewer clinical cases were included in this study, and more patients with different categories will be included in future studies. Finally, the molecular pathogenesis of lung cancer is complex. More lncRNA, miRNA and genes needed to be explored. We just explored the tip of the iceberg. There are still many mechanisms we need to explore.
In conclusion, lncRNA LINC00520 predict poor prognosis and promote progression of lung cancer by inhibiting miR-3175 expression. Thereby, it could be regarded as a novel biomarker of lung cancer diagnosis and treatment. However, there are some limitations in our study. For example, more clinical information is required and multivariable Cox’s regression analysis needs to be performed in the future.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen X, Fang J, Nie J, et al. Multivariate analysis of prognostic factors in the elderly patients with small cell lung cancer: a study of 160 patients. Chin J Lung Cancer. 2014;17(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss G, Bemis L, Sugita M, et al. MicroRNAs predicted to regulate highly expressed genes in non-small cell lung cancer (NSCLC) and small-cell lung cancer (SCLC) are localized to 19q. 2007:4517. [Google Scholar]
- 3.Sumitani M, Takifuji N, Nanjyo S, et al. Clinical relevance of sputum cytology and chest X-ray in patients with suspected lung tumors. Intern Med. 2008;47(13):1199–1205. doi: 10.2169/internalmedicine.47.0777 [DOI] [PubMed] [Google Scholar]
- 4.Kmietowicz Z. Rate of surgery for lung cancer in England and Wales rose 50% in five years. BMJ. 2013;347:f7201. doi: 10.1136/bmj.f7201 [DOI] [PubMed] [Google Scholar]
- 5.Saji H, Tsuboi M, Usuda J, Nomura M, Ohira T, Ikeda N. Abstract #2577: a gene expression signature predicts prognosis of patients with early stage non-small cell lung cancer. 2009:2577. [Google Scholar]
- 6.Simmerman EL, Dillard TA, Hao Z, Sadek RF, Khleif SN, Schroeder C. Free lung cancer screening trends toward a twofold increase in lung cancer prevalence in the underserved southeastern United States. South Med J. 2017;110(3):188. doi: 10.14423/SMJ.0000000000000619 [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi S. [Different strategies between Japan and other countries for the diagnosis and treatment of early-stage lung cancer]. Nippon Geka Gakkai Zasshi. 2008;109(6):329–332. [PubMed] [Google Scholar]
- 8.Nie Q, Zhou QH, Zhu W, et al. nm23-H1 gene inhibits lung cancer cell invasion through down-regulation of PKC signal pathway. Chin J Oncol. 2006;28(5):334–336. [PubMed] [Google Scholar]
- 9.Liao YX, Zhi-Ping Z, Jie Z, Jing-Ping L. Effects of fibronectin 1 on cell proliferation, senescence and apoptosis of human glioma cells through the PI3K/AKT signaling pathway. Cell Physiol Biochem. 2018;48:1382–1396. doi: 10.1159/000492096 [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Xu G, Chen W, et al. Detection of long-chain non-encoding RNA differential expression in non-small cell lung cancer by microarray analysis and preliminary verification. Mol Med Rep. 2015;11:1925–1932. doi: 10.3892/mmr.2014.2944 [DOI] [PubMed] [Google Scholar]
- 11.You J, Zhang Y, Liu B, et al. MicroRNA-449a inhibits cell growth in lung cancer and regulates long noncoding RNA nuclear enriched abundant transcript 1. Indian J Cancer. 2014;51(7):77. doi: 10.4103/0019-509X.154055 [DOI] [PubMed] [Google Scholar]
- 12.Wolfl S, Odyvanova L, Kroll T, Sanger J, Clement J. Screening of lung cancer samples using gene expression profiling. Nat Genet. 2001;27(4):96. doi: 10.1038/87364 [DOI] [Google Scholar]
- 13.Petersen S, Heckert C, Rudolf J, et al. Gene expression profiling of advanced lung cancer. Int J Cancer. 2000;86(4):512–517. doi: [DOI] [PubMed] [Google Scholar]
- 14.Liao Y, Cheng S, Xiang J, Luo C. lncRNA CCHE1 increased proliferation, metastasis and invasion of non-small lung cancer cells and predicted poor survival in non-small lung cancer patients. Eur Rev Med Pharmacol Sci. 2018;22:1686–1692. doi: 10.26355/eurrev_201803_14581 [DOI] [PubMed] [Google Scholar]
- 15.She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673–2680. doi: 10.3892/or.2016.5105 [DOI] [PubMed] [Google Scholar]
- 16.Qin L, Yang Y, Liu Q, Xu W, Zhao S. Down-regulated lncRNA F630028O10Rik contributes to suppress lung cancer in mice through inhibiting miR-223-3p and VEGF signaling pathway. Chest. 2016;149(4):A304. doi: 10.1016/j.chest.2016.02.317 [DOI] [PubMed] [Google Scholar]
- 17.Shen Q, Jiang Y. LncRNA NNT-AS1 promotes the proliferation, and invasion of lung cancer cells via regulating miR-129-5p expression. Biomed Pharmacother. 2018;105:176–181. doi: 10.1016/j.biopha.2018.05.123 [DOI] [PubMed] [Google Scholar]
- 18.Henry WS, Hendrickson DG, Beca F, et al. LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer. Oncotarget. 2016;7(50):81981. doi: 10.18632/oncotarget.11962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu YY, Gao W, Zhang YL, Niu M, Wang BQ. Expression and clinical significance of long non-coding RNA LINC00520 in laryngeal squamous cell carcinoma. Lin Chuang Er Bi Yan Hou Ke Za Zhi. 2018;32(2):91–95. [DOI] [PubMed] [Google Scholar]
- 20.Chen CH, Lai JM, Chou TY, et al. VEGFA upregulates FLJ10540 and modulates migration and invasion of lung cancer via PI3K/AKT pathway. PLoS One. 2009;4:e5052. doi: 10.1371/journal.pone.0005052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Schmid-Bindert G, Wang D, et al. Blocking the PI3K/AKT and MEK/ERK signaling pathways can overcome Gefitinib-resistance in non-small cell lung cancer cell lines. Adv Med Sci. 2011;56(2):275–284. doi: 10.2478/v10039-011-0043-x [DOI] [PubMed] [Google Scholar]
- 22.Liang H, Dehua L, Zhaohui L, et al. HOXB1 is a tumor suppressor gene regulated by miR-3175 in Glioma. PLoS One. 2015;10(11):e0142387. doi: 10.1371/journal.pone.0142387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi A, Han J, Jia F, Liu C. miR-3175 and miR-134 affect proliferation, invasion and apoptosis of glioma cells through PI3K/AKT signaling pathway. J BUON. 2019;24(6):2465–2474. [PubMed] [Google Scholar]
- 24.Lin L, Tu H-B, Wu L, Liu M, Jiang G. MicroRNA-21 regulates non-small cell lung cancer cell invasion and chemo-sensitivity through SMAD7. Cell Physiol Biochem. 2016;38(6):2152–2162. doi: 10.1159/000445571 [DOI] [PubMed] [Google Scholar]