Abstract
Protein coacervates serve as hubs to concentrate and sequester proteins and nucleotides and thus function as membrane-less organelles to manipulate cell physiology. We have engineered a coacervating protein to create tunable, synthetic membrane-less organelles that assemble in response to a single pulse of light. Coacervation is driven by the intrinsically disordered RGG domain from the protein LAF-1, and opto-responsiveness is coded by the protein PhoCl which cleaves in response to 405 nm light. We developed a fusion protein containing a solubilizing maltose binding protein domain, PhoCl, and two copies of the RGG domain. Several seconds of illumination at 405 nm is sufficient to cleave PhoCl, removing the solubilization domain and enabling RGG-driven coacervation within minutes in cellular-sized water-in-oil emulsions. An optimized version of this system displayed light-induced coacervation in Saccharomyces cerevisiae. The methods described here provide novel strategies for inducing protein phase separation using light.
Keywords: Biomolecular condensates, Membrane-less organelles, optogenetics, recombinant proteins, RGG, PhoCl
Recently, a subset of organelles that are not enclosed by a lipid bilayer have been identified.1,2,3 These organelles, termed membrane-less organelles or biomolecular condensates, are a result of liquid-liquid phase separation and coacervation of proteins.4 Such membrane-less organelles concentrate proteins resulting in a protein-rich phase within the organelle and a protein-poor phase outside. Many of the proteins involved in these organelles contain intrinsically disordered regions (IDRs) that are devoid of a fixed structure.5 The resulting assemblies tend to be liquid-like and dynamically exchange components with their surroundings.6 Examples of membrane-less organelles include P-bodies, Cajal bodies, stress granules, and nucleoli, amongst many others.7 These organelles serve diverse functions within the cell including stress response, gene regulation, RNA processing, and ribosome biogenesis.8 While the role of many membrane-less organelles has been elucidated, numerous questions still remain regarding their formation, regulation, and function. To answer these questions, new tools are needed for control and manipulation of membrane-less organelles. In this work, we re-engineer a protein that forms membrane-less organelles to respond to a single pulse of 405 nm light. The protein coacervates formed as a result of 405 nm light exposure do not disassemble after the light stimulus is removed. This novel opto-responsive system enables precise control over the formation of synthetic membrane-less organelles.
In addition to their biological role, biomolecular condensates have significant potential for use as a functional biomaterial. For instance, controlled release of a therapeutic peptide has been achieved by fusing the peptide to a phase separating protein and releasing it from the condensate with a protease.9,10,11 Furthermore, synthetic membrane-less organelles have been engineered with varying functions. Examples include protein coacervates designed to function as microreactors by bringing together enzymes and substrates in the coacervate,12 coacervates engineered to sequester RNA and inhibit translation,13 and coacervates encapsulated inside proto-cellular liposomes as a model organelles.14 Finally, membrane-less organelles can be used in metabolic engineering to colocalize multiple enzymes involved in a synthetic pathway.15, 16 To fully realize these applications, strategies for controlling the design and assembly of coacervates need to be developed.
Several strategies for controlling the timing and spatial location of phase separation of proteins have been implemented. Temperature can be used to control phase behavior because protein condensates generally form below or above a critical temperature that is dependent upon the sequence.17,18,19 Although temperature is a convenient control handle for the formation of protein condensates in in vitro applications, it has limited usefulness in live cells. The requirements for phase separation depend not only on protein sequence but also on post-translational modifications (PTMs);20,21 however, PTMs are less suitable for engineering applications as they can be difficult to control precisely and often require multicomponent systems. Through careful sequence design, the assembly of protein condensates can be controlled via other biomolecules such as proteases,22 RNAs,23,24 reducing agents,25,26 and crowding agents27,28. While these strategies are powerful, a biomolecule must be transported to the phase-separating protein to initiate the assembly, limiting the practicality. Finally, several strategies have been developed to control the assembly and disassembly of biomolecular condensates with light.29 Light control of organelles is desirable because light can be precisely spatially and temporally controlled experimentally and is generally orthogonal to natural cellular processes. Such optically controlled systems often rely on continuous light stimulation. In this work, we describe a single-protein system for the formation of protein coacervates in response to a single short pulse of light without the need for continuous illumination. This represents an important step towards reducing the amount of light needed to achieve stable protein coacervation.
Taking inspiration from other light-sensitive, phase-separating systems we sought to expand the capability of our previously developed, RGG-based enzyme-triggered phase separation system by creating RGG variants that are light sensitive. The intrinsically disordered domain of the Caenorhabditis elegans protein LAF-1, RGG, has been reported to be necessary and sufficient for phase separation.30 In previous work we showed that chain length of the RGG domain controls the critical temperature and concentration for phase separation. Whereas monomeric RGG is miscible at 25 °C, a fusion protein containing two RGG domains in tandem robustly condenses to form micron-size liquid coacervates. Additionally, fusion of maltose-binding protein (MBP) to tandem RGG solubilizes the protein, inhibiting phase separation at room temperature.22 Enzyme-mediated RGG coacervate assembly was achieved by creating a fusion construct wherein MBP was followed by a protease recognition site and two RGG domains. Protease treatment liberates the MBP from the RGG domains leading to RGG coacervate assembly.
We sought a genetically encoded, single-component system in which stable protein coacervates would form in response to light. Whereas previous studies have achieved protein coacervation in response to continuous illumination, we sought a system wherein coacervates assemble in response to a single light pulse without the need for continuous light stimulation. We utilized the photo-cleavable protein PhoCl in conjunction with RGG by placing PhoCl between a solubilization tag (MBP) and dimeric RGG.31 PhoCl irreversibly and spontaneously dissociates into two fragments after exposure to 405 nm light, and, when cleaved after a short pulse of 405 nm light, liberates MBP and initiates phase separation of RGG. We demonstrate light-activated coacervation with our SPLIT (stable protein coacervation using a light induced transition) system both in protocellular water droplets and in yeast cells, illustrating the versatility of this approach.
Results
To achieve single-pulse, stable, light-induced condensation of RGG into protein coacervates, we devised a composite construct containing a solubilization domain (MBP), PhoCl for photocleavage, and two RGG domains (Figure 1a). Upon 405 nm illumination, cleavage of the PhoCl domain results in two fragments: (1) an N-terminal fragment containing MBP and most of PhoCl protein and (2) a C-terminal fragment containing tandem RGG. The cleavage site of PhoCl is twelve residues from its C-terminus, meaning only these twelve residues from the PhoCl domain remain fused to tandem RGG. Upon removal of the solubilization tag, tandem RGG phase separates to form liquid coacervates.
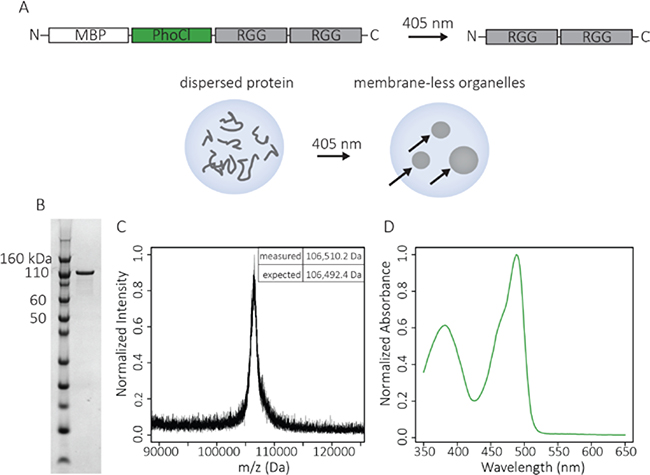
Figure 1. Characterization of the recombinant fusion protein: MBP-PhoCl-RGG-RGG.
A. Schematic of the MBP-PhoCl-RGG-RGG molecule designed to cleave upon illumination with 405 nm light to form RGG-RGG, resulting in protein condensation. Experiments were performed inside cellular-sized water droplets. Arrows indicate coacervates inside a water droplet. B. SDS-PAGE gel of MBP-PhoCl-RGG-RGG (MW = 106,492.4 Da). C. MALDI-TOF-MS spectrum of MBP-PhoCl-RGG-RGG. D. Absorbance spectrum of MBP-PhoCl-RGG-RGG showing a peak at 488 nm as a result of the folded PhoCl domain.
The protein construct, MBP-PhoCl-RGG-RGG, was produced recombinantly in Escherichia coli, and the purity and molecular weight was confirmed using gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Figure 1b-c). The absorbance spectrum of this molecule had a peak around 488 nm corresponding to the published absorbance spectrum of PhoCl (Figure 1d).31 Both to emulate a the interior of a cell and to constrain the protein to a small volume that can be wholly illuminated with a laser, the protein was encapsulated in cellular-sized water-in-oil emulsions. The water droplets containing MBP-PhoCl-RGG-RGG had uniform fluorescence at 488 nm arising from the PhoCl domain (Figure 2a) indicating that the protein had not phase separated and was disperse within the water droplets.
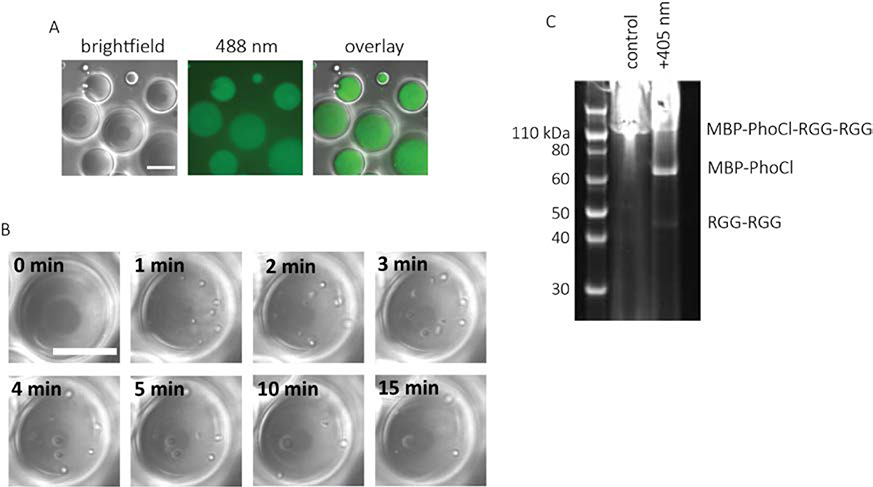
Figure 2. Light induced formation of protein coacervates.
A. Image of water-in-oil emulsions containing 2 mg/mL MBP-PhoCl-RGG-RGG. The aqueous phase contained 150 mM NaCl and 20 mM tris, pH 7.4. Image was taken in DIC and 488 nm channel. There were no visible protein coacervates before light exposure. B. Time-lapse DIC images of protein-containing emulsions after exposure to 405 nm light (5 second light pulse, 7.9 mW/cm2). Arrows indicate the location of protein condensates inside the water droplets. Scale bars: 20 μm. C. SDS-PAGE gel showing MBP-PhoCl-RGG-RGG with and without exposure to 405 nm light (5 second light pulse, 7.9 mW/cm2). The emulsions were centrifuged to recover the water phase before performing gel electrophoresis on both the light-exposed and control protein.
Photocleavage of PhoCl domain in situ was achieved using a 405 nm laser. The power was set to 7.9 mW/cm2, and the sample was illuminated for 5 seconds before imaging. Soon after illumination, numerous small protein coacervates nucleated throughout the water droplets. Because the condensed protein phase is denser than water, the protein coacervates sank to the bottom of the water droplet. The small protein coacervates also rapidly fused and grew in size (Figure 2b). To determine whether coacervate formation is a result of light-induced cleavage of MBP-PhoCl-RGG-RGG, the emulsions were recovered, and protein was analyzed by gel electrophoresis (Figure 2c & Figure S1). Prior to light exposure only one band was visible, corresponding to the full-length protein. After 5 seconds of illumination at 405 nm, three bands became visible. The band molecular weights correspond to noncleaved full-length protein and two cleavage products: MBP-PhoCl and RGG-RGG. Irreversible cleavage of the PhoCl domain to liberate MBP causes the formation of stable RGG coacervates.
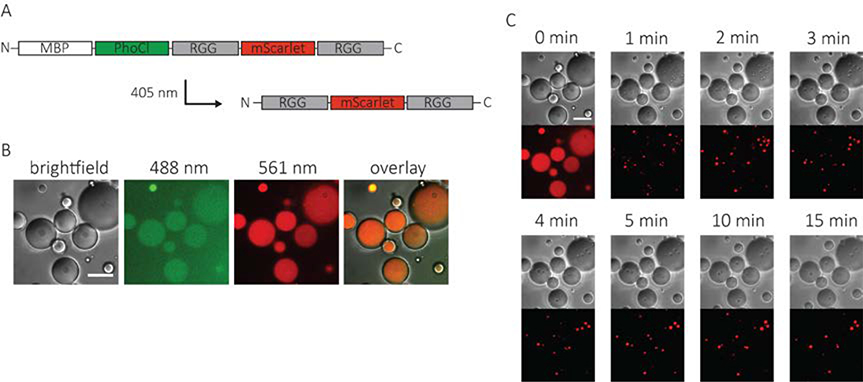
To expand the functionality of our RGG photo-coacervation system, we sought to demonstrate that a folded protein could be incorporated into the coacervates. A red fluorescent protein, mScarlet,32 was incorporated between the two RGG domains as a model protein (Figure 3a). The protein construct, MBP-PhoCl-RGG-mScarlet-RGG was contained in cellular-sized water-in-oil emulsions. The water emulsions contained no visible protein coacervates and were uniformly fluorescent in both the 488 nm (PhoCl) and 561 nm (mScarlet) channels (Figure3b). The protein-containing water droplets were exposed to 405 nm light for 5 seconds. As with the original construct, protein coacervates rapidly nucleated in the water droplets and then quickly fused and grew in size, ultimately resulting in between one and three protein coacervates in each water droplet (Figure 3c, supplemental video). Protein coacervates showed fluorescence in the 561 nm channel indicating that mScarlet was incorporated. Furthermore, the protein coacervates were also fluorescent in the 488 nm (PhoCl) channel indicating that non-cleaved MBP-PhoCl-RGG-mScarlet-RGG was also incorporated into the coacervates (Figure S2).
Figure 3. Incorporation of a folded protein into optically-regulated protein coacervates.
A. Schematic of an optically active RGG construct containing mScarlet. B. Images of water-in-oil emulsions containing 2 mg/mL MBP-PhoCl-RGG-mScarlet-RGG prior to exposure to 405 nm light. The aqueous phase contained 150 mM NaCl and 20 mM tris, pH 7.4. Image was taken in DIC, 488 nm, and 561 nm channels. C. Time-lapse DIC and 561 nm channel images of protein-containing emulsions after exposure to 405 nm light (5 second light pulse, 7.9 mW/cm2). Scale bars: 20 μm. Image intensities were background adjusted for clarity.
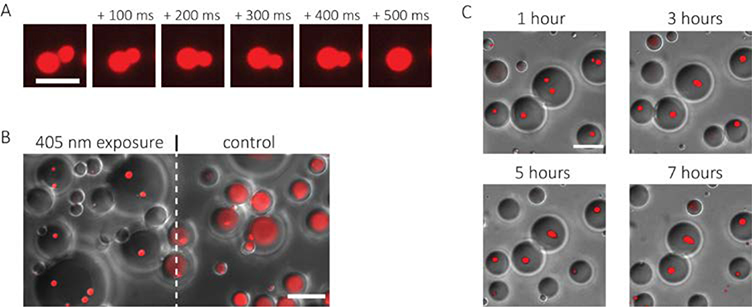
After nucleation of protein coacervates, when two coacervates came into contact, they fused and contracted to a spherical shape within hundreds of milliseconds (Figure 4a). The rapid formation of a spherical shape after fusion is expected from liquid-like coacervates stabilized by surface tension. The coacervates formed in our opto-responsive system also have excellent specificity and stability. The spatial patterning of light is highly specific; areas outside the field of illumination show no coacervate assembly. Formation of protein coacervates occurred only in water droplets that were illuminated (Figure 4b). Furthermore, the coacervates are stable for hours after induction without the need for continuous or additional illumination. The coacervates showed little change over seven hours (Figure 4c). Specificity and stability of engineered protein coacervates is critical for utility as a functional biomaterial.
Figure 4. Characterization of protein coacervates: fusion, specificity, and stability.
A. Time-lapse images showing rapid fusion of two RGG coacervates. Scale bar: 5 μm. B. Image demonstrating spatial control of coacervate formation. Area to the left of the dashed line was exposed to a 5 second 405 nm light pulse; area to the right of the dashed line was not exposed to 405 nm light. Image shows an overlay of DIC and 561 nm (mScarlet) channel images. Scale bar: 20 μm. C. Overlay of time-lapse DIC and 561 nm (mScarlet) channel images of emulsions after exposure to 405 nm light demonstrating stability over several hours. Scale bar: 20 μm.
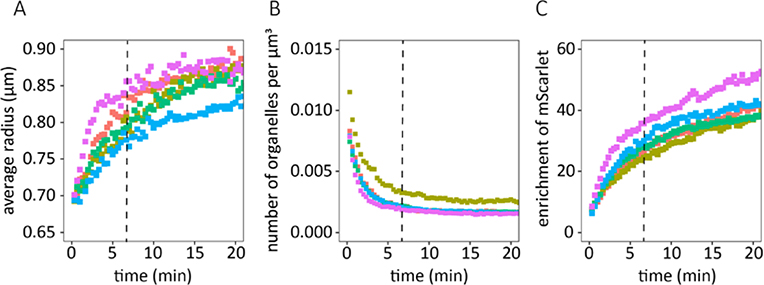
Our photo-coacervation system enables close examination of critical aspects of the nucleation and growth of RGG-based coacervates. The average coacervate radius, number density of coacervates, and enrichment of mScarlet within the coacervates were tracked with respect to time using image analysis (Figure 5a-c). We observed two distinct regimes in the growth of protein coacervates. First was a ‘fast’ regime for approximately 350 seconds following the initial 405 nm light pulse followed by a ‘slow’ regime after 350 seconds. During the fast regime, the radius of the coacervates rapidly increased and the number density of coacervates rapidly decreased. During this regime, RGG coacervates nucleated throughout the water droplet and small RGG coacervates quickly fused together to form larger coacervates. During the slow regime, the number density remained constant, meaning that we observed no new protein coacervates nucleate. The growth of the coacervates slowed during this regime, but the average radius continued to slowly increase. Furthermore, the enrichment of mScarlet appeared to continue to increase during the slow regime. This implies that, during the slow regime, the existing coacervates may continue to recruit other molecules resulting in increases in enrichment and radius.
Figure 5. Nucleation and growth of protein coacervates.
Plots of (A) average coacervate radius (μm), (B) number density of protein coacervates normalized by the volume of the water phase (1/μm3), and (C) enrichment of mScarlet ([fluorescence intensity inside coacervate]/[fluorescence intensity outside coacervate]) as a function of time (min). Each colored line represents an average taken over one field of view. Vertical dashed lines delineate the transition between the ‘fast’ and ‘slow’ coacervate growth regimes.
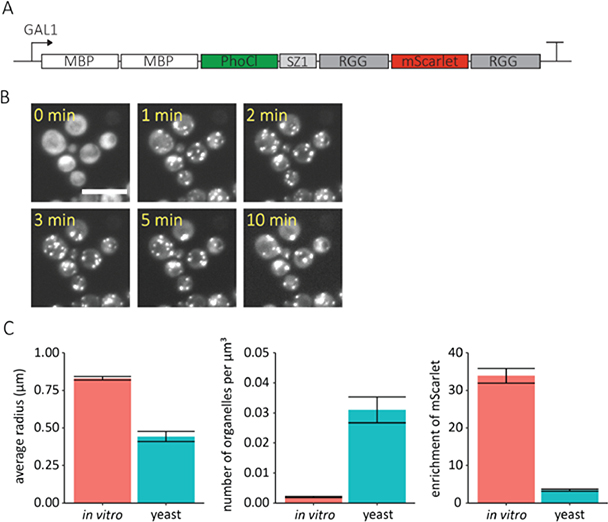
To demonstrate that photo-responsive RGG condensates can be encoded in living cells, we expressed various constructs in the model single-cell organism Saccharomyces cerevisiae (S. cerevisiae). This model cell system is broadly used for cellular engineering and our goal is to develop a platform for generating synthetic membrane-less organelles in this cell type. RGG constructs were integrated into the yeast genome and expressed from various constitutive or inducible promoters. Through optimization we found that two MBP domains, rather than one, were needed to solubilize the RGG construct in yeast. Additionally, we included a SYNZIP1[33] dimerization domain adjacent to the first RGG domain for future applications requiring modular recruitment of cargos into synthetic organelles.22 The expression of this construct was regulated by the GAL1 promoter (Figure 6a), and thus was induced via galactose. To test the photo-responsiveness of the system, yeast cells were exposed to a two minute pulse of light at 405 nm (1.6 mW/cm2). To mitigate photodamage to the cells, this light pulse was at a significantly lower wattage but longer time than the conditions used for the in vitro system. Before light exposure, we observed uniform fluorescence in the 561 nm channel, indicating that the RGG construct was dispersed within cells. Shortly after 405 nm light exposure, small RGG coacervates nucleated throughout the cells. As in the in vitro system, the small coacervates fused and grew in size over several minutes (Figure 6b). Similar behavior was observed when the construct was under control of the high expression constitutive yeast promoter, TDH3 (Figure S3).
Figure 6. Optically-regulated protein coacervates formed in a single cell eukaryote.
A. Schematic of the optically active RGG protein expressed in S. cerevisiae. B. Time-lapse, maximum intensity projection, 561 nm channel images of RGG-expressing S. cerevisiae after exposure to 405 nm light (2-minute light pulse, 1.6 mW/cm2). showing formation of RGG coacervates inside the cells. Scale bar: 10 μm. C. Average coacervate radius (μm), number density of coacervates normalized by the volume of water droplet or yeast cell (1/μm3), and enrichment of mScarlet measured 10 minutes after 405 nm light exposure. Mean ± s.e.m., n ≥ 3.
It is critical to understand the differences between condensates formed in protocellular water emulsions and condensates formed in living cells. To this end, we characterized our optically regulated RGG condensates formed in S. cerevisiae using image analysis. The average protein coacervate radius, number density of coacervates, and enrichment of mScarlet were measured 10 minutes after exposure to 405 nm light and compared to RGG condensates formed in water-in-oil emulsions (Figure 6c). We found that the coacervates formed in yeast cells were about half the size of those formed in water-in-oil emulsions. Furthermore, we found that the number density of coacervates formed in yeast was approximately 16 times higher than water-in-oil emulsions. This means that there was a much greater number of coacervates when the system was transferred to yeast. Finally, the enrichment of mScarlet, defined as the ratio of mScarlet concentration inside the coacervates to outside the coacervates, was approximately 10 times lower in yeast cells as compared to the water-in-oil emulsions. We hypothesize that these differences may be due to differences in diffusivity between water and the cellular cytoplasm, or the incorporation of a cellular proteins in droplets. The diffusivity constant of proteins has been reported to be several times greater in water than in the cellular milieu.34 The lower diffusion rates in the cytoplasm lowers the rate at which smaller coacervates come into contact and fuse leading to an overall smaller coacervate size and higher number density. Furthermore, slower diffusion of the RGG protein or the incorporation of cellular proteins could result in lower enrichment.
Discussion
In this work, we re-engineered RGG-based phase separating proteins to create an optically activated protein coacervates. The coacervates were formed from a single protein containing the intrinsically disordered RGG domain of LAF-1, the photocleavable protein, PhoCl, and MBP, which is liberated before coacervate formation. The RGG based coacervates form only after exposure to 405 nm light. Moreover, only a single short pulse of light is needed to form time-stable coacervates. The use of light to form these coacervates affords precise spatial and temporal control over their assembly. We demonstrated light induced coacervate formation inside of cellular sized water droplets as well as yeast cells. We also demonstrated that a folded protein, mScarlet, could be incorporated in the coacervates. The nucleation and growth of mScarlet-containing, RGG-based coacervates was quantified using image analysis. We found that immediate nucleation occurs broadly throughout the control volume where the protein concentration is above the critical concentration. After initial nucleation, we observed a ‘fast’ growth phase involving rapid fusion of the coacervates and a rapid increase in coacervate radius. We then observed a ‘slow’ growth phase of the coacervates via recruitment of additional RGG proteins. During the slow phase, the coacervates remained at a constant number with no new nucleation but the enrichment of mScarlet inside the coacervates continued to increase. Finally, we compared RGG coacervates formed in water emulsions to those formed in yeast cells. We found that coacervates formed in yeast were smaller, had a higher number density, and had lower enrichment of mScarlet than their in vitro counterparts. We hypothesize that these differences arise from the difference in diffusivity between water and the yeast cytoplasm.
Several other systems have been previously designed in which biomolecular condensate formation is dependent upon light. For example, OptoDroplets have been developed wherein a phase separating protein was fused to the photo-activated oligomerization domain, Cry2.35 These optoDroplets formed rapidly when exposed to blue light and were used to study physiological phase transitions. Several light sensitive systems have also been developed that utilize SspB and iLID which bind in response to blue light. For example, Corelets have been developed wherein multimeric cores containing iLID domains recruit phase separating proteins containing SspB domains.36 Furthermore, in the iPOLYMER-LI system, six tandem iLID domains were fused to YFP and six SspB domains were fused to mCherry.37 The two molecules formed aggregates in response to blue light and were used as an analogue of stress granules. In the CasDrop system, SspB was fused to an IDR and iLID was bound to dCas9.38 This system enabled blue light induced, phase separated coacervates at specific target genetic loci. Finally, the light-responsive system PixELL relies on PixD and PixE which associate to form multiunit complexes in the dark and dissociate when exposed to blue light.39 PixD and PixE were fused to phase separating proteins to create protein coacervates used to study cellular memory of spatial stimuli.
A key distinction between our constructs and these achievements are that all the previously developed techniques rely on continuous light stimulation which can have a deleterious effect on proteins and cells. In our system, coacervation follows a single short pulse of light. Furthermore, the formation of coacervates in our system relies only on a single protein without an optically active dimerization domain which facilitates the study of protein condensate nucleation and growth in the absence of triggered dimerization. Furthermore, our RGG system allows for facile incorporation of additional domains such as mScarlet. We envision the technique we developed having future applications as a tool for controlling intracellular phase transitions to understand its role in physiology and disease. Our work also sets the stage for future studies exploring the nature of synthetic membrane-less organelles formed in living cells. Finally, our opto-responsive RGG construct integrates with and expands the capability of our previously developed system for enzymatic control of RGG constructs22 and is an important step towards the creation of tunable synthetic cells.
Although, this work establishes and proof of concept for protein coacervation resulting from light induced removal of a solubilization domain, future studies will be needed to adopt this method for various practical applications. In this work we noted that very different doses of light were needed in vitro versus in S. cerevisiae; we believe this reflects the complex environment in cells in which diffusion is slower and many other proteins are present to either interfere or incorporate in droplets. Furthermore, prior work has shown that PhoCl responds to light in a dose dependent manner;31, 40 therefore, it stands to reason that light dosing should be optimized for each system. Additionally, protein concentration also must be optimized for this method. Previous work has shown that RGG coacervation is strongly dependent upon concentration.22 For practical applications one would want to know what endogenous proteins become incorporated into the synthetic organelles and what effects the synthetic organelles have on cellular functions. Also, the fate of cells after exposure to light should be studied to determine whether any deleterious effects arise from either expression of the phase separating protein or irradiation with 405 nm light. While 405 nm light has been shown to have effects on yeast cell function at sufficiently high dosages,41 we did not observe any morphological changes in the yeast after expression of the RGG construct or after exposure to light at 405 nm.
Conclusion
We developed a novel approach to trigger the formation of biomolecular condensates with a single pulse of 405 nm light which we term SPLIT (stable protein coacervation using a light induced transition). The removal of a solubilization domain from a phase-separating protein upon exposure to light causes protein coacervates to rapidly form. We demonstrate this technique both in vitro and in S. cerevisiae. The SPLIT system will have applications in synthetic protocells as an organelle mimic, in metabolic engineering, and in the study of cellular phase separation.
Methods
Gene creation and recombinant protein expression in e. coli
The RGG gene was amplified by PCR from the LAF-1 gene. The LAF-1 gene was a gift from Shana Elbaum-Garfinkle and Clifford Brangwynne. The RGG gene was further modified to append additional domains using the In-Fusion cloning kit (Takara Bio) following the manufacturer’s protocol. The PhoCl gene was a gift from was a gift from Robert Campbell (Addgene plasmid # 87697). Genes were contained in the bacterial expression vector, pBamUK, a pET derivative. All sequences were verified using Sanger sequencing (Genewiz). A 6-histidine tag was included at the C-terminus for IMAC purification.
Proteins were expressed in E. coli BL21(DE3) cells (Stratagene) following the manufacturer’s protocol. Briefly, a 5 mL starter culture was grown at 37 °C while shaking overnight in Luria broth (Sigma Aldrich) containing 50 μg/mL kanamycin (Amresco). The starter was used to inoculate 500 mL of Terrific Broth (Fisher Scientific) containing 50 μg/mL kanamycin and the culture was grown at 37 °C while shaking at 250 rpm until the optical density at 600 nm reached 0.5–0.8. Protein expression was induced with isopropyl-β-D-1-thiogalactoside (Lab Scientific) at a final concentration of 0.5 mM. The culture was then grown at 18 °C overnight in the dark. Cells were harvested by centrifugation and stored at −20 °C until purification.
Protein purification
Cells were resuspended in 15 mL of lysis buffer [20 mM tris-HCl (Bio-Rad), 500 mM NaCl, 20 mM imidazole (Thermo Fisher Scientific), 10 μg/mL DNase (Roche), complete protease inhibitor (Roche), pH 7.4]. Cells were lysed using sonication (Model 60 Sonic Dismembrator, Fisher Scientific), then the lysate was clarified by centrifugation at 15,000 g for 15–30 minutes. An Akta FPLC with a 1 mL Ni-NTA His-Trap column (GE Healthcare Life Sciences) was used for purification. First, the column was equilibrated with 5 column volumes of wash buffer (20 mM tris-HCl, 500 mM NaCl, 20 mM imidazole, pH 7.4). Then, the protein was bound and the column and washed with 10–15 column volumes of wash buffer. Finally, the protein was eluted in 1 mL fractions using elution buffer (20 mM tris-HCl, 500 mM NaCl, 500 mM imidazole, pH 7.4). Fractions containing protein were pooled and dialyzed using Slide-A-Lyzer cassettes (Thermo Fisher Scientific). Dialysis was carried out at 37 °C and into a high salt buffer (20 mM tris-HCl, 500 mM NaCl, pH 7.4). Following dialysis, the protein was aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C until use. After thawing, the protein was centrifuged at 7,000 rpm for 5 minutes to remove any aggregates. Protein concentration was determined by the absorbance at 280 nm on a NanoDrop (Thermo Fisher Scientific).
SDS-PAGE
Protein samples were denatured with NuPAGE sample buffer and heated at 70 °C for 10 minutes. Samples were run on NuPAGE Novex 4–12% Bis-Tris gels (Invitrogen) for 35 minutes at 200 V. Gels were stained with Instant Blue (Expedeon) following the manufacturer’s protocol. For protein samples recovered from emulsions, the water-in-oil emulsion was centrifuged at 6,000 RCF for several minutes and the water phase was recovered from the bottom of the tube.
MALDI-TOF-MS
Spots consisting of 1 μL of protein solution and 1 μL matrix solution (10 mg/mL sinapinic acid (Sigma Aldrich) dissolved in 50:50 acetonitrile (Fisher Scientific):water with 0.1% trifluoroacetic acid (Fisher Scientific)) were pipetted onto a MTP 384 polished steel target plate (Bruker, Billercia, MA, USA). Spectra were collected on an Ultraflextreme MALDI−TOF−MS (Bruker).
Absorbance spectrum
The absorbance spectrum of the protein was taken on a TECAN infinite 200Pro. Spectrum was taken from 350 to 650 nm in 2 nm steps.
Water-in-oil Emulsions
Cellular sized water-in-oil emulsions were created by adding 2 μL of the aqueous solution being studied to 200 μL oil containing a surfactant (50 mg/mL DPHS (Croda) dissolved in light mineral oil (Sigma)) to stabilize the water droplets and pipetting up and down approximately 50 times with a 200 μL tip. This resulted in water droplets approximately 10 to 30 μm in diameter.
Microscopy
A Leica DMi8 inverted microscope equipped with a spinning disk confocal unit was used for imaging. Imaging was conducted using a 63x, 1.4 NA plan-apochromatic oil-immersion objective and a 0.55 NA condenser and an sCMOS camera (Orca Flash 4.0, Hamamatsu Photonics). 50 μL of sample was added to chambered coverglass (No. 1.5 glass thickness, Molecular Probes). Z-stack images were taken at the coverglass.
Image Analysis
After coacervate formation was induced with a 5 second 405 nm light pulse, z-stack images of were taken in 20 second time intervals in the 561 nm channel. These images were analyzed using custom built MATLAB code. A maximum intensity projection was taken at each time point. The circular Hough algorithm was used on the image taken before light exposure to determine the location and size of the water droplets or yeast cells. Then, for each time point, the circular Hough algorithm was used to determine the location and size of the protein coacervates. The enrichment of mScarlet was defined as the ratio of average pixel intensity inside the protein coacervates to that inside the water droplets or yeast cells excluding the protein coacervates.
Gene creation and protein expression in yeast
Genes were incorporated into the yeast integrating plasmid, YIp211, using In-Fusion cloning (Takara Bio). The plasmids were transformed into the yeast strain YEF473a42 using the Frozen-EZ yeast transformation kit (Zymo Research) following the manufacturer’s protocol. The YEF473a strain was a gift from Prof. Erfei Bi. Transformants were streaked onto plates with synthetic complete media lacking uracil (6.7 g/L yeast nitrogen base (BD), dropout mix (Clontech), 2% glucose) to select for yeast in which YIp211 has been incorporated. Plates were incubated at 30 °C for 2–3 days at which point colonies were streaked onto new plates and grown for another 1–2 days before taking colonies to make patches on a third new plate. Cells were taken from the patch, suspended in YPD media, and grown at 30 °C while shaking at 300 rpm for a few hours before imaging. For constructs under the GAL1 promoter, colonies were grown in dropout media plus 2% glucose overnight, followed by 2% raffinose for eight hours, followed by 2% galactose to induce expression.
Supplementary Material
Acknowledgements
DAH and MG acknowledge the support of NSF Superseed (DMR 1720530) which supported the testing of the constructs in in vitro in synthetic compartments, NIH R01-EB028320 which supported encoding and characterization of SPLIT condensates in living cells, and a grant from the DOE Biomolecular Materials Program (DE-SC000232866) which supported initial protein design and biochemical analysis of IDP coacervates within water-in-oil emulsions. BSS acknowledges support from the NIH (F32GM119430). We thank Prof. Brian Chow for use of the microplate reader and incubator.
Footnotes
The authors declare no competing financial interest.
Supporting Information: The supporting information includes two separate files. One is a document that includes replicates of SDS-PAGE gels showing MBP-PhoCl-RGG-RGG before and after exposure to 405 nm light, microscopy images showing incorporation of non-cleaved PhoCl into protein coacervates, and microscopy images demonstrating light induced formation of protein coacervates in yeast cells with the RGG construct under control of the TDH3 promoter. A second is a video of mScarlet containing, protein coacervate formation and coalescence.
References
- [1].Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, and Hyman AA (2009) Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation, Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- [2].Gomes E, and Shorter J (2019) The molecular language of membraneless organelles, J. Biol. Chem 294, 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, and Fuxreiter M (2018) Protein Phase Separation: A New Phase in Cell Biology, Trends Cell Biol. 28, 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brangwynne, Clifford P, Tompa P, and Pappu, Rohit V (2015) Polymer physics of intracellular phase transitions, Nat. Phys 11, 899–904. [Google Scholar]
- [5].Uversky VN (2017) Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder, Curr. Opin. Struct. Biol 44, 18–30. [DOI] [PubMed] [Google Scholar]
- [6].Hyman AA, Weber CA, and Julicher F (2014) Liquid-liquid phase separation in biology, Annu. Rev. Cell Dev. Biol 30, 39–58. [DOI] [PubMed] [Google Scholar]
- [7].Banani SF, Lee HO, Hyman AA, and Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry, Nat. Rev. Mol. Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mitrea DM, and Kriwacki RW (2016) Phase separation in biology; functional organization of a higher order, Cell Commun. Signaling 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Amiram M, Luginbuhl KM, Li X, Feinglos MN, and Chilkoti A (2013) Injectable protease-operated depots of glucagon-like peptide-1 provide extended and tunable glucose control, Proc. Natl. Acad. Sci. U. S. A 110, 2792–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Amiram M, Luginbuhl KM, Li X, Feinglos MN, and Chilkoti A (2013) A depot-forming glucagon-like peptide-1 fusion protein reduces blood glucose for five days with a single injection, J. Controlled Release 172, 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sinclair SM, Bhattacharyya J, McDaniel JR, Gooden DM, Gopalaswamy R, Chilkoti A, and Setton LA (2013) A genetically engineered thermally responsive sustained release curcumin depot to treat neuroinflammation, J. Controlled Release 171, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Faltova L, Kuffner AM, Hondele M, Weis K, and Arosio P (2018) Multifunctional Protein Materials and Microreactors using Low Complexity Domains as Molecular Adhesives, ACS Nano 12, 9991–9999. [DOI] [PubMed] [Google Scholar]
- [13].Simon JR, Eghtesadi SA, Dzuricky M, You L, and Chilkoti A (2019) Engineered Ribonucleoprotein Granules Inhibit Translation in Protocells, Mol. Cell 75, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Deng NN, and Huck WTS (2017) Microfluidic Formation of Monodisperse Coacervate Organelles in Liposomes, Angew. Chem., Int. Edit 56, 9736–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hammer SK, and Avalos JL (2017) Harnessing yeast organelles for metabolic engineering, Nat. Chem. Biol 13, 823–832. [DOI] [PubMed] [Google Scholar]
- [16].Zhao EM, Suek N, Wilson MZ, Dine E, Pannucci NL, Gitai Z, Avalos JL, and Toettcher JE (2019) Light-based control of metabolic flux through assembly of synthetic organelles, Nat. Chem. Biol 15, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Quiroz FG, and Chilkoti A (2015) Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers, Nat. Mater 14, 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin YH, Forman-Kay JD, and Chan HS (2018) Theories for Sequence-Dependent Phase Behaviors of Biomolecular Condensates, Biochemistry 57, 2499–2508. [DOI] [PubMed] [Google Scholar]
- [19].Dignon GL, Zheng WW, Kim YC, and Mittal J (2019) Temperature-Controlled Liquid-Liquid Phase Separation of Disordered Proteins, ACS Cent. Sci 5, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Drino A, and Schaefer MR (2018) RNAs, Phase Separation, and Membrane-Less Organelles: Are Post-Transcriptional Modifications Modulating Organelle Dynamics?, BioEssays 40. [DOI] [PubMed] [Google Scholar]
- [21].Hofweber M, and Dormann D (2019) Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics, J. Biol. Chem 294, 7137–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schuster BS, Reed EH, Parthasarathy R, Jahnke CN, Caldwell RM, Bermudez JG, Ramage H, Good MC, and Hammer DA (2018) Controllable protein phase separation and modular recruitment to form responsive membraneless organelles, Nat. Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, and Gladfelter AS (2015) RNA Controls PolyQ Protein Phase Transitions, Mol. Cell 60, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lin Y, Protter DS, Rosen MK, and Parker R (2015) Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins, Mol. Cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Reed EH, and Hammer DA (2018) Redox sensitive protein droplets from recombinant oleosin, Soft Matter 14, 6506–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Asherie N, Pande J, Pande A, Zarutskie JA, Lomakin J, Lomakin A, Ogun O, Stern LJ, King J, and Benedek GB (2001) Enhanced crystallization of the Cys18 to Ser mutant of bovine gammaB crystallin, J. Mol. Biol 314, 663–669. [DOI] [PubMed] [Google Scholar]
- [27].Kuznetsova IM, Turoverov KK, and Uversky VN (2014) What Macromolecular Crowding Can Do to a Protein, Int. J. Mol. Sci. 15, 23090–23140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wheeler RJ, and Hyman AA (2018) Controlling compartmentalization by non-membrane-bound organelles, Philos. Trans. R. Soc, B 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nakamura H, DeRose R, and Inoue T (2019) Harnessing biomolecular condensates in living cells, J. Biochem 166, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, and Brangwynne CP (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics, Proc. Natl. Acad. Sci. U. S. A 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang W, Lohman AW, Zhuravlova Y, Lu X, Wiens MD, Hoi H, Yaganoglu S, Mohr MA, Kitova EN, Klassen JS, Pantazis P, Thompson RJ, and Campbell RE (2017) Optogenetic control with a photocleavable protein, PhoCl, Nat. Methods 14, 391–394. [DOI] [PubMed] [Google Scholar]
- [32].Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, and Gadella TW Jr. (2017) mScarlet: a bright monomeric red fluorescent protein for cellular imaging, Nat. Methods 14, 53–56. [DOI] [PubMed] [Google Scholar]
- [33].Thompson KE, Bashor CJ, Lim WA, and Keating AE (2012) SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains, ACS Synth. Biol 1, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gura Sadovsky R, Brielle S, Kaganovich D, and England JL (2017) Measurement of Rapid Protein Diffusion in the Cytoplasm by Photo-Converted Intensity Profile Expansion, Cell Rep. 18, 2795–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, and Brangwynne CP (2017) Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets, Cell 168, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bracha D, Walls MT, Wei MT, Zhu L, Kurian M, Avalos JL, Toettcher JE, and Brangwynne CP (2018) Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds, Cell 175, 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nakamura H, Lee AA, Afshar AS, Watanabe S, Rho E, Razavi S, Suarez A, Lin YC, Tanigawa M, Huang B, DeRose R, Bobb D, Hong W, Gabelli SB, Goutsias J, and Inoue T (2018) Intracellular production of hydrogels and synthetic RNA granules by multivalent molecular interactions, Nat. Mater 17, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, and Brangwynne CP (2018) Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome, Cell 175, 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dine E, Gil AA, Uribe G, Brangwynne CP, and Toettcher JE (2018) Protein Phase Separation Provides Long-Term Memory of Transient Spatial Stimuli, Cell Syst. 6, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shadish JA, Strange AC, and DeForest CA (2019) Genetically Encoded Photocleavable Linkers for Patterned Protein Release from Biomaterials, J. Am. Chem. Soc 141, 15619–15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Murdoch LE, McKenzie K, Maclean M, Macgregor SJ, and Anderson JG (2013) Lethal effects of high-intensity violet 405-nm light on Saccharomyces cerevisiae, Candida albicans, and on dormant and germinating spores of Aspergillus niger, Fungal Biol. 117, 519–527. [DOI] [PubMed] [Google Scholar]
- [42].Bi EF, and Pringle JR (1996) ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae, Mol. Cell. Biol 16, 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.